Summary
The variation of macrophage functions suggests the involvement of multiple signalling pathways in fine tuning their differentiation. Macrophages that originate from monocytes in the blood migrate to tissue in response to homeostatic or ‘danger’ signals and undergo substantial morphological and functional modifications to meet the needs of the dominant signals in the microenvironment. Wnts are secreted glycoproteins that play a significant role in organ and cell differentiation, yet their impact on monocyte differentiation is not clear. In this study, we assessed the role of Wnt1 and Wnt7a on the differentiation of monocytes and the subsequent phenotype and function of monocyte‐derived macrophages (MDMs). We show that Wnt7a decreased the expression of CD14, CD11b, CD163 and CD206, whereas Wnt1 had no effect. The Wnt7a effect on CD11b was also observed in the brain and spleen of Wnt7a−/− adult brain mouse tissue and in embryonic Wnt7a−/− tissue. Wnt7a reduced the phagocytic capacity of M‐MDMs, decreased interleukin‐10 (IL‐10) and IL‐12 secretion and increased IL‐6 secretion. Collectively, these findings demonstrate that Wnt7a generates an MDM phenotype with both pro‐inflammatory and alternative MDM cytokine profiles and reduced phagocytic capacity. As such, Wnt7a can have a significant impact on macrophage responses in health and disease.
Keywords: myeloid cell, Wnt, monocyte, macrophage, regulation/suppression
Introduction
Macrophages exists as a continuum of subtypes, each with its own unique function including phagocytosis, antigen processing and presentation, and release of tissue remodelling factors, homeostatic proteins, and pro‐ and anti‐inflammatory cytokines. Due to their importance in health and disease, understanding the signals that regulate macrophage differentiation and phenotype is paramount to gaining a better understanding of their functions as well as revealing potential therapeutic targets. Circulating blood monocytes differentiate into specific macrophage phenotypes depending on the signals they receive.1, 2 Macrophage colony‐stimulating factor (M‐CSF) with lipopolysaccharide (LPS) and interferon‐γ? (IFN‐γ) induce monocytes to differentiate into M‐1 (pro‐inflammatory) like‐monocyte‐derived macrophage (MDMs). M‐CSF with the addition of interleukin‐4 (IL‐4), IL‐10, or transforming growth factor‐β generates an alternative/regulatory‐like macrophage phenotype (M2‐MDMs). M2a‐MDMs, differentiated in the presence of IL‐4, exhibit increased expression of scavenger receptors, CD163 and CD206, and have increased phagocytic capacity.3, 4 Other morphogens can skew the differentiation of monocytes, impacting their function in tissue. In particular, emerging data point to the impact of Wnts on monocyte differentiation; specifically, Wnt5a expression is associated with monocyte‐derived dendritic cells, and in haematopoietic cells there are more monocyte or pro‐monocyte cells in the presence of either Wnt5a or Wnt11, and the appearance of macrophages was inhibited.5, 6
Wnts are ubiquitously expressed proteins that guide cell fate, development and differentiation in utero and a growing body of evidence points to their importance in cell differentiation and function post‐development.7, 8, 9, 10, 11, 12, 13, 14, 15 Wnts are a family of 19 small secreted glycoproteins that are evolutionary conserved. They bind to a seven transmembrane Frizzled receptor and one of various co‐receptors – culminating in either a β‐catenin‐dependent (canonical) or independent (non‐canonical: i.e. Planar Cell Polarity or Ca2+/Calmodulin) signalling pathway. Historically, Wnts (1, 2, 2b, 3, 3a, 4, 5a, 5b, 6, 7a, 7b, 8a, 8b, 9a, 9b, 10a, 10b, 11 and 16) were categorized on the basis of whether they induce β‐catenin‐dependent or β‐catenin‐independent signalling; however, there is an increasing recognition that this classification is no longer accurate. Wnts, depending on interaction with frizzled receptors, can mediate either pathway. For example, Wnt5a, classically, signals through the non‐canonical/β‐catenin‐independent pathway.16 However, by binding to Frizzled 4 and the LRP5 co‐receptor, Wnt5a engages the canonical/β‐catenin‐dependent pathway.16 These studies underscore an emerging paradigm of Wnt signalling, whereby the context of Wnt receptors dictates canonical versus non‐canonical Wnt pathways.16, 17
We focused here on evaluating the role of Wnt1 and Wnt7a in regulating monocyte differentiation, both phenotypically and functionally. Both Wnts engage the canonical β‐catenin signalling pathway, whereby they bind to Frizzled receptors and LRP5/6 to induce a signalling cascade that inactivates glycogen synthetase kinase‐3β to stabilize β‐catenin. β‐Catenin in turn translocates to the nucleus where it binds to the T‐cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors to regulate gene expression. Although Wnt1 predominantly induces canonical β‐catenin signalling, Wnt7a can also activate the AKT/mammalian target of rapamycin (AKT/mTOR) pathway, albeit in skeletal muscles,18 and peroxisome proliferator‐activated receptor γ (PPARγ) via activation of extracellular signal‐regulated kinase 5 (ERK5).19 Wnt7a is expressed by stromal epithelial cells;20 it is required for the development of the blood–brain barrier21, 22, 23 and has also been observed to induce tumour‐suppressive cellular senescence in the lung.24 Notably, macrophages within tumours are known regulators of tumour progression, as signals from the tumour microenvironment regulate the macrophage phenotype, which in turn regulates the phenotype and function of infiltrating immune cells.25, 26 Therefore, the effect of Wnt7a on lung cancer progression may be through its influence on macrophage phenotype. As such, due to the ability of Wnt7a to activate more than one signalling pathway that potentially influences cell phenotype and function, as well as its effect on blood–brain barrier development and lung cancer pathogenesis, we hypothesize that Wnt7a may be important in the differentiation of monocytes to MDMs in the brain and other tissue. Relative to Wnt7a, the influence of Wnt1 on various cell types and disease processes has been more widely described; where in macrophages, Wnt1 induces CD36 expression27 and prevents apoptosis in microglia.28
We report here that activated monocytes down‐regulate Wnt7a expression and that Wnt7a, but not Wnt1, significantly inhibited the expression of CD14, CD11b, CD163 and CD206 on MDMs. The aforementioned proteins are classical markers associated with myeloid cell phenotype and function, specifically; CD14 is a pathogen recognition receptor (PRR) found on human monocytes and macrophages.29 CD11b is the α subunit of the integrin heterodimer CR3 or Mac‐1; known functions include cellular migration,30 phagocytosis,31 and cellular activation.32 CD11b is primarily expressed on myeloid cells and neutrophils and is essential for their defining functions; hence, assessing the expression of this protein is important when defining monocyte and macrophage subtypes. CD163 and CD206 are scavenger receptors and mannose receptors responsible for clearing low‐density lipoproteins and glycoproteins, respectively.33, 34
In vitro observations of the Wnt7a effect on CD11b were also made in an in vivo Wnt7a−/− mouse model, where absence of Wnt7a resulted in higher CD11b expression relative to the wild‐type mice. Wnt7a also inhibited the phagocytic capacity of MDMs, inhibited the secretion of IL‐10 from alternative MDM controls, and inhibited the secretion of IL‐12 from M1‐like MDMs. Conversely, IL‐6 secretion was significantly increased relative to all MDMs, including MDMs differentiated in the presence of Wnt1. Overall, these data demonstrate that Wnt7a induces monocytes to differentiate towards a unique phenotype with both M1‐like and alternative MDM properties. As such, Wnt7a can potentially be used as a tool to influence the MDM phenotype for therapeutic intervention for example, where anti‐inflammatory macrophages dominate and inhibit appropriate immune responses.
