Abstract
Prokaryote genomes are the result of a dynamic flux of genes, with increases achieved via horizontal gene transfer and reductions occurring through gene loss. The ecological and selective forces that drive this genomic flexibility vary across species. Bacillus subtilis is a naturally competent bacterium that occupies various environments, including plant-associated, soil, and marine niches, and the gut of both invertebrates and vertebrates. Here, we quantify the genomic diversity of B. subtilis and infer the genome dynamics that explain the high genetic and phenotypic diversity observed. Phylogenomic and comparative genomic analyses of 42 B. subtilis genomes uncover a remarkable genome diversity that translates into a core genome of 1,659 genes and an asymptotic pangenome growth rate of 57 new genes per new genome added. This diversity is due to a large proportion of low-frequency genes that are acquired from closely related species. We find no gene-loss bias among wild isolates, which explains why the cloud genome, 43% of the species pangenome, represents only a small proportion of each genome. We show that B. subtilis can acquire xenologous copies of core genes that propagate laterally among strains within a niche. While not excluding the contributions of other mechanisms, our results strongly suggest a process of gene acquisition that is largely driven by competence, where the long-term maintenance of acquired genes depends on local and global fitness effects. This competence-driven genomic diversity provides B. subtilis with its generalist character, enabling it to occupy a wide range of ecological niches and cycle through them.
Keywords: Bacillus subtilis, pangenome, comparative genomics, bacterial genome evolution, lateral gene transfer, genetic competence
Introduction
Prokaryotes genomes are highly dynamic, with rates of gene gain and gene loss comparable to mutation rates (Kolstø 1997; Doolittle 1999; Koonin and Wolf 2008). The major promoter of gene gain is lateral gene transfer (LGT), which dominates the bacterial world at varying levels depending on the species biology and the ecological interactions established in local environments (Kolstø 1997; Doolittle 1999; Ochman et al. 2000; Koonin and Wolf 2008; Puigbò et al. 2014). Contradicting initial concerns (Bapteste et al. 2005; Doolittle and Bapteste 2007), the dominance of LGT has not precluded the inference of vertical phylogenies that describe the diversification process and the relationships among species (Daubin et al. 2003; Lerat et al. 2005; Puigbò et al. 2009; Hug et al. 2016). It is on the top of these phylogenies that we reconstruct the evolutionary processes that shape the microbial world and where estimates of the rates of gene gain, gene loss, and gene family expansion and regression are made. These efforts have produced the body of knowledge currently available on the diversity and dynamics of bacterial genomes in their natural habitats. Studies carried out with widely distinct bacterial groups, encompassing diverse phylogenetic depths and data set completeness, led to the view that persistent gene loss intersected by episodic events of massive gene gains through LGT dominate prokaryote genome dynamics (reviewed by Wolf and Koonin 2013). Overall, these processes are expected to lead to genome streamlining, that is, to the reduction of genome size due to relaxed selection and gene loss. Genome streamlining is well documented among obligate intracellular parasites, which typically have reduced genomes (McCutcheon and Moran 2011; Merhej and Raoult 2011), and among free-living bacteria (Luo et al. 2013; Swan et al. 2013). It is also believed that if genomes are unable to laterally acquire genes, they will face extinction, similar to asexual species that are thought to be doomed to extinction due to a lack of recombination (Moran 1996; Baltrus et al. 2008; Naito and Pawlowska 2016).
The theoretical predictions that underlie many expectations of how prokaryote genomes evolve are based on population genetic processes. Yet, much of the data that have been analyzed covers billions of years of evolution, frequently encompassing evolutionary trends across major taxonomic groups, such as different phyla (Snel et al. 2002; Kunin 2003; Charlebois and Doolittle 2004; Kettler et al. 2007; Richards et al. 2014). Eased by the increased facility of sequencing new bacterial genomes, recent efforts have been made to deeply sample many genomes within individual species with the goal of uncovering intraspecific evolutionary processes. These developments have been fundamental to our understanding of intraspecific pangenome dynamics as it has become clear that bacterial genomes are dynamic containers of essential and accessory elements, where multiple isolates are required to understand the global complexity of a single species (Tettelin et al. 2005; Touchon et al. 2009; Lefébure et al. 2010; Ahmed et al. 2012; Kaas et al. 2012; McNally et al. 2013; Bolotin & Hershberg 2015). A recent mathematical model of microbial evolution that uses intraspecific data on gene acquisition and protein-level selection proposes that the number of genes in a genome reflects an equilibrium between the benefit accrued by acquiring new genes and the cost of maintaining a larger genome (Sela et al. 2016). Puigbò et al. (2014) reconstructed the genome dynamics across many groups of closely related organisms and corroborated the rapid and variable flux of genes due to extensive gene loss and LGT. These authors, however, still group under the same name genomes belonging to different species that albeit close relatives, are not sister taxa, as for instance B. subtilis and B. amyloliquefaciens (Priest and Goodfellow 1987; Rooney et al. 2009; Connor et al. 2010). Hence, a detailed analysis of such an important organism as B. subtilis is still lacking.
B. subtilis is an endospore-forming gram-positive bacterium that belongs to the deeply rooted phylum Firmicutes. It is a model organism for many molecular processes as well as an industrial workhorse (Harwood 1992; Schallmey et al. 2004). It has received considerable interest as a promoter of plant growth and as a plant–disease control organism (Schisler et al. 2004; Deng et al. 2011; Falardeau et al. 2013). Its status as a GRAS (generally regarded as safe) organism and its ability to form endospores (hereafter, spores) have prompted several applications in biomedicine and biotechnology; these include the use of spores in probiotic formulations and as efficient platforms for surface display and vaccine delivery (Hong et al. 2005; Tavares Batista et al. 2014; Wu et al. 2015). Commonly described as a ‘soil bacterium’, B. subtilis can be sampled from a diverse set of environments that includes the invertebrate and vertebrate gut as well as several plant-associated, soil, and marine niches (Tam et al. 2006; Fan et al. 2011; Um et al. 2013). This diversity of niches and the associated diversity of social interactions raise the question of how these are achieved within a single species.
In this study, we analyze the genomic dynamics within B. subtilis and estimate the size of its pangenome by following a strictly intraspecific approach. We aim at identifying the processes that can explain its pangenome size. The first description of this species’ genomic diversity, which was obtained by performing a microarray-based comparative genomic hybridization analysis with the genome of strain 168 as reference, suggested substantial gene content diversity among B. subtilis isolates (Earl et al. 2007). Earl et al. (2007) queried for the presence of each coding sequence in the genome of strain 168 in a collection of closely related strains and identified presence/absence polymorphisms among genes involved in antibiotic production, cell wall synthesis, sporulation, and germination. An unexpectedly high-genomic diversity in gene content was found, but this diversity was likely underestimated as genes present in other strains but absent from the reference genome would have been missed. This finding led to the view that B. subtilis is a highly versatile organism, which is consistent with its presence in diverse natural settings (Earl et al. 2007). The processes leading to this diversity have, however, not been addressed.
A key feature of B. subtilis is its ability to initiate a developmental program that leads to the production of competent cells that are able to efficiently internalize and recombine exogenous DNA with no apparent sequence specificity (Haijema et al. 2001; Maamar and Dubnau 2005; Smits et al. 2005). The lack of self-specificity provides opportunities for genomic diversification through the acquisition of novel genes. However, the consequences of natural competence on the genomic composition and diversity of this species remain unclear. Competence in B. subtilis is induced transiently as cells enter the stationary phase of growth. This is a stochastic process driven by noise in the expression of an auto-regulatory transcription factor, ComK, and results in competence development in only a small fraction, typically ∼1%, of the cells in populations of natural isolates (Maamar and Dubnau 2005; Smits et al. 2005; Claverys et al. 2006; Yüksel et al. 2016). Competence may provide templates for DNA repair, defense against genomic parasites, a source of nucleotides, or a source of genetic variation (Finkel and Kolter 2001; Claverys et al. 2006; Engelmoer and Rozen 2011; Johnston et al. 2014; Ambur et al. 2016; Croucher et al. 2016). Competence can also endow B. subtilis with tolerance to certain antibiotics, as cells can enter a non-dividing state that is similar to persistence (Hahn et al. 2015; Yüksel et al. 2016). The bi-stable switch governing competence is interpreted as a bet-hedging strategy to improve fitness under adverse or variable environments (Johnsen et al. 2009). Competence raises important questions concerning its contribution to the diversity and evolution of the species’ genome and the influence of local ecological conditions on genome diversity and cohesion. It is expected that the capacity to enter a natural competence state can be beneficial as it allows the input of new and locally adapted genomic diversity into a species’ gene repertoire (Wylie et al. 2010). This capacity is of particular relevance to species such as B. subtilis that are found in a wide range of different niches and have the ability to cycle among them.
In this study, we found that B. subtilis has a large and open pangenome that results from the continuous acquisition of new genes through LGT balanced by an equivalent proportion of gene loss. We found a preponderant role for gene acquisition through competence, although other modes of acquisition, such as transduction, also carry new genes into the species pangenome. We posit that by using natural competence for genetic transformation in a wide diversity of niches, this species can potentially generate countless individual niche adaptation paths.
Materials and Methods
Bacterial Strains
Forty-three genome sequences of B. subtilis strains were downloaded (November 2014) from the NCBI genome database (supplementary table S1, Supplementary Material online). Sequences identified as plasmids in original GenBank files, laboratory strains with major chromosomal modifications, and one strain from an experimental evolution experiment were not included in our data set. We added ten genomes from nine additional species of B. subtilis close relatives: B. tequilensis, B. vallismortis, B. mojavensis, B. atrophaeus, B. amyloliquefaciens, B. sonorensis, B. licheniformis, B. pumilus, and B. cereus, the latter a well-established outgroup taxa of the B. subtilis group (Rooney et al. 2009; Connor et al. 2010; Kubo et al. 2011). In addition, we included two new strains of B. subtilis (BSP2 and BSP4) that were collected from the gut of domestic farm animals (Barbosa et al. 2005; Serra et al. 2014). These new genomes enrich the available data set of public B. subtilis genomes regarding niche-specific genome composition. Five B. subtilis genomes revealed to be misidentified (see phylogenetic history and species limits in B. subtilis in the Results section) and were therefore excluded from further analyses.
