Abstract
Background
We investigated the clinical relevance of SLC 19A1 genetic variability for high dose methotrexate (HD-MTX) related toxicities in children and adolescents with acute lymphoblastic leukaemia (ALL) and non Hodgkin malignant lymphoma (NHML).
Patients and methods
Eighty-eight children and adolescents with ALL/NHML were investigated for the influence of SLC 19A1 single nucleotide polymorphisms (SNPs) and haplotypes on HD-MTX induced toxicities.
Results
Patients with rs2838958 TT genotype had higher probability for mucositis development as compared to carriers of at least one rs2838958 C allele (OR 0.226 (0.071–0.725), p < 0.009). Haplotype TGTTCCG (H4) statistically significantly reduced the risk for the occurrence of adverse events during treatment with HD-MTX (OR 0.143 (0.023–0.852), p = 0.030).
Conclusions
SLC 19A1 SNP and haplotype analysis could provide additional information in a personalized HD-MTX therapy for children with ALL/NHML in order to achieve better treatment outcome. However further studies are needed to validate the results.
Key words: acute lymphoblastic leukaemia, genetic polymorphism, haplotype, methotrexate
Introduction
Inter-individual variability in treatment responses and treatment related toxicities is an important issue in clinical practice. It can lead to therapeutic failures or adverse drug events (ADE). Identification and characterization of genetic polymorphisms and haplotypes involved in drug metabolism, transport and mechanism of action would provide important information about individual inherited differences in drug metabolism and treatment response in order to optimize treatment outcome.1
The solute carrier 19A1 (SLC19A1), trivially referred as reduced folate carrier (RFC), located in a 40719 bp region at chromosome 21q22.3, is coding for solute folate carrier (RFC1), a major transporter of folates and antifolates into the cell. The antifolate methotrexate (MTX) is a cytotoxic drug which is used in current treatment regimens for childhood acute lymphoblastic leukaemia (ALL)2, and non-Hodgkin malignant lymphoma (NHML).3,4 The most frequently studied SLC19A1 polymorphism is rs1051266 in the second exon of the SLC19A1, which results in amino acid substitution of arginine for histidine (H27R)5 in transmembrane domain 1 (TMD1), a region implicated in substrate binding and/or translocation6,7, is expected to alter SLC transport properties. Other polymorphisms have been described to date, but the functional consequences of other single nucleotide polymorphisms (SNPs) are not known yet.8
Several studies so far investigated the association of rs1051266 with high dose MTX (HD-MTX) treatment outcome and treatment related toxicities in ALL patients.9 10 11 12 13 Some of them reported a protective role of rs1051266 AA genotype14,15, while in others either an increased risk of MTX-induced toxicities and fatal outcomes in rs1051266 A carriers9,10, or even no association between SLC19A1 genotype and MTX-induced toxicities was reported.11,13,16,17 Recently, SNPs in microRNAs (miRNAs) that are involved in regulation of SLC19A1 were investigated. SNP in miR-595 which might affect SLC19A1 regulation and could affect MTX levels in patients with paediatric B-cell ALL was reported.18
To date all studies but one19 evaluated the role of individual SLC19A1 SNPs in the MTX toxicities and outcomes of ALL treatment. The discovery of HapMap (Humane gene database) made it possible to analyse many SNPs at a time. Between the former and the latter a tagSNP approach has been introduced recently. A tagSNP is a representative SNP in a region of the genome with high linkage disequilibrium. Using the tagSNPs reduces number of SNPs needed to be genotyped to capture the common variations across the genome.20 Relatively small numbers of tagSNPs will therefore mark each of the common haplotypes within the gene region.21
In the present study we investigated a possible association between SLC19A1 haplotypes, defined by common functional variants and tagSNPs, and MTX related toxicities in 88 children and adolescents with ALL/NHML. The aim was to provide additional insight into improved, cost-effective approach for a personalized HD-MTX therapy for children with ALL/NHML based on haplotype analysis in order to achieve better treatment outcome.
Patients and methods
Study design
The study group included 88 children and adolescents (41 male, 47 female) with ALL/NHML diagnosed and treated in the period from 1990 to 2008 at the Department of oncology and haematology, University Children’s Hospital in Ljubljana, Slovenia. All the patients were Central European Caucasians and below the age of 18 years at the time of diagnosis. Clinical and demographic data were obtained from the medical records and a questionnaire by a single abstractor blinded to genotypes and are presented in Table 1. Each patient enrolled in the study received a high MTX dose (77.3% of patients received 5 g/m2 and 22.7% received 2 g/ m2 of MTX, according to treatment protocols) on 4 separate occasions and concomitant leucovorin rescue therapy to reduce MTX toxicity. Overall, 352 courses of HD-MTX applications were analysed.
Table 1.
