Abstract
Inositol pyrophosphates have emerged as important regulators of many critical cellular processes from vesicle trafficking and cytoskeletal rearrangement to telomere length regulation and apoptosis. We have previously demonstrated that 5-di-phosphoinositol pentakisphosphate, IP7, is at a high level in pancreatic β-cells and is important for insulin exocytosis. To better understand IP7 regulation in β-cells, we used an insulin secreting cell line, HIT-T15, to screen a number of different pharmacological inhibitors of inositide metabolism for their impact on cellular IP7. Although the inhibitors have diverse targets, they all perturbed IP7 levels. This made us suspicious that indirect, off-target effects of the inhibitors could be involved. It is known that IP7 levels are decreased by metabolic poisons. The fact that the inositol hexakisphosphate kinases (IP6Ks) have a high Km for ATP makes IP7 synthesis potentially vulnerable to ATP depletion. Furthermore, many kinase inhibitors are targeted to the ATP binding site of kinases, but given the similarity of such sites, high specificity is difficult to achieve. Here, we show that IP7 concentrations in HIT-T15 cells were reduced by inhibitors of PI3K (wortmannin, LY294002), PI4K (Phenylarsine Oxide, PAO), PLC (U73122) and the insulin receptor (HNMPA). Each of these inhibitors also decreased the ATP/ADP ratio. Thus reagents that compromise energy metabolism reduce IP7 indirectly. Additionally, PAO, U73122 and LY294002 also directly inhibited the activity of purified IP6K. These data are of particular concern for those studying signal transduction in pancreatic β-cells, but also highlight the fact that employment of these inhibitors could have erroneously suggested the involvement of key signal transduction pathways in various cellular processes. Conversely, IP7’s role in cellular signal transduction is likely to have been underestimated.
Abbreviations: IP7, 5-di-phosphoinositol pentakisphosphate; IP6K, inositol hexakisphosphate kinase; IR, insulin receptor; PLC, phospholipase C; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; PI4K, phosphatidylinositol 4-kinase
Keywords: Di-phosphoinositol pentakisphosphate, Inositol pyrophosphate, Inositol hexakisphosphate kinase, ATP/ADP, Pancreatic β-cell, Pharmacological inhibitors
Graphical abstract
Highlights
-
•
In pancreatic β-cells several inhibitors of signal transduction reduce IP7 levels.
-
•
There is a positive correlation between IP7 reduction and decrease in ATP/ADP.
-
•
Inhibitors deplete IP7 levels indirectly by decreasing ATP/ADP levels.
-
•
Some purportedly specific cell-signaling inhibitors directly target IP6K activity.
-
•
Caution is required in interpreting data obtained using inhibitors of inositide metabolism.
1. Introduction
Inositol pyrophosphates are water soluble molecules that are based on the structural backbone of myo-inositol. They are well conserved across eukaryotes varying from yeast to humans [1], [2]. Two well characterized inositol pyrophosphates are di-phosphoinositol pentakisphosphate, IP7, and bis-diphosphoinositol tetrakisphosphate, IP8, [1], [2], [3], [4], [5]. These highly phosphorylated molecules turn over rapidly [3], [6] and have emerged as important cellular regulators involved in many critical processes including vesicle trafficking, exocytosis, apoptosis, osmoregulation, telomere length regulation, cytoskeleton modulation and neurotrophic activation [2], [7], [8]. Both conventional allosteric regulation and novel pyrophosphorylation are responsible for these effects [4], [7], [8], [9].
Different lipid and phosphate molecules derived from myo-inositol play a crucial role in pancreatic β-cell function [4]. In particular, we showed that IP7 is critical for the exocytotic capacity of the pancreatic β-cell [10]. Others have shown that mice lacking inositol hexakisphosphate kinase 1 (IP6K1), one of the enzymes that produces IP7, have decreased plasma insulin levels [11]. IP7 levels are maintained at high concentration in β-cells (~ 5 μM), even under basal glucose conditions. The range of IP7 concentrations calculated to be present in mammalian cells and yeast varies between 0.5 and 5 μM [7], [10]. Although IP7 has been described to be involved in multiple cellular processes, the mechanisms regulating IP7 in β-cells are unknown.
Our original intention was to investigate regulatory pathways that might impact IP7 production. However, our initial studies using relevant signal transduction inhibitors, e.g., against the insulin receptor/PI3K and PLC signaling pathways, made us suspicious that IP7 was being depleted by off-target effects of these reagents. Therefore, we addressed the possible non-specific effects of the inhibitors on IP7 concentrations. Since IP7 is exquisitely dependent on the cellular ATP/ADP ratio [12], [13], we hypothesized that the inhibitors indirectly reduced IP7 concentrations via depletion of ATP/ADP. Our data demonstrate that this is generally the case. In addition, some of these compounds exhibited a direct effect on IP6K activity. This study emphasizes caution when interpreting results obtained using these inhibitors of inositide metabolism to study specific signaling pathways, especially, but not exclusively, in β-cells.
2. Materials and methods
2.1. Chemicals
All cell culture reagents were purchased from Thermofisher Scientific (Stockholm, Sweden). Inhibitors such as LY294002, TBB (4, 5, 6, 7-tetrabromobenzotriazole) and wortmannin were purchased from Sigma-Aldrich (Stockholm, Sweden). HNMPA-(AM)3 (Hydroxy-2-naphthalenylmethylphosphonic Acid Trisacetoxymethyl Ester), PAO (Phenylarsine Oxide), U73343 (1-[6-((17β-3-Methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl]-2,5-pyrrolidinedione) and U73122 (1-[6-((17β-3-Methoxyestra-1,3,5(10)-trien-17-yl)amino)hexyl]-1H–pyrrole-2,5-dione) were purchased from Merck Millipore (Solna, Sweden). HNMPA was purchased from Biomol (Hamburg, Germany).
2.2. Cell culture
The hamster derived HIT-T15 β-cell line, purchased from ATCC (Teddington, UK), was cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, as previously described [10], [14].