Sequencing of New Genomes
BSP2 and BSP4 were sequenced at GATC Biotech (Konstanz, Germany) using Illumina MiSeq 300 bp paired-end reads. For each genome, 6,00,000 read-pairs were randomly selected and assembled using a development version of MIRA 4.9.4 (Chevreux et al. 1999). The resulting contigs were then filtered by MIRA for length (≥ 500 bp) and coverage (≥ 50% of average coverage of the whole project) and defined as “large contigs,” which represented the final genome assembly. Annotation was performed with Prokka 1.10 obtained from Victorian Bioinformatics Consortium (Seemann 2014) (table 1). The Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under accession numbers LZOV00000000 (BSP2) and MAFZ00000000 (BSP4).
Table 1.
Genome Statistics on Assemblies and Annotations of Newly Sequenced Genomes
| Organism | Biosample | Bioproject | WGS | Contigs | Coverage | N50 | L50 | Level | Size (bp) | Proteins | GC% | tRNA | rRNA | ncRNAa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. subtilis str. BSP2 | SAMN05201374 | PRJNA324382 | LZOV00000000 | 85 | 80× | 133173 | 11 | Contig | 4063564 | 4176 | 43.6 | 75 | 17 | 45 |
| B. subtilis str. BSP4 | SAMN05201389 | PRJNA324391 | MAFZ00000000 | 26 | 85× | 671313 | 3 | Contig | 4151750 | 4258 | 43.3 | 88 | 15 | 95 |
Includes ncRNA, misc_RNA, and misfeature.
Orthology Mapping
We carried out two parallel strategies for gene orthology mapping (supplementary table S2, Supplementary Material online). One strategy aimed to identify a subset of single-copy core genes for phylogenetic analysis, and the other aimed to describe and characterize the pangenome of B. subtilis. Further details are described in Supplementary Materials and Methods.
Phylogenetic Analyses and Species Limits in B. subtilis
Aiming at uncovering intraspecific processes, we started the analyses by defining the species limits. The definition of what a species is can trigger controversy among taxonomists and evolutionary biologists. However, the importance of defining the species limits is not contested, even when it is necessary to impose a still classification into a dynamic process. This has important consequences in medicine, biotechnology, and defense, where often it is preferable to use a not-so-perfect set of rules over using no rules at all (Godreuil et al. 2005; Konstantinidis and Tiedje 2005; Doolittle and Papke 2006; Goris et al. 2007; Richter and Rosselló-Móra 2009). In this study, we used phylogenetics to specify the species limits. We limited the taxon B. subtilis to the most inclusive well-supported monophyletic clade in a phylogenetic analysis of the single-copy core genome that includes all described B. subtilis type strains. We used JSpecies v. 1.2.1 (Richter and Rosselló-Móra 2009) to estimate the pairwise average nucleotide identity (ANI) based on BLAST, on the data set of concatenated single-copy core genes. This allowed us to ascertain whether the diversity within species was within the levels of nucleotide diversity typically observed within species of bacteria (Godreuil et al. 2005; Konstantinidis and Tiedje 2005; Doolittle and Papke 2006; Goris et al. 2007; Richter and Rosselló-Móra 2009).
The nucleotide sequences (and protein sequences) of each gene cluster were aligned with MAFFT v. 7.154 using the G-INS-I method and default parameter values (Katoh and Standley 2013), trimmed with BMGE version 1.1 using the codon option (Criscuolo and Gribaldo 2010), and finally concatenated into a single data set. Phylogenetic analyses were carried out with RaxML v. 8.0.26 (Stamatakis 2014) with the GTR + I+G + X (LG + G+I) model of evolution, where option X specifies a maximum-likelihood (ML) estimation of base frequencies. Model choice was determined with either jModelTest version 2.1.5 (Darriba et al. 2012) or ProtTest version 3.4 (Darriba et al. 2011). Nodal support was estimated via nonparametric bootstrap analysis using an automatic frequency-based criterion (autoFC option) to determine the number of replicates.
Reconstruction of Ancestral States
We reconstructed ancestral genome sizes and estimated genomic dynamics across the genealogy of B. subtilis using the ML birth–death model implemented in the software package Count v. 10.04 (Csurös and Miklós 2009; Csurös 2010) using the phylogeny estimated with the core genome and the genome content matrix of the 42 strains. We first optimized all of the model parameters by maximizing the likelihood of the data using a gain–loss-duplication model with a Poisson distribution for gene family size at the root. We assumed Gamma-distributed rate variation across gene families with the shape parameter discretized in four classes, and we assumed a fixed gain/loss ratio across lineages as in Luo et al. (2013) and Wolf et al. (2012). Rate parameters were optimized after 100 rounds of parameter optimization. Next, we estimated the profiles of posterior probabilities of events for each branch of the tree. To obtained patterns of gain/loss, we transformed these probabilities into “likely events” using a threshold of 0.5 posterior probability.
Quantification of Core and Pangenome Size
We used the GET_HOMOLOGUES package (Contreras-Moreira and Vinuesa 2013) to estimate the core and pangenome sizes of B. subtilis. This was performed by estimating both the number of shared genes and the number of novel genes as a function of the number of n strains sequentially added. To assess the variance in the estimates, we ran this analysis for 100 random replicates. GET_HOMOLOGUES applies a fitting curve to the data by applying Tettelin et al.’s (2005) exponential decaying function to the amounts of conserved and strain-specific genes to estimate the size of the core and the pangenome, respectively. To study the distribution of the cloud genome along the chromosome, we transformed each gene middle point position (bp) into radians and used circular statistics obtained from the R package Circular v. 0.4-7 (Agostinelli and Lund 2013) to compute the maximum likelihood estimates (MLEs) for the concentration parameter (kappa) of the von Mises distribution. We generated 95% bootstrap confidence intervals to test whether the kappa parameter was significantly different from 0, where 0 indicates no preferential location of the data points around the circumference. The bootstrap confidence intervals are the α/2 and 1−alpha/2 percentiles of the 1,000 replicate MLEs computed for each resampled data set, with the confidence level (alpha) equal to 0.05. For small data sets (N < 16), the MLE of kappa was bias-corrected following Best and Fisher (1981). Finally, to test whether strains sampled in similar niches were associated with a specific pattern of gene content, we carried out hierarchical clustering using Ward’s minimum variance method (ward.D2 method of the hclust function) on a distance matrix estimated with the S4 coefficient of Gower & Legendre available from the ade4 R package (Thioulouse et al. 1997). Finally, we used PHAST (Zhou et al. 2011) to identify intact prophage sequences within B. subtilis genomes, and we used BLASTn against the NCBI nr/nt database to confirm the annotation of those sequences. We considered annotations from best significant BLAST hits (e-value < 10−5) that matched to a prophage with high query cover (>50%) and high-sequence identity. For best BLAST hits matching a prophage with query cover <50%, we considered the prophage “unknown.” Genomes fragmented into contigs that are not listed in the proper order may compromise the efficiency of PHAST to identify intact prophages. For this reason, we performed whole-genome alignments of open genomes to a closely related closed genome using MAUVE (Darling et al. 2010) and executed the PHAST analysis on the re-ordered genomes.
Results
Phylogenetic History and Species Limits in B. subtilis
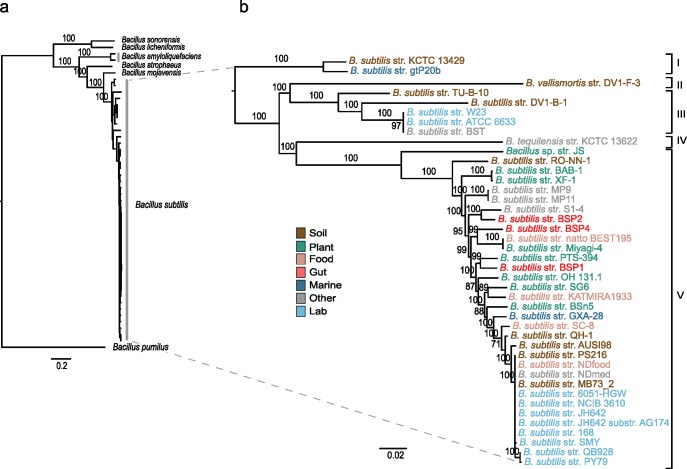
Public databases are known to sometimes include misidentified sequences (Richter and Rosselló-Móra 2009), and a preliminary analysis of the literature revealed the occasional clustering of B. vallismortis and B. tequilensis strains with strains of B. subtilis (Rooney et al. 2009; Bhandari et al. 2013). For this reason, we first determined the species limits of B. subtilis and inferred its phylogenetic history. We carried out the initial phylogenetic analysis with a data set of 398 concatenated core protein sequences (totaling 104,056 amino acids) using the genomes of all strains identified as B. subtilis in NCBI along with representative genomes of its closest relatives (supplementary table S1, Supplementary Material online). In this analysis, five genomes clustered with high-bootstrap support with the genomes of B. atrophaeus, B. amyloliquefaciens, or B. cereus type strains and were therefore considered misidentified and excluded from further analyses (supplementary fig. S1, Supplementary Material online). In subsequent phylogenetic analyses, we also dropped the most distant outgroups to avoid complications with long-branch artifacts, and we used B. pumilus to root the trees. A phylogenetic analysis of the corrected data set using 685 concatenated core genes, totaling 520,227 aligned nucleotides, recovered well-supported relationships within the B. subtilis group (fig. 1, supplementary fig. S2, Supplementary Material online). Our analyses revealed a total of 42 genomes of bona fide B. subtilis that comprised ten laboratory strains and 32 natural strains sampled from a wide diversity of sources (supplementary table S1, Supplementary Material online). We identified five major branches of diversification within B. subtilis; these branches comprise the three subspecies already described, that is, B. s. subtilis, B. s. spizizenii, and B. s. inaquosorum, as well as B. tequilensis and B. vallismortis. The marine strain B. subtilis str. gtP20b, previously classified as B. subtilis spizizenii (Fan et al. 2011) was identified in our analysis as a close relative of B. subtilis str. KCTC 13429, the type strain of the subspecies B. subtilis inaquosorum (Rooney et al. 2009). Finally, the closest outgroup of B. subtilis is B. mojavensis, followed by B. atrophaeus (supplementary fig. S2, Supplementary Material online). The core genome ANI across all B. subtilis genomes is 97.0 ± 0.09 (mean and standard error; standard deviation, 2.60), with an ANI within B. subtilis subspecies of 98.6 ± 0.31 (mean and standard error; standard deviation, 0.54; supplementary table S3, Supplementary Material online). These ANI values are within the levels of nucleotide diversity typically observed within species of bacteria (Konstantinidis and Tiedje 2005; Goris et al. 2007; Richter and Rosselló-Móra 2009). The ANI among subspecies is 94.1 ± 0.61 (mean and standard error; standard deviation, 1, 92) reflecting the phylogenetic structure in the core genome tree (fig. 1). This estimate is smaller and outside the threshold typically used to define species of bacteria (95% ANI). A species classification that favors ANI over other criteria would likely raise each subspecies to the species level. In this study, we elected to retain all of the B. subtilis type strains under the same species name, which required the inclusion of the B. tequilensis and B. vallismortis strains within the B. subtilis species limits. According to our results, the taxonomy of B. subtilis strains should be revised to account for the phylogenetic history of the core genome.
