Abstract
Renal epithelial cells are exposed to mechanical forces due to flow‐induced shear stress within the nephrons. Shear stress is altered in renal diseases caused by tubular dilation, obstruction, and hyperfiltration, which occur to compensate for lost nephrons. Fundamental in regulation of shear stress are primary cilia and other mechano‐sensors, and defects in cilia formation and function have profound effects on development and physiology of kidneys and other organs. We applied RNA sequencing to get a comprehensive overview of fluid‐shear regulated genes and pathways in renal epithelial cells. Functional enrichment‐analysis revealed TGF‐β, MAPK, and Wnt signaling as core signaling pathways up‐regulated by shear. Inhibitors of TGF‐β and MAPK/ERK signaling modulate a wide range of mechanosensitive genes, identifying these pathways as master regulators of shear‐induced gene expression. However, the main down‐regulated pathway, that is, JAK/STAT, is independent of TGF‐β and MAPK/ERK. Other up‐regulated cytokine pathways include FGF, HB‐EGF, PDGF, and CXC. Cellular responses to shear are modified at several levels, indicated by altered expression of genes involved in cell‐matrix, cytoskeleton, and glycocalyx remodeling, as well as glycolysis and cholesterol metabolism. Cilia ablation abolished shear induced expression of a subset of genes, but genes involved in TGF‐β, MAPK, and Wnt signaling were hardly affected, suggesting that other mechano‐sensors play a prominent role in the shear stress response of renal epithelial cells. Modulations in signaling due to variations in fluid shear stress are relevant for renal physiology and pathology, as suggested by elevated gene expression at pathological levels of shear stress compared to physiological shear.
Keywords: cilium, fluid flow, glycocalyx, mechanotransduction, next generation sequencing
1. INTRODUCTION
Several organs are subject to variations in fluid flow rate in response to physiological stimuli, which could be detected by different cell types via mechano‐sensing proteins or complexes. Cellular mechano‐sensitivity and mechanotransduction are essential for normal cell function, tissue development, and maintenance of organs (Goetz & Anderson, 2010; Freund, Goetz, Hill, & Vermot, 2012; Quinlan, Tobin, & Beales, 2008; Weinbaum, Duan, Satlin, Wang, & Weinstein, 2010). In the kidneys, where urinary volume, diuretics, and diet will expose the renal epithelial cells to variations in hydrodynamic forces including fluid shear stress, circumferential stretch, and drag/torque on apical cilia and probably also on microvilli (Carrisoza‐Gaytan, Carattino, Kleyman, & Satlin, 2016). Depending on the cell type and the magnitude of the hydrodynamic forces, different responses will be activated and mutations in critical components may modulate or cause (kidney) diseases (Piperi & Basdra, 2015). In addition, strong variations in hydrodynamic forces and shear stress are common in kidney diseases due to hyperfiltration, tubular dilation, and obstruction, which occur in functional nephrons, to compensate for lost glomeruli and tubules, with diabetic nephropathy and Polycystic Kidney Disease as the most common examples (Sharma, Mucino, & Ronco, 2014).
Fundamental in flow‐sensing are a number of proteins located throughout the cell membrane, cilium/ciliary base, as well as the cytoskeleton. These include ion channels, G‐protein coupled receptors (GPCRs), adherens junction proteins, focal adhesion proteins, components of the actin cytoskeleton, but also glycocalyx and lipid rafts can act as mechano‐sensors to shear stress (Curry & Adamson, 2012; Ingber, 2006; Petersen, Chung, Nayebosadri, & Hansen, 2016). Activation of aforementioned sensors upon shear stress leads to alteration of cellular signaling. Bending of the primary cilium causes ciliary influx of Ca2+, followed by an increase in cytosolic Ca2+ (DeCaen, Delling, Vien, & Clapham, 2013; Delling, DeCaen, Doerner, Febvay, & Clapham, 2013; Praetorius, Frokiaer, Nielsen, & Spring, 2003; Praetorius & Spring, 2001). It is likely that the increase in intraciliary Ca2+ does not spread to the cytosol suggesting the requirement of additional steps for amplification of the Ca2+ signal, although details are not entirely clear and under debate (Delling et al., 2013, 2016; Praetorius, 2015). Other cilia‐dependent signaling cascades affected by fluid flow include the canonical Wnt‐signaling pathway, which is restrained by fluid‐flow induced ciliary signaling in favor of non‐canonical Wnt signaling (Simons et al., 2005). Furthermore, mTOR signaling and cell‐size control, as well as STAT6/p100‐regulated transcription are thought to be negatively regulated upon flow‐induced bending of the cilium, independent from flow‐induced Ca2+ influx (Boehlke et al., 2010; Low et al., 2006; Weimbs, 2007; Zhong et al., 2016). Cilia‐independent shear‐induced alterations in renal signaling include increased Na+ and HCO3 − reabsorption and autocrine TGF‐β/ALK5 signaling (Kotsis, Boehlke, & Kuehn, 2013; Kunnen et al., 2017).
It is currently not known in detail how fluid shear stress affects cellular behavior and which signaling pathways are altered. Furthermore, gene expression and the overall cellular behavior will be the effect of an integration of the different signaling pathways, triggered by shear stress and by cytokine stimulation. In this study we set out to obtain a comprehensive overview of the transcriptome under static and shear stress conditions in renal epithelial cells to get more insight in the pathways and processes involved in the shear response. Therefore, we applied RNA‐sequencing as an unbiased means to interrogate renal epithelial cell type‐specific transcriptome alterations upon fluid shear stress. Our data indicate that genes involved in TGF‐β, MAPK, and Wnt signaling are up‐regulated by shear stress, while the JAK‐STAT related genes seems to be down‐regulated. Using ALK4/5/7 and MEK1/2 inhibitors, we showed that the shear stress‐induced signaling cascades are largely modulated by TGF‐β/ALK5 and MAPK/ERK signaling. Cilia removal abrogated shear induced gene expression of a subset of genes, but genes involved in TGF‐β, MAPK, and Wnt signaling were hardly affected, suggesting that other mechano‐sensors also play an evident role in the shear stress response of renal epithelial cells. Furthermore, altered expression of genes involved in cell‐matrix, cytoskeleton and glycocalyx remodeling, as well as amino acid, carbohydrate, and cholesterol metabolism, indicate that shear stress is regulating gene expression at several levels for cellular homeostasis. Finally, we showed that expression of several genes is elevated at pathological levels of shear stress compared to physiological controls, suggesting that variations in fluid shear stress might be relevant for the pathology in kidney diseases due to an imbalance in cellular signaling.
2. MATERIALS AND METHODS
2.1. Chemicals
ALK4/5/7 inhibitor LY‐364947 (Calbiochem; #616451) from Merck Millipore (Darmstadt, Germany), MEK1/2 inhibitor Trametinib (GSK1120212; #S2673) from Selleckchem (Bio‐Connect, Huissen, The Netherlands) and ammonium sulfate (#A‐2939) from Sigma–Aldrich (Zwijndrecht, The Netherlands) were used as previously described (Kunnen et al., 2017).
2.2. Cell culture
SV40 large T‐antigen immortalized murine proximal tubular epithelial cells (PTEC), derived from a Pkd1 lox,lox mouse, were generated and cultured as described previously (Kunnen et al., 2017; Leonhard et al., 2011). Briefly, cells were maintained at 37°C and 5% CO2 in DMEM/F‐12 with GlutaMAX (Gibco, Fisher Scientific, Landsmeer, The Netherlands; #31331‐093) supplemented with 100 U/ml Penicillin‐Streptomycin (Gibco, Life Technologies; #15140‐122), 2% Ultroser G (Pall Corporation, Pall BioSepra, Cergy St Christophe, France; #15950‐017), 1x Insulin‐Transferrin‐Selenium‐Ethanolamine (Gibco, Life Technologies; #51500‐056), 25 ng/L Prostaglandin E1 (Sigma–Aldrich; #P7527) and 30 ng/L Hydrocortisone (Sigma–Aldrich; #H0135). Cell culture was monthly tested without mycoplasma contamination using MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland; LT07‐318). New ampules were started after 15 passages.
For fluid‐flow experiments, cells were cultured on collagen‐I (Advanced BioMatrix, San Diego, CA; #5005) coated culture dishes or glass slides. Cells grown until high confluency underwent 24 hr serum starvation before the start of the treatment to exclude effects of serum‐derived growth‐factors and to synchronize cells and cilia formation.
2.3. Fluid shear stress stimulation
Cells were exposed to laminar fluid shear stress (0.25–2.0 dyn/cm2) in a cone‐plate device or parallel‐plate flow chamber as described previously (Kunnen et al., 2017). The cone‐plate device, adapted from Malek, Gibbons, Dzau, and Izumo (1993); and Malek, Ahlquist, Gibbons, Dzau, and Izumo (1995), was designed for 3.5 cm cell culture dishes (Greiner Bio‐One, Alphen aan de Rijn, The Netherlands). Cells were grown on collagen‐I coated dishes until confluence, followed by 24 hr serum starvation, before dishes were placed in the cone‐plate flow system and incubated at 37°C and 5% CO2. The confluent cell monolayer of 9.6 cm2 was subjected to fluid shear stress using 2 ml serum‐free DMEM/F‐12 medium containing penicillin‐streptomycin, with viscosity (µ) of 0.0078 dyn s/cm2 (Bacabac et al., 2005). Constant laminar (Re = 0.3) fluid‐flow was induced using a cone angle (α) of 2° and a velocity (ω) of 80 rpm, generating a fluid shear stress (τ = µω/α) of 1.9 dyn/cm2.
