Abstract
The lungs are not sterile or free from bacteria; rather, they harbor a distinct microbiome whose composition is driven by different ecological rules than for the gastrointestinal tract. During disease, there is often a shift in community composition towards Gammaproteobacteria, the bacterial class that contains many common lung-associated gram-negative “pathogens.” Numerous byproducts of host inflammation are growth factors for these bacteria. The extracellular nutrient supply for bacteria in the lungs, which is severely limited during health, markedly increases due to the presence of mucus and vascular permeability. While Gammaproteobacteria benefit from airway inflammation, they also encode molecular components that promote inflammation, potentially creating a cyclical inflammatory mechanism. In contrast, Prevotella species that are routinely acquired via microaspiration from the oral cavity may participate in immunologic homeostasis of the airways. v Areas of future research include determining for specific lung diseases (1) whether an altered lung microbiome initiates disease pathogenesis, promotes chronic inflammation, or is merely a marker of injury and inflammation, (2) whether the lung microbiome can be manipulated therapeutically to change disease progression, (3) what molecules (metabolites) generated during an inflammatory response promote cross-kingdom signaling, and (4) how the lung “ecosystem” collapses during pneumonia, to be dominated by a single pathogen.
OVERVIEW
Until very recently, it was believed that the lungs were sterile or free from bacteria.1 This would be a truly remarkable fact, if true, considering that there is virtually no environmental niche on earth which is so extreme (in oxygen, pH, hydrophobicity, temperature, salinity, predators, nutrient scarcity, etc.) that bacterial communities cannot been found.2 Furthermore, the lower airways are warm, moist surfaces centimeters past the oral and nasal cavities, which are both bacteria-rich environments over which there is a constant flow of microaersosol-generating air and fluid. Microaspiration is well-documented to occur in healthy, asymptomatic subjects.3–6 Since the first culture-independent report of a “lung microbiome” in healthy subjects,7 well over 50 published studies using molecular techniques for bacterial identification have found evidence of bacteria in the lower airways. No modern study has found evidence of a sterile lung environment. Thus, a better understanding of the nature and impact of the lung microbiome during health and disease may provide important information for diagnostic and/or therapeutic approaches.
Much more is known about the microbiome of the gastrointestinal (GI) tract than that of the lungs, largely due to non-invasive access to samples, ease of collecting serial samples and the high bacterial biomass in the samples. Thus, the principles of host-microbe interactions in the gut are sometimes presented as a universal model. While the lungs and GI tract share embryological origin and both contain mucosa-lined luminal surfaces, their anatomic as well as their macroscopic, microscopic and biochemical features are quite distinct. This results in marked differences in the composition and population dynamics of their respective microbiomes.
During health, migration of microbes in the digestive tract is from the mouth to the anus (i.e., “unidirectional”) across a variety of physical and chemical barriers. For example, orally-introduced microbes must survive the acidic pH of the stomach followed by the alkaline pH of the duodenum during transit to the cecum and colon; at which point, they must ultimately compete for resources within the densely populated microbial communities of the large intestine. The lungs are a different story. The movement of air, mucus and microorganisms in the lungs occurs with minimal physical barriers between the larynx and the most distal sites in the alveoli. This flow occurs in both directions (i.e., “bidirectional”). The end result is that the bacterial populations of the lungs are more dynamic than that those found in the lower GI tract and the opportunity to establish colonization is quite different.
The gut and lungs also have markedly different environmental conditions on their epithelial surfaces, which results in markedly different microbial communities and immune system interactions.1,8 The gastrointestinal tract is of uniform temperature (37 °C) throughout its entire length, while the epithelial surfaces of the upper and lower airways exist in a temperature gradient that ranges from ambient temperature at the point of inhalation to core body temperature in the alveoli. While the gut is anaerobic, the lungs are aerobic (during health). The epithelial surfaces of the trachea and bronchi are mucus-covered, similar to the GI tract; however, the vast majority of the lung’s surface area (the alveoli) is coated with surfactant, a lipid-rich substance that has bacteriostatic effects against select bacterial species. Luminal IgA levels are also far higher in the gut. Finally, while the epithelial and sub-epithelial tissues of the lungs and gut both harbor significant resident leukocyte populations, the extra-epithelial luminal surfaces of the gut are largely devoid of leukocytes during health. In sharp contrast, a resident phagocyte population (alveolar macrophages) actively patrol the luminal surfaces of the alveoli during health.
The emerging model for understanding the factors that control the lung microbiome during health and disease, as well as the effect of these changes on inflammatory processes and vice-versa, is that of an immunoecology “Life in Antarctica” model.8,9 In other words, similar to the story of human presence in Antarctica, there is constant viable bacterial presence in the lungs when healthy with little evidence for true long-term colonization of the lower airways. However, if the physical features of Antarctica were to change and provide for the nutritional needs of the population, long-term human colonization would be possible. Similarly, if there is a change in the physical, metabolic and predatory/defense features of the lung environment, then long-term bacterial colonization is possible. A developing question for researchers will be whether the colonization within the lung during times of stress is a cause or effect scenario and whether it contributes to the ongoing immune responses.
