Abstract
Objectives
The Ainu, the indigenous people living on the northernmost island of Japan, Hokkaido, have long been a focus of anthropological interest because of their cultural, linguistic, and physical identity. A major problem with genetic studies on the Ainu is that the previously published data stemmed almost exclusively from only 51 modern‐day individuals living in Biratori Town, central Hokkaido. To clarify the actual genetic characteristics of the Ainu, individuals who are less influenced by mainland Japanese, who started large‐scale immigration into Hokkaido about 150 years ago, should be examined. Moreover, the samples should be collected from all over Hokkaido.
Materials and methods
Mitochondrial DNA haplogroups of 94 Ainu individuals from the Edo era were successfully determined by analyzing haplogroup‐defining polymorphisms in the hypervariable and coding regions. Thereafter, their frequencies were compared to those of other populations.
Results
Our findings indicate that the Ainu still retain the matrilineage of the Hokkaido Jomon people. However, the Siberian influence on this population is far greater than previously recognized. Moreover, the influence of mainland Japanese is evident, especially in the southwestern part of Hokkaido that is adjacent to Honshu, the main island of Japan.
Discussion
Our results suggest that the Ainu were formed from the Hokkaido Jomon people, but subsequently underwent considerable admixture with adjacent populations. The present study strongly recommends revision of the widely accepted dual‐structure model for the population history of the Japanese, in which the Ainu are assumed to be the direct descendants of the Jomon people.
Keywords: ancient DNA, Jomon, Okhotsk culture, Satsumon, Siberia
1. INTRODUCTION
The Ainu, the indigenous people living on the northernmost island of Japan, Hokkaido (Figure 1), have long been a focus of anthropological and archeological interest because of their unique cultural, linguistic, and physical characteristics. From a cultural perspective, it is generally agreed that the ethnic identity of the Ainu was established in the 13th century AD. However, the origin of their biological identity remains unclear, which has been a major focus of anthropological discussion. Conventionally, based on the close morphological resemblance between the Jomon people and the Ainu (e.g., Brace & Nagai, 1982; Dodo & Ishida, 1990; Dodo & Kawakubo, 2002; Hanihara, 1991; Turner, 1976, 1990; Yamaguchi, 1963, 1982), it is generally agreed that the indigenous Hokkaido population maintained its genetic and phenetic characteristics throughout the Neolithic Jomon era to the Ainu era (Figure 2) (Dodo & Kawakubo, 2002).
Figure 1.
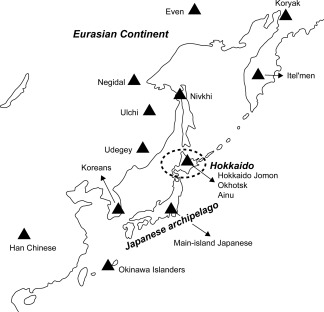
Map of Asia and the geographic locations of the populations analyzed or used for comparison in this study
Figure 2.
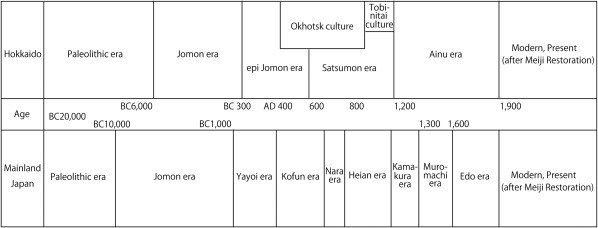
Chronological table of Hokkaido and mainland Japan
However, recent morphological and genetic studies (e.g., Hanihara, Yoshida, & Ishida, 2008; Hanihara, 2010; Ishida, Hanihara, Kondo, & Fukumine, 2009; Sato et al., 2009; Shigematsu, Ishida, Goto, & Hanihara, 2004) have indicated that the Siberian influence via the Okhotsk culture people on the Ainu significantly affected the latter's genetic structure. The Okhotsk culture people are thought to have migrated from northeastern Eurasia and been distributed in the coastal regions of northern and northeastern Hokkaido as well as southern Sakhalin during the 5th to 13th centuries AD (Amano, 2003). On Hokkaido, this culture was rapidly diminished by the invasion of Satsumon culture people at the end of the 9th century. As a result, Okhotsk culture had almost disappeared by the beginning of the 10th century. However, in the easternmost part of Hokkaido, the Okhotsk culture transformed into the Tobinitai culture under the strong influence of the Satsumon culture, and this culture continued until the beginning of the Ainu era (Segawa, 2007).
Recently, we also confirmed the considerable genetic influence of the Okhotsk culture people on the formation of the modern‐day Ainu. We found that mitochondrial DNA (mtDNA) haplogroups A, C, and Y, which are shared by the modern‐day Siberian populations, Okhotsk culture people, and the modern‐day Ainu, are not observed in the Hokkaido Jomon people (Adachi et al., 2011).
However, previous genetic studies aimed at clarifying the ethnic derivation of the Ainu have two major drawbacks. The first is that almost all of these studies (Hammer et al., 2006; Harihara, Hirai, & Omoto, 1986; Harihara et al., 1988; Horai et al., 1996; Jinam et al., 2015, 2012; Tajima et al., 2004) were based on modern‐day samples. Historically, after the Meiji government started sending settlers to Hokkaido as a national policy in 1869 (Fumoto, 2004), an enormous number of mainland Japanese migrated to Hokkaido, and their apparent genetic influence on the modern‐day Ainu was confirmed by a genome‐wide single‐nucleotide polymorphism (SNP) analysis by Jinam et al. (2012). In this context, the extent of the earlier genetic contribution of the mainland Japanese to the original formation of the Ainu cannot be evaluated.
The second problem is that the study area focused on in previous genetic studies is limited or unclear. The samples used in most studies were derived from only 51 individuals who lived in Biratori Town, central Hokkaido. Actually, Ainu people live throughout Hokkaido, so it is unclear whether these 51 individuals can represent all of the Ainu. Besides these studies, Horai et al. (1991) analyzed six early‐modern (200–300 years BP) Ainu in Hokkaido. However, unfortunately, they did not describe the region of origin of these samples.