Alternatively, cells were exposed to shear stress using a parallel plate flow chamber, as previously described (Juffer, Bakker, Klein‐Nulend, & Jaspers, 2014; Klein‐Nulend, Semeins, Ajubi, Nijweide, & Burger, 1995). Briefly, cells were grown on collagen‐I coated glass slides of 36 × 76 mm (Fisher Scientific #15178219) until confluence, followed by 24 hr serum starvation, before glass slides were place in a flow‐chamber. A confluent cell monolayer of 14.2 cm2 (24 × 59 mm) was subjected to fluid shear stress using 7.5 ml serum‐free DMEM/F‐12 medium containing penicillin‐streptomycin. Fluid was pumped at a constant flow rate (Q) of 5.5 ml/min through the chamber with 300 μm height (h), generating a constant laminar (Re = 5.0) fluid shear stress (τ = 6 µQ/h2b) of 2.0 dyn/cm2. The parallel plate flow‐chamber was placed in an incubator at 37°C and 5% CO2.
Static control cells were incubated for the same time in equal amounts of serum‐free DMEM/F12 medium containing penicillin‐streptomycin at 37°C and 5% CO2. After 4, 6, or 16 hr fluid‐flow or static (control) stimulation, cells have been harvested for mRNA isolation and gene expression analysis. In select experiments, cells were pre‐exposed to low levels of shear stress (0.25 dyn/cm2), followed by 16 hr shear stress at the same levels (physiological control) or at pathological levels of shear (2.0 dyn/cm2). ALK4/5/7 inhibitor (10 μM), MEK1/2 inhibitor (10 μM), or DMSO control (0.1%) were added 1 hr before start of fluid‐flow stimulation in the absence of medium supplements. Ammonium sulfate (AS) was used to remove primary cilia. Cells were pre‐treated with 50 mM ammonium sulfate, followed by 16 hr fluid flow in medium containing 25 mM AS, to prevent cilia restoration. Control cells were treated similarly, but without AS. Cilia formation was checked on a parallel slide by immunofluorescence using anti‐acetylated α‐tubulin antibodies (Sigma Aldrich; #T6793) as previously described (Kunnen et al., 2017).
2.4. RNA sequencing
Total RNA was isolated from fluid shear stress treated PTECs or static controls (n = 4) using TRI Reagent (Sigma–Aldrich; #T9424) and purified using Nucleospin RNA Clean‐up (Macherey‐Nagel, Düren, Germany; #740948) according to manufacturer's protocols. Next generation sequencing of mRNA was done by ServiceXS (GenomeScan, Leiden, The Netherlands) using the Illumina® HiSeq 2500 platform (San Diego, CA, USA). Illumina mRNA‐Seq Sample Prep Kit was used to process the samples according to the manufacturer's protocol. Briefly, mRNA was isolated from total RNA using the oligo‐dT magnetic beads. After fragmentation of the mRNA, a cDNA synthesis was performed. This was used for ligation with the sequencing adapters and PCR amplification of the resulting product. The quality and yield after sample preparation was measured with a DNA 1000 Lab‐on‐a‐Chip. The size of the resulting products was consistent with expected size distribution (a broad peak between 300 and 500 bp).
Clustering and cDNA sequencing using the Illumina cBot and HiSeq 2500 was performed according manufacturer's protocols. A concentration of 5.8 pM of cDNA was used. All samples were run on Pair Ends mode and 125 bp long reads. HiSeq control software HCS v2.2.38 was used. Image analysis, base calling, and quality check was performed with the Illumina data analysis pipeline RTA v1.18.61 and/or OLB v1.9 and Bcl2fastq v1.8.4. At least 87.3% of bases had a Q‐score ≥30.
Reads were aligned to mouse genome build GRCm38—Ensembl (Waterston et al., 2002) using TopHat2 version 2.0.10 (Kim et al., 2013). Gene expression was quantified using HTSeq‐Count version 0.6.1 (Anders, Pyl, & Huber, 2015), using default options (stranded = no, mode = union). Differential gene expression analysis was performed in R version 3.0.2 using DESeq (Version1.16.0). Differentially expressed genes were selected with an adjusted p‐value (corrected for multiple hypotheses testing) of <0.05. Count per million (CPM) values were calculated by dividing the read counts by total read counts of the sample, which is a measure for the abundance of the transcript. CPM > 2 was used to exclude low expressed genes.
2.5. Quantitative PCR
Gene expression analysis by quantitative PCR (qPCR) was performed as described previously (Happe et al., 2011). Briefly, cDNA synthesis of total RNA was done using Transcriptor First Strand cDNA Synthesis Kit (Roche, Almere, The Netherlands; #04897030001) according to the manufacturer's protocol. Quantitative PCR was done in triplicate on the LightCycler 480 II (Roche) using 2x FastStart SYBR‐Green Master (Roche; #04913914001) according to the manufacturer's protocol. Data was analyzed with LightCycler 480 Software, Version 1.5 (Roche). Gene expression was calculated using the 2−ΔΔCt method (Livak & Schmittgen, 2001) and normalized to the housekeeping gene Hprt, giving the relative gene expression. For primer sequences see Supplementary Table S1. Mean gene expression and standard deviation (SD) of the different treatment groups were calculated. Differences between fluid shear stress treated cells and static controls were tested using one sample t‐tests. One‐way analysis of variance (ANOVA) was used when cells were exposed for a different time or to a different flow rate. Two‐way analysis of variance (ANOVA) was used, when the shear stress response was compared to a second treatment. The ANOVA was followed by post‐hoc Fisher's LSD multiple comparison, if the overall ANOVA F‐test was significant. p < 0.05 was considered to be statistically significant.
2.6. Pathway analysis
Functional enrichment analysis was performed against the Molecular Signature Database (MSigDB: http://software.broadinstitute.org/gsea/msigdb/annotate.jsp) v5.2 (Subramanian et al., 2005) using standard hypergeometric distribution with correction for multiple hypotheses testing according to Benjamini and Hochberg. From this source we included pathway databases (KEGG, BIOCARTA, and REACTOME). Up‐ and down‐regulated genes by fluid shear stress were used as separate gene sets to discriminate between generally up‐ and down‐regulated pathways. Terms with false discovery rate (FDR) <0.01 were considered significantly enriched, giving 209 up‐regulated and 55 down‐regulated terms. Interaction networks of up‐ and down‐regulated DEG and their connecting pathways/processes were plotted using Cytoscape, version 3.4.0.
3. RESULTS
3.1. Fluid shear stress induced transcriptional changes in PTECs
To study genome wide fluid‐flow induced cellular alterations, proximal tubular epithelial cells (PTEC) were exposed to fluid shear stress of 1.9 dyn/cm2 using a cone‐plate device. Controls were similarly treated under static conditions. After 6 hr fluid shear stress or static exposure, total RNA was isolated and gene expression was analyzed using next generation sequencing (NGS) on the Illumina HiSeq 2500 platform. After quality checks the reads were aligned to mouse genome (GRCm38) and gene expression was quantified using HTSeq‐Count. Count per million (CPM) values were calculated as a measure for the abundance of the transcript (Supplementary Table S2).
A scatter plot was constructed comparing the log2 CPM values of flow vs static treated cultures, showing a substantial number of genes that are significantly (p < 0.05) up‐ or down‐regulated (Figure 1a, blue dots). Overall, RNA sequencing identified 2015 differentially expressed genes (DEG) upon shear stress exposure in PTECs (Table 1). Low expressed genes with an average counts per million (CMP) <2 were excluded, resulting in a list of 1551 DEG (Supplementary Table S3). A heat map of all 8 PTEC samples shows a clear distinction between fluid shear stress treated samples and static controls (Figure 1b). Furthermore, our genome wide RNA sequencing analysis confirmed genes known to be altered by fluid shear stress in renal epithelial cells, including Ptgs2 (Cox2), Ccl2 (Mcp1), Edn1, Egr1, Snai1, Cdh1, and Tgfb1 (Flores, Battini, Gusella, & Rohatgi, 2011; Flores, Liu, Liu, Satlin, & Rohatgi, 2012; Grabias & Konstantopoulos, 2012, 2013; Maggiorani et al., 2015; Pandit et al., 2015; Schwachtgen, Houston, Campbell, Sukhatme, & Braddock, 1998).
Figure 1.
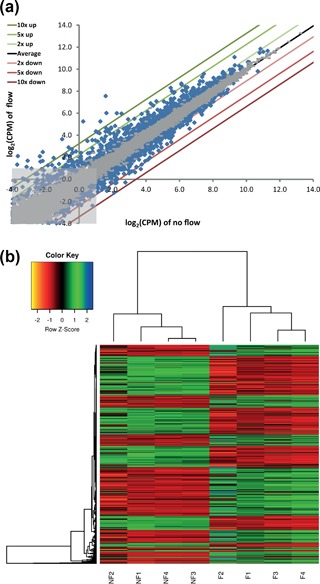
Gene expression profiling shows a strong difference between fluid shear stress treated PTECs and static controls. (a) log2 comparison of the counts per million (CPM) values of flow versus no flow treated PTEC cultures. Differentially expressed genes (DEG) are indicated by blue dots (p < 0.05). Not significant genes are indicated by gray dots. Labeled lines indicate a 2, 5, or 10 fold up‐ or down‐regulation. Black line (Average) represents equal expression for both conditions. Light‐grey box indicates the area of low expressed genes (CPM < 2). (b) Heat map showing the expression values of 1551 DEG (p < 0.05; CPM > 2) in 4 fluid shear stress treated samples (F = Flow) and 4 static controls (NF = No flow). Expression values were normalized using the Voom function in limma R package. Hierarchical clustering was applied on the samples and values were scaled by row
Table 1.
Differentially expressed genes by fluid shear stress in PTECs using next generation sequencing
| All DEG | DEG with CPM > 2 | |
|---|---|---|
| Up | 1023 | 813 |
| Down | 992 | 738 |
| Total | 2015 | 1551 |
Number of differentially expressed genes (p < 0.05) of flow versus static treated PTECs. Low expressed genes were excluded with an enrichment filter of CPM > 2. DEG, differentially expressed gene; CPM, counts per million.
