Abstract
Vernal pool clam shrimp (Eulimnadia texana) are a promising model system due to their ease of lab culture, short generation time, modest sized genome, a somewhat rare stable androdioecious sex determination system, and a requirement to reproduce via desiccated diapaused eggs. We generated a highly contiguous genome assembly using 46× of PacBio long read data and 216× of Illumina short reads, and annotated using Illumina RNAseq obtained from adult males or hermaphrodites. Of the 120 Mb genome 85% is contained in the largest eight contigs, the smallest of which is 4.6 Mb. The assembly contains 98% of transcripts predicted via RNAseq. This assembly is qualitatively different from scaffolded Illumina assemblies: It is produced from long reads that contain sequence data along their entire length, and is thus gap free. The contiguity of the assembly allows us to order the HOX genes within the genome, identifying two loci that contain HOX gene orthologs, and which approximately maintain the order observed in other arthropods. We identified a partial duplication of the Antennapedia complex adjacent to the few genes homologous to the Bithorax locus. Because the sex chromosome of an androdioecious species is of special interest, we used existing allozyme and microsatellite markers to identify the E. texana sex chromosome, and find that it comprises nearly half of the genome of this species. Linkage patterns indicate that recombination is extremely rare and perhaps absent in hermaphrodites, and as a result the location of the sex determining locus will be difficult to refine using recombination mapping.
Keywords: genomics, genome assembly, invertebrate genetics, sex chromosomes, genome biology, HOX genes
Introduction
The clam shrimp Eulimnadia texana is a desert vernal pool shrimp found in the southwestern United States. It is a relative of the other vernal pool branchiopods such as the Triops tadpole shrimp and the Anostraca fairy shrimp, and shares many of its unique traits with them (Weeks et al. 2009). Eulimnadia texana has, along with these other vernal pool shrimp, been noted for its unique sex determining system (Sassaman and Weeks 1993), its rare (in Metazoa) requirement to reproduce via desiccated diapaused eggs (Sassaman and Weeks 1993), and its unique habitat. This androdioecious (Sassaman and Weeks 1993) species has three common arrangements of sex alleles (Sassaman and Weeks 1993) or “proto-sex chromosomes” (Weeks et al. 2010). Males are always homozygous for the “Z” male allele, while hermaphrodites may be “ZW” or “WW,” with WW hermaphrodites only capable of producing hermaphrodite offspring. Much effort ( Weeks et al. 2010) has gone into attempting to identify the E. texana sex locus because of this unique arrangement; this, coupled with the fact that close relatives of the species have ordinary male–female sexual dimorphism (Weeks et al. 2009), makes the Eulimnadia clade, and E. texana in particular, an excellent study system for understanding the genetic changes that underlie changes in sex determination. The fact that, unlike in most animals, both the “Z” and “W” sex determination alleles are capable of being homozygous is interesting as a comparator for testing the hypothesis that the lack of recombination in “Y” and “W” alleles drives degradation of sex chromosomes. The ability of eggs to remain in diapause for years at a time (Brendonck 1996) is especially valuable to geneticists because very few macroscopic animals exist in which populations can be archived for long periods without changes occurring in the genetics of the population (genetic drift, loss of linkage disequilibrium, etc.). Furthermore, clam shrimp live in desert vernal pools; naturally limited migration from pool to pool makes them well suited to the study of populations evolving in relative genetic isolation.
Genome assembly of nonmodel organisms was financially unrealistic until the advent of high-throughput next generation sequencing. Unfortunately, next generation sequencing methods such as Illumina are limited to short read sequencing, which is not ideal for genome assembly; assemblies produced using Illumina-type short read data tend to have low contiguity (Treangen and Salzberg 2011). This problem can be overcome by using PacBio (Eid et al. 2009), Oxford Nanopore (Laver et al. 2015), or other long read sequencing technologies to supplement or replace Illumina sequencing. A hybrid approach to sequencing and assembly using both short and long reads has been shown to produce highly contiguous assemblies in Drosophila-sized genomes (Chakraborty et al. 2016). Genome annotation of de novo assemblies is routinely performed using RNAseq data (Wang et al. 2009), and tools for that purpose are already available (Stanke and Waack 2003; Grabherr et al. 2011).
Here, we lay out our attempt to extend genetic research on E. texana into the world of whole genome sequence analysis using the latest genomics techniques. We used a combination of short read Illumina (Shen et al. 2005) and long read PacBio (Eid et al. 2009) sequencing to generate a high-quality draft genome assembly and performed an annotation of genes using RNAseq (Wang et al. 2009). We generated a genome assembly for a WW hermaphrodite clam shrimp strain consisting of 112 contigs totaling 120 Mb in length with a contig N50 of 18 Mb. Using RNAseq data we annotate 17,667 genes, of which ∼99% of hermaphrodite transcripts are placed into our assembly. This assembly is the most contiguous assembly of a crustacean genome of which we are aware. By comparison, Daphnia pulex has a scaffold N50 of 494 kb (Ye et al. 2017).
Materials and Methods
Shrimp Collection and Rearing
Clam shrimp (fig. 1) used here were initially sampled from New Mexico and Arizona, then inbred in the laboratory (Weeks and Zucker 1999). We reared the clam shrimp in the laboratory until day 10 of their life cycles, then extracted DNA and RNA from them. Clam shrimp populations were reared in 50×30×8 cm disposable aluminum foil catering trays (Catering Essentials, full size steam table pan). In each pan, we mixed 500 ml of soil with 6 l of water purified via reverse osmosis. 0.3 g of aquarium salt (API aquarium salt, Mars Fishcare North America, Inc.) were added to each tray to ensure that necessary nutrients were available to the shrimp. Trays were checked daily for nonclam shrimp, especially the carnivorous Triops longicaudatus, and all nonclam shrimp were immediately removed from trays. We identified the following nonclam shrimp: T. longicaudatus, D. pulex, and an unknown species of Anostraca fairy shrimp. An inbred population of clam shrimp, here referred to by its numerical title JT4(4)5, was derived from the JT4 wild population and used for Illumina sequencing for the genome assembly. We generated this population by collecting a set of JT4 monogenic hermaphrodites and raising them in the laboratory for six generations (Weeks 2004). Because monogenic hermaphrodites cannot interbreed and can only produce hermaphroditic offspring, the resulting population was the exclusive product of selfing for six generations. Although diversity may exist between individuals in this population, each individual is highly homozygous. We sampled a single hermaphrodite from this population and expanded it to obtain the isohermaphrodite line (JT4(4)5-L) and used the line for sequencing.
Fig. 1.
—A male clam shrimp (left), and a hermaphrodite clam shrimp (right). Both are exemplars of the E. texana species. Note the presence of clasping arms on the male—these are required for nonself-fertilized sex, and the presence of a brood pouch along the dorsal surface of the hermaphrodite.
Library Preparation and Sequencing
Illumina Library for Genome Assembly
DNA for Illumina sequencing was extracted from 50 inbred monogenic hermaphrodites from the JT4(4)5-L strain. We generated the inbred, isohermaphrodite shrimp population JT4(4)5-L from the inbred JT4(4)5 population generated by Weeks (2004). The JT4(4)5-L population has been inbred in the laboratory (full selfing) for six generations, and was used for all gDNA sequencing. We performed the Illumina Truseq library preparation protocol. We chose this method over Nextera library preparation for the library for genome assembly for two reasons: first, Nextera library preparation has been shown to produce a bias in coverage that can cause problems during genome assembly (Lan et al. 2015); second, the Covaris shearing used in the Truseq protocol allowed us to control the fragment length of the DNA to produce pseudo long reads obtained by joining overlapping read pairs (we refer to these a “pontigs” for paired-contigs). In order to produce an average pontig fragment length of 150 bp, we used the following Covaris shearing settings: 60 s × 6 at 10% duty cycle, 5 intensity, 200 cycles per burst. We size selected the final library on an agarose gel to get the desired 150 bp read length. We ran one lane of paired-end 100 bp Illumina sequencing on an Illumina HiSeq 2500, producing 124.9 Gb of sequence data.
