Abstract
Female post-mating behaviors are regulated by complex factors involving males, females, and the environment. In insects, plant secondary compounds that males actively forage for, may indirectly modify female behaviors by altering male behavior and physiology. In the tephritid fruit fly, Bactrocera tryoni, females mated with males previously fed on plant-derived phenylpropanoids (=“lures” based on usage in tephritid literature), have longer mating refractoriness, greater fecundity, and reduced longevity than females mated with non-lure fed males. This system thus provides a model for studying transcriptional changes associated with those post-mating behaviors, as the genes regulating the phenotypic changes are likely to be expressed at a greater magnitude than in control females. We performed comparative transcriptome analyses using virgin B. tryoni females, females mated with control males (control-mated), and females mated with lure-fed males (lure-mated). We found 331 differentially expressed genes (DEGs) in control-mated females and 80 additional DEGs in lure-mated females. Although DEGs in control-mated females are mostly immune response genes and chorion proteins, as reported in Drosophila species, DEGs in lure-mated females are titin-like muscle proteins, histones, sperm, and testis expressed proteins which have not been previously reported. While transcripts regulating mating (e.g., lingerer) did not show differential expression in either of the mated female classes, the odorant binding protein Obp56a was down-regulated. The exclusively enriched or suppressed genes in lure-mated females, novel transcripts such as titin and histones, and several taxa-specific transcripts reported here can shed more light on post-mating transcriptional changes, and this can help understand factors possibly regulating female post-mating behaviors.
Keywords: polyandry, sterile insect technique, methyl eugenol, zingerone, Tephritidae, indirect genetic effects
Introduction
Mating can induce profound physiological changes and behavioral switches in females, including changes in oviposition patterns, mating refractoriness, and longevity (Thornhill and Alcock, 1983; Fowler and Partridge, 1989; Miyatake et al. 1999). These changes have a wide range of fitness impacts, and hence evolutionary implications (Andersson 1994; Elgar 1998; Arnqvist and Nilsson 2000; Jennions and Petrie 2000; Mays and Hill 2004): the intensity and tendency of these changes have been linked to sexual selection (Birkhead and Pizzari 2002), kin selection (Hughes et al. 2008), sexual conflict (Parker 1979; Chapman et al. 2003; Parker 2006), and speciation (Martin and Hosken 2003; Ritchie 2007). For example, reduced female remating receptivity is considered to directly benefit the male partner; whereas for females, reception of quality sperm from the first male confers indirect genetic benefits (e.g., in quality offspring) in mate selection scenarios (Arnqvist and Nilsson 2000; Fedorka and Mousseau 2002).
Female post-mating changes can be mediated by a complex of factors including the male, the female herself, and the environment (e.g., resource availability, operational sex ratio) (Thornhill and Alcock 1983; Andersson 1994; Aluja et al. 2009). In insects, plants with which they interact may also play a significant role in mediating those behaviors, additional to their “simple” role as a food source. For example, male euglossine bees utilize secondary plant compounds from orchids for their sex pheromone communication, and the plant compounds thus indirectly modify female behaviors (Landolt and Phillips 1997; Zimmermann et al. 2009; Weber et al. 2016). Similarly, in frugivorous fruit flies of the genus Bactrocera (Diptera: Tephritidae), plant secondary compounds such as methyl eugenol, raspberry ketone, and zingerone modify sexual calling and mediate male mating success (Shelly 2010). These compounds are known commonly as “male lures” or simply “lures” in the tephritid literature, because it is the males which predominantly respond to and feed upon these compounds, and the chemicals are used as lures in monitoring and pest management: this generic terminology is used in this paper hereafter.
Unlike the condition-dependent preferences and sexual selection recorded in many systems (Andersson 1986; Cotton et al. 2006), female preference for lure-fed males in Bactrocera is not due to specific conditions as lures do not provide any direct dietary benefits (Raghu et al. 2002). Further, lures are not an essential component of mating in Bactrocera, but they do very commonly modify mate selection (Raghu 2004; Shelly 2010; Kumaran et al. 2014a). The lure-mediated male mating success is apparent in several Bactrocera species (Shelly and Villalobos 1995; Shelly and Nishida 2004; Wee et al. 2007; Shelly et al. 2010, Obra and Resilva 2013; Haq et al. 2014), although not ubiquitous across all species (Raghu and Clarke 2003; Shelly 2017). Although there are well documented direct, lure-induced male behavioral changes in most species studied, female changes after mating with a lure-fed male are far less studied and the results are contradictory. Shelly (2000) and Shelly and Nishimoto (2016) found no evidence for female post-mating changes in Bactrocera dorsalis and Zeugodacus curcurbitae, respectively, after they had mated with lure-fed males. However, in B. tryoni, mating with a lure-fed male indirectly suppressed female receptivity for remating and induced greater life-time fecundity over a shorter period (Kumaran et al. 2013).
The mechanisms mediating the female post-mating changes in B. tryoni are not known; but they must be indirect through modified male pheromone compounds and/or other lure-modified male physiological traits (Hee and Tan 1998; Kumaran et al. 2014a, 2014b). Lures induce expression of numerous energy metabolic genes and pathways in males, and empirical evidence suggests that lure-fed males become physically fitter than non-lure fed males (Kumaran et al. 2014b). Additionally, several accessory gland protein (Acp) transcripts are differentially expressed in lure-fed B. tryoni compared with non-lure fed males (Kumaran et al. 2014b), and this could possibly mediated the female post-mating phenotypic changes observed. Generally, Acps or sperm that males transfer to females during copulation are regarded as the proximate mechanisms mediating phenotypic changes occurring in females after mating (Chen et al. 1988; Herndon and Wolfner 1995; Wolf et al. 1998; Chapman et al. 2003; Gillott 2003; Wigby et al. 2009; Sitnik et al. 2016). For instance, in knock-down studies using D. melanogaster, it was confirmed that female receptivity for remating is regulated by male produced sex peptide (Acp70a) (Chapman et al. 2003; Liu and Kubli 2003), and in tephritid fruit flies, B. tryoni and Ceratitis capitata, Acps were found to induce sexual inhibition (Radhakrishnan and Taylor 2008) and oviposition (Jang 1995; Scolari et al. 2012).
Transcriptome profiling of females post-mating offers insights to understand the mechanisms regulating female post-mating behaviors (e.g., Lawniczak and Begun 2004; McGraw et al. 2004; Bono et al. 2011). For example, McGraw et al. (2008) identified >2,500 genes and Lawniczak and Begun (2004) found 38 differentially expressed genes (DEGs) in D. melanogaster females post-mating. Among the DEGs were several genes of known function, including Odorant binding protein 99a, immune response genes, yolk proteins, and chorion proteins. Studying transcriptional changes exclusively in the reproductive tracts of Drosophila mojavensis, Bono et al. (2011) found 12 female-origin genes out of 18 DEGs including Obp93A and immune response transcripts. In female C. capitata post-mating, 34 DEGs including Obp19d and chemoreception transcripts were recorded (Gomulski et al. 2012), and in Ostrinia nibulalis 978 DEGs including peptidases, immune response genes and hormone receptors were recorded (Al-Wathiqui et al. 2014). Despite such transcriptome studies, female post-mating changes are still poorly understood because of significant variation in the genes regulated, often low levels of differential expression in those genes which are differentially expressed, and/or the low number of functional genes regulated. The varied results and lack of information on functional genes necessitates that more studies in different systems are needed.
The B. tryoni-lure system, where lures regulate many genes in males (Kumaran et al. 2014b) and strongly promote female post-mating effects (Kumaran et al. 2013), provides a model system to capture deeper and behavior-specific transcriptional changes in females post-mating. Since lures modify refractoriness, fecundity and longevity in B. tryoni, it is likely that genes regulating those phenotypic changes will be expressed at a greater magnitude. To this end, we used RNA-seq analyses to carry out comparative transcriptome analysis of B. tryoni virgin females with females mated with non-lure fed males (=control-mated) and lure-fed males (=lure-mated). Mapping of genetic changes in females post-mating, especially in a pest species, is not only of importance to develop deeper understanding of mechanisms influencing female post-mating behaviors, but may also help with the pest management options, such as the Sterile Insect Technique (Klassen 2005).
Materials and Methods
Mating in B. tryoni
Bactrocera tryoni is a dusk mating species. In combined male and female aggregations, males commence rapid wing movements associated with dispersion of a pheromone at dusk when light intensity drops below a critical level, which elicits female response (Tychsen 1978; Ekanayake et al. 2017). Receptive females are subsequently mounted by males and copulation ensues. Males are mostly polygynous, whereas females were regarded as monandrous until recent empirical evidence confirmed that multiple mating, although not inevitable, is prevalent (Song et al. 2007; Radhakrishnan et al. 2009; Chinajariyawong et al. 2010). Although polyandry and polygyny is prevalent, mating occurs only once a day given the strict mating window.
Insect Source and Experimental Groups
Bactrocera tryoni were obtained as pupae from the rearing facility at the [Queensland Government] Department of Agriculture Fisheries and Forestry, Brisbane. Adults emerging from pupae were provided with water, sugar, and protein hydrolysate ad libitum and maintained at 27 °C and 70% Relative Humidity in a room illuminated with natural light, in addition to fluorescent lighting between 0700 and 1,600 h every day.
