Summary
Invariant natural killer T (iNKT) cells are adaptive T cells with innate‐like characteristics including rapid cytokine production and a proliferative response to stimulation. Development of these cells in the thymus is dependent on expression of the microRNA (miRNA) processing enzyme Dicer, indicating that iNKT cells probably have distinct miRNA requirements for gene regulation during development. The miRNA miR‐155 has previously been shown to have numerous roles in T cells, including regulation of proliferation and differentiation, and positive modulation of interferon‐γ expression. We examined the role of miR‐155 in the development and function of iNKT cells. Using germline‐deficient miR‐155 mice, we showed that loss of miR‐155 resulted in unchanged iNKT cell frequency and cell number. Although miR‐155 was up‐regulated in iNKT cells upon activation with α‐galactosylceramide, loss of miR‐155 did not affect cytokine production or proliferation by iNKT cells. Hence, cytokine production occurs in iNKT cells independently of miR‐155 expression.
Keywords: cytokine, development, invariant natural killer T cells, microRNAs
Introduction
Invariant natural killer T (iNKT) cells are a versatile T‐cell population, expressing a semi‐invariant T‐cell receptor (TCR) (Vα14‐Jα18 in mice) and the ability to rapidly respond to their cognate glycolipid to produce an array of cytokines and rapidly mobilize an immune response.1 Although the unique microRNA (miRNA) profile associated with iNKT cells during activation is currently unclear, there is precedence for studying the role of miRNAs in iNKT cell development, lineage differentiation and function. Conditional loss of Dicer (the RNAaseII enzyme responsible for miRNA processing) led to a significant reduction in iNKT cells during development and in peripheral tissue, strongly indicating that miRNAs play a role in iNKT cell development.2, 3, 4 Substantial evidence also exists for a function for individual miRNAs in the development of iNKT cells. Mir‐150 is required for iNKT cell maturation and its loss results in increased interferon‐γ (IFN‐γ) production,5 miR‐181 is essential for the early development of iNKT cells6 and Let‐7 miRNAs specifically target Promyelocytic leukemia zinc finger protein (PLZF) expression during iNKT cell development in the thymus.7 Recently, it was shown that the miRNA cluster miR17~92 regulates iNKT cell development through regulation of transforming growth factor‐β.8
miR‐155 was one of the first miRNAs discovered in mammals and is expressed by multiple cells of the immune system. In B cells, over‐expression of miR‐155 led to the transformation and development of lymphoma, suggesting a role for miR‐155 in regulating cell proliferation.9 In T cells, miR‐155 appears to have multiple unique functions. Initially, T cells from mice with germline miR‐155 deficiency were shown to skew toward the T helper type 2 lineage, possibly due to loss of miR‐155 negative regulation of the transcription factor c‐Maf (a positive regulator of interleukin‐4).10, 11 miR‐155 has been shown to regulate both the CD8+ effector T‐cell response and the natural killer cell response to viral infection, through effects on the proliferation of these cells, although miR‐155 appears to target different mRNAs during proliferation depending on cell type.12, 13, 14, 15 In one study, survival of CD8+ effector T cells was also impaired in the absence of miR‐155.12 miR‐155 has been reported to regulate cell lineage decisions, with central memory CD8+ T cells expressing higher levels of miR‐155 than effector memory subsets.16, 17 A role for miR‐155 in the survival of regulatory T cells through its action on SOCS1 has also been observed.18, 19 Aside from a function in proliferation and survival, miR‐155 can also influence cytokine production by immune cells. Indeed, miR‐155 can positively regulate IFN‐γ production by CD4 and CD8 T cells through its repression of the phosphatase SHIP1, a negative regulator of IFN‐γ expression.20, 21
We examined a role for miR‐155 in the development and function of iNKT cells. Using germline‐deficient miR‐155 mice, we showed that loss of miR‐155 subtly affected the frequency of iNKT cells in the thymus but not in peripheral tissues. Loss of miR‐155 also resulted in marginally more CD44+ NK1.1+ stage 3 cells in the thymus. Although miR‐155 functions to regulate conventional T‐cell proliferation and cytokine production in peripheral tissue, these parameters were unaffected by the loss of miR‐155 in peripheral iNKT cells. We therefore concluded that IFN‐γ production by iNKT cells is regulated by a mechanism independent of miR‐155.