3.2. Pathway analysis of RNA sequencing data
We used functional enrichment analysis of the MSigDB (Subramanian et al., 2005) as the tool to identify biological pathways or processes associated with fluid‐shear stress in PTECs. The list of 1551 DEG (Supplementary Table S3) was split into up‐regulated (813) and down‐regulated (738) genes in order to get pathways that are generally up‐ or down‐regulated. The 209 up‐regulated and 55 down‐regulated biological annotations in flow‐stimulated cells are presented in Supplementary Tables S4 and S5, respectively. We subdivided the biological pathways in core signal transduction, as well as cell–cell/matrix interaction, metabolism, cytokine signaling, other cellular processes and diseases. These processes show many connections as indicated by interaction networks of genes with the annotated pathways (Supplementary Figure S1). Cell–cell/matrix interactions are clearly affected by fluid‐flow (Supplementary Table S4). This is revealed by increased gene expression of cytoskeletal components (Actb, Actg1, Actn1, Flna), cadherins (Cdh10, Cdh11), tight junction molecule (Cldn4), cell adhesion molecules (Cadm1, Cadm3, Epcam, Ncam1, Vcam1), extracellular matrix components (Col1a1, Col5a1, Fn1, Lamc1, Lamc2), and integrins (Itgav, Itga2, Itga5, Itgb1, Itgb3, Itgb4, Itgb5). Furthermore, we see a shear stress enhanced expression of genes involved in glycosaminoglycan and carbohydrate metabolism, including proteoglycans (Gpc1, Sdc1, Sdc2, Sdc3, Cd44), heparan sulfate, carbohydrate or uronyl sulfotransferases (Hs2st1, Hs3st3b1, Hs6st1, Chst7, Chst11, Ust). Genes involved in apoptosis and cell cycle activity are increased by shear stress, including pro‐apoptotic (Trp53, Bid, Fas, Pmaip1) as well as pro‐survival (Bcl2, E2f3, Ctnnb1, Myc) and cell cycle arrest (Gadd45, Sfn, Cdkn2b) genes, while key players in apoptosis (Bad, Bak, Bax, and caspases) and cell cycle (cyclins and CDKs) were not altered in gene expression (Supplementary Table S2–S5). Pathways involved in cytokine signaling and other cellular processes and diseases are up‐regulated as well and have broad overlap with the core signal transduction pathways. Of those, the most prominently up‐regulated pathways by fluid flow include MAPK, TGF‐β, Wnt, PDGF, and p53 signaling (Table 2). We previously reported changes in TGF‐β signaling, involving genes encoding proteins relaying the signal from cell membrane toward the nucleus, i.e. the ligands Tgfb1‐3 and the receptor Alk5 (Tgfbr1), as well as down‐stream targets, that is, Pai1 (Serpine1), Fn1, Col1a1, and Snai1 (Kunnen et al., 2017). Our gene expression profile now also shows increased expression of genes encoding proteins involved in TGF‐β ligand activation (Furin, Thbs1) or ligand inhibition (Ltbp2), the transcription factor Smad3, but also the inhibitors Smad7, Smurf1, Skil, and Tgif1, all critical components of the pathway (Table 2).
Table 2.
Core signaling pathways affected by fluid shear stress—up‐regulated genes
| Pathway | Pathways description | Database | k/K | FDR | Genes |
|---|---|---|---|---|---|
| MAPK | MAPK signaling pathway | KEGG | 39/267 | 3.1E‐23 | MAP2K1; TGFB1; PAK1; RRAS; CACNB3; CACNA1G; PDGFA; PDGFB; PRKCA; FGF1; FGF9; TGFB3; TP53; TGFBR1; MYC; FAS; FOS; RAP1B; FLNA; PLA2G4A; PPP3CA; NFATC2; NFATC4; RASA1; ZAK; NR4A1; GADD45A; GADD45B; GADD45G; MAP2K3; DUSP4; DUSP6; DUSP7; SRF; DUSP1; HSPB1; MAP4K4; DUSP9; RELB |
| MAP kinase activation in TLR cascade | REACTOME | 8/50 | 2.9E‐05 | MAP2K1; FOS; MAP2K3; DUSP4; DUSP6; DUSP7; PPP2R1B; IRAK2 | |
| MAPKinase signaling pathway | BIOCARTA | 10/87 | 4.5E‐05 | MAP2K1; TGFB1; PAK1; TGFB3; TGFBR1; MYC; FOS; MAP2K3; MAP4K4; MAPK6 | |
| TGF‐β | Signaling by TGF‐beta Receptor Complex | REACTOME | 15/63 | 7.6E‐12 | TGFB1; TGFBR1; MYC; SMAD3; FURIN; NCOR2; CDKN2B; SERPINE1; SMURF1; SMAD7; UBE2D1; JUNB; SKIL; TGIF1; PMEPA1 |
| TGF‐beta signaling pathway | KEGG | 15/86 | 1.3E‐10 | TGFB1; TGFB3; TGFBR1; MYC; SMAD3; CDKN2B; THBS1; PPP2R1B; SMURF1; SMAD7; BMPR2; ACVR1; INHBA; NOG; ID1 | |
| TGF beta signaling pathway | BIOCARTA | 7/19 | 2.6E‐06 | MAP2K1; TGFB1; TGFB3; TGFBR1; SMAD3; SMAD7; SKIL | |
| Signaling by BMP | REACTOME | 6/23 | 2.9E‐05 | SMURF1; SMAD7; UBE2D1; BMPR2; NOG; FSTL1 | |
| Wnt | Wnt signaling pathway | KEGG | 17/151 | 6.6E‐09 | PRKCA; TP53; MYC; PPP3CA; NFATC2; NFATC4; CTNNB1; SMAD3; TCF7; FZD7; FZD8; WNT7A; WNT7B; PPP2R1B; CSNK1E; FOSL1; PORCN |
| PDGF | Signaling by PDGF | REACTOME | 16/122 | 1.0E‐08 | MAP2K1; PDGFA; PDGFB; PRKCA; RASA1; NR4A1; FOXO1; COL1A1; COL5A1; COL4A3; FURIN; PDGFC; THBS1; ITPR3; ADCY7; PHLPP1 |
| PDGF signaling pathway | BIOCARTA | 6/32 | 1.8E‐04 | MAP2K1; PDGFA; PRKCA; FOS; RASA1; SRF | |
| p53 | p53 signaling pathway | KEGG | 12/69 | 1.3E‐08 | TP53; FAS; GADD45A; GADD45B; GADD45G; BID; THBS1; SERPINE1; IGFBP3; PMAIP1; SFN; CCNG2 |
| p53 signaling pathway | BIOCARTA | 4/16 | 7.6E‐04 | TP53; GADD45A; BCL2; TIMP3 | |
| VEGF | VEGF signaling pathway | KEGG | 10/76 | 3.6E‐06 | MAP2K1; PRKCA; PLA2G4A; PPP3CA; NFATC2; NFATC4; HSPB1; PTK2; PTGS2; SPHK1 |
| HGF | Signaling of Hepatocyte Growth Factor Receptor | BIOCARTA | 8/37 | 6.1E‐06 | MAP2K1; PAK1; FOS; RAP1B; RASA1; ITGB1; PTK2; PTK2B |
| ETS | ETS pathway | BIOCARTA | 5/18 | 1.4E‐04 | FOS; NCOR2; ETS1; CSF1; ETS2 |
| FGF | Signaling by FGFR | REACTOME | 10/112 | 2.2E‐04 | MAP2K1; PRKCA; FGF1; FGF9; NR4A1; FOXO1; CBL; ITPR3; ADCY7; PHLPP1 |
Pathway analysis (KEGG, Reactome and Biocarta) done on 813 significantly up‐regulated genes upon fluid shear stress in PTECs using MSigDB. The most significantly altered core signaling pathways are shown and ordered together, followed by the lowest false discovery rate (FDR). K, number of genes in pathway database; k, number of genes in overlap. For the complete list of the pathway analysis of up‐regulated genes upon fluid shear stress see Supplementary Table S4.
However, the most prominently activated signaling pathway is the mitogen‐activated protein kinase (MAPK) pathway (Tables 2 and S4). The MAPK pathway is a set of intracellular signal transduction cascades that regulate a wide variety of stimulated cellular processes, including proliferation, differentiation, apoptosis and stress responses. The canonical cascades identified in mammalians are extracellular signal‐regulated kinase 1 and 2 (ERK1/2), c‐Jun N‐terminal kinase (JNK), p38, and ERK5, responding to different mitogens or forms of stress. Consequently, the MAPK pathway comprises a large number of molecules. Increased expression by fluid shear was observed for several MAP kinases (i.e., Map2k1, Map2k3, Map4k4, Mapk6, Map3k20 = Zak) as well as dual‐specific phosphatases (Dusp1, 4, 6, 7, and 9), which negatively regulate members of the MAP kinase superfamily. The classical MAP kinase (ERK1/2) pathway, activating proliferation and differentiation, shows increased expression of Ras (Rras), MEK1 (Map2k1) and c‐Fos (Fos). Upstream mitogens, PDGF (Pdgfa, b, and c), HB‐EGF (Hbegf) and FGF (Fgf1,9) are increased by fluid shear as well. Also the stress‐mitogen pathway is modified, including increased mRNA levels of TNFα and TNFα‐receptors (Tnfaip2, Tnfaip3, C1qtnf3, Fas, Tnfrsf1b, Tnfrsf12a, Tnfrsf23, Relt). Furtheremore, CXC, CX3C, and CC chemokines and receptors are increased by shear (Cxcl10, Cxcl14, Cxcl16, Cx3cl1, Ccl2, Cxcr4). Cooperation between MAPK pathway and NFAT proteins integrates two important signaling pathways that are altered by shear stress, the MAPK‐pathway and calcium signaling. This involves elevated expression of Nfatc2 and Nfatc4, as well as expression of several calcium channels (Cacnb3, Cacna1g) and calcium/calmodulin dependent proteins (Camk2n1, Ccbe1, Ncs1, Carhsp1). Other transcription factors that are reported to be regulated by MAPK/ERK are Ets1 and Ets2 (Foulds, Nelson, Blaszczak, & Graves, 2004), which are both increased by fluid shear stress as well (Table 2).
