Abstract
The development of new computational tools has recently opened up the study of natural behaviors at a precision that was previously unachievable. These tools permit a highly quantitative analysis of behavioral dynamics at timescales that are well matched to the timescales of neural activity. Here we examine how combining these methods with established techniques for estimating an animal’s sensory experience presents exciting new opportunities for dissecting the sensorimotor transformations performed by the nervous system. We focus this review primarily on examples from Caenorhabditis elegans and Drosophila melanogaster – for these model systems, computational approaches to characterize behavior, in combination with unparalleled genetic tools for neural activation, silencing, and recording, have already proven instrumental for illuminating underlying neural mechanisms.
Introduction
Behavior is dynamic, complex, and seemingly noisy – this presents challenges for both quantifying it and connecting it with the underlying neural activity that generates it [1]. One challenge with quantifying behavior is defining it – while neural activity can be measured in spikes or fluctuations of membrane voltage, scientists have yet to agree on a definition of what constitutes ‘behavior’. For example, a survey of ethologists [2] yielded largely inconsistent definitions. The respondents agreed with incompatible statements such as both ‘only animals behave’ and ‘algae chemotaxis is behavior’, ‘behavior is always executed through muscular activity’ but ‘sponges behave’ (without muscles), and ‘a person deciding to do nothing is behaving’ (without using muscles). One way to make progress on this question may then be to simply quantify as much as possible about what an animal does (what we here refer to as ‘behavior’). Thanks to developments in computer science and the increasing availability of high resolution, high-frame rate cameras, we are now able to capture and quantify orders of magnitude more data about animal movements and actions than was possible even a decade ago. Faced with these new large datasets, there has been a recent explosion of interest in developing algorithms to automate the classification of behavior. Rather than focus on summary statistics, these algorithms provide the ability to precisely measure individual-level variation in behavior, as it evolves over time. In addition, these methods represent a powerful alternative to human classification, which is typically slow, difficult to reproduce, and often introduces unwanted biases [3,4]. These automated tools further provide consistency when analyzing phenotypes that result from genetic and neural perturbations [4].
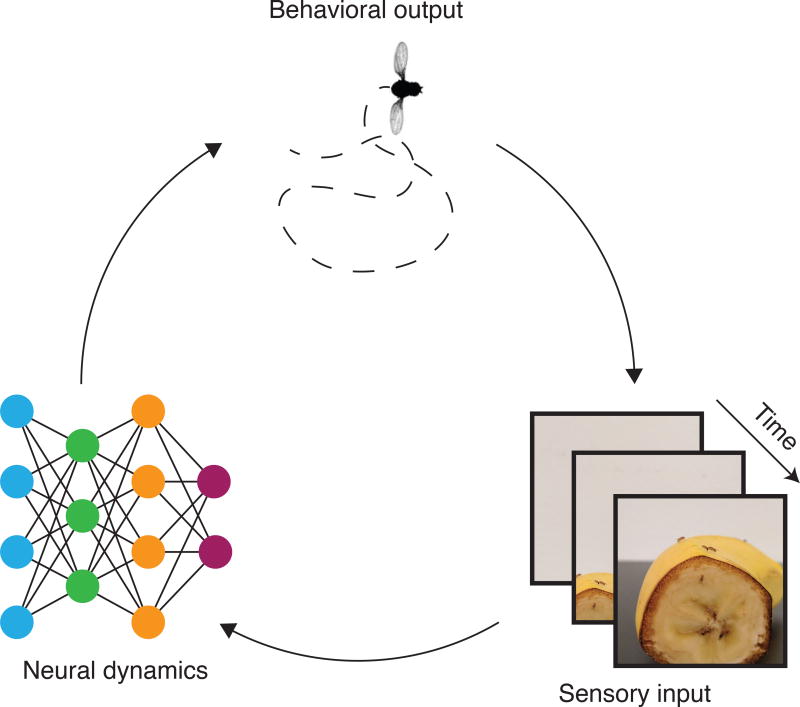
In parallel, new computational frameworks now allow scientists to estimate the sensory experience of the animal under study. That is, both the input and output space of the nervous system can be extensively sampled. These data can then be used to build models that not only predict behavior (keeping in mind that an animal’s own behavior affects its sensory experience), but also identify the internal computations that the nervous system performs. These capabilities become even more powerful when combined with a simple nervous system and genetic toolkit that facilitate testing predictions from models via targeted neural manipulations and recordings (e.g., determining the specific neurons involved in a given behavior and over what timescales). We therefore focus our review on new methods for behavioral quantification in worms and flies, delving into how to use quantitative behavioral analysis in combination with genetic tools to map full sensorimotor pathways, from neurons that process sensory information all the way to neurons that coordinate behavior (Figure 1).
Figure 1. Mapping full sensorimotor pathways.
Solving the sensorimotor transformations that nervous systems perform requires quantification of sensory inputs, neural dynamics and behavioral outputs. Sensory inputs influence neural activity which drives behavior, which in turn can change the sensory input that an animal receives (image of fly adapted from Muijres et al 2014).