Materials and methods
Mice
All experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. The miR‐155 knockout (KO) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and bred to C57BL/6 mice to generate wild‐type (WT) littermate controls. Mice were bred and housed in specific pathogen‐free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of Pittsburgh. αGalCer (2 μg, Avanti Polar Lipids, Alabaster, AL) was administered intraperitoneally.
Flow cytometry
Single‐cell suspensions were prepared from thymus and spleen. Bone marrow was prepared by isolating lymphocytes from the femurs of mice and preparing a single‐cell suspension. Liver single‐cell suspensions and adipose single‐cell suspensions were prepared as previously described.23 Lung single‐cell suspensions were prepared by mincing lung, incubating minced tissue with collagenase IV at 37° before filtration. The following antibodies were used: anti‐TCR‐β FITC (clone H57‐597), anti‐CD4 FITC (L3T4), anti‐CD24 (M1/69), CD1d Tetramer Alexa 488 [NIH Tetramer Core Facility, Atlanta, GA (mCD1d/PBS57)], CD1d Tetramer phycoerythrin (PE) [NIH Tetramer Core Facility (mCD1d/PBS57)], anti‐TCR‐β PE (H57‐597), anti‐PLZF PE (Mags.21F7), anti‐NK1.1 Peridinin chlorophyll protein‐Cy5.5 (PK136), anti‐NK1.1 allophycocyanin (APC) (PK136), anti‐TCR‐β APC (H57‐597), anti‐TBET APC (4B10), CD1d Tetramer Alexa 647 [NIH Tetramer Core Facility (mCD1d/PBS57)], Ki‐67 Pacific Blue (SolA15) and Annexin V PE (BD Pharmingen, San Diego, CA, USA). All antibodies were purchased from eBioscience (San Diego, CA, USA) unless otherwise specified. Samples were collected on a FACS LSRII, FACS Fortessa or FACS Aria (BD Biosciences, San Jose, CA, USA) and were analysed with flowjo software (TreeStar, Ashland, OR, USA). TCR‐β + CD1d Tetramer+ iNKT cells were sorted with a FACS Aria to at least 98% purity.
Quantitative PCR
For miR‐155, samples were reverse transcribed using a TaqMan™ MicroRNA Reverse Transcription Kit (P/N 4366597; Applied Biosystems, Foster City, CA, USA). Mouse MiR155 TaqMan™ MicroRNA assays were purchased from Applied Biosystems (P/N 4427975). Sno202 was used to normalize.
Statistical analysis
Two‐group comparisons were assessed with an unpaired Student's t‐test. graph‐pad prism software (version 5; GraphPad, San Diego, CA, USA) was used for statistical analyses. P‐values of less than 0·05 were considered significant.
Results
miR‐155 deficiency results in increased iNKT cell frequency in the thymus
Using miR‐155 germline‐deficient (miR‐155 KO) mice, we first characterized conventional T‐cell development in the thymus of these animals. We observed that total thymocyte frequency and cell number were not significantly different between WT and miR‐155 KO mice (Fig. 1a). To characterize the functional importance of miR‐155 in the development of iNKT cells, we determined the frequency and absolute number of iNKT cells in the thymus of WT and miR‐155 KO mice. Although the frequency of iNKT cells was significantly increased in the absence of miR‐155, we observed no substantial difference in absolute iNKT cell number in the thymus between WT and miR‐155 KO mice (Fig. 1b).
Figure 1.