THE NASAL MICROBIOME
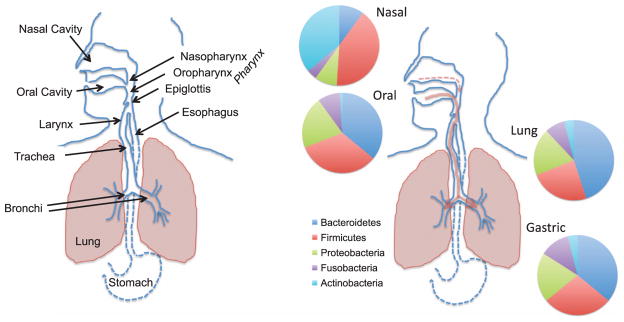
The resident nasal bacterial microbiome in healthy adults is largely distinct from that found in the oral cavity and shares membership with that of the skin microbiome (Figure 1).10–13 In the culture-independent 16S rRNA amplicon sequencing surveys of 300 healthy adults performed by the Human Microbiome Project, as well as similar sequencing studies from our group and others,11,12,14 the most commonly found and dominant members of the nasal microbiota were drawn from the following genera: Propionibacteria, Corynebacteria, Staphylococcus, and Moraxella. This is consistent with decades of culture-based analyses of the nasal microbiota. The Human Microbiome Project metagenomic studies could further speciate the major colonizing organisms as P. acnes, C. accolens, C. kropenstedtii, S. aureus, and S. epidermitidis.11 In studies by Charlson et al., the bacterial communities in the nasopharynx (as well as the oropharynx) were significantly different in smokers compared with non-smokers, both in terms of membership and distribution.12 Aging and disease processes, such as chronic rhinosinusitis, can also markedly alter the nasal microbiota.15,16 Bisgaard et al. reported that blood eosinophil counts and total IgE at 4 years of age were significantly increased in children colonized neonatally with S. pneumoniae, M. catarrhalis, H. influenzae, or a combination of these organisms.17 Furthermore, the prevalence of asthma and the reversibility of airway resistance after β2-agonist administration at 5 years of age were significantly increased in the children colonized neonatally with these organisms as compared with the children without such colonization.
Figure 1.
Comparison of the anatomy and bacterial microbiomes of the upper regions of the aerodigestive tract in humans. Bacterial composition is shown at the phylum-level. Gammaproteobacteria are a class in the Proteobacteria phylum. Arrows depict flow of air and microaerosols, with a greater contribution in most individuals during health coming from the oral cavity.
THE ORAL MICROBIOME
While not discussed in this review, the human oral cavity contains a number of different microbiological habitats that range from the teeth and gingival pockets to the tongue, cheeks and tonsils as well as the hard and soft palates.18 Numerous culture-independent studies have shown that the oral microbiome is comprised of hundreds of different species, with distinct clusters of microbes predominating in different oral habitats. The major bacterial inhabitants of the oral cavity are Prevotella, Veillonella, Streptococcus, Haemophilus, Fusobacterium, Neisseria, and Corynebacteria species.11,12,14,18,19 Similar to the nasal cavity, smoking, aging and disease processes can all markedly alter the oral microbiota.
UPPER AIRWAY ANATOMY
The upper and lower airways comprise an anatomically contiguous surface that begins in the mouth and nose and terminates in the lungs (Figure 1). A healthy human inhales 12–15 times per minute at rest, on average, with ~20% of the air in the lungs being exchanged on each breath. Both of these parameters change during disease. Air that enters the lungs passes through the nasal and oral cavity and across the microbe-laden mucosal surfaces of these sites. All regions of the upper airways are mucus-covered epithelium, with a flow of mucus, resident microbes, trapped particulates and inhaled microbes down the aerodigestive tract (the vast majority moving with salivary flow into the gastrointestinal tract). In addition to air passing through oropharynx, there is a constant flow of saliva carrying resident and ingested oral microbes (~2 liters per day). Further along, in the laryngopharynx, food, fluid, saliva, and mucus is diverted into the esophagus by the epiglottis, a flap of elastic cartilage, while air is diverted into the opening of the larynx and trachea. However, this constant movement of fluid and air through this cavity also creates a biologically significant amount of aerosols, even in healthy individuals. Microaspiration of these aerosols is responsible for the vast majority of the constant microbial seeding of the lower airways from the oral cavity during health and both nasal and oral cavity when nasal mucus flow increases (discussed below).