PacBio Library for Genome Assembly
We followed the general protocol outlined in Chakraborty et al. (2016) to generate the PacBio library used here. We homogenized 265 inbred monozygotic hermaphrodites from the JT4(4)5-L strain in liquid nitrogen using a mortar and pestle. We then extracted DNA using the Qiagen Blood and Cell culture DNA Midi Kit (Qiagen, Valencia, CA, USA). We made two modifications to the protocol: first, we incubated the tissue powder in the mixture of G2 buffer, RNaseA, and protease for 18 h, rather than the 2 h listed in the protocol; second, we doubled the RNaseA added from 19 up to 38 µl, and halved the protease added from 500 to 250 µl. We made these changes based on the presence of RNA in earlier attempts to use this kit. After gDNA extraction, we sheared the gDNA using a 1.5-in., 24-gauge blunt tipped needle for 20 strokes. We visualized both the original gDNA and the sheared DNA using field inversion gel electrophoresis as in Chakraborty et al. (2016). We size selected the DNA using a 15–50 kb cutoff using the BluePippin gel electrophoresis platform (Sage Science, Beverly, MA, USA). We prepared the sequencing library using 5 µg of this product, and then size selected again using a 15–50 kb cutoff on the BluePippin gel electrophoresis platform. This produced a total of 0.149 nmol of library. We sequenced this library using 10 SMRTcells on the PacBio RS II sequencer, producing 6.7 Gb of sequence data and a read length N50 of 15.2 kb.
RNA Sequencing
Male clam shrimp for RNA sequencing came from the outbred WAL population (Weeks and Zucker 1999). This is a natural population, raised for only a single generation in the laboratory. Hermaphrodites came from the JT4(4)5-L population used for gDNA sequencing. Adult males and hermaphrodites were sequenced separately. RNA extraction was performed using Trizol (Chomczynski and Sacchi 1987). We cleaned the RNA using RNeasy Mini columns (74104, Qiagen) following the manufacturer’s protocols, and then used this RNA to generate Illumina TruSeq RNAseq libraries according to the standard Illumina protocol. The male and hermaphrodite libraries were sequenced using one lane each of paired end 100 bp Illumina sequencing. We generated 23 Gb of sequence data for males and 23 Gb of sequence data for hermaphrodites.
k-Mer Counting
We generated k-mers using Jellyfish, v. 1.1.6 (Marçais and Kingsford 2011). We counted all 25-mers in the joined, but uncorrected, pontigs, then identified a local maximum coverage of 76×, then computed the genome size using the following formula:
where T = 15.7 Gb = total basepairs of pontig data, L = 112.7 = mean read length, M = 24 = mer length – 1, and C = 76 = coverage (cf. Lamichhaney et al. 2016). This produced a genome size estimate of 144 Mb. We use this genome size estimate throughout this work.
Genome Assembly
Hybrid Assembly
Genome assembly was performed according to the protocol established in Chakraborty et al. (2016). We first generated “pontigs” from the PE100 reads obtained from the 150 bp insert library by assembling individual read pairs. There is some evidence (cf. read joining with a third read in Gnerre et al. 2011) that such long, contiguous, error-free reads are slightly better for genome assembly than trimmed paired reads. We generated pontigs using the fq-join function in ea-utils (Aronesty 2013), and then used Quake (Kelley et al. 2010) to error correct the pontigs. We then assembled the corrected pontigs using Platanus (Kajitani 2014), a De Bruijn graph assembler, with its default settings. This produced an assembly with an N50 of 5.2 kb. We input this assembly, plus the raw PacBio reads, into DBG2OLC (Ye et al. 2016). The input data set producing the highest contiguous assembly was identified via a set of hybrid assemblies using a range of quality cutoffs—we tested every whole numbered quality cutoff from 82% to 92%, and, in keeping with (Chakraborty et al. 2016), downsampled each PacBio data set down to the longest 30×. The 85% cutoff produced the highest N50 of 1.92 Mb and an assembly size of 120 Mb. All N50s are summarized in supplementary table 1, Supplementary Material online.
PacBio-Only Assembly
We used Celera 8.2, release candidate 3 (Myers et al. 2000), to generate the PacBio-only assembly, using the specfile listed in the supplementary text, Supplementary Materials online. The assembly had an N50 of 3.4 Mb, and a genome size of 126 Mb.
Assembly Merging
We used Quiver (Chin et al. 2013) to correct both the hybrid assembly and the PacBio assembly, then performed merging using quickmerge (Chakraborty et al. 2016). We used the following command line settings:
python merge_wrapper.py -pre merged_quivered_shrimp_assemblies -hco 5.0 -c 1.5/path/to/quivered/hybrid/path/to/quivered/pbonly
Here, -hco refers to the stringency with which seed high confidence overlaps are filtered, and -c refers to the stringency with which other HCOs are filtered. After merging, we corrected the resultant assembly by using Quiver again. In keeping with the Quiver standard practices, we ran Quiver on this assembly one more time, and then quantified differences between the assemblies using MUMmer (Kurtz et al. 2004). We noted a decrease in the number of SNPs and indels identified between the final two Quiver runs, so we took the final quivered assembly as our final assembly.
Annotation
We used Trinity (Grabherr et al. 2011) and Augustus (Stanke and Waack 2003) to generate an annotation of the genome assembly. We ran Trinity three times: once for the male RNAseq data, once for the hermaphrodite RNAseq data, and once for the combination of both males and hermaphrodites. We used a custom script to convert Augustus data into a generic gff3 file, and another custom script to identify 4-fold degenerate sites based on the same annotation. We used BLAST (Altschul et al. 1990) to align the entire Drosophila melanogaster proteome against the Augustus-generated shrimp CDS and vice versa. Mutual best hits with an e-value below 10−5 were considered significant. We tentatively assert that these genes are correctly annotated, and that they are orthologous or paralogous to genes in D. melanogaster.
Differential Expression Analysis
We identified differences in expression between males and hermaphrodites using Tophat (Trapnell et al. 2009) and the DESeq 1 package (Love et al. 2014). Tophat was used for transcript counting, while DESeq was used for differential expression analysis. Because we did not have replicated RNAseq data, we used the “blind” method to estimate dispersion using the following R code:
We then identified differences between the base means of the “male” and “herm” groups using the modified binomial test featured in DESeq, using the following R code:
BLAST Annotation
We annotated all gene functions using blastp to align the E. texana genes to the D. melanogaster NCBI protein database, and vice versa. We regard the mutual best hits (those pairs that had e-values below 10−5 in both directions, and that paired in both BLAST directions) as the annotations in which we were most confident. In the 13 peaks of high interest discussed below, we annotated the genes that did not have mutual best BLAST hits in D. melanogaster by taking the most significant BLAST hit for each gene (identified using blastp against the D. melanogaster nr protein database) and assigning that putative identity to the gene of interest.