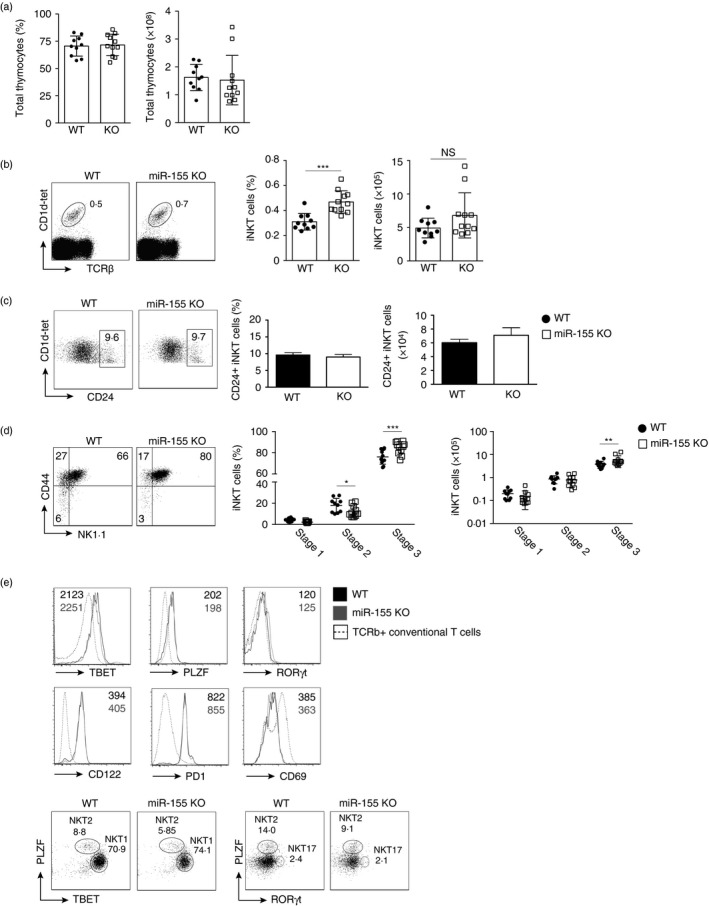
Loss of miR‐155 promotes maturation of thymic invariant natural killer T (iNKT) cells. T cells from the thymus of wild‐type (WT) and miR‐155 germline‐deficient (miR‐155 KO) mice were characterized. (a) Graphs indicating frequency and total thymocytes from WT and miR‐155 KO (KO) mice. (b) Flow cytometric analysis of iNKT cell frequency in WT and miR‐155 KO gated lymphocytes from the thymus. Numbers indicate percentage of cells within the drawn gate. Graphs indicate the average frequency and absolute number (± SEM) of CD1d‐tet+ TCR‐β + gated iNKT cells from the indicated genotype. (c) Flow cytometry plots indicating CD24 expression on gated iNKT cells in the thymus. Graphs indicate the average frequency and absolute number (± SEM) of CD24+ gated iNKT cells from the indicated genotype. (d) Flow cytometric analysis of CD44+ and NK1.1+ expression by gated iNKT lymphocytes from the indicated genotype. Graphs indicate average frequency and absolute number (± SD) of maturation stages as defined by CD44 and NK1.1 expression on CD1d‐tet+ TCR‐β + gated iNKT cells from the indicated genotype. (e) Histograms showing TBET, PLZF, ROR γt, CD122, PD1 and CD69 expression by gated iNKT cells from WT and miR‐155 KO mice. Numbers indicate the median fluorescence intensity (MFI) from the indicated genotype with WT shown in black and miR‐155 KO shown in grey. The dotted line indicates expression by conventional T cells for comparison. Flow cytometry plots indicate TBET versus PLZF and PLZF versus ROR γt expression by gated iNKT cells in the thymus from the indicated genotype. Data are representative of three independent experiments with two to four mice per group per experiment. Statistical significance was evaluated with unpaired Student's t‐test, where n.s., not significant, *P < 0·05, **P < 0·01 and ***P < 0·001.
Similar to conventional T cells, iNKT cells go though sequential stages of development in the thymus. The first, stage 0, is characterized by high CD24 expression. Gating on CD1d Tetramer+ CD24+ iNKT cells, we noted no difference in the frequency of absolute number of iNKT cells expressing CD24 in the thymus. We concluded that progenitor stage 0 iNKT cells were unaffected by the absence of miR‐155 (Fig. 1c). After stage 0, iNKT cells down‐regulate CD24 becoming CD44− NK1.1− stage 1 iNKT cells before up‐regulation of CD44 (stage 2) and NK1.1 (stage 3). Closer analysis of iNKT cell stage development in the thymus revealed that loss of miR‐155 resulted in marginally decreased stage 2 (CD44+ NK1.1−) and increased stage 3 (CD44+ NK1.1+) iNKT cells (Fig. 1d). Stage 1 (CD44− NK.1−) cells were unaffected by loss of miR‐155 expression (Fig. 1d). We conclude that miR‐155 deficiency results in increased frequency of stage 3 CD44+ NK1.1+ iNKT cells in the thymus.