LOWER AIRWAY ANATOMY
Connecting the larynx to the bronchial trees of the lungs is the trachea, which is connected to the two primary bronchi of the lungs (Figure 1). This Y-shaped split of the conducting airways is called the carina and recent studies from our laboratory have shown that the bacterial communities of the carina are more similar to those found in the oral cavity than at any other point below the epiglottis (Dickson et al., submitted). Moving deeper into the lower airways, each bronchus is further divided into smaller, secondary bronchi, which continue to successively branch off (forming tertiary bronchi) that further divide into terminal bronchioles. The surface and wall temperatures of the upper portion of the trachea and bronchial tree are typically lower than core body temperature due to the movement of colder ambient air into the lungs. This is a factor that has not been historically considered when discussing the human microbiome and/or airway colonization. During disease, such as asthma and chronic obstructive pulmonary disease (COPD), airflow markedly changes in the lower bronchi and bronchioles due to a number of structural and inflammatory factors, all of which can markedly affect the dynamics of microbial immigration into the airways.
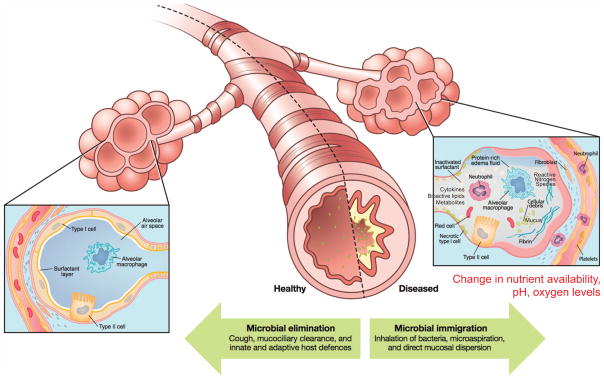
The bronchial tree is a mucosal surface and is the only section of the aerodigestive tract where mucus flow is backward (toward the oropharynx). Diseases or activities that destroy ciliary function, reduce the cough reflex, increase lower airway mucus secretion or increase the viscosity of the mucus, can markedly affect the dynamics of microbial elimination from the airways, increase airway temperatures and also create anaerobic zones for microbial growth in the lungs. However, the cellular architecture and function of the lungs markedly changes as you advance past the terminal bronchioles into the alveoli. Overall, the total surface area of the alveoli in an adult is estimated to be around 70 m2. The alveoli are not lined with the type of epithelium found in the bronchial tree; rather, the alveoli contain a thin single-cell squamous epithelial layer (type 1 alveolar epithelial cells) that are coated with a thin layer of surfactant (not mucus). Pulmonary surfactant is a lipid-rich lipoprotein complex (phospholipoprotein) formed by type 2 alveolar epithelial cells, which contains a number of free fatty acids, as well as phosphatidylcholine-containing lipids (primarily dipalmitoyl phosphatidylcholine) and sphingomyelins. It also contains a number of proteins, some of which have potent anti-bacterial or opsonization activity, especially for Gram-negative bacteria. Free fatty acids are bacteriocidal to many Gram positive bacteria. Thus, the large surface area of the alveoli is an aerobic lipid-covered surface, making it a markedly distinct ecological habitat from those found in the upper airways and GI tract. As described below, disease and inflammatory processes that introduce anaerobic zones, cause serum leak or mucus accumulation in the alveoli, or result in alveolar collapse (atalectasis) can markedly change the microbiome of the lungs, which in turn, has the potential to further drive inflammatory processes.
Finally, another critical difference in the alveolar surface compared with that of the bronchi, trachea, upper airways and GI tract is the presence of a host-derived microbial “predator” as a resident of the microbiome during health: the alveolar macrophage. This is the only “externally-exposed” surface of the body in which a leukocyte resides on the surface during health, rather than in the epithelium or sub-epithelial layers. Functioning much as an ameba might function in a pond environment, alveolar macrophages are highly active phagocytes. Thus, microbes that have developed virulence strategies based on competition with ameba in aqueous environments (e.g. Legionella, Pseudomonas, etc.) can often use those same mechanisms for surviving predatory alveolar macrophages.
THE IMMUNOECOLOGY OF THE RESPIRATORY TRACT MICROBIOME
In the “Life in Antarctica” model of the lung microbiome, the balance of three factors determine and shape the lung microbiome (Figure 2):9 (1) microbial immigration into the airways, (2) elimination of microbes from the airways, and (3) the relative reproduction rates of the microbes found in the airways, which is determined by the growth conditions of the region. Any change in the lung microbial communities, within an individual or as viewed across disease states, must be the result of a change in one of these three factors. Evidence from our lab and others strongly points to subclinical microaspiration as the primary source of microbial immigration5,14,20–23 (Dickson et al., submitted). Subclinical microaspiration of pharyngeal secretions among healthy subjects is well-documented. Other contributing factors can include the inhalation of bacteria from the air and direct migration along airway mucosal surfaces. Microbial elimination is accomplished by a combination of mucociliary clearance, cough, and host immune defenses (both innate and adaptive).