Hox Gene Annotation
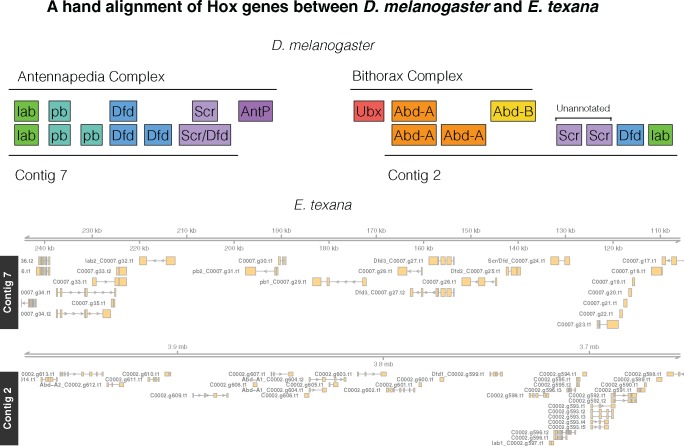
We identified an initial set of HOX genes using mutual best hit BLAST and found six apparent HOX genes spread across two contigs (C0002 and C0007, fig. 2). We then used a protein–protein BLAST (BLASTP, cutoff = 10−5) of all E. texana annotated genes onto all D. mel annotated genes, and identified five more genes that BLASTed to the D. mel HOX region. We aligned all protein sequences with Clustal-Omega (Goujon et al. 2010; Sievers et al. 2011; McWilliam et al. 2013, default settings), and then built a tree using MEGA v. 7.0.26 (Kumar et al. 2016). Our MEGA settings were maximum likelihood tree, using only conserved residues, 300-iteration bootstrap consensus. We called any E. texana gene with only one D. mel HOX gene in its sister clade as an ortholog of the D. mel HOX gene. Finally, we ran a tBLASTx of the D. mel HOX genes against the E. texana genome to identify possible unannotated HOX genes (cutoff = 10−5). We identified Scr as the ortholog of the two unannotated E. texana genes by aligning their genomic regions, and all E. texana and D. mel HOX CDS sequences in Clustal-Omega, then calling them orthologs using the same criterion as above.
Fig. 2.
—Top: The D. melanogaster HOX regions hand-aligned against the E. texana HOX regions. The ortholog identities of the E. texana HOX genes are established via bootstrap consensus maximum likelihood trees in MEGA. Note the similarity between the Antennapedia complex and Contig 7, and note that Contig 2 appears to be a combination of a copy of the Antennapedia complex and a portion of the Bithorax complex. Bottom: a visualization of the genome regions identified above. In this bottom panel, genes have been renamed for clarity. Genes that correspond to a hox gene are renamed in the figure as “DrosophilaName_E.texana name” with the Drosophila gene name prefixed to the E. texana gene name. Each instance of a given Drosophila name is numbered. To extract the correspond gene from E texana annotation files the Drosophila prefix should be removed.
Results
Genome Assembly
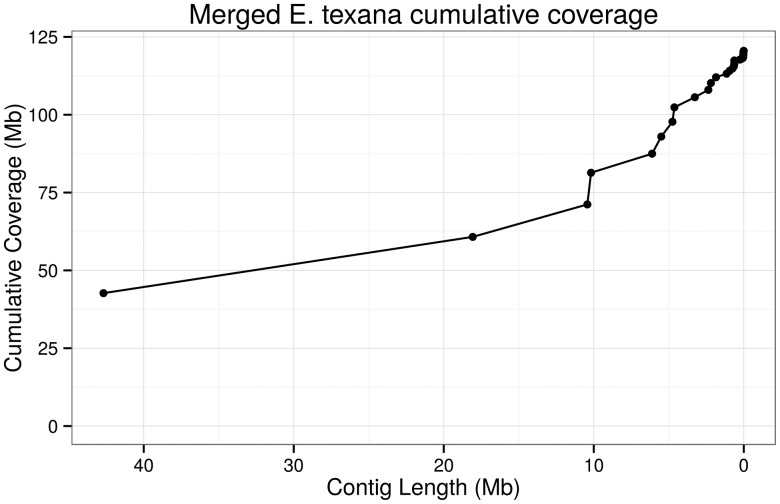
We assembled the genome using both the hybrid approach suggested by DBG2OLC (Ye et al. 2016) and the PacBio-only approach used in PBcR (Berlin et al. 2015), and then merged the two assemblies using quickmerge (Chakraborty et al. 2016) to produce the final assembly. The genome assembled into 112 contigs totaling 120 Mb. These contigs had an N50 of 18 Mb. A plot of cumulative coverage versus contig length (fig. 3) demonstrates that a substantial portion (85%) of the genome is contained in the eight largest contigs. The largest contig is 41 Mb in length. This level of contiguity is a dramatic improvement for vernal pool research: the highest quality vernal pool species currently assembled is D. pulex, with a genome size of 153 Mb and a scaffold N50 of 494 kb (Ye et al. 2017). Other major invertebrate genomes include the honey bee (Apis mellifera, contig N50 = 46 kb, scaffold N50 = 997 kb, Elsik et al. 2014), the Tribolium beetle (contig N50 = 41 kb, scaffold n50 = 992 kb, Richards et al. 2008), the argentine ant (Linepithena humile, contig N50 = 35 kb, scaffold N50 = 1.3 Mb, Smith et al. 2011), and the amphipod shrimp Parhyale hawaiensis (contig N50 = 81 kb, Kao et al. 2016). Note that scaffold N50 differs from contig N50 in that scaffolds are inferred by joining contigs with gaps, while contigs are gapless; thus, the difference between the assemblies is more dramatic than the numbers seem to indicate.
Fig. 3.
—A plot of cumulative genome coverage of the E. texana genome assembly by contig. As the plot progresses from left to right, the contig lengths are added to the cumulative coverage in order from largest to smallest. A high-quality assembly should achieve a high cumulative coverage with a small number of contigs. Here ∼80% of the assembly is contained in contigs larger than ∼5 Mb.
The observation that the estimated genome size is 144 Mb, and the final assembly size is 120 Mb, indicates that some portions of the genome were not assembled. This is ordinary in genome assembly, as highly repetitive heterochromatin regions tend to be impossible to assemble with current technology. For instance, the D. melanogaster genome is estimated to be 175 Mb in size (Ellis et al. 2014), yet the D. melanogaster assembled genome (easily among the best higher eukaryote assemblies) is “only” 143 Mb (dos Santos et al. 2015).
Two lines of evidence lead us to have confidence in this genome assembly: the quality of other genome assemblies produced using similar data and the same bioinformatics pipeline, and empirical evidence of the quality of this assembly. The genome assembly pipeline used in Chakraborty et al. (2016) has been thoroughly evaluated under a variety of genome size and coverage circumstances, and the genome size and coverage of these test assemblies match very closely to the genome size and coverage of our E. texana assembly. In particular, the Chakraborty (2016) assembly that used 39× of coverage to assemble a 140 Mb genome had an assembly N50 of 6.7 Mb, only 3,194 misassemblies, and 12.25 mismatched bases per 100 kb. Empirical evidence of the quality of a never-before-assembled genome is difficult to acquire, but we can report on the fraction of the Trinity-assembled (Grabherr et al. 2011; detailed below) RNAseq-derived transcripts that are present within the final assembly. We find that, if we use transcripts assembled entirely from RNA from hermaphrodites of the reference strain JT4(4)5-L, 98.9% of the transcripts align with above 92% identity, according to BLAT (Kent 2002). Interestingly, using the entire RNAseq data set, which contained both the hermaphrodites from the reference strain and males from the WAL strain, produced 95.5% successful alignment, which opens the possibility that some genes are present only in some male fraction of the genome not sampled in our WW hermaphrodite. Unfortunately, this difference could alternatively be strain-specific, rather than male-specific, with no simple way to differentiate those possibilities without further experimentation.
Repeatmasker identified 624 SINEs, 16,044 LINEs, 2,302 LTRs, 24,817 DNA elements, and 88,928 unclassified elements, together making up 26.4% of the genome. This contrasts with the relatively low rate of repetitive elements in D. melanogaster, at 3.9% (Kaminker et al. 2002). That said a large portion of this repetitive sequence is “unclassified”; if we remove the unclassified repeats from the count, only 9.8% of the genome consists of interspersed repeats. Other (noninterspersed) repeats make up 5.1% of the genome.