Materials and methods
Ethics statement
Research involving human subjects was conducted in accordance with institutional (IRBL06080703) and U.S. Government guidelines on human research.
Animal care
Animal experiments were conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by respective institutional Animal Care and Use (IACCU) committees. Adult animals were housed in the Biologic Resources Laboratory (BRL), UIC under protocol (15‐264). Wnt7a −/− embryos (e13.5) (Parr and McMahon, 1995) (JAX 004715) were from the Frederick National Laboratory in Fredrick, MD, under IACUC protocol (ASP 14‐078).
Wnt7a−/− mice
Wnt7a −/− mice (Parr and McMahon, 1995) were used as frozen embryos (e13.5) and 11‐ to 12‐week‐old adults. Wnt7a−/− adult mice and their wild‐type (WT) littermates are inbred strains on a B6/129S1 genetic background (The Jackson Laboratory, Bar Harbor, ME; Stock number 004715). Wnt7a−/− mice were bred until congenic in C57BL/6J strain. As the homozygous Wnt7a−/− mice were sterile, we made use of heterozygotes for colony maintenance. The genetic background of the mice was determined using PCR of DNA from tail biopsies. For embryos; after removing a limb for genotyping, the embryos were briefly fixed in 2% paraformaldehyde in PBS before being embedded in optimal cutting temperature compound (OCT) and snap frozen. Adult mice were killed using CO2 and perfused with PBS, before removal of brain and spleen. Half of the adult brain was fixed overnight in 2% paraformaldehyde followed by 30% sucrose and fixed in OCT blocks before cryo‐sectioning. Mouse spleens were crushed and incubated with red blood cell lysis buffer (Clontech Laboratories, Mountain View, CA) before cell culture experiments.
Generation of monocyte‐derived macrophages
Monocyte populations were enriched directly from healthy human whole blood by negative immune‐selection (STEMCELL Technologies, Vancouver, BC, Canada). Approximately 0·5 × 106 cells/ml were cultured in RPMI‐1640 supplemented with 10% fetal bovine serum (FBS) for 7 days in the presence of 50 ng/ml M‐CSF (R&D Systems, Minneapolis, MN). To obtain M1‐like and M2‐like MDM cells, 50 ng/ml IFN‐γ (R&D Systems) and 1 μg/ml LPS (Sigma‐Aldrich, St Louis, MO, USA) or 10 ng/ml IL‐4 (R&D Systems), respectively, were added. Experimental groups were treated with 50 ng/ml M‐CSF in addition to 300 ng/ml of either Wnt1 or Wnt7a (Abcam, Cambridge, UK) active full‐length human recombinant proteins. Growth factors, cytokines and Wnt proteins were added every other day, whereas IL‐4 was added during the last 48 hr of the experiment, IFN‐γ was added 48 hr before, followed by stimulation with LPS 24 hr before ending the experiment. Mouse MDMs were obtained by culturing splenocytes overnight in complete macrophage medium. Briefly, complete macrophage medium consisted of Dulbecco's modified Eagle's medium with high glucose and pyridoxine hydrochloride (Gibco, Waltham, MA), heat‐inactived fetal calf serum, modified Eagle's medium essential amino acids (Gibco), l‐glutamine (Gibco), HEPES buffer (Gibco), penicillin and streptomycin, and 50 mm 2‐mercaptoethanl. After overnight activation and adherence of monocytes, suspension cells were removed and adherent cells were washed twice with Dulbecco's PBS (Corning, Manassas, VA) before replacing complete macrophage medium. Cells were cultured for 7 days followed by overnight incubation with Wnt7a and subsequent flow cytometric analysis.
Multiplex assay
Secretion of IL‐6, IL‐1β, IL‐10, IL‐12p70 and tumour necrosis factor‐α (TNF‐α) from differentiated MDMs were measured with a human cytokine magnetic 5‐Plex Panel (EMD Millipore, St Louis, MO) according to the manufacturer's protocol.
Phagocytosis assay
Phagocytosis was assessed on 7‐day differentiated MDMs using pHrodo Staphylococcus aureus bioparticles (Life Technologies, Carlsbad, CA) following the manufacturer's protocol scaled to a 12‐well format. Briefly, differentiated macrophages were cultured with pHrodo S. aureus bioparticles for 90 min, washed with PBS and fluorescence signal was assessed by microscopy and by flow cytometry to calculate median fluorescence intensity to identify phagocytic cells.
Flow cytometry
Differentiated macrophages were assessed by flow cytometry for extracellular markers: lineage marker CD3/CD19/CD20/CD56 (allophycocyanin; Biolegend, San Diego, CA), CD14 (BV786; Biolegend), CD16 (Peridinin chlorophyll protein‐Cy7; BD Biosciences, San Jose, CA), CD11b (allophycocyanin‐Cy7; BD Biosciences), HLA‐DR (Peridinin chlorophyll protein; BD Biosciences), CD163 (PeTexas Red, BD Biosciences) and CD206 (phycoerythrin, FITC; BD Biosciences). Data were collected on the BD Fortessa Flow Cytometer with bd facs diva software (BD Biosciences) and analysed using flow jo flow Analysis Software (Treestar Inc., Ashland, OR).