Wnt signaling is activated when secreted Wnt ligands bind to specific Frizzled (FzD) receptors on the surface of target cells to trigger the canonical (Wnt/β‐catenin) or non‐canonical (β‐catenin‐independent) pathways. Particularly, canonical Wnt signaling seems activated by fluid shear. Expression of both Wnt7a and Wnt7b is increased, as well as Porcupine (Porcn), required for Wnt secretion. Also expression of FzD receptors (Fzd7 and 8) is up‐regulated (although the co‐receptor Lrp6 is down‐regulated) as well as the key players β‐catenin (Ctnnb1) and Tcf7, which are regulating the expression of down‐stream target genes (Wisp1, Fosl1, Myc).
Overall, less core signaling pathways were identified that were down‐regulated by fluid shear stress. The most prominently down‐regulated pathway is JAK/STAT or Interferon signaling (Table 3), with reduced expression of receptors (Ifngr1, Il6st, Il5ra, and Lifr), signal transducers (Jak2, Stat1, Stat5a and Irf7, 8, 9) as well as target genes (Socs2 and Gbp6, 7). Other down‐regulated pathways include Rho, PDGF, Hedgehog, and Insulin signaling, as well as different metabolic pathways (Tables 3 and S5). This also includes PI3K/AKT related signaling, which is not included as core signaling pathway from KEGG in the MSigDB.
Table 3.
Core signaling pathways affected by fluid shear stress—down‐regulated genes
| Pathway | Pathways description | Database | k/K | FDR | Genes |
|---|---|---|---|---|---|
| JAK‐STAT | Cytokine Signaling in Immune system Interferon signaling Interferon alpha/beta signaling Immune system Interferon gamma signaling | REACTOME REACTOME REACTOME REACTOME REACTOME | 25/270 19/159 13/64 37/933 8/63 | 2.3E‐10 9.9E‐10 2.2E‐09 4.5E‐06 2.9E‐04 | STAT1; IRF7; IRF9; H2‐M3; IRF8; ISG15; USP18; IFIT1; MX2; IFI27; IFI35; IFIT3; XAF1; JAK2; IFNGR1; GBP7; DDX58; TRIM25; UBA7; PIK3R1; STAT5A; MAP2K6; IL6ST; SOCS2; BLNK; PTEN; CDH1; RASGRP2; PPP2R5A; IFIH1; DHX58; RAP1GAP2; RAP1GAP; DUSP3; ICOSL; KLHL13; FBXO44 |
| Jak‐STAT signaling pathway | KEGG | 10/155 | 2.7E‐03 | STAT1; IRF9; JAK2; IFNGR1; PIK3R1; STAT5A; IL6ST; SOCS2; PIK3R5; LIFR | |
| Rho | Signaling by Rho GTPases | REACTOME | 9/113 | 1.6E‐03 | NGEF; ARHGEF9; FAM13A; CHN2; RHOF; ARHGAP24; ARHGAP19; ARHGAP18; ARHGAP29 |
| PDGF | Signaling by PDGF | REACTOME | 9/122 | 2.5E‐03 | STAT1; PIK3R1; STAT5A; PTEN; GRB7; PRKAR2B; PRKCE; COL4A5; ADCY9 |
| Hedgehog | Hedgehog signaling pathway | KEGG | 6/56 | 3.9E‐03 | LRP2; WNT16; WNT6; PTCH1; BMP7; GAS1 |
| Insulin | Insulin signaling pathway | KEGG | 9/137 | 4.2E‐03 | PIK3R1; SOCS2; PIK3R5; PPARGC1A; LIPE; PRKAR2B; MKNK2; SORBS1; PPP1R3C |
Pathway analysis (KEGG, Reactome and Biocarta) done on 738 significantly down‐regulated genes upon fluid shear stress in PTECs using MSigDB. The most significantly altered core signaling pathways are shown and ordered together, followed by the lowest false discovery rate (FDR). K, number of genes in pathway database; k, number of genes in overlap. For the complete list of the pathway analysis of down‐regulated genes upon fluid shear stress see Supplementary Table S5.
Expression of a selected set of genes was validated by quantitative PCR using a parallel plate flow‐chamber (Kunnen et al., 2017) and confirmed fluid‐shear induced expression of Ccbe1, Prune2, Wisp1, Fbln5, Plk2, Junb, Gsto1, Hbefg, Map3k20 (Zak), Wnt7b, Tes, Runx1, Ets1, Map4k4, Itgb1, and Itgav, while Jak2 and Stat1 expression was decreased by fluid shear stress (Figure 2). After 16 hr gene expression was significantly increased for all tested genes (Supplementary Figure S2). While several genes reached significance already at 6 hr, others did not. Furthermore, we investigated if the changes in gene expression by shear stress were reversible, by doing a static post incubation of 8 hr, after removal of shear. For several genes, shear stress induced gene expression returned to levels close to the static controls, while other genes showed similar or higher expression levels after post incubation without shear (Supplementary Figure S3), indicating that in time genes can respond differently to variations in fluid shear stress.
Figure 2.
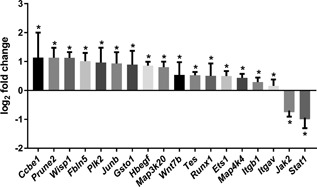
qPCR validation of RNA sequencing results. Gene expression (log2 fold change) of selected target genes is altered upon 16 hr fluid shear stress, as measured by quantitative PCR. Parallel plate flow‐chamber induced fluid shear stress at 2.0 dyn/cm2 in PTECs; n = 13 per condition; Hprt served as housekeeping gene to correct for cDNA input; data were normalized to static controls (log2 fold change = 0). *Indicates significantly altered expression by flow versus no flow (p < 0.05) using a one sample t‐test
3.3. Fluid shear stress response in PTECs is dominated by TGF‐β/ALK5 and MAPK/ERK pathways
We previously showed shear stress induced TGF‐β/ALK5 dependent SMAD2/3 signaling and target gene expression (Kunnen et al., 2017). In addition to increased expression of canonical SMAD2/3 targets, we see shear stress induced expression of other genes known to be induced by TGF‐β signaling, including Junb and Fbln5 (Figure 2) (Lee, Hong, & Bae, 2002; Schiemann, Blobe, Kalume, Pandey, & Lodish, 2002; Topalovski, Hagopian, Wang, & Brekken, 2016). Our results indicate that shear stress induced Junb and Fbln5 expression was ALK4/5/7 dependent (Figure 3a). In addition, genes involved in other (core) signaling pathways, like MAPK (Map3k20 and Map4k4), Wnt (Wisp1), ETS (Ets1), and other pathways (Plk2, Prune2), were strongly repressed by the ALK4/5/7 inhibitor (Figure 3a), suggesting that TGF‐β/ALK5 signaling is interacting with more pathways than the canonical TGF‐β pathway alone. In contrast, fluid shear stress induced down‐regulation of Stat1 and Jak2 was not altered upon ALK4/5/7 inhibition, although Jak2 basal levels were already higher with the ALK4/5/7 inhibitor (Figure 3a).
Figure 3.
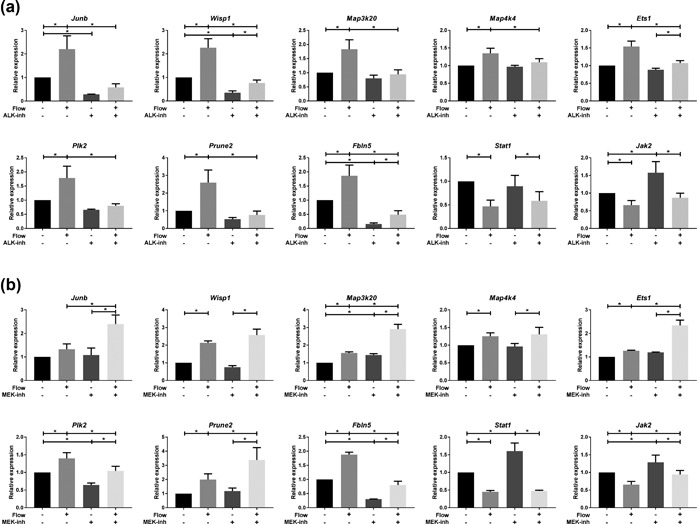
Shear stress response in PTECs is modulated by ALK4/5/7 and MEK1/2 inhibitors. Relative expression of selected genes upon 16 hr fluid shear stress exposure, as measured by quantitative PCR. (a) ALK4/5/7 inhibitor (10 μM LY‐364947) significantly reduces shear stress increased expression of Junb, Wisp1, Map3k20, Map4k4, Ets1, Plk2, Prune2, and Fbln5, while shear stress induced down‐regulation of Jak2 and Stat1 was not altered. (b) MEK1/2 inhibition (10 μM Trametinib) significantly reduces shear stress increased expression of Plk2 and Fbln5, while fluid‐flow increased expression of Junb, Map3k20, Ets1, and Prune2 is further elevated. Wisp1 and Map4k4 expression was not altered upon MEK inhibition. Jak2 and Stat1 expression was still down‐regulated by shear stress upon MEK inhibition, although basal levels were slightly higher. (a, b) Parallel plate flow‐chamber induced fluid shear stress at 2.0 dyn/cm2 in PTECs; t = 16 hr; qPCR, Hprt served as housekeeping gene to correct for cDNA input; data normalized to unstimulated controls (fold change); n = 3–5 per condition. *Indicates p < 0.05 by two‐way ANOVA, followed by post‐hoc Fisher's LSD multiple comparison. ALK‐inh = ALK4/5/7 inhibitor (LY‐364947). MEK‐inh = MEK1/2 inhibitor (Trametinib, GSK1120212)
Since SMAD2/3 mediated gene transcription can be either restrained or induced by ERK1/2 signaling, as shown before (Hough, Radu, & Dore, 2012; Kretzschmar, Doody, Timokhina, & Massague, 1999; Kunnen et al., 2017), we also investigated the involvement of MAPK/ERK signaling in the shear stress response. Our data indicate that only Plk2 and Fbln5 induction by fluid‐shear is lowered using MEK1/2 inhibitors (Figure 3b), although the flow response is still present. In contrast, Junb, Map3k20 (Zak), Ets1, and Prune2 expression was further elevated using MEK inhibitors, which was also seen for many canonical SMAD2/3 targets (Kunnen et al., 2017), while the shear stress response of Wisp1 and Map4k4 was not significantly changed upon MEK inhibition (Figure 3b). Fluid shear stress induced down‐regulation of Jak2 and Stat1 is still present upon MEK inhibition (Figure 3b), although basal levels were slightly higher with MEK1/2 inhibitors. In conclusion, our data suggest complex regulation of the fluid shear stress response in PTECs, which is largely modulated by TGF‐β/ALK5 and MAPK/ERK pathways.