Flies were sexed within 2–3 days of emergence, when still sexually immature, and the sexes were then housed separately in Perspex cages (30 × 30 × 30 cm). Three groups of females were maintained: mature virgin females (=“virgin females” hereafter), females mated with control males (=“control-mated females” hereafter) and females mated with males previously fed on the plant-derived secondary compound zingerone [4-(3-methoxy-4-hydroxyphenyl)-butan-2-one] (=“lure-mated females” hereafter). To obtain zingerone-fed males, flies (14 days old) were provided with 1.5 mL of zingerone (10 µg/µl of 95% ethanol, Sigma-Aldrich, CHEME, GmbH, Germany, >96% purity) on a cotton wick placed on an inverted petri dish for 2 h from 0800 to 1000 h on the day of mating. Our previous observations showed that males process zingerone within 3 h after feeding (Kumaran et al. 2014a). The concentrations, dilutions and presentation methods are based on previous studies on other Bactrocera flies (Shelly and Villalobos 1995; Hee and Tan 1998; Kumaran et al. 2013). To obtain mated females 50 females (14 days old) were housed in Perspex cages (30 × 30 × 30 cm) and either 50 control or lure-fed males were released at 1500 h, approximately 2 h before mating commences. Once mating commenced between 1700 and 1730 h, the mating pairs were transferred to new cages while ensuring that the flies remain paired. For virgin females, 50 females without males were maintained. Two cages per female group were maintained.
RNA Isolation, Library Preparation, Assembly, and Annotation
From each of three groups (virgin females, control-mated females, and lure-mated females), totally 40 females were collected approximately 10–12 h after mating and snap-frozen in liquid nitrogen. Females were mated only once given the strict mating window. Although females in the cages were not monitored individually, our previous studies showed that lures do not modify copula duration (Kumaran et al. 2013). Total RNA from whole body was extracted using TRIzol and purified with a Qiagen RNeasy kit following the manufacturer’s instructions. Two replicates, each with RNA from 20 females pooled together, were maintained for each of the female groups. The quality of RNA was tested on 1.5% agarose gel as well as on a Agilent Bioanalyzer 2100, and only samples that had a RNA integrity number of >8 were used. Detailed methodology on RNA isolation, library construction, assembly and annotation are presented in our previous studies (Arthofer et al. 2014; Kumaran et al. 2014b; van der Burg et al. 2016; Stewart et al. 2017). Low quality leading and trailing bases (<Q30) were trimmed and adapter sequences removed from each sequence read using Trimmomatic (Bolger et al. 2014). The final transcriptome was assess for completeness using BUSCO (Simão et al. 2015). The raw sequence files are deposited in the Sequence Read Archive accessions SRR5927915 (virgin 1), SRR5927916 (virgin 2), SRR5927913 (normal-mated 1), SRR5927914 (normal-mated 2), SRR5927911 (lure-mated 1), and SRR5927912 (lure-mated 2) in NCBI.
Analysis of Differentially Expressed Genes
To determine differentially expressed genes (DEGs), sequencing reads were mapped to contigs using bowtie2 and Fragments Per kb per Million fragments (FPKM) values were used to determine expression levels and differential gene expression for all the comparisons viz., virgin versus control-mated, virgin versus lure-mated, and control-mated versus lure-mated using the trinity platform (Haas et al. 2013) using version 3.4.0 of the BioConductor package edgeR (Robinson et al. 2010). Transcripts with an FPKM value of zero were excluded from downstream analysis. A False discovery rate (FDR) of ≤0.001 and log two fold change of ≥2 was used to determine statistically significant differential expression in virgin versus control-mated versus lure-mated females. We undertook gene set enrichment analysis to determine whether particular Gene Ontology (GO) categories were overrepresented in up- and down-regulated DEGs using GOSeq (Young et al. 2010).
Results
Sequence Assembly Statistics
The total number of reads generated for all libraries was 490,218, 344. The number of reads generated per library was as follows: lure-mated 1 = 74,256, 386; lure-mated 2 = 53,599,704; control-mated 1 = 59,549,082; control-mated 2 = 90,701,413; virgin 1 = 66,654,335; virgin 2 = 65,411,462. The raw reads are deposited in the SRA accessions SRR5927912-SRR5927916 under BioProject PRJNA397485 and BioSample SAMN07459442 in NCBI. The total number of contigs generated in the combined assembly (before reducing redundancy and bias with CD-HIT) was 108,313. After CD-HIT, 67,492 contigs (transcripts) remained with a total of 58,291 trinity genes. The average contig length was 765.3 bp, the contig N50 was 1,420 and the percentage GC content was 38.8%. Our transcriptome was largely complete with >95% of the 1,658 core insect genes present as full length copies in the assembly (BUSCO benchmarking = C: 95.5%[S: 63.1%, D: 32.4%], F: 2.3%, M: 2.2%, n: 1,658).
DEGs in Control-Mated Females Versus Virgin Females
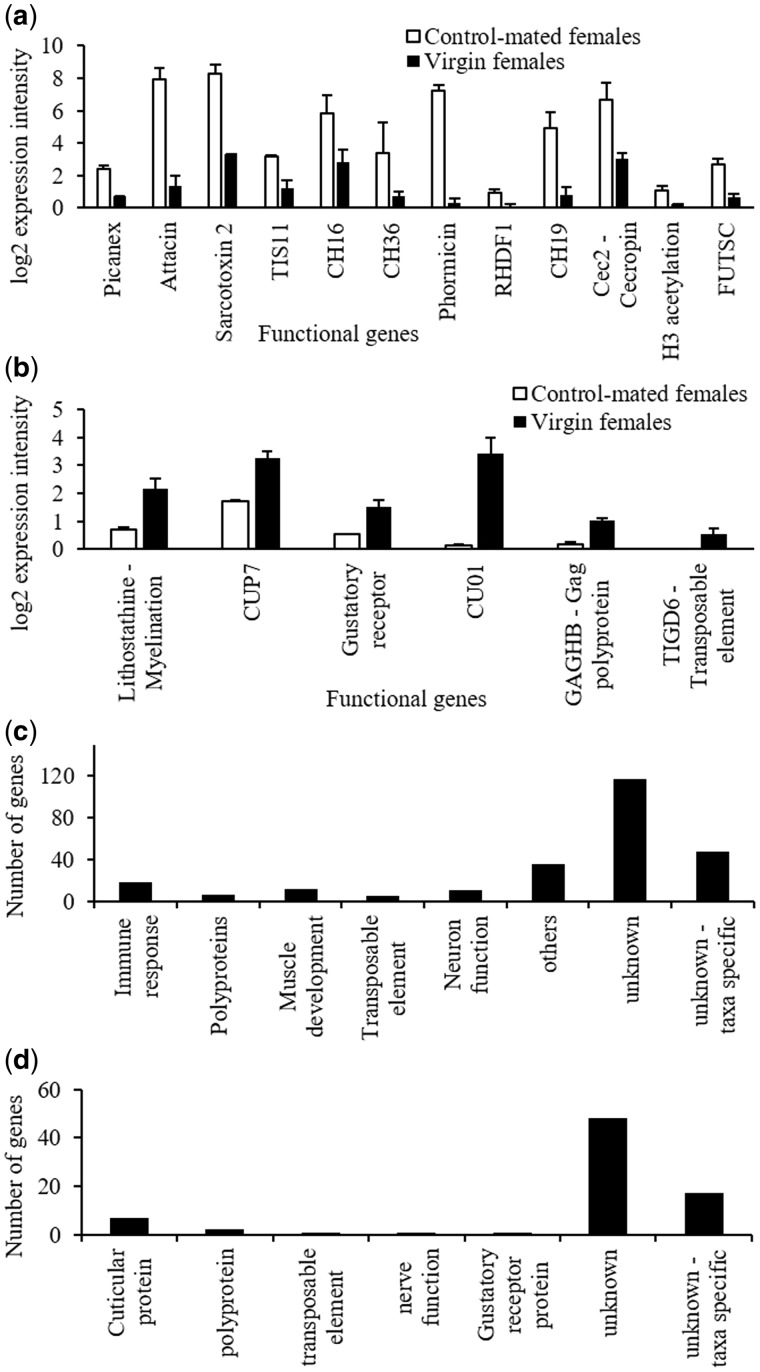
Three-hundred and thirty-one genes were differentially expressed (DEGs) (254 up-regulated and 77 down-regulated) in control-mated females when compared with virgin females (supplementary tables S1 and S2, Supplementary Material online). Among the functional transcripts up-regulated, immune response transcripts were highly enriched followed by genes encoding chorion proteins, transposable elements, titin-like muscle proteins and histone proteins (fig. 1a). The most significantly down-regulated genes in mated females encoded cuticle proteins, gustatory receptor transcript, polyprotein and a transposable element (fig. 1b). The majority of DEGs (165 out of 254 up-regulated and 60 out of 77 down-regulated) had unknown functions; and approximately 30% of the up-regulated and 25% of the down-regulated genes are known only from tephritid fruit flies (B. dorsalis and/or C. capitata) (fig. 1c and d). The difference in expression intensity in DEGs was up to 11 fold for up- and down-regulated transcripts (supplementary fig. S1, Supplementary Material online).