Immunofluorescence
Mouse embryos and adult brain cryosections (20 μm) from Wnt7a −/− and WT mice were stained with F4/80 (1 : 100, anti‐rabbit Cy3; Abcam), CD11b (1 : 100, anti‐rat AF647; Abcam), isolectin b4 (1 : 100, AF488; Life Technologies), and DAPI (Molecular Probes, Eugene, OR) and visualized using an LSM 710 or LSM 700 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Quantitative real‐time RT‐PCR
Total RNA was extracted using the RNeasy mini prep kit (Qiagen, Hilden, Germany), followed by incubation with DNase (Sigma‐Aldrich) and retrotranscription (cDNA qScript; Quantabio, Beverly, MA). Quantitative real‐time RT‐PCR was performed using Ssofast Evagreen Supermix with a Low Rox kit (Bio‐Rad, Hercules, CA) in a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) using 7500 software v2.0.1. Melting curve analysis was performed to ensure the amplification of a single product. The change in binding was calculated by relative quantification using the comparative threshold cycle (CT) method, with results reported as fold change relative to control (ΔCT = CT Target – CT control; ΔΔCt = ΔCT Target – ΔCT experimental control; fold change relative to control = 2−ΔΔCt). Primers were designed using IDTDNA (San Diego, CA) primerquest design software. They are: Wnt1 Forward GAA CGC TCT CTT CCA GTT CTC, Wnt1 Reverse GGA GAG ATG GAT CGC TAT GAA C; Wnt2 Forward 5′‐TAC TGT ATC AGG GAC CGA GAG, Wnt2 Reverse GCC TCT CCC ACA ACA CAT AA; Wnt2b Forward 5′‐GAT GGA TTT AGG AGG CTG AGT G‐3′, Wnt2b Reverse GAG GAT GAG CCT TGA CTT AGT G; Wnt3 Forward CTG GAC CAC ATG CAC CTA AA, Wnt3 Reverse CGT ACT TGT CCT TGA GGA AGT C; Wnt3a Forward GGC AGC TGT GAA GTG AAG A, Wnt3a Reverse GGT GTT TCT CTA CCA CCA TCT C; Wnt4 Forward CAG TGG AGA ACT GGA GAA GTG, Wnt4 Reverse TCC ACA AAG GAC TGT GAG AAG; Wnt5a Forward GCT AGA GAA AGG GAA CGA ATC C, Wnt5a Reverse CCA GAC ACT CCA TGA CAC TTA C; Wnt5b Forward CGA GAG CGT GAG AAG AAC TTT, Wnt5b Reverse GGC GAC ATC AGC CAT CTT AT; Wnt6 Forward TCA GTT CCA GTT CCG TTT CC, Wnt6 Reverse GAA AGC TGT CTC TCG GAT GTC; Wnt7a Forward GGT GCG AGC ATC ATC TGT AA, Wnt7a Reverse CAT TTG GGA GCC TTC TCC TAT G; Wnt7b Forward GGC AGT GTG GAT GGA TGT T, Wnt7b Reverse GGG TGT CCT CAA ATA GGG TTA G; Wnt8a Forward GTG TCA TGG CAT CTC AGG AA, Wnt8a Reverse GCG GTC ATA CTT GGC CTT TA; Wnt8b Forward CTA ACC GGG AGA CAG CAT TT, Wnt8b Reverse GAG TCA TCA CAG CCA CAG TT; Wnt9a Forward CAG TAC CAG TTC CGC TTT GA, Wnt9a Reverse GAA GAG ATG GCG TAG AGG AAA G; Wnt9b Forward GAA GCA GTG TGA CCT ACT GAA G, Wnt9b Reverse GCC TGA ACT GGA ACT GAC AT; Wnt10a Forward ACT CCG ACC TGG TCT ACT TT; Wnt10b Forward GAA TGC GGA TCC ACA ACA AC; Wnt11 Forward GGG CTT CAA AGG AAA CTG ATA GG, Wnt11 Reverse ATC CCT GCC CTG ACT TCT T; Wnt16 Forward GTC CTG CTA GCC ATG AAT CTA C, Wnt16 Reverse GGC AGT CCA CAG ACA TTA ACT; GAPDH Forward TGACTTCAACAGCGACACCCACT, GAPDH Reverse ACCACCCTGTTGCTGTAGCCAAAT.
Statistical analyses
When the data were distributed normally, analysis of variance and post‐hoc tests were used. When the data were not normally distributed, non‐parametric analysis was performed. All tests assumed a two‐sided significance level of 0·05 using graphpad instat 3 Software (San Diego, CA) for data analysis.
Results
Wnt7a mRNA expression decreases after monocyte activation
We evaluated mRNA expression of the 19 Wnts 24 hr after monocytes were cultured in the presence of M‐CSF (Table 1). At 24 hr, both Wnt2 and Wnt7a were significantly decreased, whereas the expression of Wnt1 and Wnt5a was significantly increased in comparison to freshly isolated monocytes. These data suggest that Wnts are differentially expressed upon monocyte activation at the early stages of differentiation.
Table 1.
Differential expression of Wnts mRNAs after monocyte activation
| Wnt | Fold change rel. to T = 0 hr |
|---|---|
| Wnt1 | 49·035a |
| Wnt2 | 0·194a |
| Wnt2b | 2·757 |
| Wnt3 | 7·117 |
| Wnt3a | 22·651 |
| Wnt4 | 5·267 |
| Wnt5a | 56·482a |
| Wnt5b | 18·389 |
| Wnt6 | 1·560 |
| Wnt7a | 0·118a |
| Wt7b | 8·641 |
| Wnt8a | 17·467 |
| Wnt8b | 35·426 |
| Wnt9a | 2·552 |
| Wnt9b | 19·885 |
| Wnt10a | 2·716 |
| Wnt10b | 5·179 |
| Wnt11 | 4·366 |
| Wnt16 | 2·516 |
mRNA expression of 19 Wnts measured from freshly isolated whole blood human monocytes and at 24 hr post M‐CSF stimulation. RNA was measured by RT‐qPCR using 19 custom designed Wnt primer pairs. C t values were normalized to the C t for GAPDH, followed by calculations of ΔC t and ΔΔC t and fold change calculated relative to T = 0. Data are representative of three donors
P < 0·05.
