Abstract
Pathologic complete response (pCR) is a predictor for favorable outcome after neoadjuvant treatment in early breast cancer. Modulation of gene expression may also provide early readouts of biological activity and prognosis, offering the possibility for timely response‐guided treatment adjustment. The role of early transcriptional changes in predicting response to neoadjuvant chemotherapy plus bevacizumab was investigated. One‐hundred‐and‐fifty patients with large, operable and locally advanced HER2‐negative breast cancer received epirubicin and docetaxel, with the addition of bevacizumab. Patients underwent tumor biopsies at baseline, after Cycle 2 and at the time of surgery. The primary end point, pCR, and its relation with the secondary endpoints event‐free survival (EFS), overall survival (OS) and gene expression profiles, are reported. The pCR rate was 13% (95% CI 8.6–20.2), with significantly more pCRs among triple‐negative [28% (95% CI 14.8–45.4)] than among hormone receptor positive (HR+) tumors [9% (95% CI 4.6–16.3); (OR = 3.9 [CI = 1.5–10.3])]. pCR rates were not associated with EFS or OS. PAM50 subtypes significantly changed after Cycle 2 (p = 0.03) and an index of absolute changes in PAM50 correlations between these time‐points was associated with EFS [HR = 0.62 (CI = 0.3–1.1)]. In univariable analyses, signatures for angiogenesis, proliferation, estrogen receptor signaling, invasion and metastasis, and immune response, measured after Cycle 2, were associated with pCR in HR+ tumors. Evaluation of changes in molecular subtypes and other signatures early in the course of neoadjuvant treatment may be predictive of pCR and EFS. These factors may help guide further treatment and should be considered when designing neoadjuvant trials.
Keywords: Neoadjuvant, breast cancer, Phase 2 trial, pathological complete response, PAM50, AIMS
Short abstract
What's new?
It's a good sign for a patient's prognosis if, after pre‐operative chemotherapy, no breast cancer cells survive. But this metric isn't perfect, and varies depending on the tumor's molecular subtype. Here, the authors analyzed changes in gene expression brought on by chemotherapy. They analyzed molecular markers in biopsies from 150 breast cancer patients at three time points: baseline, after 2 cycles of neoadjuvant therapy, and right before surgery. Neoadjuvant chemotherapy significantly changed the tumor's gene expression profile, they found, and these changes could have predictive value: a bigger change between baseline and Cycle 2 correlated with longer event‐free survival.
Neoadjuvant chemotherapy is widely used to treat early‐stage breast cancer and the resulting increase in breast conservation1 has led to more patients with early breast cancer being offered neoadjuvant therapy. The underlying rationale is that response to neoadjuvant treatment in the loco‐regional area reflects the effect on micro‐metastatic disease. Patients achieving a pathologic complete response (pCR) to preoperative chemotherapy are expected to experience a favorable prognosis, especially those with highly proliferative tumors.2, 3 The extent and properties of residual disease in the breast also impact on prognosis.4, 5 The effect of adding experimental treatments for patients not achieving a pCR after neoadjuvant chemotherapy, that is, for patients with an adverse prognosis, is being investigated in several studies (e.g., NCT02032823 and NCT01772472), and has been reported to be beneficial in a large Phase III study, CREATE‐X.6
The neoadjuvant treatment setting also offers the possibility to study the effect of treatment on the primary tumor, and the modulation of tissue biomarkers over time. In the GeparTrio study7, 8, 9 a clinical response‐guided chemotherapy regimen was found to be significantly better in prolonging disease‐free (DFS) and overall survival (OS) than a non‐individualized approach with a fixed number of cycles, specifically among patients with hormone receptor (HR)‐positive tumors. In patients with estrogen receptor (ER) positive disease treated with neoadjuvant endocrine treatment, Ki67 expression after 2 weeks of treatment was a better predictor of long‐term outcome than a baseline measurement of the same marker (reviewed in Ref. 10). Further, Varadan et al. recently investigated this brief‐exposure paradigm at the molecular level, uncovering a TGF‐β gene signature predictive of pCR after neoadjuvant anti‐angiogenic therapy.11 Clearly, to exploit the full potential of neoadjuvant chemotherapy, better understanding of the effect of a specific therapy on predictive biomarkers during the course of such treatment is needed. Sampling of tumor tissue at an early time‐point, enabling the evaluation of changes in, for example, molecular subtypes, may provide valuable information in predicting long‐term outcome.
The molecular subtypes of breast cancer display differential responses to conventional neoadjuvant chemotherapy,3, 12 complicating the investigation of the predictive value of biomarkers. Patients with basal and HER2‐positive disease have the highest pCR rates, while the luminal subtypes less frequently achieve a pCR.2, 13, 14 In addition, pCR is not a perfect surrogate marker for long‐term survival,15 since many patients with residual disease after neoadjuvant therapy still have a good prognosis, particularly those with luminal tumors. The caveats raised when translating the prognostic value of pCR on the individual level to the assumption that this will lead to an improved prognosis on a group level16 emphasizes the need to identify biomarkers predictive of both short and long‐term treatment outcomes.