Demographic and clinical characteristics of children with ALL/NHML and MTX treatment toxicity
| Characteristic | ALL patients |
|---|---|
| (N = 88) | |
| Gender (%) | |
| Male | 41 (46.6) |
| Female | 47 (53.4) |
| Median age at diagnosis, years (range) | 4.58 (0.3–16.6) |
| Median body surface area, m2 (range) | 0.73 (0.4–1.9) |
| Leukaemia subtype (%); | |
| B-cell | 72 (81.8) |
| T-cell | 13 (14.7) |
| Undetermined | 3 (3.4) |
| Median No. leukocytes at diagnosis, 109 cells/L (range) | 9.35 (1–1650) |
| Median No. thrombocytes at diagnosis, 109 cells/L (range) | 72 (3–553) |
| Median haemoglobin conc., g/l (range) | 84 (43–148) |
| Median % of blasts at diagnosis (range) | 23.5 (0–97) |
| Median MTX dose, mg (range) | 3300 (540–9200) |
| MTX response | Present, N (%) |
| Relapse | 8 (9.2)** |
| Exitus | 4 (4.6)** |
| All MTX-induced ADEs* | 48 (54.5) |
| Leukopenia | 33 (37.5) |
| Thrombocytopenia | 16 (18.2) |
| Mucositis | 15 (17.0) |
| Neurotoxicity | 7 (8.0)** |
data is missing for one patient
data was collected after the first cycle of MTX
All patients and/or their parents or legal guardians gave their written informed consent to participate in the study. The study was approved by the Republic of Slovenia National Medical Ethics Committee and was carried out according to the Helsinki Declaration.
Outcome measurement
Adverse drug events (ADE) were recorded retrospectively from patients’ files throughout the course of the HD-MTX treatment. ADEs were classified as (1) leukopenia, defined as white blood cells count less than 5*109 per litre, (2) thrombocytopenia, defined as thrombocyte count less than 140*109 per litre, (3) mucositis, determined and graded according to the WHO oral toxicity scale classification22 or based on physician’s descriptions in medical records, need for analgesia and/ or total parenteral nutrition and (4) neurotoxicity, determined and graded according to National Cancer Institute Common Toxicity Criteria Version 1 and included patients with seizures (regardless of grade) and patients with grade 3 or 4.23 The data on relapse and exitus were also recorded. All the reported ADEs were used for the evaluation of MTX toxicity.
SLC19A1 SNP selection
A total of seven SNPs (rs2838951, rs2838956, rs2838958, rs1051266, rs1131596, rs2838451 and rs17004785) in SLC19A1 spanning 40 kbp across the chromosomal 21q22.3 region were selected. SNP selection was based on known function from the literature and/or National Center for Biotechnology Information SNP database (NCBI) and block-tagging ability. Among all, two SNPs were a functional variant (rs1051266 and rs1131596), while five (rs2838951, rs2838956, rs2838958, rs2838451, rs17004785) were tagging SNPs, selected based on International HapMap Project CEPH data (phase II, March 2008) with the following criteria: r2 > 0.8, and the frequency of minor allele greater than 5%. Tagging SNP selection was done using HaploView software version 4.1 (Supplementary Table 1).24
SLC19A1 genotyping
In total 5 ml of peripheral blood was collected into tubes with sodium citrate and stored for short term at -20°C until DNA isolation. Genomic DNA was isolated from peripheral blood leukocytes using a Qiagen FlexiGene kit (Qiagen GmbH, Germany). Genotyping was performed using a fluorescence-based competitive allele-specific (KASPar) assay.25 Real time PCR was performed on the ABI 7900HT in a 4 μL reaction mix containing 0.055 μL of KASPar allele specific PCR assay, 2 μL of KASPar PCR Master Mix (2x concentration, containing KTaq polymerase and ROX), 2.2 mM MgCl2 and 10 ng of DNA.
Statistical analysis
SNP based analyses
Binary logistic regression analysis with the addition of independent variables, such as sex, age at time of diagnosis and body surface area (BSA) was done using SPSS for Windows version 16.0.1 software (Statistical Package for the Social Sciences, Chicago, IL). The risk for toxicity of MTX treatment was expressed as odds ratio (OR) with 95% confidence intervals (CI). The level of significance was set to 0.05. Deviation from Hardy-Weinberg equilibrium (HWE) was calculated for each SNP using Chi-square test. The level of deviation was set to 0.05. Power calculations were done using G*Power software version 3.1.0. Our study had an 80% statistical power to detect medium effect sizes (0.331) with an alpha level of 0.05. The correction for false-positive findings was done using a method for detecting false-positive report probability (FPRP) described by Wacholder et al.26
The cut-off for noteworthy results was set as 0.5. The expected ORs were based on the previous reports of an association of SNPs in SLC19A1 with MTX-toxicity.9,10,11,14,15 Prior probability was set to high (π = 0.1) due to well documented association of SLC19A1 and MTX transport.27
Haplotype based analysis
SLC19A1 haploblocks and linkage disequilibrium (LD) between the selected SNPs were visualized using HaploView software version 4.1.24 SLC19A1 haplotype construction was performed using Tagger pairwise method.
Results
Patient characteristics
Of the 88 patients recruited, 48 (54.5%) patients suffered from MTX-induced adverse events. Specific MTX-induced ADEs after the first cycle of MTX and their frequencies are presented in Table 1.