Annotation and Differential Expression
We collected one lane of Illumina RNAseq data from 25 male clam shrimp from the WAL wild population, and another lane from 25 inbred monozygotic females from the JT4(4)5-L population (the reference population used for the assembly). We used a combination of Trinity (Grabherr et al. 2011) and Augustus (Stanke and Waack 2003) to generate an annotation. We did three runs of Trinity—one run using only the males, one run using only the hermaphrodites, and one run using both together. The combined run produced 85,721 transcripts, while the male and hermaphrodite runs produced 77,257 and 55,845 transcripts, respectively. We ran Augustus using the combined run to generate gene predictions for E. texana. This generated a total of 17,667 genes and 23,965 transcripts. Of these genes, 5,438 were found to be mutual best hits with known D. melanogaster genes.
Phylogeny and the Genome
Crustaceans are a diverse group with highly variable genomes. Genome sizes range from the very small 160 Mb genome of the branchiopod water flea Scapholeberis kingii (Beaton 1988) to the huge 63 Gb genome of the arctic amphipod Ampelisca macrocephala (Rees et al. 2007). The number of genes in crustacean genomes appears to be less variable, and is not necessarily connected to genome size. Daphnia pulex (genome size ∼200 Mb) has the most genes of any known animal at 31,000 (Colbourne 2011), and the amphipod P. hawaiensis (genome size 3.6 Gb) was annotated as having 28,000 genes (Kao et al. 2016). By comparison, E. texana appears to have a genome size of 144 Mb, with ∼17,667 genes. This makes E. texana the smallest crustacean genome on record. In addition, E. texana’s gene count is substantially lower than known E. texana relatives. It is difficult to say conclusively that 17,000 is the true gene number of E. texana, but BUSCO (Simão et al. 2015), a program for estimating completeness of a gene set, successfully finds 88% of its expected ortholog sets in the E. texana proteome, indicating ∼88% completeness of the gene set.
Eulimnadia texana is a member of the clam shrimp order Spinicaudata, which is a member of the Branchiopoda class of crustaceans (Weeks et al. 2009). Branchiopoda is one of the major classes of the subphylum Crustaceae, and is believed to be the sister taxon to Multicrustacea, which includes copepods, malacostracans, and thecostracans (Regier et al. 2010). The closest relatives of the clam shrimp are fellow branchiopods such as the Triops tadpole shrimp and the Anostraca fairy shrimp, but neither of these has had a thorough genomic analysis. As discussed above, another branchiopod, D. pulex, is the closest thoroughly sequenced relative of E. texana, but the amount of differentiation between them is substantial. Fossil evidence indicates that E. texana, a member of Spinicaudata, and D. pulex, a member of Cladocera, diverged in the Silurian period, 443MYA–419MYA, but genetic evidence indicates a much more recent divergence in the Jurassic period (201MYA–145MYA) (Sun et al. 2016). Either way, the divergence time of the two species is substantial. Both E. texana and D. pulex have small genomes, with E. texana’s assembled genome at 120 Mb, and D. pulex’s at 153 Mb. C-value measurment of other Spinicaudatans and Cladocerans (Beaton 1988) indicate that closely related species have similar-sized genomes. For example, the clam shrimp Lynceus brachyurus has a genome size of 290 Mb (Beaton 1988), and the water flea Daphnia magna also has a genome size of 290 Mb (Jalal et al. 2013). Strangely, the closest measured relative of the clade containing E. texana and D. pulex, Anostraca, contains species with vertebrate-sized genomes. The measured anostracan genome sizes (respectively, Artemia salina, Branchinecta paludosa, and Artemiopsis stephanssoni) are 2.8, 2.7, and 850 Mb (Rheinsmith et al. 1974; Beaton 1988).
HOX Gene Annotation
In order to validate our annotation and assembly, we attempted to identify HOX genes in the clam shrimp genome, and compare their order to that of the HOX genes in D. melanogaster. HOX genes are an interesting test case as they are important in development, they are believed to cluster in two different chromosomal regions in invertebrates, their order tends to be conserved across all animals, and that order reflects where they are expressed along the anterior/posterior axis (reviewed in Duboule 2007). We identified HOX genes using mutual best hit BLAST and found six apparent HOX genes spread across two contigs (C0002 and C0007, fig. 2). We then used a protein–protein BLAST of all E. texana annotated genes onto all D. mel annotated genes, and identified five more genes that BLASTed to the D. mel HOX region. We removed one of these genes (C0002.g600) from the analysis because, upon multiple alignment with Clustal-Omega (Goujon et al. 2010; Sievers et al. 2011; McWilliam et al. 2013), there was no evidence that it contained a HOX motif. Finally, we ran a tBLASTx of the D. mel HOX genes against the E. texana genome to identify possible unannotated HOX genes in the region, and found two more candidates. Although we cannot confirm the reason that these putative HOX genes were not annotated in our genome, it may be that they are only expressed in the larval stages of the E. texana life cycle, which we did not sequence. Because we based our annotation on RNAseq data, any genes not expressed in adults would not be annotated. BLAST was unable to identify orthologs of the D. mel HOX-associated miRNAs miR-iab-4, miR-iab-8, miR-10, or miR-993. We took this collection of 12 genes, found orthologs between E. texana and D. mel using Clustal-Omega and MEGA v. 7.0.26 (supplementary figs. 1 and 2, Supplementary Material online; see also Kumar et al. 2016), and hand-ordered them relative to D. mel. The identity of these genes is not certain, but from our results, it appears that nearly all genes are grouped spatially with their orthologs, and the rough order of the orthologous gene groups is conserved between D. mel and E. texana, especially when comparing the D. mel Antennapedia complex to the E. texana genome (of the D. mel Bithorax complex, only Abd-A orthologs were identified in E. texana) (fig. 2). In addition, Contig 2 appears to contain a partial duplication of the Antennapedia locus from D. mel.
The identified HOX genes are divided across two contigs (contigs 2 and 7). We identified six putative HOX orthologs on Contig 2, and another six on contig 7. Because we do not have access to linkage information, it is possible that these contigs are each part of the same chromosome. Regardless, based on the distance from each HOX cluster to its respective contig edge, the clusters must be a minimum of 3.8 Mb away from each other. The clusters are similar in size at 214 kb for the contig 2 cluster, and 98 kb for the contig 7 cluster. The two HOX clusters contain a number of genes that are interspersed between the HOX genes: 10 in the contig 2 cluster, and 3 in the contig 7 cluster. In the contig 7 cluster, there are several runs of multiple HOX genes in a row. C0007.g24 and C0007.g25 correspond, respectively, to (1) either Scr or Dfd (the Clustal-Ω tree is unresolved here) and (2) Dfd. These genes are adjacent and collinear, as are C0007.g31 and C0007.g32, which correspond to pb and lab, respectively.