Fig. 1.
—The recent diversification of B. subtilis. (A) Phylogenetic analysis of B. subtilis strains and their closest relatives and (B) of the B. subtilis clade alone. ML (GTR + I+G + X) and bootstrap analyses were carried out on a concatenated data set of 685 core genes (520,227 nt). The tree was rooted following the results provided in supplementary figure S2, Supplementary Material online. Numbers on the nodes indicate bootstrap support, and the scale bar indicates the expected number of nucleotide substitutions per site. Due to the scale of the tree, some branches are too small for their statistical support to be shown; the cladogram in figure 3 distinguishes the branches with bootstrap support higher than 80%. The Roman numerals on the right indicate the five major branches of diversification within B. subtilis. The suggested taxonomic nomenclature for these branches is as follows: I B. subtilis inaquosorum, II B. subtilis vallismortis, III B. subtilis spizizenii, IV B. subtilis tequilensis, and V B. subtilis subtilis. Taxa are colored to indicate the niche at the site of sampling.
A Large, Dispersed, Open Pangenome
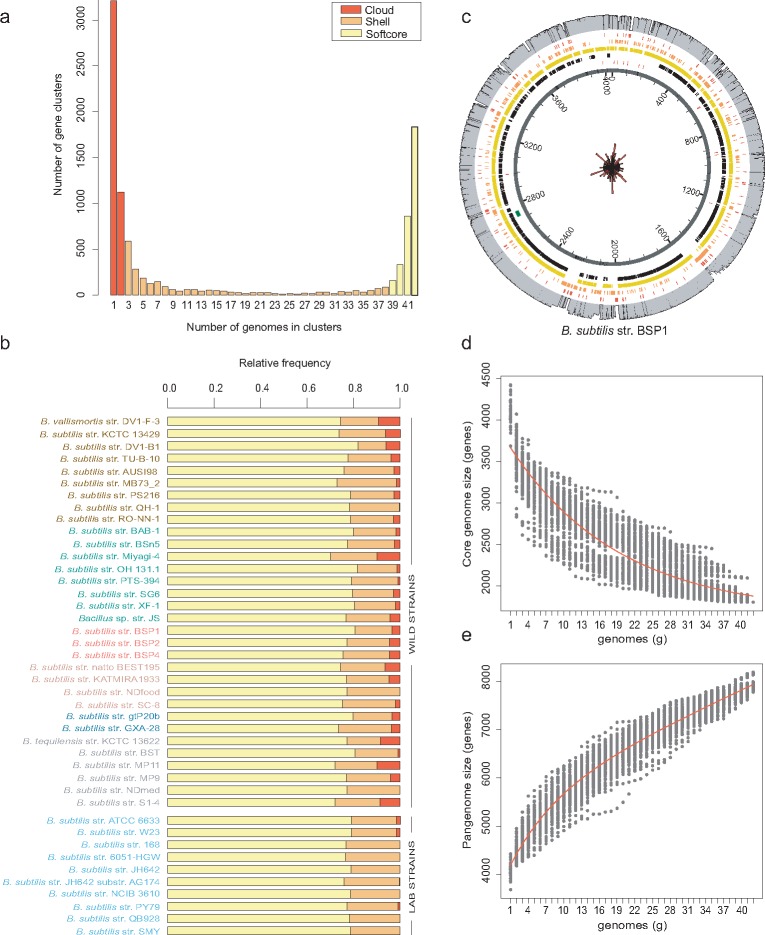
Comparative analysis of the B. subtilis genomes indicated that the total genome size can vary from 3.88 to 4.45 Mb, with the number of predicted protein-coding sequences ranging from 3,608 to 4,538 (supplementary table S1, Supplementary Material online). Orthology mapping of the 42 genomes of B. subtilis retrieved 10,137 homologous protein clusters (supplementary data set, Supplementary Material online). Depending on their frequency among the sampled genomes (size of the cluster), homologous clusters were classified as core (present in all 42 genomes), softcore (present in 39–42 genomes), shell (present in 3–38 genomes), or cloud (present in only one or two genomes). This classification resulted in 1,834 strict core genes, 3,193 softcore genes, 2,610 shell genes, and 4,334 cloud genes (fig. 2A). The u-shape distribution indicates that most genes in the B. subtilis pangenome either exist at very low frequencies or are found in almost all genomes, with only 25.7% of the pangenome being present at intermediate frequencies. The relative proportions of these polymorphic classes per genome are relatively constant, and each genome of B. subtilis has on average (mean and standard error) 77.1% ± 0.4% of softcore genes, 19.7% ± 0.4% of shell genes, and 3.2% ± 0.5% of cloud genes (fig. 2B). Thus, the ∼43% of the total pangenome that is present in only one to two genomes represents, on average, no more than 3% of each B. subtilis genome, a percentage that lowers to 0.5% among the laboratory strains (fig. 2B). The inclusion of open genomes in the analysis adds noise to the data set, as a small proportion of genes might not have been sequenced or might have been truncated and not annotated. However, and importantly, we found no significant differences in the numbers of shell and cloud genes between wild strains with open genomes and wild strains with closed genomes (t = 2.02, P-value = 0.052, and t = 1.42, P-value = 0.167, respectively). This indicates the absence of strong bias in the data set due to genome assembly quality.
Fig. 2.
—B. subtilis has a large, dispersed, and open pangenome. (A) B. subtilis pangenome gene cluster frequency spectrum. The color coding indicates the partitioning of the pangenome matrix (10,137 gene clusters) into classes of genomic polymorphism: cloud (one to two genomes), shell (3–38 genomes), and softcore (39–42 genomes). (B) Relative frequencies of each polymorphic class per genome. The cloud genome accounts for 43% of the species pangenome but represents on average only 3% of each genome. Cloud genes are almost absent from the genomes of laboratory strains, which likely reflects the axenic conditions under which these strains are typically cultivated. (C) Genome atlas of B. subtilis str. BSP1, indicating that the variable pangenome is distributed throughout the chromosome. Circles from the center represent the location of the following elements: coding sequences (dark gray), tRNA (red) and prophages (intact: dark green; questionable: green; incomplete: light green); core genome (black); softcore genome (yellow); shell genome (orange); cloud genome (red). The outer circle in gray represents the relative frequency of each gene cluster in the data set. The radial histogram at the center is a rose diagram showing the dispersion of cloud genes around the genome after transforming each gene middle-point position into radians. The genome was divided into 100 bins, and the radii of the sectors are equal to the square root of the relative frequencies of the cloud genome, making the area of each sector proportional to its frequency. The rose diagram was scaled to the inner circle of the genome atlas. Quantification of the core genome (D) and the pangenome (E) of B. subtilis after 100 random samples of 42 genomes. The numbers of shared (D) and novel (E) genes are plotted against the number of n strains sequentially added. The red line is the fitted curve following the exponential function of Tettelin (2005). Bacillus subtilis has an open pangenome that translates into 1,659 (SE 216.77) core genes and an average number of new genes asymptotically predicted for further genome sequencing of 57.
The asymptotic properties of the distribution of conserved and strain-specific genes in B. subtilis indicate a core genome of 1,659 genes (residual standard error (SE) of 216.77) and an extrapolated pangenome growth rate of 57 new genes (residual standard error of 223.93) per new genome added (see Eqs. 1 and 2 in Supplementary Material online). By reaching a nonzero asymptotic value for the rate of growth, this model predicts that the B. subtilis pangenome is unbounded, or open. The high proportion of variable pangenome is not a result of the domestication and manipulation of laboratory strains. The estimates of core and pangenome sizes of B. subtilis wild strains do not differ substantially from the ones estimated with the whole data set, extrapolating a core genome of 1,803 genes (residual SE 225.20) and an asymptotic value of 73 new genes (residual SE 202.77) for every new genome sequenced (supplementary fig. S3, Supplementary Material online).