Association between individual SNPs and MTX toxicity
The characteristics and the minor allele frequencies (MAF) of the investigated SNPs in SLC19A1 are presented in Supplementary Table 1. In one patient genotyping failed for all SNPs, while genotyping for rs2838951 and rs17004785 failed in two patients. The observed frequencies of the studied SNPs in ALL/NHML patients did not deviate from HWE (data not shown).
With regard to specific ADEs, univariate logistic regression analysis revealed only the association of rs2838958 with the occurrence of mucositis development (p = 0.009). We observed that patients with rs2838958 TT genotype had higher probability for mucositis development as compared to carriers of at least one polymorphic rs2838958 C allele (OR 0.226 (0.071–0.725), p < 0.009) (Table 2).
Table 2.
The influence of rs2838958 genotype on mucositis development - univariate logistic regression analysis
| Degree of moscositis | rs2838958 genotype | OR(95%CI) | P | |
|---|---|---|---|---|
| TT (%) | TC + CC (%) | |||
| 0 | 18 (66.7) | 53 (89.8) | reference | |
| 1 | 4 (14.8) | 4 (6.8) | 0.340 (0.077–1.500) | 0.154 |
| 2 | 4 (14.8) | 2 (3.4) | 0.170 (0.029–1.007) | 0.051 |
| 4 | 1 (3.7) | 0 (0.0) | -* | |
| 1–4 | 9 (33.3) | 6 (10.2) | 0.226 (0.071–0.725) | 0.009 |
| Mean value ± STD | 0.59 | 0.14 | 0.007 | |
Calculation was not possible
We then investigated the possible association between SLC19A1 polymorphisms and MTX-induced ADEs with the multivariate analysis using an additive model (per allele), but a statistically significant relationship could be established neither for overall nor for specific ADEs (Table 3). A marginally significant impact on the risk of mucositis was observed for rs2838951 (OR 2.160 (0.909–5.129), p = 0.081).
Table 3.
The impact of SLC19A1 genotypes on the occurrence of any and specific MTX-induced adverse events using multivariate analysis – an additive model
| SNP ID | Any adverse events | Leukopenia | Thrombocytopenia | Mucositis | Neurotoxicity | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | P | OR | P | OR | P | OR | P | OR | P | |
| (CI 95%) | (CI 95%) | (CI 95%) | (CI 95%) | (CI 95%) | ||||||
| rs1131590 | 0.719 | 0.360 | 0.609 | 0.150 | 1.170 | 0.696 | 1.292 | 0.579 | 0.542 | 0.361 |
| (0.354–1.458) | (0.310–1.197) | (0.532–2.576) | (0.525–3.170) | (0.146–2.016) | ||||||
| rs1051200 | 1.392 | 0.360 | 1.643 | 0.150 | 0.855 | 0.696 | 0.775 | 0.579 | 0.542 | 0.361 |
| (0.686–2.824) | (0.836–3.230) | (0.388–1.881) | (0.316–1.906) | (0.146–2.016) | ||||||
| rs2838958 | 1.221 | 0.583 | 1.684 | 0.136 | 0.556 | 0.172 | 0.476 | 0.130 | 0.453 | 0.236 |
| (0.599–2.490) | (0.849–3.339) | (0.240–1.291) | (0.182–1.243) | (0.122–1.677) | ||||||
| rs2838956 | 1.411 | 0.349 | 1.742 | 0.113 | 0.689 | 0.374 | 0.705 | 0.454 | 0.596 | 0.434 |
| (0.686–2.899) | (0.878–3.458) | (0.303–1.566) | (0.283–1.759) | (0.163–2.182) | ||||||
| rs17004785 | 0.849 | 0.772 | 1.393 | 0.545 | 0.854 | 0.821 | 0.672 | 0.606 | 1.226 | 0.800 |
| (0.280–2.572) | (0.476–4.079) | (0.218–3.349) | (0.148–3.055) | (0.255–5.899) | ||||||
| rs12483377 | 1.475 | 0.478 | 1.825 | 0.238 | 1.688 | 0.368 | 0.930 | 0.914 | 1.061 | 0.939 |
| (0.504–4.318) | (0.672–4.956) | (0.540–5.280) | (0.249–3.473) | (0.232–4.861) | ||||||
| rs2838951 | 1.261 | 0.483 | 0.740 | 0.351 | 1.652 | 0.185 | 2.160 | 0.081 | 1.693 | 0.337 |
| (0.666–2.412) | (0.392–1.394) | (0.786–3.473) | (0.909–5.129) | (0.577–4.967) | ||||||
The estimates for the odds ratio (OR) were adjusted to the following variables: age, sex, and treatment regimen.
We also performed haplotype analysis to ascertain whether the combination of polymorphisms that define a specific haplotype impacts the occurrence of ADEs. As shown in Figure 1 the investigated polymorphisms were lying in the same haploblock.
Figure 1.