This sampling bias precludes genetic evaluation of possible regional differences of the Ainu as suggested by previous morphological studies (e.g., Dodo, Kawakubo, Sawada, & Ishida, 2012; Hanihara et al., 2008; Hanihara, 2010; Ito, 1967; Kondo, 1995; Ossenberg, Dodo, Maeda, & Kawakubo, 2006; Shigematsu et al., 2004; Yamaguchi, 1981). Therefore, to clarify the process of formation of the Ainu and their regional characteristics, individuals who were not influenced by the large‐scale immigration of mainland Japanese must be examined. In addition, regional bias of the samples must be corrected.
In the current study, we thus present the mtDNA data of Ainu skeletons excavated from all over Hokkaido. All of these skeletons are dated to the Edo era (1603 to 1868 AD), a period for which no distinct proof of an intensive genetic influence of mainland Japanese on the Ainu has been reported. A detailed analysis of the coding and control regions of mtDNA was used to address the questions regarding the matrilineal genetic characteristics of the Ainu, as well as those about the relationship between the Ainu and the ancient and modern‐day populations in eastern Eurasia. Moreover, regional differences of the Ainu were discussed.
2. MATERIALS AND METHODS
2.1. Samples
We sampled the teeth from 115 Ainu skeletons from the Edo era (Edo Ainu, hereafter), which were housed at Sapporo Medical University and Date City Institute of Funkawan Culture, Hokkaido, Japan. Only well‐preserved teeth were selected from each individual. The use of these samples was approved by the Ainu Association of Hokkaido and Educational Committee of Hokkaido. Moreover, this study was approved by the ethics committee of the Faculty of Medicine of the University of Yamanashi. This study was performed against the background of discussions being held among the Ainu Association of Hokkaido, the Anthropological Society of Nippon, and the Japan Archaeological Association together with the Ministry of Education, Culture, Sports, Science and Technology, Japan, about ways of studying Ainu skeletal remains and burial goods; this work forms a part of such study.
2.2. Authentication methods
To minimize the risk of prelaboratory contamination (Sampietro et al., 2006), the tooth samples were collected only by N.A. and K.S. while wearing a face mask, laboratory coat, cap, and gloves. DNA‐based studies were performed in a laboratory dedicated exclusively to ancient DNA analysis. In the current study, we employed standard precautions to avoid contamination, such as the separation of pre‐ and post‐PCR experimental areas, use of disposable laboratory wares and filter‐plugged pipette tips, treatment with DNA contamination removal solution (DNA Away; Molecular Bio Products, San Diego, CA), UV irradiation of equipment and benches, and negative extraction and PCR controls.
2.3. DNA extraction and purification
DNA was extracted from the skeletal samples in accordance with slightly modified versions of previously published protocols (Adachi, Sawada, Yoneda, Kobayashi, & Itoh, 2013) at the University of Yamanashi by N.A. In brief, tooth samples were immersed in a 13% bleach solution (Nacalai Tesque Inc., Kyoto, Japan) for 15 min, rinsed several times with DNase/RNase‐free distilled water (Thermo Fisher Scientific, Waltham, MA), and allowed to air‐dry. Furthermore, the outer surface of the samples was removed to a depth of 1 mm by using a dental drill. Next, the samples were rinsed with DNase/RNase‐free distilled water once again and allowed to air‐dry under irradiation in a UV crosslinker for 90 min, during which time they were turned over twice. The samples were then encased in Exafine silicone rubber (GC, Tokyo, Japan) following the protocol described by Gilbert et al. (2003). After approximately 5 min, or once the rubber was hard, the tip of the root of the tooth sample was removed with a horizontal cut and then powdered using a mill (Multi‐beads Shocker®; Yasui Kikai, Osaka, Japan). Thereafter, the dentin around the dental cavity and the dental pulp was powdered and removed through the root tip by using a dental drill. The powdered samples were then decalcified with 0.5 M EDTA (pH 8.0) (Thermo Fisher Scientific) at 56°C overnight. The EDTA buffer was then replaced with a fresh solution, and the samples were decalcified for a further 36 hr at 56°C. Decalcified samples were lysed in 1,000 μL of Genomic Lyse buffer (Genetic ID, Fairfield, IA) with 50 μL of 20 mg/mL proteinase K (Thermo Fisher Scientific) overnight at 56°C. Finally, the lysate was extracted twice with 1,500 μL of phenol‐chloroform‐isoamyl alcohol (25:24:1) (Thermo Fisher Scientific) and 1,500 μL of chloroform (Wako Pure Chemical Industries, Osaka, Japan). DNA was extracted from the lysate by using a FAST ID DNA Extraction Kit (Genetic ID) in accordance with the manufacturer's instructions. Finally, 130 μL of DNA extract was obtained from each sample.
2.4. Sequencing and SNP typing of mtDNA
Sequencing and SNP typing of mtDNA were performed at the University of Yamanashi by N.A., T.K., and R.T. For all samples, we followed slightly modified versions of previously described protocols (Adachi et al., 2013) to amplify and sequence the segments of mtDNA. The segments examined covered parts of the tRNAPro gene, hypervariable segment (HVS) 1 (nucleotide positions [np] 15,999–16,141, 16,121–16,238, and 16,209–16,366, relative to the revised Cambridge reference sequence [rCRS: Andrews et al., 1999]), and HVS 2 (128–256).
Amplifications were carried out in a total reaction volume of 20 μL containing a 2‐μL aliquot of DNA extract, 0.2 μM of each primer, and reagents of a Multiplex PCR kit (QIAGEN, Hilden, Germany). The PCR conditions were as follows: initial denaturation at 95°C for 15 min; followed by 36 cycles at 94°C for 30 s, 57°C for 90 s, and 72°C for 30 s; and final extension at 72°C for 10 min. Following PCR, the products were subjected to direct sequencing by using the same primers and the BigDye® Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on a 310 DNA Sequencer (Applied Biosystems).
To accurately assign mtDNAs under study to the relevant haplogroups, haplogroup‐diagnostic polymorphisms, especially those in coding regions, were also analyzed. This analysis was performed by using a multiplex system for analyzing mitochondrial single‐nucleotide polymorphisms (mtSNPs), which relies on a novel amplified product‐length polymorphism (APLP) method that uses inosine‐flapped primers and is specifically designed for the detailed haplogrouping of degraded mtDNA (Kakuda et al., 2016). This analysis was performed only for the samples for which the sequence analysis was successful. Sequence and APLP analyses were performed at least twice to confirm reproducibility.