2.3. Static incubation
To measure IP7, HIT-T15 cells were labeled for 120 h in a custom made RPMI medium containing 10 μCi/ml [3H] myo-inositol (American radiolabeled chemicals, St. Louis, MO, USA) [10], [14]. On the day of the experiment, cells (~ 1.6 × 105 cells/cm2) were incubated for 30 min or 2 h or 4 h with inhibitor or vehicle (DMSO) in a modified KREBS buffer containing 0.1 mM glucose, 119 mM NaCl, 2 mM CaCl2, 1 mM MgSO4, 4.6 mM KCl, 0.15 mM Na2HPO4, 0.4 mM KH2PO4, 5 mM NaHCO3, 0.5 mg/ml BSA, 20 mM HEPES and pH 7.4. For experiments involving insulin stimulation, cells were incubated for 30 min in modified KREBS buffer followed by stimulation with the same buffer containing 5 mU/ml insulin (Actrapid®, Novo Nordisk, Denmark) for 3 min. The buffer was removed and cells were lysed by 5% ice-cold trichloroacetic acid (TCA) followed by extraction (see below).
2.4. IP7 measurements
The acid lysed cells were incubated on ice for 30 min after addition of 0.5 M EDTA, pH 8.0, to a final concentration of 2 mM. Inositol phosphates, ATP and ADP were extracted essentially as previously described [12], [15]. The extracts were neutralized by 0.5 M EDTA, pH 8.0. Ion-exchange HPLC containing a SAX column (250 × 4.6 mm Partisphere 5 μm, Hichrom, Berkshire, United Kingdom) with mobile phases, 1 mM EDTA (buffer A) and 1.25 M (NH4)2HPO3 in 1 mM EDTA, pH 4.4 with H3PO4 (buffer B) was used to separate inositol phosphates as previously described [14] with a modified elution profile (0% B - 0 to 5 min; B linearly increased to 50% - 5 to 20 min; 50% B - 20 to 40 min; B linearly increased to 70% - 40 to 55 min; 70% B - 55 to 75 min; B linearly increased to 85% - 75 to 77 min; 85% B - 77 to 90 min; B linearly increased to 100% - 90 to 92 min; 100% B – 92 to 98 min). Separated fractions were mixed with scintillation solution (Perkin Elmer, The Netherlands) and radioactivity was measured using a liquid scintillation counter (Perkin Elmer). IP7 and IP6 data are expressed as fold change normalized to average of controls.
2.5. ATP/ADP measurements
For ATP/ADP measurements, cells were cultured in RPMI 1640 medium for 2 days. Static incubation and extraction were then performed as described above. An aliquot of the acid extracted sample was used for ATP/ADP measurements as previously described [12] or using the ApoSENSOR Kit (Biovision, Milpitas, California) following manufacturer's instructions. Data are expressed as fold change normalized to average of controls.
2.6. Expression and purification of IP6K1
The cDNA of HsIP6K1 (residues 9 to 441) was subcloned into the pDest-566 vector. This vector encodes a 6 × His tag, a maltose binding protein tag and TEV protease cleavage site at the N-terminus. Lemo21 (DE3) Competent E. coli cells (New England Biolabs, Massachusetts, USA) were transformed with the resultant plasmid. An overnight culture of the transformed E. coli cells was inoculated into nutrient-rich 2xYT medium supplemented with 0.6 mM l-rhamnose at pH 7.5 and grown at 37 °C to A595 = 0.7. Isopropyl β-d-thiogalactopyranoside (0.1 mM) was then added and cultures were continued at 15 °C for 2 days. The cells were disrupted using a constant cell disruption system (Constant Systems Ltd., Northants, UK) under 20 KPsi. Recombinant HsIP6K1 was purified with a Ni-NTA agarose column (Qiagen, Germantown, MD, USA) followed by a HiTrap™ Heparin HP column (GE Healthcare, Pittsburgh, USA). The purity was estimated to be > 80% as judged by SDS-PAGE. The purified proteins were concentrated and stored at − 80 °C.
2.7. IP6K activity measurements
Purified IP6K1 protein (150 ng) with inhibitors mentioned in Table 1 or vehicle [DMSO; final concentration = 1% (v/v)] was added to 100 μl assay buffer comprising: 100 mM KCl, 3 mM MgSO4, 1 mM Na2ATP, 1 mM Na2EDTA, 2 mg/ml BSA, 20 mM HEPES pH 7.2 with KOH, approximately 20,000 d.p.m. [3H] IP6 (Perkin Elmer) and 10 μM IP6 (Merck Millipore). Assays were quenched at 15 min with 100 μl ice-cold assay buffer followed immediately by 40 μl ice-cold 2 M HClO4 plus 0.2 mg/ml IP6. Samples were neutralized and analyzed by Partsiphere SAX HPLC as previously described [16]. The IP6K inhibitor TNP [N2-(m-Trifluorobenzyl), N6-(p-nitrobenzyl)purine] (Merck Millipore) was used as a positive control in the kinase assay [17].
Table 1.
Effect of inhibitors on the IP6K1 activity.
| Inhibitor | IP6K1 activity (% of control) |
|---|---|
| DMSO control | 100 ± 7.74 (4) |
| Wortmannin, 10 μM | 90.13 ± 2.88 (4) |
| HNMPA, 75 μM | 89.66 ± 11.97 (3) |
| U73343, 10 μM | 86.31 ± 1.16 (3) |
| TBB, 10 μM | 79.62 ± 4.96 (4) |
| LY294002, 25 μM | 75.08 ± 4.03 (4)* |
| PAO, 1 μM | 8.94 ± 0.45 (3)*** |
| U73122, 10 μM | 0.46 ± 0.46 (3)*** |
| TNP, 20 μM | 7.61 ± 1.23 (3)*** |
Purified IP6K1 was incubated with various inhibitors in IP6K assay buffer, as described in Materials and methods. Treatment with LY 294002 (PI3K inhibitor), PAO (PI4K inhibitor), U73122 (PLC inhibitor) and TNP (IP6K inhibitor, positive control) decreased IP6K activity significantly. Data are expressed as Means ± SEM (number of independent experiments), *p < 0.05, ***p < 0.001, one-way ANOVA followed by Tukey's multiple comparison test.
2.8. Gene expression analysis by real-time RT-PCR
Total RNA was isolated from HIT-T15 cells (~ 1.6 × 105 cells/cm2) using the RNeasy Mini Kit (Qiagen, Sollentuna, Sweden). Briefly, cells were lysed with RLT lysis buffer (Qiagen). Lysates were applied to a spin column and the subsequent RNA isolation and DNase treatment were carried out according to manufacturer's recommendations. Total RNA was reverse transcribed at 37 °C with the Applied Biosystems High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). Gene expression was measured by real-time PCR with PowerUp™ SYBR® Green Master Mix (ThermoFisher Scientific) on an ABI7300 instrument (Applied Biosystems). Reactions were performed with equal amount of cDNA (equivalent to 10 ng of RNA) in a total volume of 40 μL. The forward and reverse primers were purchased from Sigma-Aldrich (Supplemental Table S1) and used at the final concentration of 0.25 μM. The initial step at 50 °C for 2 min was followed by 1 cycle at 95 °C for 10 min and 40 cycles of tandem 95 °C for 15 s and 60 °C for 1 min. The PCR melting curve produced one single peak corresponding to a single amplified product. Gapdh was used as an endogenous reference gene for the normalization of the Ct values.
