Abstract
Background
Tuberculosis Immune Reconstitution Inflammatory Syndrome (TB-IRIS) remains incompletely understood. Neutrophils are implicated in tuberculosis pathology but detailed investigations in TB-IRIS are lacking. We sought to further explore the biology of TB-IRIS and in particular the role of neutrophils.
Setting
Two observational, prospective cohort studies in HIV/TB co-infected patients starting antiretroviral therapy, one to analyze gene expression and subsequently one to explore neutrophil biology.
Methods
nCounter gene expression analysis was performed in TB-IRIS patients (n=17) versus antiretroviral-treated HIV/TB co-infected controls without IRIS (n=17) in Kampala, Uganda. Flow cytometry was performed in TB-IRIS patients (n=18) and controls (n=11) in Cape Town, South Africa to determine expression of neutrophil surface activation markers, intracellular cytokines and Human Neutrophil Peptides (HNP). Plasma neutrophil Elastase and HNP1-3 were quantified using ELISA. Lymph node immunohistochemistry was performed on three further TB-IRIS cases.
Results
There was a significant increase in gene expression of S100A9 (p=0.002), NLRP12 (p=0.018), COX-1 (p=0.025) and IL-10 (p=0.045) two weeks after ART initiation in Ugandan TB-IRIS patients versus controls, implicating neutrophil recruitment. IRIS patients in both cohorts demonstrated increases in blood neutrophil count, plasma HNP and elastase concentrations from ART initiation to week two. CD62L (L-selectin) expression on neutrophils increased over 4 weeks in South African controls while IRIS patients demonstrated the opposite. Intense staining for the neutrophil marker CD15 and IL-10 was seen in necrotic areas of TB-IRIS patients’ lymph nodes.
Conclusion
Neutrophils in TB-IRIS are activated, recruited to sites of disease and release granule contents, contributing to pathology.
Keywords: Tuberculosis, HIV-1, neutrophils, immune reconstitution inflammatory syndrome, IRIS
Introduction
When patients with HIV-associated TB begin Antiretroviral Therapy (ART), approximately 18% develop Tuberculosis-associated Immune Reconstitution Inflammatory Syndrome (TB-IRIS) [1]. TB-IRIS is an exaggerated immune response to M. tuberculosis (MTB) antigens associated with reconstitution of the immune system. It is characterized by excessive inflammatory responses and deterioration in clinical status [1, 2].
According to the International Network for the Study of HIV associated IRIS (INSHI) case definitions, two forms of TB-IRIS exist: ‘paradoxical’ (clinical worsening of a patient on TB treatment after starting ART) and ‘unmasking’ (undiagnosed TB becoming apparent after starting ART) [3].
TB-IRIS has been associated with perturbations in both the adaptive and innate immune systems [4, 5]. These include increased secretion of neutrophil-associated mediators such as S100A8/A9 and matrix metalloproteinases (MMPs) [6–8], perforin and granzyme B by CD4+ T cells [9], higher expression and imbalance of C1Q and C1-inhibitor (complement system) [10], activation of monocytes [11], inflammasome and Toll-like receptor signaling [12, 13] as well as elevated chemokine and cytokine production [14–16] with a particular role for the IL-10 family [17]. Although rapid changes in CD4+ T cell count have long been associated with all forms of IRIS, recent research has focused on these latter phenomena of inflammasome activation and release of soluble mediators from innate cells [4, 12]. However, the clinical syndromes associated with TB-IRIS, especially suppurative lymphadenitis and abscess formation, implicate neutrophils as critical effector cells mobilized by these inflammatory signals.
To gain further understanding into the biology of TB-IRIS, we recruited and prospectively followed patients with HIV-associated tuberculosis (HIV+TB+) at risk of developing IRIS at two clinical sites, in Uganda and South Africa. First, we conducted an assessment of gene expression in putative pathways. On the basis of previous research summarized above, we chose to study the T-cell receptor, cytokine genes including the IL-10 pathway [17] and the inflammasome [12, 13]. Subsequently, in a separate cohort, we performed functional assays chosen on the basis of genes that were over-expressed in IRIS patients versus controls: these experiments focused on neutrophils which, although implicated [6], have not been extensively studied before in TB-IRIS.
Materials and Methods
Patient recruitment and study visits
Cohort 1: Patients with a confirmed diagnosis of both HIV and TB, on TB treatment (for a median [IQR] of 40 [24-59] days) and who were eligible for ART initiation according to the July 2008 Ugandan national treatment guidelines (CD4 count <250 cells/μL), were recruited in 2009 at Mulago National Tuberculosis and Leprosy clinic and the Infectious Diseases Institute in Kampala for gene expression studies, as previously described [18]; see Supplementary Table 1. Patients were reviewed at week 0 (before ART initiation), week 2 and months 1-12 (after ART initiation). Patients who developed TB-IRIS (cases) were defined according to the INSHI clinical case definitions [3] and were matched by age (<10 years difference between patients), CD4 cell count before ART initiation (mean (SD) difference, 5.3 (6.8) cells/μL) and sex with those that did not develop TB-IRIS (non-IRIS controls). Sampling at the IRIS time-point was performed before patients received corticosteroids. All patients provided written informed consent. The Uganda National Council of Science and Technology, Makerere Faculty of Medicine Ethics Committee (IRB-Makerere-05_2007), Infectious Disease Scientific Review Committee, University of Antwerp Ethics Committee and the Institute of Tropical Medicine, Antwerp, Belgium (CME_UZA_7/29/157) approved the study.
Cohort 2: Recruitment of patients for neutrophil studies took place in Cape Town, South Africa as part of the longitudinal Tissue Destruction in Tuberculosis 2 (TDTB2) study (Supplementary Table 1). Patients were recruited in 2013 at Ubuntu clinic, a primary care HIV treatment clinic in Site B, Khayelitsha. HIV-infected patients at high risk of developing TB-IRIS (CD4 count <200 cells/μL at enrolment) were followed up during anti-tuberculosis treatment and initiation of ART until twelve weeks of ART. Samples for neutrophil studies were collected at ART initiation (week 0), week two and week four of ART. TB-IRIS diagnosis was made retrospectively after week 12 by a consensus panel using the INSHI case definition; controls (non-IRIS) were those patients who were also sampled at ART initiation and Week 2 / Week 4 follow-up visits but did not develop the syndrome [3]. At the IRIS/week 2 time point, two TB-IRIS and one non-IRIS control were receiving corticosteroids. Ethical approval was obtained from the Faculty of Health Sciences Human Research Ethics Committee, University of Cape Town (HREC REF: 516/2011); all patients provided written informed consent.
Samples for detailed analysis were available from 34 patients in Cohort 1 (17 cases and 17 controls) and 29 patients in Cohort 2 (18 cases and 11 controls). Supplementary Figure 1 summarises the study design.