Wnt7a inhibits the expression of CD14, CD11b, CD163 and CD206
Isolation of monocytes, flow cytometry gating strategy/culture purity, generation of MDM, M1‐MDMs and M2a‐MDMs, and the treatment paradigm with Wnt1 or Wnt7a are described in the Supplementary material (Fig. S1). MDM, M1‐MDMs or M2a‐MDMs were treated with either Wnt1 or Wnt7a and expression of CD14, CD11b, CD163 and CD206 was evaluated by flow cytometry. CD14 and CD11b expression distinguish macrophages from other cell types. These proteins are involved in response to microbial products, adhesion and migration, and are therefore important for macrophage function.35, 36 CD163 and CD206 are scavenger and mannose receptors, respectively, often associated with alternative macrophage phenotypes and phagocytic function.37 Wnt7a significantly reduced CD14 expression on M‐MDM, M1‐MDM and M2a‐MDM in comparison with untreated cultures and in comparison with Wnt1‐treated cultures (Fig. 1a). Wnt7a also significantly inhibited the expression of CD11b on M‐MDM, M1‐MDM and M2a‐MDM (Fig. 1b) and reduced CD163 expression on M1‐MDM (P < 0·05), but not on M‐MDMs or M2a‐MDMs (Fig. 1c). Wnt7a reduced CD206 expression on both M‐MDM and M1‐MDM cells (P < 0·05) but not on M2a‐MDMs (Fig. 1d). The M2a‐MDM control expressed higher levels of CD163 and CD206 relative to the M‐MDM and M1‐MDM controls and Wnt7a addition to M2a‐MDMs did not decrease CD163 or CD206 expression (data not shown). Wnt5a, which was enhanced at the transcriptional level after monocyte activation (Table 1), did not show significant effects on these surface markers and as such was excluded from further analysis (data not shown). These data demonstrate that Wnt7a inhibits the expression of classical markers of macrophage differentiation and polarization in MDMs differentiated in vitro.
Figure 1.
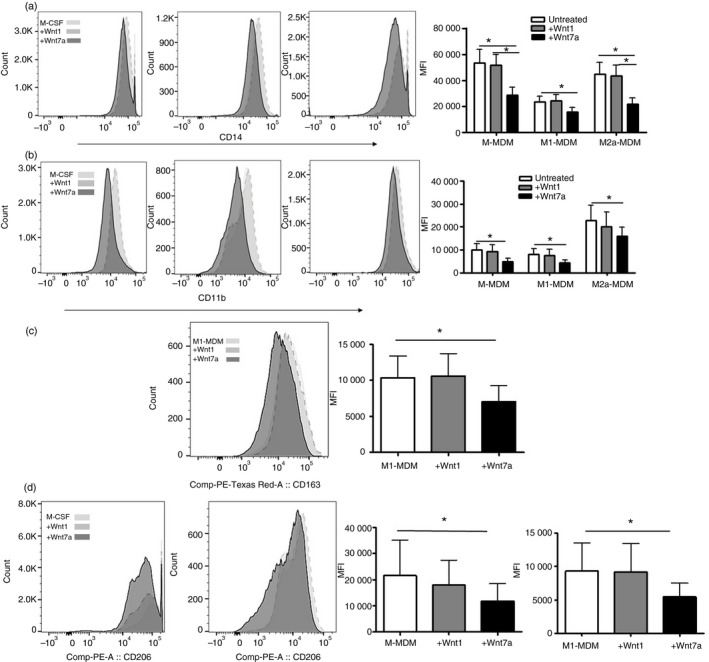
Wnt7a reduces CD14, CD11b, CD163, and CD206 expression on monocyte‐derived macrophages in vitro. Representative histogram for median fluorescence intensity (MFI), and cumulative data of CD14 (a), CD11b (b), CD163 (c), and CD206 (d) expression on human M‐MDM, M1‐MDM, M2a‐MDM with or without Wnt1 and Wnt7a treatments as described in the Supplementary material (Fig. S1) (n = 6 or n = 7 using four different donors, *P < 0·05 in comparison with monocyte‐derived macrophages (MDMs) or between M1‐MDM versus M1‐MDM+7a, as indicated.
Wnt7a−/− mice express higher levels of CD11b
To assess whether these in vitro observations translate to in vivo conditions, we assessed CD11b and F4/80 (a mouse macrophage marker) expression on Wnt7a−/− e13.5 mouse embryos and 11‐ to 12‐week‐old mice by immunofluorescence. Wnt7a−/− embryos had higher levels of F4/80 and CD11b expression than WT controls (Fig. 2a). To assess whether the difference observed in embryos also occurs in adult Wnt7a−/− mice, we obtained whole brain tissue from WT and Wnt7a −/− mice for immunofluorescence and flow cytometry analysis of the macrophage population. Adult brain tissue from Wnt7a −/− mice had a significantly higher frequency of CD11b+ staining relative to WT controls (Fig. 2b) as well as significantly higher median fluorescence intensity (Fig. 2c). Finally, to determine if exposure of macrophages from Wnt7a −/− mice to recombinant Wnt7a would further reduce CD11b expression, we differentiated monocytes from mouse splenocytes into macrophages in macrophage‐conditioned media then incubated the differentiated cells for 48 hr with Wnt7a protein. Macrophages differentiated from Wnt7a −/− animals showed significant reduction in CD11b expression after Wnt7a treatment (Fig. 2d). These data demonstrate that Wnt7a down‐regulates CD11b expression on monocytes both in vitro and in vivo.
Figure 2.