2.5. Haplogroup assignment
Haplogroup assignment was performed by N.A. and T.K. Each Edo Ainu mtDNA was assigned to the relevant haplogroup according to the HVS and coding region data, by using Phylotree, the updated comprehensive phylogenetic tree of global human mtDNA variation (www.phylotree.org; mtDNA tree Build 17) (Van Oven & Kayser, 2009), so that each mtDNA was allocated to the smallest named haplogroup. Since several segments of the same mtDNA sample were analyzed independently, meticulous care was taken to avoid artificial recombination of the results.
2.6. Population, statistical, and phylogenetic analyses
These analyses were performed by H.K‐K. and K.S. To clarify the biological relationships between the Edo Ainu and other populations, their haplogroup frequencies were compared with those of 14 ancient and modern‐day East Asian and Siberian populations obtained from previously published data (Adachi et al., 2011; Lee et al., 2006; Maruyama, Minaguchi, & Saitou, 2003; Sato et al., 2009; Schurr, Sukernik, Starikovskaya, & Wallace, 1999; Sekiguchi et al., 2008; Starikovskaya, Sukernik, Schurr, Kogelnik, & Wallace, 1998; Starikovskaya et al., 2005; Sukernik et al., 2012; Tajima et al., 2004; Xu & Hu, 2015; Yao, Kong, Bandelt, Kivisild, & Zhang, 2002). The approximate locations of these populations are shown in Figure 1.
To investigate the relationship between the Edo Ainu and the other populations, neighbor‐joining (NJ) trees based on haplogroup frequency‐based pairwise Fst were constructed in the program Mega 7.0.18 (Kumar, Tamura, & Nei, 2004).
2.7. Comparison between regions
As described earlier, previous morphological studies have indicated possible regional differences in the Ainu people. These studies divided Hokkaido Island in several different ways. Among them, Dodo et al. (2012) explicitly explained their particular system of division as follows. The area that is southwest of Ishikari Low Land (southwestern Hokkaido, classified by the dashed line in Figure 3) often shared the same culture as the northern part of Honshu since the Jomon era. Moreover, the influence of the Kofun culture of northern Honshu, which played an important role in the formation of the Satsumon culture, extended into this area. Therefore, this area is expected to have been genetically more influenced by the mainland Japanese than the other areas of Hokkaido. On the other hand, the area along the coast of the Sea of Okhotsk was intensively inhabited by the Okhotsk culture people; therefore, their influence is expected to be more evident in this area. The other area is considered to be less influenced by mainland Japanese and the Okhotsk people, and is expected to have retained the characteristics of the indigenous Jomon people. We adopted this classification and analyzed the observed haplogroups in each region and their frequencies to elucidate the regional differences of the Ainu. However, the number of samples from along the coast of the Sea of Okhotsk that were successfully analyzed was small (seven individuals from four sites, see Figure 3), so we combined this area with the central area and analyzed this as “northeastern/central Hokkaido.”
Figure 3.
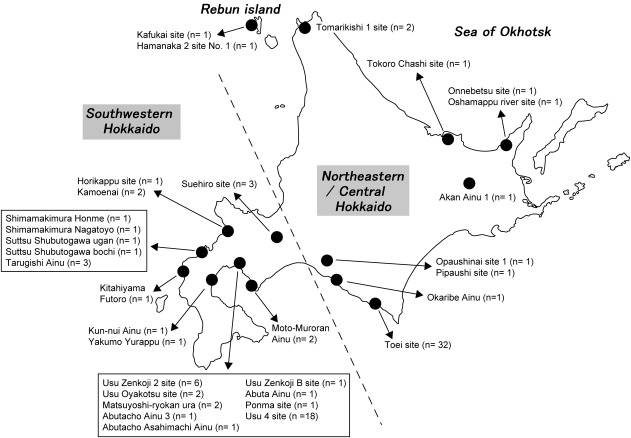
Regional classification of the population sample sites. Geographic locations of the sites are indicated by black dots. Numbers in parentheses denote the numbers of samples from each site that were successfully analyzed
Aside from the Hokkaido Jomon and the Okhotsk culture people, the populations inhabiting the neighboring area of Hokkaido are expected to have contributed to the establishment of the Ainu. Therefore, to elucidate the regional differences of the haplogroups observed in the Edo Ainu, we classified them into four types as follows: (1) Hokkaido Jomon type (the haplogroups that are observed in the Hokkaido Jomon); (2) Okhotsk type (the haplogroups that are observed in the Okhotsk culture people, but are absent from the Hokkaido Jomon); (3) mainland Japanese type (the haplogroups that are absent from the Hokkaido Jomon and the Okhotsk culture people, but are observed in modern‐day mainland Japanese at higher frequencies than in modern‐day native Siberians); and (4) Siberian type (the haplogroups that are absent from the Hokkaido Jomon and the Okhotsk culture people, but are observed in modern‐day native Siberians at higher frequencies than in modern‐day mainland Japanese). Based on the frequencies of these four haplogroup types, regional differences of the Ainu between southwestern Hokkaido and northeastern/central Hokkaido were examined by Fisher's exact probability test by using R version 3.4.1 (R Core Team, 2017).
3. RESULTS
3.1. Authenticity of the mtDNA data
Both negative extraction and PCR controls consistently showed negative results throughout the experiment. Of the 115 samples, 3 were fragile and broke down during the decontamination process, and 7 did not yield PCR products in the sequence analysis. Of the remaining 105 samples that yielded PCR products, 11 were excluded from APLP analysis due to the lack of reproducibility and/or ambiguity of the sequence results. As a result, 94 samples were successfully sequenced. APLP analysis was performed for these 94 samples, generating reproducible and unambiguous results (Supporting Information Table S1 and Figure 3).
All mtDNAs that were successfully analyzed fully represented the haplogroup motifs; therefore, we confidently assigned them to the relevant haplogroups. This means that the reliability of the data obtained from the samples was supported by the established mtDNA phylogenetic system. We have submitted these 94 mtDNA sequences to the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/index-e.html) with the accession numbers LC219528‐LC219567, LC219569‐LC219594, LC219596‐LC219663, LC219665‐LC219690, LC219692‐LC219719.