Sample collection and processing
For Cohort 1, venous blood (30–40 ml) was collected in EDTA tubes (BD Pharmingen, Franklin Lakes, New Jersey, USA) at week 0 and week 2 after initiation of ART. Peripheral Blood Mononuclear Cells (PBMC) were isolated by Ficoll-Hypaque gradient centrifugation and cryopreserved for further processing (see below). For Cohort 2, blood samples (30–40 ml) were collected in sodium heparin vacutainers (BD Pharmingen) at weeks 0, 2 and 4 after initiation of ART and were processed for plasma generation within two hours of collection; an aliquot (1 ml) of blood was removed for functional assays as described below.
nCounter gene expression analysis
RNA was extracted from PBMC using standard techniques (Supplementary Methods). ProbeSet sequences for the gene sets of interest (T-cell receptors, the inflammasome, IL-10 pathway and cytokines; 148 genes in total) are shown in Supplementary Table 2.
Determination of neutrophil activation and degranulation
We investigated neutrophil activation in whole blood by flow cytometry, measuring cell surface expression of CD11b, CD16, CD62L, CD66a,c,e [19] and IL-8RA. An aliquot of whole blood was stained on ice with CD11b-PE-Cy7, CD16-APC-H7, CD62L-FITC, CD66a,c,e-PE, IL-8 RA-APC (BD Pharmingen) and viability dye (eFluor 450, eBiosciences; San Diego, California, USA or ViViD, Invitrogen; Carlsbad, California, USA). After washing, the stained sample was fixed in 2% paraformaldehyde and acquired on a Becton Dickinson Fortessa flow cytometer (BD Biosciences). Data analysis was performed with FlowJo software (FlowJo 10.1r5, Tree Star, Ashland, OR) using the gating strategy in Supplementary Figure 2.
Determination of neutrophil elastase and Human Neutrophil Peptides (HNP1-3) in plasma
Neutrophil elastase and Human Neutrophil Peptides (HNP1-3) plasma concentrations were quantified using ELISA according to the manufacturer’s instructions (Hycult Biotech; Uden, The Netherlands). Assays were performed in duplicate. The sensitivity for neutrophil elastase was 0.67 ng/ml and for HNP1-3 was 4.25 pg/ml. The elastase assay detects both free and complexed elastase.
Immunohistochemistry (IHC) staining of lymph nodes
Patient selection, lymph node (LN) preparation and immunohistochemistry were carried out as previously described [20] and summarized in Supplementary Methods.
Statistical analysis
Comparison between the two groups was performed using t tests (unpaired for IRIS vs non-IRIS comparisons, paired for within-group comparisons between ART initiation and later time points), the Mann-Whitney U test or Wilcoxon test for continuous variables and Fisher exact tests for categorical variables. Statistics were performed using GraphPad Prism Version 7.0 (La Jolla, California, USA) and Qlucore Omics explorer version 3.2. (Lund, Sweden) Significance was inferred below a two-tailed p-value of 0.05.
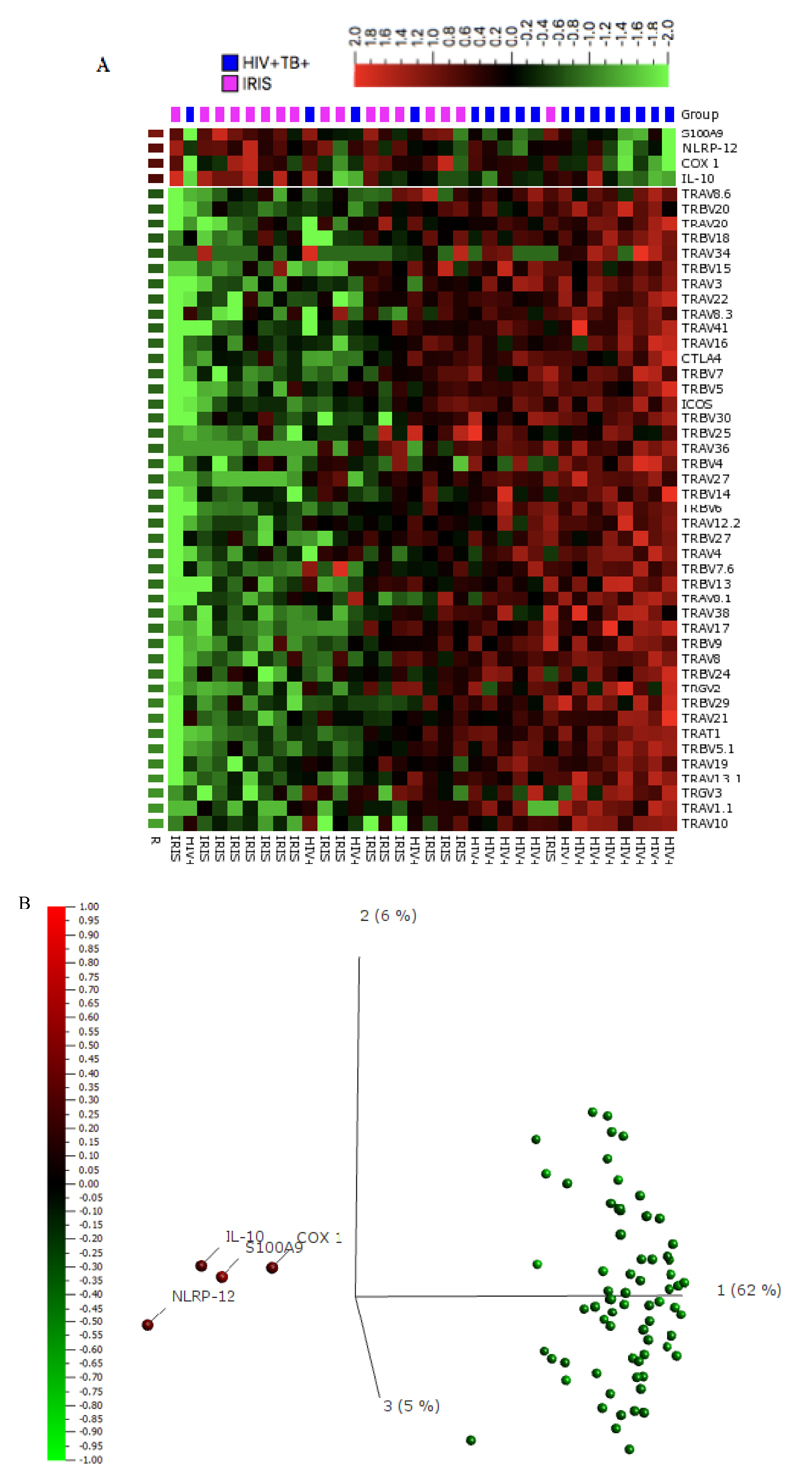
Gene expression analysis to identify discriminating transcripts between the groups (based on p-value <0.05 and q value (False Discovery Rate-adjusted p-value) <0.1) was performed using Qlucore Omics explorer and displayed on a heatmap. The IRIS (pink) and non-IRIS (blue) patients (columns) and genes (rows) were ordered using principal component analysis (PCA) and R statistic respectively. Gene expression at the week two time point on the heatmap was classified as high or low (relative to the entire cohort) if colored red and green respectively. A PCA plot, with the projection score and variance filtering set at 0.38 and 0.43 respectively, was used to detect strong signals within the data on gene transcript abundance. Principal Component Analysis identifies the major vectors (‘components’) which differentiate multi-parameter data sets. The genes were colored according to their R statistic with green and red if higher in non-IRIS controls or IRIS patients respectively, and the distance between individual genes reflects their correlation coefficient.