Fig. 1.
—Differential expression in control-mated females compared with virgin females. (a) Example up-regulated transcripts and their expression levels; (b) Example down-regulated functional transcripts and their expression levels; (c) Up-regulated genes within functional classes differentially expressed; and (d) Down-regulated genes within functional classes differentially expressed.
Gene Ontology (GO) analysis revealed that up-regulated DEGs were represented in 194 biological processes (supplementary table S3, Supplementary Material online), 33 molecular function (supplementary table S4, Supplementary Material online) and 51 cellular component GO terms (supplementary table S5, Supplementary Material online). Most significantly enriched GO terms corresponded with DEGs and comprised mostly of defence (immune) response transcripts and muscle functions (table 1). The suppressed DEGs were represented in 98 GO terms in control-mated females when compared with virgin females (supplementary table S6, Supplementary Material online). Most significantly suppressed GO terms included transcripts encoding metabolic processes, biosynthetic processes, transporter activities, and catalytic activities (table 2).
Table 1.
Most Significant Up-Regulated GO Terms in Control-Mated Females Compared with Virgin Females (full list appended as supplementary file, Supplementary Material online)
| GO Id. | GO Term | Total Numbers in GO Term | Numbers Differentially Expressed | P Value |
|---|---|---|---|---|
| Biological Processes | ||||
| GO: 0007062 | Sister chromatid cohesion | 35 | 10 | 0 |
| GO: 0007076 | Mitotic chromosome condensation | 37 | 10 | 0 |
| GO: 0007522 | Visceral muscle development | 19 | 10 | 0 |
| GO: 0016203 | Muscle attachment | 37 | 10 | 0 |
| GO: 0042742 | Defense response to bacterium | 184 | 19 | 2.97E-17 |
| GO: 0009617 | Response to bacterium | 200 | 19 | 1.26E-16 |
| GO: 0045087 | Innate immune response | 257 | 21 | 7.23E-16 |
| GO: 0045214 | Sarcomere organization | 60 | 14 | 5.93E-15 |
| GO: 0098542 | Defense response to other organism | 241 | 19 | 1.08E-14 |
| GO: 0051707 | Response to other organism | 342 | 21 | 3.47E-14 |
| Molecular function | ||||
| GO: 0008307 | Structural constituent of muscle | 40 | 12 | 1.69E-12 |
| GO: 0003779 | Actin binding | 195 | 15 | 1.63E-09 |
| GO: 0008092 | Cytoskeletal protein binding | 395 | 17 | 1.6E-07 |
| GO: 0005198 | Structural molecule activity | 520 | 17 | 2.59E-07 |
| GO: 0016709 | Oxidoreductase activity | 11 | 3 | 0.000172 |
| GO: 0001872 | (1→3)-beta-D-glucan binding | 5 | 2 | 0.000343 |
| GO: 0043914 | NADPH: sulfur oxidoreductase activity | 3 | 2 | 0.001285 |
| GO: 0004521 | Endoribonuclease activity | 137 | 5 | 0.001533 |
| GO: 0004540 | Ribonuclease activity | 156 | 5 | 0.002875 |
| GO: 0004523 | RNA-DNA hybrid ribonuclease activity | 105 | 4 | 0.003011 |
| Cellular Processes | ||||
| GO: 0000794 | Condensed nuclear chromosome | 31 | 10 | 0 |
| GO: 0005859 | Muscle myosin complex | 23 | 11 | 0 |
| GO: 0005863 | Striated muscle myosin thick filament | 17 | 10 | 0 |
| GO: 0016460 | Myosin II complex | 28 | 11 | 0 |
| GO: 0031674 | I band | 23 | 11 | 0 |
| GO: 0032982 | Myosin filament | 30 | 11 | 0 |
| GO: 0036379 | Myofilament | 26 | 11 | 0 |
| GO: 0005576 | Extracellular region | 790 | 29 | 1.67E-15 |
| GO: 0030017 | Sarcomere | 40 | 13 | 1.93E-14 |
| GO: 0044449 | Contractile fiber part | 122 | 17 | 6.96E-12 |
Table 2.
Most Significant Down-Regulated GO Terms in Control-Mated Females Compared with Virgin Females (full list appended as supplementary file, Supplementary Material online)
| GO Id. | GO Term | Total Numbers in GO Term | Numbers Differentially Expressed | P Value |
|---|---|---|---|---|
| Biological Processes | ||||
| GO: 0051246 | Regulation of protein metabolic process | 807 | 0 | 0.000975 |
| GO: 0031323 | Regulation of cellular metabolic process | 2,804 | 9 | 0.00132 |
| GO: 0032268 | Regulation of cellular protein metabolic process | 756 | 0 | 0.001448 |
| GO: 0044248 | Cellular catabolic process | 870 | 0 | 0.002026 |
| GO: 0044711 | Single-organism biosynthetic process | 903 | 0 | 0.002672 |
| GO: 0080090 | Regulation of primary metabolic process | 2,660 | 9 | 0.002726 |
| GO: 0009056 | Catabolic process | 1,118 | 1 | 0.003689 |
| GO: 0019538 | Protein metabolic process | 2,370 | 8 | 0.004911 |
| GO: 0033036 | Macromolecule localization | 901 | 1 | 0.005483 |
| GO: 0006811 | Ion transport | 784 | 0 | 0.006089 |
| Molecular Function | ||||
| GO: 0003674 | Molecular function | 11,752 | 62 | 7.42E-07 |
| GO: 0022892 | Substrate-specific transporter activity | 883 | 0 | 0.002882 |
| GO: 0005215 | Transporter activity | 1,072 | 1 | 0.005253 |
| GO: 0022891 | Transmembrane transporter activity | 791 | 0 | 0.006853 |
| GO: 0015075 | Ion transmembrane transporter activity | 696 | 0 | 0.012545 |
| GO: 0003824 | Catalytic activity | 5,755 | 28 | 0.015134 |
| GO: 0022857 | Transmembrane transporter activity | 915 | 1 | 0.020015 |
| GO: 0043167 | Ion binding | 4,881 | 25 | 0.021984 |
| GO: 0005509 | Calcium ion binding | 444 | 0 | 0.026681 |
| GO: 0005488 | Binding | 8,977 | 54 | 0.037584 |
| Cellular Component | ||||
| GO: 0016020 | Membrane | 3,753 | 12 | 0.000259 |
| GO: 0043227 | Membrane-bounded organelle | 5,567 | 23 | 0.000416 |
| GO: 0044464 | Cell part | 10,245 | 57 | 0.00056 |
| GO: 0043231 | Intracellular membrane-bounded organelle | 5,261 | 23 | 0.002085 |
| GO: 0005886 | Plasma membrane | 1,655 | 3 | 0.002407 |
| GO: 0031982 | Vesicle | 992 | 1 | 0.003442 |
| GO: 0005737 | Cytoplasm | 2,727 | 10 | 0.004576 |
| GO: 0031988 | Membrane-bounded vesicle | 933 | 1 | 0.005007 |
| GO: 0005634 | Nucleus | 3,432 | 13 | 0.005385 |
| GO: 0005739 | Mitochondrion | 763 | 0 | 0.006402 |
DEGs in Lure-Mated Females Versus Virgin Females
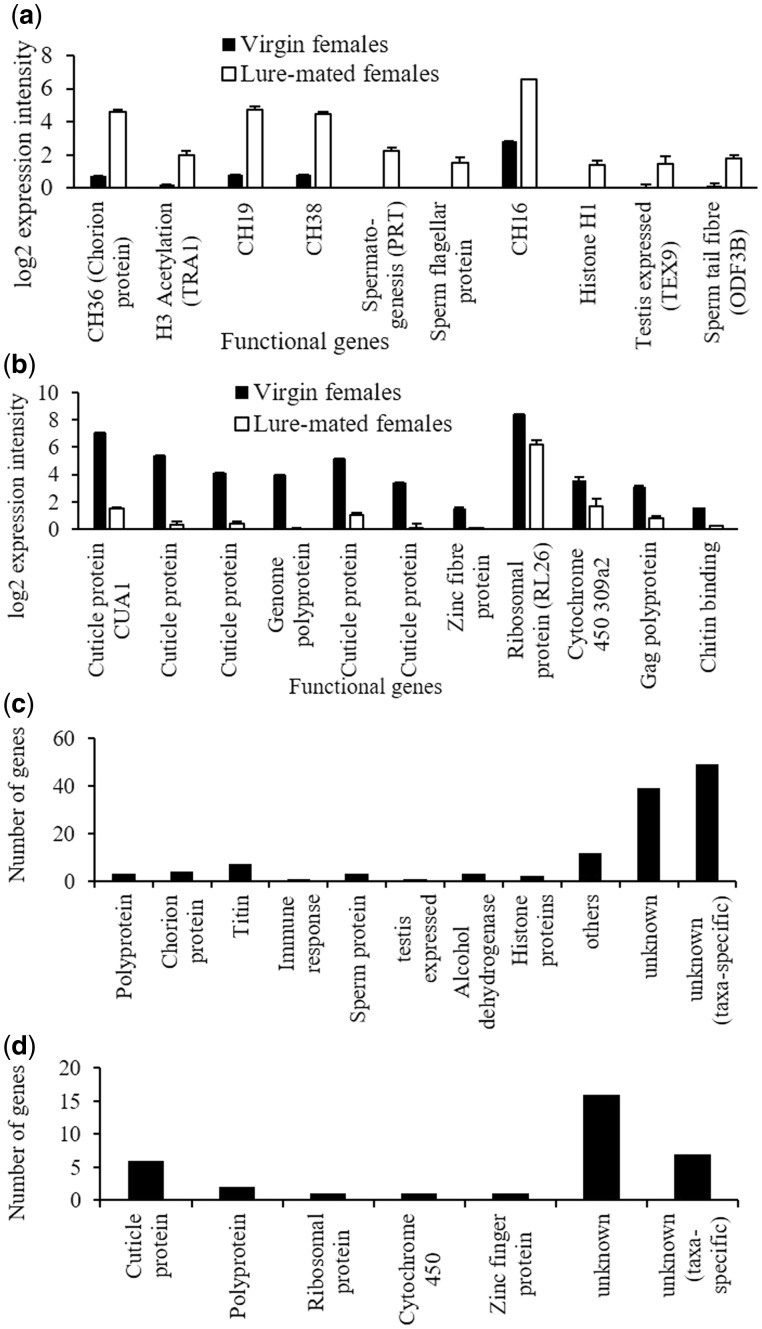
We found 124 genes up-regulated and 34 genes down-regulated in lure-mated females when compared with virgin females (supplementary tables S7 and S8, Supplementary Material online), with several fold difference in expression intensities (supplementary fig. S2, Supplementary Material online). The DEGs were dissimilar to that of control-mated females, with 67 new up-regulated transcripts identified. In addition to genes encoding chorion and titin proteins, DEGs in lure-mated females comprised testis expressed proteins, sperm proteins, and a different set of histone proteins (fig. 2a). Further, fewer immune response genes were over-expressed in lure-mated females than in control-mated females: of the 124 up-regulated genes, only two genes were immune related compared with 16 (out of 254) in control-mated females. Additionally, 13 genes were switched off only in lure-mated females: these were not differentially expressed in control-mated females. The down-regulated transcripts that encoded proteins with known functions were cuticle proteins, polyproteins, ribosomal protein (RL26), and cytochrome 450 309a2 (fig. 2b). Several taxa-specific transcripts with unknown functions were also differentially expressed in lure-mated females (fig. 2c and d).