2.9. Quantification of IP6K1 protein levels using immunoblotting
HIT-T15 cells were treated with inhibitors or vehicle as described under Static incubation. Then, cells were lysed with RIPA buffer (150 mM NaCl, 1% NP-40, 0.1% sodium deoxycholate, 1 mM EDTA, 0.1% SDS, 50 mM Tris, pH 7.6) supplemented with complete mini protease inhibitor cocktail and PhosSTOP phosphatase inhibitors (Roche Diagnostics, Stockholm, Sweden). Cell lysates were sonicated and centrifuged at 11,400 g for 15 min to remove cell debris. Protein content was measured by Pierce™ BCA protein assay kit (Thermofisher Scientific). Equal amounts of protein extracts (10 μg/lane) were separated using pre-cast NuPAGE™ 10% Bis-Tris gel (Thermofisher Scientific) and transferred to nitrocellulose membranes. Membranes were blocked with 5% skim milk in Tris-buffered saline containing 150 mM NaCl, 0.1% Tween-20, 20 mM Tris base, pH 7.4 (TBST) for 1 h and then incubated overnight with the primary antibody (rabbit-anti IP6K1, Sigma-Aldrich HPA040825, dilution 1: 1000) diluted in TBST containing 5% BSA at 4 °C. After repeated washing with TBST, membranes were incubated with the HRP-conjugated donkey anti-rabbit antibody (NA934, GE Healthcare, Uppsala, Sweden, dilution 1: 5000) diluted in TBST containing 5% skim milk for 1 h at room temperature. The blots were then washed with TBST and developed with Amershan ECL Prime Western Blotting Detection Reagent (GE Healthcare). The chemiluminescence signal was detected by ChemiDoc™ Imaging System and analyzed by densitometry using Image Lab™ (Bio-Rad, Solna, Sweden). The membranes were then washed repeatedly with TBST, probed with HRP-conjugated mouse anti-β-actin (Sigma-Aldrich®A3854, dilution 1: 40,000) for 1 h, washed and developed as described above.
2.10. Statistical analysis
Data are expressed as Mean ± SEM. Data analyses were performed with GraphPad Prism 5.0 using the statistical tests mentioned in the figure legends. A p-value < 0.05 was considered significant.
3. Results and discussion
3.1. Inhibitors against pathways downstream of insulin signaling decrease IP7 levels
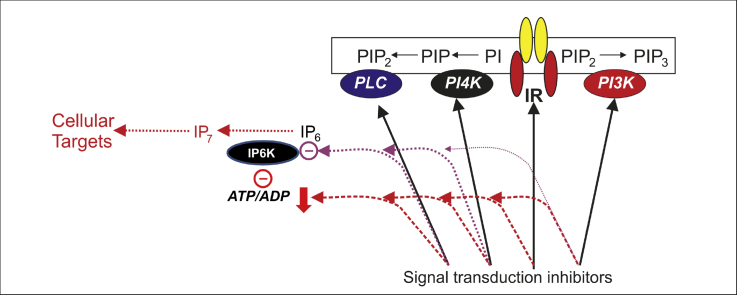
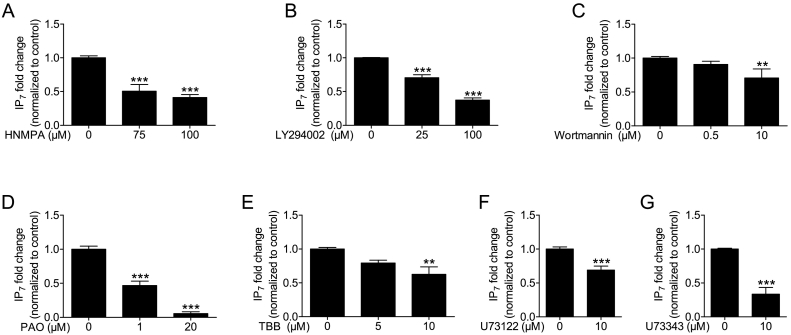
The pancreatic β-cell has a unique feedback mechanism operating even under basal conditions, namely autocrine stimulation of insulin receptors (IRs) by secreted insulin and thereby activation of downstream insulin signaling pathways, particularly through PI3K [18], [19]. It is reasonable that this might control β-cell IP7 levels, since insulin stimulation in other tissues was shown to increase the levels of the inositol pyrophosphate [20]. Thus we interrogated IR signaling as a basis for the high IP7 levels in β-cells. Among cell lines that are a model for insulin secretion, hamster HIT-T15 cells are particularly useful for the current study because they recapitulate the unusually high IP7 level in primary β-cells [10], [21]. Blocking insulin receptor signaling using an inhibitor of IR tyrosine kinase activity, HNMPA-(AM)3, decreased IP7 levels significantly in insulin secreting HIT-T15 cells. We used 75 μM and 100 μM HNMPA-(AM)3 [22], [23], and both these concentrations decreased IP7 almost by 50% (Fig. 1A). These data could be interpreted to suggest that IP7 may be regulated by insulin feedback. To further investigate this, we looked into the activity of PI3Ks, key signaling kinases that are downstream of the IR, and their involvement in IP7 regulation. PI3Ks were blocked by using two different inhibitors, LY294002 and wortmannin. LY294002 at concentrations of 25 and 100 μM [21] decreased IP7 levels (Fig. 1B), whereas a more specific inhibitor, wortmannin [24], at a concentration 0.5 μM [25], did not (Fig. 1C). This discrepancy questioned the involvement of PI3K in IP7 production. A higher dose of wortmannin, 10 μM, was able to decrease IP7 levels by 29% (Fig. 1C). At this high concentration, wortmannin is known to block PI4K [26] and thus PI4K rather than PI3K activity might be implied. Therefore, we used a PI4K inhibitor, PAO to see if PI4K was involved in IP7 regulation. PAO at a concentration of 1 and 20 μM [27], [28] decreased IP7 levels dramatically by 53 and 95%, respectively (Fig. 1D). It is also possible that the effect of LY294002 was due to an off target effect, as LY294002 has been reported to inhibit CK2 [29]. Therefore, we used the CK2 inhibitor TBB and examined its effect on IP7. TBB at a concentration of 5 and 10 μM [30] decreased IP7 levels by 21% and 38%, respectively (Fig. 1E).