Results
Patient characteristics
Supplementary Table 1 summarizes demographic and basic laboratory data for both cohorts. At ART initiation, there were no statistical differences in patient characteristics between those who subsequently developed IRIS and those who did not. The median [IQR] time to IRIS presentation across both studies was 14 [10-15] days.
RNA analysis reveals higher expression of genes implicated in neutrophilic inflammation in TB-IRIS patients compared to controls
We used NanoString nCounter technology to ascertain gene expression in PBMC of IRIS and non-IRIS patients at the IRIS time-point (median of 14 days) or after 2 weeks of ART in controls. The nCounter gene expression values obtained were log 2 transformed pre-analysis to normalize data as per standard transcriptomic analytical pathways; a false discovery rate (q-value) of 0.1 was applied to account for multiple comparisons. A heatmap to visualize the pattern of transcript abundance in IRIS patients and non-IRIS controls revealed over 70 discriminating transcripts with modest clustering of IRIS cases (pink) and non-IRIS controls (blue); there was generally lower gene expression (green) in the IRIS patients compared to the non-IRIS controls (Figure 1A). On the contrary, Cyclooxygenase-1 (COX-1), Interleukin-10 (IL-10), Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) Family Pyrin Domain Containing 12 (NLRP12 / Pypaf-7), and S100 calcium-binding protein A9 (S100A9) were significantly more abundant in the IRIS cases than in the non-IRIS controls at two weeks of ART.
Figure 1. Gene expression analysis in PBMCs from patients with HIV-associated TB-IRIS and HIV/TB co-infected controls without clinical IRIS:
A. 100 ng of total RNA was used to obtain values for gene expression analysis using nCounter technology. Unsupervised hierarchical clustering of transcript abundance data from TB-IRIS (pink) (n = 17) and non-IRIS (blue) (n = 17) patients at week two/IRIS-time point was performed using a heatmap in Qlucore Omics explorer v3.2. The columns represent patients while the rows are genes identified as discriminatory (p<0.05, q<0.1). Relative gene expression compared to the entire cohort was classified as low (green) and high (red) respectively. Genes were ordered according to their R statistic between IRIS and non-IRIS patients. B. Discriminatory genes were visualized on a PCA plot. The genes (variables) were colored according to their R statistic; green for the lowest (implying greater abundance in non-IRIS vs IRIS) and red if the highest (implying greater abundance in IRIS vs non-IRIS). The genes with the highest expression in IRIS were COX-1, IL-10, NLRP-12 and S100A9.
Abbreviations: ASC; Apoptosis-associated speck-like protein containing a Caspase Recruitment Domain (CARD); CD, Cluster of Differentiation; COX-1/PTGS, Cyclooxygenase-1/prostaglandin-endoperoxide synthase; CTLA4, Cytotoxic T Lymphocyte-associated protein 4 (CD152); GATA3, Glycine, Alanine, Thymine, Alanine binding protein 3; ICOS, Inducible T-cell costimulator; IFN-ϒ, Interferon gamma; IL, Interleukin; IL-7R, Interleukin-7 receptor; ITK, Interleukin-2-inducible T-cell kinase; pypaf-7, PYRIN-containing Apaf-1-like proteins; S100A9, S100 calcium-binding protein A9; Tbet, T-box transcription factor; TRAC, T-cell Receptor alpha constant; TRAV, T-cell Receptor alpha variable; TRBC, T-cell Receptor beta constant; TRBV, T-cell Receptor beta variable; TRDV, T-cell Receptor delta variable; TRGC, T-cell Receptor gamma constant; TRGV; T-cell Receptor gamma variable.
PCA was then used to detect correlation patterns within the discriminating transcripts. The four genes (COX-1, δ=0.96, fc=1.9, R=0.38, p=0.025, q=0.051; IL-10, δ=0.75, fc=1.7, R=0.35, p=0.045, q=0.077; NLRP12, δ=1.27, fc=2.4, R=0.40, p=0.018, q=0.042; and S100A9, δ=1.10, fc=2.1, R=0.52, p=0.002, q=0.018) which were more abundant in IRIS cases versus non-IRIS controls clearly correlated with each other and separated from the other transcripts (Figure 1B).
Next, we quantitatively analyzed these four transcripts using the log2 transformed nCounter gene expression values. As shown in Supplementary Figure 3, S100A9 expression significantly increased at the two-week time point in the IRIS patients (median log2 expression, 16.07; IQR, 15.15–16.35) from ART initiation (median, 14.59; IQR, 14.06–15.22) and was higher at 2 weeks compared to the controls (median, 15.05; IQR, 14.12–15.50; p=0.002). NLRP-12 expression also significantly increased from ART initiation (median, 5.66; IQR, 4.12–6.77) to the two-week time point in TB-IRIS patients (median, 6.94; IQR, 6.23–7.68), when it was higher compared to the controls (median, 6.15; IQR, 5.44–6.93; p=0.016). IL-10 significantly decreased in controls from ART initiation (median, 7.56; IQR, 6.42–7.73) to two weeks (median, 6.41; IQR, 5.38–7.02; p=0.005), and significantly greater IL-10 expression was seen in the IRIS cases (median, 6.83; IQR, 6.33–8.02) versus controls (median, 6.41; IQR, 5.38–7.02; p=0.049) at two weeks. Significantly higher COX-1 expression was also seen in the IRIS group (median, 8.93; IQR, 7.87-9.51) versus the non-IRIS controls (median, 7.94; IQR, 6.95-8.81; p=0.049) at the two-week time point.
TB-IRIS is characterized by neutrophilia
The most up-regulated gene in TB-IRIS identified in our expression analysis was S100A9, which is implicated in neutrophil accumulation in tuberculosis [21]. Similarly, NLRP12 (Pypaf-7) is crucial for neutrophil recruitment in other models of infection [22], including to the lungs [23], while (among its other actions) COX-1 generates eicosanoids which activate neutrophils [24]. We have also shown that neutrophil markers strongly co-localise with IL-10 in human tuberculous granulomas [20]. Our gene expression data therefore suggested a role for neutrophils in TB-IRIS pathogenesis and we examined this in another patient cohort, subsequently recruited in Cape Town. Supplementary Table 1 details participant characteristics.