Fig. 2.
—Differential expression in lure-mated females compared with virgin females. (a) Example up-regulated transcripts and their expression levels; (b) Example down-regulated functional transcripts and their expression levels; (c) Up-regulated genes within functional classes differentially expressed; and (d) Down-regulated genes within functional classes differentially expressed.
One-hundred and seventy-nine GO terms in the biological processes category were significantly enriched (P < 0.05) with muscle development, DNA packaging, and chromosome condensation being the most significantly enriched terms (table 3 and supplementary table S9, Supplementary Material online). In the cellular component category, 38 GO terms showed significant enrichment and the most significantly enriched terms were muscle filament, sarcomere, myosin filament, and condensed chromosome (table 3 and supplementary table S10, Supplementary Material online). There were 28 GO terms enriched in the molecular function category, which included muscle structural constituents, actin binding, and enzymatic activities (table 3 and supplementary table S11, Supplementary Material online). Forty GO terms were down-regulated in lure-mated females with regulation of metabolic processes, signal transduction, and negative regulation of biological processes being the most significantly down-regulated GO terms (table 4 and supplementary table S12, Supplementary Material online).
Table 3.
Most Significantly Up-Regulated GO Terms in Lure-Mated Females Compared with Virgin Females
| GO Id. | GO Term | Total Numbers in GO Term | Numbers Differentially Expressed | P Value |
|---|---|---|---|---|
| Biological processes | ||||
| GO: 0007522 | Visceral muscle development | 19 | 8 | 2.38E-12 |
| GO: 0030261 | Chromosome condensation | 54 | 9 | 2.06E-11 |
| GO: 0006323 | DNA packaging | 55 | 9 | 2.19E-11 |
| GO: 0016203 | Muscle attachment | 37 | 8 | 1.26E-10 |
| GO: 0007062 | Sister chromatid cohesion | 35 | 8 | 1.34E-10 |
| GO: 0007076 | Mitotic chromosome condensation | 37 | 8 | 1.58E-10 |
| GO: 0007520 | Myoblast fusion | 56 | 8 | 7.51E-10 |
| GO: 0007519 | Skeletal muscle tissue development | 54 | 8 | 8.58E-10 |
| GO: 0035206 | Regulation of hemocyte proliferation | 54 | 8 | 1.35E-09 |
| GO: 0000768 | Syncytium formation by plasma membrane fusion | 65 | 8 | 1.56E-09 |
| Cellular Component | ||||
| GO: 0005863 | Striated muscle myosin thick filament | 17 | 8 | 0 |
| GO: 0030017 | Sarcomere | 40 | 10 | 0 |
| GO: 0005859 | Muscle myosin complex | 23 | 8 | 4.85E-11 |
| GO: 0031674 | I band | 23 | 8 | 5.07E-11 |
| GO: 0032982 | Myosin filament | 30 | 8 | 5.75E-11 |
| GO: 0036379 | Myofilament | 26 | 8 | 7.57E-11 |
| GO: 0000794 | Condensed nuclear chromosome | 31 | 8 | 1.47E-10 |
| GO: 0016460 | Myosin II complex | 28 | 8 | 2.15E-10 |
| GO: 0000793 | Condensed chromosome | 39 | 8 | 4.00E-10 |
| GO: 0044449 | Contractile fiber part | 122 | 12 | 3.58E-09 |
| Molecular Function | ||||
| GO: 0008307 | Structural constituent of muscle | 40 | 8 | 1.57E-08 |
| GO: 0003779 | Actin binding | 195 | 10 | 1.39E-07 |
| GO: 0005198 | Structural molecule activity | 520 | 10 | 4.41E-06 |
| GO: 0016763 | Transferase activity, transferring pentosyl groups | 27 | 3 | 1.73E-05 |
| GO: 0008092 | Cytoskeletal protein binding | 395 | 10 | 1.91E-05 |
| GO: 0003796 | Lysozyme activity | 12 | 2 | 0.000157 |
| GO: 0004568 | Chitinase activity | 29 | 2 | 0.001118 |
| GO: 0017061 | S-methyl-5-thioadenosine phosphorylase activity | 3 | 1 | 0.004963 |
| GO: 0002060 | Purine nucleobase binding | 3 | 1 | 0.005099 |
| GO: 0004731 | Purine-nucleoside phosphorylase activity | 3 | 1 | 0.005099 |
Table 4.
Most Significant GO Terms Down-Regulated in Lure-Mated Females Compared with Virgin Females
| GO Id. | GO Term | Total Numbers in GO Term | Numbers Differentially Expressed | P Value |
|---|---|---|---|---|
| Biological Processes | ||||
| GO: 0031323 | Regulation of cellular metabolic process | 2,804 | 1 | 0.000548 |
| GO: 0060255 | Regulation of macromolecule metabolic process | 2,773 | 1 | 0.000557 |
| GO: 0080090 | Regulation of primary metabolic process | 2,660 | 1 | 0.00085 |
| GO: 0007165 | Signal transduction | 1,883 | 0 | 0.00187 |
| GO: 0048519 | Negative regulation of biological process | 2,116 | 1 | 0.002111 |
| GO: 0019222 | Regulation of metabolic process | 3,216 | 3 | 0.002131 |
| GO: 0048523 | Negative regulation of cellular process | 1,911 | 1 | 0.004281 |
| GO: 0010468 | Regulation of gene expression | 2,204 | 1 | 0.006921 |
| GO: 0009653 | Anatomical structure morphogenesis | 1,407 | 0 | 0.008501 |
| GO: 0051171 | Regulation of nitrogen compound metabolic process | 2,127 | 1 | 0.008567 |
| Cellular Component | ||||
| GO: 0016020 | Membrane | 3,753 | 4 | 0.006526 |
| GO: 0043227 | Membrane-bounded organelle | 5,567 | 8 | 0.006829 |
| GO: 0043231 | Intracellular membrane-bounded organelle | 5,261 | 8 | 0.016174 |
| GO: 0044425 | Membrane part | 3,809 | 5 | 0.038189 |
| GO: 0005886 | Plasma membrane | 1,655 | 1 | 0.048329 |
| Molecular Function | ||||
| GO: 0043169 | Cation binding | 3,480 | 3 | 0.004204 |
| GO: 0046872 | Metal ion binding | 3,420 | 3 | 0.004734 |
| GO: 0043167 | Ion binding | 4,881 | 8 | 0.019285 |
DEGs in Lure-Mated Females Versus Control-Mated Females
When lure-mated females were compared with control-mated females, 89 DEGs were detected: 70 up-regulated and 19 down-regulated (supplementary tables S13 and S14, Supplementary Material online). The expression intensity was low with up to 7-fold difference (supplementary figs. S3 and S4, Supplementary Material online), compared with 11-fold difference recorded in control-mated versus virgin or lure-mated versus virgin categories. Testis expressed, sperm, farnesol dehydrogenase, reverse transcriptase, and histone 1 to histone 5 linker genes were up-regulated (supplementary fig. S5a, Supplementary Material online); odorant binding (Obp56A), occluding homology, binding, nuclear transport, and transposable elements were the down-regulated transcripts (supplementary fig. S5b, Supplementary Material online). A number of differentially expressed transcripts with unknown function were detected and most of these were taxa-specific, having no significant BLAST hit to any other species in the current databases (supplementary fig. S5c and d, Supplementary Material online).