It has been observed in several crustaceans that AntP and Ubx are bicistronic—they are present on a single transcribed region of the genome (Shiga et al. 2006). Despite the high quality of the clam shrimp assembly, we failed to detect both AntP and Ubx. It is of note, however, that while the HOX gene orthologs are easily detected due to the presence of a HOX motif in each one, the identity of each ortholog is difficult to confirm. Indeed, different BLAST search schemes associated a given HOX ortholog with up to five different HOX genes; hence, our reliance upon multiple alignment and tree building to determine orthology. The upshot of this is that it is possible that AntP and Ubx orthologs actually are included in our HOX gene set, but are mis-annotated. It is also possible that these genes are truly absent in clam shrimp. This carries over to another point: overall, the HOX genes of E. texana appear atypical for a crustacean, and certainly seem to be worthy of further investigation. An organism with no Ubx, AntP, or Abd-B, but two copies of lab and pb, and three to four copies of Scr and Dfd, could reveal a great deal about the relationship between segment development and HOX gene expression. Still, based only on the data available here, it is difficult to precisely determine which E. texana HOX genes are orthologous to which D. melanogaster HOX genes, so some sort of independent confirmation of these HOX gene identities is needed before strong conclusions can be drawn. Here, then, are tentative comparisons to known arthropods with atypical HOX gene configurations. There are several arthropods with vastly reduced HOX gene sets, including C. elengans nematodes, which are missing Pb, zen, Dfd, Antp, Ubx, and abd-A (Aboobaker and Blaxter 2003), and tardigrades, which are missing Pb, Scr, AntP, Ubx, and abd-A (Smith et al. 2016). It is interesting to note that both of these organisms have lost both AntP and Ubx, and both have body plans that differ markedly from the average arthropod. This brings up the possibility that loss of AntP and Ubx, along with other changes in Hox gene organization, may be central to reorganization of development in arthropod species with highly divergent body plans.
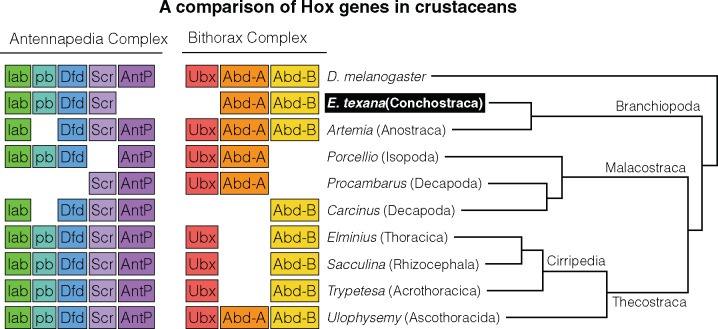
Several studies have attempted to identify patterns in the rearrangements and losses of the known HOX genes across the arthropods. In contrast to vertebrate development, crustacean development apparently does not require HOX genes to be collinear, transcribe in the same direction, or match the gene order to the order of segments as in Drosophila (Dressler and Gruss 1989). There are 10 canonical HOX gene ancestors from which all arthropod HOX orthologs are apparently descended (Akam et al. 1994), and duplication and loss of these genes is fairly common across the arthropod tree (Deutsch and Mouchel-Vielh 2003). Our annotation allowed us to identify orthologs of 6 out of these 10 genes (fig. 4), and we were not able to locate AntP, Ubx, zen, or ftz. The closest relative of E. texana with thoroughly investigated Hox genes, the Anostracan shrimp Artemia, is missing pb and zen, meaning that, assuming both annotations are complete, the overlap of missing Hox genes between these two species is limited to zen. In addition, the E. texana genome is apparently lacking orthologs for the Antp and Ubx genes, which are rarely lost (except in the case of the Decapod Carcinus, which is missing Ubx). As noted above, due to the challenge of correctly assigning the HOX orthologs, it is difficult to say with certainty that the Antp and Ubx gene orthologs are truly lost in E. texana; we recommend further inquiry into this topic in future studies. Finally, there is a known relationship between Ubx and Antp expression and the presence of maxillipeds on the thorax in crustaceans. In most crustaceans, Ubx and Abd-a expression in a thoracic segment correspond with loss of maxillipeds at that segment (Deutsch and Mouchel-Vielh 2003). Clam shrimp are lacking maxillipeds across their thoracic region. Thus, it must either be the case that Ubx is present but unannotated in the clam shrimp genome or that Ubx is not required for maxilliped development. With currently available data, it is difficult to determine which of these hypotheses is correct, though Deutsch and Mouchel-Vielh (2003) note, based on segment expression data, that the removal of the Ubx protein might be necessary but not sufficient for maxilliped development. Thus, current knowledge about Ubx does allow for the possibility that clam shrimp may have lost Ubx without gaining maxillipeds.
Fig. 4.
—An illustration of Hox gene loss in the crustaceans. Tree branch lengths are not informative. As shown here, the loss of AntP and Ubx is uncommon in crustaceans.
Immunity Gene Annotation
Recent work has produced a thorough catalogue of the arthropod genes that relate to immune system function (Waterhouse et al. 2007). Immunity genes are believed to be among the most rapidly evolving in insects (Sackton et al. 2007), and it stands to reason that this will hold true in other arthropods. We identified orthologs of these immunity genes in E. texana with BLAST. We extracted the complete list of known arthropod genes from ImmunoDB (Waterhouse et al. 2007) and BLASTed it against the E. texana protein set. Although only 106 genes were identified as orthologs by mutual best hit BLAST, 279 of the 346 ImmunoDB genes either BLASTed the collection of E. texana proteins, or were BLASTed by the collection of E. texana proteins, with an e-value below 10−5, implicating a total of 1,184 E. texana genes in possible immune activity. Still, this analysis focuses on the 106 mutual best hit genes in order to remain conservative. In a given immunity gene family, the percentage of genes that successfully hit orthologs could vary dramatically, from 0% in the cases of the AMP and PGRP families to 100% in the JAKSTAT family (table 1).
Table 1.
A List of Gene Counts for Drosophila melanogaster Immune-Related Genes (from ImmunoDB) and the Count and Fraction of Orthologs Detected in the Eulimnadia texana Proteome
| Immune Gene Families with Orthologs in E. texana | |||
|---|---|---|---|
| Gene Family | D. melanogaster Genes | E. texana Orthologs | Fraction Identified |
| ML | 10 | 1 | 0.10 |
| CLIP | 47 | 12 | 0.26 |
| TOLLPATH | 5 | 4 | 0.80 |
| IAP | 4 | 3 | 0.75 |
| CTL | 34 | 7 | 0.21 |
| AMP | 21 | 0 | 0.00 |
| IMDPATH | 8 | 5 | 0.63 |
| CAT | 2 | 1 | 0.50 |
| SPZ | 6 | 5 | 0.83 |
| GALE | 7 | 2 | 0.29 |
| PRDX | 20 | 11 | 0.55 |
| SRPN | 29 | 3 | 0.10 |
| SCR | 21 | 10 | 0.48 |
| PPO | 3 | 1 | 0.33 |
| PGRP | 22 | 0 | 0.00 |
| FREP | 14 | 4 | 0.29 |
| TOLL | 9 | 2 | 0.22 |
| JAKSTAT | 3 | 3 | 1.00 |
| CASP | 7 | 1 | 0.14 |
| CASPA | 6 | 1 | 0.17 |
| APHAG | 23 | 13 | 0.57 |
| LYS | 13 | 1 | 0.08 |
| REL | 3 | 2 | 0.67 |
| SRRP | 13 | 9 | 0.69 |
| BGBP | 3 | 1 | 0.33 |
| SOD | 4 | 2 | 0.50 |
| TEP | 10 | 3 | 0.30 |
Comparison of these results to those of other species reveals broad trends of gene family gain and loss. Analyses of insect genomes (specifically, D. melanogaster, Anopheles Gambiae, and Aedes Aegypti—Waterhouse et al. 2007) reveals that specific gene families are highly conserved and are likely to have common orthologs between species, while other gene families tend to have species-specific genes with no detectable orthology. In particular, AMP family genes tend to be highly species-specific, CTL family genes tend to be intermediate, and the IAP, SOD, and SCR gene families tend to be conserved. This is reflected in our results—if we rank the E. texana gene families by the fraction of D. melanogaster genes with detected E. texana orthologs, we find that AMP < CTL < IAP, SOD, SCR. Thus, E. texana data seem consistent with Waterhouse et al. (2007)’s conclusion that AMP and CTL are relatively fast-evolving gene families. On a similar note, recent sequencing of the shrimp P. hawaiensis allowed for the observation that the PGRP family, which is present in most arthropods, does not seem to occur in Parhyale, and therefore may not exist in crustaceans; on the other hand, Toll-like receptors were found in Parhyale. Clam shrimp reveal the same pattern: mutual best hit BLAST did not reveal any PGRP orthologs in E. texana, but two of nine D. melanogaster Toll-like receptors were found to have E. texana orthologs. It is not likely that the undetectability of PGRP is simply a problem with the low power of mutual hit BLAST, as BLAST failed to identify a single blast hit either to or from PGRP. It is possible that the PGRP family’s orthologs cannot be identified because PGRP is a rapidly evolving gene family, like AMP and CTL above, but Waterhouse et al. (2007) actually indicates that PGRP is a relatively highly conserved family. In the absence of other evidence, it seems that total loss of PGRP genes from the crustacean genome is a reasonable hypothesis.