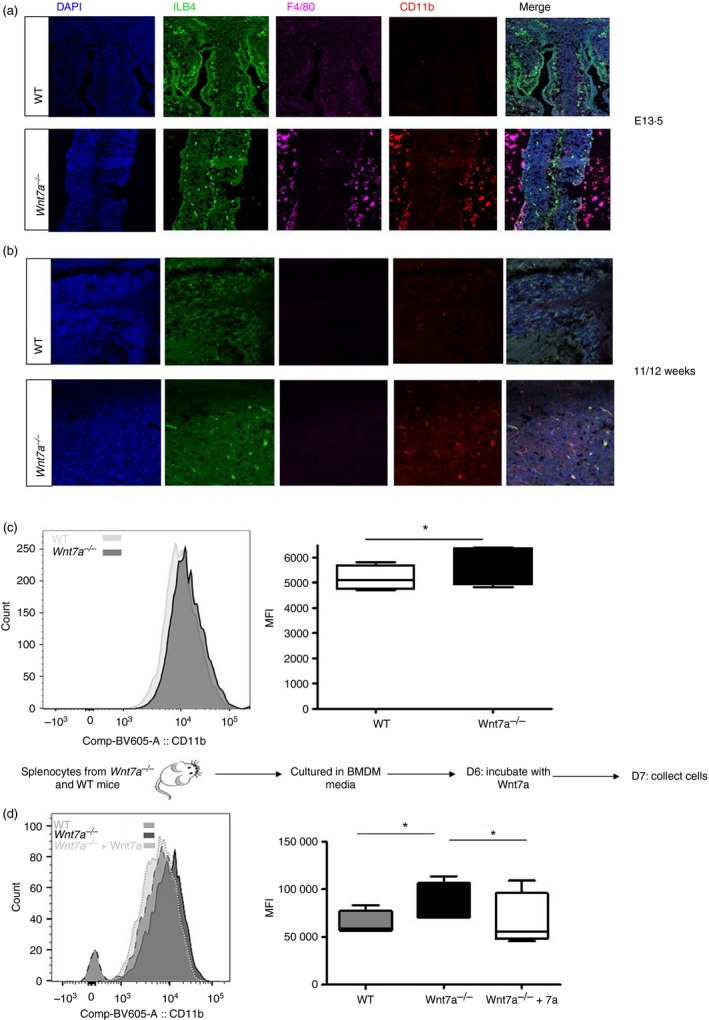
Wnt7a−/− mice have a higher expression of CD11b. Anti‐CD11b and mouse F4/80 antibodies were used to stain for mouse macrophages/microglia in E13.5 mouse embryo telencephalon (a) and in 11‐ to 12‐week‐old mouse brain cortex (b) for both wild‐type (WT) and Wnt7a−/− mice. Green: Isolectin B4, blue: DAPI, red: CD11b, magenta: F4/80. (c) Flow cytometric analysis of CD11b expression on macrophages/microglia isolated from the brain of Wnt7a WT and Wnta7a−/− mice represented as median fluorescence intensity (MFI). Data are representative of four mice in each group, P < 0·05. (d) Splenocytes isolated from Wnt7a−/− and WT mice were used to culture macrophages as indicated in the experimental paradigm then cells were stained for CD11b expression (n = 4 for each experimental group; *P < 0·05; WT versus Wnt7a−/− and Wnt7a−/− versus Wnt7a−/− +Wnt7a).
Wnt7a inhibits phagocytosis
Given that Wnt7a reduced the expression of two molecules associated with macrophage phagocytic capability, CD11b and CD206, we assessed the impact of Wnt7a on MDM phagocytic function. MDMs, after in vitro differentiation, were incubated with pHrodo S. aureus bioparticles for 90 min. M‐MDMs differentiated in the presence of Wnt7a showed a significantly reduced ability to take up foreign particles (Fig. 3a,b). Wnt1 or Wnt7a had no significant effect on the phagocytic capacity of M1‐MDMs or M2a‐MDMs. These data demonstrate that Wnt7a reduces the phagocytic potential of M‐MDM.
Figure 3.
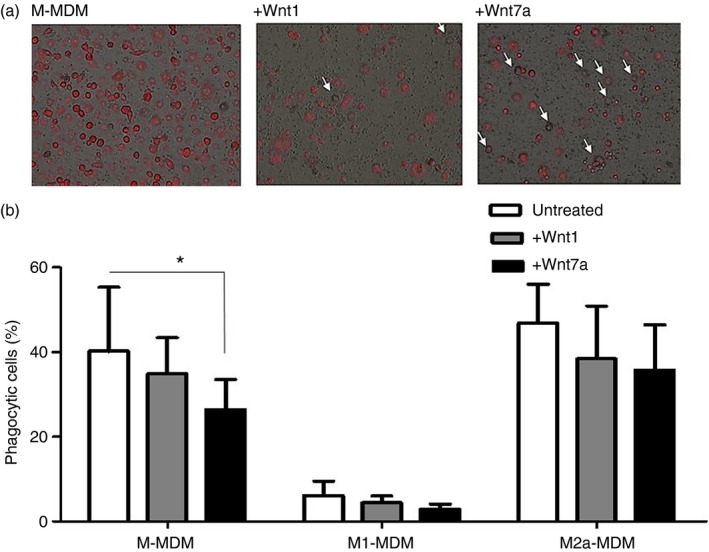
Wnt7a reduces phagocytic ability of monocyte‐derived macrophages (MDMs). (a) Phagocytic potential of human M‐MDM with or without Wnt1 or Wnt7a treatment was assessed using immune fluorescence (a) of pHrodo® Red Staphylococcus aureus Bioparticles®, white arrows indicate non‐phagocytic cells (n = 3) and by flow cytometry (b) for PE (excitation 585 nm) MFI of experimental group in comparison with control MDMs (n = 3, *P < 0·05 for M‐MDM versus M‐MDM + Wnt7a).
Wnt7a influences the cytokine profile of MDMs
To assess the impact of Wnt7a on cytokine secretion of MDMs, we collected cell culture supernatant of fully differentiated cells for analysis with a human cytokine magnetic 5‐Plex Panel for the Luminex™ for IL‐1β, TNF‐α, IL‐10, IL‐6 and IL‐12p70. In the presence of Wnt7a, there was a decrease in IL‐10 relative to the M‐MDM and M2a‐MDM controls (p < 0·05), but not relative to the M1‐MDM control (Fig. 4a). There was also a reduction in IL‐12p70 in the presence of Wnt7a, but not Wnt1, in the M1‐MDM control, and IL‐12p70 was undetectable in M‐MDM and M2a‐MDMs (Fig. 4b). Interleukin‐6 secretion was significantly increased in the presence of Wnt7a relative to all MDM controls (Fig. 4c). Neither IL‐1β nor TNF‐α levels were significantly altered by Wnt7a (Fig. 4d,e). These data demonstrate that Wnt7a influences cytokine secretion from MDMs, particularly in a manner that does not promote classical polarization of cytokine responses associated with M1 or M2 phenotypes.
Figure 4.