Most of the DEGs in lure-mated females (e.g., sperm proteins, testis expressed proteins) were only differentially expressed (67 out of 70 up-regulated and 13 out of 19 down-regulated) in lure-mated females; those genes were not differentially expressed in control-mated females when compared with virgin females. There were 54 unknown function transcripts up-regulated only in lure-mated females, and 31 of them were specific to tephritids (supplementary fig. S4, Supplementary Material online). The genes suppressed only in lure-mated females included Obp56A, occudin homology domain, carboxylate reductase, and binding nuclear protein, with nine unknown function transcripts (supplementary fig. S4, Supplementary Material online). The remaining six down-regulated genes and the three up-regulated genes showed reversal in expression, that is, those six genes were up-regulated and the three genes down-regulated, respectively, in control-mated females (fig. 3).
Fig. 3.
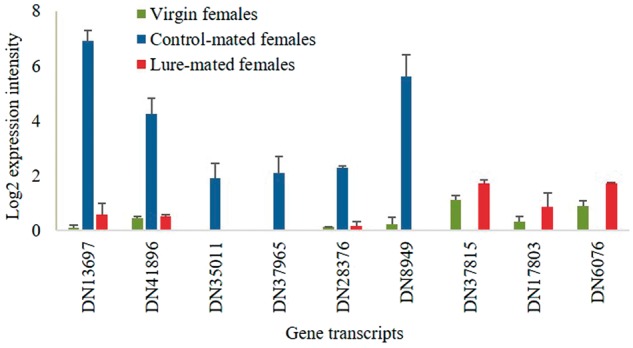
—The functional transcripts showed reversal in expression pattern in lure-mated females. There were six genes enriched after control-mating but suppressed in lure-mated females, and there were three genes suppressed after control-mating but enriched in lure-mated females.
One-hundred and twenty-six GO terms were enriched within biological processes category with metabolic processes, biosynthetic processes, and cellular metabolic compound salvage being the most significantly enriched terms (table 5 and supplementary table S15, Supplementary Material online). Within cellular component category, six GO terms showed significant enrichment with the most significant being nucleosome, DNA bending, and DNA packing complex (table 5). There were 37 GO terms enriched within molecular function category with transferase activity, transferring pentosyl groups, and enzymatic activities most significantly enriched among other GO terms (table 5 and supplementary table S16, Supplementary Material online). Thirteen GO terms with nine biological processes, three cellular component, and one molecular function category were significantly down-regulated in lure-mated females (table 6).
Table 5.
Most Significant Up-Regulated GO Terms in Lure-Mated Females Compared with Control-Mated Females (full list appended as supplementary file, Supplementary Material online)
| GO Id. | GO Term | Total Numbers in GO Term | Numbers Differentially Expressed | P Value |
|---|---|---|---|---|
| Biological Processes | ||||
| GO: 0043101 | Purine-containing compound salvage | 15 | 2 | 0.00013 |
| GO: 0046128 | Purine ribonucleoside metabolic process | 120 | 3 | 0.000338 |
| GO: 0042278 | Purine nucleoside metabolic process | 126 | 3 | 0.000389 |
| GO: 0072522 | Purine-containing compound biosynthetic process | 128 | 3 | 0.000416 |
| GO: 0009119 | Ribonucleoside metabolic process | 142 | 3 | 0.000558 |
| GO: 0009116 | Nucleoside metabolic process | 156 | 3 | 0.00073 |
| GO: 0043094 | Cellular metabolic compound salvage | 38 | 2 | 0.000847 |
| GO: 1901657 | Glycosyl compound metabolic process | 170 | 3 | 0.000932 |
| GO: 0019523 | L-idonate metabolic process | 1 | 1 | 0.001442 |
| GO: 0046176 | Aldonic acid catabolic process | 1 | 1 | 0.001442 |
| Molecular Function | ||||
| GO: 0016763 | Transferase activity, transferring pentosyl groups | 27 | 2 | 0.000447 |
| GO: 0017061 | S-methyl-5-thioadenosine phosphorylase activity | 3 | 1 | 0.00338 |
| GO: 0002060 | Purine nucleobase binding | 3 | 1 | 0.003383 |
| GO: 0004731 | Purine-nucleoside phosphorylase activity | 3 | 1 | 0.003383 |
| GO: 0042301 | Phosphate ion binding | 4 | 1 | 0.004507 |
| GO: 0002054 | Nucleobase binding | 4 | 1 | 0.004648 |
| GO: 0008422 | Beta-glucosidase activity | 9 | 1 | 0.010515 |
| GO: 0004553 | Hydrolase activity, hydrolyzing O-glycosyl compounds | 147 | 2 | 0.012497 |
| GO: 0003796 | Lysozyme activity | 12 | 1 | 0.013456 |
| GO: 0016798 | Hydrolase activity, acting on glycosyl bonds | 162 | 2 | 0.015102 |
| Cellular Component | ||||
| GO: 0000786 | Nucleosome | 17 | 2 | 0.000192 |
| GO: 1990104 | DNA bending complex | 17 | 2 | 0.000192 |
| GO: 0044815 | DNA packaging complex | 22 | 2 | 0.000352 |
| GO: 0032993 | Protein-DNA complex | 33 | 2 | 0.0007 |
| GO: 0045261 | Proton-transporting ATP synthase complex, catalytic core F(1) | 12 | 1 | 0.01361 |
| GO: 0033178 | Proton-transporting two-sector ATPase complex, catalytic domain | 33 | 1 | 0.037039 |
Table 6.
Most Significant Down-Regulated GO Terms in Lure-Mated Females Compared with Control-Mated Females (full list appended as supplementary file, Supplementary Material online)
| GO Id. | GO Term | Total Numbers in GO Term | Numbers Differentially Expressed | P Value |
|---|---|---|---|---|
| GO: 0050789 | Regulation of biological process (BP) | 5,387 | 1 | 0.00232 |
| GO: 0050794 | Regulation of cellular process (BP) | 5,044 | 1 | 0.00422 |
| GO: 0065007 | Biological regulation (BP) | 5,805 | 2 | 0.007141 |
| GO: 0005515 | Protein binding (MF) | 3,223 | 0 | 0.009313 |
| GO: 0019222 | Regulation of metabolic process (BP) | 3,216 | 0 | 0.010203 |
| GO: 0044464 | Cell part (CC) | 10,245 | 8 | 0.014812 |
| GO: 0031323 | Regulation of cellular metabolic process (BP) | 2,804 | 0 | 0.020083 |
| GO: 0060255 | Regulation of macromolecule metabolic process (BP) | 2,773 | 0 | 0.020886 |
| GO: 0080090 | Regulation of primary metabolic process (BP) | 2,660 | 0 | 0.025073 |
| GO: 0032502 | Developmental process (BP) | 3,787 | 1 | 0.030784 |
| GO: 0005575 | Cellular_component (CC) | 11,673 | 11 | 0.036444 |
| GO: 0044424 | Intracellular part (CC) | 8,928 | 7 | 0.041381 |
| GO: 0044767 | Single-organism developmental process (BP) | 3,523 | 1 | 0.0436 |
Across all three conditions (virgin, control-mated, and lure-mated), 434 genes were differentially expressed (supplementary fig. S6, Supplementary Material online); among, 80,214 and 83 genes were uniquely up-regulated in virgin, control-mated and lure-mated females respectively. There were 69 DEGs found in both lure-mated and control-mated females with 47 up-regulated and 20 down-regulated transcripts. Up-regulated functional genes included titin like muscle proteins, chorion proteins, Nesprin1, replicase polyprotein, and H3 acetylation. Down-regulated genes included adult cuticle protein, genome polyprotein, zinc finger protein, and chitin binding protein.
Discussion
Summary
Our study has revealed 331 DEGs in control-mated females when compared with virgin females. Up-regulated genes were mostly transcripts governing immune response functions, as previously documented in D. melanogaster and D. mojavensis (Lawniczak and Begun 2004; McGraw et al. 2004; Bono et al. 2011). Within tephritids, post-mating up-regulation of several immune genes (cecropin, sapecin, attacin, defensin, and diptericin) was reported in B. dorsalis (Wei et al. 2016; Zheng et al. 2016), but none were reported for C. capitata (Gomulski et al. 2012). It is possible that the female immune system has responded to the reception of Acps, sperm, and associated contaminants from males. Genes encoding chorion proteins were the next dominant transcripts up-regulated in mated females, and this result contradicts McGraw et al. (2008) who found suppression of CH36 and CH38 in D. melanogaster.