3.4. Primary cilia only play a role in a part of the shear stress response in PTECs
Since defects in cilia formation and function have profound effects on the development and physiology of kidneys and other organs (Goetz & Anderson, 2010; Quinlan et al., 2008), we investigated the shear stress response in PTECs after cilia removal by ammonium sulfate. Expression of Plk2, Prune2, and Ets1 were clearly cilia dependent, since the shear stress induced response was completely lost after cilia ablation (Figure 4). In contrast, genes involved in TGF‐β, Wnt, MAPK, and JAK/STAT signaling, that is, Junb, Fbln5, Wisp1, Map3k20, Map4k4 as well as Stat1, were only slightly or not affected in the shear stress response upon cilia removal (Figure 4). Although shear induced down‐regulation of Jak2 was abrogated, Jak2 expression in static cells was already reduced upon ammonium sulfate treatment (Figure 4). Our data suggests that shear stress regulated gene expression in PTECs is only partially cilia dependent and other mechano‐sensors are involved as well.
Figure 4.
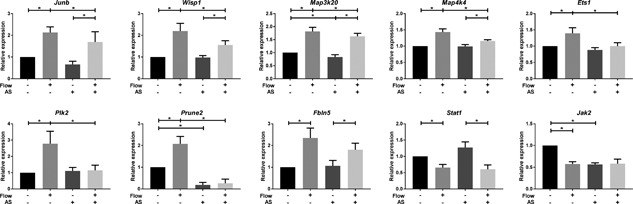
Shear stress altered gene expression in PTECs is partially cilia dependent. Relative expression of selected genes upon 16 hr fluid shear stress exposure in controls and cells treated with 50 mM ammonium sulfate (AS), as measured by quantitative PCR. Shear stress induced expression of Ets1, Plk2, and Prune2 was abrogated after cilia ablation. Junb, Wisp1, Map3k20, Map4k4, Fbln5, and Stat1 expression was only slightly or not affected in the shear stress response upon cilia removal. Shear induced down‐regulation of Jak2 was abrogated, since Jak2 expression in static cells was already reduced upon ammonium sulfate treatment. Parallel plate flow‐chamber induced fluid shear stress at 2.0 dyn/cm2 in PTECs; Hprt served as housekeeping gene to correct for cDNA input; data were normalized to static controls (fold change); n = 5 per condition. *Indicates p < 0.05 by two‐way ANOVA, followed by post‐hoc Fisher's LSD multiple comparison
3.5. Shear stress induced gene expression in PTECs is flow rate dependent
Thus far we applied fluid shear stress of 2.0 dyn/cm2, which is known to be an increased physio‐pathological shear stress (Essig, Terzi, Burtin, & Friedlander, 2001; Grabias & Konstantopoulos, 2012, 2013; Weinbaum et al., 2010). To compare the gene expression to physiological levels of shear, we exposed the cells to a shear stress range of 0.25–2.0 dyn/cm2. Expression of Wisp1, Map3k20, Map4k4, and Ets1 was clearly flow rate dependent and this trend was also visible for Junb, Plk2, Prune2, and Fbln5 (Figure 5a). To mimic the induction of hyperfiltration, PTECs were pre‐exposed to physiological levels of shear (0.25 dyn/cm2) for 4 hr, followed by 16 hr shear stress at the same physiological level or at pathological levels of shear (2.0 dyn/cm2). Expression of Wisp1, Map3k20, Map4k4, Ets1, and Fbln5 was significantly higher at pathological levels of shear compared to physiological levels, while this trend was also visible for Junb and Plk2 (Figure 5b). For the downregulated genes, Stat1 and Jak2, there was no difference in expression between physiological and pathological shear. So, our data indicate that higher levels of shear and a switch from physiological to pathological shear, result in increased gene expression, at least for most the genes analyzed in this experiment.
Figure 5.
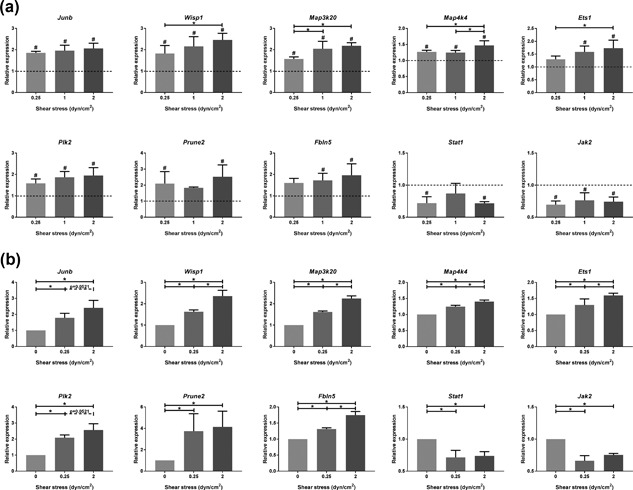
Shear stress altered gene expression in PTECs is flow rate dependent. Relative expression of selected genes upon different levels of fluid shear stress exposure (0.25–2.0 dyn/cm2) for 16 hr, as measured by quantitative PCR. (a) After starvation under static conditions, cells were directly exposed to the indicated level of fluid shear stress. (b) Cells were first pre‐exposed for 4 hr to low levels of shear stress (0.25 dyn/cm2), followed by 16 hr shear stress exposure at indicated levels. Expression of all genes was significantly increased by shear stress compared to static controls (dashed line in a) and was flow rate dependent for most genes. (a, b) Parallel plate flow‐chamber induced fluid shear stress of 0.25–2.0 dyn/cm2 in PTECs; t = 16 hr (a) or t = 4 + 16 hr (b); qPCR, Hprt served as housekeeping gene to correct for cDNA input; data normalized to unstimulated controls (fold change); n = 3 per condition. #Significant difference compared to unstimulated control (dashed line in a) or *significant difference between treatment groups (p < 0.05 by one‐way ANOVA, followed by post‐hoc Fisher's LSD multiple comparison)
4. DISCUSSION
In this study we used RNA sequencing to get a comprehensive overview of the transcriptome alterations upon fluid shear stress in proximal tubular epithelial cells. Physiological shear stress in renal epithelial cells is ranging from 0.05–1.0 dyn/cm2, where proximal tubular cells experience the highest range of shear stress (Essig et al., 2001; Grabias & Konstantopoulos, 2012, 2013; Weinbaum et al., 2010). We applied a fluid shear stress of 2.0 dyn/cm2, which is known to be an increased physio‐pathological shear stress, mimicking hyperfiltration after renal mass reduction or during progression of renal disease. Our genome wide RNA sequencing data confirmed previously reported fluid flow‐induced changes in gene expression of Cox2 (Ptgs2), Ccl2 (Mcp1), Edn1, Egr1, Snai1, and Cdh1 in renal epithelial cells (Flores et al., 2011, 2012; Maggiorani et al., 2015; Pandit et al., 2015; Schwachtgen et al., 1998). Furthermore, our data reveal >1,500 other genes to be altered by fluid shear stress in PTECs. We validated a subset of genes by qPCR and showed that the shear stress response was time dependent within the first 16 hr. Furthermore, after removal of the shear, the shear‐induced gene expression was reversible for some of the genes, while other genes showed similar or higher differential gene expression upon static post incubation. Differences in signaling and cytokine production upon shear may explain the different responses as well as differences in transcriptional activation and stability of transcripts. For example, Fn1 is a very long transcript, which requires more time for transcription and degradation, while Pai1 (Serpine1) has an faster turn‐over (Kunnen et al., 2017; 't Hoen et al., 2011).
Pathway analysis indicated increased expression of cell–cell/cell‐matrix interaction genes, including cytoskeletal components, cell adhesion and tight junction molecules, extracellular matrix components and integrins. This suggests strengthening of epithelial cells and their surroundings to resist (increased) physiological shear stress (Duan et al., 2008; Essig et al., 2001; Jang et al., 2013). Another study showed loss of epithelial cell morphology during high pathological shear stress of 5 dyn/cm2 (Maggiorani et al., 2015). Long‐term high shear exposure therefore might also lead to fibrotic deposition and tubulointerstitial lesions, which is commonly seen after renal mass reduction or during progression of renal diseases (Essig & Friedlander, 2003; Essig et al., 2001; Grabias & Konstantopoulos, 2014; Rohatgi & Flores, 2010; Venkatachalam et al., 2010). Pro‐apoptotic as well as pro‐survival and cell cycle arrest genes were induced by shear stress, while key players in apoptosis (Bad, Bak, Bax, and caspases) and cell cycle (cyclins and CDKs) were not altered in gene expression. This suggests that apoptosis and cell cycle related gene expression are not dramatically altered during shear exposure.