Figure 2.
Model of the environmental, microbial, structural, and immunologic factors that control the composition of the lung microbiome during health and disease.
Inflammation plays a key role in the third ecological factor that shapes the lung microbiome, largely by affecting the regional growth conditions (e.g. nutrient availability, temperature, pH, oxygen tension). Inflammation can increase vascular leak into the airways, providing critical nutrient elements such as carbon sources, amino acids, vitamins, and iron. Epithelial cell damage can create exposed zones of basement membrane matrix, which facilitates bacterial adherence. Damaged epithelial cells also produce key innate cytokines, such as TSLP, IL-25, and IL-33, in response to bacterial components.24 These mediators can induce activation of ILC2s that produce IL-5 and IL-13 leading to additional inflammation, such as eosinophils, and expansion of goblet cells.25 Goblet cell hyperplasia, seen in allergies and other states of chronic IL-13 production, can lead to excessive mucus levels deep in the airways providing anaerobic niches that inhibit phagocytosis and further enhance bacterial colonization.26 Inflammatory cells can also be sources of catecholamines that could modulate bacterial virulence.27,28 Finally, as discussed in more detail below, the production of reactive nitrogen species can provide the necessary terminal electron acceptors to grow in airways that have become anaerobic due to excessive mucus production, atalectasis or inflammatory consolidation. When healthy, the regional growth conditions generally do not support robust bacterial growth, resulting in relatively little bacterial growth.
THE ORAL MICROBIOME AS THE PRIMARY SOURCE OF THE BACTERIAL MICROBIOTA IN HUMAN LUNGS DURING HEALTH
The end result of this interplay of these three factors is that in the vast majority of healthy individuals, the microbiome of human lungs during health more closely resembles that of the oropharynx than it does inhaled air, the nasopharynx or the lower gastrointestinal tract.14,19,29,30 The dominant bacteria genera found in the lower airways are Prevotella, Veillonella, and Streptococcus but also include Fusobacterium and Haemophilus. The nasal microbiome contributes little to lung communities during health in most, but not all, individuals.14,29 This is likely due the low volume of nasal secretions relative to salivary flow. However, liquids applied to the nares do appear in the lungs3 and rhinorrhea can cause the lung and nasal microbial communities to look more similar. Nasal colonizers such as S. aureus and M. catarrhalis are both also well-documented respiratory pathogens during pneumonia and various other inflammatory respiratory diseases. While the oral microbiome is the primary source of the bacterial microbiota in human lungs during health, it remains to be determined what are the factors that additionally shape the lung microbiome in those healthy individuals with enhanced similarity between their nasal and lung microbiomes.14
THE LUNG MICROBIOME CHANGES DURING DISEASE
Acute and chronic lung diseases can dramatically change the ecological determinants of the lung microbiome - immigration, elimination and regional growth conditions, resulting in markedly different microbial communities.1,8 Surprisingly, smoking does not change the lung microbiome, although it does change the nasal microbiome as well as immune function in the airways. A major feature of the microbiome in diseased lower airways is a shift in community composition away from the Bacteroidetes phylum that dominates the healthy lung microbiome towards Gammaproteobacteria, the class that contains many common lung-associated gram-negative “pathogens.” Changes in the lung microbiota are just beginning to be associated with important clinical features of chronic lung disease such as exacerbation frequency in bronchiectasis,31 mortality in idiopathic pulmonary fibrosis,32 and responsiveness to corticosteroids and antibiotics in asthma.33,34
THE LUNG MICROBIOME AND INFLAMMATION: A TWO-WAY STREET
Inflammation is most often viewed as a process that generates activated leukocytes and antimicrobial molecules to destroy invading microbes. However, it is now appreciated that inflammation plays a key role in wound repair35 and unregulated inflammation is an underlying cause of many chronic diseases. More recently, it has been proposed that certain taxonomic groups of bacteria, especially those found in the Gammaproteobacteria class, encode the metabolic capacity to utilize inflammatory byproducts to survive and prosper under anaerobic and low oxygen conditions.22,36,37 During chronic inflammation these Gammaproteobacteria can markedly increase in relative abundance in a site (i.e., “bloom”) and outcompete bacteria that lack the metabolic capacity to benefit from inflammation. This connection between Gammaproteobacteria blooms and inflammation and has been largely studied for the gastrointestinal (GI) tract. However, the same mechanisms are likely at play in the respiratory tract in chronic respiratory diseases such as asthma and chronic obstructive pulmonary disease, with the most intensely studied bacteria being Pseudomonas aeruginosa outgrowth during cystic fibrosis.38