Fig. 1.
Inhibitors against inositide and related signaling pathways decrease IP7 levels. HIT-T15 cells, labeled with 10 μCi/ml [3H] myo-inositol, were incubated with respective inhibitor for 30 min. (A) Blocking IR signaling using HNMPA-(AM)3 decreased IP7 levels significantly (n = 6, for control; n = 3, for 75 μM; n = 4, for 100 μM). (B) Blocking PI3K by LY294002 (n = 9, for control; n = 7, for 25 μM; n = 6, for 100 μM) decreased IP7 levels dose dependently, whereas (C) wortmannin (n = 8, for control; n = 4, for 0.5 μM; n = 3, for 10 μM), did not decrease IP7 levels at 0.5 μM, the most commonly used concentration. At high concentration, 10 μM, wortmannin also decreased IP7 levels. (D) PI4K inhibitor, PAO, strongly decreased IP7 levels dose dependently (n = 5, for control; n = 5, for 1 μM; n = 3, for 20 μM). (E) CK2 inhibitor, TBB, also reduced IP7 levels both at 5 and 10 μM (n = 5, for control; n = 3, for 5 μM; n = 4, for 10 μM). (A- E) Data are presented as Means ± SEM, ***p < 0.001, **p < 0.01 compared to controls, one-way ANOVA followed by Tukey's multiple comparison test. (F) PLC inhibitor, U73122, decreased IP7 levels by 31% (n = 7, for control; n = 5, for 10 μM). (G) Whereas, the negative control for PLC inhibitor, U73343, decreased IP7 levels more potently by 66% (n = 6, for control; n = 5, for 10 μM). (F and G) Data are presented as Means ± SEM, ***p < 0.001, Student's t-test.
An interpretation of the above results is that PI3K signaling is not involved in the regulation of IP7 levels in β-cells. It also throws doubts on the role of the IR. This is consistent with the fact that direct stimulation of these cells with insulin at concentrations previously determined to activate insulin receptor signaling [31], did not lead to an increase in IP7 (Supplemental Fig. S1). Therefore, the observed IP7 changes are likely due to off-target effects of the inhibitors. Nonetheless, this data suggest the potential involvement of other signaling pathways in the establishment of IP7 in β-cells.
3.2. Blocking PLC by U73122 indirectly decreases IP7 levels
PLC is a key enzyme which breaks down phosphatidylinositol 4,5-bisphosphate (PIP2) to inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). The availability of IP3 is the critical step in the formation of higher inositol phosphates, including IP7 [32], [33]. Furthermore, since PI4K activity is important for PIP2 formation and inhibition of this kinase resulted in dramatic IP7 loss, the impact of PI4K inhibition may be to remove an important precursor. Thus, we examined the possible involvement of PLC in the generation of IP7. To test this idea we used a PLC inhibitor and its negative control [34]. Fig. 1F and G shows that the negative control of PLC inhibitor, U73343 (Fig. 1G), decreased IP7 levels more potently than the actual inhibitor, U73122, itself (Fig. 1F). Thus the observed decrease in IP7 by these reagents is difficult to interpret. The negative control data suggest the involvement of off-target effects on IP7 levels.
Pharmacological inhibitors are convenient and often employed tools for the elucidation of biochemical pathways, but the specificity of these compounds is a known concern [29], [35]. Inhibitors have also been reported to non-specifically affect cellular ATP levels [36], [37]. This is important because the IP6Ks that are responsible for IP7 generation have a high Km for ATP [38], [39] and compromising ATP/ADP levels has been shown to affect IP7 levels [12], [13]. Given the above scenario, we investigated the hypothesis that these inhibitors affect IP7 levels in β-cells indirectly. We therefore examined the ATP/ADP ratios after treating HIT-T15 cells with the above broad spectrum of inhibitors.
3.3. Inhibitors against the insulin signaling pathway reduce cellular ATP/ADP levels
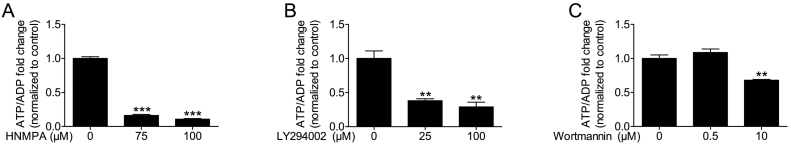
HIT-T15 cells were incubated with HNMPA-(AM)3, the IR inhibitor, for 30 min. HNMPA-(AM)3 treatment at concentrations used to measure IP7 levels, 75 μM and 100 μM, decreased ATP/ADP levels by 84 and 89%, respectively (Fig. 2A). Thus the IP7 decrease observed upon blocking the insulin receptor is likely to be mediated by a reduction in ATP/ADP levels. We then tested if this was also the case for the other inhibitors that block the PI3K pathway downstream of the IR.
Fig. 2.
Effect of insulin signaling pathway inhibitors on cellular ATP/ADP levels. HIT-T15 cells were incubated for 30 min in the presence of each inhibitor and ATP/ADP levels were measured. (A) IR inhibitor, HNMPA-(AM)3, reduced ATP/ADP levels at 75 and 100 μM (n = 4). (B) PI3K inhibitor, LY294002 (n = 3), also decreased ATP/ADP levels at both concentrations, 25 and 100 μM, whereas, (C) wortmannin (n = 4) decreased ATP/ADP only at 10 μM and not at 0.5 μM, following its effect on IP7 (Fig. 1C). Data are presented as Means ± SEM, ***p < 0.001, **p < 0.01 compared to controls, one-way ANOVA followed by Tukey's multiple comparison test.
Measuring ATP/ADP upon inhibiting PI3K by LY294002 and wortmannin showed that the decrease in IP7 levels using these inhibitors also reflected reduced ATP/ADP levels. In addition to decreasing IP7 levels (Fig. 1B), LY294002 decreased ATP/ADP levels at 25 μM and 100 μM (Fig. 2B). In contrast, only 10 μM wortmannin decreased ATP/ADP levels (Fig. 2C) in keeping with its impact on IP7 levels (Fig. 1C), In summary, these data suggest that the observed decrease in IP7 levels were mediated by the inhibitor compromising ATP/ADP levels.