Automated Methods to Quantify Behavior
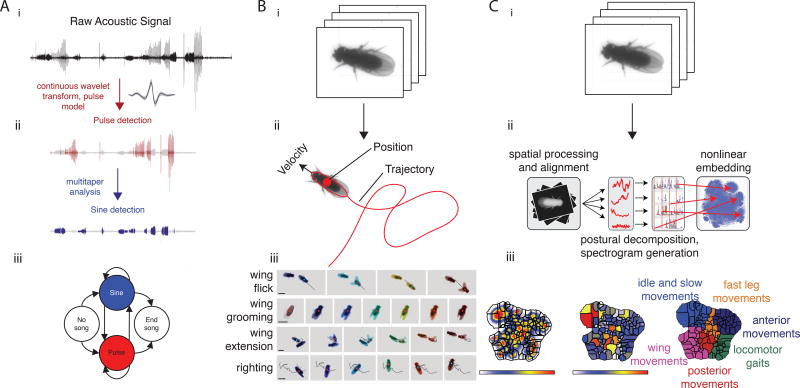
The most widely used automated methods take raw recordings – consisting of video, audio, or other measurements – and segment them into behaviors that are pre-defined by the experimenter (Figure 2Aii and 2Bii). For example, tracking software can find the location of animals in raw video frames by identifying pixels that differ from the background. The trajectory of each individual animal is then tracked across time, providing additional features of the animal’s behavior such as its velocity and changes in orientation [5–8]. More complex species-specific behaviors can be identified by adding additional assumptions. For instance, a fruit fly has two wings that can be recognized by virtue of being more transparent than the body – quantification of wing position permits identification of, for example, aggressive wing threats[9]. Other forms of raw data - such as acoustic recordings - can also be automatically segmented, for example by using a pre-defined template to identify when an animal is emitting a specific acoustic signal[10] (Figure 2A). While effective at classifying specific behaviors, these methods are not flexible. In contrast, the Janelia Automatic Animal Behavior Annotator (JAABA)[11] starts with video clips segmented using some of the aforementioned tools, but then allows the user to select video frames that correspond to a particular behavior (e.g., wing grooming) (Figure 2B). The program automatically ‘learns’ to identify all other frames with similar features. This class of machine learning algorithm can be easily trained on almost any type of data.
Figure 2. Automated algorithms for quantifying behavior.
(A) A fully supervised algorithm for segmenting acoustic behavior begins by (i) taking raw audio recordings, (ii) using an algorithm to identify salient features - here, ‘pulse’ and ‘sine’ types of song - and then inferring (iii) the longer-term bout structure (consisting of alternating trains of pulse and sine song) (modified from Arthur et al. 2013). (B) Starting with video data (i), other supervised algorithms (ii) find salient features such as center of mass (position), velocity, trajectory, and so on. (iii) A semi-supervised machine learning algorithm (JAABA) can use these features to identify discrete actions defined by the experimenter (modified from Kabra et al. 2013). (C) Largely unsupervised methods attempt to identify all behaviors from raw video data (i). (ii) One such method takes a set of aligned images from movies of flies and decomposes the dynamics into a low-dimensional basis set. Time series are produced by projecting the original pixel values onto this basis set, and these trajectories are then embedded into two dimensions (using t-SNE). (iii) Each position in the behavioral map corresponds to a unique set of postural dynamics, with nearby points representing similar motions (modified from Berman et al 2014). Maps are built by computing the probability of being embedded in this point in 2D space (left), then clustered with a watershed algorithm into discrete actions (middle) before identifying what general behaviors large regions of space belong to (right).
Ultimately though, one would like to identify all the actions that an animal performs (e.g. singing, running, or reaching) without needing the experimenter to painstakingly define one after another. The first step in this process is to identify some simpler representation of an animal’s posture. The power of this analysis was first shown in C. elegans, an animal that can be described as a curved 1-dimensional line. Performing a dimensionality reduction technique known as principal component analysis (PCA) revealed that only four dimensions account for the overwhelming majority of a worm’s postures [12]. How the animal’s posture changes in each of these dimensions is sufficient to explain a wide variety of behaviors, from foraging to thermotaxis[12,13], and has led to the discovery of new behaviors [14] as well as identified novel roles for genes in behavior[15]. Similar approaches that use automated tools to more closely quantify posture have been used to characterize larval zebrafish [16] and larval Drosophila [17] behavior.
While the aforementioned methods provide a description of an animal’s posture at any given moment in time, they lack information about the context in which those postures were used. Converting postures (e.g., ‘straight forearm’ or ‘extended elbow’) into actions (e.g., ‘reaching’) requires some way of clustering together stereotyped sequences of postures that are repeatedly seen with only minor variations. Methods to accomplish this range from embedding postures into a low-dimensional space to find ‘clumps’ of similar sequences[18] to identifying when one posture is predictable from previous postures[19], though other methods exist[15,17,20,21]. What is exciting about these ‘unsupervised’ algorithms is that they offer not only ever-more-precise quantification of what an animal is doing at each moment in time, but also reveal the underlying structure of behavior (e.g., which behaviors are sub-programs of other behaviors or which behaviors co-occur), and offer the potential for discovering completely new behaviors. These methods are not limited to single animals either: additional behaviors may fall out of clustering data from multiple animals engaged in social behaviors[22,23]. In addition, one can ascend another level of timescales and cluster actions into ‘sequences of actions’. Doing so in Drosophila reveals long timescale structure in behavior that does not fall out of the simple transition probabilities between actions[24].
These methods provide the foundation for dissecting the underlying neural circuit activity. With sophisticated tools for behavioral quantification in hand, we can ask which neurons are necessary and which sufficient to drive a particular behavior by using the powerful genetic toolkits available for C. elegans and Drosophila [25,26]. Even single neurons and cell types can be targeted, providing a level of circuit dissection simply unavailable in larger organisms. For example, one study screened the individual behaviors of more than 37,000 Drosophila larvae, each expressing Channelrhodopsin in one of roughly 1,000 different neural populations [27]. Using a behavioral tracker, the authors identified behaviors driven by activation of a neural population and then built maps connecting specific neurons to each behavior. Rapidly screening so many animals would not have been possible without automated methods. A more recent study expanded this approach to adult Drosophila, automatically cataloging the neural correlates of behaviors using 400,000 flies and targeting 2,204 different neuronal populations for activation [28]. Other studies in adult flies have used similar automated strategies to identify, for example, the connection between specific visual neurons and behavior [29] or that the same subset of neurons can induce different behaviors (courtship vs. aggression), depending on levels of neural activation [30]. In C. elegans, automated behavioral classification has been used to distinguish between modes of foraging behavior, and then connect these modes with specific neuromodulatory pathways [31], or to identify the specific neural subsets involved in discrete aspects of random search behavior [32]. Automated behavioral analysis can also lead to a more precise manipulation of neural activity [33].