In the GI tract, inflammation changes the metabolic environment of the mucosa by providing the terminal electron acceptors needed for anaerobic respiration by many Gamma-proteobacteria, thereby allowing them to outgrow the resident anaerobes that primarily utilize fermentation for their energy needs (fermentation generates less ATP than respiration).22,36,37 Production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) are two of the primary antimicrobial effectors of inflammatory cells. Reactive nitrogen species are produced via inducible nitric oxide synthase (iNOS) in myeloid cells and even other leukocytes.39 O2− and NO can react together to produce peroxynitrite (ONOO−), which breaks down into nitrate (NO3−) and nitrite (NO2−). This extracellular nitrate can be used as a terminal electron acceptor to support anaerobic respiration and outgrowth of Gamma-proteobacteria in vivo via denitrification.21,22,37,40–44 Thus, iNOS/RNS can also promote the colonization of inflamed mucosal surfaces by providing a selective advantage for denitrifying facultative anaerobic bacteria such as E. coli, S. typhimurium, and K. pneumoniae. Most Gammaproteobacteria, including Pseudomonas aeruginosa, encode these pathways. P. aeruginosa has long been held to be an obligate aerobic bacterium; however, recent studies have highlighted that this is not true. P. aeruginosa has a highly branched respiratory system for growth under both microaerophilic and anaerobic conditions.45 Inflammation is a common feature of chronic respiratory diseases such as asthma and chronic obstructive pulmonary disease. It has been observed in a number of studies of humans and mice that the relative proportion of Gammaproteobacteria in the lungs increases during disease.1,38,46 We have recently reviewed the role of inflammation in promoting the growth of P. aeruginosa in cystic fibrosis and inflamed airways.38 Thus, one of the consequences of the upregulated host inflammatory response on mucosal surfaces is the production of terminal electron acceptors for anaerobic respiration that allow P. aeruginosa and other Gammaproteobacteria to grow, persist and outcompete all other microbial members on that surface.
Gammaproteobacteria will not only benefit from mucosal inflammation, they also encode molecular components that promote inflammation. Microbe-associated molecular patterns (MAMPs) on the surface of bacteria interact with receptors on immune cells to drive inflammation, including lipopolysaccharide (LPS), which interacts with TLR4 on immune cells.47 LPS that is hexa-acylated is 100-fold more immunostimulatory when bound to TLR4 then LPS that is penta-aceylated..48 While penta-aceylated LPS is found in most Gram-negative bacteria, Gammaproteobacteria both penta- and hexa-aceylated LPS. The difference in inflammatory potential between commensal oral Prevotella sp. in the lungs and COPD/asthma-associated Gammaproteobacteria is likely partly due to differences in LPS structure.47,49 Many Gammaproteobacteria also encode TLR5-binding flagellins, as well as TLR9-binding unmethylated CpG motifs.50,51 Thus, a feedback loop can develop where bacteria that contain the most immunostimulatory microbe-associated molecular patterns are also the ones that benefit most metabolically from an inflammatory environment.
LUNG MICROBIOTA AND IMMUNITY
The lung and airway microbiome can directly impacts immunity and disease or can be an effect of altered local immunity/inflammation during the development of disease. Early and severe viral infections, such as respiratory syncytial virus (RSV), influenza, and even rhinovirus, can alter the long-term immune response of the lung, especially in infants that may not yet have a stable lung microbiome. Numerous correlative studies have suggested that changes in the lung microbiome may accompany disease progression, including allergic disease, but other than in confirmed cases of bacterial pneumonia, it has yet to be definitively established whether specific members of the lung microbiota provide a direct stimuli that leads to disease susceptibility or protection.
However, there is emerging mechanistic data in this area, most notably on the role of Prevotella spp. in the airways. As described earlier, Gram-negative Prevotella spp. are often found in the airways of healthy individuals14,19,29,30 and exhibit differences in LPS structure compared with Gram-negative Gammaproteobacteria.47,49 In one study, human monocyte-derived dendritic cells (Hu mDCs) were cultured with either Prevotella spp. or Haemophilus spp. and activation markers and cytokine expression were monitored. While all bacteria could induce CD83, CD40 and CD86, Haemophilus spp. induced higher levels of IL-23, IL-12p70 and IL-10. Furthermore, co-culture experiments found that Prevotella spp. were able to reduce Haemophillus influenzae-induced IL-12p70 in Hu mDCs.49 Similar responses were observed in mice for both in vitro and in vivo assays.47 In another set of studies from 112 patients post-lung transplantation, it was observed that a shift away from a Prevotella-dominant airway microbiome correlated with the development of inflammatory or remodeling profiles in macrophages.46 Prevotella-high and -low community profiles have been noted in healthy subjects,14,30 with the Prevotella-high profile being associated with enhanced “subclinical” lung inflammation, that is notable for enhanced expression of inflammatory cytokines and elevated Th-17 lymphocytes.30,52 However, association studies do not determine the direction of causality and this data would be very consistent with a model where subclinical airway inflammation leads to higher levels of micro-aspirated Prevotella. In support of this model is the observation in cystic fibrosis patients that colonization with P. aeruginosa, a pro-inflammatory bacteria, significantly increased the likelihood that Prevotella will be present.53 However, as described earlier, chronic airway inflammation is ultimately associated with the emergence of Gammaproteobacteria dominated bacterial communities in the airways.1,38,46 The inflammation-microbiome connection is a two-way street in that pulmonary inflammation in LPS/ elastase-treated mice (a model of COPD) has been reported to increase Pseudomonas spp. and reduce Prevotella spp. and intranasal transfer of bronchoalveolar lavage fluid that contains the lung microbiota from these diseased mice enhances IL-17A production in the lungs of antibiotic-treated or germ-free recipients.54 This is consistent with the findings that targeting of the lung microbiota by innate immunity, as a result of pIgR/ sIgA deficiency, results in progressive small airway remodeling and emphysema.55 Collectively, these observations support an emerging model that exposure of the pulmonary immune system to oral-derived microbiota (notably Prevotella spp.) during health is part of the central homeostatic processes that regulate pulmonary inflammatory responses.