Differential Expression
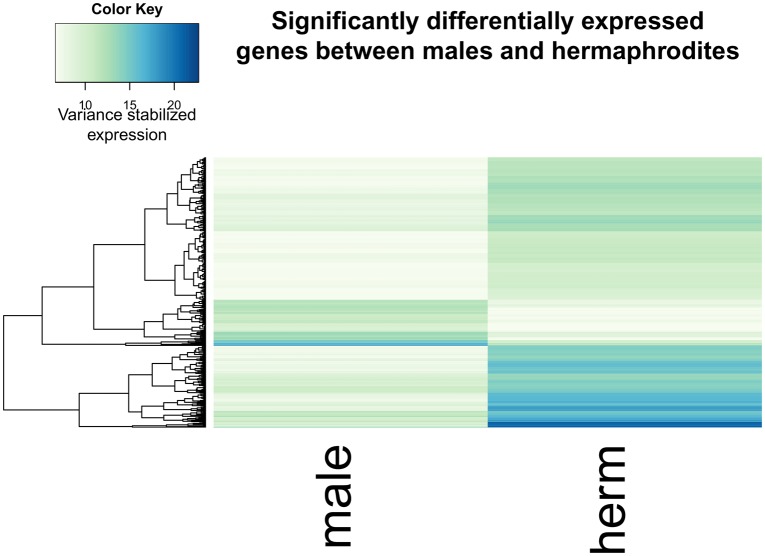
We next compared the RNAseq data from males and hermaphrodites to identify differentially expressed genes. We found 486 differentially expressed genes (Benjamini–Hochberg–Yekutieli [Benjamini and Yekutieli 2001] adjusted P-value <0.05) (fig. 5) out of the 17,667 genes identified by Augustus. Forty of these genes are among the genes with D. melanogaster orthologs. Gene ontology enrichment analysis with GOrilla (Eden et al. 2009) indicates an enrichment of the following GO terms based on the rank order of significance of differential expression (GO terms with a Benjamini–Hochberg corrected P-value <0.05 are listed): structural constituent of cuticle, chitin binding, structural constituent of chitin-based larval cuticle, structural constituent of chitin-based cuticle, carboxypeptidase activity, chitin deacetylase activity, and association with the condensin complex, extracellular region, and DNA packaging complex (supplementary table 2, Supplementary Material online). Hermaphrodites have both testes and ovaries, while males have only testes; additionally, hermaphrodites typically store up to several hundred large eggs in their carapace prior to ovipositioning (Weeks et al. 1997). These two large phenotypic differences between males and females are likely to drive many of the observed expression differences.
Fig. 5.
—A heat map of expression for genes differentially expresses between males and hermaphrodites (adjusted P<0.05). Note the small portion of genes that have nearly zero expression in males, and high expression in hermaphrodites.
Sex Locus Localization
The quality of the clam shrimp genome assembly allows us to identify the contig harboring the sex-determining locus of E. texana. Previous analyses of allozymes and microsatellites (Weeks 2004; Weeks et al. 2010) indicate the sex determining locus is linked to several markers, with at least three markers so tightly linked that they can be used to genotype the sex locus status of individuals (ZZ vs. ZW vs. WW) with relatively high accuracy. We used BLAST (Altschul et al. 1990) to align the sequences of the four best such markers (the allozyme Fum and microsatellites CS8, CS11, and CS15) to the E. texana assembly (supplementary fig. 3, Supplementary Material online). We found that the three microsatellite loci aligned to our largest (41 Mb; contig 1) contig, while the allozyme Fum aligned to a smaller 1 Mb contig (that we speculate would join contig 1 in a more contiguous assembly). The order (in the assembly) of the three microsatellite markers that map to contig 1 does not agree with the order inferred genetically in Weeks et al. (2010; see figure), indicating a problem with either the mapping or the assembly. We are relatively confident in the quality of our assembly, and there is reason to think that the mapping could be incorrect. Weeks et al. (2010) found a very high rate of recombination between three microsatellites when looking at male meiosis. Specifically, he observed recombination distances of 94 and 73 cM; recombination fractions indistinguishable from free recombination. In contrast, in hermaphrodites, adjacent markers were separated by a very small number of recombinants with only approximately 5 total crossover events inferred in 170 individuals. We posit that recombination does not occur in amphigenic hermaphrodites, or occurs very seldom, and that much of the inference of marker order may actually be due to a low (∼1%) rate of mis-genotyping of the microsatellite markers. Our highly contiguous genome allows for future experiments to determine if indeed amphigenic hermaphrodites experience recombination in E. texana.
It is important to note that the genome assembly was produced using data from WW hermaphrodites. Thus, the male version of the sex-determining locus is not expected to be present in the genome assembly. This may make detection of the sex-determining locus more difficult, depending on the divergence of the “Z” and “W” versions of the sex locus. If the two loci are highly diverged, they may not align to each other; on the other hand, if they are not highly diverged, they may align to each other, but show a signal of increased polymorphism. A future de novo assembly of a male will help elucidate the location of the sex determination locus.
Residual Variation and Assembly Errors
We aligned the Illumina data from the inbred JT4(4)5-L line used for the genome assembly to the reference genome and observed SNPs at a rate of 0.00018 per bp. This indicates, as expected, a very low SNP rate within the inbred strain we sequenced (supplementary fig. 4, Supplementary Material online). If JT4(4)5-L was not fully inbred then we expect runs of heterozygous sites, whereas isolated SNPs are likely assembly errors. Consistent with this prior belief, there are notable differences in patterns of heterozygosity amongst the contigs. The largest three contigs are almost completely free of heterozygosity (0.000024 SNPs per bp), reflecting a very low assembly error rate at with respect to point mutations. In contrast, the fourth contig and several others generally have higher levels of heterozygosity (contig 4: 0.00021 SNPs per bp). 64% of the heterozygosity in the genome is contained in the 26 most SNP-dense contigs, which account for only 5.6 Mb of the genome. Thus, most of the genome is nearly heterozygosity free with blocks of residual heterozygosity. We speculate that these small contigs with high levels of heterozygosity could be mis-assembled, leading to incorrect read mapping that appears as heterozygosity, or regions that did not become homozygous following inbreeding that then failed to assemble adequately because of the heterozygosity therein.