Linking Sensation to Action
Although these new methods for behavioral quantification offer a greater understanding of what the animal is doing, they are in general agnostic to the sensory experience of the animal. Bringing together highly quantitative readouts of both behavior and dynamic sensory stimuli will enable mapping complete sensorimotor transformations [34].
One way to begin is to simply activate sensory neurons directly and quantify which behaviors are produced as a result [29,35–37] (Figure 3A). By varying the activity of sensory neurons and observing the motor output, one can directly compute the mapping the animal uses for behavior. However, animals that are naturally behaving receive a diverse range of sensory inputs. Establishing which inputs drive behavior can be challenging as natural sensory spaces not only contain both temporal and spatial correlations, they can also be altered by an animal’s own behavior. This means that simply examining the stimuli that precede or co-occur with a particular behavior is insufficient. Instead, newer computational techniques are needed to estimate relevant sensory features in spite of these correlations, by finding those most linked to the output[38–40]. These methods, in general, fit a linear filter that processes an input before being passed through a nonlinear function [41]. The linear filter (Figure 3, column iii) describes which temporal features of the inputs drive the output. Though these methods were initially developed to predict neural activity from complex sensory stimuli, they can be extended to predict behavior. This has now been done in a few cases using C. elegans and Drosophila, highlighted below.
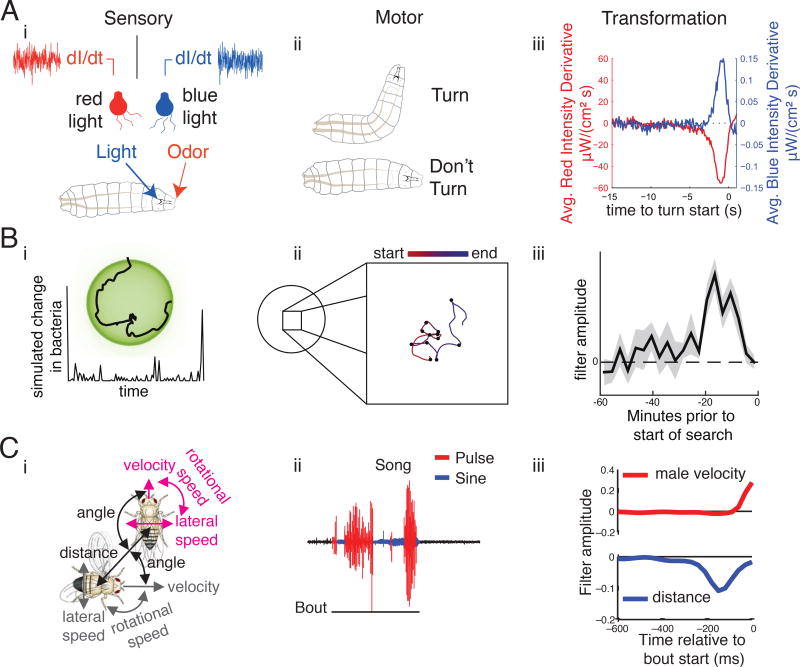
Figure 3. Inferring sensorimotor transformations from behavioral data.
(A) Visual and olfactory pathways can be driven by two different colors of light use Gaussian white noise patterns (i). On the basis of this activation, Drosophila larvae will decide whether or not to turn (ii). (iii) A reverse-correlation analysis finds the filters that transform sensory neuron activation into behavioral choices (modified from Gepner et al 2015). (B) (i) C. elegans that are on a food patch will explore (insert) and experience changes in food concentration. (ii) When they are exploring off-food, they will explore a small area by emitting a certain number of large-angle turns (represented here as black dots) to interrupt their forward locomotion and keep them in a small area. (iii) This sensory experience and turning output can be linked by a linear filter that keeps track of ~25 minutes of experience (modified from Calhoun et al 2015). (C) (i) Drosophila males will court a female by observing features such as the distance between the male and female and her velocity and (ii) will produce a dynamic song via wing vibration. (iii) The type of song that is emitted can be predicted based on the male’s own velocity and the inter-fly distance, for instance (modified from Coen et al 2014).
For example, when C. elegans leave a patch of food they will forage for new food by searching a local area before switching to a more global search (Figure 3B). How does this search strategy - this particular sequence of actions - change based on the animal’s sensory experience? The sensory environment can be estimated by measuring the concentration of food and position of the animal’s nose, while the motor output can be measured using automated tools described above to track the worm’s trajectory[42,43] (similar measurements have been made using Drosophila larvae in an olfactory environment [44]). Sensorimotor filters obtained from these data reveal a role for long timescale integration (~25 minutes) in guiding foraging decisions. Identifying these transformations enables finding the neurobiological underpinnings. In this case, the temporal width of the filter is determined by the concentration of the CREB molecule in a single pair of interneurons.
Social interactions add another layer of complexity, where the behavioral output of one animal serves as input to the sensorimotor processes of another animal. Drosophila courtship was previously thought to be comprised of a static sequence of actions, but measurement of the movements of two flies along with the song that the male produces identified a novel role for sensory feedback (e.g., the movements of the female) in song patterning decisions (e.g., choosing which song mode to produce at each moment in time) [45] (Figure 3C). Although courtship involves a dynamic feedback loop between two animals, the loop can be ‘broken’ by presenting controlled artificial stimuli. For example, the motion of an apparent female can modify the amplitude of song pulses produced by a tethered, walking male[46]. Similar analyses of the sensory cues that drive behavior can also be performed in flying animals, though this has the added complexity of requiring more degrees of freedom to explain sensorimotor transformations in a 3D environment[47–49]. These studies that rely on the precise measurement of behavior reveal that events that were previously considered ‘random’ or ‘noisy’ are, in fact, carefully orchestrated sensorimotor processes (though note that this careful orchestration may also include some beneficial stochasticity, for example in creating variable behavioral responses to reliable sensory stimuli [50,51]).