3.2. Genetic characteristics of the Ainu mtDNAs
Twenty‐one haplogroups and their subhaplogroups were identified in 94 Edo Ainu individuals (Supporting Information Table S1). As described earlier, conventionally, the Ainu are considered to be descended from the Hokkaido Jomon people, with little admixture with other populations. Among the haplogroups observed in the Hokkaido Jomon (N9b1, N9b4, N9b*, D4h2, G1b*, M7a2, M7a*; Adachi et al., 2011), haplogroups N9b1, G1b*, and M7a2 are also observed in the Edo Ainu. Above all, haplogroup N9b1, which is the most frequently observed haplogroup in the Hokkaido Jomon people (55.6%, 30 of 54 individuals; Adachi et al., 2011), is also observed at a relatively high frequency (20.2%, 19 of 94 individuals) in the Edo Ainu. These findings indicate the genetic continuity between the Hokkaido Jomon and the Ainu. This possible genetic continuity is corroborated by Y chromosome DNA analysis of the modern Ainu. Y chromosomal DNA haplogroup D1b, which is considered to be a strong candidate for the Jomon paternal lineage, was observed at high frequency in the modern Ainu (Hammer et al., 2006; Tajima et al., 2004).
However, there is a crucial difference between the Hokkaido Jomon people and the Ainu. Four haplogroups (N9b4, N9b*, D4h2, M7a*) are missing in the Edo Ainu, whereas they have 19 haplogroups that are not observed in the Hokkaido Jomon. This raises questions about the conventional hypothesis that the Ainu are the direct descendants of the Hokkaido Jomon people.
To clarify the matrilineal genetic relationship between the Edo Ainu and the other populations, pairwise Fst values between each pair of populations (Supporting Information Table S2) were calculated from the mtDNA haplogroup frequencies of the Edo Ainu and the 14 ancient and modern‐day East Asian and Siberian populations (Table 1, Figure 1). In the frequency‐based clustering of compared populations determined by neighbor‐joining based on the Fst values described earlier, the Ainu of the Edo era was located almost in the median center between the Hokkaido Jomon/Udegey cluster and the Lower Amur region cluster including the Okhotsk culture people (Figure 4). This confirmed our hypothesis (Adachi et al., 2011) that the genetic characteristics of the Ainu are based on the Hokkaido Jomon people and the subsequent input of Lower Amur region Siberian genes through the Okhotsk culture people. However, when examining in detail the haplogroups observed in the Ainu examined in the present study, haplogroups M7a1, M7b1a1a1, D4 (except for D4h2), M8a, Z1a, M9a, F1b, N9a, A5a, and A5c are observed neither in the Hokkaido Jomon nor in the Okhotsk culture people. This suggests the presence of populations other than the Hokkaido Jomon and the Okhotsk culture people that contributed to the formation of the Ainu. Mainland Japanese and the native Siberians are considered to be the major candidates for the origin of these haplogroups because of the proximity of their distributions to Hokkaido.
Table 1.
mtDNA haplogroup frequencies in the Edo Ainu and the ancient and present‐day East Eurasian populations
| Haplogroup | Edo Ainu (n = 94) | Hokkaido Jomon (n = 54) | Okhotsk (n = 37) | Modern Ainu (n = 51) | Mainland Japanese (n = 211) | Okinawa Islanders (n = 156) | Koreans (n = 694) | Han Chinese (n = 471) | Ulchi (n = 160) | Even (n = 87) | Nivkhi (n = 56) | Udegey (n = 46) | Negidal (n = 33) | Koryak (n = 147) | Itel'men (n = 46) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N9b | 20.2 | 64.8 | 10.8 | 7.8 | 1.9 | 2.6 | 0.3 | 0.0 | 4.4 | 0.0 | 0.0 | 30.4 | 0.0 | 0.0 | 0.0 |
| M7a1 | 2.1 | 0.0 | 0.0 | 15.7 | 9.5 | 28.2 | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M7a2 | 2.1 | 1.9 | 5.4 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 | 0.0 | 19.6 | 0.0 | 0.0 | 0.0 |
| M7a* | 0.0 | 5.6 | 0.0 | 0.0 | 0.0 | 0.6 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| G1a1 | 0.0 | 0.0 | 0.0 | 5.9 | 5.7 | 0.0 | 2.3 | 0.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| G1b | 9.6 | 11.1 | 24.3 | 15.7 | 0.0 | 0.0 | 0.0 | 0.0 | 7.5 | 10.3 | 5.4 | 0.0 | 27.3 | 42.9 | 69.5 |
| G2a | 0.0 | 0.0 | 0.0 | 3.9 | 3.3 | 1.9 | 3.9 | 2.8 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| G3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 1.6 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Y1 | 31.9 | 0.0 | 43.2 | 19.6 | 0.5 | 0.0 | 1.3 | 0.8 | 43.1 | 5.7 | 66.1 | 8.7 | 21.2 | 8.8 | 4.3 |
| C1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| C4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 1.3 | 2.4 | 2.5 | 5.0 | 28.7 | 0.0 | 17.4 | 9.1 | 23.8 | 0.0 |
| C5a | 3.2 | 0.0 | 5.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.9 | 2.3 | 0.0 | 0.0 | 0.0 | 12.2 | 13.0 |
| D4o1 | 2.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 | 3.6 | 0.0 | 0.0 | 0.0 | 0.0 |
| D4h2 | 0.0 | 16.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