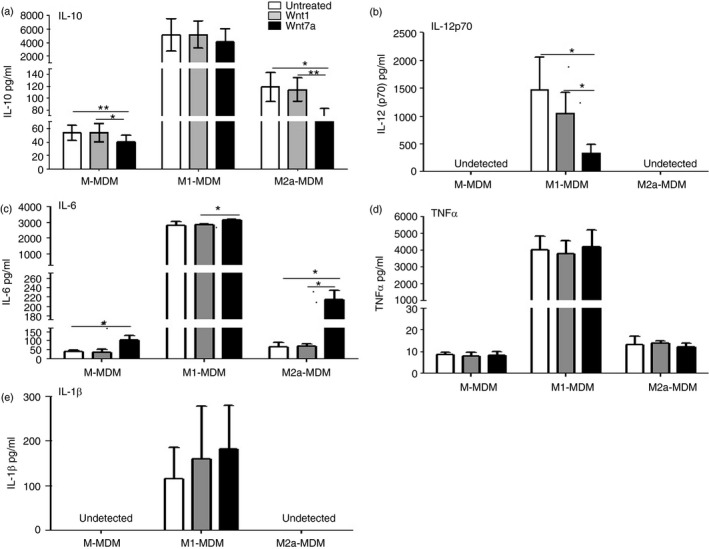
Wnt7a regulates cytokine expression in monocyte‐derived macrophages (MDMs). Interleukin‐10 (IL‐10) (a), IL‐12p70 (b), IL‐1β (c), tumour necrosis factor‐α (TNF‐α) (d) and IL‐6 (b) secreted protein levels of untreated human MDMs, and Wnt1‐ or Wnt7a‐treated MDMs was measured by Luminex assay (n = 4, *P < 0·05, **P < 0·01).
Discussion
Monocyte‐derived macrophages are exceptionally plastic, a trait that allows them to alter their function to meet the requirements of their microenvironment. Therefore, it is important to define physiologically relevant factors that fine‐tune MDM phenotype and function, including their rate of development, the proteins they secrete, and the receptors they express. While skewing of monocytes to M‐1 or M2a‐MDM phenotypes is well characterized in response to classical stimulation (e.g. M‐CSF/LPS/IFN‐γ or M‐CSF+IL‐4/IL‐10/or transforming growth factor β, respectively), little is known about the role of Wnts in skewing monocyte differentiation.
To date, the role of Wnt3a and Wnt5a has been evaluated in the context of macrophage functions. Wnt5a through CamMK11 signalling induced inflammatory cytokines of macrophages38 and it was also reported to induce immunosuppressive macrophages.39 These rather dichotomous reports may be context dependent. Further, Wnt5a was reported to stimulate phagocytosis, although this was based on mouse macrophage studies rather than human macrophages.40 Wnt3a also induced pro‐inflammatory cytokines by mouse macrophages, but led to sensitization of macrophage responses to subsequent Toll‐like receptor 2 and 9 stimulation.41 We show here that Wnt7a induces monocytes to differentiate towards a unique MDM phenotype with both M1‐like and alternative‐like MDM properties. As such, levels of Wnt7a in various tissues can significantly alter monocyte differentiation, regulating the phenotype and function of MDMs.
Specifically, we show that activated monocytes down‐regulate Wnt7a expression and that Wnt7a but not Wnt1 significantly inhibited the expression of CD14, CD11b, CD163 and CD206 on MDMs. For CD11b, this inhibition was also observed in vivo in a Wnt7a−/− mouse model. Wnt7a also inhibited phagocytic capacity of MDMs, which is probably due to Wnt7a‐mediated down‐regulation of CD206 and CD11b, two molecules associated with phagocytosis.37, 42
Wnt7a inhibited the secretion of IL‐10 from alternative MDM controls and inhibited the secretion of IL‐12 from M1‐like MDM controls. Conversely, IL‐6 secretion was significantly increased relative to all MDM controls as well as MDMs differentiated in the presence of Wnt1. IL‐6 has been shown to skew MDMs toward an alternative phenotype;43, 44 however, IL‐6 is notoriously pleiotropic and needs to be assessed in combination with other factors to determine its role in each context. In this case, cells primed with IFN‐γ and stimulated with LPS secreted significantly higher amounts of IL‐6 than cells that were differentiated in just M‐CSF or M‐CSF with IL‐4. The cells that secreted more IL‐6 also down‐regulated CD206 and CD163 expression and had significantly higher expression of IL‐12p70, TNF‐α and IL‐1β relative to the M‐CSF and alternative controls. The significant decrease in IL‐12p70 expression as well as the limited effect on IL‐1β and TNF‐α suggests that the Wnt7a MDMs may not be completely pro‐inflammatory. Further, Wnt7a‐mediated down‐regulation of IL‐12p70 and IL‐10 simultaneously underscores the heterogeneity of MDMs in vivo to display both pro‐ and anti‐inflammatory characteristics simultaneously.45, 46
The finding that Wnt7a skews monocyte differentiation is particularly relevant to HIV invasion in the central nervous system. HIV invades the central nervous system within weeks of infection through both trafficking of T cells and also monocytes that harbour HIV DNA, yet are not productively infected unless they differentiate into macrophages.47 Studies from our laboratory demonstrated that robust β‐catenin is expressed in monocytes and blocks active HIV transcription, yet as these monocytes differentiate into macrophages they support productive infection. As such, Wnt7a secreted by endothelial cells and neurons48, 49, 50, 51 can potentially regulate monocyte differentiation in the brain, as the data suggest, in a way that may inhibit their differentiation, and subsequent susceptibility to HIV infection. Further, Wnt7a‐responsive macrophages are likely to be less phagocytic and as such may not contribute significantly to controlling neuroinflammation by way of clearing debris and dying cells that can perpetuate inflammation. However, because microglia and astrocytes have both demonstrated phagocytic capacity,52, 53, 54 it remains to be seen whether Wnt7a‐mediated reduction in phagocytic capacity has a significant impact on neuroinflammation. These findings underscore the complexity of monocyte/macrophage differentiation in response to a variety of morphogens and highlight the complexity of these responses as context dependent and not necessarily as just M‐1 and M‐2 like macrophages. Indeed, Wnt7a, and similar physiological signals, may serve to regulate inflammatory responses, resulting in reduced cell damage that can occur from chronic inflammation. Simultaneous reduction in IL‐10 also suggests that Wnt7a‐MDM may not inhibit the response of other immune cells, but rather serves as a regulator that modulates excessive inflammatory or anti‐inflammatory responses. An intermediate MDM phenotype may be favourable in disease and homeostasis; in neuroAIDS inflammatory M1 macrophages perpetuate neuroinflammation,55 whereas alternative macrophage phenotypes are a marker of a worsened prognosis due to the inability of macrophages to promote anti‐viral CD8+ T‐cell responses.56 Additionally, macrophages inhibit would healing,57, 58 while anti‐inflammatory macrophages are notorious allies in tumour development through their ability to inhibit anti‐tumour responses from other immune cells.59 Future experiments should assess the relevance of Wnt7a on MDM function in neuroAIDS by gain and loss of function studies using in vitro and in vivo Wnt7a knockout and over‐expression models. Ultimately, understanding how monocyte/macrophages respond to signals in their microenvironment, particularly from proteins such as Wnts that demonstrated ubiquitous expression and ability to influence cell fate and function, can serve as novel approaches to regulate immune responses in particular diseases and/or tissues.