Dissecting Circuits for Sensorimotor Behaviors
How exactly does sensation lead to changes in behavior? An early example of solving this problem comes from the careful measurement of turning responses to visual motion [52,53]. These measurements suggested models with a clear prediction: motion should be detected via correlated activation of two nearby units coupled by a delay (such that activation of one unit followed by activation of the other would appear as motion). Building on these models, recent studies using Drosophila and its genetic and neural circuit toolkit, have identified pairs of visual neurons that implement precisely this algorithm: one pair for correlated increases in luminance and one pair for correlated decreases [54]. Further quantification of the sensory environment that flies experience in nature has revealed more accurate descriptions of both visuomotor behavior and the underlying circuitry [55,56]. These examples highlight the value of starting with a detailed model of a sensorimotor process inspired by quantitative behavioral data in order to solve neural circuit mechanisms.
For some behaviors, it may not be as obvious which neural pathways are involved – in this case, genetic and neural circuit manipulations combined with quantitative models of behavior can provide insights. For example, prior to the quantification of Drosophila courtship described in the previous section, there was little reason to examine neural coding in visual pathways when considering the neural basis for courtship song patterning [45]. However, the modeling made the key prediction that distance between two individuals is a major predictor of song choice (pulse versus sine), which was confirmed with mutations that rendered the male fly blind. Similarly, in a separate study, high-resolution behavioral tracking of groups of Drosophila presented with an aversive odor identified a social component to odor avoidance. By silencing and activating leg mechanosensory neurons, the authors found a surprising role for touch receptors in collective responses to odor stimuli [57].
Beyond identifying which neurons contribute to behavior, we would also like to identify the patterns of neural activity that are responsible for driving such dynamic behavior. To gain cellular-resolution access to the nervous system, it is often necessary to head-fix or immobilize the animal – while this limits behavior, it is still possible to identify neural correlates of sensorimotor processing from such recordings using either electrophysiology or imaging of calcium sensors [58]. For example, worms are typically restrained in a microfluidic chip for calcium imaging [59]. In such a preparation, whole-brain imaging revealed that the neural dynamics are highly constrained to only a few main modes of activity, each of which corresponds to distinct actions [60]. In contrast, head-fixed flies can be allowed to fly [61] or to walk on an air-supported treadmill [62]. Stimuli can then be presented in closed-loop with the fly’s motion, and the nervous system can be imaged at high resolution in such virtual environments. Employing such a setup has already revealed many of the sensorimotor processes that underlie spatial navigation in Drosophila [63,64]. Recording neural activity during behavior also facilitates finding neural responses that co-vary with behavior on short timescales (such as within a single trial). For instance, visual cells in the Drosophila eye responsive to wide-field horizontal or vertical motion also encode information about the velocity of the animal’s self-motion to compensate for the expected effect on the incoming visual signal [65,66].
Remarkably, we are already able to record from the majority of the neurons, at cellular resolution, in two animals during free behavior, C. elegans [67,68] and another invertebrate, Hydra [69], though progress is being made toward similar capabilities in adult Drosophila [70,71], larval Drosophila [72], and larval zebrafish [73]. For C. elegans, real-time quantification of movement drives the motion of a motorized stage that re-positions the worm directly under the neural imaging scope [67,68]. Such experiments can connect population neural activity with natural behaviors, such as forward or backward motion, or turning, and can even track activity over long-term behavioral state changes [74]. Due to the complexity of the neural dynamics, dimensionality reduction techniques such as PCA can be used to identify subnetworks that co-vary in their activity and are implicated in certain behaviors.
In addition to worms and flies, the larval zebrafish also offers a compact nervous system along with an advanced genetic toolkit to facilitate studies of sensorimotor processes [75]. Automated behavioral tools (though fully supervised) have also been used to track fish and study locomotion [76], prey capture [77], or social behaviors [78]. Most importantly, facilitated by its transparency, methods have been developed to rapidly image neural activity at cellular-resolution from the entire brain during fictive behavior [79]. This has led to breakthroughs in, for example, dissecting the circuits underlying the optomotor response [80] or locomotor learning [81] in the zebrafish. The investigation of whole-brain activity is arguably more mature in zebrafish than in either C. elegans or Drosophila. The next few years should hopefully lead to a convergence between studies of these organisms, with the behavioral quantification tools developed in the latter species applied to the former and the circuit dissection philosophy of zebrafish brought to bear on worms and flies.
Future Directions
As outlined above, recent advances in behavioral quantification facilitate solving the underlying neural circuit mechanisms. However, several challenges remain. First, the methods described above require animals to be placed in artificial environments in order to cleanly segment videos. However, placing animals in richer environments would provide better access to the computations that brains evolved to solve. Methods that make ‘unsupervised’ behavioral classification more compatible with recordings taken in natural settings would expand the dictionary of behaviors that can be identified. Second, most behavioral quantification methods capture video of animals from a single vantage point, but characterization of the full repertoire of animal behavior will likely require combining data from multiple cameras and from other types of recordings (e.g., muscle activity [82] or leg kinematics [83]). Third, as we advocate in this review, behavioral quantification should be intertwined with studies of the sensory feedback that drives or modulates behavior. One current limitation is that experimenters pre-define the features of the sensory space to feed into models. It should be possible to identify relevant sensory features in an ‘unsupervised’ fashion [84], similar to new methods for behavioral analysis. Fourth, current models of sensorimotor transformations often assume an animal does not switch between behavioral states despite the fact that we know that internal state plays an important role in behavior [85–87]. Models that can capture longer timescales in behavior or ‘hidden states’ (e.g., using hidden Markov models) could identify the influence of these internal states and will facilitate finding their neural correlates. Finally, many of the tools described in this review characterize the relationships between animal postures and their environments as linear, when in fact they may be highly nonlinear. The use of machine learning techniques, including deep learning, to introduce nonlinearities into the relationships between postures should produce more precise descriptions of behavior [88–90].