|
D4 (excl. D4o1 and D4h2) |
11.7 | 0.0 | 0.0 | 13.7 | 36.0 | 25.6 | 27.1 | 14.4 | 16.3 | 39.1 | 25.0 | 0.0 | 24.2 | 1.4 | 0.0 |
| D5 | 0.0 | 0.0 | 0.0 | 3.9 | 5.2 | 2.6 | 5.3 | 8.0 | 0.6 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M7b1a1a1 | 3.2 | 0.0 | 0.0 | 3.9 | 2.8 | 7.1 | 3.5 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| M7c | 0.0 | 0.0 | 0.0 | 0.0 | 0.5 | 0.6 | 2.9 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| F1 | 2.1 | 0.0 | 0.0 | 2.0 | 5.2 | 3.2 | 6.9 | 9.8 | 3.1 | 5.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| F (excl. F1) | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.4 | 5.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| N9a | 1.1 | 0.0 | 0.0 | 0.0 | 1.9 | 3.2 | 6.3 | 4.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| B4 | 0.0 | 0.0 | 0.0 | 2.0 | 7.6 | 7.1 | 10.5 | 11.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| B5 | 0.0 | 0.0 | 2.7 | 0.0 | 3.3 | 0.6 | 4.3 | 3.4 | 0.6 | 0.0 | 0.0 | 0.0 | 12.1 | 0.0 | 0.0 |
| A5a | 1.1 | 0.0 | 0.0 | 0.0 | 5.2 | 0.6 | 3.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| A5c | 6.4 | 0.0 | 0.0 | 0.0 | 0.5 | 2.6 | 0.4 | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| A8 | 0.0 | 0.0 | 8.2 | 2.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 | 6.5 |
| Z1a | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | 2.3 | 0.0 | 0.0 | 0.0 | 5.4 | 6.5 |
| M8a | 1.1 | 0.0 | 0.0 | 0.0 | 0.5 | 0.6 | 1.0 | 4.0 | 3.8 | 0.0 | 0.0 | 15.2 | 0.0 | 0.0 | 0.0 |
| M9a | 1.1 | 0.0 | 0.0 | 2.0 | 0.9 | 1.9 | 1.6 | 2.1 | 0.6 | 0.0 | 0.0 | 8.7 | 0.0 | 0.0 | 0.0 |
| Others | 0.0 | 0.0 | 0.0 | 2.0 | 8.1 | 9.0 | 12.2 | 24.8 | 1.9 | 2.3 | 0.0 | 0.0 | 6.1 | 2.7 | 0.0 |
Frequencies of mtDNA haplogroups are inferred from published data as follows: Hokkaido Jomon (Adachi et al., 2011); Okhotsk (Sato et al., 2009); Modern Ainu (Tajima et al., 2004);.
Mainland Japanese (Maruyama et al., 2003); Ryukyu Islanders (Sekiguchi et al., 2008); Koreans (Lee et al., 2006); Han Chinese (Xu & Hu, 2015; Yao et al., 2002);.
Nivkhi, Udegey, Negidal (Starikovskaya et al., 2005); Ulchi, Even (Sukernik et al., 2012); Koryak, Itel'men (Schurr et al., 1999). All populations except for the Edo Ainu, Okhotsk, and.
Hokkaido Jomon are modern ones. Haplogroups of the modern Ainu and Ryukyu Islanders were inferred from the sequence of HVS 1 of mtDNA.
Figure 4.
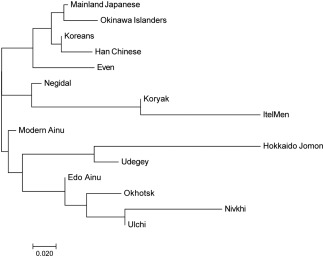
Frequency‐based clustering of 15 ancient and modern East Asian and Siberian populations determined by neighbor‐joining using the Fst values based on haplogroup frequencies
3.3. Regional differences of the Ainu inferred from mtDNA data
When comparing the frequencies of the four haplogroup types observed in southwestern and northeastern/central Hokkaido (Table 2), their regional differences were statistically significant at the 5% level (p‐value = .03194). As described earlier, the results of mtDNA analysis of the Edo Ainu suggested the multiple origins of this population. This possible multiplicity of origins of the Ainu is considered to be an important factor behind their regional differences.
Table 2.
Frequencies of the four types of mtDNA haplogroups (%) observed in the Edo Ainu in each region
| Hokkaido Jomon type |
Total (n = 94) |
Southwestern Hokkaido (n = 51) |
Northeastern/Central Hokkaido (n = 43) |
|---|---|---|---|
| N9b1 | 20.2 (n = 19) | 13.7 (n = 7) | 27.9 (n = 12) |
| M7a2 | 2.1 (n = 2) | 2.0 (n = 1) | 2.3 (n = 1) |
| G1b* | 8.5 (n = 8) | 3.9 (n = 2) | 14.0 (n = 6) |
| Total | 30.9 (n = 29) | 19.6 (n = 10) | 44.2 (n = 19) |
| Okhotsk type | |||
| Y1 | 31.9 (n = 30) | 29.4 (n = 15) | 34.9 (n = 15) |
| C5a2b | 3.2 (n = 3) | 5.9 (n = 3) | 0 |
| Total | 35.1 (n = 33) | 35.3 (n = 18) | 34.9 (n = 15) |
| Mainland Japanese type | |||
| D4 (exc. D4o1) | 11.7 (n = 11) | 17.6 (n = 9) | 4.7 (n = 2) |
| M7b1a1a1 | 3.2 (n = 3) | 5.9 (n = 3) | 0 |
| F1b1a | 2.1 (n = 2) | 3.9 (n = 2) | 0 |
| N9a | 1.1 (n = 1) | 2.0 (n = 1) | 0 |
| M7a1a7 | 2.1 (n = 2) | 2.0 (n = 1) | 2.3 (n = 1) |
| A5a | 1.1 (n = 1) | 2.0 (n = 1) | 0 |
| A5c | 6.4 (n = 6) | 3.9 (n = 2) | 9.3 (n = 4) |
| Total | 27.7 (n = 26) | 37.3 (n = 19) | 16.3 (n = 7) |
| Siberian type | |||
| D4o1 | 2.1 (n = 2) | 3.9 (n = 2) | 0 |
| G1b1 | 1.1 (n = 1) | 0 | 2.3 (n = 1) |
| Z1a | 1.1 (n = 1) | 2.0 (n = 1) | 0 |
| M8a | 1.1 (n = 1) | 0 | 2.3 (n = 1) |
| M9a | 1.1 (n = 1) | 2.0 (n = 1) | 0 |
| Total | 6.4 (n = 6) | 7.8 (n = 4) | 4.7 (n = 2) |
4. DISCUSSION
4.1. Ethnic derivation of the Ainu inferred from mtDNA data
As widely recognized, mtDNA provides only matrilineal information of the populations. Moreover, it covers only a tiny portion of genetic variation of the individual and is very prone to genetic drift. Therefore, many of the population parameters examined in the present study must be interpreted cautiously. We tried to shed as much light as possible on the genetic features of the Ainu despite these restrictions.