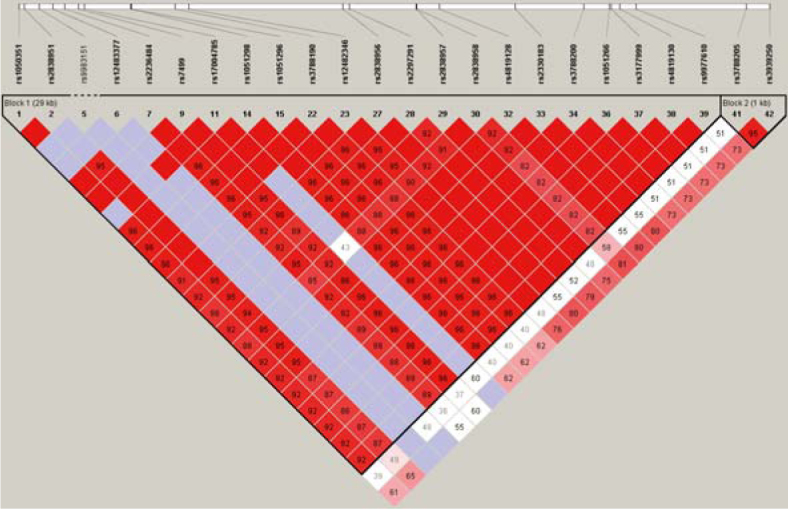
Graphic presentation of haploblocks and selected SNPs in SLC19A1.
The frequencies of SLC19A1 haplotypes in patients with ALL and NHML that were treated with HD-MTX are shown in Table 4. The Haplotype TGTTCCC (H2), which has wild type alleles present on all loci, was selected as the reference haplotype for the analysis. By frequency, it is the second most common haplotype in the patient test group with a frequency of 33.4%. The haplotype CACCCCG (H1) had polymorphic alleles present on the first four and on the last locus, with wild type alleles present on the fifth and sixth loci. The haplotype TGTTGTC (H3) had wild type alleles present on the first four and on the last locus, with polymorphic alleles present on the fifth and sixth. The haplotype TGTTCCG (H4) had wild type alleles present at all loci, except for the last one, while haplotype CATTCCC (H5) had polymorphic alleles present on the first, second, fifth and sixth locus, with wild type alleles on the remaining three.
Table 4.
SLC19A1 haplotype frequencies in patients with ALL and NHML
| Haplotype* | Frequency | Standard error |
|---|---|---|
| H1 = CACCCCG | 0.396 | 0.037 |
| H2 = TGTTCCC | 0.334 | 0.036 |
| H3 = TGTTGTC | 0.094 | 0.022 |
| H4 = TGTTCCG | 0.053 | 0.017 |
| H5 = CATTCCC | 0.017 | 0.010 |
| Other | 0.106 |
SNP rs1131596, rs1051266, rs2838958, rs2838956, rs17004785, rs12483377, rs2838951 from 5’ to 3’, respectively
H1 was the most frequent haplotype in the patient test group with a frequency of 39.6%. Together, H1, H2, and H3 haplotypes accounted for 82.4% of all haplotypes. In patients with ALL and ML, the haplotypes H4 and H5 were more rarely represented, with the frequency of a single haplotype between 1 to 5%. Therefore haplotype H5 and the other very rare haplotypes have been grouped together as ‘rare’ for further statistical analysis.
We examined whether there is a link between the SLC19A1 haplotypes and ADEs in patients treated with HD-MTX. We included the age and sex of the patient, and the treatment regimen as co-variables in the model. We found a statistically significant association of haplotype H4 in the SLC19A1 gene and the treatment regimen on the occurrence of any possible MTX-induced ADEs. Patients who were treated with ALL-BFM 95 were at an increased risk for the occurrence of any MTX-induced ADEs (OR 2.050 (1.065–3.943), p = 0.031), while those with haplotype H4 had a reduced risk for the occurrence of any possible ADE after treatment with HD-MTX (OR 0.143 (0.023–0.852), p = 0.030) (Table 5). The impact of the SLC19A1 haplotypes on specific MTX-induced ADEs was not observed, probably due to the small number of individual toxic events (Table 5).
Table 5.