Recent reports have indicated that transcription factor expression can define iNKT cell subsets.24 Gating on iNKT cells and examining TBET, PLZF and RORγt expression, we could determine no substantial difference in expression of these transcription factors by WT and miR‐155 KO cells (Fig. 1e). Additionally, cell surface proteins expressed by iNKT cells including CD122, PD1 and CD69, were not differentially expressed by miR‐155 KO cells (Fig. 1e). Gating on thymic iNKT cells and examination of NKT1, NKT2 and NKT17 cells by transcription factor expression, we noted a slight decrease in NKT2 cells and an increase in NKT1 cells (Fig. 1e) consistent with the subtle decrease we observed in stage 2 and increase in stage 3 cells (Fig. 1d). NKT17 cell frequency remained unchanged in the absence of miR‐155 expression (Fig. 1e).
Loss of miR‐155 does not affect peripheral iNKT cells
We next determined whether the modest increase in the frequency of stage 3 iNKT cells that we observed in miR‐155 KO thymocytes was consistent in peripheral iNKT cells. We compared the frequency and absolute number of iNKT cells in the spleens of WT and miR‐155 KO mice. Here we observed there was no difference in the frequency and absolute number of iNKT cells in the spleen between WT mice and those with germline loss of miR‐155 expression (Fig. 2a). When we examined iNKT cell stages, we also noted that there was no significant difference between stage 1 (CD44lo NK1.1lo), 2 (CD44hi NK1.1lo) and 3 (CD44hi NK1.1hi) cells in the periphery of WT and miR‐155 KO mice (Fig. 2b). To investigate cell death, we examined Annexin V and 7‐AAD expression by WT and miR‐155 KO iNKT cells and could determine no difference in expression (Fig. 2c). As in the thymus, the expression of the transcription factors TBET, PLZF and RORγ was unaffected by loss of miR‐155 (Fig. 2d). Finally, in other peripheral lymphoid tissues such as the liver, bone marrow, lung and adipose tissue, we did not observe a significant difference in the frequency of iNKT cells (Fig. 2e). Hence, we concluded that the increased frequency of iNKT cells, and specifically stage 3 iNKT cells, that we observed in the thymus was lost as the cells migrated from the thymus and matured in peripheral tissue.
Figure 2.
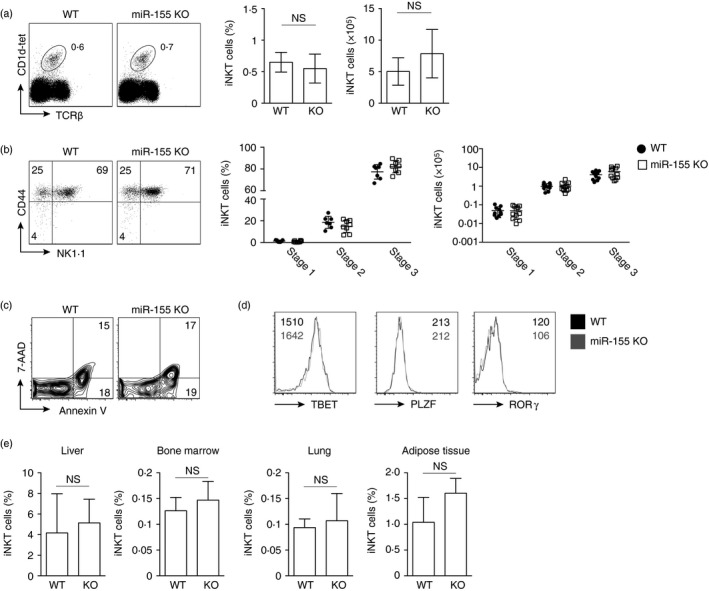
miR‐155 expression is dispensable for invariant natural killer T (iNKT) cell homeostasis in peripheral tissue. Wild‐type (WT) and miR‐155 germline‐deficient (miR‐155 KO) mice were characterized. (a) Flow cytometric analysis of iNKT cell frequency in WT and miR‐155 KO lymphocytes from the spleen. Numbers indicate percentage of cells within the drawn gate. Graphs indicate the average frequency and absolute number (± SEM) of CD1d‐tet+ TCR‐β + gated iNKT cells from the indicated genotype. (b) Flow cytometric analysis of CD44+ and NK1.1+ expression by gated iNKT lymphocytes from the indicated genotype. Graphs indicate average frequency and absolute number (± SEM) of