Disclosure
The authors declare no competing interests.
Supporting information
Figure S1. Gating strategy for monocytes, generation of monocyte‐derived macrophages and treatment paradigm.
Acknowledgements
This work is supported by National Institutes of Health grant 5R01NS060632 (LA), R01MH00628 (LA), R01CA138528 (RW) and CCR, part of NCI's intramural research program, NIH, Department of Health and Human Services (DHHS).
References
- 1. Bain CC, Bravo‐Blas A, Scott CL, Gomez Perdiguero E, Geissmann F, Henri S et al Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol 2014; 15:929–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Olingy CE, San Emeterio CL, Ogle ME, Krieger JR, Bruce AC, Pfau DD et al Non‐classical monocytes are biased progenitors of wound healing macrophages during soft tissue injury. Sci Rep 2017; 7:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci 2008; 1:453–61. [DOI] [PubMed] [Google Scholar]
- 4. Rey‐Giraud F, Hafner M, Ries CH. In vitro generation of monocyte‐derived macrophages under serum‐free conditions improves their tumor promoting functions. PLoS ONE 2012; 7:e42656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lehtonen A, Ahlfors H, Veckman V, Miettinen M, Lahesmaa R, Julkunen I. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol 2007; 82:710–20. [DOI] [PubMed] [Google Scholar]
- 6. Brandon C, Eisenberg LM, Eisenberg CA. WNT signaling modulates the diversification of hematopoietic cells. Blood 2000; 96:4132–41. [PubMed] [Google Scholar]
- 7. Wong C, Chen C, Wu Q, Liu Y, Zheng P. A critical role for the regulated wnt‐myc pathway in naive T cell survival. J Immunol 2015; 194:158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khoo MLM, Carlin SM, Lutherborrow MA, Jayaswal V, Ma DDF, Moore JJ. Gene profiling reveals association between altered Wnt signaling and loss of T‐cell potential with age in human hematopoietic stem cells. Aging Cell 2014; 13:744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kvell K, Fejes AV, Parnell SM, Pongracz JE. Active Wnt/β‐catenin signaling is required for embryonic thymic epithelial development and functionality ex vivo . Immunobiology 2014; 219:644–52. [DOI] [PubMed] [Google Scholar]
- 10. Ferrando‐Martınez S, Ruiz‐Mateos E, Dudakov JA, Velardi E, Grillari J, Kreil DP et al Wnt signaling suppression in the senescent human thymus. J Gerontol A Biol Sci Med Sci 2015; 70:273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gattinoni L, Zhong X‐S, Palmer DC, Ji Y, Hinrichs CS, Yu Z et al Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med 2009; 15:808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu B, Crampton SP, Hughes CCW. Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity 2007; 26:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lin JC, Chang RL, Chen YF, Yang JJ, Baskaran R, Chung LC et al β‐Catenin overexpression causes an increase in inflammatory cytokines and NF‐κB activation in cardiomyocytes. Cell Mol Biol (Noisy‐le‐grand) 2016; 63:17–22. [DOI] [PubMed] [Google Scholar]
- 14. Liu XZ, Fan J, Qi K, Liu SP, Xu WD, Gao Y et al Dishevelled2 promotes apoptosis and inhibits inflammatory cytokine secretion in rheumatoid arthritis fibroblast‐like synoviocytes through crosstalk with the NF‐κB pathway. Oncotarget 2017; 8:12649–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jang J, Jung Y, Chae S, Chung S‐I, Kim S‐M, Yoon Y. WNT/β‐catenin pathway modulates the TNF‐α‐induced inflammatory response in bronchial epithelial cells. Biochem Biophys Res Commun 2017; 484:442–9. [DOI] [PubMed] [Google Scholar]
- 16. Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits β‐catenin–TCF signaling depending on receptor context. PLoS Biol 2006; 4:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Amerongen R, Fuerer C, Mizutani M, Nusse R. Wnt5a can both activate and repress Wnt/β‐catenin signaling during mouse embryonic development. Dev Biol 2012; 369:101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Von Maltzahn J, Bentzinger CF, Rudnicki MA. Wnt7a‐Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat Cell Biol 2011; 14:186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Winn RA, Scoyk MV, Hammond M, Rodriguez K, Crossno JT Jr, Heasley LE et al Antitumorigenic effect of Wnt 7a and Fzd 9 in non‐small cell lung cancer cells is mediated through ERK‐5‐dependent activation of peroxisome proliferator‐activated receptor c. J Biol Chem 2006; 281:26943–50. [DOI] [PubMed] [Google Scholar]
- 20. Pongracz J, Hare K, Harman B, Anderson G, Jenkinson EJ. Thymic epithelial cells provide WNT signals to developing thymocytes. Eur J Immunol 2003; 33:1949–56. [DOI] [PubMed] [Google Scholar]
- 21. Daneman R, Agalliu D, Zhou L, Kuhnert F, Kuo CJ, Barres BA. Wnt/β‐catenin signaling is required for CNS, but not non‐CNS, angiogenesis. Proc Natl Acad Sci U S A 2009; 106:641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Posokhova E, Shukla A, Seaman S, Volate S, Hilton MB, Wu B et al GPR124 functions as a WNT7‐specific coactivator of canonical β‐catenin signaling. Cell Rep 2015; 10:123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Y, Nathans J. Gpr124 controls CNS angiogenesis and blood–brain barrier integrity by promoting ligand‐specific canonical wnt signaling. Dev Cell 2014; 31:248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bikkavilli RK, Avasarala S, Van Scoyk M, Arcaroli J, Brzezinski C, Zhang W et al Wnt7a is a novel inducer of β‐catenin‐independent tumor‐suppressive cellular senescence in lung cancer. Oncogene 2015; 34:5317–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mantovani A. Tumor‐associated macrophages. Curr Opin Immunol 1989; 2:689–92. [DOI] [PubMed] [Google Scholar]
- 26. Mantovani A, Bottazzi B, Colotta F, Sozzani S, Ruco L. The origin and function of tumor‐associated macrophages. Immunol Today 1992; 13:265–70. [DOI] [PubMed] [Google Scholar]
- 27. Wang S, Sun Z, Zhang X, Li Z, Wu M, Zhao W et al Wnt1 positively regulates CD36 expression via TCF4 and PPAR‐γ in macrophages. Cell Physiol Biochem 2015; 35:1289–302. [DOI] [PubMed] [Google Scholar]