The models described in this review are also largely descriptive in nature. However, models that are generative, or able to generate simulated behavior, can provide a set of underlying rules that the nervous system may implement [91]. These rules could then be used to develop a theory of why behaviors are generated in the way that they are [1]. Such a comprehensive theory will lead to better measures of behavior that can ultimately be related back to the neural activity that generates it. Nonetheless, the recent explosion of methods for behavioral quantification, combined with the unparalleled genetic and neural circuit toolkits of worms and flies, has yielded exciting results with regard to the cellular mechanisms underlying both sensorimotor transformations and the generation of behavior. These studies reveal the promise of these model systems in providing a sophisticated understanding of the fundamental principles that shape behavior.
Highlights.
New automated methods permit characterization of the full repertoire of an animal’s behavior.
Quantification of dynamic sensory stimuli in combination with behavioral analysis will facilitate identification of the computations the nervous system performs to transform sensory responses into motor outputs.
Extensive genetic tools and a simple nervous system make worms and flies attractive model systems for harnessing the full power of these new computational methods to solve the mechanisms underlying sensorimotor transformations.
Acknowledgments
We thank Andrew Leifer, David Schoppik, and the entire Murthy Lab for helpful comments. AJC was funded by an NIH T32 and a Simons Collaboration on the Global Brain postdoctoral fellowship and MM was funded by an NIH New Innovator Award, an NSF CAREER award, an NSF BRAIN Initiative EAGER award, and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krakauer JW, Ghazanfar AA, Gomez-Marin A, MacIver MA, Poeppel D. Neuroscience Needs Behavior: Correcting a Reductionist Bias. Neuron. 2017;93:480–490. doi: 10.1016/j.neuron.2016.12.041. [DOI] [PubMed] [Google Scholar]
- 2.Levitis DA, Lidicker WZ, Jr, Freund G. Behavioural biologists do not agree on what constitutes behaviour. Animal Behaviour. 2009;78:103–110. doi: 10.1016/j.anbehav.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson DJ, Perona P. Toward a science of computational ethology. Neuron. 2014;84:18–31. doi: 10.1016/j.neuron.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Gomez-Marin A, Paton JJ, Kampff AR, Costa RM, Mainen ZF. Big behavioral data: psychology, ethology and the foundations of neuroscience. Nat. Neurosci. 2014;17:1455–1462. doi: 10.1038/nn.3812. [DOI] [PubMed] [Google Scholar]
- 5.Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nat. Methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramot D, Johnson BE, Berry TL, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Branson K, Robie AA, Bender J, Perona P, Dickinson MH. High-throughput ethomics in large groups of Drosophila. Nat. Methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng Y, Coen P, Sun M, Shaevitz JW. Efficient multiple object tracking using mutually repulsive active membranes. PLoS ONE. 2013;8:e65769. doi: 10.1371/journal.pone.0065769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dankert H, Wang L, Hoopfer ED, Anderson DJ, Perona P. Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods. 2009;6:297–303. doi: 10.1038/nmeth.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthur BJ, Sunayama-Morita T, Coen P, Murthy M, Stern DL. Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biol. 2013;11:11. doi: 10.1186/1741-7007-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabra M, Robie AA, Rivera-Alba M, Branson S, Branson K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods. 2013;10:64–67. doi: 10.1038/nmeth.2281. **The authors developed a powerful method to identify and categorize behaviors using semi-supervised machine learning algorithms. These methods do not require tuning the software to track specific behaviors but rather automate the process of identifying behaviors based on user-defined examples. [DOI] [PubMed] [Google Scholar]
- 12.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput. Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. **The authors developed a new ‘unsupervised’ and automated pipeline for clustering sequences of postures (from movies of walking flies) into discrete, stereotyped actions. They observed that flies exhibit approximately 100 stereotyped behaviours that are interspersed with frequent bouts of non-stereotyped behaviours – this framework is powerful for identifying behaviors and behavioral transitions that can ultimately be linked to dynamic neural activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. From modes to movement in the behavior of Caenorhabditis elegans. PLoS ONE. 2010;5:e13914. doi: 10.1371/journal.pone.0013914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broekmans OD, Rodgers JB, Ryu WS, Stephens GJ. Resolving coiled shapes reveals new reorientation behaviors in C. elegans. eLife. 2016;5:428. doi: 10.7554/eLife.17227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AEX, Yemini EI, Grundy LJ, Jucikas T, Schafer WR. A dictionary of behavioral motifs reveals clusters of genes affecting Caenorhabditis elegans locomotion. Proc. Natl. Acad. Sci. U.S.a. 2013;110:791–796. doi: 10.1073/pnas.1211447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girdhar K, Gruebele M, Chemla YR. The Behavioral Space of Zebrafish Locomotion and Its Neural Network Analog. PLoS ONE. 2015;10:e0128668. doi: 10.1371/journal.pone.0128668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szigeti B, Deogade A, Webb B. Searching for motifs in the behaviour of larval Drosophila melanogaster and Caenorhabditis elegans reveals continuity between behavioural states. Journal of the Royal Society Interface. 2015;12:20150899. doi: 10.1098/rsif.2015.0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berman GJ, Choi DM, Bialek W, Shaevitz JW. Mapping the stereotyped behaviour of freely moving fruit flies. Journal of the Royal Society Interface. 