The impact of gender, age, treatment regimen and SLC19A1 haplotypes on the occurrence of any and specific (thrombocytopenia, mucositis, and neurotoxicity) MTX-induced adverse events
| All | Thrombocytopenia | Mucositis | Neurotoxicity | |||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Gender | 2.385 | 0.078 | 1.615 | 0.413 | 1.523 | 0.520 | 0.531 | 0.475 |
| (0.908–6.259) | (5.134–5.076) | (0.423–5.472) | (0.093–3.024) | |||||
| Age | 1.028 | 0.588 | 9.526 | 0.440 | 1.054 | 0.396 | 1.088 | 0.289 |
| (0.931–1.134) | (8.422–1.078) | (0.935–1.190) | (0.931–1.272) | |||||
| Treatment regimen | 2.050 | 0.031* | 9.839 | 0.962 | 1.676 | 0.255 | 1.282 | 0.678 |
| (1.065–3.943) | (4.620–2.095) | (0.689–4.075) | (0.397–4.139) | |||||
| H1 = CACCCCG | 1.000 | 0.943 | 6.615 | 0.352 | 0.419 | 0.096 | 0.309 | 0.120 |
| (0.472–2.117) | (2.769–1.579) | (0.150–1.167) | (0.070–1.364) | |||||
| H3 = TGTTGTC | 1.110 | 0.844 | 9.220 | 0.891 | 0.593 | 0.515 | 0.951 | 0.934 |
| (0.335–3.618) | (2.254–3.750) | (0.122–2.877) | (0.175–5.150) | |||||
| H4 = TGTTCCC | 0.143 | 0.030* | 3.617 | 0.379 | 0.005 | 0.985 | 0.014 | 0.992 |
| (0.023–0.852) | (4.568–2.541) | (0.0–NC) | (0.0–NC) | |||||
| Rare haplotypes | 2.002 | 0.275 | 9.113 | 0.871 | 2.002 | 0.275 | 2.002 | 0.275 |
| (0.580–6.775) | (2.607–3.171) | (0.580–6.775) | (0.580–6.775) | |||||
p < 0.05; H2 was the reference haplotype
Discussion
In the present study we have used haplotype based approach to investigate the association of genetic variability in SLC19A1 gene with the high-dose MTX-induced ADEs in paediatric ALL and NHML patients. SLC19A1 is one of the main influx transporters for folates and antifolates, however the reports on the impact on genetic variability on the MTX treatment outcome are conflicting. Protective role of rs1051266 AA genotype14,15, as well as increased risk for MTX-induced ADEs10 or no association of rs1051266 was reported.11,13,16
Among the so far published studies on the role of SLC19A1 in the treatment of ALL with MTX all studies but one19 have only investigated the most common functional SNP rs1051266 (SLC19A1 G80A), which may alter the transport characteristics of SLC19A1.28 By using a haplotype based approach we were able to investigate the impact of genetic variability of SNPs that are in linkage disequilibrium within the broader region within of the SLC19A1 on the occurrence of MTX-induced ADEs and disease outcome and effectiveness of treatment with HD-MTX in children and adolescents with ALL/NHML.
Among the investigated SLC19A1 polymorphisms univariate logistic regression analysis showed that the rs2838958 TT genotype increases the risk of developing mucositis. This association remained borderline significant in the multivariate analysis using the additive model. The other SLC19A1 polymorphisms were not associated with the occurrence of MTX-induced ADEs.
In the haplotype based analysis we found that H4 haplotype (TGTTCCG), which, excluding the last locus (rs2838951), has wild type alleles present, statistically significantly reduces the risk for the occurrence of ADEs during treatment with HD-MTX. Since the frequency is low in our sample it is difficult to ascertain its clinical relevance given the number of patients and the small number of individual toxic events.
Haplotype based approaches have been rarely used when investigating HD-MTX induced adverse events in ALL and NHML patients. One study investigated the influence of MTHFR SNPs and haplotypes on treatment response29 and HD-MTX toxicity30,31, while the other studies focused on DNA repair and cell cycle processes on risk for leukaemia development using haplotype based approach.32-36
Only study of Lopez-Lopez focused on MTX transporters. This study examined 384 SNPs in 12 transporter genes, including SLC19A1, and their relationship to MTX plasma levels and MTX-related toxicities.19 They found no association between SLC19A1 haplotypes and MTX plasma levels. However they confound the results of Treviño and colleagues on association between SLCO1B1 rs11045879 polymorphism and toxicity and observed for the first time a significant association with MTX plasma levels and rs9516519 in ABCC4, rs3740065 and haplotype GCGGG in ABCC2.19
However, haplotype based approach has been used in analysing the influence of the polymorphisms in the broader region of SLC19A1 on the occurrence of low dose MTX-induced ADEs in rheumatoid arthritis (RA) patients in order to facilitate the design of personalized low dose MTX treatment.37,38 Bohanec Grabar et al. reported that individual SNP and haplotype analysis suggest that rs1051266 could be a functional variant altering MTX toxicity; rs1051266 and rs1131956 were significantly associated with protection against discontinuation of treatment owing to MTX toxicity. Rs2838956 was significantly associated with protection against skin ADEs, while two common SLC19A1 haplotypes carrying rs1051266 and rs1131596 minor alleles had a protective effect towards MTX induced ADEs. Significant association was also found for rs1051266 and rs1131596 with infection.37 Lima et al. reported that SLC19A1 and genotypes and haplotypes may help to identify RA patients with increased risk of MTX-related gastrointestinal toxicity since protective effect of carriers of wild type allele of rs1051266 and rs2838956 regarding MTX induced gastrointestinal toxicity as well as association between GGAG haplotype for SLC19A1 rs7499, rs1051266, rs2838956 and rs3788200 with MTX gastrointestinal toxicity was established.38
The results of the haplotype based analysis indicate, that other polymorphisms that are in linkage disequilibrium with rs1051266 and rs1131596, may impact the functioning of SLC19A1 via, for example, alterations in SLC19A1 splicing.39,40 As well, other genes coding for proteins that are involved in the transport and metabolism of MTX can also impact pharmacokinetics and the occurrence of MTX-induced AE, such as SLCO1B1 SNPs that are associated with lower clearance of MTX and SLCO1A2 as well.41 Furthermore, SNP in miR-595 which might affect SLC19A1 regulation and could affect MTX levels in patients with paediatric B-cell ALL was recently reported.18
The results of our study show that SLC19A1 genotyping could be one of the factors that could contribute to safer and more effective treatment with HD-MTX, especially for identifying patient groups at risk of mucositis development in Slovenian patients. As the occurrence of ADEs and the treatment outcome could be affected by other genetic factors involved in the transport and metabolism of MTX as well as by concomitant supportive therapy and the current pathophysiological condition of the patient the results of our study should be confirmed in a larger number of patients.