Nowadays, among the theories explaining the population history of the Japanese, the dual‐structure model proposed by Hanihara (1991) is the most widely accepted. This model assumes that the first occupants of the Japanese archipelago came from somewhere in Southeast Asia and they gave rise to the Jomon people. Thereafter, the second wave of migration from Northeast Asia took place in and after the Aeneolithic Yayoi age. These two populations gradually mixed with each other and brought the Japanese people into existence. In this model, the Ainu are considered to be descended from the Jomon people, with little admixture with other populations.
However, the results of our study suggest that the later admixture of Ainu with other populations than Jomon people was more considerable than it was proposed until today.
First, our results showed that the genetic influence of the Okhotsk culture people on the Ainu is significant. The proportion of Okhotsk‐type haplogroups in the Edo Ainu was 35.1%, which is as high as that of the Jomon‐type haplogroups (30.9%). This suggests that the Okhotsk culture people were one of the main genetic contributors to the formation of the Ainu.
Moreover, intriguingly, a genetic contribution of the mainland Japanese to the Edo Ainu is evident (28.1%), which is almost as considerable as those of the Jomon and the Okhotsk culture people. As referred to above, conventionally, the genetic influence of the mainland Japanese on the Ainu is considered to have been limited until the Meiji government started sending settlers to Hokkaido as a national policy in 1869. However, our findings cast doubt on this accepted notion.
In addition, Siberian‐type haplogroups are observed in the Edo Ainu. Although their frequency is low (7.3%), as described earlier, the existence of these haplogroups may hint at the continuity of the genetic relationship between the Ainu and native Siberians even after the Okhotsk culture disappeared from Hokkaido. However, the number of Okhotsk people who were genetically analyzed is still small (n = 37; Sato et al., 2009), so it is possible that these haplogroups will be identified in the Okhotsk people in further study.
4.2. Regional differences of the Ainu
By classifying the mtDNA haplogroups into four types as described earlier, regional differences of the Ainu people were highlighted. Judging from the data shown in Table 2, the high frequencies of Jomon‐type haplogroups in northeastern/central Hokkaido (44.2%) and the high frequencies of mainland Japanese‐type haplogroups in southwestern Hokkaido (37.3%) might be plausible reasons for these regional differences.
This result is consistent with the result of a morphological analysis by Ossenberg et al. (2006). They described that, among the Ainu in Hokkaido, individuals in southeastern Hokkaido (this area is contained within our category of “northeastern/central Hokkaido”) are the closest to the Jomon people, whereas the individuals in western Hokkaido (this area is included within our category of “southwestern Hokkaido”) are the closest to mainland Japanese. This result is considered reasonable, given the geographical proximity of southwestern Hokkaido to the main island of Japan.
However, surprisingly, there were no regional differences in the frequencies of the Okhotsk‐type haplogroups (35.3% in southwestern Hokkaido and 34.9% in northeastern/central Hokkaido). This indicates that the genetic influence of the Okhotsk culture people diffused rapidly in the fledgling Ainu.
As described earlier, Segawa (2007) stated that the invasion of the Satsumon culture people into the areas inhabited by the Okhotsk culture people and the subsequent decline of Okhotsk culture occurred rapidly. The Okhotsk culture people were considered to have been assimilated rapidly into the Satsumon culture during this process, and became part of the basis of the Ainu.
5. CONCLUDING REMARKS
In the current study, we clarified that the Ainu were established from the Hokkaido Jomon people and subsequently underwent considerable admixture with adjacent populations. The present study strongly recommends review of the widely accepted dual‐structure model regarding the population history of the Japanese, in which the Ainu are assumed to be the direct descendants of the Jomon people. However, the causes of the regional differences in the genetic influence of the Okhotsk culture people on the Ainu remain unresolved by mtDNA analysis. Nuclear genome analysis using next‐generation sequencing is expected to be helpful to resolve this issue.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Supporting Information Table S1
Supporting Information Table S2
ACKNOWLEDGMENTS
We are grateful to the Ainu Association of Hokkaido, Dr. Mineko Fujimiya and Dr. Hirofumi Matsumura of Sapporo Medical University, Japan, and Dr. Naoyuki Ohshima of Date City Institute of Funkawan Culture, Japan, for granting permission to genetically analyze the skeletons. We also express our gratitude to Mr. Ken Nagata of the Department of Marine Bio‐diversity Research, Japan Agency for Marine‐Earth Science and Technology (JAMSTEC), for his help in writing this article. We also thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript. This study was supported by Grants‐in‐aid for Scientific Research from the Japan Society for the Promotion of Science (Nos. 25251043, 26440257, 15K01136, 16H01954, 17H02019, 17H03737), and JSPS Core to Core Program (Advanced Research Networks) “Advanced Core Research Center for the History of Human Ecology in the North.”
Adachi N, Kakuda T, Takahashi R, Kanzawa‐Kiriyama H, Shinoda K‐I. Ethnic derivation of the Ainu inferred from ancient mitochondrial DNA data. Am J Phys Anthropol. 2018;165:139–148. https://doi.org/10.1002/ajpa.23338
Funding information Japan Society for the Promotion of Science, Grant/Award Numbers: 25251043, 26440257, 15K01136, 16H01954, 17H02019, 17H03737; JSPS Core to Core Program (Advanced Research Networks) “Advanced Core Research Center for the History of Human Ecology in the North.”