The analysis performed to infer the presence of intact prophages within the genomes indicated that B. subtilis has on average two intact prophages, ranging from one to four in the marine strain GXA-28 and reaching six in strain S1–4, which was isolated from a chicken feather (supplementary fig. S4 and table S4, Supplementary Material online). The PBSX phage-like element is the most common prophage present in the genomes analyzed. This phage is induced by the SOS response and results in cell lysis (Krogh et al. 1996). The SPβ prophage and the prophage-like skin element, a defective prophage, are mostly restricted to strains of the subspecies B. s. subtilis (supplementary fig. S4, table S4, Supplementary Material online). Both these elements are inserted within the coding region of sporulation-specific genes. SPβ interrupts the spsM gene (Abe et al. 2014), whereas skin is inserted into the sigK gene (Stragier et al. 1989). SPβ and skin have to be excised by site-specific recombinases during sporulation for the reconstitution of functional spsM and sigK genes. Note, however, that excision is restricted to the mother cell, which is a terminal cell line that lyses at the end of sporulation to release the mature spore into the environment (Abe et al. 2014; Stragier et al., 1989). Thus, in the genome of the spore, SPβ and skin remain intact and are transmitted to the next generation. These elements are also found in strains that branch in parts of the phylogenetic tree that do not include the subspecies B. s. subtilis; specifically, the skin element was detected in strains DV1-B1 and S1–4, and the SPβ prophage was found in strain GXA-28 (supplementary fig. S4 and table S4, Supplementary Material online). This may indicate ancestral acquisition with posterior elimination from the genome of many strains or an evolutionary trajectory that includes both vertical and horizontal inheritance. The pairwise alignment statistics (% query cover and % identity) of best BLAST hits (supplementary table S4, Supplementary Material online) also indicate high polymorphism in the sequence content and in the nucleotide diversity of homologous prophages and skin elements. This genomic mosaicism with regions of high similarity interspersed with segments that show no homology has been described previously and may result from recombination between phages (Hendrix et al. 1999; Bobay et al. 2013). We also detected events of prophage degradation. For example, the laboratory strain PY79 has a highly fragmented SPβ prophage distributed through different parts of its genome, precluding its identification by PHAST. Other intact prophages were detected, but searches of the NCBI databases did not produce good identifications; therefore these prophages were considered “unknown.” For example, strain TU-B-10 has two intact and identical prophages inserted in tandem near a cluster of tRNA genes, distant from sigK. Nevertheless, the best BLAST hit of these prophage sequences matches a skin element, although with 36% coverage and 88% identity from the canonical sequence. For the purposes of this study, intact but “unknown” prophages were considered functional, but their precise identification and adaptive value require further investigation.
We tested whether the distribution of the cloud genome along the chromosome was particularly enriched within prophages. For the large majority of strains, we found no significant association of the cloud genome with prophages, which reflects the frequency at which prophages PBSX and SPβ and the skin element were detected among B. subtilis genomes (supplementary table S5, Supplementary Material online). Since skin is a defective prophage (above), it does not function as a vector of LGT. Importantly, however, our conclusions do not change when we exclude the skin element from the list of intact phages (supplementary table S5, Supplementary Material online). We did find a group of 12 strains in which the cloud genome is particularly enriched within prophages, particularly low-frequency prophages for which we found no identification (the “unknown” class, above). Among this group, it is noteworthy strains GXA-28, AUSI98, and S1–4 with large proportions of the cloud genome concentrated within intact prophages (44.7%, 45.4%, and 26.9%, respectively; supplementary table S5, Supplementary Material online). However, because the remaining cloud genes are distributed throughout the rest of the genome, circular statistics applied to the variable pangenome indicated the absence of an aggregated distribution, as indicated by a concentration parameter of the von Mises distribution not significantly different from zero (fig. 2C, supplementary fig. S5 and table S6, Supplementary Material online). From these analyses, we conclude that for the large majority of B. subtilis genomes, there are no hotspot regions, that is, regions more prone to harbor the most recently acquired genes (cloud genome). We also estimated the concentration parameter (kappa) of the von Mises distribution for the shell genome. Several strains have confidence intervals that do not include zero, suggesting a clumped distribution (supplementary table S6, Supplementary Material online). However, the shell genome can be derived from gene acquisitions that spread vertically, or horizontally, across strains, and gene losses. All of these processes occurring in one class can lead to very complex patterns.
Dynamics of Genome Content
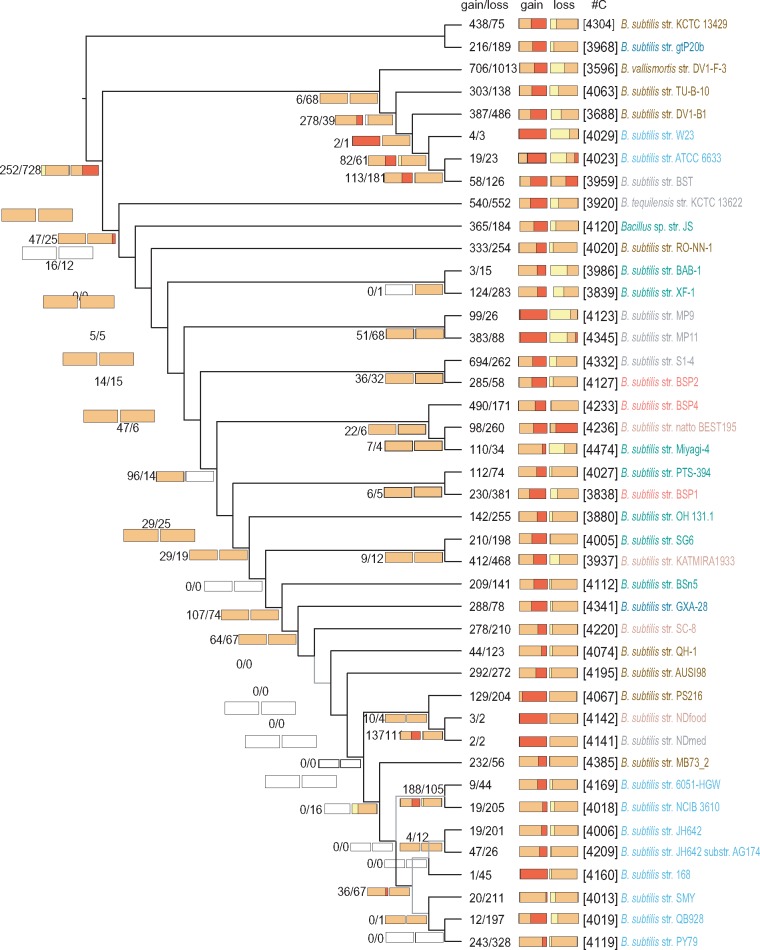
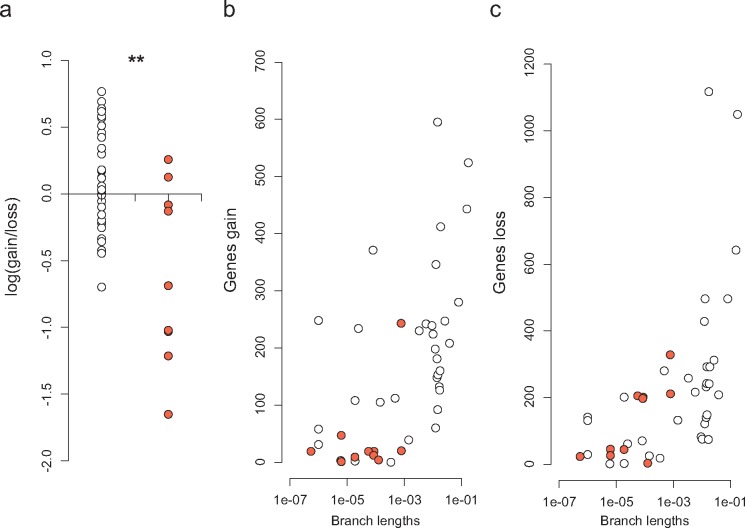
To analyze the genome dynamics responsible for the observed distribution of gene frequencies, we applied a birth–death likelihood model to reconstruct ancestral genome sizes and the dynamics of genome content across the genealogy of B. subtilis. Current distributions can either reflect the genealogical process, implying a minimal number of transition events, or result from an intricate series of gene gains and losses that likely reflect local selective pressures and social interactions. As in other studies (Kettler et al. 2007; Luo et al. 2013; Puigbò et al. 2014), our analyses show that the majority of gene gains and losses occur at the terminal branches. This finding suggests that the diversity in genome content within the B. subtilis pangenome is mainly due to recent events that are not shared by many strains (fig. 3). In general, longer terminal branches show more events, as apparent in the branches leading to strains DV1-B-1, B. tequilensis and B. vallismortis (fig. 3). But this relationship is not linear as high gene gains and losses also occur in some short branches, for example, the branches leading to strains PS216, AUSI198, and PY79 (fig. 3). We found no general prevalence of gene loss relative to gene gain among terminal branches leading to genomes of wild strains (fig. 4), and the variation observed does not reflect differences in the niche at the site of sampling. Laboratory strains show gene losses comparable to those of wild strains with similar terminal branch lengths but show marginal gene gains with almost no cloud genome, likely reflecting their history in axenic cultures (fig. 4). Strain PY79, with an estimated number of 243 gene gains and 328 gene losses, stands out as the exception among laboratory strains, which otherwise show gene gains and losses of the same order of magnitude (fig. 4). This result explains why strain PY79, which has a core genome almost identical to those of other laboratory strains (close relationship and small branch lengths on the core genome tree, fig. 1), has a distinct pangenome composition that is visible in the clustering of the pangenome tree of figure 5A (see also below).
Fig. 3.
—B. subtilis genome dynamics. Estimated events of genome content evolution are mapped onto the genealogy estimated with the core genome (here shown without branch lengths for clarity). Numbers below the branches and at terminal nodes represent the estimated numbers of gene gain and loss events (gain/loss). The two horizontal bars are stacked histograms that represent the relative proportions of softcore (yellow), shell (orange) and cloud (red) gene gain and loss dynamics inferred for each node. Numbers in squared brackets (#C) are the total numbers of gene clusters per genome. Light gray branches indicate phylogenetic relationships with bootstrap support lower than 80% (see fig. 1).