Among the DEGs in lure-mated females, 50% (67 up-regulated and 13 down-regulated) were unique transcripts differentially expressed only in lure-mated females, and these were not differentially expressed when control-mated females were compared with virgin females. This suggests that mating with zingerone-fed males is indirectly modulating the expression pattern of a set of genes not observed in previous studies. Some of the unique transcripts in lure-mated females encode histone proteins, testis expressed proteins and sperm proteins, with very few immune response genes and no immune response GO terms. Such an observation may indicate that mating with lure-fed males does not elicit a strong immune response in females and may help to explain female preference for lure-fed males as “immunity” can be under sexual selection (Lawniczak et al. 2007). While it is unknown if lures reduced contamination in male Acps or ejaculates, lure-fed males are physically fitter than unfed males (Kumaran et al. 2014b), and it is yet to be investigated whether lures help male B. tryoni to burn unwanted fats and associated contaminations. With respect to sperm and testis expressed genes, expression is possibly from the sperm stored in the spermatheca as B. tryoni females store sperm for at least 15 days after mating (Perez‐Staples et al. 2007) and this observation perhaps explains the reduced refractoriness observed in lure-mated females (Kumaran et al. 2013). However, the reason for the expression of male-biased testis expressed genes in mated females remains unclear.
We a priori expected DEGs detected in both control-mated and lure-mated females (78 out of 158 in lure-mated females) to differ in their expression intensities (i.e., with higher expression in lure-mated females), which we hoped would shed light on the female specific genes mediating fecundity and refractoriness. However, no difference in expression intensity was observed except for nine genes which, paradoxically, showed reversal in expression (fig. 3). Overall, our results indicate the possibility of the previously unobserved, but differentially expressed genes to be involved in mediating fecundity and refractoriness. It is highly likely that the DEGs found in the mated females were regulated exclusively by mating; however, the possibility that the lure-fed males could have modified the close-range courtship behaviors (e.g., harassment and intrasex competition) and underlying transcription factors in females (during precopulatory interactions) cannot be excluded.
The number of DEGs detected following mating (331 from control-mated females plus 80 new DEGs from lure-mated females) were substantially greater than the 32 transcripts reported in C. capitata (Gomulski et al. 2012) and the 83 in B. dorsalis (Zheng et al. 2016), but only approximately one-fifth of the number reported in Drosophila (McGraw et al. 2004). Although the magnitude of DEGs we detected was greater (up to an 11-fold expression difference) compared with a 2-fold differences reported in D. melanogaster and C. capitata, these differences can be attributed to analytical methodology used (RNA-seq vs. microarray) rather than a difference among species. In addition, although our replicates are pooled samples of 20 females, it is possible that two replicates still could have underestimated the number of DEGs in B. tryoni (Schurch et al. 2016). Despite the methodological differences, the additional unique transcripts recorded in B. tryoni can help understand the female factors possibly mediating the post-mating changes. In addition to the already reported immune genes, sperm proteins, and testis expressed genes, our study has revealed expression of several titin like muscle proteins and histone proteins and we elaborate on these below.
Transcript Homologues Regulating Mating and/or Remating
Mating response homologues takeout, Ca(2+)/calmodulin-responsive adenylate cyclase, protein yellow, ejaculatory bulb specific protein, lingerer, sarah, fruitless, and sex peptide receptor did not show differential expression in either control-mated or lure-mated females. We expected these genes to be suppressed in lure-mated females at a greater magnitude compared with control-mated females, since we found greater refractoriness in lure-mated females (Kumaran et al. 2013). It is possible that these genes are only transcribed during the narrow temporal window of dusk, when copulation occurs in B. tryoni (Ekanayake et al. 2017). The other possible explanation is that there are sex specific differences in the expression of these genes, with greater representation of mating transcripts in males compared with females: most of the mating related genes listed above were over-represented in male B. tryoni (Kumaran et al. 2014b). These mating response homologues were also not reported as DEGs in female Drosophila and other species which supports the sex specific expression hypothesis (McGraw et al. 2008; Gomulski et al. 2012; Zheng et al. 2016).
Odorant binding protein Obp56a was down-regulated in mated B. tryoni females, as noticed in D. melanogaster (McGraw et al. 2004), with greater suppression in lure-mated females (supplementary fig. S8, Supplementary Material online). It is possible that down regulation of odorant binding proteins suppress female attraction or receptiveness towards courting males, ultimately controlling remating frequency.
Titin-like Muscle Proteins
We recorded several other transcripts up-regulated after mating that are unique to this study, including titin-like proteins and histone proteins. Titin (=connectin) was not reported previously either in Drosophila or tephritids, although a few muscle related proteins were differentially expressed in Ostrinia nubilalis (Al-Wathiqui et al. 2014). Titin is found primarily in skeletal muscles and is involved in sarcomere related functions (Greaser 2001). It contains a protein kinase domain positioned to sense mechanical load and it is found that the kinase domain interacts with the zinc-finger proteins to respond to mechanical stimuli in humans (Lange et al. 2005). The role of these muscle proteins in mated female B. tryoni is not known; given that males remain mounted on females during several hours of copulation (Kumaran et al. 2014b), perhaps those proteins were activated to hold the male weight. Since there was no previous record in insects, it is also possible that titin encode differential functions unlike in humans.
Histone Proteins
There were two transcripts encoding histone protein analogues (H1 and H5 linker) overexpressed only in lure-mated females, and a H3 acetylation transcript was overexpressed in both female types. There was a greater expression of H3 acetylation in lure-mated females, which along with the expression of H1 and H5 linker, suggests possible epigenetic changes (histone modifications) post-mating in B. tryoni. Differential expression of histone transcripts has not been reported previously in any model organisms or tephritids; however, targeted epigenetic studies have identified histone modifications in females post-mating (Zhou et al. 2014). Histones function to package DNA into nucleosomes and it is a main protein within chromatin. Since DNA wraps around histones, they play a role in gene regulation by altering chromatin structure (Grunstein 1997). Acetylation of histone H3 occurs at several different lysine positions in the histone tail and is performed by the enzymes called histone acetyltransferases (HATs) (Bannister and Kouzarides 2011). For instance, H3 acetylation localized to discrete sites in the mammalian genome mediate distinct chromatin functions that dictate transgene expression or silencing (Yan and Boyd 2006). Epigenetic variations are reported to mediate individual differences in behavior and such variations have often been traced through several generations (Jensen 2013; Dias et al. 2015). In B. tryoni males, the “lure foraging” trait was found to be passed onto offspring sired by males fed on phytochemicals (Kumaran and Clarke 2014) suggesting possible phytochemical mediated epigenetic changes.
Taxa-Specific Transcripts
There were a great number of transcripts with unknown functions and most of these are predicted proteins that have only previously been identified within other tephritid fruit fly genomes. The reason for the failure to assign functions is perhaps because female specific transcriptome resources are generally lacking for most tephritid species, or that they are restricted to this group for which genomic resources have only recently become available. The functional role of taxa-specific transcripts needs to be investigated as they may be involved in mediating post-mating behaviors such as oviposition and remating.
Female-Specific Factors of Post-mating Behaviors
Although Acps are the proximate mechanisms of female postmating changes, we strongly believe that females are also likely to control their post-mating patterns to some extent for the following reasons: 1) behaviorally, females choose whether to mate or not, and with whom in most of the species (Andersson and Simmons 2006); and 2) mating, egg production, and oviposition is physiologically costly (Baer and Schmid‐Hempel 2001; Colegrave et al. 2002; Wigby and Chapman 2005), thus selection should act on females to utilise their resources optimally to maximize reproductive success. This hypothesis does not compete with the effect of male Acps in mediating female behaviors, but insist additional factors need to be studied to fully understand complex female post-mating behaviors (Immonen and Ritchie 2012). The transcriptomic changes found in B. tryoni and other systems will be highly useful resources for future studies targeting female-specific factors.
Conclusion
This study presents a wide range of functional transcripts differentially regulated in mated females. Transcripts such as those encoding titin-like muscle proteins, histone proteins and a number of unknown genes in mated females suggest their possible role in mediating post-mating changes and warrants further research on female-specific genetic changes. Exclusively enriched or suppressed genes in lure-mated females and taxa-specific transcripts suggest complex factors, additional to male Acps and sperm, contribute to female post-mating behaviors. Further targeted studies on differentially expressed genes, not regulated by male Acps, could shed more light on understanding the evolutionary implications of post-mating changes and female factors mediating those changes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We acknowledge the help of Dr. Kevin Dudley and Mrs. Sahana Manoli (Molecular Genetics Research Facility, QUT) with sequencing. Discussions with Drs. Mark Schutze, Matt Krosch, Paul Cunningham, and Stuart Wigby (Oxford University) helped generate ideas and discussions. We are grateful to three anonymous reviewers for their helpful comments on the manuscript. This study was supported by Horticulture Innovation Australia Ltd (HIA) as project AI14002 of the HIA Transformational Postdoctoral Fellowship scheme. We acknowledge the support of HIA, which included funds it manages on behalf of the Australian Government.