Discussion
On Nonmodel Organisms and Genome Assembly
One of the long standing assumptions in genomics is that high-quality whole-genome genetic analysis is not possible with nonmodel organisms because of the lack of genetics resources available for such systems, such as genome assemblies and annotations. Before the advent of high-throughput sequencing (i.e., Illumina sequencing), nonmodel genome assembly was prohibitively expensive. The human genome project cost approximately $3 billion, while the Celera human genome assembly was seen as comparatively affordable at $300 million. The advent of Illumina sequencing and De Bruijn graph assembly dropped the cost of genome assembly to on the order of $10,000—depending on the genome size and complexity—but the contiguity of these assemblies tended to be low because of the short length of Illumina-type reads. Thus, most arthropods, with the exception of D. melanogaster, have had low contiguity genome assemblies when they have assemblies at all. One of the most studied insects, the Heliconius melpomene butterfly, is a representative example. Its 454 and Illumina-based assembly, published in 2012 by a large consortium, had an N50 of 277 kb (Heliconius Genome Consortium 2012), which was considered very respectable contiguity for a nonmodel assembly at that time. In 2016, PacBio sequencing and linkage analysis was used to bring the N50 of H. melpomene to 2.1 Mb, highlighting the advances possible with long read sequencing technology (Davey et al. 2016). Still, outside of the insects, high-quality genome assemblies are rare. Daphnia pulex, which has been used as a model organism for many years, has an assembly with a scaffold N50 of 470 kb (Colbourne et al. 2005). We have now generated what is, to our knowledge, the most contiguous crustacean assembly ever completed. Here, we demonstrate that the generation of a genome for a new model organism is not necessarily difficult or costly. Modern sequencing techniques (i.e., PacBio) allow for de novo genome assembly of a ∼200 Mb genome for ∼$10 K USD. A preliminary genome annotation using RNAseq for a handful of tissues can be accomplished for ∼$3 K USD. This combination of factors makes genomics in nonmodel systems an attractive target for evolutionary biologists.
We present here a de novo whole genome assembly for E. texana with an N50 of 18 Mb. This genome will be a useful resource for the vernal pool research community, and will elevate the status of clam shrimp as an emerging model organism. In addition, we present a draft annotation of the genome that allows for accurate identification of genic, intergenic, etc., regions, as well as homology-based comparisons with genes in other species. Finally, we carried out an initial analysis of differential gene expression between males and hermaphrodites and identify some gene ontology terms that seem to be associated with differential expression between males and hermaphrodites.
The Reduced E. texana Genome
The small genome size of E. texana (144 Mb) and the small size of the E. texana proteome (17,667 genes) relative to other crustaceans, seem to indicate an overall reduction in the E. texana genome compared to its best sequenced relatives. Based on this minimal information, we can only speculate as to the reason for this reduction. One major difference between E. texana and the extremely gene-rich D. pulex is the fact that Daphnia can switch between sexual and asexual reproduction. Colbourne (2011) suggests that this switching, and other phenotypic changes driven by environment in Daphnia, may require genes that are not needed in other organisms. It is also possible that gene number is over-estimated in fragmented genome assemblies, as was the case in humans before a complete draft assembly was completed. Early in the millennium, gene count betting pools predicted up to 100,000 genes in the human genome (Begley 2003). The E. texana gene count is remarkably similar to Drosophila and C. elegans, suggesting to us that gene counts are likely over-estimated in other arthropods. These are possible explanations for the difference in gene number between E. texana and Daphnia, but it does not explain the difference in genome size. Genome size is often believed to be inversely correlated with effective population size (Lynch and Conery 2003). Given that clam shrimp are substantially larger than Daphnia, it would be a surprise if they had a larger effective population size than Daphnia; on the other hand, E. texana is obligately sexual with selfing, and Daphnia is facultatively sexual, so the effective population sizes of the two organisms should not be assumed without a thorough genomic analysis.
The Proto-Sex Chromosome
Much effort has gone into identifying the structure of the sex locus in individuals with recently derived sex chromosomes (Zhou and Bachtrog 2012; Charlesworth 2013). Eulimnadia texana is androdioecious, but is believed to be descended from a dioecious ancestor that was ancestral to the entire Eulimnadia clade (Weeks et al. 2009). Linkage analysis has indicated that the sex-determining region is likely to be a large autosomal linkage group or a “proto-sex” chromosome. We identified a single contig that contained all but one of the previously identified sex-linked markers. This contig likely harbors the sex determining linkage group. Linked genetic markers were spread across the entire 42-Mb contig, and the order of the markers differed from the order predicted by linkage mapping. It is not clearly relevant to the evolution of sex chromosomes, but it is an interesting observation that the sex chromosome represents roughly a third of the clam shrimp genome. We were unable to identify the sex-determining locus within this chromosome, since it is possible that hermaphrodites do not recombine, as is the case in D. melanogaster (Lenormand 2003) and other organisms. A lack of recombination in hermaphrodites would make linkage-mapping the sex-determining locus impossible. Our genome assembly should allow for new experiments using SNP markers to confirm or refute the existence of recombination in hermaphrodites and perhaps map the sex-determining locus. Alternatively, a second male specific assembly, in concert with GWAS-type approaches, may allow the sex determining region to be identified. In addition, we cannot rule out the possibility that the entire chromosome, rather than a narrow locus is involved in sex determination.
We mapped RNAseq derived transcripts from hermaphrodites and males back to the genome assembly. Despite our ability to map ∼99% of hermaphrodite transcripts back to the reference genome, ∼4% of the male transcripts failed to map. Thus, there are transcripts present in males that are too distinct to map to the hermaphrodite derived genome assembly. This suggests one of three possibilities: first, there may be a genomic region that only occurs in males, which is absent from our current assembly; second, there is a region present in both male and hermaphrodite versions of the genome, but that the male and hermaphrodite alleles are too diverged from one another for male derived transcripts to map back to hermaphrodite alleles; or third, some male transcripts do not map back to the reference simply due to polymorphism segregating in this species. We note that the RNAseq data were obtained from two different strains, with the hermaphrodite strain being the same one from which the assembly is derived. A further study could elucidate which of these hypotheses is correct by generating a whole genome assembly of a male genome (or, although less informative, aligning hermaphrodite specific transcripts from the WAL strain back to the reference genome).
Conclusions
We generated a highly contiguous, annotated genome assembly with an N50 of 18 Mb for the clam shrimp E. texana. This genome assembly allowed us to identify numerous genes with homology to genes in D. melanogaster, and we identified a subset of these genes as being differentially expressed between males and females.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank the UC Irvine genomics core facility and the University of Kansas genomics facility, as well as Stuart Macdonald, for assistance in library preparation and sequencing. This work was supported by NIH grants AI126037, GM115562, and OD10974 to A.D.L., and NSF grant DEB-9628865 to S.C.W.