Fig. 4.
—Gene gain and loss events at terminal branches of the B. subtilis phylogeny. There is no bias of gene gain to gene loss among natural strains (white), whereas laboratory strains (red) have a bias toward gene loss (A), even when contrasted with natural strains with similar terminal branch lengths (B, C). Ratios were log-transformed with the log1p function and tested for significant differences using the Mann–Whitney–Wilcoxon test in R (W = 261; P-value = 0.002). Branch lengths are in units of expected numbers of substitutions per nucleotide site.
Fig. 5.
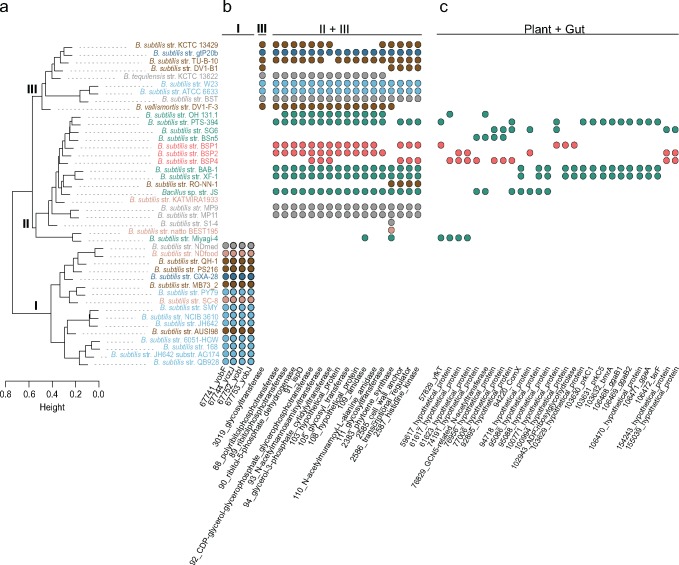
—Hierarchical cluster analysis of B. subtilis pangenome diversity and niche-specific genome content. (A) Dendrogram of the pairwise distances estimated with protein genome content; colors indicate niche at the site of sampling as in figure 1. Roman numbers on the nodes indicate the three major clusters of diversity; I and II include genomes of B. s. subtilis, and cluster III contains genomes from all other B. subtilis subspecies. On the right are phylogenetic profiles for selected gene clusters. In (B) are gene clusters distributed exclusively to clusters I, III, and II + III. Cluster II is more heterogeneous than the other clusters, and we found no gene with a distribution exclusive to this group of strains. In (C) are gene clusters identified only among genomes of strains sampled in plants or in the gut. These are putative examples of LGT associated with niche occupancy.
Cloud genes exist in only one or two strains and are likely acquired through LGT. It is unknown whether the current distributions of the variable pangenome reflect the genealogical process, indicating strictly vertical inheritance after initial acquisition. We used the results of the previous analysis, shown in figure 3, to estimate the number of state transitions (presence to absence and vice versa) for each gene cluster in the data set. Our results indicate that genes from the shell genome generally have an evolutionary history characterized by multiple gains and losses, with the median number of state transitions being 4 (first and third quartiles are Q1 = 3 and Q3 = 6). In contrast, the number of transitions is 1 for softcore genes (Q1 = 1; Q3 = 2) and 1 (Q1 = 1; Q3 = 3) for cloud genes. Interestingly, of the 2610 shell genes in the data set, only 130, corresponding to 5.0%, were acquired only once with no subsequent loss. The relative proportions of each polymorphic class in the gains and loss events inferred on the core genome tree are shown in the histograms of figure 3. Not surprisingly, gains of cloud genes, indicated in red on the left-hand stacked-bar histograms of figure 3, occur mainly at the terminal branches, as do the losses of softcore genes, represented in yellow on the right-hand stacked-bar histograms. Note that losses of strain-specific genes at terminal branches cannot be detected, making the gene losses of cloud genes necessarily underestimated in this type of analysis.
In summary, gene gains and losses are balanced in the evolution of B. subtilis genomes, with most events occurring at terminal branches, particularly along long branches. Unlike softcore and cloud genes, shell genes are the result of complex histories characterized by frequent events of gains and losses.
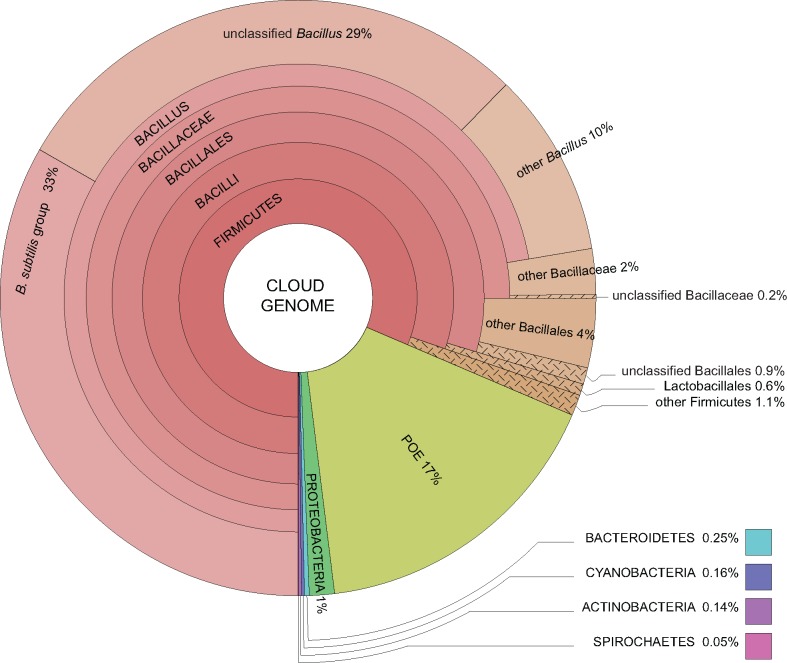
Origin of the Cloud Genome
To identify putative donor species of the cloud genome, we carried out a BLASTP search for each protein against the NCBI nr database excluding B. subtilis entries and using an e-value of 10−5. The taxonomy of the best BLAST hits indicates that 72.3% of the cloud genome has a close homologue within the genus Bacillus and another 9.1% has a close homologue within the phylum Firmicutes (fig. 6). These results show that recently acquired genes are likely transfers from closely related species. In the absence of a selective mechanism that interferes with exogenous DNA integration, this result must reflect the species diversity at the ecological niche. LGT from distant taxa was identified for 85 genes, corresponding to ∼2.0% of the cloud genome. These genes have their closest homologues in other phyla, particularly in the Proteobacteria (59 genes), the Bacteroidetes (11 genes), the Cyanobacteria (7 genes), and the Actinobacteria (6 genes). We recovered no significant BLAST hits for 720 genes (16.6% of the cloud genome). These are B. subtilis genes with no homologues in the NCBI non-redundant database; they might correspond to newly formed genes, annotation errors, or genes present in taxa that have yet to be sequenced.
Fig. 6.
—Recently acquired genes from the cloud genome of B. subtilis have homologous copies within closely related groups. Taxonomic characterization of best BLAST hits of each protein from the cloud genome of B. subtilis indicates that 72.3% of these genes have a close homologue within the genus Bacillus and an additional 9.1% within the phylum Firmicutes. Eighty-five genes (2.0%) have best BLAST hits with species from other phyla, such as Proteobacteria and Bacteroidetes, which is interpreted as evidence of long-distance LGT. POE (16.6%) represents putative orphan (or error) genes; these are B. subtilis genes with no homologues in other species represented in the NCBI nr database. These results were obtained after a BLASTP search for each protein from the cloud genome against the NCBI nr database excluding B. subtilis entries using an e-value threshold of 10−5.
Niche-Specific Signals in Pangenome Diversity
We surveyed the literature for niche characterization at the site of collection (supplementary table S7, Supplementary Material online). Based on this survey, we classified the strains into seven categories: Plant, Soil, Gut, Food (fermented food products), Marine, Lab, and Other. Mapping the niche classifications onto the core genome tree of figure 1 revealed that niche occupancy occurs independently of the genealogy and that strains with different genealogical histories can be sampled in similar environments. In particular, the three strains sampled in the vertebrate gut have close relatives in other niches, and the strains sampled in the soil or in association with plants are distributed throughout the core genome tree without clustering by niche. Hence, niche occupancy by B. subtilis was not achieved by population fragmentation and specialization; rather, it occurs repeatedly throughout the evolution of the species.
If niche occupancy were achieved due to the acquisition of a particular group of genes, then strains sampled in similar niches would have similar pangenomes. Otherwise, similarity in genome composition should reflect the genealogical history. A hierarchical cluster analysis on the pairwise whole genome differences (pangenome or gene-content tree) can thus allow inferences of niche-specific signals whenever strains from different lineages cluster in the pangenome tree following the niche classification. In our analysis, the deepest split in the pangenome tree does not follow the phylogeny, as it segregates the strains of the subspecies B. s. subtilis into two clusters, clusters I and II (fig. 5), with strains from cluster II being more similar to strains from all other subspecies (cluster III) than to strains from cluster I. This is a surprising result given the short evolutionary time that can be inferred for B. s. subtilis from the core genome tree (fig. 1). By searching for genes that sustain the branching of the pangenome tree, we identified four genes that are present in all strains of cluster I but are absent from clusters II and III (fig. 5B; see supplementary data set 3, Supplementary Material online for a full list of genes exclusive to cluster I). These genes, yobF, yozJ, yobI, and yobJ, coding for proteins of unknown functions, are located 80 Kb upstream of prophage SPβ, which is also exclusive to this group. These genes represent a DNA acquisition that has been maintained through vertical inheritance in the lineage of B. s. subtilis. We found no genes exclusive to cluster II (supplementary data set 4, Supplementary Material online) and only one gene exclusive to cluster III, coding for a putative glycosyltransferase (NCBI ProteinID: YP_003865706.1; supplementary data set 5, Supplementary Material online). The association of cluster II with cluster III is not sustained on core genes absent from cluster I but rather on 668 genes present at various frequencies among strains from both clusters II and III but absent from cluster I. Examples include the tarIJKL genes involved in the synthesis of poly(ribitol phosphate), a cell wall teichoic acid found in some strains of B. subtilis (Lazarevic et al. 2002) (gene clusters 89, 90, 91, and 92 in fig. 5 and supplementary table S8, Supplementary Material online; full data set is provided in supplementary data set 6, Supplementary Material online). Several of the genes exclusive to clusters II and III have a paralogue copy in cluster I that is on average 40% divergent from the copy in II and III. An example of these genes is the lytABC genes, which are annotated with the same names in strain 168 (supplementary table S8, Supplementary Material online).