Literature Cited
- Aluja M, Rull J, Sivinski J, Trujillo G, Pérez-Staples D.. 2009. Male and female condition influence mating performance and sexual receptivity in two tropical fruit flies (Diptera: Tephritidae) with contrasting life histories. J Insect Physiol. 55(12):1091–1098. [DOI] [PubMed] [Google Scholar]
- Al-Wathiqui N, Lewis SM, Dopman EB.. 2014. Using RNA sequencing to characterize female reproductive genes between Z and E Strains of European Corn Borer moth (Ostrinia nubilalis). BMC Genomics 15(1):189–201.http://dx.doi.org/10.1186/1471-2164-15-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. 1986. Evolution of condition‐dependent sex ornaments and mating preferences: sexual selection based on viability differences. Evolution 40:804–816. [DOI] [PubMed] [Google Scholar]
- Andersson MB. 1994. Sexual selection. New Jersey: Princeton University Press. [Google Scholar]
- Andersson M, Simmons LW.. 2006. Sexual selection and mate choice. Trends Ecol Evol. 21(6):296–302.http://dx.doi.org/10.1016/j.tree.2006.03.015 [DOI] [PubMed] [Google Scholar]
- Arnqvist G, Nilsson T.. 2000. The evolution of polyandry: multiple mating and female fitness in insects. Anim Behav. 60(2):145–164.http://dx.doi.org/10.1006/anbe.2000.1446 [DOI] [PubMed] [Google Scholar]
- Arthofer W, et al. 2014. Genomic resources notes accepted 1 December 2013–31 January 2014. Mol Ecol Res. 14(3):664–665.http://dx.doi.org/10.1111/1755-0998.12255 [DOI] [PubMed] [Google Scholar]
- Baer B, Schmid‐Hempel P.. 2001. Unexpected consequences of polyandry for parasitism and fitness in the bumblebee, Bombus terrestris. Evolution 55(8):1639–1643. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T.. 2011. Regulation of chromatin by histone modifications. Cell Res. 21(3):381–395.http://dx.doi.org/10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead TR, Pizzari T.. 2002. Postcopulatory sexual selection. Nat Rev Genet. 3(4):262–273.http://dx.doi.org/10.1038/nrg774 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120.http://dx.doi.org/10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JM, Matzkin LM, Kelleher ES, Markow TA.. 2011. Postmating transcriptional changes in reproductive tracts of con-and heterospecifically mated Drosophila mojavensis females. Proc Natl Acad Sci U S A. 108(19):7878–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, et al. 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci U S A. 100(17):9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PS, et al. 1988. A male accessory gland peptide that regulates reproductive behavior of female Dropsophila melanogaster. Cell 54(3):291–298.http://dx.doi.org/10.1016/0092-8674(88)90192-4 [DOI] [PubMed] [Google Scholar]
- Chinajariyawong A, Drew RAI, Meats A, Balagawi S, Vijaysegaran S.. 2010. Multiple mating by females of two Bactrocera species (Diptera: Tephritidae: Dacinae). Bull Entomol Res. 100(03):325–330. [DOI] [PubMed] [Google Scholar]
- Colegrave N, Kotiaho JS, Tomkins JL.. 2002. Mate choice or polyandry: reconciling genetic compatibility and good genes sexual selection. Evol Ecol Res. 4:911–917. [Google Scholar]
- Cotton S, Small J, Pomiankowski A.. 2006. Sexual selection and condition-dependent mate preferences. Curr Biol. 16(17):R755–R765. [DOI] [PubMed] [Google Scholar]
- Dias BG, Maddox SA, Klengel T, Ressler KJ.. 2015. Epigenetic mechanisms underlying learning and the inheritance of learned behaviors. Trends Neurosci. 38(2):96–107.http://dx.doi.org/10.1016/j.tins.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake WMTD, Jayasundara MSH, Peek T, Clarke AR, Schutze MK.. 2017. The mating system of the true fruit fly Bactrocera tryoni and its sister species, Bactrocera neohumeralis. Insect Sci. 24(3):478–490. [DOI] [PubMed] [Google Scholar]
- Elgar MA. 1998. Sperm competition and sexual selection in spiders In: Birkhead TR, Moller AP, editors. Sperm competition and sexual selection. London: Academic press; p. 307–339. [Google Scholar]
- Fedorka KM, Mousseau TA.. 2002. Material and genetic benefits of female multiple mating and polyandry. Anim Behav. 64(3):361–367.http://dx.doi.org/10.1006/anbe.2002.3052 [Google Scholar]
- Fowler K, Partridge L.. 1989. A cost of mating in female fruit flies. Nature 338(6218):760–761.http://dx.doi.org/10.1038/338760a0 [Google Scholar]
- Gillott C. 2003. Male accessory gland secretions: modulators of female reproductive physiology and behavior. Annu Rev Entomol. 48:163–184.http://dx.doi.org/10.1146/annurev.ento.48.091801.112657 [DOI] [PubMed] [Google Scholar]
- Gomulski LM, et al. 2012. Transcriptome profiling of sexual maturation and mating in the Mediterranean fruit fly, Ceratitis capitata. PLoS One 7(1):e30857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaser M. 2001. Identification of new repeating motifs in titin. Proteins 43(2):145–149.http://dx.doi.org/10.1002/1097-0134(20010501)43:2<145::AID-PROT1026>3.0.CO;2-B [DOI] [PubMed] [Google Scholar]
- Grunstein M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389(6649):349–352.http://dx.doi.org/10.1038/38664 [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 8(8):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq I, Vreysen MJ, Cacéres C, Shelly TE, Hendrichs J.. 2014. Methyl eugenol aromatherapy enhances the mating competitiveness of male Bactrocera carambolae Drew & Hancock (Diptera: Tephritidae). J Insect Physiol. 68:1–6. [DOI] [PubMed] [Google Scholar]
- Hee AKW, Tan KH.. 1998. Attraction of female and male Bactrocera papayae to conspecific males fed with methyl eugenol and attraction of females to male sex pheromone components. J Chem Ecol. 24:753–764.http://dx.doi.org/10.1023/A:1022302605357 [Google Scholar]
- Herndon LA, Wolfner MF.. 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc Nat Acad Sci U S A. 92(22):10114–10118.http://dx.doi.org/10.1073/pnas.92.22.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WO, Oldroyd BP, Beekman M, Ratnieks FL.. 2008. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320(5880):1213–1216.http://dx.doi.org/10.1126/science.1156108 [DOI] [PubMed] [Google Scholar]
- Immonen E, Ritchie MG.. 2012. The genomic response to courtship song stimulation in female Drosophila melanogaster. Proc Biol Sci. 279(1732):1359–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang EB. 1995. Effects of mating and accessory gland injections on olfactory-mediated behavior in the female Mediterranean fruit fly, Ceratitis capitata. J Insect Physiol. 41(8):705–710.http://dx.doi.org/10.1016/0022-1910(95)00015-M [Google Scholar]
- Jennions MD, Petrie M.. 2000. Why do females mate multiply? A review of the genetic benefits. Biol Rev. 75(1):21–64.http://dx.doi.org/10.1017/S0006323199005423 [DOI] [PubMed] [Google Scholar]
- Jensen P. 2013. Transgenerational epigenetic effects on animal behaviour. Prog Biophys Mol Biol. 113(3):447–454. [DOI] [PubMed] [Google Scholar]
- Klassen W. 2005. Area-wide integrated pest management and the sterile insect technique In: Dyck VA, Hendrichs J, Robinson AS, editors. Sterile insect technique. The Netherlands: Springer; pp. 36–98. [Google Scholar]
- Kumaran N, Balagawi S, Schutze MK, Clarke AR.. 2013. Evolution of lure response in tephritid fruit flies: phytochemicals as drivers of sexual selection. Anim Behav. 85(4):781–789.http://dx.doi.org/10.1016/j.anbehav.2013.01.024 [Google Scholar]
- Kumaran N, Clarke AR.. 2014. Indirect effects of phytochemicals on offspring performance of Queensland fruit fly, Bactrocera tryoni (Diptera: Tephritidae). J App Entomol. 138(5):361–367. [Google Scholar]
- Kumaran N, Hayes RA, Clarke AR.. 2014a. Cuelure but not zingerone make the sex pheromone of male Bactrocera tryoni (Tephritidae: Diptera) more attractive to females. J Insect Physiol. 68:36–43. [DOI] [PubMed] [Google Scholar]
- Kumaran N, Prentis PJ, Mangalam KP, Schutze MK, Clarke AR.. 2014b. Sexual selection in true fruit flies (Diptera: Tephritidae): transcriptome and experimental evidences for phytochemicals increasing male competitive ability. Mol Ecol. 23:4645–4657. [DOI] [PubMed] [Google Scholar]
- Landolt PJ, Phillips TW.. 1997. Host plant influences on sex pheromone behavior of phytophagous insects. Annu Rev Entomol. 42:371–391.http://dx.doi.org/10.1146/annurev.ento.42.1.371 [DOI] [PubMed] [Google Scholar]
- Lange S, et al. 2005. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308(5728):1599–1603.http://dx.doi.org/10.1126/science.1110463 [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, et al. 2007. Mating and immunity in invertebrates. Trends Ecol Evol. 22(1):48–55.http://dx.doi.org/10.1016/j.tree.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, Begun DJ.. 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47(5):900–910.http://dx.doi.org/10.1139/g04-050 [DOI] [PubMed] [Google Scholar]