- 28. Shang YC, Chong ZZ, Hou J, Maiese K. Wnt1, FoxO3a, and NF‐κB oversee microglial integrity and activation during oxidant stress. Cell Signal 2010; 22:1317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM et al CD14 controls the LPS‐induced endocytosis of Toll‐like receptor 4. Cell 2011; 147:868–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meerschaert J, Furie MB. The adhesion molecules used by monocytes for migration across endothelium include CD11a/CD18, CD11b/CD18, and VLA‐4 on monocytes and ICAM‐1, VCAM‐1, and other ligands on endothelium. J Immunol 1995; 154:4099–112. [PubMed] [Google Scholar]
- 31. Le Cabec V, Carréno S, Moisand A, Bordier C, Maridonneau‐Parini I. Complement receptor 3 (CD11b/CD18) mediates type I and type II phagocytosis during nonopsonic and opsonic phagocytosis, respectively. J Immunol 2002; 169:2003–9. [DOI] [PubMed] [Google Scholar]
- 32. Shappell SB, Toman C, Anderson DC, Taylor AA, Entman ML, Smith CW. Mac‐1 (CD11b/CD18) mediates adherence‐dependent hydrogen peroxide production by human and canine neutrophils. J Immunol 1990; 144:2702–11. [PubMed] [Google Scholar]
- 33. Nielsen MJ, Andersen CBF, Moestrup SK. CD163 binding to haptoglobin‐hemoglobin complexes involves a dual‐point electrostatic receptor‐ligand pairing. J Biol Chem 2013; 288:18834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee SJ, Evers S, Roeder D, Parlow AF, Risteli J, Risteli L et al Mannose receptor‐mediated regulation of serum glycoprotein homeostasis. Science 2002; 295:1898–901. [DOI] [PubMed] [Google Scholar]
- 35. Gabrilovich DI, Ostrand‐Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 2012; 12:253–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005; 5:953–64. [DOI] [PubMed] [Google Scholar]
- 37. Rőszer T. Understanding the Mysterious M2 Macrophage through activation markers and effector mechanisms. Mediators Inflamm 2015; 18:816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pereira C, Schaer DJ, Bachli EB, Kurrer MO, Schoedon G. Wnt5A/CaMKII signaling contributes to the inflammatory response of macrophages and is a target for the antiinflammatory action of activated protein C and interleukin‐10. Arterioscler Thromb Vasc Biol 2008; 28:504–10. [DOI] [PubMed] [Google Scholar]
- 39. Bergenfelz C, Medrek C, Ekström E, Jirström K, Janols H, Wullt M et al Wnt5a induces a tolerogenic phenotype of macrophages in sepsis and breast cancer patients. J Immunol 2012; 188:5448–58. [DOI] [PubMed] [Google Scholar]
- 40. Maiti G, Naskar D, Sen M. The Wingless homolog Wnt5a stimulates phagocytosis but not bacterial killing. Proc Natl Acad Sci U S A 2012; 109:16600–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu C‐H, Nguyen TTK, Irvine KM, Sweet MJ, Frazer IH, Blumenthal A. Recombinant Wnt3a and Wnt5a elicit macrophage cytokine production and tolerization to microbial stimulation via Toll‐like receptor 4. Eur J Immunol 2014; 44:1480–90. [DOI] [PubMed] [Google Scholar]
- 42. Fossati‐Jimack L, Ling GS, Cortini A, Szajna M, Malik TH, McDonald JU et al Phagocytosis is the main CR3‐mediated function affected by the lupus‐associated variant of CD11b in human myeloid cells. PLoS ONE 2013; 8:e57082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mauer J, Chaurasia B, Goldau J, Vogt MC, Ruud J, Nguyen KD et al Signaling by IL‐6 promotes alternative activation of macrophages to limit endotoxemia and obesity‐associated resistance to insulin. Nat Immunol 2014; 15:423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fernando MR, Reyes JL, Iannuzzi J, Leung G, McKay DM. The pro‐inflammatory cytokine, interleukin‐6, enhances the polarization of alternatively activated macrophages. PLoS ONE 2014; 9:e94188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gordon S, Plüddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev 2014; 262:36–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas . J Clin Invest 2012; 122:787–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aljawai Y, Richards MH, Seaton MS, Narasipura SD, Al‐Harthi L. β‐Catenin/TCF‐4 signaling regulates susceptibility of macrophages and resistance of monocytes to HIV‐1 productive infection. Curr HIV Res 2014; 12:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stenman JM, Rajagopal J, Carroll TJ, Ishibashi M, McMahon J, McMahon AP. Canonical Wnt signaling regulates organ‐specific assembly and differentiation of CNS vasculature. Science 2008; 322:1247–50. [DOI] [PubMed] [Google Scholar]
- 49. Qu Q, Sun G, Murai K, Ye P, Li W, Asuelime G et al Wnt7a regulates multiple steps of neurogenesis. Mol Cell Biol 2013; 33:2551–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N et al The Wnt/b‐catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 2004; 131:2791–801. [DOI] [PubMed] [Google Scholar]
- 51. Ciani L, Boyle KA, Dickins E, Sahores M, Anane D, Lopes DM et al Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca2+/Calmodulin‐dependent protein kinase II. Proc Natl Acad Sci U S A 2011; 108:10732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hanisch U‐K, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 2007; 10:1387–94. [DOI] [PubMed] [Google Scholar]
- 53. Block ML, Zecca L, Hong J‐S. Microglia‐mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci 2007; 8:57–69. [DOI] [PubMed] [Google Scholar]
- 54. Wyss‐Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F et al Adult mouse astrocytes degrade amyloid‐β in vitro and in situ . Nat Med 2003; 9:453–7. [DOI] [PubMed] [Google Scholar]
- 55. Burdo TH, Lackner A, Williams KC. Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev 2013; 254:102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kumar A, Herbein G. The macrophage: a therapeutic target in HIV‐1 infection. Mol Cell Ther 2014; 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sindrilaru A, Peters T, Wieschalka S, Baican C, Baican A, Peter H et al An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest 2011; 121:985–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med 2011; 11:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sica A, Schioppa T, Mantovani A, Allavena P. Tumour‐associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti‐cancer therapy. Eur J Cancer 2006; 42:717–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Gating strategy for monocytes, generation of monocyte‐derived macrophages and treatment paradigm.