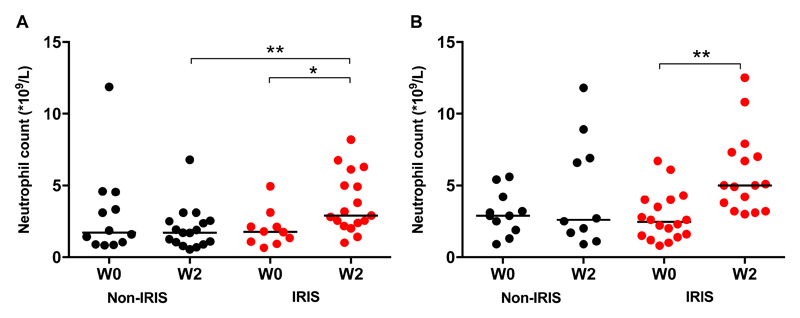
The IRIS cases in both cohorts demonstrated an increase in peripheral neutrophil counts from ART initiation to the IRIS time-point / week 2 (Cohort 1 median [IQR] 1.77 [1.04–2.37] x109/L to 2.91 [2.29–5.56] x109/L, p=0.049, Figure 2A; Cohort 2 median [IQR] 2.45 [1.48–4.00] x109/L to 5.00 [3.35–7.23] x109/L, p=0.001, Figure 2B). There were no changes in non-IRIS controls from ART initiation to two weeks. At two weeks, IRIS patients in Cohort 1 had significantly higher neutrophil counts versus the controls (median [IQR] 2.91 [2.29–5.56] x109/L) and median [IQR] 1.70 [0.97–2.52] x109/L respectively, p=0.003, Figure 2A).
Figure 2. TB-IRIS patients exhibit a rise in neutrophil count after two weeks of ART.
A: Neutrophil counts from TB-IRIS (n = 10 at ART initiation, n = 17 at Week 2 (W2)) and non-IRIS (n=12 at ART initiation, n = 17 at W2) patients (Cohort 1) are presented at ART initiation and at the Week 2 (W2) time point. B: Neutrophil counts from TB-IRIS (n =18 at ART initiation, n = 16 at W2) and non-IRIS (n =11 at ART initiation, n = 10 at W2) patients (Cohort 2) are presented at initiation of ART and at Week 2 (W2). Mann Whitney and Wilcoxon tests were used (* p < 0.05, ** p < 0.01).
There were no differences between IRIS patients and controls’ total lymphocyte or monocyte counts at either baseline or at the two week / IRIS time point.
TB-IRIS patients demonstrate activation of neutrophils, as defined by surface marker expression
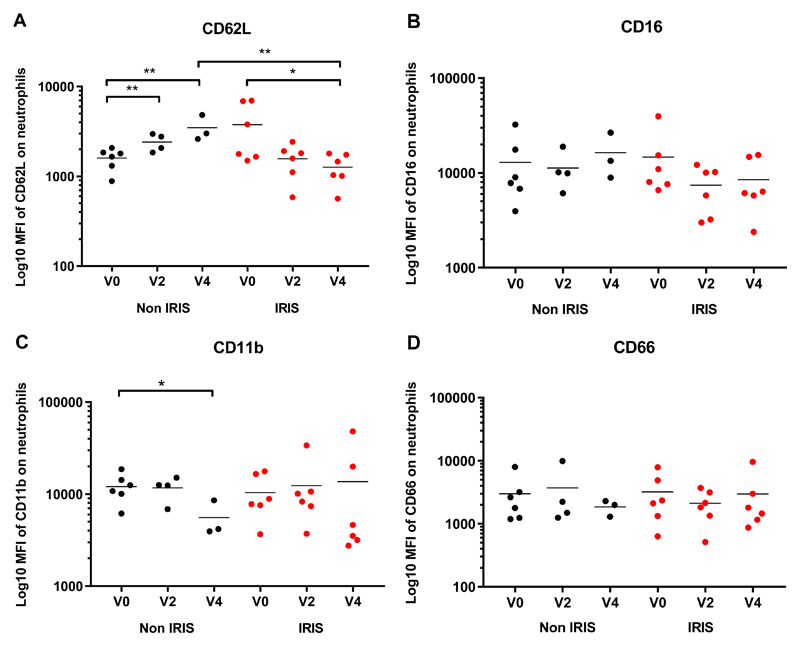
Neutrophil cell surface activation markers (CD11b, CD16, CD62L and CD66a,c,e) were analyzed in whole blood from a subset of patients in Cohort 2 (n=6 per group) using flow cytometry. There was a significant linear trend towards decreased expression of CD62L, as defined by median fluorescence intensity, on TB-IRIS patients’ neutrophils over the first four weeks from ART initiation (p=0.014), with a significant difference between neutrophil CD62L expression at ART initiation (mean, 3881; SD, 2746) versus four weeks (mean, 1229; SD, 483; p=0.042; Figure 3A). Significantly higher expression of CD62L was observed in non-IRIS controls (mean, 3422; SD, 1196) compared to TB-IRIS cases (mean, 1269; SD, 483; p=0.005; Figure 3A) at week four, consistent with significantly increased CD62L expression on non-IRIS controls’ neutrophils from ART initiation (mean, 1596; SD, 427) to two weeks (mean, 2387; SD, 517; p=0.003) and further to four weeks (mean, 3422; SD, 1196; p=0.009; Figure 3A). Supplementary Figure 2B presents representative CD62L MFI at the Week 2 / IRIS time point.
Figure 3. Neutrophil activation in TB-IRIS patients and Non-IRIS controls:
The Median Fluorescence Intensity of CD62L (A), CD16 (B), CD11b (C) and CD66a,c,e (D) on neutrophils in fresh whole blood is shown for TB-IRIS patients (red, n=6) and non-IRIS controls (black, n=6 at ART initiation (Week (W) 0), n = 4 at W2, n = 3 at W4). Lines represent means and p-values (* p < 0.05, ** p < 0.01) were derived from unpaired and paired t tests.
A similar pattern was seen for CD16 expression (Figure 3B) although comparisons did not reach statistical significance. Median fluorescence intensity of CD11b decreased in the control group from ART initiation (mean, 12130; SD, 4253) to Week 4 (mean, 5562; SD, 2584; p=0.047; Figure 3C) but no difference was seen in the IRIS group. No differences were seen in CD66a,c,e expression (Figure 3D), nor in IL-8 RA (data not shown).