Core signaling pathways altered by shear stress comprise MAPK and TGF‐β signaling. Even more, TGF‐β/ALK5‐induced target gene expression in renal epithelial cells is partially restrained by MEK1/2‐mediated signaling (Kunnen et al., 2017). Using ALK4/5/7 inhibitors, we showed that many genes, but not all genes, are dependent on shear induced TGF‐β/ALK5 signaling, including genes involved in other core signaling pathways like MAPK and Wnt signaling. The role of TGF‐β as a master regulator of the shear response is related to the TGF‐β/ALK5 interaction since we previously showed that also TGF‐β neutralizing antibodies inhibit the response (Kunnen et al., 2017). It is conceivable that under flow conditions TGF‐β processing and binding of the active ligand is enhanced. Interestingly, a recent publication showed that TGF‐β can be released from its latency‐associated peptide (LAP) by shear stress, probably by forces exerted on αv‐β6‐integrins via the actin cytoskeleton (Dong et al., 2017; Ha, 2017). We also noticed an increase in gene expression of several integrins during shear stress, including integrin αv (Itgav). In addition, there are several connections between TGF‐β and MAPK signaling (Hough et al., 2012; Kretzschmar et al., 1999; Lee et al., 2007; Muthusamy et al., 2015), thereby modulating the response to shear. Our data show that the shear stress response of a subset of genes is attenuated upon MEK1/2 inhibition, while other genes showed an enhanced response. Since there are multiple interactions between TGF‐β and MAPK/ERK signaling pathways, the integration of these pathways is complex and biological context dependent, and therefore difficult to predict (Kunnen et al., 2017).
In addition to TGF‐β signaling, increased expression of other cytokines observed in our study suggests attraction and activation of macrophages and inflammatory cells upon shear in vivo. This is a common phenomenon during development of kidney diseases, where shear stress is fluctuating due to changes in glomerular filtration rate, tubular hyperfiltration and obstruction (Akchurin & Kaskel, 2015). Altered expression of other growth factors or cytokine signaling pathways include FGF, HB‐EGF, PDGF, CXC, and other cytokines. FGF, HB‐EGF, and PDGF can bind to tyrosine kinase receptors that upon activation stimulate the Ras/Raf/ERK (MAPK) pathway and/or the PI3K/AKT pathway (up‐regulated upon shear stress) and/or STAT‐signaling (down‐regulated upon shear stress) (Pileri & Piccaluga, 2012; Turner & Grose, 2010). At several levels these pathways can be amplified or negatively modulated, and they can interact with each other as well. Multiple ligand isoforms can bind to the receptors with different affinities. Upon fluid flow, transcript levels of several ligands is increased (Fgf1, Fgf9, Hbegf, Pdgfa, Pdgfb, Pdgfc), but not the receptors. Whether increased signaling is related to endocrine/paracrine loops, as seen for TGF‐β, needs more extensive investigation. Interestingly, we also observed altered expression of proteoglycans, like syndecans and glypican, as well as modifying enzymes involved in glycosaminoglycan, heparan‐sulphate or chondroitin‐sulphate metabolism, which are all involved in glycocalyx remodeling (Reitsma, Slaaf, Vink, van Zandvoort, & oude Egbrink, 2007). Cell‐surface‐associated heparan sulfate proteoglycans have been shown to be essential for FGF signal transduction and, more general, the glycocalyx is able to significantly modify the cellular response to growth factors including PDGF and FGF. It has been shown that the glycocalyx plays an important role in mechanotransduction of shear stress in endothelial cells. It is required for the cytoskeleton to respond to shear stress and acts as a signaling platform integrating shear stress, growth factor, chemokine and cytokine signaling (Ebong, Lopez‐Quintero, Rizzo, Spray, & Tarbell, 2014; Thi, Tarbell, Weinbaum, & Spray, 2004; Zeng, 2017; Zeng & Liu, 2016). So, our data indicate that fluid shear stress induce genes involved in glycocalyx remodeling in PTECs, although it has to be further investigated whether the glycocalyx is equally involved in mechano‐sensing upon shear stress in renal epithelial cells.
The shear stress response in PTECs can be regulated by a variety of mechano‐sensors at different sub‐cellular locations (Curry & Adamson, 2012; Ingber, 2006; Petersen et al., 2016). We investigated the role of primary cilia, since defects in cilia formation and function are associated with developmental disorders and (kidney) diseases (Goetz & Anderson, 2010; Quinlan et al., 2008). Our results indicate that fluid shear stress induced Plk2, Prune2, and Ets1 expression is cilia dependent, since “removal” of the cilium by ammonium sulphate completely abolished the shear stress response. Genes involved in TGF‐β, MAPK, and Wnt signaling were not or only slightly reduced upon ammonium sulphate treatment, suggesting that mechano‐sensors at other cellular locations are also contributing to the shear stress response in PTECs.
The main shear stress down‐regulated pathway is JAK/STAT signaling. However, this is largely related to reduced expression of components of the interferon signaling pathway since only a few STAT1 target genes are differentially expressed (Irf7, Irf9, Ifi35, Ifi27, Trim25) (Satoh & Tabunoki, 2013). Interferon itself is not expressed in our in vitro system (Supplementary Table S2), but reduced expression of components of the signaling pathway support a study in endothelial cells, describing attenuation of IFNγ‐induced responses by laminar flow, via the suppression of STAT1 activation (Tsai et al., 2007). We show that reduced Stat1 expression by shear stress was ALK4/5/7, MEK1/2 as well as cilium independent, although there was slightly higher expression when using the MEK1/2 inhibitors in static cells. A similar pattern was observed for Jak2 expression, with the notification that ammonium sulphate treatment already reduced expression of Jak2 as much as shear stress. For another Stat‐family member, STAT6, reduced expression of target genes has been reported. During fluid flow both STAT6 and the transcriptional co‐activator p100 locate in the primary cilia, while at static conditions these proteins translocate to the nucleus (Low et al., 2006).
Other down‐regulated genes by shear stress include genes involved in amino acid, carbohydrate, fatty acid, ketone body and cholesterol metabolism (Supplementary Figure S1B, Table S5). Also in endothelial cells shear stress exposure decreased expression of genes involved in glycolysis (Doddaballapur et al., 2015; Kim, Lee, Kawata, & Park, 2014), lipid metabolism (Fisslthaler & Fleming, 2009; Mun, An, Park, Jo, & Boo, 2008; Yamamoto & Ando, 2013) and cholesterol biosynthesis (Fisslthaler, Fleming, Keseru, Walsh, & Busse, 2007; Yamamoto & Ando, 2015). This was dependent on AMPK, which is an important kinase in energy metabolism (Carling, 2004; Fisslthaler & Fleming, 2009) and plays a central role in fluid flow induced primary cilium bending and down‐regulation of mTORC1 activity in renal epithelial cells (Boehlke et al., 2010; Zhong et al., 2016). Overall, the data show that increased shear stress reduces metabolic activity in renal epithelial cells.
This in vitro study gives a comprehensive overview of fluid shear stress altered gene expression in renal epithelial cells, but is not fully representative for the in vivo situation, since several other cells types and cytokines in the nephrons are involved. Nevertheless, our results give an overview of genes and pathways that are modulated by shear stress in renal epithelial cells, which could help us to understand relevant biological processes involved in mechano‐sensing. Several of the shear regulated processes are altered in kidney diseases as well, including TGF‐β, Wnt, and JAK‐STAT signaling (Gewin, Zent, & Pozzi, 2017). We hypothesize that large variations in shear stress, occurring in kidney diseases, might contribute to the disease phenotype. This hypothesis is supported by our data showing that the expression of several genes involved in TGF‐β, MAPK, and Wnt signaling is further elevated upon switching from physiological to pathological levels of shear.
In conclusion, this study provides a comprehensive profile of genes altered upon shear stress in PTECs. Both cell cycle activity and apoptosis are not dramatically altered and molecular alterations are more related to cell remodeling, involving cell–cell and cell‐matrix interactions, cytoskeleton and glycocalyx remodeling, as well as glycolysis and cholesterol metabolism. MAPK/ERK and TGF‐β signaling are master regulators of shear‐induced gene expression, since inhibitors modulate other signaling pathways as well. Nevertheless, altered JAK/STAT signaling, the main core signaling pathways down‐regulated upon shear stress, is independent of MAPK/ERK and TGF‐β. Our results indicate that different mechano‐sensors are involved in shear stress sensing in PTECs, because cilia ablation did only affect expression of a subset of shear modulated genes. Imbalance in cellular signaling due to variations in fluid shear stress are probably relevant for renal physiology and pathology as suggested by elevated expression of genes at pathological levels of shear stress compared to physiological controls. At this moment only a limited number of genes have been annotated to pathways and transcriptional target genes are hardly included, thereby limiting the interpretation of data to what is currently known. In the future the use of gene‐specific targeting, high‐throughput RNA‐sequencing, and connectivity maps will probably reveal additional information on shear induced signaling and how shear stress regulated processes influence epithelial cell integrity and cellular plasticity in renal disease.
CONFLICTS OF INTEREST
The authors declare no competing or financial interests.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Figure S1. Interaction network of genes regulated by fluid shear stress.
Figure S1. Shear stress induced expression in PTECs in time.
Figure S1. Shear stress induced expression in PTECs is partially reversible after removal of shear.
Table S1. Primer sequences used for qPCR.
Table S2. Counts per million values of all samples analyzed by RNA sequencing.
Table S3. Fold change of differentially expressed genes by fluid shear stress in PTECs.
Table S4. Pathway analysis of up‐regulated genes by fluid shear stress in PTECs.
Table S5. Pathway analysis of down‐regulated genes by fluid shear stress in PTECs.
ACKNOWLEDGMENTS
We thank Ron Wolterbeek (Medical Statistics, LUMC) for support with statistical analysis and Prof. Jenneke Klein‐Nuland for facilitating the research with the parallel‐plate fluid flow system. This work was supported by funding from the Netherlands Organization for Scientific Research (NWO) [grant number 820.02.016] and from the People Program (Marie Curie Actions) of the European Union's Seventh Framework Program FP7/2077‐2013 [REA grant agreement no. 317246].