maturation stages as defined CD44 and NK1.1 expression on CD1d‐tet+ TCR‐β + gated iNKT cells from the indicated genotype. Graphs are the average of 10 mice from three independent experiments. (c) Flow cytometry plots indicating Annexin V and 7‐AAD expression on gated iNKT lymphocytes from the indicated genotype. (d) Histograms showing TBET, PLZF and ROR γt expression by gated iNKT cells from WT and miR‐155 KO mice. Numbers indicate the median fluorescence intensity (MFI) from the indicated genotype. (e) Graphs indicating the frequency of iNKT cells from WT and miR‐155 KO (KO) mice from the indicated lymphoid organ. Statistical significance was evaluated with unpaired Student's t‐test, where n.s., not significant.
Loss of miR‐155 does not affect iNKT cell cytokine production
Previous data have indicated that miR‐155 can indirectly positively regulate IFN‐γ expression in conventional T cells, through targeting of the src homology 2 domain‐containing inositol phosphatase 1 (SHIP1) for degradation. Therefore, we determined whether miR‐155 could affect cytokine production by peripheral iNKT cells. Interferon‐γ and interleukin‐4 production can be detected by iNKT cells 1–3 hr after activation with their cognate ligand α‐galactosylceramide (α‐GC). First, we analysed expression of miR‐155 in WT iNKT cells after α‐GC treatment. Sorting iNKT cells from WT mice after in vivo activation, we observed up‐regulation of miR‐155 ~2‐fold after 3 hr and ~30‐fold after 3 days of activation relative to naive iNKT cells (Fig. 3a). Activating WT and miR‐155 KO mice with α‐GC for 3 hr, we could detect no difference in frequency or absolute number of splenic iNKT cells (Fig. 3b). When we examined IFN‐γ and interleukin‐4 production by WT and miR‐155 KO iNKT cells at 3 hr after activation in vivo using intracellular staining, we observed no difference in production of these cytokines between the WT and miR‐155 KO iNKT cells (Fig. 3c). Hence, we conclude that miR‐155, despite its increased expression within 3 hr after iNKT cell activation, is dispensable for production of canonical cytokines by iNKT cells.
Figure 3.
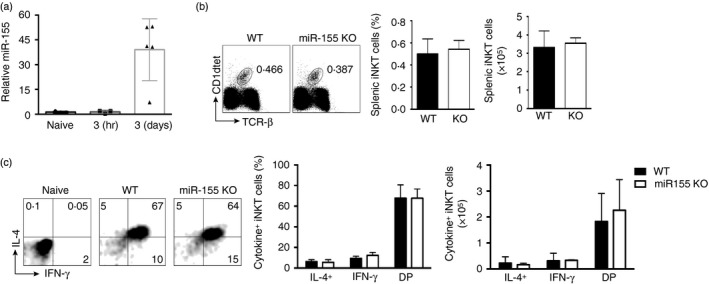
miR‐155 is up‐regulated in activated invariant natural killer T (iNKT) cells. (a) Quantitative PCR indicating relative expression of miR‐155 by sorted CD1d‐tet+ TCR‐β + iNKT cells from naive mice or mice injected with 2 μg α‐galactosylceramide (α‐GC) for 3 hr or 3 days. (b) Flow cytometric analysis of iNKT cell frequency in the spleen after intraperitoneal injection with 2 μg α‐GC for 3 hr. Graphs indicate the average frequency and absolute number (± SEM) of gated iNKT cells from the indicated genotype. (c) Flow cytometry and bar graphs indicate frequency and cell number of interferon‐γ (IFN‐γ) and interleukin‐4 (IL‐4) expression by gated splenic iNKT lymphocytes from the indicated genotype. DP, double positive for IFN‐γ and IL‐4. Graphs are representative of two experiments with two mice per group.