TB-IRIS patients exhibit increased Neutrophil Elastase and Human Neutrophil Peptide 1-3 plasma concentrations
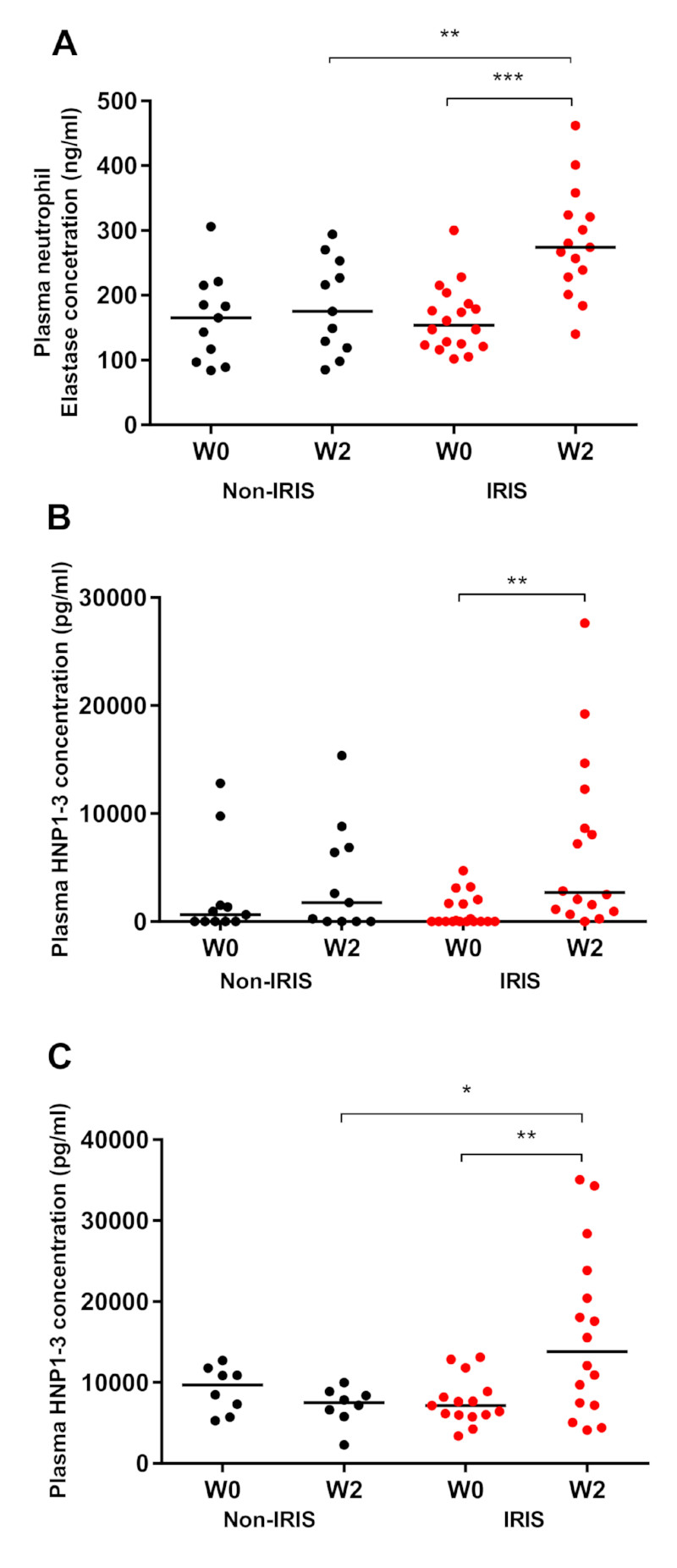
Neutrophil elastase is implicated in inflammation and tissue damage [25], and we measured this marker in plasma samples from Cohort 2. Neutrophil elastase concentration increased significantly in TB-IRIS patients between ART initiation (median 154 ng/mL; IQR, 122.5–191.3) and week two (median 274 ng/mL; IQR, 228–324; p=0.0004; Figure 4A). At two weeks after ART initiation, there was a significantly higher plasma neutrophil elastase concentration in TB-IRIS patients compared to non-IRIS controls (median, 274 ng/mL; IQR, 228–324 versus median, 175 ng/mL; IQR, 119–253 p=0.005; Figure 4A).
Figure 4. Analysis of plasma levels of neutrophil elastase and HNP1-3 in patients with TB-IRIS and non-IRIS controls.
A. Neutrophil Elastase (TB-IRIS patients (red, n = 18 at ART initiation, n = 15 at W2) and non-IRIS controls (black n = 11)) plasma concentrations were quantified using ELISA in Cohort 2. B. Human Neutrophil Peptide (HNP) 1-3 (TB-IRIS patients (red, n = 18 at ART initiation, n = 16 at W2) and non-IRIS controls (black n = 11)) plasma concentrations were quantified using ELISA in Cohort 2. C. Human Neutrophil Peptide (HNP) 1-3 plasma concentrations were quantified using ELISA in Cohort 1 (TB-IRIS patients (n =15 at ART initiation, n = 16 at W2) and non-IRIS controls (n = 8)). Lines represent medians and p-values (** p < 0.01, *** p < 0.001) were derived from Mann-Whitney and Wilcoxon tests.
Analysis of plasma Human Neutrophil Peptide (HNP) 1-3 concentrations in Cohort 2 revealed an increase in TB-IRIS patients from ART initiation (median, 0 pg/mL; IQR, 0–1775) to the week two-time point (median, 2675 pg/mL; IQR, 990–11353; p=0.005; Figure 4B). In Cohort 1, HNP1-3 concentrations also increased from week 0 (median, 7153 pg/mL; IQR, 5998–8896) to week two (median, 13821 pg/mL; IQR, 7271–22975; p=0.001), when they were higher compared to controls (median, 7510 pg/mL; IQR, 6007–8751; p=0.038; Figure 4C).
Analysis of a wider cohort recruited identically in Uganda confirmed significant differences in HNP concentration between TB-IRIS patients and non-IRIS controls at the IRIS time-point / Week 2, with resolution of these differences by later time points (Supplementary Figure 4).