Wortmannin has been held as a more specific PI3K inhibitor than LY294002 [24] and our results underscore that finding. Only at wortmannin concentrations higher than usually employed, did the inhibitor make any impact on either IP7 or ATP/ADP. Nevertheless, the LY294002 concentration range we have used are typical for inhibiting PI3K activity in other publications [40], [41], [42]. Thus, our data raise the possibility that the drop in IP7 might have contributed to the biological effects reported upon blocking PI3K by LY294002.
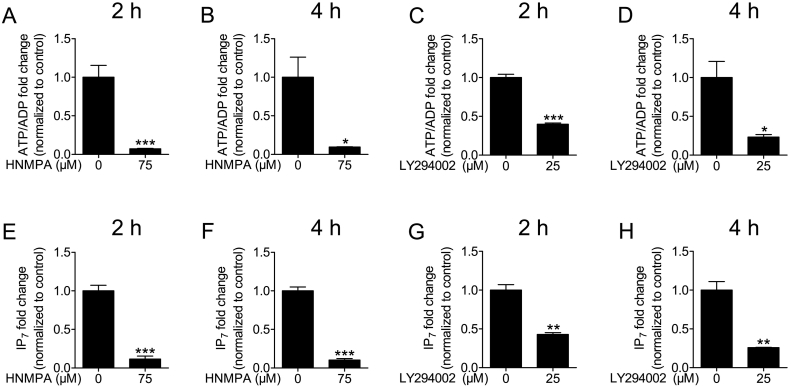
We have also evaluated the consequences of long-term treatment with inhibitors comparing HNMPA-(AM)3 and LY294002. The non-specific effects of HNMPA-(AM)3 and LY294002 upon cellular bioenergetics and IP7 levels, observed at 30 min, were confirmed after 2 h and 4 h incubations (Fig. 3). There was a considerable reduction in both ATP/ADP and IP7 levels after 4 h treatment compared to 30 min.
Fig. 3.
Effect of long term treatment with HNMPA-(AM)3 and LY294002 on ATP/ADP and IP7 levels. HIT-T15 cells, labeled with 10 μCi/ml [3H] myo-inositol, were incubated with respective inhibitor for 2 h (A, C, E, G) or 4 h (B, D, F, H). Treatment with HNMPA-(AM)3, IR signaling blocker, or LY294002, PI3K inhibitor, decreased ATP/ADP ratios (A–D, n = 3) and IP7 levels (E–H, n = 4) significantly. Data are presented as Means ± SEM, *p < 0.05 **p < 0.01, ***p < 0.001, Student's t-test.
3.4. Effect of PI4K and CK2 inhibitors on cellular ATP/ADP levels
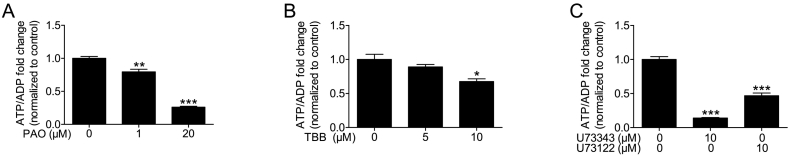
We then tested the effect of PI4K and CK2 inhibitors on ATP/ADP levels. PAO decreased ATP/ADP levels dose dependently (Fig. 4A). At 1 μM and 20 μM PAO, when the inhibitor reduced IP7 by 53% and 95% (Fig. 1D), ATP/ADP levels dropped by 21% and 74%, respectively (Fig. 4A). At 10 μM, TBB decreased ATP/ADP levels by 32% (Fig. 4B). At the same concentration IP7 levels were also reduced by 38% (Fig. 1E). This again suggests that the observed IP7 drop using PAO and TBB could depend on the decrease in cellular ATP/ADP levels.
Fig. 4.
Effect of PAO, TBB, U73343 and U73122 on cellular ATP/ADP levels. HIT-T15 cells were incubated for 30 min in the presence of each inhibitor before ATP/ADP measurements. (A) PI4K inhibitor, PAO, decreased ATP/ADP levels dose dependently. (B) CK2 inhibitor, TBB, reduced ATP/ADP levels significantly at 10 μM. (C) Both PLC inhibitor negative control, U73343, and PLC inhibitor, U73122, reduced ATP/ADP levels. PLC inhibitor negative control, U73343, strongly decreased ATP/ADP levels, an effect that was more pronounced than that obtained by the actual PLC inhibitor, U73122. Data are presented as Means ± SEM, n = 3, ***p < 0.001, **p < 0.01 compared to controls, one-way ANOVA followed by Tukey's multiple comparison test.
3.5. Effect of the PLC inhibitor, U73122 and its negative control, U73343 on cellular ATP levels
The negative control of PLC inhibitor (U73343) decreased ATP/ADP levels more potently (by 86%) than U73122 (by 53%), the authentic PLC inhibitor (Fig. 4C). Also in this case, the ATP/ADP reduction detected using these inhibitors are well matched to the lower IP7 levels observed in Fig. 1F and G.
3.6. Direct effects of inhibitors on the IP6K
The above results show that all the inhibitors at concentrations affecting IP7 levels impacted ATP/ADP ratios. Although this association indicates the involvement of ATP/ADP in IP7 synthesis, the contribution of other factors such as IP6K activity and IP6K protein levels cannot be excluded. In order to investigate this further, we screened all the inhibitors, at the minimum concentration in which they impacted IP7 levels, against purified IP6Ks. In pancreatic β-cells from mouse, both cell lines and primary islets, there was a dominant expression of IP6K1 and IP6K2 [10]. The same is the case for HIT-T15 cells derived from the Syrian hamster (Supplemental Fig. S2). Thus use of IP6K1 is a reasonable surrogate for assessing impact on HIT-T15 cell IP6K activity. PI4K inhibitor (PAO) and PLC inhibitor (U73122) potently inhibited IP6K1 activity as efficiently as the well characterized IP6K inhibitor TNP (Table 1). There was also a minor (25%) decrease in IP6K1 activity upon treatment with LY294002 (Table 1). Thus the disruption of IP6K activity upon using these three inhibitors can contribute to the decrease in IP7 levels observed in cells. However, the kinase inhibition observed in the test-tube setting may not be directly translated to the cellular situation due to compound-specific interactions with other cellular components. The data obtained from the PLC inhibitor and its negative control can be used as an example to understand this. Since the negative control U73343 and the actual PLC inhibitor U73122 are structurally similar their uptake and availability in an intact cell should be alike. U73343, that did not decrease IP6K1 activity, had a more profound effect in decreasing IP7 and ATP/ADP levels than U73122, which had 95% inhibitory effect in the enzymatic assay. This reemphasizes the fact that the degree of IP6K inhibition observed in the cellular context is lower, compared to the test-tube setting.