Here, we report the clinical and transcriptional profiling outcomes of a multi‐center, Phase II trial (PROMIX) of neoadjuvant anthracycline (epirubicin) plus taxane (docetaxel)‐based chemotherapy with the addition of bevacizumab (Avastin®) for patients with HER2‐negative breast cancer. The overall aim was to evaluate the sensitivity of molecular markers and functional imaging to detect response to neoadjuvant treatment at an early time‐point and to identify tumor characteristics and treatment‐induced changes in tumor characteristics predictive of long‐term prognosis. Here, we focus on evaluating whether transcriptional changes induced by short‐term exposure to chemotherapy can predict short‐term (pCR) and long‐term (event‐free survival, EFS) outcomes. We report that neoadjuvant treatment‐induced changes in molecular subtypes and other gene signatures may be predictive of pCR rates and EFS, and propose an index, ΔPAM50, based on the overall change in PAM50 subtype correlations between baseline and Cycle 2, predictive of EFS. These results suggest that an early readout of response after brief exposure to therapy may minimize the exposure to ineffective toxic treatments and may also help guide decision‐making regarding adjustment of treatment strategies targeting the residual tumor burden. In addition to breast and lymph node imaging, response evaluation tools reflecting gene expression profiles before and after a brief exposure to chemotherapy should be included in the development of future neoadjuvant treatment strategies in breast cancer.
Material and Methods
Study design and treatment
Between September 2008 and November 2011, 150 women with large, operable and locally advanced HER2‐negative breast cancer were enrolled in the multi‐center single‐arm Phase II clinical trial, PROMIX (ClinicalTrials.gov NCT00957125). Eligible patients were women aged ≥18 years with localized HER2‐negative primary breast cancer suitable for primary medical treatment with or without regional lymph node metastases, with adequate bone marrow, renal, hepatic and cardiac functions, no other uncontrolled medical or psychiatric disorders and with an ECOG performance status of 0–1. The main exclusion criteria were distant metastases, other malignancy in the past two years (except for radically treated basal or squamous cell carcinoma of the skin or carcinoma in situ of the cervix), HER2 amplification and pregnancy or lactation.
The patients were scheduled to receive six cycles of epirubicin and docetaxel, at doses of 75 mg/m2 i.v. each once every three‐week cycle, with the addition of bevacizumab (15 mg/kg i.v.) on Day 1 during cycles 3–6 in the absence of a clinical complete response (cCR), followed by surgery.
Clinical and radiological response evaluations were performed after two, four and six courses of treatment. All patients underwent core needle biopsies, which were collected free‐hand or with ultra‐sound guidance at baseline and after Cycle 2, while post‐treatment residual tumor biopsy samples were collected during surgery. Patients post‐surgery were treated according to the Swedish national guidelines.17 The CONSORT diagram in Figure 1 illustrates the clinical trial and number of patients and tumor samples included. Pathology review was performed locally. ER and progesterone receptor status were determined by immunohistochemistry and was considered positive if ≥10% of tumor cells were positive. The clinicopathological data at baseline are summarized in Table 1. The study was approved by the Ethics Committee at Karolinska University Hospital, 2007/1529–31/2 and patients provided written informed consent to participate. For more details about the trial, see ClinicalTrials.gov and Supporting Information Materials.
Figure 1.
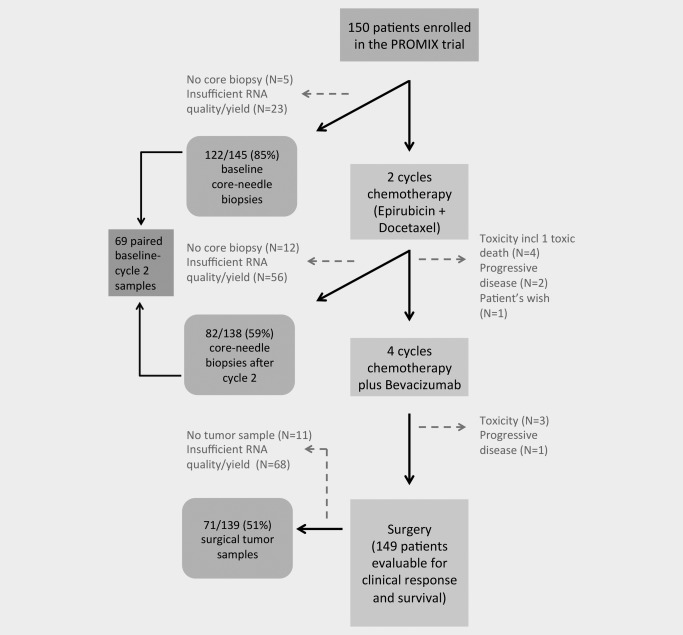
CONSORT diagram illustrating the PROMIX clinical trial, depicting patient enrollment and tumor biopsy retrieval for gene expression profiling.
Table 1.
Baseline clinicopathological characteristics for the patients included in the PROMIX trial
| Factor | N (%) |
|---|---|
| Age (years) | |
| Mean | 50 |
| Range | 28–71 |
| ER status | |
| Positive | 107 (73%) |
| Negative | 40 (27%) |
| NA | 2 |
| PR status | |
| Positive | 83 (57%) |
| Negative | 63 (43%) |
| NA | 3 |
| Histology | |
| Ductal | 107 (73%) |
| Lobular | 22 (15%) |
| Other | 18 (12%) |
| NA | 2 |
| Nodal status | |
| Positive | 89 (60%) |
| Negative | 60 (40%) |
| NA | 0 |
| Tumor size (mm) | |
| Mean | 58.9 |
| Range | 20–180 |
| Clinical subtype | |
| HR positive | 111 (76%) |
| TNBC | 36 (24%) |
| NA | 2 |
| PAM50 subtype | |
| Luminal A | 25 (21%) |
| Luminal B | 55 (45%) |
| HER2‐enriched | 6 (5%) |
| Basal | 26 (21%) |
| Normal‐like | 10 (8%) |
| NA | 27 |
| AIMS subtype | |
| Luminal A | 19 (16%) |
| Luminal B | 29 (24%) |
| HER2‐enriched | 19 (16%) |
| Basal | 30 (24%) |
| Normal‐like | 25 (20%) |
| NA | 27 |
NA: not available; ER: estrogen receptor; PR: progesterone receptor; HR: hormone receptor; TNBC: triple‐negative breast cancer; PAM50: prediction analysis of microarrays 50; AIMS: Absolute Intrinsic Molecular Subtyping.