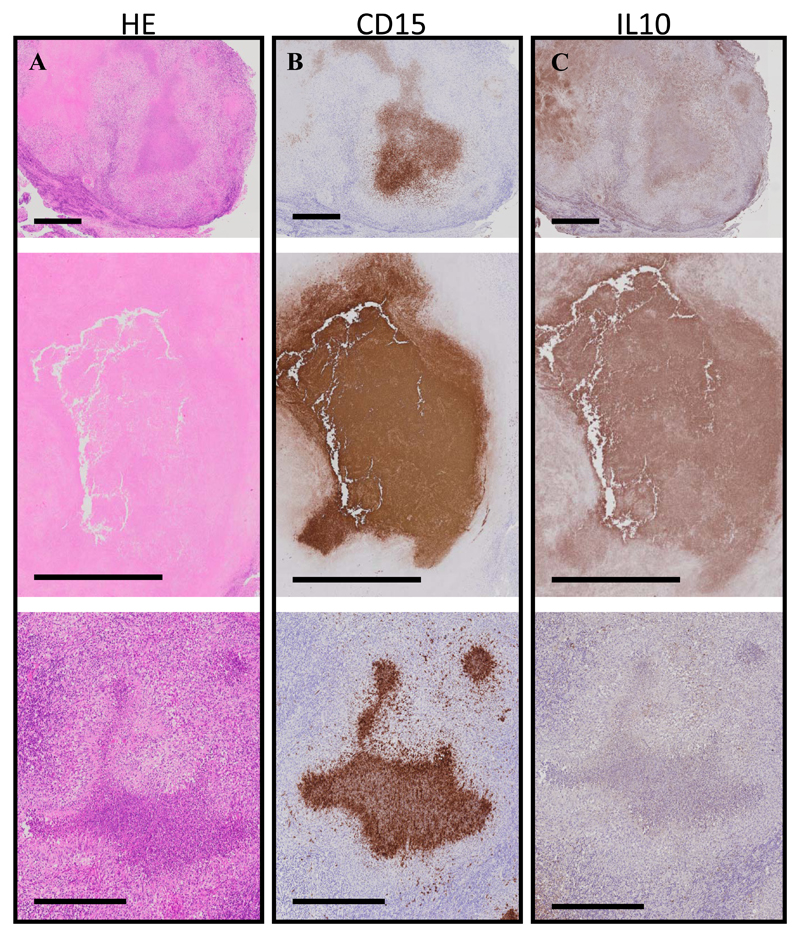
Lymph node granulomas from IRIS patients show significant neutrophil infiltration and IL-10 production
We proceeded to characterize neutrophil infiltration and accumulation in lymph nodes of TB-IRIS patients in situ, using immunohistochemistry. There was intense staining in the centre of the biopsies for the neutrophil marker CD15, correlating with areas of significant necrosis (Figure 5). Lymph nodes from patients with TB-IRIS also stained strongly for IL-10, largely correlating with neutrophils, as previously shown [20].
Figure 5. Neutrophil infiltration in the lymph nodes of TB-IRIS patients.
Caseous granulomas from consecutive cross-sectional lymph node sections of TB-IRIS patients (n = 3) that were stained with Hematoxylin and Eosin (H&E) (A), CD15 (neutrophils, B), or IL-10 (C). Intense neutrophil staining localizes within most of these caseous granulomas. IL-10 staining was diffuse but did localize within and near caseous granulomas. Black bars represent 200 μm.
Discussion
TB-IRIS immunopathogenesis remains incompletely defined and a lack of predictive markers makes its diagnosis and treatment complex. Given the temporal association of IRIS with reconstitution of CD4+ T lymphocyte numbers on antiretroviral therapy, many studies have focused on Th1 cells [26, 27]. However, TB-IRIS is not explained simply by a change in CD4 numbers, and innate cells are also implicated in the syndrome [5, 12]. Neutrophils are increasingly recognised in tuberculosis pathology [28–30], as we have previously described in TB-meningitis IRIS [6], but they had not previously been studied in this detail.
We recruited HIV+TB+ patients at risk of developing IRIS (Cohort 1) and investigated transcript abundance of genes relating to inflammasome, T-cell receptor, cytokines and their receptors. The gene transcripts that were most abundant in IRIS patients versus non-IRIS controls, and clearly discriminatory on a PCA plot, were S100A9, IL-10, NLRP-12 and COX-1. Increased expression of inflammasome and neutrophil-associated genes in TB-IRIS is consistent with previous results [12, 31], but the lower abundance of TCR-associated genes in TB-IRIS patients was unexpected and deserves further analysis. This may reflect poor reconstitution of normal T cell function in TB-IRIS and again supports the concept that the phenomenon is driven by innate inflammation without an orchestrated acquired immune response.
Among the more abundant transcripts, S100A9 contributes to inflammation in tuberculosis due to its role in neutrophil recruitment [6, 21, 32] and it has been proposed as a promising biomarker for TB diagnosis [33, 34]. NLRP-12 also plays an important role in neutrophil recruitment [22, 23]. We have reported increased levels of the IL-10 cytokine family in IRIS [17] and observed significant IL-10 staining in tuberculous granulomas where it associates with neutrophil markers and necrosis [20]. The source of IL-10 in TB-IRIS remains unclear, with conflicting data on whether regulatory T cell populations are expanded (reviewed in [4]). Again, it may be that innate cells are responsible for the production of immunosuppressive cytokines. Gene expression data therefore suggested a role of neutrophils in the development of TB-IRIS and we recruited a further cohort to perform neutrophil functional assays.
In both cohorts, we first demonstrated that patients meeting INSHI criteria for IRIS exhibited an increase in neutrophil count from ART initiation. We observed that neutrophils accumulate intensely at sites of pathology in TB-IRIS and associate with areas of necrosis. IRIS patients’ neutrophils were activated, shedding their CD62L/L-Selectin over time with a significant drop from ART initiation to four weeks (despite the initiation of corticosteroids in three patients); the reverse pattern being observed in controls. A similar trend to CD62L was seen for CD16. We have previously shown that at ART initiation, neutrophils in antiretroviral-naïve HIV-infected patients are activated, rapidly undergo cell death and their ability to kill M. tuberculosis is impaired compared to HIV-uninfected controls [18]. Our data confirms that abnormal activation is reversed on ART in patients with an uncomplicated clinical course (undergoing protective immune reconstitution), while in IRIS the neutrophil dysfunction becomes exaggerated (these patients undergo pathogenic immune reconstitution).
We did not see differences between the groups in other activation markers, including CD11b and CD66a,c,e. However, loss of CD16 and CD62L occurs preferentially as neutrophils progress to cell death [35]. Collectively, these data suggest that neutrophil activation and presumably early cell death is a hallmark of TB-IRIS [28, 30]. Increased neutrophil influx and death at disease sites will lead to release of cytotoxic granule contents causing local tissue damage and amplifying inflammatory responses [29, 36], consistent with necrotic abscesses and lymphadenopathy often observed in TB-IRIS.
Compatible with this conclusion, we found an increased neutrophil elastase concentration in the plasma of TB-IRIS patients versus non-IRIS controls two weeks after initiation of ART in cohort 2. There was also an increase from ART initiation in the South African TB-IRIS patients’ elastase concentration, and an increase in HNP 1-3 in both cohorts. The difference in neutrophil elastase concentration between IRIS patients and controls was seen despite no significant difference in absolute neutrophil count in Cohort 2, suggesting that plasma concentrations of this granule product might represent more than simply a higher number of circulating neutrophils.
Notably, some activation parameters in the patients developing IRIS tended to be less abnormal at ART initiation. This is consistent with observations by others [14, 37, 38] that TB-IRIS may be heralded by lower cytokine concentrations at ART initiation but subsequent large magnitude changes.
Limitations of our study include relatively small group sizes. We were unable to perform neutrophil functional assays including phagocytosis, mycobacterial killing and cell death in sufficient numbers, as few samples met our stringent pre-specified neutrophil purity and viability criteria of >90%. Differences in HNP concentrations between the cohorts might be due to differences in pre-analytical handling; in Cohort 1 blood was collected in Uganda and assays performed in Belgium, whereas South African samples were analysed locally. We also note a difference in neutrophil and CD4 counts between the two cohorts, likely to reflect the clinical realities of treating HIV-TB co-infection in Uganda in 2009 compared to South Africa in 2013, as well as differences in analysis platforms and racial background. However, the fact that we could demonstrate a role for neutrophils in two geographically different cohorts increases the generalizability of our findings.