2014;11:20140672–20140672. doi: 10.1098/rsif.2014.0672. **The authors developed an ‘unsupervised’ method (distinct from Berman et al. 2014) to classify mouse behavior from depth imaging cameras. This method uses dimensionality reduction followed by computational modeling (an autoregressive (AR) hidden Markov model (HMM)) to identify behavioral modules and to capture the transition probabilities between modules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiltschko AB, Johnson MJ, Iurilli G, Peterson RE, Katon JM, Pashkovski SL, et al. Mapping Sub-Second Structure in Mouse Behavior. Neuron. 2015;88:1121–1135. doi: 10.1016/j.neuron.2015.11.031. **The authors investigate several ‘unsupervised’ methods for classifying Drosophila behavior, comparing several alternative methods for dimensionality reduction and cluster assignment. They find that principal components analysis (as opposed to t-SNE, see Berman et al. 2014) followed by Gaussian mixture modeling outperforms other methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd JG, Kain JS, de Bivort B. Systematic exploration of unsupervised methods for mapping behavior. bioRxiv. 2016:051300. doi: 10.1101/051300. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Marin A, Stephens GJ, Brown AEX. Hierarchical compression of Caenorhabditis eleganslocomotion reveals phenotypic differences in the organization of behaviour. Journal of the Royal Society Interface. 2016;13:20160466. doi: 10.1098/rsif.2016.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klibaite U, Berman G, Cande J, Stern D, Shaevitz J. An unsupervised method for quantifying the behavior of paired animals. Phys Biol. 2017 doi: 10.1088/1478-3975/aa5c50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Escudero A, Vicente-Page J, Hinz RC. idTracker: tracking individuals in a group by automatic identification of unmarked animals. Nature. 2014 doi: 10.1038/nmeth.2994. [DOI] [PubMed] [Google Scholar]
- 24.Berman GJ, Bialek W, Shaevitz JW. Predictability and hierarchy in Drosophila behavior. Proc. Natl. Acad. Sci. U.S.a. 2016;113:11943–11948. doi: 10.1073/pnas.1607601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boulin T, Hobert O. From genes to function: the C. elegans genetic toolbox. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1:114–137. doi: 10.1002/wdev.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogelstein JT, Park Y, Ohyama T, Kerr RA, Truman JW, Priebe CE, et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science. 2014;344:386–392. doi: 10.1126/science.1250298. *The authors used ‘unsupervised’ methods to classify Drosophila larval behavior. These methods were then used to screen a library of more than 1,000 genetic enhancer lines, each driving a neural activator in a different population of neurons. This paper demonstrates the power of automated behavioral methods for connecting identifiable neurons with their associated behaviors. A similar approach was taken by Robie et al. in adult flies. [DOI] [PubMed] [Google Scholar]
- 28.Robie AA, Hirokawa J, Edwards AW, Umayam LA, Lee A, Phillips ML, et al. Mapping the Neural Substrates of Behavior. Cell. 2017;170:393–406. e28. doi: 10.1016/j.cell.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 29.Wu M, Nern A, Williamson WR, Morimoto MM, Reiser MB, Card GM, et al. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife. 2016;5:7587. doi: 10.7554/eLife.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoopfer ED, Jung Y, Inagaki HK, Rubin GM, Anderson DJ. P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. eLife. 2015;4:2700. doi: 10.7554/eLife.11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, Bargmann CI. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell. 2013;154:1023–1035. doi: 10.1016/j.cell.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts WM, Augustine SB, Lawton KJ, Lindsay TH. A stochastic neuronal model predicts random search behaviors at multiple spatial scales in C. elegans. eLife. 2016 doi: 10.7554/eLife.12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bath DE, Stowers JR, Hörmann D, Poehlmann A, Dickson BJ, Straw AD. FlyMAD: rapid thermogenetic control of neuronal activity in freely walking Drosophila. Nat. Methods. 2014;11:756–762. doi: 10.1038/nmeth.2973. [DOI] [PubMed] [Google Scholar]
- 34.Clark DA, Freifeld L, Clandinin TR. Mapping and cracking sensorimotor circuits in genetic model organisms. Neuron. 2013;78:583–595. doi: 10.1016/j.neuron.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez-Nunez L, Belina J, Klein M, Si G, Claus L, Carlson JR, et al. Reverse-correlation analysis of navigation dynamics in Drosophila larva using optogenetics. eLife. 2015;4:e06225. doi: 10.7554/eLife.06225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gepner R, Skanata MM, Bernat NM, Kaplow M, Gershow M, Calabrese RL. Computations underlying Drosophila photo-taxis, odor-taxis, and multi-sensory integration. eLife. 2015;4:e06229. doi: 10.7554/eLife.06229. *Directly stimulating sensory neurons during free behavior is a powerful way to identify a sensorimotor transformation. The authors used optical stimulation of Drosophila larval visual and olfactory neurons to identify how sensory information is integrated within the nervous system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulze A, Gomez-Marin A, Rajendran VG, Lott G, Musy M, Ahammad P, et al. Dynamical feature extraction at the sensory periphery guides chemotaxis. eLife. 2015;4:1129. doi: 10.7554/eLife.06694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharpee T, Rust NC, Bialek W. Analyzing neural responses to natural signals: maximally informative dimensions. Neural Computation. 2004;16:223–250. doi: 10.1162/089976604322742010. [DOI] [PubMed] [Google Scholar]
- 39.Fitzgerald JD, Sincich LC, Sharpee TO. Minimal models of multidimensional computations. PLoS Comput. Biol. 2011;7:e1001111. doi: 10.1371/journal.pcbi.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pillow JW, Shlens J, Paninski L, Sher A, Litke AM, Chichilnisky EJ, et al. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature. 2008;454:995–999. doi: 10.1038/nature07140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clemens J, Murthy M. The use of computational modeling to link sensory processing with behavior in Drosophila. 2017 doi: 10.7287/peerj.preprints.2720v1. [DOI] [Google Scholar]