Endpoints
The primary endpoint of the trial was pCR, defined as the absence of invasive cancer in the breast and lymph nodes at surgery; presence of residual non‐invasive DCIS was allowed. Secondary endpoints included EFS (time from surgery to breast cancer metastasis or death), OS (time from surgery to death), and transcriptional changes after brief exposure to neoadjuvant therapy.
A power analysis was performed prior to initiation of the trial based on the assumption that 120 patients would undergo surgery after the sixth treatment cycle, and that 25% would achieve a pCR. The power for detection of a true difference in, for example, molecular markers of 1 SD between patients with pCR compared with those without was >99%, with α = 0.05.
Gene expression analysis
Core biopsies (at baseline and after Cycle 2) and surgical biopsies (after Cycle 6) were snap frozen immediately. Total RNA was extracted from fresh frozen tumor samples using the Allprep DNA/RNA mini kit (QIAGEN, Valencia, CA) in a QIAcube (QIAGEN) according to the manufacturer's instructions. RNA was quantified using a NanoDrop ND‐1000 (NanoDrop Products, Wilmington, DE) and the RNA integrity was assessed on a Caliper LabChip (Advanced Molecular Vision, Lincolnshire, UK). Global transcriptional profiles were determined using Illumina Human HT‐12 v4.0 Expression BeadChips (Illumina, San Diego, CA) in three randomized batches at the SCIBLU Genomics Center at Lund University, Sweden. Three samples were included in all batches to control and manage potential technical related biases. The data were normalized using BioArray Software Environment (BASE)18 and R.19 Data were background corrected and quantile normalized and probes with p‐values >0.01, >30% missing values and/or a variance <0.15 were filtered out. Furthermore, the Illumina probes were re‐annotated using Re‐annotation and Mapping for Oligonucleotide Array Technologies (ReMOAT),20 and only probes annotated as “good” or “perfect” quality were selected for further analyses. The remaining probes were log2 transformed and mean centered across tumors. Gene expression data are available through GEO (accession number: GSE87455). A principal component analysis (PCA) was performed and associations between principal components and technical and biological annotations were evaluated; no serious batch effect or technical bias requiring correction was detected (Supporting Information Fig. S1).
All tumors were centroid classified into the five intrinsic subtypes using the PAM50 gene signature.21 To confirm the robustness of the subtype determination, different methods of computing the PAM50 subtypes were applied: (i) subtyping after exclusion of surgical biopsies (which were mainly normal‐like and could bias the subtype assignments); (ii) normalization of all samples in the PROMIX cohort with a large cohort of 2,000 primary (untreated) breast cancers,22 followed by subtyping; and (iii) subtyping using the recently developed single sample predictor Absolute Intrinsic Molecular Subtyping (AIMS).23 The agreement between the PAM50 subtype calls between the different cohorts (all PROMIX only, PROMIX excluding surgical biopsies, and all PROMIX with 2,000 primary breast cancers) was very high (Kappa >0.9 for all comparisons) and the agreement between the PAM50 versus AIMS for all PROMIX samples was moderate (Kappa = 0.6).
To explore the significance of global subtype changes over time (i.e., during neoadjuvant treatment), an index, delta‐PAM50 (ΔPAM50), was computed by adding the absolute values for each of the intrinsic PAM50 subtype correlations for each tumor and calculating the change in this index between baseline and Cycle 2. This ΔPAM50 index reflects the change in all five subtype correlations upon neoadjuvant treatment, providing a numeric value corresponding to the global changes in subtype correlations. Scores, ranging from 0.116 to 4.159, were ranked and plotted to display the distribution (Supporting Information Fig. S2a), whereby the data were divided into tertiles to explore the long‐term (EFS) prognostic value of the ΔPAM50 index. To confirm the robustness of the index, the deltas for all the different methods of computing subtypes were also tested and very high correlations (r > 0.9 for all comparisons) were observed between all the delta values, irrespective of the method used. Highly similar survival distributions were obtained regardless of method used (Supporting Information Figs. S2b–S2e).
Finally, the activity of previously defined gene signatures related to hypoxia,24 vascular invasion,25 stromal PDGF signaling26 and seven previously described modules representing biological processes related to breast cancer,27 was calculated for each tumor as described in the respective original publications.
Statistical analyses
Correlative analyses investigating associations between outcome (pCR and EFS) and tumor pathological factors, and treatment‐induced changes in gene signatures and outcome, were evaluated by uni and multivariable logistic regression or Cox proportional hazards models, where applicable. Gene signatures were analyzed on a continuous scale. Kaplan–Meier plots were generated to visualize the survival difference between patients stratified by conventional prognostic variables (ER status, nodal status, proliferation, molecular subtype) and pCR (using a landmark approach) and the log‐rank test was used to check for statistically significant differences. p‐values from two‐sided statistical tests are reported and p < 0.05 was considered significant.