Furthermore, other inhibitors such as HNMPA, wortmannin, TBB and U73343 do not inhibit IP6K1 directly (Table 1), so their action against IP7 can be ascribed mostly to the effect on cellular ATP/ADP levels. We have also investigated a possible effect of all the inhibitors on the IP6K1 protein levels in HIT-T15 cells. Using immunoblotting, we were able to verify that the reduction in IP7 is not due to a decrease in IP6K1 protein (Supplemental Fig. S3). Our attempts to measure IP6K2 protein levels ended in vain due to the lack of antibody specific for hamster IP6K2.
3.7. Correlation between ATP/ADP and IP7 levels
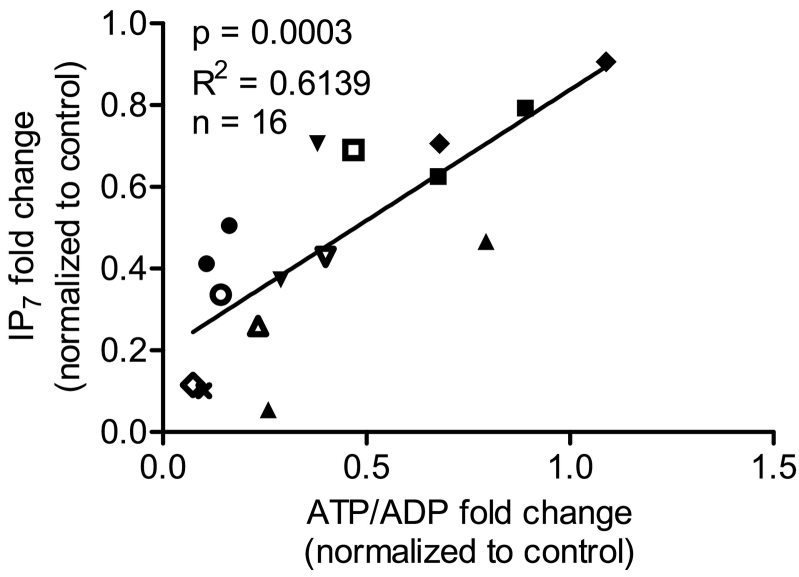
Since the drop in IP7 was mostly accompanied by decreased ATP/ADP levels, we pooled all the data obtained with different inhibitors to examine whether there was an overall correlation between ATP/ADP and IP7. Fig. 5 shows that there was a positive correlation (R = 0.7835, R2 = 0.6139, p = 0.0003). Yet, the correlation improved (R = 0.9129, R2 = 0.8334, p = 0.0006) when we excluded inhibitors that directly inhibited IP6K activity. Therefore, these data suggest that in the case of PAO, U73122 and LY294002 both direct kinase inhibition and reduced ATP/ADP contribute to the overall IP7 depletion in intact cells.
Fig. 5.
Correlation between ATP/ADP and IP7. Correlation analysis was performed using both IP7 and ATP/ADP data obtained from all the inhibitors. IP7 levels show a significant positive correlation with cellular ATP/ADP levels, R = 0.7835, R2 = 0.6139, *p = 0.0003, n = 16 using Pearson's correlation. ● HNMPA-(AM)3 30 min, ♢ HNMPA-(AM)3 2 h, × HNMPA-(AM)3 4 h, ■ TBB 30 min, ♦ wortmannin 30 min, ▲ PAO 30 min, □ U73122 30 min, ○ U73343 30 min, ▼ LY294002 30 min, ▽ LY294002 2 h and △ LY294002 4 h. When we excluded the inhibitors that affect the kinase activity (PAO, U73122 and LY294002), the correlation improved, R = 0.9129, R2 = 0.8334, *p = 0.0006, n = 9 using Pearson's correlation.
It should be noted that the off-target effect was quite specific to IP7, for example, with 30 min or 2 h or 4 h treatment with the inhibitor there was no significant decrease in other higher inositol phosphates like its immediate precursor, IP6 (Supplemental Table S2 and S3). However, at longer time incubations (4 h), particularly with HNMPA-(AM)3, which dramatically reduced the ATP/ADP ratio, there was a tendency for a reduction in IP6 too (Supplemental Table S3). These data speak against the use of at least HNMPA-(AM)3 in longer term experiments in pancreatic β-cells. The difference in sensitivity of IP6 vs. IP7 production to ATP/ADP reduction reflects the different Km of the kinases involved. IP5Ks have a Km for ATP in the micromolar range [43]. In contrast, as we mentioned previously, IP6Ks have an unusually high Km for ATP, i.e. at least an order of magnitude higher (around 1 mM) [39]. This makes them uniquely sensitive to cellular ATP changes in pancreatic β-cells, whose free concentration of ATP is around 1 mM [44].
3.8. Conclusions
The above results show that most of the protein kinase- and lipase inhibitors used decreased intracellular IP7 levels largely by reducing ATP/ADP ratios and to an extent by a direct impact on IP6K activity. These data support the view that IP7 levels are strongly dependent on the bioenergetics status of the β-cell. Hence, a compromised energy metabolism, irrespective of the cause, will affect IP7 and consequently the multitude of processes that this molecule is involved in [1], [3], [4], [5], [45]. This is something we are currently investigating further. In addition, the strong direct inhibition of IP6K activity by PAO and U73122 suggests that these reagents must be considered as effective IP6K inhibitors, thus potentially changing the interpretation of data generated by their use. The particular action of PAO may relate to its chemical property of attacking vicinal cysteines in proteins [46]. IP6K, for example, has been reported to possess cysteines important for catalytic activity [47]. Although these results are of a particular concern for those studying signal transduction in pancreatic β-cells, they also highlight the fact that the use of the described inhibitors could have erroneously suggested the involvement of key signal transduction pathways in various cellular processes. Another implication of our data is that inositol pyrophosphates may be involved in several cellular processes currently ascribed to unrelated signaling pathways.
Given the broad spectrum of inhibitors that affect IP7 levels, it is possible that many other inhibitors, as yet untested, could be having a similar impact on IP7 levels. Whilst many laboratories do not have the resources or equipment to investigate whether their inhibitor of interest is indirectly depleting IP7, a reasonable strategy that we recommend would be to test the impact of their inhibitor of choice on the ATP/ADP ratio using a commercially available kit. In this way at least some of the potential problems with interpretation could be quickly exposed.