Kunnen SJ, Malas TB, Semeins CM, Bakker AD, Peters DJM. Comprehensive transcriptome analysis of fluid shear stress altered gene expression in renal epithelial cells. J Cell Physiol. 2018;233: 3615–3628. https://doi.org/10.1002/jcp.26222
REFERENCES
- Akchurin, O. M. , & Kaskel, F. (2015). Update on inflammation in chronic kidney disease. Blood Purification, 39, 84–92. [DOI] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq‐a Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacabac, R. G. , Smit, T. H. , Cowin, S. C. , Van Loon, J. J. , Nieuwstadt, F. T. , Heethaar, R. , & Klein‐Nulend, J. (2005). Dynamic shear stress in parallel‐plate flow chambers. Journal of Biomechanics, 38, 159–167. [DOI] [PubMed] [Google Scholar]
- Boehlke, C. , Kotsis, F. , Patel, V. , Braeg, S. , Voelker, H. , Bredt, S. , … Kuehn, E. W. (2010). Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nature Cell Biology, 12, 1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling, D. (2004). The AMP‐activated protein kinase cascade‐a unifying system for energy control. Trends in Biochemical Sciences, 29, 18–24. [DOI] [PubMed] [Google Scholar]
- Carrisoza‐Gaytan, R. , Carattino, M. D. , Kleyman, T. R. , & Satlin, L. M. (2016). An unexpected journey: Conceptual evolution of mechanoregulated potassium transport in the distal nephron. American Journal of Physiology Cell Physiology, 310, C243–C259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry, F. E. , & Adamson, R. H. (2012). Endothelial glycocalyx: Permeability barrier and mechanosensor. Annals of Biomedical Engineering, 40, 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaen, P. G. , Delling, M. , Vien, T. N. , & Clapham, D. E. (2013). Direct recording and molecular identification of the calcium channel of primary cilia. Nature, 504, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling, M. , DeCaen, P. G. , Doerner, J. F. , Febvay, S. , & Clapham, D. E. (2013). Primary cilia are specialized calcium signalling organelles. Nature, 504, 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling, M. , Indzhykulian, A. A. , Liu, X. , Li, Y. , Xie, T. , Corey, D. P. , & Clapham, D. E. (2016). Primary cilia are not calcium‐responsive mechanosensors. Nature, 531, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddaballapur, A. , Michalik, K. M. , Manavski, Y. , Lucas, T. , Houtkooper, R. H. , You, X. , … Boon, R. A. (2015). Laminar shear stress inhibits endothelial cell metabolism via KLF2‐mediated repression of PFKFB3. Arteriosclerosis, Thrombosis, and Vascular Biology, 35, 137–145. [DOI] [PubMed] [Google Scholar]
- Dong, X. , Zhao, B. , Iacob, R. E. , Zhu, J. , Koksal, A. C. , Lu, C. , … Springer, T. A. (2017). Force interacts with macromolecular structure in activation of TGF‐beta. Nature, 542, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. , Gotoh, N. , Yan, Q. , Du, Z. , Weinstein, A. M. , Wang, T. , & Weinbaum, S. (2008). Shear‐induced reorganization of renal proximal tubule cell actin cytoskeleton and apical junctional complexes. Proceedings of the National Academy of Sciences of the United States of America, 105, 11418–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebong, E. E. , Lopez‐Quintero, S. V. , Rizzo, V. , Spray, D. C. , & Tarbell, J. M. (2014). Shear‐induced endothelial NOS activation and remodeling via heparan sulfate, glypican‐1, and syndecan‐1. Integrative Biology (Camb), 6, 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essig, M. , & Friedlander, G. (2003). Tubular shear stress and phenotype of renal proximal tubular cells. Journal of the American Society of Nephrology, 14, S33–S35. [DOI] [PubMed] [Google Scholar]
- Essig, M. , Terzi, F. , Burtin, M. , & Friedlander, G. (2001). Mechanical strains induced by tubular flow affect the phenotype of proximal tubular cells. American Journal of Physiology Renal Physiology, 281, F751–F762. [DOI] [PubMed] [Google Scholar]
- Fisslthaler, B. , & Fleming, I. (2009). Activation and signaling by the AMP‐activated protein kinase in endothelial cells. Circulation Research, 105, 114–127. [DOI] [PubMed] [Google Scholar]
- Fisslthaler, B. , Fleming, I. , Keseru, B. , Walsh, K. , & Busse, R. (2007). Fluid shear stress and NO decrease the activity of the hydroxy‐methylglutaryl coenzyme A reductase in endothelial cells via the AMP‐activated protein kinase and FoxO1. Circulation Research, 100, e12–e21. [DOI] [PubMed] [Google Scholar]
- Flores, D. , Battini, L. , Gusella, G. L. , & Rohatgi, R. (2011). Fluid shear stress induces renal epithelial gene expression through polycystin‐2‐dependent trafficking of extracellular regulated kinase. Nephron Physiology, 117, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, D. , Liu, Y. , Liu, W. , Satlin, L. M. , & Rohatgi, R. (2012). Flow‐induced prostaglandin E2 release regulates Na and K transport in the collecting duct. American Journal of Physiology Renal Physiology, 303, F632–F638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds, C. E. , Nelson, M. L. , Blaszczak, A. G. , & Graves, B. J. (2004). Ras/mitogen‐activated protein kinase signaling activates Ets‐1 and Ets‐2 by CBP/p300 recruitment. Molecular and Cellular Biology, 24, 10954–10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund, J. B. , Goetz, J. G. , Hill, K. L. , & Vermot, J. (2012). Fluid flows and forces in development: Functions, features and biophysical principles. Development, 139, 1229–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewin, L. , Zent, R. , & Pozzi, A. (2017). Progression of chronic kidney disease: Too much cellular talk causes damage. Kidney International, 91, 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz, S. C. , & Anderson, K. V. (2010). The primary cilium: A signalling centre during vertebrate development. Nature Reviews Genetics, 11, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabias, B. M. , & Konstantopoulos, K. (2012). Epithelial‐mesenchymal transition and fibrosis are mutually exclusive reponses in shear‐activated proximal tubular epithelial cells. FASEB Journal, 26, 4131–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabias, B. M. , & Konstantopoulos, K. (2013). Notch4‐dependent antagonism of canonical TGF‐beta1 signaling defines unique temporal fluctuations of SMAD3 activity in sheared proximal tubular epithelial cells. American Journal of Physiology Renal Physiology, 305, F123–F133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabias, B. M. , & Konstantopoulos, K. (2014). The physical basis of renal fibrosis: Effects of altered hydrodynamic forces on kidney homeostasis. American Journal of Physiology Renal Physiology, 306, F473–F485. [DOI] [PubMed] [Google Scholar]
- Ha, T. (2017). Structural biology: Growth factor rattled out of its cage. Nature, 542, 40–41. [DOI] [PubMed] [Google Scholar]
- Happe, H. , van der Wal, A. M. , Leonhard, W. N. , Kunnen, S. J. , Breuning, M. H. , De Heer, E. , & Peters, D. J. (2011). Altered Hippo signalling in polycystic kidney disease. The Journal of Pathology, 224, 133–142. [DOI] [PubMed] [Google Scholar]
- Hough, C. , Radu, M. , & Dore, J. J. (2012). Tgf‐beta induced Erk phosphorylation of smad linker region regulates smad signaling. PLoS ONE, 7, e42513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber, D. E. (2006). Cellular mechanotransduction: Putting all the pieces together again. FASEB Journal, 20, 811–827. [DOI] [PubMed] [Google Scholar]
- Jang, K. J. , Mehr, A. P. , Hamilton, G. A. , McPartlin, L. A. , Chung, S. , Suh, K. Y. , & Ingber, D. E. (2013). Human kidney proximal tubule‐on‐a‐chip for drug transport and nephrotoxicity assessment. Integrative Biology (Camb), 5, 1119–1129. [DOI] [PubMed] [Google Scholar]
- Juffer, P. , Bakker, A. D. , Klein‐Nulend, J. , & Jaspers, R. T. (2014). Mechanical loading by fluid shear stress of myotube glycocalyx stimulates growth factor expression and nitric oxide production. Cell Biochemistry and Biophysics, 69, 411–419. [DOI] [PubMed] [Google Scholar]
- Kim, B. , Lee, H. , Kawata, K. , & Park, J. Y. (2014). Exercise‐mediated wall shear stress increases mitochondrial biogenesis in vascular endothelium. PLoS ONE, 9, e111409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. , & Salzberg, S. L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology, 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein‐Nulend, J. , Semeins, C. M. , Ajubi, N. E. , Nijweide, P. J. , & Burger, E. H. (1995). Pulsating fluid flow increases nitric oxide (NO) synthesis by osteocytes but not periosteal fibroblasts‐correlation with prostaglandin upregulation. Biochemical and Biophysical Research Communications, 217, 640–648. [DOI] [PubMed] [Google Scholar]
- Kotsis, F. , Boehlke, C. , & Kuehn, E. W. (2013). The ciliary flow sensor and polycystic kidney disease. Nephrology, Dialysis, Transplantation, 28, 518–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar, M. , Doody, J. , Timokhina, I. , & Massague, J. (1999). A mechanism of repression of TGFbeta/ Smad signaling by oncogenic Ras. Genes & Development, 13, 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnen, S. J. , Leonhard, W. N. , Semeins, C. , Hawinkels, L. J. A. C. , Poelma, C. , Ten Dijke, P. , … Peters, D. J. M. (2017). Fluid shear stress‐induced TGF‐beta/ALK5 signaling in renal epithelial cells is modulated by MEK1/2. Cellular and Molecular Life Sciences, 74, 2283–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. S. , Hong, S. H. , & Bae, S. C. (2002). Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor‐beta and bone morphogenetic protein. Oncogene, 21, 7156–7163. [DOI] [PubMed] [Google Scholar]