Results
Biopsy sample retrieval and RNA extraction
Core needle biopsy samples were successfully collected from 145/150 (97%) patients at baseline, 138 (92%) after Cycle 2 and tissue samples from 139 (93%) at surgery. The mRNA quality and yield was adequate for the generation of high quality gene expression data from 122/145 samples (85%) at baseline, 82/138 (59%) after Cycle 2 and 71/139 (51%) at surgery (Fig. 1). There were paired baseline–Cycle 2 data for 69 patients.
One hundred and thirty‐nine of 150 patients (93%) received all six cycles of chemotherapy–bevacizumab combination treatment. Treatment was prematurely disrupted in 11 patients (7%; Fig. 1). All patients were evaluable for response, except for one patient who died due to toxic colitis after the first course of treatment and was excluded from further analyses.
At baseline, 24% of evaluable tumors were triple‐negative breast cancers (TNBCs) and 76% were HR+ (Table 1).
Clinical endpoints
Objective response
No patient achieved a clinical complete response (cCR) after two cycles of chemotherapy. In total, 20 patients [13% (95% CI 8.6–20.2)] achieved a pCR and 100 patients [67% (95% CI 58.8–74.5)] experienced partial response. Among the 20 patients achieving a pCR, 16 had no residual tumor (ypT0 ypN0), and four displayed residual DCIS (ypT0/Tis ypN0).
pCR by subtype at baseline
The pCR rate was significantly higher among TNBCs [28% (95% CI 14.8–45.4)] compared with the HR+ tumors [9% (95% CI 4.6–16.3)]; [OR = 3.9 (CI = 1.5–10.3), Supporting Information Table S1]. Seventeen patients who achieved pCR had baseline gene expression data. pCR rates varied significantly between the PAM50 subtypes; 8% (2/25) of luminal A, 6% (3/55) of luminal B, 17% (1/6) of HER2‐enriched, 20% (2/10) of normal‐like and 35% (9/26) of basal tumors. Importantly, although 53% (9/17) of the evaluable patients achieving a pCR had basal tumors at baseline, the specificity of the basal subtype for predicting pCR was only 35% (9/26). Nonetheless, the odds of a patient with a basal tumor achieving a pCR was significantly higher than that of a patient with a luminal A tumor [OR = 6.1 (CI = 1.2–32)]. Using the single sample predictor AIMS, the pCR rates followed a similar pattern, with 0, 4, 10, 20 and 30% for luminal A, luminal B, HER2‐enriched, normal‐like and basal tumors, respectively. Likewise, patients with basal tumors had a significantly higher likelihood of achieving a pCR compared with luminal B tumors [OR = 12.0 (CI = 1.4–102)]. HR status and molecular subtype were however not independently predictive of pCR when included in the same multivariable logistic regression model. Taken together, although correlations between HR status/molecular subtype and pCR were observed, their sensitivity and specificity for predicting pCR was only modest.
Event‐free and overall survival
The cut‐off for survival data presented was March 9, 2016. The median follow‐up for EFS and OS was 48.3 (IQR 35–60) and 49.0 (IQR 41–60) months, respectively. Forty patients (27%) had a recurrence and 29 (19%) died from disseminated breast cancer. There was no statistically significant difference in EFS and OS for patients with pCR vs. no pCR in the whole cohort (Figs. 2 a and 2 b), but a trend toward inferior EFS and OS was noted among patients with a pCR relative to those without. As most patients experiencing a pCR had basal tumors, a subtype conferring a poor prognosis, a post hoc survival analysis restricted to basal tumors was conducted. Inferior EFS and OS was observed for patients without a pCR in this subgroup, although this was not significant (Figs. 2 c and 2 d).
Figure 2.
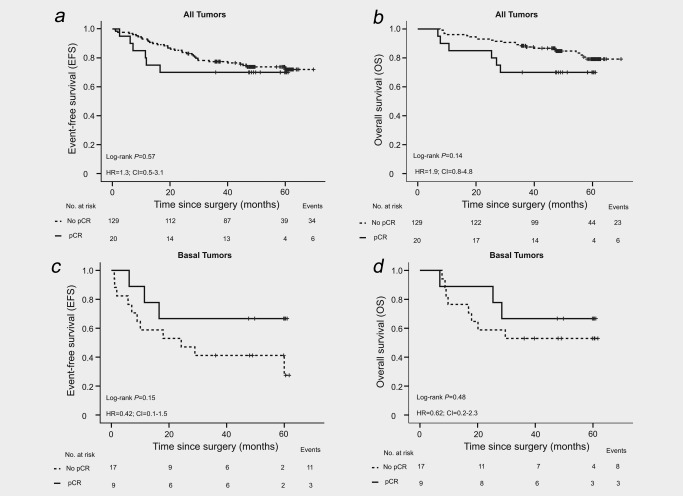
Kaplan–Meier survival analyses. (a) EFS and (b) OS for all patients in the trial (N = 149). (c) EFS and (d) OS for patients with basal tumors (N = 26).
Due to the poor performance of pCR in predicting long‐term prognosis in this cohort, we investigated the ability of conventional prognostic factors in predicting long‐term outcome. As expected, HR negativity, positive nodal status, high tumor proliferation index (Ki67) and the basal subtype were all significantly prognostic for an inferior EFS and OS in univariable analyses (p < 0.05 for all factors, Supporting Information Table S2).
The non‐significant trend for better long‐term outcome for patients with basal tumors experiencing a pCR indicates that even within this subtype, pCR is not a precise surrogate marker for long‐term survival.