miR‐155 is dispensable for the iNKT cell response at day 3 after activation
miR‐155 was previously reported to affect proliferation of conventional CD8+ effector T cells.12, 13, 14 Furthermore, we observed ~30‐fold increase in miR‐155 expression in iNKT cells upon activation with α‐GC at 3 days after activation (Fig. 3a). To determine a role for miR‐155 in iNKT cells 3 days after activation, we compared WT and miR‐155 KO iNKT cells at this time‐point. We examined iNKT cells in the spleen, liver and bone marrow and all trended towards reduced frequency of iNKT cells in the absence of miR‐155 (Fig. 4a). However, iNKT cell absolute number in these tissues was unaffected by loss of miR‐155 expression (Fig. 4a). Moreover, we could determine no differences in NK1.1 and CD44 expression in iNKT cells from miR155 KO mice relative to WT, although as previously reported, NK1.1 down‐regulation after activation with α‐GC was observed in both WT and miR‐155 KO iNKT cells (Fig. 4b). Previous reports have indicated that PD1 is up‐regulated on activated iNKT cells.25 Therefore, we examined PD1 expression on WT and miR155 KO iNKT cells 3 days after activation. As with overall iNKT cell frequency and stages, we observed no differences in PD1 expression on day 3 activated iNKT cells in the absence of miR‐155 (Fig. 4c). To determine proliferation, we examined Ki‐67 expression in WT and miR‐155 KO iNKT cells 3 days after activation with α‐GC. In all tissues examined, a subset of iNKT cells were dividing (Ki‐67+) (Fig. 4d). Importantly, loss of miR‐155 in iNKT cells did not affect cell division by these cells in peripheral tissue after activation (Fig. 4d). Finally, we examined transcription factor expression and could detect no difference in expression of TBET, PLZF or RORγ in WT or miR‐155 KO iNKT cells after activation (Fig. 4e). Hence, we have established that despite high miR‐155 expression in iNKT cells at day 3 after activation, miR‐155 is dispensable for the proliferation of iNKT cells in peripheral tissue.
Figure 4.
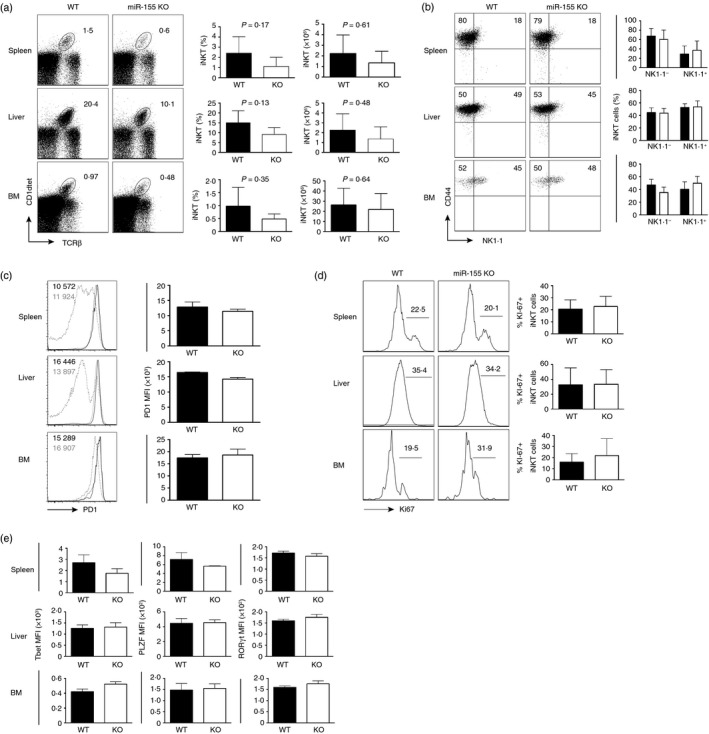
Loss of miR‐155 does not affect invariant natural killer T (iNKT) cells at 3 days after activation. (a) Flow cytometric analysis of iNKT cell frequency in the spleen, liver and bone marrow (BM) at 3 days post intraperitoneal injection with 2 μg α‐galactosylceramide (α‐GC). Bar graphs indicate iNKT cell frequency and absolute number (± SEM) in the indicated tissue. (b) Flow cytometric analysis of CD44+ and NK1.1+ expression by gated iNKT lymphocytes from the indicated genotype and tissue. Graphs indicate average frequency (± SEM) of NK1.1− CD44+ or NK1.1+ CD44+ CD1d‐tet+ TCR‐β + gated iNKT cells from the indicated genotype and tissue. (c) Flow cytometric analysis and median fluorescence intensity (MFI) of PD1 expression by gated iNKT lymphocytes from the indicated genotype and tissue. Black line indicates wild‐type (WT), grey line indicates miR‐155‐deficient (miR‐155 KO) and dotted line indicates conventional T‐cell control for comparison. (d) Histograms indicating Ki‐67 expression in iNKT cells from the indicated genotype and tissue at day 3 after activation. Graphs indicate average frequency (± SEM) of Ki‐67+ CD1d‐tet+ TCR‐β + gated iNKT cells from the indicated genotype and tissue. (e) Graphs showing TBET, PLZF and ROR γt MFI by gated iNKT cells from WT and miR‐155 KO mice after 3 days activation with α‐GC. Graphs are representative of four to six mice from two independent experiments.