A strength of our analysis was the inclusion of both peripheral blood and lymph node samples, although longitudinal analyses were conducted exclusively in peripheral blood which may not be representative of the tissue environment. However, as peripheral blood does exhibit significant perturbations in TB-IRIS, is easily accessible for serial measurements and contains many components of both the innate and acquired immune systems, we believe that analysis of this compartment is informative.
In conclusion, our data suggest that TB-IRIS is characterized by aberrant immunological recovery with inflammasome activation and neutrophil recruitment instead of reconstitution of normal T cell receptor function. Within the context of local and systemic inflammation, recruited neutrophils are activated, are likely to undergo rapid cell death and will release cytotoxic granule contents. This drives tissue damage and further inflammation, paradoxically associated with immunosuppressive IL-10 release which may compromise host control of any remaining viable mycobacteria. As neutrophils are likely to be key effector cells mediating pathological damage in TB-IRIS, it seems logical to consider host-directed therapies to reduce neutrophil recruitment (eg CXCR2 inhibitors [39] and anti-C5a inhibitors [40]) or to promote neutrophil apoptosis (eg statins [41]): these questions require further research.
Supplementary Material
Acknowledgements
We would like to thank Prof Jon Friedland and Prof Paul Elkington for their expert input into the design and supervision of the TDTB2 study in Cape Town, from which our patients were recruited.
Funding
This work was supported by: EC FP6 Specific Targeted Research Project (STREP) (LSHP-CT-2007-037659-TBIRIS); the European and South African HIV co-infection research consortium (PIRSES-GA-2011-295214 to J.K.N); the Netherlands Organization for Scientific Research – WOTRO Science for Global Development (NACCAP W 07.05.20100); the Infectious Diseases Network for Treatment and Research in Africa (INTERACT); the individual PhD scholarship of the Institute of Tropical Medicine (ITM), supported by the Directorate General for Development (to J.K.N); the Wellcome Trust (087754 to D.M.L, 094000 to N.F.W, 098316 to G.M., 104803 & 203135 to R.J.W., FC00110218 to Francis Crick Institute); British Federation of Women Graduates (Ruth Bowden Scholarship to N.F.W); Cancer Research UK (FC00110218 to Francis Crick Institute); Medical Research Council UK (FC00110218 to Francis Crick Institute); European Union (FP7-HEALTH-F3-2012-305578 to R.J.W.); National Research Foundation of South Africa (96841 to R.J.W.)
References
- 1.Namale PE, Abdullahi LH, Fine S, et al. Paradoxical TB-IRIS in HIV-infected adults: a systematic review and meta-analysis. Future Microbiol. 2015;10:1077–1099. doi: 10.2217/fmb.15.9. [DOI] [PubMed] [Google Scholar]
- 2.Bana TM, Lesosky M, Pepper DJ, et al. Prolonged tuberculosis-associated immune reconstitution inflammatory syndrome: characteristics and risk factors. BMC Infect Dis. 2016;16:518. doi: 10.1186/s12879-016-1850-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meintjes G, Lawn SD, Scano F, et al. Tuberculosis-associated immune reconstitution inflammatory syndrome: case definitions for use in resource-limited settings. Lancet Infect Dis. 2008;8:516–523. doi: 10.1016/S1473-3099(08)70184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai RP, Meintjes G, Wilkinson RJ. HIV-1 tuberculosis-associated immune reconstitution inflammatory syndrome. Semin Immunopathol. 2016;38:185–198. doi: 10.1007/s00281-015-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai RP, Nakiwala JK, Meintjes G, et al. The immunopathogenesis of the HIV tuberculosis immune reconstitution inflammatory syndrome. Eur J Immunol. 2013;43:1995–2002. doi: 10.1002/eji.201343632. [DOI] [PubMed] [Google Scholar]
- 6.Marais S, Wilkinson KA, Lesosky M, et al. Neutrophil-associated central nervous system inflammation in tuberculous meningitis immune reconstitution inflammatory syndrome. Clin Infect Dis. 2014;59:1638–1647. doi: 10.1093/cid/ciu641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravimohan S, Tamuhla N, Kung SJ, et al. Matrix Metalloproteinases in Tuberculosis-Immune Reconstitution Inflammatory Syndrome and Impaired Lung Function Among Advanced HIV/TB Co-infected Patients Initiating Antiretroviral Therapy. EBioMedicine. 2016;3:100–107. doi: 10.1016/j.ebiom.2015.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tadokera R, Meintjes GA, Wilkinson KA, et al. Matrix metalloproteinases and tissue damage in HIV-tuberculosis immune reconstitution inflammatory syndrome. Eur J Immunol. 2014;44:127–136. doi: 10.1002/eji.201343593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson KA, Walker NF, Meintjes G, et al. Cytotoxic mediators in paradoxical HIV-tuberculosis immune reconstitution inflammatory syndrome. J Immunol. 2015;194:1748–1754. doi: 10.4049/jimmunol.1402105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran HT, Van den Bergh R, Loembe MM, et al. Modulation of the complement system in monocytes contributes to tuberculosis-associated immune reconstitution inflammatory syndrome. AIDS. 2013;27:1725–1734. doi: 10.1097/QAD.0b013e328361648b. [DOI] [PubMed] [Google Scholar]
- 11.Andrade BB, Singh A, Narendran G, et al. Mycobacterial antigen driven activation of CD14++CD16- monocytes is a predictor of tuberculosis-associated immune reconstitution inflammatory syndrome. PLoS Pathog. 2014;10:e1004433. doi: 10.1371/journal.ppat.1004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai RP, Meintjes G, Wilkinson KA, et al. HIV-tuberculosis-associated immune reconstitution inflammatory syndrome is characterized by Toll-like receptor and inflammasome signalling. Nat Commun. 2015;6:8451. doi: 10.1038/ncomms9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan HY, Yong YK, Shankar EM, et al. Aberrant Inflammasome Activation Characterizes Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome. J Immunol. 2016;196:4052–4063. doi: 10.4049/jimmunol.1502203. [DOI] [PubMed] [Google Scholar]
- 14.Goovaerts O, Jennes W, Massinga-Loembe M, et al. LPS-binding protein and IL-6 mark paradoxical tuberculosis immune reconstitution inflammatory syndrome in HIV patients. PLoS One. 2013;8:e81856. doi: 10.1371/journal.pone.0081856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tadokera R, Meintjes G, Skolimowska KH, et al. Hypercytokinaemia accompanies HIV-tuberculosis immune reconstitution inflammatory syndrome. Eur Respir J. 2011;37:1248–1259. doi: 10.1183/09031936.00091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan HY, Yong YK, Andrade BB, et al. Plasma interleukin-18 levels are a biomarker of innate immune responses that predict and characterize tuberculosis-associated immune reconstitution inflammatory syndrome. AIDS. 2015;29:421–431. doi: 10.1097/QAD.0000000000000557. [DOI] [PubMed] [Google Scholar]