Translational endpoints
Molecular subtype changes in relation to outcome
The distribution of the molecular subtypes changed during treatment (Fig. 3 a, Supporting Information Table S3). The comparison of matched baseline and Cycle 2 gene expression profiles suggested that luminal B to luminal A conversions were common and a shift to the normal‐like subtype was predominant compared with a change to the other subtypes. Of the 10 patients achieving a pCR with available paired baseline and Cycle 2 data, seven changed PAM50 subtype; six (86%) of these changed to the normal‐like subtype (Fig. 3 b). By contrast, among the 59 patients not achieving a pCR with paired baseline and Cycle 2 data, only 46% changed PAM50 subtype; 30% of these changed to the normal‐like subtype (Fig. 3 c). Very similar shifts were seen for AIMS (Supporting Information Fig. S3, Supporting Information Table S3).
Figure 3.
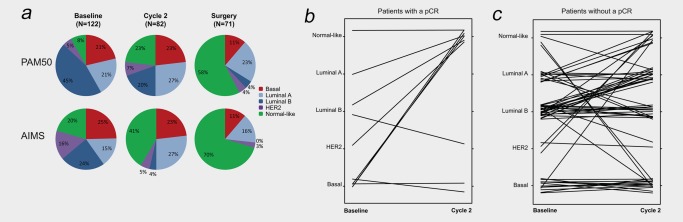
Molecular subtypes. (a) Distribution of PAM50 and AIMS subtypes during treatment. (b) PAM50 subtype switches between baseline and after Cycle 2 for patients with a pCR (N = 10). (c) PAM50 subtype switches between baseline and after Cycle 2 for patients without a pCR (N = 59). [Color figure can be viewed at wileyonlinelibrary.com]
The changes in the distribution of subtypes between baseline and Cycle 2 and baseline and surgery were statistically significant (McNemar's p < 0.05 for all comparisons). Of note, patients with basal tumors at baseline who did not achieve a pCR were also less likely to change subtype. However, subtype after Cycle 2 was not a significant predictor of the likelihood of achieving a pCR (PAM50, p = 0.5; AIMS, p = 1.0), but a statistically significant association was noted between subtype after Cycle 2 and both EFS (Log‐rank p = 0.003 for PAM50; p < 0.001 for AIMS) and OS (Log‐rank p = 0.006 for PAM50; p < 0.001 for AIMS). Inferior survival was associated with basal tumors, as expected. These findings suggest that a change in subtype early in the course of treatment provides pertinent information regarding long‐term outcome.
As discrepant results have been reported regarding the concordance of the molecular subtypes between matched pre‐ and post‐treatment exposure biopsy samples,11, 28, 29 we investigated if the observed subtype conversions were associated with a true treatment response or whether they reflect tumor heterogeneity. First, we examined the expression of the proliferation‐associated gene module (AURKA), which is most likely to be affected in response to chemotherapy, and the ESR1 and ERBB2 modules, which are less likely to be altered by this treatment,27 and found a significant decrease of the AURKA module, but no significant change in the scores of the ESR1 and ERBB2 modules between the paired baseline and Cycle 2 samples (Supporting Information Fig. S4).
To further investigate the correlation between subtype conversions and outcome an index, ΔPAM50, representing the sum of the absolute changes in all PAM50 subtype correlations between baseline and Cycle 2, was computed for each patient. We then tested the hypothesis that the value of this index was associated with EFS. Intriguingly, patients whose tumors changed subtype between baseline and Cycle 2 had a better prognosis than those whose tumors remained stable (Fig. 4 a–d, Supporting Information Fig. S5), and an increase in the absolute ΔPAM50 index correlated with prolonged EFS [HR = 0.62 (CI = 0.3–1.1); Fig. 4 e], irrespective of pCR status on completion of neoadjuvant chemotherapy. Importantly, this finding was robust, as the same results were obtained irrespective of the subtyping method (Supporting Information Fig. S2). These exploratory analyses suggest that treatment‐induced shifts in molecular subtypes, measured as early as after two cycles of treatment, may be clinically relevant, although the role of intratumoral heterogeneity due to sampling or treatment requires further study.
Figure 4.
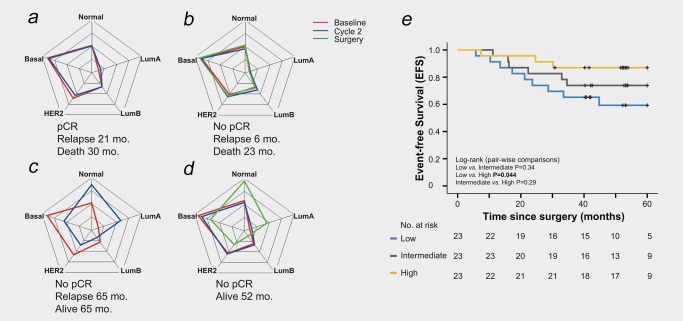
PAM50 subtype correlations between baseline and Cycle 2 in relation to outcome. (a–d) Examples of spider plots (correlations from −1 in the center, to 1 at each node) showing correlations to all PAM50 subtypes in representative patients with basal tumors with pCR status and survival time indicated. (e) Kaplan–Meier survival plot showing the association between the absolute change in subtype correlation (ΔPAM50 index) and EFS (N = 69). [Color figure can be viewed at wileyonlinelibrary.com]
Predicting outcome using pre‐defined gene signatures
Finally, we investigated if pre‐ or on‐treatment sampling specifically modified the associations between nine gene signatures24, 25, 26, 27 and outcome in the clinical subtypes (HR+ and TNBC). Supporting Information Figure S6 depicts the activity of each signature at baseline and after Cycle 2 in relation to pCR and EFS. Overall, differential expression of signatures after Cycle 2 displayed a better association with outcome compared to baseline levels. Specifically, among HR+ tumors, pCR was significantly associated with the baseline activity of only three signatures (high apoptosis and immune response and low ER signaling scores) compared with the Cycle 2 activity of six signatures (low angiogenesis, proliferation, ER signaling and activated stroma scores, and high invasion and metastasis and immune response scores; Figs. 5 a and 5 b). The predictive potential of the angiogenesis, apoptosis, invasion and metastasis, proliferation, and immune response signatures remained statistically significant when the change in the signature activity between baseline and Cycle 2 was considered for pCR (Supporting Information Fig. S7a). By contrast, only the baseline activity of the activated stroma signature was significantly associated with EFS among HR+ tumors (Fig. 5 c).