Numerous studies have also demonstrated that the composition of the gut microbiome has an impact on the development of pulmonary immune responses, such as allergic airway disease.56–60 For example, in the study by Trompette et al., mice fed a high-fiber diet had increased circulating levels of short-chain fatty acids (SCFAs) and were protected against allergic inflammation in the lung, whereas a low-fiber diet decreased levels of short-chain fatty acids and increased allergic airway disease.59 The mechanism underlying this immune modulation involved alterations in bone marrow hematopoiesis that were characterized by enhanced generation of macrophage and dendritic cell (DC) precursors and subsequent seeding of the lungs by DCs with high phagocytic capacity but an impaired ability to promote Th2 cell effector functions. Despite observations in mouse models over a decade ago, the mechanisms that underlie the gut-lung axis of immune regulation still emain to be completely elucidated.
While the PAMP distribution from various organisms likely contributes to the nature of innate immune responses that can be pathogenic, the organisms themselves produce intermediates that can alter the subsequent immune responses. The ability of bacteria-derived metabolites to affect the nature of the immune response has been outlined in several studies in the gut microbiome. For example, a link between diet and microbiome for regulation of immune responses has been established with dietary fiber and Clostridium species leading to production of short-chain fatty acids and development of Treg cells.61–65 Other microbial members of the micrbiota may provide protective mediators, including anti-inflammatory omega-3 polyunsaturated fatty acids (ω-3 PUFA), as well as immune modulating tryptophan metabolites.66,67 Thus, by understanding the cohorts of micro-organisms that colonize the lung during health and disease, and the metabolites that they generate, a clearer picture of the immune environment and modulated response can be identified.
CONCLUSIONS AND FUTURE DIRECTIONS
There is much to be explored and proven about the concept that the relationship between inflammation and changes in the lung microbiome is a two-way street. The luminal surfaces of the lungs are significantly different from those of the GI tract and ecological principles are the driving force of lung microbiome-immmune system interactions (“immunoecology”). Numerous byproducts of the host inflammatory response are known growth factors for select bacterial species, including catecholamines,68 inflammatory cytokines,69–71 increased temperature,72,73 and free ATP.72,73 The extracellular nutrient supply for bacteria in the airways, which is severely limited during health, is abruptly increased by the presence of mucus and vascular permeability. Areas of future research include determining for specific lung diseases (1) whether an altered lung microbiome initiates disease pathogenesis, promotes chronic inflammation, or is merely a marker of injury and inflammation, (2) whether the lung microbiome can be manipulated therapeutically to change disease progression, (3) what other molecules (metabolites) generated during an inflammatory response promote cross-kingdom signaling and augmentation of virulence, and (4) how the diverse bacterial community composition and dynamic homeostasis of the lung “ecosystem” collapses during pneumonia, to be dominated by a single dominant pathogen.9
Acknowledgments
We have been supported in part by the following grants: NHLBI ROI-HL114447 (G.B.H.), NHLBI U01-HL098961 (G.B.H.), Nesbitt Program for CF Research at the University of Michigan (G.B.H.), the University of Michigan Host-Microbiome Initiative (G.B.H. & R.P.D.), NHLBI K23-HL130641 (R.P.D.), UL1TR000433 the Michigan Institute for Clinical & Health Research (R.P.D.), the University of Michigan Center for Integrative Research in Critical Care (R.P.D.), NIAID R01-AI036302 (N.W.L.), and NHLBI R01-HL114858 (N.W.L.). G.B.H. and N.W.L. are also supported by funds from the Mary H. Weiser Food Allergy Center at the University of Michigan.