- 17.Tadokera R, Wilkinson KA, Meintjes GA, et al. Role of the interleukin 10 family of cytokines in patients with immune reconstitution inflammatory syndrome associated with HIV infection and tuberculosis. J Infect Dis. 2013;207:1148–1156. doi: 10.1093/infdis/jit002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Worodria W, Menten J, Massinga-Loembe M, et al. Clinical spectrum, risk factors and outcome of immune reconstitution inflammatory syndrome in patients with tuberculosis-HIV coinfection. Antivir Ther. 2012;17:841–848. doi: 10.3851/IMP2108. [DOI] [PubMed] [Google Scholar]
- 19.Lowe DM, Bangani N, Goliath R, et al. Effect of Antiretroviral Therapy on HIV-mediated Impairment of the Neutrophil Antimycobacterial Response. Ann Am Thorac Soc. 2015;12:1627–1637. doi: 10.1513/AnnalsATS.201507-463OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diedrich CR, O'Hern J, Gutierrez MG, et al. Relationship between HIV-1 co-infection, IL-10, and M. tuberculosis in human lymph node granulomas. J Infect Dis. 2016;214:1309–1318. doi: 10.1093/infdis/jiw313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopal R, Monin L, Torres D, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am J Respir Crit Care Med. 2013;188:1137–1146. doi: 10.1164/rccm.201304-0803OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai S, Batra S, Del Piero F, et al. NLRP12 modulates host defense through IL-17A-CXCL1 axis. Mucosal Immunol. 2016;9:503–514. doi: 10.1038/mi.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulland TK, Jain N, Hornick EE, et al. Nlrp12 mutation causes C57BL/6J strain-specific defect in neutrophil recruitment. Nat Commun. 2016;7:13180. doi: 10.1038/ncomms13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinz C, Aldrovandi M, Uhlson C, et al. Human Platelets Utilize Cycloxygenase-1 to Generate Dioxolane A3, a Neutrophil-activating Eicosanoid. J Biol Chem. 2016;291:13448–13464. doi: 10.1074/jbc.M115.700609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gehrig S, Duerr J, Weitnauer M, et al. Lack of neutrophil elastase reduces inflammation, mucus hypersecretion, and emphysema, but not mucus obstruction, in mice with cystic fibrosis-like lung disease. Am J Respir Crit Care Med. 2014;189:1082–1092. doi: 10.1164/rccm.201311-1932OC. [DOI] [PubMed] [Google Scholar]
- 26.Bourgarit A, Carcelain G, Samri A, et al. Tuberculosis-associated immune restoration syndrome in HIV-1-infected patients involves tuberculin-specific CD4 Th1 cells and KIR-negative gammadelta T cells. J Immunol. 2009;183:3915–3923. doi: 10.4049/jimmunol.0804020. [DOI] [PubMed] [Google Scholar]
- 27.Meintjes G, Wilkinson KA, Rangaka MX, et al. Type 1 helper T cells and FoxP3-positive T cells in HIV-tuberculosis-associated immune reconstitution inflammatory syndrome. Am J Respir Crit Care Med. 2008;178:1083–1089. doi: 10.1164/rccm.200806-858OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe DM, Bandara AK, Packe GE, et al. Neutrophilia independently predicts death in tuberculosis. Eur Respir J. 2013;42:1752–1757. doi: 10.1183/09031936.00140913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe DM, Redford PS, Wilkinson RJ, et al. Neutrophils in tuberculosis: friend or foe? Trends Immunol. 2012;33:14–25. doi: 10.1016/j.it.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Ong CW, Elkington PT, Brilha S, et al. Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis. PLoS Pathog. 2015;11:e1004917. doi: 10.1371/journal.ppat.1004917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marais S, Lai RP, Wilkinson KA, et al. Inflammasome activation underlies central nervous system deterioration in HIV-associated tuberculosis. J Infect Dis. 2017;215:677–686. doi: 10.1093/infdis/jiw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruenster M, Kurz AR, Chung KJ, et al. Extracellular MRP8/14 is a regulator of beta2 integrin-dependent neutrophil slow rolling and adhesion. Nat Commun. 2015;6:6915. doi: 10.1038/ncomms7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J. Early diagnosis of pulmonary tuberculosis using serum biomarkers. Proteomics. 2015;15:6–7. doi: 10.1002/pmic.201400532. [DOI] [PubMed] [Google Scholar]
- 34.Xu D, Li Y, Li X, et al. Serum protein S100A9, SOD3, and MMP9 as new diagnostic biomarkers for pulmonary tuberculosis by iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics. 2015;15:58–67. doi: 10.1002/pmic.201400366. [DOI] [PubMed] [Google Scholar]
- 35.Hart SP, Ross JA, Ross K, et al. Molecular characterization of the surface of apoptotic neutrophils: implications for functional downregulation and recognition by phagocytes. Cell Death Differ. 2000;7:493–503. doi: 10.1038/sj.cdd.4400680. [DOI] [PubMed] [Google Scholar]
- 36.Korbel DS, Schneider BE, Schaible UE. Innate immunity in tuberculosis: myths and truth. Microbes Infect. 2008;10:995–1004. doi: 10.1016/j.micinf.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 37.Ravimohan S, Tamuhla N, Steenhoff AP, et al. Immunological profiling of tuberculosis-associated immune reconstitution inflammatory syndrome and non-immune reconstitution inflammatory syndrome death in HIV-infected adults with pulmonary tuberculosis starting antiretroviral therapy: a prospective observational cohort study. Lancet Infect Dis. 2015;15:429–438. doi: 10.1016/S1473-3099(15)70008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goovaerts O, Jennes W, Massinga-Loembe M, et al. Lower Pre-Treatment T Cell Activation in Early- and Late-Onset Tuberculosis-Associated Immune Reconstitution Inflammatory Syndrome. PLoS One. 2015;10:e0133924. doi: 10.1371/journal.pone.0133924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Soyza A, Pavord I, Elborn JS, Smith D, Wray H, Puu M, et al. A randomised, placebo-controlled study of the CXCR2 antagonist AZD5069 in bronchiectasis. Eur Respir J. 2015;46:1021–1032. doi: 10.1183/13993003.00148-2015. [DOI] [PubMed] [Google Scholar]
- 40.Woodruff TM, Nandakumar KS, Tedesco F. Inhibiting the C5-C5a receptor axis. Mol Immunol. 2011;48:1631–1642. doi: 10.1016/j.molimm.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Thomson NC. Novel approaches to the management of noneosinophilic asthma. Ther Adv Respir Dis. 2016;10:211–234. doi: 10.1177/1753465816632638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.