Figure 5.

Forest plots showing associations between outcome and gene signatures in HR+ (a–d) and TNBC (e–h) tumors, respectively. Odds ratios for pCR for each signature at baseline (a, e) and after Cycle 2 (b, f). Hazard ratios for EFS for each signature at baseline (c, g) and after Cycle 2 (d, h). *p < 0.05.
Within the TNBC subset, none of the signatures were significantly associated with pCR (Figs. 5 e and 5 f), while a significant association was noted between disease recurrence and high Cycle 2 activity of the angiogenesis and hypoxia signatures as well as low ER signaling after Cycle 2 (Fig. 5 h). Moreover, a consistent and significant association was observed between high tumor proliferation activity and increased incidence of metastasis both after Cycle 2 and for the change between baseline and Cycle 2 (Fig. 5 h, Supporting Information Fig. S7d).
Taken together, these data suggest that brief exposure to chemotherapy may modify the associations between certain gene signatures and outcome, lending support to the proposal that a biopsy sample obtained during neoadjuvant treatment may improve the potential to predict both short‐term (pCR) and long‐term (EFS) outcome, albeit to a varying extent between HR+ and TNBC subtypes.
Discussion
The potential advantages of neoadjuvant systemic chemotherapy are several, including higher rates of breast conserving surgery, allowing for the possibility of measuring in vivo therapy response early and thereby enabling adjustment of treatment strategies and unique opportunities for individualization of treatment strategies.9 To leverage these potentials, we have evaluated the sensitivity of molecular signatures and biomarkers from early on‐treatment samples to predict response to neoadjuvant treatment in different breast cancer subtypes in the PROMIX trial.
The overall pCR rate was 13%, which is lower than the 18–59% rates reported for similar trials evaluating the addition of bevacizumab to neoadjuvant anthracycline–taxane‐based chemotherapy in HER2‐negative breast cancer.30, 31, 32, 33 Inter‐trial differences in the distribution of patients in the clinical and molecular subtypes, tumor stage at entry, type, dose, design and timing of chemotherapy regimens, numbers of bevacizumab cycles and in the definitions of pCR, with or without centralized review of tumor‐pathological factors, may account for these differences. Altered vasculature may also contribute to treatment resistance. Nonetheless, our study confirmed the notion that pCR rates differ between subtypes, with the highly proliferating TNBC subset and, specifically, the basal tumors attaining the highest pCR rates.3 It is becoming increasingly clear that molecular features, including molecular subtypes, need to be taken into consideration when recruiting patients to neoadjuvant trials and when interpreting pCR rates.2 As a consequence, treatment studies are preferentially restricted to specific subsets of breast cancers today.
In 2012, the FDA approved pCR as a valid predictor of long‐term clinical benefit, such as DFS and OS, in neoadjuvant settings.34 However, assessment of pCR as a surrogate end‐point for long‐term prognosis following neoadjuvant chemotherapy is more relevant for HER2‐positive and TNBC, less valid in luminal B (HER2‐negative or positive) tumors and probably less informative for luminal A tumors.2, 3 In a two‐step multi‐variable analysis, excluding the interaction between subtype and pCR, Bonnefoi et al. showed that pCR contributed to the prediction of EFS across all subtypes, although this was not significant for the less proliferative subtypes.16 These assertions are corroborated by the present study and by others.3 In this study, patients with basal tumors, with or without a pCR, still experienced a significantly worse outcome than patients with luminal tumors, but post hoc analyses among basal tumors suggested that patients achieving a pCR may expect a better EFS and OS. These findings continue to emphasize the need for better therapies for women with basal tumors, and for additional factors to predict long‐term outcome, particularly in patients, such as those with luminal subtypes, who may not achieve pCR but who may still derive benefit from a neoadjuvant therapeutic approach. Several models for the evaluation of prognosis in relation to residual disease burden after neoadjuvant chemotherapy have been developed in order to refine the evaluation beyond pCR/non‐pCR. Such models have proven useful for predicting prognosis, but obviously lack the possibility to guide treatment modifications early during the treatment course. In this context, Symmans and colleagues recently reported that a residual cancer burden (RCB) index was prognostic for long‐term survival after neoadjuvant chemotherapy in HR‐positive/HER2‐negative, HER2‐positive and TNBC subsets of breast cancer,35 emphasizing the importance of assessing tumor characteristics after exposure to treatment.
Due to the single‐arm design of the PROMIX study, the specific contribution of bevacizumab to outcome remains unclear. Nonetheless, the repeated finding from previous trials36, 37 and this study that increased pCR rates may not translate into more favorable outcomes remains a concern.