The main limitation of our study, which is probably also one of the reasons for the inconsistent results among studies published so far, is the limited sample size. Since ALL is a rare disease in children, the number of patients that could be as comparable as possible in the entire treatment process is limited. We therefore also included five patients with NHML in the study, who were treated according to the same protocol as patients with ALL.
The great advantage of our study is that all subjects come from a small and very homogeneous population42, that the patient group is clinically well defined and very homogeneous given the type and course of treatment, centralized treatment of patients with an established treatment and supportive regimen. The group was treated by the same medical team in a single centre according to standard criteria.
For genotyping, we used a method that would be suitable for genetic testing in clinical practice, as it is easy to perform, fast, affordable, and enables simultaneous analysis of multiple samples and different polymorphisms if they are run under the same conditions, and also require small amounts of DNA for analysis.
Employing a haplotype based approach we tried to examine the impact of genetic variability in the entire region of the studied gene and also included the possible functional polymorphisms that are in linkage disequilibrium with the selected SNPs. With this approach an even stronger influence of genetic variability of SLC19A1 with MTX-induced ADEs could be shown as opposed to taking into account only individual SNPs.
The observations of our study provide additional information to the ongoing discussion about the most suitable biological markers to be evaluated during treatment with HD-MTX in order to achieve more efficient, safer and more rational treatment of children and adolescents with ALL. Additional prospective pharmacogenetic studies on higher sample sizes are needed to further evaluate a possible impact of genetic variability of the MTX transporters that have an impact on pharmacokinetics of MTX and enzymes involved in MTX metabolism that could help us to identify patients that are threatened by serious ADEs during HD-MTX treatment.
Footnotes
Disclosure: No potential conflicts of interest were disclosed.
Supplementary Material Details
References
- 1.Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE. Pharmacogenomics and individualized medicine: Translating science into practice. Clin Pharmacol Ther. 2012;92:467–75. doi: 10.1038/clpt.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV, Rodman JH, Crom WR, Boyett JM, Pui CH. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. N Engl J Med. 1998;338:499–505. doi: 10.1056/NEJM199802193380803. [DOI] [PubMed] [Google Scholar]
- 3.Murphy SB, Fairclough DL, Hutchison RE, Berard CW. Non-Hodgkin’s lymphomas of childhood: an analysis of the histology, staging, and response to treatment of 338 cases at a single institution. J Clin Oncol. 1989;7:186–93. doi: 10.1200/JCO.1989.7.2.186. [DOI] [PubMed] [Google Scholar]
- 4.Anderson JR, Jenkin RD, Wilson JF, Kjeldsberg CR, Sposto R, Chilcote RR. et al. Long-term follow-up of patients treated with COMP or LSA2L2 therapy for childhood non-Hodgkin’s lymphoma: a report of CCG-551 from the Childrens Cancer Group. J Clin Oncol. 1993;11:1024–32. doi: 10.1200/JCO.1993.11.6.1024. [DOI] [PubMed] [Google Scholar]
- 5.Shaw GM, Lammer EJ, Zhu HP, Baker MW, Neri E, Finnell RH. Maternal periconceptional vitamin use, genetic variation of infant reduced folate carrier (A80G), and risk of spina bifida. Am J Med Genet. 2002;108:1–6. doi: 10.1002/ajmg.10195. [DOI] [PubMed] [Google Scholar]
- 6.Assaraf YG. The role of multidrug resistance efflux transporters in antifolate resistance and folate homeostasis. Drug Resist Updat. 2006;9:227–46. doi: 10.1016/j.drup.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Zeng H, Chen ZS, Belinsky MG, Rea PA, Kruh GD. Transport of methotrexate (MTX) and folates by multidrug resistance protein (MRP) 3 and MRP1: effect of polyglutamylation on MTX transport. Cancer Res. 2001;61:7225–32. [PubMed] [Google Scholar]
- 8.Pharmacogenomics. Knowledge. Implementation. 2017 Feb 06 www.pharmgkb.org The Pharmacogenomics Knowledgebase. [Google Scholar]
- 9.Laverdiere C, Chiasson S, Costea I, Moghrabi A, Krajinovic M. Polymorphism G80A in the reduced folate carrier gene and its relationship to methotrexate plasma levels and outcome of childhood acute lymphoblastic leukemia. Blood. 2002;100:3832–4. doi: 10.1182/blood.V100.10.3832. [DOI] [PubMed] [Google Scholar]