Conflict of interest
P-OB is CEO of the biotech company Biocrine AB, and CJB is a consultant with this company.
Acknowledgments
Acknowledgements
We acknowledge the help of Andreas Nagel in the original establishment of the ATP assay. We thank Dr. Michael Tekle and Karin Åvall for their help in setting up the PCR analysis. This study was supported by the Swedish Research Council, the Novo Nordisk Foundation, Karolinska Institutet, the Swedish Diabetes Association, The Family Knut and Alice Wallenberg Foundation, Diabetes Research and Wellness Foundation (677_2015 PG), Berth von Kantzow's Foundation, The Skandia Insurance Company Ltd., STINT and CAPES program and grants 2014/05214-1, 2006/54691-0, 2010/02272-0 from FAPESP, grant 301617/2016-3 from CNPq (PQ-1D), Strategic Research Programme in Diabetes at Karolinska Institutet, ERC-2013-AdG 338936-BetaImage, the Stichting af Jochnick Foundation and the Family Erling-Persson Foundation. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellsig.2017.10.008.
Contributor Information
Per-Olof Berggren, Email: per-olof.berggren@ki.se.
Christopher J. Barker, Email: chris.barker@ki.se.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Thota S.G., Bhandari R. The emerging roles of inositol pyrophosphates in eukaryotic cell physiology. J. Biosci. 2015;40(3):593–605. doi: 10.1007/s12038-015-9549-x. [DOI] [PubMed] [Google Scholar]
- 2.Bennett M., Onnebo S.M., Azevedo C., Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell. Mol. Life Sci. 2006;63(5):552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson M.S., Livermore T.M., Saiardi A. Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 2013;452(3):369–379. doi: 10.1042/BJ20130118. [DOI] [PubMed] [Google Scholar]
- 4.Barker C.J., Berggren P.-O. New horizons in cellular regulation by inositol polyphosphates: insights from the pancreatic β-cell. Pharmacol. Rev. 2013;65(2):641–669. doi: 10.1124/pr.112.006775. [DOI] [PubMed] [Google Scholar]
- 5.Wundenberg T., Mayr G.W. Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol. Chem. 2012;393(9):979–998. doi: 10.1515/hsz-2012-0133. [DOI] [PubMed] [Google Scholar]
- 6.Menniti F., Miller R., Putney J., Shears S. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 1993;268(6):3850–3856. [PubMed] [Google Scholar]
- 7.Shears S.B., Gokhale N.A., Wang H., Zaremba A. Diphosphoinositol polyphosphates: what are the mechanisms? Adv. Enzym. Regul. 2011;51(1):13–25. doi: 10.1016/j.advenzreg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas M.P., Potter B.V. The enzymes of human diphosphoinositol polyphosphate metabolism. FEBS J. 2014;281(1):14–33. doi: 10.1111/febs.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty A., Kim S., Snyder S.H. Inositol pyrophosphates as mammalian cell signals. Sci. Signal. 2011;4(188) doi: 10.1126/scisignal.2001958. (re1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illies C., Gromada J., Fiume R., Leibiger B., Yu J., Juhl K., Yang S.-N., Barma D.K., Falck J.R., Saiardi A. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic β cells. Science. 2007;318(5854):1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- 11.Bhandari R., Juluri K.R., Resnick A.C., Snyder S.H. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc. Natl. Acad. Sci. 2008;105(7):2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagel A., Barker C.J., Berggren P.O., Illies C. Diphosphosinositol polyphosphates and energy metabolism: assay for ATP/ADP ratio. Methods Mol. Biol. 2010;645:123–131. doi: 10.1007/978-1-60327-175-2_8. [DOI] [PubMed] [Google Scholar]
- 13.Wundenberg T., Grabinski N., Lin H., Mayr G.W. Discovery of InsP6-kinases as InsP6-dephosphorylating enzymes provides a new mechanism of cytosolic InsP6 degradation driven by the cellular ATP/ADP ratio. Biochem. J. 2014;462(1):173–184. doi: 10.1042/BJ20130992. [DOI] [PubMed] [Google Scholar]
- 14.Larsson O., Barker C.J., Sjöholm Å., Carlqvist H., Michell R.H., Bertorello A., Nilsson T., Honkanen R.E., Mayr G.W., Zwiller J. Inhibition of phosphatases and increased Ca2 + channel activity by inositol hexakisphosphate. Science. 1997;278(5337):471–474. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- 15.Barker C.J., Illies C., Berggren P.O. HPLC separation of inositol polyphosphates. Methods Mol. Biol. 2010;645:21–46. doi: 10.1007/978-1-60327-175-2_2. [DOI] [PubMed] [Google Scholar]
- 16.Safrany S.T., Ingram S.W., Cartwright J.L., Falck J., McLennan A.G., Barnes L.D., Shears S.B. The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 1999;274(31):21735–21740. doi: 10.1074/jbc.274.31.21735. [DOI] [PubMed] [Google Scholar]
- 17.Padmanabhan U., Dollins D.E., Fridy P.C., York J.D., Downes C.P. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 2009;284(16):10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leibiger I.B., Leibiger B., Berggren P.-O. Insulin signaling in the pancreatic β-cell. Annu. Rev. Nutr. 2008;28:233–251. doi: 10.1146/annurev.nutr.28.061807.155530. [DOI] [PubMed] [Google Scholar]
- 19.Yu J., Berggren P.-O., Barker C.J. An autocrine insulin feedback loop maintains pancreatic β-cell 3-phosphorylated inositol lipids. Mol. Endocrinol. 2007;21(11):2775–2784. doi: 10.1210/me.2006-0473. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty A., Koldobskiy M.A., Bello N.T., Maxwell M., Potter J.J., Juluri K.R., Maag D., Kim S., Huang A.S., Dailey M.J. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143(6):897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leibiger B., Leibiger I.B., Moede T., Kemper S., Kulkarni R.N., Kahn C.R., de Vargas L.M., Berggren P.-O. Selective insulin signaling through A and B insulin receptors regulates transcription of insulin and Glucokinase genes in pancreatic β cells. Mol. Cell. 2001;7(3):559–570. doi: 10.1016/s1097-2765(01)00203-9. [DOI] [PubMed] [Google Scholar]