In this trial, we were able to retrieve tumor material from a substantial fraction of patients at baseline, during treatment (Cycle 2) and at surgery. Predicting response to neoadjuvant chemotherapy based on analyzing baseline samples alone has so far been unsuccessful, especially among HR+ tumors.7, 8, 9 In our analyses, short‐term exposure to chemotherapy significantly modulated the gene expression profiles of many tumors. Tumors from patients achieving a pCR generally changed to the normal‐like subtype already after two cycles of chemotherapy. Importantly, a higher absolute subtype correlation difference between baseline and Cycle 2 (ΔPAM50 index) was associated with prolonged EFS, indicating the robust prognostic importance of the molecular subtypes. Similar shifts toward the normal‐like subtype were observed among 32 women receiving neoadjuvant chemotherapy in the population‐based SCAN‐B study,38 in which 50% of tumors changed subtype, one‐third of which became normal‐like. These findings suggest that molecular subtype shifts may reflect a plasticity of some tumors regardless of subtype, a feature which may be a useful indicator of outcome after neoadjuvant therapy; however, the role of other factors, such as stromal alterations, intratumoral heterogeneity, and tumor proliferation changes needs to be adequately investigated before firm conclusions can be drawn.
The interim molecular evaluation investigated as part of the PROMIX trial also revealed that the ability to predict outcome by nine gene signatures was greatly improved when an early‐treatment sample was used, especially among HR+ tumors, a subgroup for which biomarkers of pCR or long‐term prognosis after neoadjuvant therapy are seriously lacking. Although no gene signature was found to be associated with pCR among TNBCs in the present study, proliferation rate and ER signaling after Cycle 2 were significantly associated with EFS. By using baseline gene expression profiles, Prat et al. also showed that proliferation and luminal signatures were significantly associated with pCR and DFS, although this was specifically in basal tumors and not TNBCs per se.39 The PROMIX trial was not powered to specifically study TNBC/basal tumors. The modest success rate for gene expression profiling could reflect a possible confounding effect of tumor heterogeneity, possibly enhanced by the large size of the tumors. This also underpowered the statistical analyses, warranting caution when interpreting these exploratory analyses and underlining the need for further studies to validate and extend our results.
In conclusion, we suggest that evaluation of gene expression changes in samples taken early in the course of neoadjuvant treatment can provide predictive information that may be used to inform further treatment strategies and may provide the opportunity for truly personalized medicine. Based on these findings, we propose that the coming generation of neoadjuvant trials should include sampling of residual tumor during treatment and at surgery for assessment of, for example, ΔPAM50 to guide adjuvant treatment in the controlled clinical trial setting.
Supporting information
Supporting Information
Acknowledgements
The authors are grateful to the women who participated in the PROMIX trial. We thank Anna Ebbesson for technical assistance.
Financial disclosures: J. Bergh: Research grants from Astra Zeneca, Amgen, Bayer, Merck, Pfizer, Roche and Sanofi‐Aventis to Karolinska Institutet and University Hospital; payment from UpToDate to Asklepios Medicine HB for a book chapter on breast cancer diagnostics. T. Hatschek: Research grants from Roche and Pfizer; Translational Advisory Board, Pfizer. No other financial interests or relationships of commercial nature that might be construed as a conflict of interest are disclosed by any of the authors.
Contributor Information
Thomas Hatschek, Email: thomas.hatschek@ki.se.
Ingrid Hedenfalk, Email: Ingrid.Hedenfalk@med.lu.se.
References
- 1. van der Hage JA, van de Velde CJ, Mieog JS. Preoperative chemotherapy for women with operable breast cancer (Review). The Cochrane Library 2009;(2):CD005002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. von Minckwitz G, Untch M, Blohmer JU, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 2012;30:1796–804. [DOI] [PubMed] [Google Scholar]
- 3. Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long‐term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164–72. [DOI] [PubMed] [Google Scholar]
- 4. Symmans WF, Peintinger F, Hatzis C, et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 2007;25:4414–22. [DOI] [PubMed] [Google Scholar]
- 5. von Minckwitz G, Schmitt WD, Loibl S, et al. Ki67 measured after neoadjuvant chemotherapy for primary breast cancer. Clin Cancer Res. 2013;19:4521–31. [DOI] [PubMed] [Google Scholar]
- 6. Toi M, Lee SJ, Ohtani S, et al. A phase III trial of adjuvant capecitabine in breast cancer patients with HER2‐negative pathologic residual invasive disease after neoadjuvant chemotherapy (CREATE‐X, JBCRG‐04). Proceedings from the San Antonio Breast Cancer Symposium, December 8–12, 2015; San Antonio, TX [Google Scholar]
- 7. Huober J, von Minckwitz G, Denkert C, et al. Effect of neoadjuvant anthracycline‐taxane‐based chemotherapy in different biological breast cancer phenotypes: overall results from the GeparTrio study. Breast Cancer Res Treat 2010;124:133–40. [DOI] [PubMed] [Google Scholar]
- 8. von Minckwitz G, Kummel S, Vogel P, et al. Intensified neoadjuvant chemotherapy in early‐responding breast cancer: phase III randomized GeparTrio study. J Natl Cancer Inst 2008;100:552–62. [DOI] [PubMed] [Google Scholar]
- 9. Untch M, Konecny GE, Paepke S, et al. Current and future role of neoadjuvant therapy for breast cancer. Breast 2014;23:526–37. [DOI] [PubMed] [Google Scholar]