Discussion
We show that miR‐155 has a minor role in the development of iNKT cells in the thymus. We demonstrate how loss of miR‐155 resulted in increased stage 3 cells with a corresponding decrease in stage 2 cells. Although thymic development of iNKT cells was slightly perturbed in the absence of miR‐155, this defect was not apparent in peripheral tissue. Loss of miR‐155 resulted in normal development and cytokine production by iNKT cells in the periphery and activation was unaffected by loss of miR‐155 expression. Moreover, miR‐155 expression, although high in iNKT cells at day 3 after activation, was dispensable for their proliferation at this time‐point.
Importantly, our data are largely consistent with a previously published report in which miR‐155 was over‐expressed using an Lck‐miR‐155 transgene.26 Here it was shown that over‐expression of miR‐155 resulted in an increase in stage 2 cells.26 In this miR‐155 over‐expression study, peripheral iNKT cell frequency and cell number were perturbed,26 in contrast to our findings here in which peripheral iNKT cells lacking expression of miR‐155 were unaffected. It is possible that loss of miR‐155 expression is compensated for by other microRNAs or even by other proteins in peripheral iNKT cells, which would account for normal iNKT cell homeostasis and activation observed in the periphery in the absence of miR‐155.
Previous work has also indicated that miR‐155 can indirectly regulate IFN‐γ expression, mediated in part through its repression of SHIP1, in conventional CD4+ and CD8+ T cells.20 Invariant NKT cells can produce IFN‐γ within a matter of hours after activation, a process that was unaffected by loss of miR‐155, indicating that activation‐induced IFN‐γ production is independent of miR‐155 in these cells upon activation. Overall, we find that miR‐155 has a moderate effect on the development of iNKT cells in the thymus but that loss of miR‐155 does not affect peripheral homeostasis, cytokine production or activation of iNKT cells in peripheral tissues.
Disclosure
The authors declare they have no commercial or financial conflicts of interest.
Acknowledgements
We would like to thank all members of the D'Cruz laboratory and Drs Lawrence Kane and Amanda Poholek for critical reading of this manuscript. ABF performed the experiments and wrote the manuscript, HMB performed experiments, AN performed experiments, LMD designed the study and wrote the manuscript. This work was supported by grants from the NIH including R21 AI 121744 to LMD and T32 AI089443 to ABF and HMB. We would like to thank Dewayne Falkner, Aarika Yates and Hiroshi Yano for assistance with cell sorting.