- 10.Imanishi H, Okamura N, Yagi M, Noro Y, Moriya Y, Nakamura T. et al. Genetic polymorphisms associated with adverse events and elimination of methotrexate in childhood acute lymphoblastic leukemia and malignant lymphoma. J Hum Genet. 2007;52:166–71. doi: 10.1007/s10038-006-0096-z. [DOI] [PubMed] [Google Scholar]
- 11.Kishi S, Griener J, Cheng C, Das S, Cook EH, Pei D. et al. Homocysteine, pharmacogenetics, and neurotoxicity in children with leukemia. J Clin Oncol. 2003;21:3084–91. doi: 10.1200/JCO.2003.07.056. [DOI] [PubMed] [Google Scholar]
- 12.Shimasaki N, Mori T, Samejima H, Sato R, Shimada H, Yahagi N. et al. Effects of methylenetetrahydrofolate reductase and reduced folate carrier 1 polymorphisms on high-dose methotrexate-induced toxicities in children with acute lymphoblastic leukemia or lymphoma. J Pediatr Hematol Oncol. 2006;28:64–8. doi: 10.1097/01.mph.0000198269.61948.90. [DOI] [PubMed] [Google Scholar]
- 13.Faganel Kotnik B, Dolzan V, Grabnar I, Jazbec J. Relationship of the reduced folate carrier gene polymorphism G80A to methotrexate plasma concentration, toxicity, and disease outcome in childhood acute lymphoblastic leukemia. Leuk Lymphoma. 2010;51:724–6. doi: 10.3109/10428191003611402. [DOI] [PubMed] [Google Scholar]
- 14.Shimasaki N, Mori T, Torii C, Sato R, Shimada H, Tanigawara Y. et al. Influence of MTHFR and RFC1 polymorphisms on toxicities during maintenance chemotherapy for childhood acute lymphoblastic leukemia or lymphoma. J Pediatr Hematol Oncol. 2008;30:347–52. doi: 10.1097/MPH.0b013e318165b25d. [DOI] [PubMed] [Google Scholar]
- 15.Faganel Kotnik B, Grabnar I, Bohanec Grabar P, Dolžan V, Jazbec J. Association of genetic polymorphism in the folate metabolic pathway with methotrexate pharmacokinetics and toxicity in childhood acute lymphoblastic leukaemia and malignant lymphoma. Eur J Clin Pharmacol. 2011;67:993–1006. doi: 10.1007/s00228-011-1046-z. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Tissing WJ, de Jonge R, van Zelst BD, Pieters R. Polymorphisms in folate-related genes: association with side effects of high-dose methotrexate in childhood acute lymphoblastic leukemia. Leukemia. 2008;22:1798–800. doi: 10.1038/leu.2008.66. [DOI] [PubMed] [Google Scholar]
- 17.Zgheib NK, Akra-Ismail M, Aridi C, Mahfouz R, Abboud MR, Solh H. et al. Genetic polymorphisms in candidate genes predict increased toxicity with methotrexate therapy in Lebanese children with acute lymphoblastic leukemia. Pharmacogenet Genomics. 2014;24:387–96. doi: 10.1097/FPC.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 18.Iparraguirre L, Gutierrez-Camino A, Umerez M, Martin-Guerrero I, Astigarraga I, Navajas A. et al. MiR-pharmacogenetics of methotrexate in childhood B-cell acute lymphoblastic leukemia. Pharmacogenet Genomics. 2016;26:517–25. doi: 10.1097/FPC.0000000000000245. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Lopez E, Ballesteros J, Piñan MA, Sanchez de Toledo J, Garcia de Andoin N, Garcia-Miguel P. et al. Polymorphisms in the methotrexate transport pathway: a new tool for MTX plasma level prediction in pediatric acute lymphoblastic leukemia. Pharmacogenet Genomics. 2013;23:53–61. doi: 10.1097/FPC.0b013e32835c3b24. [DOI] [PubMed] [Google Scholar]
- 20.Ribas G, González-Neira A, Salas A, Milne RL, Vega A, Carracedo B. et al. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet. 2006;118:669–79. doi: 10.1007/s00439-005-0094-9. [DOI] [PubMed] [Google Scholar]
- 21.Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G. et al. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–7. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 22.Handbook for reporting results of cancer treatment. Geneva: 1979. pp. 15–22. World Health Organization. [Google Scholar]
- 23.Common Toxicity Criteria Version 1. 2017 Feb 06 http://ctep.cancer.gov/reporting/ctc.html National Cancer Institute. [Google Scholar]
- 24.HaploView software version 4.1. 2017 Feb 06 http://www.broad.mit.edu/haploview/haploview-downloads Broad Institute. [Google Scholar]
- 25.Science for a safer world. 2017 Feb 06 http://www.kbioscience.co.uk LGC. [Google Scholar]
- 26.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;1796:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscow JA, Gong M, He R, Sgagias MK, Dixon KH, Anzick SL. et al. Isolation of a gene encoding a human reduced folate carrier (RFC1) and analysis of its expression in transport-deficient, methotrexate-resistant human breast cancer cells. Cancer Res. 1995;55:3790–4. [PubMed] [Google Scholar]