- 10. Yeo B, Dowsett M. Neoadjuvant endocrine therapy: Patient selection, treatment duration and surrogate endpoints. Breast 2015;24: S78–83. [DOI] [PubMed] [Google Scholar]
- 11. Varadan V, Kamalakaran S, Gilmore H, et al. Brief‐exposure to preoperative bevacizumab reveals a TGF‐beta signature predictive of response in HER2‐negative breast cancers. Int J Cancer 2016;138:747–57. [DOI] [PubMed] [Google Scholar]
- 12. Esserman LJ, Berry DA, DeMichele A, et al. Pathologic complete response predicts recurrence‐free survival more effectively by cancer subset: results from the I‐SPY 1 TRIAL–CALGB 150007/150012, ACRIN 6657. J Clin Oncol 2012;30:3242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Houssami N, Macaskill P, von Minckwitz G, et al. Meta‐analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur J Cancer 2012;48:3342–54. [DOI] [PubMed] [Google Scholar]
- 14. Kong X, Moran MS, Zhang N, et al. Meta‐analysis confirms achieving pathological complete response after neoadjuvant chemotherapy predicts favourable prognosis for breast cancer patients. Eur J Cancer 2011;47:2084–90. [DOI] [PubMed] [Google Scholar]
- 15. Berruti A, Amoroso V, Gallo F, et al. Pathologic complete response as a potential surrogate for the clinical outcome in patients with breast cancer after neoadjuvant therapy: a meta‐regression of 29 randomized prospective studies. J Clin Oncol 2014;32:3883–91. [DOI] [PubMed] [Google Scholar]
- 16. Bonnefoi H, Litiere S, Piccart M, investigators EBS , et al. Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: a landmark and two‐step approach analyses from the EORTC 10994/BIG 1‐00 phase III trial. Ann Oncol 2014;25:1128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.SweBCG, Nationella Riktlinjer for behandling av bröstcancer, 2013. Available at: http://www.swebcg.se/vardprogram/. Accessed May 2017.
- 18. Vallon‐Christersson J, Nordborg N, Svensson M, et al. BASE–2nd generation software for microarray data management and analysis. BMC Bioinformatics 2009;10:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Team RDC. The R Project for Statistical Computing, www.r-project.org. Available at: https://www.r-project.org. Accessed December 2016.
- 20. Barbosa‐Morais NL, Dunning MJ, Samarajiwa SA, et al. A re‐annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res 2010;38:e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paquet ER, Hallett MT. Absolute assignment of breast cancer intrinsic molecular subtype. J Natl Cancer Inst 2015;107:357 [DOI] [PubMed] [Google Scholar]
- 24. Buffa FM, Harris AL, West CM, et al. Large meta‐analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer 2010;102:428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mannelqvist M, Stefansson IM, Bredholt G, et al. Gene expression patterns related to vascular invasion and aggressive features in endometrial cancer. Am J Pathol 2011;178:861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frings O, Augsten M, Tobin NP, et al. Prognostic significance in breast cancer of a gene signature capturing stromal PDGF signaling. Am J Pathol 2013;182:2037–47. [DOI] [PubMed] [Google Scholar]
- 27. Desmedt C, Haibe‐Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res 2008;14:5158–65. [DOI] [PubMed] [Google Scholar]
- 28. Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 29. Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol 2016;34:542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. von Minckwitz G, Eidtmann H, Rezai M, et al. Neoadjuvant chemotherapy and bevacizumab for HER2‐negative breast cancer. N Engl J Med 2012;366:299–309. [DOI] [PubMed] [Google Scholar]
- 31. Sikov WM, Berry DA, Perou CM, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once‐per‐week paclitaxel followed by dose‐dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple‐negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 2015;33:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Earl HM, Hiller L, Dunn JA, et al. Efficacy of neoadjuvant bevacizumab added to docetaxel followed by fluorouracil, epirubicin, and cyclophosphamide, for women with HER2‐negative early breast cancer (ARTemis): an open‐label, randomised, phase 3 trial. Lancet Oncol 2015;16:656–66. [DOI] [PubMed] [Google Scholar]
- 33. Bear HD, Tang G, Rastogi P, et al. Bevacizumab added to neoadjuvant chemotherapy for breast cancer. N Engl J Med 2012;366:310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.FC MY, Pathologic Complete Response in Neoadjuvant Treatment of High‐Risk Early‐Stage Breast Cancer: Use as an Endpoint to Support Accelerated Approval, 2012.
- 35. Symmans WF, Wei C, Gould R, et al. Long‐term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol 2017; 35:1049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nahleh ZA, Barlow WE, Hayes DF, et al. SWOG S0800 (NCI CDR0000636131): addition of bevacizumab to neoadjuvant nab‐paclitaxel with dose‐dense doxorubicin and cyclophosphamide improves pathologic complete response (pCR) rates in inflammatory or locally advanced breast cancer. Breast Cancer Res Treat 2016;158:485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von Minckwitz G, Loibl S, Untch M, groups GA‐Bs , et al. Survival after neoadjuvant chemotherapy with or without bevacizumab or everolimus for HER2‐negative primary breast cancer (GBG 44‐GeparQuinto). Ann Oncol 2014;25:2363–72. [DOI] [PubMed] [Google Scholar]
- 38. Saal LH, Vallon‐Christersson J, Hakkinen J, et al. The Sweden Cancerome Analysis Network ‐ Breast (SCAN‐B) initiative: a large‐scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med 2015;7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prat A, Lluch A, Albanell J, et al. Predicting response and survival in chemotherapy‐treated triple‐negative breast cancer. Br J Cancer 2014;111:1532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