References
- 1. Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol 2007; 25:297–336. [DOI] [PubMed] [Google Scholar]
- 2. Fedeli M, Napolitano A, Wong MP, Marcais A, de Lalla C, Colucci F et al Dicer‐dependent microRNA pathway controls invariant NKT cell development. J Immunol 2009; 183:2506–12. [DOI] [PubMed] [Google Scholar]
- 3. Seo KH, Zhou L, Meng D, Xu J, Dong Z, Mi QS. Loss of microRNAs in thymus perturbs invariant NKT cell development and function. Cell Mol Immunol 2010; 7:447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG et al Tie2cre‐induced inactivation of the miRNA‐processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci USA 2009; 106:10266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng Q, Zhou L, Mi QS. MicroRNA miR‐150 is involved in Vα14 invariant NKT cell development and function. J Immunol 2012; 188:2118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henao‐Mejia J, Williams A, Goff LA, Staron M, Licona‐Limon P, Kaech SM et al The microRNA miR‐181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity 2013; 38:984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pobezinsky LA, Etzensperger R, Jeurling S, Alag A, Kadakia T, McCaughtry TM et al Let‐7 microRNAs target the lineage‐specific transcription factor PLZF to regulate terminal NKT cell differentiation and effector function. Nat Immunol 2015; 16:517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fedeli M, Riba M, Garcia Manteiga JM, Tian L, Vigano V, Rossetti G et al miR‐17 approximately 92 family clusters control iNKT cell ontogenesis via modulation of TGF‐β signaling. Proc Natl Acad Sci USA 2016; 113:E8286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N et al Pre‐B cell proliferation and lymphoblastic leukemia/high‐grade lymphoma in Eµ‐miR155 transgenic mice. Proc Natl Acad Sci USA 2006; 103:7024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR et al Requirement of bic/microRNA‐155 for normal immune function. Science 2007; 316:608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y et al Regulation of the germinal center response by microRNA‐155. Science 2007; 316:604–8. [DOI] [PubMed] [Google Scholar]
- 12. Dudda JC, Salaun B, Ji Y, Palmer DC, Monnot GC, Merck E et al MicroRNA‐155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity 2013; 38:742–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J et al The microRNA miR‐155 controls CD8+ T cell responses by regulating interferon signaling. Nat Immunol 2013; 14:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lind EF, Elford AR, Ohashi PS. Micro‐RNA 155 is required for optimal CD8+ T cell responses to acute viral and intracellular bacterial challenges. J Immunol 2013; 190:1210–6. [DOI] [PubMed] [Google Scholar]
- 15. Zawislak CL, Beaulieu AM, Loeb GB, Karo J, Canner D, Bezman NA et al Stage‐specific regulation of natural killer cell homeostasis and response against viral infection by microRNA‐155. Proc Natl Acad Sci USA 2013; 110:6967–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Almanza G, Fernandez A, Volinia S, Cortez‐Gonzalez X, Croce CM, Zanetti M. Selected microRNAs define cell fate determination of murine central memory CD8 T cells. PLoS ONE 2010; 5:e11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsai CY, Allie SR, Zhang W, Usherwood EJ. MicroRNA miR‐155 affects antiviral effector and effector Memory CD8 T cell differentiation. J Virol 2013; 87:2348–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR‐155 contributes to the development of regulatory T cells. J Immunol 2009; 182:2578–82. [DOI] [PubMed] [Google Scholar]
- 19. Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K et al Foxp3‐dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009; 30:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huffaker TB, Hu R, Runtsch MC, Bake E, Chen X, Zhao J et al Epistasis between microRNAs 155 and 146a during T cell‐mediated antitumor immunity. Cell Rep 2012; 2:1697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR‐155. Proc Natl Acad Sci USA 2009; 106:7113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol 2010; 11:240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fonseca DM, Hand TW, Han SJ, Gerner MY, Glatman Zaretsky A, Byrd AL et al Microbiota‐dependent sequelae of acute infection compromise tissue‐specific immunity. Cell 2015; 163:354–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady‐state production of IL‐4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 2013; 14:1146–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL‐10‐producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest 2014; 124:3725–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burocchi A, Pittoni P, Tili E, Rigoni A, Costinean S, Croce CM et al Regulated expression of miR‐155 is required for iNKT cell development. Front Immunol 2015; 6:140. [DOI] [PMC free article] [PubMed] [Google Scholar]