Footnotes
AUTHOR CONTRIBUTION
All authors contributed to the conceptual framework of this review. G.B.H. wrote the initial draft of the manuscript and all authors were involved in the editing, discussion and revisions of the draft.
DISCLOSURE
The authors declare no conflict of interest.
References
- 1.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horikoshi K, Grant WD. Extremophiles: Microbial Life In Extreme Environments. Vol. 20. Wiley-Liss; 1998. [Google Scholar]
- 3.Gleeson K, Eggli DF, Maxwell SL. Quantitative aspiration during sleep in normal subjects. Chest. 1997;111:1266–1272. doi: 10.1378/chest.111.5.1266. [DOI] [PubMed] [Google Scholar]
- 4.Huxley EJ, Viroslav J, Gray WR, Pierce AK. Pharyngeal aspiration in normal adults and patients with depressed consciousness. Am J Med. 1978;64:564–568. doi: 10.1016/0002-9343(78)90574-0. [DOI] [PubMed] [Google Scholar]
- 5.Quinn LH, Meyer OO. The relationship of sinusitis and bronchiectasis. Arch Otolaryngol—Head Neck Surg. 1929;10:152. [Google Scholar]
- 6.Amberson JB. A clinical consideration of abscesses and cavities of the lung. Bull Johns Hopkins Hosp. 1954;94:227–237. [PubMed] [Google Scholar]
- 7.Hilty M, et al. Disordered microbial communities in asthmatic airways. PLOS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLOS Pathog. 2015;11:e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Resp Med. 2014;2:238–246. doi: 10.1016/S2213-2600(14)70028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlson ES, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen TT, Kirkeby LP, Poulsen K, Reinholdt J, Kilian M. Resident aerobic microbiota of the adult human nasal cavity. APMIS. 2000;108:663–675. doi: 10.1034/j.1600-0463.2000.d01-13.x. [DOI] [PubMed] [Google Scholar]
- 14.Bassis CM, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam K, Schleimer R, Kern RC. The etiology and pathogenesis of chronic rhinosinusitis: a review of current hypotheses. Curr Allergy Asthma Rep. 2015;15:41. doi: 10.1007/s11882-015-0540-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelan FJ, et al. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thorac Soc. 2014;11:513–521. doi: 10.1513/AnnalsATS.201310-351OC. [DOI] [PubMed] [Google Scholar]
- 17.Bisgaard H, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med. 2007;357:1487–1495. doi: 10.1056/NEJMoa052632. [DOI] [PubMed] [Google Scholar]
- 18.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris A, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickson RP, et al. Spatial variation in the healthy human lung microbiome and the adapted island model of lung biogeography. Ann Am Thorac Soc. 2015;12:821–830. doi: 10.1513/AnnalsATS.201501-029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivera-Chavez F, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLOS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7:245–257. doi: 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartemes KR, Kita H. Dynamic role of epithelium-derived cytokines in asthma. Clin Immunol. 2012;143:222–235. doi: 10.1016/j.clim.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlon ST, McKenzie AN. Type 2 innate lymphoid cells: new players in asthma and allergy. Curr Opin Immunol. 2012;24:707–712. doi: 10.1016/j.coi.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Lai HY, Rogers DF. Mucus hypersecretion in asthma: intracellular signalling pathways as targets for pharmacotherapy. Curr Opin Allergy Clin Immunol. 2010;10:67–76. doi: 10.1097/ACI.0b013e328334643a. [DOI] [PubMed] [Google Scholar]
- 27.Flierl MA, et al. Phagocyte-derived catecholamines enhance acute inflammatory injury. Nature. 2007;449:721–725. doi: 10.1038/nature06185. [DOI] [PubMed] [Google Scholar]
- 28.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkataraman A, et al. Application of a neutral community model to assess structuring of the human lung microbiome. MBio. 2015;6:e02284–14. doi: 10.1128/mBio.02284-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal LN, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers GB, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc. 2014;11:496–503. doi: 10.1513/AnnalsATS.201310-335OC. [DOI] [PubMed] [Google Scholar]
- 32.Molyneaux PL, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang YJ, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. e371–373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goleva E, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188:1193–1201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med. 2011;13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winter SE, Baumler AJ. Why related bacterial species bloom simultaneously in the gut: principles underlying the ‘Like will to like’ concept. Cell Microbiol. 2014;16:179–184. doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter SE, Baumler AJ. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes. 2014;5:71–73. doi: 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scales BS, Dickson RP, Huffnagle GB. A tale of two sites: how inflammation can reshape the microbiomes of the gut and lungs. J Leuk Biol. 2016;100:943–950. doi: 10.1189/jlb.3MR0316-106R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36:161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Lopez CA, Rivera-Chavez F, Byndloss MX, Baumler AJ. The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar typhimurium during colitis. Infect Immun. 