REFERENCES
- Adachi, N. , Sawada, J. , Yoneda, M. , Kobayashi, K. , & Itoh, S. (2013). Mitochondrial DNA analysis of the human skeleton of the initial Jomon phase excavated at the Yugura Cave site, Nagano, Japan. Anthropological Science, 121, 137–143. [Google Scholar]
- Adachi, N. , Shinoda, K. , Umetsu, K. , Kitano, T. , Matsumura, H. , Fujiyama, R. , … Tanaka, M. (2011). Mitochondrial DNA analysis of Hokkaido Jomon skeletons: Remnants of archaic maternal lineages at the southwestern edge of former Beringia. American Journal of Physical Anthropology, 146, 346–360. [DOI] [PubMed] [Google Scholar]
- Amano, T. (2003). Okhotsk bunka to ha nanika In Nomura T. & Utagawa H. (Eds.), Shin Hokkaido no kodai 2: Zoku‐jomon, Okhotsk bunka (pp. 110–133). Sapporo, Japan: Hokkaido Shinbunsha (in Japanese; ). [Google Scholar]
- Andrews, R. M. , Kubacka, I. , Chinnery, P. F. , Lightowlers, R. N. , Turnbull, D. M. , & Howell, N. (1999). Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genetics, 23, 147. [DOI] [PubMed] [Google Scholar]
- Behar, D. , Van Oven, M. , Rosset, S. , Metspalu, M. , Loogväli, E. L. , Silva, N. M. , … Villems, R. (2012). A “Copernican” reassessment of the human mitochondrial DNA tree from its root. American Journal of Human Genetics, 90, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace, C. L. , & Nagai, M. (1982). Japanese tooth size: Past and present. American Journal of Physical Anthropology, 59, 399–411. [DOI] [PubMed] [Google Scholar]
- Dodo, Y. , & Ishida, H. (1990). Population history of Japan as viewed from cranial nonmetric variation. Anthropological Science, 98, 269–287. [Google Scholar]
- Dodo, Y. , & Kawakubo, Y. (2002). Cranial affinities of the Epi‐Jomon inhabitants in Hokkaido, Japan. Anthropological Science, 110, 1–32. [Google Scholar]
- Dodo, Y. , Kawakubo, Y. , Sawada, J. , & Ishida, H. (2012). The Ainu and their neighbors as seen from the perspective of nonmetric cranial trait variation II. Regional differences among the Ainu peoples in Hokkaido and Sakhalin. Anthropological Science (Japanese Series), 120, 135–149. in Japanese). [Google Scholar]
- Fumoto, S. (2004). Kindai‐teki kaitaku no rosen In Tabata H. (Eds.), Kaidou no nihon‐shi 2: Ezo‐chi kara hokkaido he (pp. 84–111). Tokyo, Japan: Yoshikawa Kobunkan (in Japanese; ). [Google Scholar]
- Gilbert, M. T. P. , Willerslev, E. , Hansen, A. J. , Barnes, I. , Rudbeck, L. , Lynnerup, N. , & Cooper, A. (2003). Distribution patterns of postmortem damage in human mitochondrial DNA. American Journal of Human Genetics, 72, 32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, M. F. , Karafet, T. M. , Park, H. , Omoto, K. , Harihara, S. , Stoneking, M. , & Horai, S. (2006). Dual origins of the Japanese: Common ground for hunter‐gatherer and farmer Y chromosomes. Journal of Human Genetics, 51, 47–58. [DOI] [PubMed] [Google Scholar]
- Hanihara, K. (1991). Dual structure model for the population history of Japanese. Japan Review, 2, 1–33. [Google Scholar]
- Hanihara, T. , Yoshida, M. , & Ishida, H. (2008). Craniometric variation of the Ainu: An assessment of differential gene flow from Northeast Asia to northern Japan, Hokkaido. American Journal of Physical Anthropology, 137, 283–293. [DOI] [PubMed] [Google Scholar]
- Hanihara, T. (2010). Metric and nonmetric dental variation and the population structure of the Ainu. American Journal of Human Biology, 22, 163–171. [DOI] [PubMed] [Google Scholar]
- Harihara, S. , Hirai, M. , & Omoto, K. (1986). Mitochondrial DNA polymorphism in Japanese living in Hokkaido. Japanese Journal of Human Genetics, 31, 73–83. [DOI] [PubMed] [Google Scholar]
- Harihara, S. , Saitou, N. , Hirai, M. , Gojobori, T. , Park, K. S. , Misawa, S. , … Omoto, K. (1988). Mitochondrial DNA polymorphism among five Asian populations. American Journal of Human Genetics, 43, 134–143. [PMC free article] [PubMed] [Google Scholar]
- Horai, S. , Kondo, R. , Murayama, K. , Hayashi, S. , Koike, H. , & Nakai, N. (1991). Phylogenetic affiliation of ancient and contemporary humans inferred from mitochondrial DNA. Philosophical Transactions of the Royal Society B, 333, 409–417. [DOI] [PubMed] [Google Scholar]
- Horai, S. , Murayama, K. , Hayasaka, K. , Matsubayashi, S. , Hattori, Y. , Fucharoen, G. , … Pan, I. H. (1996). mtDNA polymorphism in East Asian populations, with special reference to the peopling of Japan. American Journal of Human Genetics, 59, 579–590. [PMC free article] [PubMed] [Google Scholar]
- Ishida, H. , Hanihara, T. , Kondo, O. , & Fukumine, T. (2009). Craniometric divergence history of the Japanese populations. Anthropological Science, 117, 147–156. [Google Scholar]
- Ito, S. (1967). Local differences on Ainu skulls‐Metrical observations. Bulletin of Institute for Study of North Eurasian Culture, Hokkaido University, 2, 191–238. [Google Scholar]
- Jinam, T. , Kanzawa‐Kiriyama, H. , Inoue, I. , Tokunaga, K. , Omoto, K. , & Saitou, N. (2015). Unique characteristics of the Ainu population in Northern Japan. Journal of Human Genetics, 60, 565–571. [DOI] [PubMed] [Google Scholar]
- Jinam, T. , Nishida, N. , Hirai, M. , Kawamura, S. , Oota, H. , Umetsu, K. , … Saitou, N. (2012). The history of human populations in the Japanese archipelago inferred from genome‐wide SNP data with a special reference to the Ainu and the Ryukyuan populations. Journal of Human Genetics, 57, 787–795. [DOI] [PubMed] [Google Scholar]
- Kakuda, T. , Shojo, H. , Tanaka, M. , Nambiar, P. , Minaguchi, K. , Umetsu, K. , & Adachi, N. (2016). Multiplex APLP system for haplogrouping extremely degraded East‐Asian mtDNAs. PLoS One, 11, e0158463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, O. (1995). An analysis of Ainu population structure, based on cranial morphology. Anthropological Science, 103, 369–384. [Google Scholar]