- 42.Calhoun AJ, Tong A, Pokala N, Fitzpatrick JAJ, Sharpee TO, Chalasani SH. Neural Mechanisms for Evaluating Environmental Variability in Caenorhabditis elegans. Neuron. 2015;86:428–441. doi: 10.1016/j.neuron.2015.03.026. **C. elegans off-food search was thought to be a purely stochastic sequence of turning events. However, this study used automated methods to quantify the time-varying behavior and sensory environment of the animal to uncover a precise control of the turning events. The authors used this knowledge to identify the neural circuitry and molecular mechanisms that support this sensorimotor behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calhoun AJ, Chalasani SH, Sharpee TO. Maximally informative foraging by Caenorhabditis elegans. eLife. 2014;3:8616. doi: 10.7554/eLife.04220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomez-Marin A, Stephens GJ, Louis M. Active sampling and decision making in Drosophila chemotaxis. Nature Communications. 2011;2:441. doi: 10.1038/ncomms1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coen P, Clemens J, Weinstein AJ, Pacheco DA, Deng Y, Murthy M. Dynamic sensory cues shape song structure in Drosophila. Nature. 2014;507:233–237. doi: 10.1038/nature13131. **Drosophila courtship was long thought to be a sequence of fixed action patterns. However, this study used automated measurements of interactions between animals to identify how sensory cues dynamically modulate the song patterns males produce. The authors were able to predict which type of song the male produced during song bouts, based on the sensory feedback he received from the female. [DOI] [PubMed] [Google Scholar]
- 46.Coen P, Xie M, Clemens J, Murthy M. Sensorimotor Transformations Underlying Variability in Song Intensity during Drosophila Courtship. Neuron. 2016;89:629–644. doi: 10.1016/j.neuron.2015.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Censi A, Straw AD, Sayaman RW, Murray RM, Dickinson MH. Discriminating external and internal causes for heading changes in freely flying Drosophila. PLoS Comput. Biol. 2013;9:e1002891. doi: 10.1371/journal.pcbi.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mischiati M, Lin H-T, Herold P, Imler E, Olberg R, Leonardo A. Internal models direct dragonfly interception steering. Nature. 2015;517:333–338. doi: 10.1038/nature14045. [DOI] [PubMed] [Google Scholar]
- 49.Lin H-T, Leonardo A. Heuristic Rules Underlying Dragonfly Prey Selection and Interception. Curr. Biol. 2017;0 doi: 10.1016/j.cub.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Gordus A, Pokala N, Levy S, Flavell SW, Bargmann CI. Feedback from network states generates variability in a probabilistic olfactory circuit. Cell. 2015;161:215–227. doi: 10.1016/j.cell.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell HC. Behavioral Variability in the Service of Constancy. International Journal of Comparative Psychology. 2014;27 [Google Scholar]
- 52.Hassenstein V, Reichardt W. System theoretical analysis of time, sequence and sign analysis of the motion perception of the snout-beetle Chlorophanus. Z Naturforsch B. 1956 [Google Scholar]
- 53.Barlow HB, Levick WR. The mechanism of directionally selective units in rabbit's retina. The Journal of Physiology. 1965;178:477–504. doi: 10.1111/(ISSN)1469-7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Behnia R, Clark DA, Carter AG, Clandinin TR, Desplan C. Processing properties of ON and OFF pathways for Drosophila motion detection. Nature. 2014;512:427–430. doi: 10.1038/nature13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leonhardt A, Ammer G, Meier M, Serbe E, Bahl A, Borst A. Asymmetry of Drosophila ON and OFF motion detectors enhances real-world velocity estimation. Nat. Neurosci. 2016;19:706–715. doi: 10.1038/nn.4262. [DOI] [PubMed] [Google Scholar]
- 56.Arenz A, Drews MS, Richter FG, Ammer G, Borst A. The Temporal Tuning of the Drosophila Motion Detectors Is Determined by the Dynamics of Their Input Elements. Curr. Biol. 2017;27:929–944. doi: 10.1016/j.cub.2017.01.051. [DOI] [PubMed] [Google Scholar]
- 57.Ramdya P, Lichocki P, Cruchet S, Frisch L, Tse W, Floreano D, et al. Mechanosensory interactions drive collective behaviour in Drosophila. Nature. 2015;519:233–236. doi: 10.1038/nature14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. Ultra-sensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chalasani SH, Chronis N, Tsunozaki M, Gray JM. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007 doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 60.Kato S, Kaplan HS, Schrödel T, Skora S, Lindsay TH, Yemini E, et al. Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell. 2015;163:656–669. doi: 10.1016/j.cell.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 61.Maimon G, Straw AD, Dickinson MH. Active flight increases the gain of visual motion processing in Drosophila. Nat. Neurosci. 2010;13:393–399. doi: 10.1038/nn.2492. [DOI] [PubMed] [Google Scholar]
- 62.Seelig JD, Chiappe ME, Lott GK, Dutta A, Osborne JE, Reiser MB, et al. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat. Methods. 2010;7:535–540. doi: 10.1038/nmeth.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seelig JD, Jayaraman V. Neural dynamics for landmark orientation and angular path integration. Nature. 2015;521:186–191. doi: 10.1038/nature14446. *The authors combined imaging of neural activity with simultaneous measurement of walking behavior to uncover the computations performed by neurons in the Drosophila central complex. These neurons are involved in the sensorimotor transformation from visual feedback to changes in walking orientation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green J, Adachi A, Shah KK, Hirokawa JD, Magani PS, Maimon G. A neural circuit architecture for angular integration in Drosophila. Nature. 2017;546:101–106. doi: 10.1038/nature22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujiwara T, Cruz TL, Bohnslav JP, Chiappe ME. A faithful internal representation of walking movements in the Drosophila visual system. Nat. Neurosci. 2017;20:72–81. doi: 10.1038/nn.4435. [DOI] [PubMed] [Google Scholar]