- Lee, M. K. , Pardoux, C. , Hall, M. C. , Lee, P. S. , Warburton, D. , Qing, J. , … Derynck, R. (2007). TGF‐beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO Journal, 26, 3957–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhard, W. N. , van der Wal, A. , Novalic, Z. , Kunnen, S. J. , Gansevoort, R. T. , Breuning, M. H. , … Peters, D. J. M. (2011). Curcumin inhibits cystogenesis by simultaneous interference of multiple signaling pathways: In vivo evidence from a Pkd1‐deletion model. American Journal of Physiology Renal Physiology, 300, F1193–F1202. [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Low, S. H. , Vasanth, S. , Larson, C. H. , Mukherjee, S. , Sharma, N. , Kinter, M. T. , … Weimbs, T. (2006). Polycystin‐1, STAT6, and P100 function in a pathway that transduces ciliary mechanosensation and is activated in polycystic kidney disease. Developmental Cell, 10, 57–69. [DOI] [PubMed] [Google Scholar]
- Maggiorani, D. , Dissard, R. , Belloy, M. , Saulnier‐Blache, J. S. , Casemayou, A. , Ducasse, L. , … Buffin‐Meyer, B. (2015). Shear stress‐induced alteration of epithelial organization in human renal tubular cells. PLoS ONE, 10, e0131416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek, A. M. , Ahlquist, R. , Gibbons, G. H. , Dzau, V. J. , & Izumo, S. (1995). A cone‐plate apparatus for the in vitro biochemical and molecular analysis of the effect of shear stress on adherent cells. Methods in Cell Science, 17, 165–176. [Google Scholar]
- Malek, A. M. , Gibbons, G. H. , Dzau, V. J. , & Izumo, S. (1993). Fluid shear stress differentially modulates expression of genes encoding basic fibroblast growth factor and platelet‐derived growth factor B chain in vascular endothelium. The Journal of Clinical Investigation, 92, 2013–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun, G. I. , An, S. M. , Park, H. , Jo, H. , & Boo, Y. C. (2008). Laminar shear stress inhibits lipid peroxidation induced by high glucose plus arachidonic acid in endothelial cells. American Journal of Physiology. Renal Physiology, 295, H1966–H1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy, B. P. , Budi, E. H. , Katsuno, Y. , Lee, M. K. , Smith, S. M. , Mirza, A. M. , … Derynck, R. (2015). ShcA protects against epithelial‐Mesenchymal transition through compartmentalized inhibition of TGF‐beta‐Induced smad activation. PLoS Biology, 13, e1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, M. M. , Inscho, E. W. , Zhang, S. , Seki, T. , Rohatgi, R. , Gusella, L. , … Kohan, D. E. (2015). Flow regulation of endothelin‐1 production in the inner medullary collecting duct. American Journal of Physiology Renal Physiology, 308, F541–F552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, E. N. , Chung, H. W. , Nayebosadri, A. , & Hansen, S. B. (2016). Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nature Communications, 7, 13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri, S. A. , & Piccaluga, P. P. (2012). New molecular insights into peripheral T cell lymphomas. American Journal of Physiology. Renal Physiology, 122, 3448–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperi, C. , & Basdra, E. K. (2015). Polycystins and mechanotransduction: From physiology to disease. World Journal of Experimental Medicine, 5, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetorius, H. A. (2015). The primary cilium as sensor of fluid flow: New building blocks to the model. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. American Journal of Physiology Cell Physiology, 308, C198–C208. [DOI] [PubMed] [Google Scholar]
- Praetorius, H. A. , Frokiaer, J. , Nielsen, S. , & Spring, K. R. (2003). Bending the primary cilium opens Ca(2+)‐sensitive intermediate‐Conductance K+ channels in MDCK cells. Journal of Membrane Biology, 191, 193–200. [DOI] [PubMed] [Google Scholar]
- Praetorius, H. A. , & Spring, K. R. (2001). Bending the MDCK cell primary cilium increases intracellular calcium. Journal of Membrane Biology, 184, 71–79. [DOI] [PubMed] [Google Scholar]
- Quinlan, R. J. , Tobin, J. L. , & Beales, P. L. (2008). Modeling ciliopathies: Primary cilia in development and disease. Current Topics in Developmental Biology, 84, 249–310. [DOI] [PubMed] [Google Scholar]
- Reitsma, S. , Slaaf, D. W. , Vink, H. , van Zandvoort, M. A. , & oude Egbrink, M. G. (2007). The endothelial glycocalyx: Composition, functions, and visualization. Pflugers Archiv, 454, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi, R. , & Flores, D. (2010). Intratubular hydrodynamic forces influence tubulointerstitial fibrosis in the kidney. Current Opinion in Nephrology and Hypertension, 19, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, J. , & Tabunoki, H. (2013). A comprehensive profile of chIP‐Seq‐Based STAT1 target genes suggests the complexity of STAT1‐mediated gene regulatory mechanisms. Gene Regulation and Systems Biology, 7, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann, W. P. , Blobe, G. C. , Kalume, D. E. , Pandey, A. , & Lodish, H. F. (2002). Context‐specific effects of fibulin‐5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin‐5 is induced by transforming growth factor‐beta and affects protein kinase cascades. The Journal of Biological Chemistry, 277, 27367–27377. [DOI] [PubMed] [Google Scholar]
- Schwachtgen, J. L. , Houston, P. , Campbell, C. , Sukhatme, V. , & Braddock, M. (1998). Fluid shear stress activation of egr‐1 transcription in cultured human endothelial and epithelial cells is mediated via the extracellular signal‐related kinase 1/2 mitogen‐activated protein kinase pathway. The Journal of Clinical Investigation, 101, 2540–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A. , Mucino, M. J. , & Ronco, C. (2014). Renal functional reserve and renal recovery after acute kidney injury. Nephron Clinical Practice, 127, 94–100. [DOI] [PubMed] [Google Scholar]
- Simons, M. , Gloy, J. , Ganner, A. , Bullerkotte, A. , Bashkurov, M. , Kronig, C. , … Walz, G. (2005). Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nature Genetics, 37, 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo, P. , Mootha, V. K. , Mukherjee, S. , Ebert, B. L. , Gillette, M. A. , … Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge‐based approach for interpreting genome‐wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi, M. M. , Tarbell, J. M. , Weinbaum, S. , & Spray, D. C. (2004). The role of the glycocalyx in reorganization of the actin cytoskeleton under fluid shear stress: A “bumper‐car” model. Proceedings of the National Academy of Sciences of the United States of America, 101, 16483–16488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 't Hoen, P. A. , Hirsch, M. , de Meijer, E. J. , de Menezes, R. X. , van Ommen, G. J. , & den Dunnen, J. T. (2011). MRNA degradation controls differentiation state‐dependent differences in transcript and splice variant abundance. Nucleic Acids Research, 39, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalovski, M. , Hagopian, M. , Wang, M. , & Brekken, R. A. (2016). Hypoxia and transforming growth factor beta cooperate to induce fibulin‐5 expression in pancreatic cancer. The Journal of Biological Chemistry, 291, 22244–22252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, Y. C. , Hsieh, H. J. , Liao, F. , Ni, C. W. , Chao, Y. J. , Hsieh, C. Y. , & Wang, D. L. (2007). Laminar flow attenuates interferon‐induced inflammatory responses in endothelial cells. Cardiovascular Research, 74, 497–505. [DOI] [PubMed] [Google Scholar]
- Turner, N. , & Grose, R. (2010). Fibroblast growth factor signalling: From development to cancer. Nature Reviews Cancer, 10, 116–129. [DOI] [PubMed] [Google Scholar]
- Venkatachalam, M. A. , Griffin, K. A. , Lan, R. , Geng, H. , Saikumar, P. , & Bidani, A. K. (2010). Acute kidney injury: A springboard for progression in chronic kidney disease. American Journal of Physiology Renal Physiology, 298, F1078–F1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R. H. , Lindblad‐Toh, K. , Birney, E. , Rogers, J. , Abril, J. F. , Agarwal, P. , … Lander, E. S . (2002). Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- Weimbs, T. (2007). Polycystic kidney disease and renal injury repair: Common pathways, fluid flow, and the function of polycystin‐1. American Journal of Physiology Renal Physiology, 293, F1423–F1432. [DOI] [PubMed] [Google Scholar]
- Weinbaum, S. , Duan, Y. , Satlin, L. M. , Wang, T. , & Weinstein, A. M. (2010). Mechanotransduction in the renal tubule. American Journal of Physiology Renal Physiology, 299, F1220–F1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K. , & Ando, J. (2013). Endothelial cell and model membranes respond to shear stress by rapidly decreasing the order of their lipid phases. Journal of Cell Science, 126, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K. , & Ando, J. (2015). Vascular endothelial cell membranes differentiate between stretch and shear stress through transitions in their lipid phases. American Journal of Physiology Renal Physiology, 309, H1178–H1185. [DOI] [PubMed] [Google Scholar]
- Zeng, Y. (2017). Endothelial glycocalyx as a critical signalling platform integrating the extracellular haemodynamic forces and chemical signalling. Journal of Cellular and Molecular Medicine, 21, 1457 –1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Y. , & Liu, J. (2016). Role of glypican‐1 in endothelial NOS activation under various steady shear stress magnitudes. Experimental Cell Research, 348, 184–189. [DOI] [PubMed] [Google Scholar]
- Zhong, M. , Zhao, X. , Li, J. , Yuan, W. , Yan, G. , Tong, M. , … Jiang, Y. (2016). Tumor suppressor folliculin regulates mTORC1 through primary cilia. The Journal of Biological Chemistry, 291, 11689–11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Figure S1. Interaction network of genes regulated by fluid shear stress.
Figure S1. Shear stress induced expression in PTECs in time.
Figure S1. Shear stress induced expression in PTECs is partially reversible after removal of shear.
Table S1. Primer sequences used for qPCR.
Table S2. Counts per million values of all samples analyzed by RNA sequencing.
Table S3. Fold change of differentially expressed genes by fluid shear stress in PTECs.
Table S4. Pathway analysis of up‐regulated genes by fluid shear stress in PTECs.
Table S5. Pathway analysis of down‐regulated genes by fluid shear stress in PTECs.