- Kumar, S. , Tamura, K. , & Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics, 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Lee, H. Y. , Yoo, J. E. , Park, M. J. , Chung, U. , Kim, C. Y. , & Shin, K. J. (2006). East Asian mtDNA haplogroup determination in Koreans: Haplogroup‐level coding region SNP analysis and subhaplogroup‐level control region sequence analysis. Electrophoresis, 27, 4408–4418. [DOI] [PubMed] [Google Scholar]
- Mabuchi, T. , Susukida, R. , Kido, A. , & Oya, M. (2007). Typing the 1.1 kb control region of human mitochondrial DNA in Japanese individuals. Journal of Forensic Sciences, 52, 355–363. [DOI] [PubMed] [Google Scholar]
- Maruyama, S. , Minaguchi, K. , & Saitou, N. (2003). Sequence polymorphisms of the mitochondrial DNA control region and phylogenetic analysis of mtDNA lineages in the Japanese populations. International Journal of Legal Medicine, 117, 218–225. [DOI] [PubMed] [Google Scholar]
- Ossenberg, N. S. , Dodo, Y. , Maeda, T. , & Kawakubo, Y. (2006). Ethnogenesis and craniofacial change in Japan from the perspective of nonmetric traits. Anthropological Science, 114, 99–115. [Google Scholar]
- R Core Team . (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- Sampietro, M. L. , Gilbert, M. T. P. , Lao, O. , Caramelli, D. , Lari, M. , Bertranpetit, J. , & Lalueza‐Fox, C. (2006). Tracking down human contamination in ancient human teeth. Molecular Biology and Evolution, 23, 1801–1807. [DOI] [PubMed] [Google Scholar]
- Sato, T. , Amano, T. , Ono, H. , Ishida, H. , Kodera, H. , Matsumura, H. , … Masuda, R. (2009). Mitochondrial DNA haplogrouping of the Okhotsk people based on analysis of ancient DNA: An intermediate of gene flow from the continental Sakhalin people to the Ainu. Anthropological Science, 117, 171–180. [Google Scholar]
- Schurr, T. G. , Sukernik, R. I. , Starikovskaya, E. B. , & Wallace, D. C. (1999). Mitochondrial DNA variation in Koryaks and Itel'men: Population replacement in the Okhotsk Sea‐Bering Sea region during Neolithic. American Journal of Physical Anthropology, 108, 1–39. [DOI] [PubMed] [Google Scholar]
- Segawa, T. (2007). Ainu no rekishi: Umi to takara no nomad. Tokyo, Japan: Koudansha (in Japanese; ). [Google Scholar]
- Sekiguchi, K. , Imaizumi, K. , Fujii, K. , Mizuno, N. , Ogawa, Y. , Akutsu, T. , … Kasai, K. (2008). Mitochondrial DNA population data of HV 1 and HV 2 sequences from Japanese individuals. Legal Medicine, 10, 284–286. [DOI] [PubMed] [Google Scholar]
- Shigematsu, M. , Ishida, H. , Goto, M. , & Hanihara, T. (2004). Morphological affinities between Jomon and Ainu: Reassessment based on nonmetric cranial traits. Anthropological Science, 112, 161–172. [Google Scholar]
- Starikovskaya, E. B. , Sukernik, R. I. , Derbeneva, O. A. , Volodko, N. V. , Ruiz‐Pesini, E. , Torroni, A. , … Wallace, D. C. (2005). Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Annals of Human Genetics, 69, 67–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starikovskaya, E. B. , Sukernik, R. I. , Schurr, T. G. , Kogelnik, A. , & Wallace, D. C. (1998). Mitochondrial DNA diversity in Chukchi and Siberian Eskimos: Implications for the genetic history of ancient Beringia and the peopling of the New World. American Journal of Human Genetics, 63, 1473–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukernik, R. I. , Volodko, N. V. , Mazunin, I. O. , Eltsov, N. P. , Dryomov, S. V. , & Starikovskaya, E. B. (2012). Mitochondrial gene diversity in the Tubalar, Even, and Ulchi: Contribution to prehistory of Native Siberians and their affinities to Native Americans. American Journal of Physical Anthropology, 148, 123–138. [DOI] [PubMed] [Google Scholar]
- Tajima, A. , Hayami, M. , Tokunaga, K. , Juji, T. , Matsuo, M. , Marzuki, S. , … Horai, S. (2004). Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages. Journal of Human Genetics, 49, 187–193. [DOI] [PubMed] [Google Scholar]
- Turner, C. G. (1976). Dental evidence on the origins of the Ainu and Japanese. Science, 193, 911–913. [DOI] [PubMed] [Google Scholar]
- Turner, C. G. (1990). Major features of Sundadonty and Sinodonty, including suggestions about East Asian microevolution, population history, and late Pleistocene relationships with Australian Aboriginals. American Journal of Physical Anthropology, 82, 295–317. [DOI] [PubMed] [Google Scholar]
- Van Oven, M. , & Kayser, M. (2009). Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Human Mutation, 30, E386–E394. http://www.phylotree.org. [DOI] [PubMed] [Google Scholar]
- Xu, K. , & Hu, S. (2015). Population data of mitochondrial DNA HVS‐I and HVS‐II sequences for 208 Henan Han Chinese. Legal Medicine, 17, 287–294. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, B. (1963). The human skeletal remains from Bozuyama in central Hokkaido. Journal of Anthropological Society Nippon, 71, 55–71. in Japanese). [Google Scholar]
- Yamaguchi, B. (1981). Human skeletal remains of prehistoric times in Hokkaido In Ogata T. (Eds.), The Japanese I (anthropology 5) (pp. 137–156). Tokyo, Japan: Yuzankaku (in Japanese). [Google Scholar]
- Yamaguchi, B. (1982). A review of the osteological characteristics of the Jomon population in prehistoric Japan. Journal of Anthropological Society Nippon, 90, 77–90. [Google Scholar]
- Yao, Y. G. , Kong, Q. P. , Bandelt, H. J. , Kivisild, T. , & Zhang, Y. P. (2002). Phylogenetic differentiation of mitochondrial DNA in Han Chinese. American Journal of Human Genetics, 70, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Supporting Information Table S1
Supporting Information Table S2