- 66.Kim AJ, Fitzgerald JK, Maimon G. Cellular evidence for efference copy in Drosophila visuomotor processing. Nat. Neurosci. 2015;18:1247–1255. doi: 10.1038/nn.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen JP, Shipley FB, Linder AN, Plummer GS, Liu M, Setru SU, et al. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.a. 2016;113:E1074–81. doi: 10.1073/pnas.1507110112. *The authors performed whole-brain calcium imaging at cellular resolution in freely behaving C. elegans. This was accomplished via extremely fast behavioral tracking in order to keep the head directly under the objective at all times. This method will be critical for characterizing the activity that underlies sensorimotor transformations during natural behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Venkatachalam V, Ji N, Wang X, Clark C, Mitchell JK, Klein M, et al. Panneuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.a. 2016;113:E1082–8. doi: 10.1073/pnas.1507109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dupre C, Yuste R. Non-overlapping Neural Networks in Hydra vulgaris. Curr. Biol. 2017 doi: 10.1016/j.cub.2017.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grover D, Katsuki T, Greenspan RJ. Flyception: imaging brain activity in freely walking fruit flies. Nat. Methods. 2016;13:569–572. doi: 10.1038/nmeth.3866. [DOI] [PubMed] [Google Scholar]
- 71.Aimon S, Katsuki T, Grosenick L, Broxton M, Deisseroth K, Sejnowski TJ, et al. Linking stimuli and behavior with fast near-whole brain recordings in adult Drosophila. bioRxiv. 2016:033803. doi: 10.1101/033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, et al. Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms. Nature Photonics. 2015;9:113–119. doi: 10.1038/nphoton.2014.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cong L, Wang Z, Chai Y, Hang W, Shang C, Yang W, et al. Rapid Whole Brain Imaging Of Neural Activities In Freely Behaving Larval Zebrafish. bioRxiv. 2017:131532. doi: 10.1101/131532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nichols ALA, Eichler T, Latham R, Zimmer M. A global brain state underlies<i>C. elegans</i> sleep behavior. Science. 2017;356:eaam6851. doi: 10.1126/science.aam6851. [DOI] [PubMed] [Google Scholar]
- 75.Orger MB, de Polavieja GG. Zebrafish Behavior: Opportunities and Challenges. 2015;40 doi: 10.1146/annurev-neuro-071714-033857. Https://Doi.org/10.1146/Annurev-Neuro-071714-033857. annurev–neuro–071714–033857. [DOI] [PubMed] [Google Scholar]
- 76.Dunn TW, Mu Y, Narayan S, Randlett O, Naumann EA. Brain-wide mapping of neural activity controlling zebrafish exploratory locomotion. eLife. 2016 doi: 10.7554/eLife.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Semmelhack JL, Donovan JC, Thiele TR, Kuehn E, Laurell E, Baier H. A dedicated visual pathway for prey detection in larval zebrafish. eLife. 2014;3:17968. doi: 10.7554/eLife.04878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hinz RC, de Polavieja GG. Ontogeny of collective behavior reveals a simple attraction rule. Proc. Natl. Acad. Sci. U.S.a. 2017;114:2295–2300. doi: 10.1073/pnas.1616926114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013 doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 80.Naumann EA, Fitzgerald JE, Dunn TW, Rihel J, Sompolinsky H, Engert F. From Whole-Brain Data to Functional Circuit Models: The Zebrafish Optomotor Response. Cell. 2016;167:947–960. e20. doi: 10.1016/j.cell.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawashima T, Zwart MF, Yang C-T, Mensh BD, Ahrens MB. The Serotonergic System Tracks the Outcomes of Actions to Mediate Short-Term Motor Learning. Cell. 2016;167:933–946. e20. doi: 10.1016/j.cell.2016.09.055. [DOI] [PubMed] [Google Scholar]
- 82.Lindsay T, Sustar A, Dickinson M. The Function and Organization of the Motor System Controlling Flight Maneuvers in Flies. Current Biology. 2017;27:345–358. doi: 10.1016/j.cub.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 83.Mendes CS, Bartos I, Akay T, Márka S, Mann RS. Quantification of gait parameters in freely walking wild type and sensory deprived Drosophila melanogaster. eLife. 2013 doi: 10.7554/eLife.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xin, Chen, Beck JM, Pearson JM. Neuron's Eye View: Inferring Features of Complex Stimuli from Neural Responses. 2015 doi: 10.1371/journal.pcbi.1005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim SM, Su C-Y, Wang JW. Neuromodulation of Innate Behaviors in Drosophila. Annu. Rev. Neurosci. 2017;40 doi: 10.1146/annurev-neuro-072116. annurev–neuro–072116–031558. [DOI] [PubMed] [Google Scholar]
- 86.Jourjine N, Mullaney BC, Mann K, Scott K. Coupled Sensing of Hunger and Thirst Signals Balances Sugar and Water Consumption. Cell. 2016;166:855–866. doi: 10.1016/j.cell.2016.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Corrales-Carvajal VM, Faisal AA, Ribeiro C. Internal states drive nutrient homeostasis by modulating exploration-exploitation trade-off. eLife. 2016;5:119. doi: 10.7554/eLife.19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marblestone AH, Wayne G, Kording KP. Toward an Integration of Deep Learning and Neuroscience. Frontiers in Computational Neuroscience. 2016;10:406. doi: 10.3389/fncom.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benjamin AS, Fernandes HL, Tomlinson T, Ramkumar P, VerSteeg C, Miller L, et al. Modern machine learning far outperforms GLMs at predicting spikes. bioRxiv. 2017:111450. doi: 10.1101/111450. [DOI] [Google Scholar]
- 90.Viejo G, Cortier T, Peyrache A. Resolving neuronal population code and coordination with gradient boosted trees. bioRxiv. 2017:148643. doi: 10.1101/148643. [DOI] [Google Scholar]
- 91.Gomez-Marin A, Oron E, Gakamsky A, Dan Valente, Benjamini Y, Golani I. Generative rules of Drosophila locomotor behavior as a candidate homology across phyla. Scientific Reports. 2016;6:27555. doi: 10.1038/srep27555. [DOI] [PMC free article] [PubMed] [Google Scholar]