- 28.Whetstine JR, Gifford AJ, Witt T, Liu XY, Flatley RM, Norris M. et al. Single nucleotide polymorphisms in the human reduced folate carrier: characterization of a high-frequency G/A variant at position 80 and transport properties of the His(27) and Arg(27) carriers. Clin Cancer Res. 2001;7:3416–22. [PubMed] [Google Scholar]
- 29.Aplenc R, Thompson J, Han P, La M, Zhao H, Lange B. et al. Methylenetetrahydrofolate reductase polymorphisms and therapy response in pediatric acute lymphoblastic leukemia. Cancer Res. 2005;65:2482–7. doi: 10.1158/0008-5472.CAN-04-2606. [DOI] [PubMed] [Google Scholar]
- 30.Kałuzna E, Strauss E, Zając-Spychała O, Gowin E, Świątek-Kościelna B, Nowak J. et al. Functional variants of gene encoding folate metabolizing enzyme and methotrexate-related toxicity in children with acute lymphoblastic leukemia. Eur J Pharmacol. 2015;769:93–9. doi: 10.1016/j.ejphar.2015.10.058. [DOI] [PubMed] [Google Scholar]
- 31.Liao QC, Li XL, Liu ST, Zhang Y, Li TY, Qiu JC. [Association between the methylenetetrahydrofolate reductase gene polymorphisms and haplotype with toxicity response of high dose methotrexate chemotherapy]. [Chinese] Zhonghua Liu Xing Bing Xue Za Zhi. 2012;33:735–9. [PubMed] [Google Scholar]
- 32.Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, Macias-Gomez N, Ayala-Madrigal ML, Barros-Nuñez P. et al. XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res. 2009;8:1451–8. doi: 10.4238/vol8-4gmr687. [DOI] [PubMed] [Google Scholar]
- 33.Chokkalingam AP, Bartley K, Wiemels JL, Metayer C, Barcellos LF, Hansen HM. et al. Haplotypes of DNA repair and cell cycle control genes, X-ray exposure, and risk of childhood acute lymphoblastic leukemia. Cancer Causes Control. 2011;22:1721–30. doi: 10.1007/s10552-011-9848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosor M, Ziółkowska I, Januszkiewicz-Lewandowska D, Nowak J. Polymorphisms and haplotypes of the NBS1 gene in childhood acute leukaemia. Eur J Cancer. 2008;44:2226–32. doi: 10.1016/j.ejca.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 35.Semsei AF, Erdélyi DJ, Ungvári I, Kámory E, Csókay B, Andrikovics H. et al. Association of some rare haplotypes and genotype combinations in the MDR1 gene with childhood acute lymphoblastic leukaemia. Leuk Res. 2008;32:1214–20. doi: 10.1016/j.leukres.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Pakakasama S, Sirirat T, Kanchanachumpol S, Udomsubpayakul U, Mahasirimongkol S, Kitpoka P. et al. Genetic polymorphisms and haplotypes of DNA repair genes in childhood acute lymphoblastic leukaemia. Pediatr Blood Cancer. 2007;48:16–20. doi: 10.1002/pbc.20742. [DOI] [PubMed] [Google Scholar]
- 37.Bohanec Grabar P, Leandro-García LJ, Inglada-Pérez L, Logar D, Rodríguez-Antona C. Dolžan V. Genetic variation in the SLC19A1 gene and methotrexate toxicity in rheumatoid arthritis patients. Pharmacogenomics. 2012;13:1583–94. doi: 10.2217/pgs.12.150. [DOI] [PubMed] [Google Scholar]
- 38.Lima A, Bernardes M, Azevedo R, Monteiro J, Sousa H, Medeiros R, Seabra V. SLC19A1, SLC46A1 and SLCO1B1 polymorphisms as predictors of methotrexate-related toxicity in Portuguese rheumatoid arthritis patients. Toxicol Sci. 2014;142:196–209. doi: 10.1093/toxsci/kfu162. [DOI] [PubMed] [Google Scholar]
- 39.Williams FM, Flintoff WF. Structural organization of the human reduced folate carrier gene: evidence for 5’ heterogeneity in lymphoblast mRNA. Somat Cell Mol Genet. 1998;24:143–56. doi: 10.1023/b:scam.0000007117.50428.63. [DOI] [PubMed] [Google Scholar]
- 40.Whetstine JR, Flatley RM, Matherly LH. The human reduced folate carrier gene is ubiquitously and differentially expressed in normal human tissues: identification of seven non-coding exons and characterization of a novel promoter. Biochem J. 2002;367:629–40. doi: 10.1042/BJ20020512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trevino LR, Shimasaki N, Yang W, Panetta JC, Cheng C, Pei D. et al. Germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–8. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidan-Jeras B, Jurca B, Dolzan V, Jeras M, Breskvar K, Bohinjec M. Gjertson DW, Terasaki PI. Caucasian Slovenian normal. Lenexa: American society for histocompability and immunogenetics; 1998. 1998. pp. 180–1. HLA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