- Liu H, Kubli E.. 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc Nat Acad Sci U S A. 100(17):9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin OY, Hosken DJ.. 2003. The evolution of reproductive isolation through sexual conflict. Nature 423(6943):979–982.http://dx.doi.org/10.1038/nature01752 [DOI] [PubMed] [Google Scholar]
- Mays HL, Hill GE.. 2004. Choosing mates: good genes versus genes that are a good fit. TREE 19(10):554–559. [DOI] [PubMed] [Google Scholar]
- McGraw LA, Clark AG, Wolfner MF.. 2008. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics 179(3):1395–1408.http://dx.doi.org/10.1534/genetics.108.086934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF.. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 14(16):1509–1514.http://dx.doi.org/10.1016/j.cub.2004.08.028 [DOI] [PubMed] [Google Scholar]
- Miyatake T, Chapman T, Partridge L.. 1999. Mating-induced inhibition of remating in female Mediterranean fruit flies Ceratitis capitata. J Insect Physiol. 45(11):1021–1028.http://dx.doi.org/10.1016/S0022-1910(99)00083-9 [DOI] [PubMed] [Google Scholar]
- Obra GB, Resilva SS.. 2013. Influence of adult diet and exposure to methyl eugenol in the mating performance of Bactrocera philippinensis. J Appl Entomol. 137:210–216.http://dx.doi.org/10.1111/j.1439-0418.2011.01678.x [Google Scholar]
- Parker GA. 1979. Sexual selection and sexual conflict In: Blum M, Blum N, editors. Sexual selection and reproductive competition in insects. New York: Academic Press; pp. 123–166. [Google Scholar]
- Parker GA. 2006. Sexual conflict over mating and fertilization: an overview. Philos Trans R Soc Lond B Biol Sci. 361(1466):235–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Staples D, Harmer AMT, Taylor PW.. 2007. Sperm storage and utilization in female Queensland fruit flies (Bactrocera tryoni). Physiol Entomol. 32(2):127–135. [Google Scholar]
- Radhakrishnan P, Taylor PW.. 2008. Ability of male Queensland fruit flies to inhibit receptivity in multiple mates, and the associated recovery of accessory glands. J Insect Physiol. 54: 421–428. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan P, Pérez-Staples D, Weldon CW, Taylor PW.. 2009. Multiple mating and sperm depletion in male Queensland fruit flies: effects on female remating behaviour. Anim Behav. 78: 839–846. [Google Scholar]
- Raghu S. 2004. Functional significance of phytochemical lures to dacine fruit flies (Diptera: Tephritidae): an ecological and evolutionary synthesis. Bull Entomol Res. 94(5):385–399. [DOI] [PubMed] [Google Scholar]
- Raghu S, Clarke AR.. 2003. Sexual selection in a tropical fruit fly: role of a plant derived chemical in mate choice. Entomol Exp Appl. 108(1):53–58. [Google Scholar]
- Raghu S, Clarke AR, Yuval B.. 2002. Investigation of the physiological consequences of feeding on methyl eugenol by Bactrocera cacuminata (Diptera: Tephritidae). Environ Entomol. 31(6):941–946.http://dx.doi.org/10.1603/0046-225X-31.6.941 [Google Scholar]
- Ritchie MG. 2007. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 38(1):79–102.http://dx.doi.org/10.1146/annurev.ecolsys.38.091206.095733 [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140.http://dx.doi.org/10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurch NJ, et al. 2016. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use?. RNA 22(10):839–851.http://dx.doi.org/10.1261/rna.053959.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolari F, et al. 2012. Transcriptional profiles of mating-responsive genes from testes and male accessory glands of the Mediterranean fruit fly, Ceratitis capitata. PLoS One 7(10):e46812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly TE. 2000. Fecundity of female oriental fruit flies (Diptera: Tephritidae): effects of methyl eugenol-fed and multiple mates. Ann Entomol Soc Am. 93:559–564.http://dx.doi.org/10.1603/0013-8746(2000)093[0559:FOFOFF]2.0.CO;2 [Google Scholar]
- Shelly TE. 2017. Zingerone and the mating success and field attraction of male melon flies (Diptera: Tephritidae). J Asia Pac Entomol. 20(1):175–178.http://dx.doi.org/10.1016/j.aspen.2016.12.013 [Google Scholar]
- Shelly TE, Nishida R.. 2004. Larval and adult feeding on methyl eugenol and the mating success of male oriental fruit flies, Bactrocera dorsalis. Entomol Exp Appl. 112(2):155–158. [Google Scholar]
- Shelly TE, Nishimoto J.. 2016. Does female mate choice confer direct fitness benefits? results from a tephritid fruit fly. Ann Entomol Soc Am. 110:204–211. [Google Scholar]
- Shelly TE, Villalobos EM.. 1995. Cue lure and the mating behavior of male melon flies (Diptera: Tephritidae). Fla Entomol. 78(3):473–482.http://dx.doi.org/10.2307/3495532 [Google Scholar]
- Shelly TE, Edu J, McInnis D.. 2010. Pre-release consumption of methyl eugenol increases the mating competitiveness of sterile males of the oriental fruit fly, Bactrocera dorsalis, in large field enclosures. J Insect Sci. 10(8):1–16.http://dx.doi.org/10.1673/031.010.0801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly TE. 2010. Effects of methyl eugenol and raspberry ketone/cue lure on the sexual behavior of Bactrocera species (Diptera: Tephritidae). Appl Entomol Zool. 45(3):349–361.http://dx.doi.org/10.1303/aez.2010.349 [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3232. [DOI] [PubMed] [Google Scholar]
- Sitnik JL, Gligorov D, Maeda RK, Karch F, Wolfner MF.. 2016. The female post-mating response requires genes expressed in the secondary cells of the male accessory gland in Drosophila melanogaster. Genetics 202(3):1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SD, Drew RA, Hughes JM.. 2007. Multiple paternity in a natural population of a wild tobacco fly, Bactrocera cacuminata (Diptera: Tephritidae), assessed by microsatellite DNA markers. Mol Ecol. 16(11):2353–2361.http://dx.doi.org/10.1111/j.1365-294X.2007.03277.x [DOI] [PubMed] [Google Scholar]
- Stewart ZK, Pavasovic A, Hock DH, Prentis PJ.. 2017. Transcriptomic investigation of wound healing and regeneration in the cnidarian Calliactis polypus. Sci Rep. 7:41458.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill R, Alcock J.. 1983. The evolution of insect mating systems. Cambridge: Harvard University Press. [Google Scholar]
- Tychsen PH. 1978. Mating behaviour of the Queensland fruit fly, Dacus tryoni (Diptera: Tephritidae), in field cages. Aust J Entomol. 16(4):459–465.http://dx.doi.org/10.1111/j.1440-6055.1977.tb00139.x [Google Scholar]
- van der Burg CA, Prentis PJ, Surm JM, Pavasovic A.. 2016. Insights into the innate immunome of actiniarians using a comparative genomic approach. BMC Genomics 17(1):850–864.http://dx.doi.org/10.1186/s12864-016-3204-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MG, Mitko L, Eltz T, Ramírez SR.. 2016. Macroevolution of perfume signalling in orchid bees. Ecol Lett. 19(11):1314–1323. [DOI] [PubMed] [Google Scholar]
- Wee SL, Tan KH, Nishida R.. 2007. Pharmacophagy of methyl eugenol by males enhances sexual selection of Bactrocera carambolae. J Chem Ecol. 33(6):1272–1282.http://dx.doi.org/10.1007/s10886-007-9295-0 [DOI] [PubMed] [Google Scholar]
- Wei D, et al. 2016. Transcriptome analysis to identify genes for peptides and proteins involved in immunity and reproduction from male accessory glands and ejaculatory duct of Bactrocera dorsalis. Peptides 80:48–60.http://dx.doi.org/10.1016/j.peptides.2015.08.007 [DOI] [PubMed] [Google Scholar]
- Wigby S, Chapman T.. 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr Biol. 15(4):316–321.http://dx.doi.org/10.1016/j.cub.2005.01.051 [DOI] [PubMed] [Google Scholar]
- Wigby S, et al. 2009. Seminal fluid protein allocation and male reproductive success. Curr Biol. 19(9):751–757.http://dx.doi.org/10.1016/j.cub.2009.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Brodie III ED, Cheverud JM, Moore AJ, Wade MJ.. 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol Evol. 13(2):64–69.http://dx.doi.org/10.1016/S0169-5347(97)01233-0 [DOI] [PubMed] [Google Scholar]
- Yan C, Boyd DD.. 2006. Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol. 26(17):6357–6371.http://dx.doi.org/10.1128/MCB.00311-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A.. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11(2):14..http://dx.doi.org/10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Luo D, Wu F, Wang J, Zhang H.. 2016. RNA sequencing to characterize transcriptional changes of sexual maturation and mating in the female oriental fruit fly Bactrocera dorsalis. BMC Genomics 17:194.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Mackay TF, Anholt RR.. 2014. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genomics 15:927.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann Y, Ramírez SR, Eltz T.. 2009. Chemical niche differentiation among sympatric species of orchid bees. Ecology 90(11):2994–3008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.