Competence Allows Laterally Acquired Genes to Propagate Across B. subtilis Strains that Share the Same Environment
Analyses of the pangenome tree indicate a tighter clustering of Plant and Gut strains than the clustering recovered from core genome tree. We queried the pangenome of B. subtilis for genes of the shell genome that could explain this aggregation. Figure 5C shows phylogenetic profiles for selected examples (full results are provided on supplementary data set 7, Supplementary Material online). All selected examples were manually curated. These genes are at relatively low frequencies in our data set and some are syntenic, suggesting common events of transference, although none overlap with intact prophages and thus must rely on another mode of transmission, such as competence. Gene acquisitions through LGT are a primordial mode by which bacteria can promote local adaptation by expansion of their metabolic repertoire; however, many of these genes have unknown functions still requiring a full functional characterization. Others, however, are LGT events that provide xenologous copies of genes that were previously present in the genome. Supplementary figure S6, Supplementary Material online, shows the evolutionary history of the bmrA gene, which codes for a multidrug resistance ABC transporter ATP-binding protein. The evolutionary history of the bmrA gene is characterized by frequent LGT between gram-positive bacteria. Within B. subtilis alone, we identified three homologous copies, of which one is from the softcore genome (NCBI ProteinID: NP_391362.1) and has been transmitted vertically within the B. subtilis group. The other two copies belong to the shell (NCBI ProteinID: AGE62337) and cloud (NCBI ProteinID: ADV95129) genomes and were recently acquired by B. subtilis strains from unrelated organisms. These xenologous copies do not overlap with any of the described prophages. Importantly, since their initial acquisition, these genes have been transferred across strains sampled from Plant niches. This observation strongly argues in favor of a mode of transmission, such as natural competence, that does not rely on a phage.
The presence of highly divergent homologous copies in the genomes of B. subtilis is clear demonstrations of the pervasive influence that LGT has had on the diversity of gene content in this species. Whether these laterally acquired xenologous copies maintain the same function as the vertically inherited copy is unclear and will require a full functional characterization to determine. In any event, we have shown here that genes acquired from unrelated species can propagate across B. subtilis strains that share environments (examples are genes presented in fig. 5C) or be transmitted vertically, as with the genes of cluster I that are plotted in figure 5B. In both cases, their maintenance in the B. subtilis pangenome likely reflects adaptation to the environment.
Discussion
We show that B. subtilis has a large, open and dynamic pangenome that largely results from the continuous acquisition of genes from closely related species along with extensive gene loss. This pattern of genome dynamics suggests a key role for natural competence coupled with a wide niche breadth in the creation and maintenance of the species’ pangenome diversity. We corroborate previous results on the evolution of bacterial genomes, showing that this evolution is a highly dynamic process. Importantly, we show that this dynamicity does not lead to major differences among strains. On average, 77.1% of all genes in each genome are shared among all strains, and 43% of the species pangenome that exists in only one or two strains represents no more than 3% of the genome of natural strains and 0.5% of that of laboratory strains. Consistent with the results of previous studies (reviewed by Wolf and Koonin 2013), we infer that the main determinants in the evolution of bacterial genomes are gene gain through LGT and gene loss. The net result of these processes shape and maintain a species-specific genome cohesion by streamlining the genomes along with the acquisition of a new and diverse genomic repertoire. At the terminals of the core genome tree, we do not recover the gene loss bias that has been widely documented in the literature (Kunin 2003; Makarova et al. 2006; Csurös and Miklós 2009; Wolf et al. 2012; Puigbò et al. 2014). Instead, we found high and ubiquitous gene gain in the evolution of wild strains, suggesting this to be a fairly frequent rather than episodic mechanism of genome diversity acquisition. Similar dynamics were observed in the genome evolution of Escherichia coli but were attributed to phage-related genes or transposable elements (Touchon et al. 2009). Bolotin and Hershberg (2015, 2016) suggest that a gene-loss bias in intraspecific genome dynamics is mainly a feature of highly clonal species. However, in species with higher rates of recombination, such as B. subtilis, the two rates are expected to balance out. More generally, Puigbò et al. (2014) suggest that gene loss bias may not manifest at short evolutionary scales. Losses of strain-specific genes cannot be detected, imposing an underestimation of the gene loss at these branches. It is also possible that stochastically acquired genes could temporarily accumulate in the genomes if neutral or slightly deleterious, similar to the inflated number of non-synonymous to synonymous substitutions estimated in data sets of intraspecific genomes (Rocha et al. 2006).
The high rates of gene gain and gene loss that we recover for the tips of the tree coupled with the ability of B. subtilis to uptake exogenous DNA without self-specificity of sequence identification are highly suggestive of an evolutionary model in which cloud genes are the result of stochastic events of gene acquisitions. The long-term maintenance of these genes in the genome repertoire of the species should thus depend on their fitness effects given the genomic context (epistatic interactions) and the specificities of the biotic and abiotic environment (Graham and Istock 1979; Berg and Kurland 2002; Johnsen et al. 2009). Several studies have addressed the fate of acquired genes, and the results suggest that gene loss of recently acquired genes is pervasive in many bacterial groups (van Passel et al. 2008; Lo et al. 2015; Kuo and Ochman 2009). Simulations under a birth-and-death model of prokaryote genome evolution suggest that neutral or nearly neutral gene acquisitions in microbial populations are expected to generate a large diversity of transient gene content, where only sequences that are under strong selection, globally or in individual patches, are expected to persist (Berg and Kurland 2002). An example of strong local episodic selection promoting genome diversity was previously proposed for B. subtilis. In this species, competence develops in non-dividing cells in an otherwise growing population, imposing a short-term fitness cost on the competent cells. Johnsen et al. (2009) show that this impairment can be overcome if episodic stresses, such as antimicrobials, preferentially affect the dividing cells, an advantage that is highly augmented if selection favors the competent cells that have acquired new DNA (Johnsen et al. 2009).
It is tempting to speculate that through competence, B. subtilis stochastically surveys the environment for new genes, potentiating a dynamic process of niche adaptation in which each organism can have its own evolutionary trajectory, as proposed by Gogarten et al. (2002). A population could simultaneously express diverse phenotypes in an extension of bet-hedging strategies, as documented for genetically identical cells (Ackermann et al. 2008; Leisner et al. 2008; Veening et al. 2008; Beaumont et al. 2009). Bet-hedging is important for survival during the rapid expansion of sub-populations in a rapidly changing environment or when an organism frequently transits between niches, as it is likely the case in B. subtilis (Kussell and Leibler 2005; Tam et al. 2006; Wolf et al. 2005; Wylie et al. 2010). The maintenance of different genomes in a local population might also foster strategies for the division of labor and the evolution of cooperative behaviors (Morris et al. 2012; Mas et al. 2016), both of which are well documented in B. subtilis (Lopez et al. 2009; Shank et al. 2011; van Gestel et al. 2015).
Nevertheless, transduction by the integration of phage DNA into the bacterial chromosome certainly plays a role in the acquisition of new genes, but it is not the prevailing mode of LGT in B. subtilis. Only a few strains show evidence of having prophages enriched in cloud genes, and in those strains, a large proportion of the recently acquired cloud genome is distributed throughout the entire genome and does not map to prophages (fig. 2C, supplementary fig. S5 and table S5, Supplementary Material online). We have not analyzed the diversity present in plasmids. We note, however, that although genes in replicative plasmids would likely further increase the pangenome size of B. subtilis, the integration of plasmids or other integrative and conjugative elements into the chromosome is expected to generate a clumped distribution of laterally transferred genes (Wozniak and Waldor 2010), which is clearly not the dominant pattern observed here (fig. 2C, supplementary fig. S5, Supplementary Material online).
Natural competence is not expected to play a similar role in every organism. For instance, both Streptococcus pneumoniae and Haemophilus influenzae are naturally competent species, but both have pangenome sizes that are much smaller than the one estimated here for B. subtilis (Hiller et al. 2007; Hogg et al. 2007). These species have a narrower niche breadth that is typically restricted to the human respiratory tract, in which they can become highly pathogenic. Streptococcus pneumonia, unlike B. subtilis, lacks a specific DNA damage repair mechanism (Charpentier et al. 2012), and it might rely on competence to overcome DNA damage caused by the host immune system (Claverys et al. 2006). In contrast, in H. influenza, the diversity of the genes acquired through transformation is clearly constrained by the need for a sequence-specific identifier that limits DNA binding and uptake to intraspecific DNA (Mell and Redfield 2014).