- 22.Beith J.L., Alejandro E.U., Johnson J.D. Insulin stimulates primary β-cell proliferation via Raf-1 kinase. Endocrinology. 2008;149(5):2251–2260. doi: 10.1210/en.2007-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jozic I., Blanco G., Barbieri M.A. Inhibition of Rab5 activation during insulin receptor-mediated endocytosis. Curr. Cell. Biochem. 2011;1(1):20–32. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao W., Qi J., Wang L., Zhang M., Wang P., Gao C. LY294002 inhibits TLR3/4-mediated IFN-β production via inhibition of IRF3 activation with a PI3K-independent mechanism. FEBS Lett. 2012;586(6):705–710. doi: 10.1016/j.febslet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Kaminski A., Welters Hannah J., Kaminski Edward R., Morgan Noel G. Human and rodent pancreatic β-cells express IL-4 receptors and IL-4 protects against β-cell apoptosis by activation of the PI3K and JAK/STAT pathways. Biosci. Rep. 2010;30(3):169–175. doi: 10.1042/BSR20090021. [DOI] [PubMed] [Google Scholar]
- 26.Xu J.-X., Si M., Zhang H.-R., Chen X.-J., Zhang X.-D., Wang C., Du X.-N., Zhang H.-L. Phosphoinositide kinases play key roles in norepinephrine-and angiotensin II-induced increase in phosphatidylinositol 4, 5-bisphosphate and modulation of cardiac function. J. Biol. Chem. 2014;289(10):6941–6948. doi: 10.1074/jbc.M113.527952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue J., Liu J., Shen X. Inhibition of phosphatidylinositol 4-kinase results in a significant reduced respiratory burst in formyl-methionyl-leucyl-phenylalanine-stimulated human neutrophils. J. Biol. Chem. 2001;276(52):49093–49099. doi: 10.1074/jbc.M101328200. [DOI] [PubMed] [Google Scholar]
- 28.Sorensen S.D., Linseman D.A., McEwen E.L., Heacock A.M., Fisher S.K. A role for a wortmannin-sensitive phosphatidylinositol-4-kinase in the endocytosis of muscarinic cholinergic receptors. Mol. Pharmacol. 1998;53(5):827–836. [PubMed] [Google Scholar]
- 29.Davies S.P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351(1):95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarno S., Reddy H., Meggio F., Ruzzene M., Davies S.P., Donella-Deana A., Shugar D., Pinna L.A. Selectivity of 4, 5, 6, 7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (‘casein kinase-2’) FEBS Lett. 2001;496(1):44–48. doi: 10.1016/s0014-5793(01)02404-8. [DOI] [PubMed] [Google Scholar]
- 31.Leibiger B., Moede T., Uhles S., Barker C.J., Creveaux M., Domin J., Berggren P.-O., Leibiger I.B. Insulin-feedback via PI3K-C2α activated PKBα/Akt1 is required for glucose-stimulated insulin secretion. FASEB J. 2010;24(6):1824–1837. doi: 10.1096/fj.09-148072. [DOI] [PubMed] [Google Scholar]
- 32.York J.D., Odom A.R., Murphy R., Ives E.B., Wente S.R. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285(5424):96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 33.Tsui M.M., York J.D. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzym. Regul. 2010;50(1):324–337. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banfic H., Bedalov A., York J.D., Visnjic D. Inositol pyrophosphates modulate S phase progression after pheromone-induced arrest in Saccharomyces cerevisiae. J. Biol. Chem. 2013;288(3):1717–1725. doi: 10.1074/jbc.M112.412288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garuti L., Roberti M., Bottegoni G. Non-ATP competitive protein kinase inhibitors. Curr. Med. Chem. 2010;17(25):2804–2821. doi: 10.2174/092986710791859333. [DOI] [PubMed] [Google Scholar]
- 36.Choi K., Mollapour E., Choi J.H., Shears S.B. Cellular energetic status supervises the synthesis of bis-diphosphoinositol tetrakisphosphate independently of AMP-activated protein kinase. Mol. Pharmacol. 2008;74(2):527–536. doi: 10.1124/mol.107.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerriero C.J., Weisz O.A. N-WASP inhibitor wiskostatin nonselectively perturbs membrane transport by decreasing cellular ATP levels. Physiology. 2007;292(4):C1562–C1566. doi: 10.1152/ajpcell.00426.2006. [DOI] [PubMed] [Google Scholar]
- 38.Azevedo C., Szijgyarto Z., Saiardi A. The signaling role of inositol hexakisphosphate kinases (IP6Ks) Adv. Enzym. Regul. 2011;51(1):74–82. doi: 10.1016/j.advenzreg.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Shears S.B. Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 2009;76(2):236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimoto M., Suda Y., Vernhettes S., Nakano A., Ueda T. Phosphatidylinositol 3-kinase and 4-kinase have distinct roles in intracellular trafficking of cellulose synthase complexes in Arabidopsis thaliana. Plant Cell Physiol. 2015;56(2):287–298. doi: 10.1093/pcp/pcu195. [DOI] [PubMed] [Google Scholar]
- 41.Blommaart E.F., Krause U., Schellens J.P., Vreeling-Sindelárová H., Meijer A.J. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243(1–2):240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 42.Chan C., Paulmurugan R., Reeves R., Solow-Cordero D., Gambhir S. Molecular imaging of phosphorylation events for drug development. Mol. Imaging Biol. 2009;11(3):144–158. doi: 10.1007/s11307-008-0187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ives E.B., Nichols J., Wente S.R., York J.D. Biochemical and functional characterization of inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinases. J. Biol. Chem. 2000;275(47):36575–36583. doi: 10.1074/jbc.M007586200. [DOI] [PubMed] [Google Scholar]
- 44.Kennedy H.J., Pouli A.E., Ainscow E.K., Jouaville L.S., Rizzuto R., Rutter G.A. Glucose generates sub-plasma membrane ATP microdomains in single islet β-cells. Potential role for strategically located mitochondria. J. Biol. Chem. 1999;274(19):13281–13291. doi: 10.1074/jbc.274.19.13281. [DOI] [PubMed] [Google Scholar]
- 45.Wild R., Gerasimaite R., Jung J.-Y., Truffault V., Pavlovic I., Schmidt A., Saiardi A., Jessen H.J., Poirier Y., Hothorn M. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016;352(6288):986–990. doi: 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- 46.He X., Ma Q. Critical cysteine residues of Kelch-like ECH-associated protein 1 in arsenic sensing and suppression of nuclear factor erythroid 2-related factor 2. J. Pharmacol. Exp. Ther. 2010;332(1):66–75. doi: 10.1124/jpet.109.160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Onnebo S.M.N., Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 2009;423(1):109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.