2015;83:3470–3478. doi: 10.1128/IAI.00351-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bliska JB, van der Velden AW. Salmonella “sops” up a preferred electron receptor in the inflamed intestine. MBio. 2012;3:e00226–00212. doi: 10.1128/mBio.00226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez CA, et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio. 2012;3:e00143–12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez-Torres A, Baumler AJ. Nitrate, nitrite and nitric oxide reductases: from the last universal common ancestor to modern bacterial pathogens. Curr Opin Microbiol. 2015;29:1–8. doi: 10.1016/j.mib.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spees AM, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio. 2013;4:e00430–13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabra W, Kim EJ, Zeng AP. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology. 2002;148:3195–3202. doi: 10.1099/00221287-148-10-3195. [DOI] [PubMed] [Google Scholar]
- 46.Bernasconi E, et al. Airway microbiota determines innate cell inflammatory or tissue remodeling profiles in lung transplantation. Am J Respir Crit Care Med. 2016;194:1252–1263. doi: 10.1164/rccm.201512-2424OC. [DOI] [PubMed] [Google Scholar]
- 47.Larsen JM, Musavian HS, Butt TM, Ingvorsen C, Thysen AH, Brix S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp. promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology. 2015;144:333–342. doi: 10.1111/imm.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brix S, Eriksen C, Larsen JM, Bisgaard H. Metagenomic heterogeneity explains dual immune effects of endotoxins. J Allergy Clin Immunol. 2015;135:277–280. doi: 10.1016/j.jaci.2014.09.036. [DOI] [PubMed] [Google Scholar]
- 49.Larsen JM, et al. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLOS ONE. 2012;7:e31976. doi: 10.1371/journal.pone.0031976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 51.Honko AN, Mizel SB. Effects of flagellin on innate and adaptive immunity. Immunol Res. 2005;33:83–101. doi: 10.1385/IR:33:1:083. [DOI] [PubMed] [Google Scholar]
- 52.Segal LN, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. doi: 10.1038/nmicrobiol.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tunney MM, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am J Respir Crit Care Med. 2008;177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 54.Yadava K, et al. Microbiota promotes chronic pulmonary inflammation by enhancing IL-17A and autoantibodies. Am J Respir Crit Care Med. 2016;193:975–987. doi: 10.1164/rccm.201504-0779OC. [DOI] [PubMed] [Google Scholar]
- 55.Richmond BW, et al. Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat Commun. 2016;7:11240. doi: 10.1038/ncomms11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herbst T, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 57.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73:30–38. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun. 2004;72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 60.Fujimura KE, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci USA. 2014;111:805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol. 2015;36:3–12. doi: 10.1016/j.it.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kosiewicz MM, Dryden GW, Chhabra A, Alard P. Relationship between gut microbiota and development of T cell associated disease. FEBS Lett. 2014;588:4195–4206. doi: 10.1016/j.febslet.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 64.Geuking MB, McCoy KD, Macpherson AJ. Metabolites from intestinal microbes shape Treg. Cell Res. 2013;23:1339–1340. doi: 10.1038/cr.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Memari B, et al. Engagement of the aryl hydrocarbon receptor in mycobacterium tuberculosis-infected macrophages has pleiotropic effects on innate immune signaling. J Immunol. 2015;195:4479–4491. doi: 10.4049/jimmunol.1501141. [DOI] [PubMed] [Google Scholar]
- 67.Zelante T, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Freestone PP, et al. Pseudomonas aeruginosa-catecholamine inotrope interactions: a contributory factor in the development of ventilator-associated pneumonia? Chest. 2012;142:1200–1210. doi: 10.1378/chest.11-2614. [DOI] [PubMed] [Google Scholar]
- 69.Kanangat S, et al. Effects of cytokines and endotoxin on the intracellular growth of bacteria. Infect Immun. 1999;67:2834–2840. doi: 10.1128/iai.67.6.2834-2840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaza SK, McClean S, Callaghan M. IL-8 released from human lung epithelial cells induced by cystic fibrosis pathogens Burkholderia cepacia complex affects the growth and intracellular survival of bacteria. Int J Med Microbiol. 2011;301:26–33. doi: 10.1016/j.ijmm.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 71.Porat R, Clark BD, Wolff SM, Dinarello CA. Enhancement of growth of virulent strains of Escherichia coli by interleukin-1. Science. 1991;254:430–432. doi: 10.1126/science.1833820. [DOI] [PubMed] [Google Scholar]
- 72.Marks LR, Davidson BA, Knight PR, Hakansson AP. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. MBio. 2013;4:e00438–13. doi: 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt A, et al. Neutrophil elastase-mediated increase in airway temperature during inflammation. J Cyst Fibros. 2014;13:623–631. doi: 10.1016/j.jcf.2014.03.004. [DOI] [PubMed] [Google Scholar]