The diversity of the B. subtilis pangenome reflects both the vertical inheritance of genes (the genealogical process) and convergent evolution related to the occupancy of similar environments. Overall, there is not a strong pangenome signature for niche occupancy, as we did not recover niche-specific genes. We note, however, that strains from cluster II (fig. 5A) can be traced to an environment in which host–microbe interactions are likely. This cluster contains all Plant- and Gut-associated strains as well as two strains sampled from fermented food, one strain sampled from a chicken feather, and two strains isolated from the abdomen and nest of fungus-growing termites. Microbes that cycle through environments could benefit from common (in addition to specific) adaptation strategies in such cross-kingdom host colonizations (Wiedemann and Virlogeux-Payant 2015). In addition, fecal contamination of the soil by animals can expose plants to gut bacteria, creating a cycle of transmission that closes when animals are fed with plants and fermented (probiotic) foods, exposing the animal gut to plant-adapted and fermented bacteria (Tam et al. 2006; Barak and Schroeder 2012; Melotto et al. 2014; Serra et al. 2014). The lack of a strong signature for niche adaptation could reflect a sampling problem if the strains were isolated as spores, which can be easily dispersed, and not as growing cells. It might also be explained by the influences of the several environments through which the strains cycled or by the ability of bacteria to adopt a multitude of different strategies to adapt to the same environment.
Two genomes can be similar if they share a common evolutionary history of gene gains and losses or if they evolve convergently. However, when there is no tendency for closely related strains to occupy similar niches, as appears to be the case for B. subtilis, a common history in the genomic dynamics of vertically inherited genes would be shared, at least transiently, between strains sampled in different environments. For this reason, it is not always easy to differentiate common ancestry from convergent evolution in a pangenome-wide analysis. A lack of genes with a distribution that closely follows the niche classification most likely reflects a species that has not specialized into living in a particular habitat, that is, a species that is a generalist microorganism, of which B. subtilis might be a paradigm.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Eduardo Rocha and Luis Teixeira for critically reading earlier versions of this manuscript. This study was supported by “Fundação para a Ciência e a Tecnologia” (FCT) through grants PTDC/EBB-BIO/119006/2010 to J.P.L. and PEst-OE/EQB/LA0004/2011 to A.O.H. This study was also financially supported by Project LISBOA-01-0145-FEDER-007660 (“Microbiologia Molecular, Estrutural e Celular”) funded by FEDER funds through COMPETE2020—“Programa Operacional Competitividade e Internacionalização” (POCI), by national funds through the FCT to A.O.H. P.H.B. acknowledges a post-doctoral fellowship (SFRH/BPD/89907/2012) and C.S. received a PhD fellowship (SFRH/BD/29397/06), both from the FCT.
Literature Cited
- Abe K. 2014. Developmentally-regulated excision of the SPβ prophage reconstitutes a gene required for spore envelope maturation in Bacillus subtilis Viollier, PH, editor. PLoS Genet. 10:e1004636.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M, et al. 2008. Self-destructive cooperation mediated by phenotypic noise. Nature 454(7207):987–990.http://dx.doi.org/10.1038/nature07067 [DOI] [PubMed] [Google Scholar]
- Agostinelli C, Lund U.. 2013. R package ‘circular’: circular statistics (version 0.4-7). URL: https://r-forger-projectorg/projects/circular/. https://r-forger-projectorg/projects/circular/.
- Ahmed A, et al. 2012. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol. 194(15):3922–3937.http://dx.doi.org/10.1128/JB.00056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambur OH, Engelstädter J, Johnsen PJ, Miller EL, Rozen DE.. 2016. Steady at the wheel: conservative sex and the benefits of bacterial transformation. Philos Trans R Soc B: Biol Sci. 371(1706):20150528.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Guillemin K, Phillips PC.. 2008. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62(1):39–49. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. 2005. Do orthologous gene phylogenies really support tree-thinking? BMC Evol Biol. 5:33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak JD, Schroeder BK.. 2012. Interrelationships of food safety and plant pathology: the life cycle of human pathogens on plants. Annu Rev Phytopathol. 50:241–266.http://dx.doi.org/10.1146/annurev-phyto-081211-172936 [DOI] [PubMed] [Google Scholar]
- Barbosa TM, Serra CR, La Ragione RM, Woodward MJ, Henriques AO.. 2005. Screening for bacillus isolates in the broiler gastrointestinal tract. Appl Environ Microbiol. 71(2):968–978.http://dx.doi.org/10.1128/AEM.71.2.968-978.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB.. 2009. Experimental evolution of bet hedging. Nature 462(7269):90–93. [DOI] [PubMed] [Google Scholar]
- Berg OG, Kurland CG.. 2002. Evolution of microbial genomes: sequence acquisition and loss. Mol Biol Evol. 19(12):2265–2276.http://dx.doi.org/10.1093/oxfordjournals.molbev.a004050 [DOI] [PubMed] [Google Scholar]
- Best DJ, Fisher NI.. 1981. The BIAS of the maximum likelihood estimators of the von mises-fisher concentration parameters. Commun Stat: Simul Comput. 10(5):493–502.http://dx.doi.org/10.1080/03610918108812225 [Google Scholar]
- Bhandari V, Ahmod NZ, Shah HN, Gupta RS.. 2013. Molecular signatures for Bacillus species: demarcation of the Bacillus subtilis and Bacillus cereus clades in molecular terms and proposal to limit the placement of new species into the genus Bacillus. Int J Syst Evol Microbiol. 63(Pt 7):2712–2726. [DOI] [PubMed] [Google Scholar]
- Bobay L-M, Touchon M, Rocha EPC.. 2013. Manipulating or superseding host recombination functions: a dilemma that shapes phage evolvability, Casadesús, J, editor. PLoS Genet. 9:e1003825.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin E, Hershberg R.. 2015. Gene loss dominates as a source of genetic variation within clonal pathogenic bacterial species. Genome Biol Evol. 7(8):2173–2187.http://dx.doi.org/10.1093/gbe/evv135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin E, Hershberg R.. 2016. Bacterial intra-species gene loss occurs in a largely clocklike manner mostly within a pool of less conserved and constrained genes. Sci Rep. 6(1):35168..http://dx.doi.org/10.1038/srep35168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF.. 2004. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14(12):2469–2477.http://dx.doi.org/10.1101/gr.3024704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier X, Polard P, Claverys J-P.. 2012. Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS?. Curr Opin Microbiol. 15(5):570–576.http://dx.doi.org/10.1016/j.mib.2012.08.001 [DOI] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S.. 1999. Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB), pp. 45–56.
- Claverys J-P, Prudhomme M, Martin B.. 2006. Induction of competence regulons as a general response to stress in gram-positive Bacteria. Annu Rev Microbiol. 60:451–475.http://dx.doi.org/10.1146/annurev.micro.60.080805.142139 [DOI] [PubMed] [Google Scholar]
- Connor N, et al. 2010. Ecology of speciation in the genus Bacillus. Appl Environ Microbiol. 76(5):1349–1358.http://dx.doi.org/10.1128/AEM.01988-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Moreira B, Vinuesa P.. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 79(24):7696–7701.http://dx.doi.org/10.1128/AEM.02411-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S.. 2010. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Mostowy R, Wymant C, Turner P.. 2016. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict. PLoS Biol. doi: 10.1371/journal.pbio.1002394.s020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csuos M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinfo Appl Note 26(15):1910–1912. CrossRef] [DOI] [PubMed] [Google Scholar]
- Csurös M, Miklós I.. 2009. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol Biol Evol. 26(9):2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5(6):e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9(8):772..http://dx.doi.org/10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165.http://dx.doi.org/10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Moran NA, Ochman H.. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301(5634):829–832.http://dx.doi.org/10.1126/science.1086568 [DOI] [PubMed] [Google Scholar]
- Deng Y, et al. 2011. Complete genome sequence of Bacillus subtilis BSn5, an endophytic bacterium of Amorphophallus konjac with antimicrobial activity for the plant pathogen Erwinia carotovora subsp. carotovora. J Bacteriol. 193(8):2070–2071.http://dx.doi.org/10.1128/JB.00129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 1999. Lateral genomics. Trends Cell Biol. 9(12):M5–M8. [PubMed] [Google Scholar]
- Doolittle WF, Bapteste E.. 2007. Pattern pluralism and the tree of life hypothesis. Proc Natl Acad Sci USA. 104(7):2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Papke RT.. 2006. Genomics and the bacterial species problem. Genome Biol. 7(9):116..http://dx.doi.org/10.1186/gb-2006-7-9-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Losick R, Kolter R.. 2007. Bacillus subtilis genome diversity. J Bacteriol. 189(3):1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmoer DJP, Rozen DE.. 2011. Competence increases survival during stress in Streptococcus pneumoniae. Evolution 65(12):3475–3485.http://dx.doi.org/10.1111/j.1558-5646.2011.01402.x [DOI] [PubMed] [Google Scholar]
- Falardeau J, Wise C, Novitsky L, Avis TJ.. 2013. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J Chem Ecol. 39(7):869–878.http://dx.doi.org/10.1007/s10886-013-0319-7 [DOI] [PubMed] [Google Scholar]
- Fan L, et al. 2011. Genome sequence of Bacillus subtilis subsp. spizizenii gtP20b, isolated from the Indian Ocean. J Bacteriol. 193(5):1276–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE, Kolter R.. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J Bacteriol. 183(21):6288–6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godreuil S, Cohan F, Shah H, Tibayrenc M.. 2005. Which species concept for pathogenic bacteria? An E-debate. Infect Genet Evol. 5(4):375–387.http://dx.doi.org/10.1016/j.meegid.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG.. 2002. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 19(12):2226–2238.http://dx.doi.org/10.1093/oxfordjournals.molbev.a004046 [DOI] [PubMed] [Google Scholar]
- Goris J, et al. 2007. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 57(Pt 1):81–91. [DOI] [PubMed] [Google Scholar]
- Graham JP, Istock CA.. 1979. Gene exchange and natural selection cause Bacillus subtilis to evolve in soil culture. Science 204(4393):637–639.http://dx.doi.org/10.1126/science.107592 [DOI] [PubMed] [Google Scholar]
- Hahn J, Tanner AW, Carabetta VJ, Cristea IM, Dubnau D.. 2015. ComGA–RelA interaction and persistence in the Bacillus subtilis K-state. Mol Microbiol 97(3):454–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema BJ, Hahn J, Haynes J, Dubnau D.. 2001. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol 40(1):52–64.http://dx.doi.org/10.1046/j.1365-2958.2001.02363.x [DOI] [PubMed] [Google Scholar]
- Harwood CR. 1992. Bacillus subtilis and its relatives: molecular biological and industrial workhorses. Trends Biotechnol. 10(7):247–256.http://dx.doi.org/10.1016/0167-7799(92)90233-L [DOI] [PubMed] [Google Scholar]
- Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF.. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 96(5):2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, et al. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol. 189(22):8186–8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JS, et al. 2007. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 8(6):R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong HA, Duc LH, Cutting SM.. 2005. The use of bacterial spore formers as probiotics. FEMS Microbiol Rev. 29(4):813–835.http://dx.doi.org/10.1016/j.femsre.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Hug LA, et al. 2016. A new view of the tree of life. Nat Microbiol. 1:16048.. [DOI] [PubMed] [Google Scholar]
- Johnsen PJ, Dubnau D, Levin BR.. 2009. Episodic selection and the maintenance of competence and natural transformation in Bacillus subtilis. Genetics 181(4):1521–1533.http://dx.doi.org/10.1534/genetics.108.099523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C, Martin B, Fichant G, Polard P, Claverys J-P.. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 12(3):181–196. [DOI] [PubMed] [Google Scholar]
- Kaas RS, Friis C, Ussery DW, Aarestrup FM.. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverseEscherichia coli genomes. BMC Genomics 13(1):1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.http://dx.doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettler GC, et al. 2007. Patterns and implications of gene gain and loss in the evolution of prochlorococcus. PLoS Genet. 3(12):e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolstø AB. 1997. Dynamic bacterial genome organization. Mol Microbiol. 24(2):241–248. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM.. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 102(7):2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI.. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36(21):6688–6719.http://dx.doi.org/10.1093/nar/gkn668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh S, O’Reilly M, Nolan N, Devine KM.. 1996. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology 142:2031–2040. [DOI] [PubMed] [Google Scholar]
- Kubo Y, et al. 2011. Phylogenetic analysis of Bacillus subtilis strains applicable to Natto (fermented soybean) production. Appl Environ Microbiol. 77(18):6463–6469.http://dx.doi.org/10.1128/AEM.00448-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Ouzounis CA.. 2003. The balance of driving forces during genome evolution in Prokaryotes. Genome Res. 13(7):1589–1594.http://dx.doi.org/10.1101/gr.1092603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-H, Ochman H.. 2009. The fate of new bacterial genes. FEMS Microbiol Rev 33(1):38–43.http://dx.doi.org/10.1111/j.1574-6976.2008.00140.x [DOI] [PubMed] [Google Scholar]
- Kussell E, Leibler S.. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309(5743):2075–2078.http://dx.doi.org/10.1126/science.1114383 [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Abellan F-X, Möller SB, Karamata D, Mauël C.. 2002. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology (Reading, Engl) 148(3):815–824. [DOI] [PubMed] [Google Scholar]
- Lefébure T, Bitar PDP, Suzuki H, Stanhope MJ.. 2010. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol Evol. 2(0):646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner M, Stingl K, Frey E, Maier B.. 2008. Stochastic switching to competence. Curr Opin Microbiol. 11(6):553–559.http://dx.doi.org/10.1016/j.mib.2008.09.020 [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA.. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3(5):e130.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W-S, Gasparich GE, Kuo C-H.. 2015. Found and lost: the fates of horizontally acquired genes in arthropod-symbiotic spiroplasma. Genome Biol Evol. 7(9):2458–2472.http://dx.doi.org/10.1093/gbe/evv160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R.. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 33(1):152–163.http://dx.doi.org/10.1111/j.1574-6976.2008.00148.x [DOI] [PubMed] [Google Scholar]
- Luo H, Csurös M, Hughes AL, Moran MA.. 2013. Evolution of divergent life history strategies in marine alphaproteobacteria. MBio 4(4):e00373-13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Dubnau D.. 2005. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol 56(3):615–624.http://dx.doi.org/10.1111/j.1365-2958.2005.04592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, et al. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 103(42):15611–15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas A, Jamshidi S, Lagadeuc Y, Eveillard D, Vandenkoornhuyse P.. 2016. Beyond the black queen hypothesis. ISME J. 10(9):2085–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- McNally A, Cheng L, Harris SR, Corander J.. 2013. The evolutionary path to extraintestinal pathogenic, drug-resistant Escherichia coli is marked by drastic reduction in detectable recombination within the core genome. Genome Biol Evol. 5(4):699–710.http://dx.doi.org/10.1093/gbe/evt038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mell JC, Redfield RJ.. 2014. Natural competence and the evolution of DNA uptake specificity. J Bacteriol 196(8):1471–1483. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Panchal S, Roy D.. 2014. Plant innate immunity against human bacterial pathogens. Front Microbiol. 5:411.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Raoult D.. 2011. Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc. 86(2):379–405.http://dx.doi.org/10.1111/j.1469-185X.2010.00151.x [DOI] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 93(7):2873–2878.http://dx.doi.org/10.1073/pnas.93.7.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER.. 2012. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio 3(2):e00036-12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M, Pawlowska TE.. 2016. Defying Muller's Ratchet: ancient heritable endobacteria escape extinction through retention of recombination and genome plasticity. MBio 7(3):e02057-15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA.. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405(6784):299–304.http://dx.doi.org/10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- Priest FG, Goodfellow M, Shute LA, Berkeley RCW.. 1987. Bacillus amyloliquefaciens sp. nov., nom. rev. Int J Syst Bacteriol. 37(1):69–71. [Google Scholar]
- Puigbò P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV.. 2014. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 12:66.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P, Wolf YI, Koonin EV.. 2009. Search for a ‘Tree of Life’ in the thicket of the phylogenetic forest. J Biol. 8(6):59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards VP, et al. 2014. Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol Evol. 6(4):741–753.http://dx.doi.org/10.1093/gbe/evu048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 106(45):19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, et al. 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 239(2):226–235. [DOI] [PubMed] [Google Scholar]
- Rooney AP, Price NPJ, Ehrhardt C, Swezey JL, Bannan JD.. 2009. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int J Syst Evol Microbiol. 59(10):2429–2436. [DOI] [PubMed] [Google Scholar]
- Schallmey M, Singh A, Ward OP.. 2004. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 50(1):1–17.http://dx.doi.org/10.1139/w03-076 [DOI] [PubMed] [Google Scholar]
- Schisler DA, Slininger PJ, Behle RW, Jackson MA.. 2004. Formulation of Bacillus spp. for biological control of plant diseases. Phytopathology 94(11):1267–1271. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069.http://dx.doi.org/10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sela I, Wolf YI, Koonin EV.. 2016. Theory of prokaryotic genome evolution. Proc Natl Acad Sci USA. 113(41):11399–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra CR, Earl AM, Barbosa TM, Kolter R, Henriques AO.. 2014. Sporulation during growth in a gut isolate of Bacillus subtilis. J Bacteriol. 196(23):4184–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank EA, et al. 2011. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci USA. 108(48):19107–19108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, et al. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol 56(3):604–614.http://dx.doi.org/10.1111/j.1365-2958.2005.04488.x [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA.. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 12(1):17–25.http://dx.doi.org/10.1101/gr.176501 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313.http://dx.doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Kunkel B, Kroos L, Losick R.. 1989. Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243(4890):507–512.http://dx.doi.org/10.1126/science.2536191 [DOI] [PubMed] [Google Scholar]
- Swan BK, et al. 2013. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 110(28):11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam NKM, et al. 2006. The intestinal life cycle of Bacillus subtilis and close relatives. J Bacteriol. 188(7):2692–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares Batista M, et al. 2014. Gut adhesive Bacillus subtilis spores as a platform for mucosal delivery of antigens. Infect Immun. 82(4):1414–1423.http://dx.doi.org/10.1128/IAI.01255-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial ‘pan-genome’. Proc Natl Acad Sci USA. 102(39):13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thioulouse J, Chessel D, Dolédec S, Olivier JM.. 1997. ADE-4: a multivariate analysis and graphical display software. Stat Comput. 7(1):75–83. [Google Scholar]
- Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5(1):e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um S, Fraimout A, Sapountzis P, Oh D-C, Poulsen M.. 2013. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci Rep. 3:3250.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gestel J, Vlamakis H, Kolter R.. 2015. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate, Laub, MT, editor. PLoS Biol. 13:e1002141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MW, Marri PR, Ochman H.. 2008. The emergence and fate of horizontally acquired genes in Escherichia coli. 4:e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J-W, Smits WK, Kuipers OP.. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol. 62:193–210.http://dx.doi.org/10.1146/annurev.micro.62.081307.163002 [DOI] [PubMed] [Google Scholar]
- Wiedemann A, Virlogeux-Payant I, Chausse A-M, Schikora A, Velge P.. 2015. Interactions of Salmonella with animals and plants. Front Microbiol. 5:45–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DM, Vazirani VV, Arkin AP.. 2005. Diversity in times of adversity: probabilistic strategies in microbial survival games. J Theor Biol. 234(2):227–253. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Koonin EV.. 2013. Genome reduction as the dominant mode of evolution. BioEssays 35(9):829–837.http://dx.doi.org/10.1002/bies.201300037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Makarova KS, Yutin N, Koonin EV.. 2012. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol Direct 7:46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RAF, Waldor MK.. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 8(8):552–563.http://dx.doi.org/10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Wu I-L, et al. 2015. A versatile nano display platform from bacterial spore coat proteins. Nat Commun. 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CS, Trout AD, Kessler DA, Levine H.. 2010. Optimal strategy for competence differentiation in Bacteria. PLoS Genet. 6(9):e1001108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yüksel M, Power JJ, Ribbe J, Volkmann T, Maier B.. 2016. Fitness trade-offs in competence differentiation of Bacillus subtilis. Front Microbiol. 7:3110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS.. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39(Web Server issue):W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.