Literature Cited
- Aboobaker A, Blaxter M.. 2003. Hox gene evolution in nematodes: novelty conserved. Curr Opin Genet Dev. 13(6):593–598.http://dx.doi.org/10.1016/j.gde.2003.10.009 [DOI] [PubMed] [Google Scholar]
- Akam M, et al. 1994. The evolving role of Hox genes in arthropods. Development 1994:209–215. [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinform J. 7(1):1–8. [Google Scholar]
- Beaton MJ. 1988. Genome size variation in the cladocera. MSc thesis, University of Windsor, Windsor, Ontario, Canada
- Begley S. 2003. How many genes does it take to make a human? Wanna bet? Wall Street Journal May 23 http://www.wsj.com/articles/SB105363997823088600, last accessed December 19, 2017. [Google Scholar]
- Benjamini Y, Yekutieli D.. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 29(4):1165–1188. [Google Scholar]
- Berlin K, et al. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 33(6):623–630. [DOI] [PubMed] [Google Scholar]
- Brendonck L. 1996. Diapause, quiescence, hatching requirements: what we can learn from large freshwater branchiopods (Crustacea: Branchiopoda: Anostraca, Notostraca, Conchostraca). Hydrobiologia 320(1–3):85–97.http://dx.doi.org/10.1007/BF00016809 [Google Scholar]
- Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ.. 2016. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 44:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. 2013. Plant sex chromosome evolution. J Exp Bot. 64(2):405–420.http://dx.doi.org/10.1093/jxb/ers322 [DOI] [PubMed] [Google Scholar]
- Chin C-S, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10(6):563–569.http://dx.doi.org/10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N.. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 162(1):156–159. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, et al. 2011. The ecoresponsive genome of Daphnia pulex. Science 331(6017):555–561.http://dx.doi.org/10.1126/science.1197761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Singan VR, Gilbert DG.. 2005. wFleaBase: the Daphnia genome database. BMC Bioinformatics 6:45.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, et al. 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda) 6(3):695–708.http://dx.doi.org/10.1534/g3.115.023655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch JS, Mouchel-Vielh E.. 2003. Hox genes and the crustacean body plan. Bioessays 25(9):878–887.http://dx.doi.org/10.1002/bies.10319 [DOI] [PubMed] [Google Scholar]
- dos Santos G, et al. 2015. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucl. Acids Res. 43:D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR, Gruss P.. 1989. Anterior boundaries of Hox gene expression in mesoderm-derived structures correlate with the linear gene order along the chromosome. Differentiation 41(3):193–201.http://dx.doi.org/10.1111/j.1432-0436.1989.tb00747.x [DOI] [PubMed] [Google Scholar]
- Duboule D. 2007. The rise and fall of Hox gene clusters. Development 134(14):2549–2560.http://dx.doi.org/10.1242/dev.001065 [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z.. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323(5910):133–138.http://dx.doi.org/10.1126/science.1162986 [DOI] [PubMed] [Google Scholar]
- Ellis LL, et al. 2014. Intrapopulation genome size variation in D. melanogaster reflects life history variation and plasticity. PLOS Genet. 10(7):e1004522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, et al. 2014. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15:86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. 1995. Statistical properties of segregating sites. Theor Popul Biol. 48(2):172–197http://dx.doi.org/10.1006/tpbi.1995.1025 [DOI] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. PNAS 108(4):1513–1518.http://dx.doi.org/10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M, et al. 2010. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38(Web Server issue):W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 29(7):644–652.http://dx.doi.org/10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliconius Genome Consortium. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal M, Wojewodzic MW, Laane CMM, Hessen DO, Bainard J.. 2013. Larger Daphnia at lower temperature: a role for cell size and genome configuration? Genome 56(9):511–519. [DOI] [PubMed] [Google Scholar]
- Kajitani R. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395.http://dx.doi.org/10.1101/gr.170720.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3:research0084.1–84.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao D, et al. 2016. The genome of the crustacean Parhyale hawaiensis, a model for animal development, regeneration, immunity and lignocellulose digestion. eLife Sci. 5:e20062.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Schatz MC, Salzberg SL.. 2010. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 11(11):R116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12(4):656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.http://dx.doi.org/10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, et al. 2016. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat Genet. 48(1):84–88.http://dx.doi.org/10.1038/ng.3430 [DOI] [PubMed] [Google Scholar]
- Lan JH, et al. 2015. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum Immunol. 76(2–3):166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver T, et al. 2015. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif. 3:1–8..http://dx.doi.org/10.1016/j.bdq.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T. 2003. The evolution of sex dimorphism in recombination. Genetics 163(2):811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 302(5649):1401–1404. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C.. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, et al. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41(Web Server issue):W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, et al. 2000. A whole-genome assembly of Drosophila. Science 287(5461):2196–2204.http://dx.doi.org/10.1126/science.287.5461.2196 [DOI] [PubMed] [Google Scholar]
- Rees DJ, Dufresne F, Glémet H, Belzile C.. 2007. Amphipod genome sizes: first estimates for Arctic species reveal genomic giants. Genome 50(2):151–158. [DOI] [PubMed] [Google Scholar]
- Regier JC, et al. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463(7284):1079–1083.http://dx.doi.org/10.1038/nature08742 [DOI] [PubMed] [Google Scholar]
- Rheinsmith EL, Hinegardner R, Bachmann K.. 1974. Nuclear DNA amounts in Crustacea. Compar Biochem Physiol B: Compar Biochem. 48(3):343–348.http://dx.doi.org/10.1016/0305-0491(74)90269-7 [DOI] [PubMed] [Google Scholar]
- Richards S, et al. 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452(7190):949–955.http://dx.doi.org/10.1038/nature06784 [DOI] [PubMed] [Google Scholar]
- Sackton TB, et al. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 39(12):1461.http://dx.doi.org/10.1038/ng.2007.60 [DOI] [PubMed] [Google Scholar]
- Sassaman C, Weeks SC.. 1993. The genetic mechanism of sex determination in the conchostracan shrimp Eulimnadia texana. Am Nat. 141(2):314–328. [DOI] [PubMed] [Google Scholar]
- Shen R, et al. 2005. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 573(1–2):70–82. [DOI] [PubMed] [Google Scholar]
- Shiga Y, et al. 2006. Transcriptional readthrough of Hox genes Ubx and Antp and their divergent post-transcriptional control during crustacean evolution. Evol Dev 8(5):407–414.http://dx.doi.org/10.1111/j.1525-142X.2006.00114.x [DOI] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith CD, et al. 2011. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). PNAS 108(14):5673–5678.http://dx.doi.org/10.1073/pnas.1008617108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FW, et al. 2016. The compact body plan of tardigrades evolved by the loss of a large body region. Curr Biol. 26(2):224–229.http://dx.doi.org/10.1016/j.cub.2015.11.059 [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S.. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl 2):ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Sun X-Y, Xia X, Yang Q.. 2016. Dating the origin of the major lineages of Branchiopoda. Palaeoworld 25:303–317.http://dx.doi.org/10.1016/j.palwor.2015.02.003 [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111.http://dx.doi.org/10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL.. 2011. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 13(1):36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M.. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 10(1):57–63.http://dx.doi.org/10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316(5832):1738–1743.http://dx.doi.org/10.1126/science.1139862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks SC. 2004. Levels of inbreeding depression over seven generations of selfing in the androdioecious clam shrimp, Eulimnadia texana. J Evol Biol. 17(3):475–484.http://dx.doi.org/10.1111/j.1420-9101.2004.00712.x [DOI] [PubMed] [Google Scholar]
- Weeks SC, Benvenuto C, Sanderson TF, Duff RJ.. 2010. Sex chromosome evolution in the clam shrimp, Eulimnadia texana. J Evol Biol. 23(5):1100–1106.http://dx.doi.org/10.1111/j.1420-9101.2010.01963.x [DOI] [PubMed] [Google Scholar]
- Weeks SC, Chapman EG, Rogers DC, Senyo DM, Hoeh WR.. 2009. Evolutionary transitions among dioecy, androdioecy and hermaphroditism in limnadiid clam shrimp (Branchiopoda: Spinicaudata). J Evol Biol. 22(9):1781–1799. [DOI] [PubMed] [Google Scholar]
- Weeks SC, Marcus V, Alvarez S.. 1997. Notes on the life history of the clam shrimp, Eulimnadia texana. Hydrobiologia 359:191–197.http://dx.doi.org/10.1023/A:1003106702451 [Google Scholar]
- Weeks SC, Zucker N.. 1999. Rates of inbreeding in the androdioecious clam shrimp Eulimnadia texana. Can J Zool. 77(9):1402–1408.http://dx.doi.org/10.1139/z99-103 [Google Scholar]
- Ye C, Hill CM, Wu S, Ruan J, Ma Z. (Sam). 2016. DBG2OLC: efficient assembly of large genomes using long erroneous reads of the third generation sequencing technologies. Sci Rep. 6(1):31900.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, et al. 2017. A new reference genome assembly for the microcrustacean Daphnia pulex. G3 (Bethesda) 7:1405–1416.http://dx.doi.org/10.1534/g3.116.038638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D.. 2012. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337(6092):341–345.http://dx.doi.org/10.1126/science.1225385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.