Abstract
In this research we aim to demonstrate that an ontology-based system can categorize potential drug-drug interaction (PDDI) evidence items into complex types based on a small set of simple questions. Such a method could increase the transparency and reliability of PDDI evidence evaluation, while also reducing the variations in content and seriousness ratings present in PDDI knowledge bases. We extended the DIDEO ontology with 44 formal evidence type definitions. We then manually annotated the evidence types of 30 evidence items. We tested an RDF/OWL representation of answers to a small number of simple questions about each of these 30 evidence items and showed that automatic inference can determine the detailed evidence types based on this small number of simpler questions. These results show proof-of-concept for a decision support infrastructure that frees the evidence evaluator from mastering relatively complex written evidence type definitions.
Keywords: Drug Interactions, Ontologies, Artificial Intelligence
Introduction
While medication therapies are generally beneficial to a patient’s health, they can also result in harm. A recent review of epidemiologic studies published over a 14 year period found that 3.5% of hospital admissions are the result of an adverse drug reaction [1]. The United States Department of Health and Human Services recently stated that reducing the rate of adverse drug events should be a national priority [2]. Research on the safety of any given drug starts during its development and continues after marketing. During these phases, studies examine the potential for one drug to alter the pharmacokinetic properties (absorption, distribution, metabolism, or distribution) or clinical effect (pharmacodynamics) of another drug. The results of these studies can suggest PDDIs that might lead to preventable harm to patients without proper management [3].
The spectrum of study types used to research PDDIs is broad and complex. These include in vitro experiments, population pharmacokinetic analyses, randomized controlled clinical trials, and observational epidemiologic studies [4]. Data mining adverse event reports and case report evaluation can also be included because these are research activities that generate PDDI hypotheses [5]. Not only are there many different types of PDDI studies, there are also numerous considerations that can influence the validity of a given study [2,4]. The range and complexity of study designs and the resulting evidence types can make it very difficult to evaluate a body of evidence to determine if a PDDI exists. This difficulty might be an important factor influencing the large differences in content found in PDDI knowledge bases that are designed to help clinicians make management decisions [6,7]. Indeed, a more systematic approach to evaluating evidence for the existence of PDDIs was one of the recommendations put forth by a recent expert consensus conference series [8].
We believe that an ontology-based system can categorize PDDI evidence items into complex types based on a small set of simple questions, thereby reducing the cognitive load experienced by evidence evaluators. Our approach is to have a computer program infer the specific evidence type of a PDDI study based on simple data provided by the evidence evaluator. Here, we report our progress creating an infrastructure for this kind of decision support. We demonstrate a proof-of-concept for using an off the shelf OWL reasoner and formal evidence type definitions so that an evidence base curator only has to identify the study type at a high level (e.g., clinical study) and provide some simple design features (e.g., ‘randomization’). With this information, the OWL reasoner can efficiently and consistently infer the specific evidence type with which to tag the study (see Fig. 1).
Figure 1.
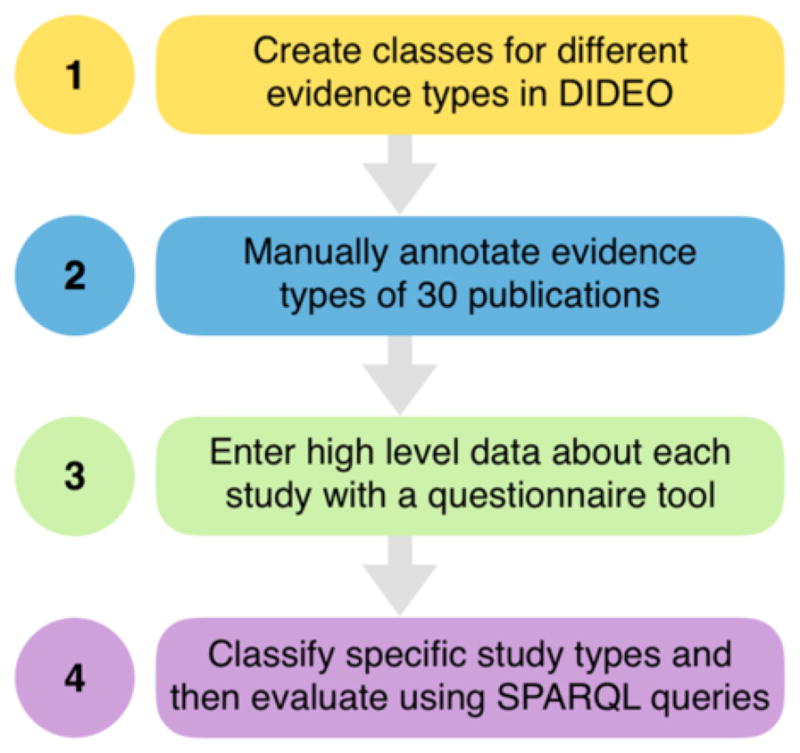
The steps taken to build the proof-of-concept for categorizing PDDI evidence.
Methods
DIDEO (https://github.com/DIDEO) is an OWL ontology developed to facilitate managing information about PDDIs from multiple sources (clinical studies, case studies, in vitro experiments, etc.). It is freely accessible from http://purl.obolibrary.org/obo/dideo.owl. The fundamental ontological commitments of DIDEO have been described in detail [9]. One key contribution of DIDEO is to provide OWL classes for evidence types (see Fig. 2).
Figure 2.
The evidence information types from DIDEO. All subclasses of “evidence information from clinical study” currently existing in DIDEO are displayed.
The process creating the textual definitions for each evidence type has been previously reported [10]. For Step 1 of the current project, we wrote necessary and sufficient conditions (i.e., OWL equivalent class axioms) for each textual definition using entities defined or imported into DIDEO (see Table 1). We then ran an OWL reasoner (e.g. HermiT (http://www.hermit-reasoner.com)) to categorize the evidence types into a multilevel hierarchy.
Table 1.
(Step 1) Clinical trial evidence types represented in DIDEO, their textual definitions, and their necessary and sufficient conditions.
| Evidence type (IRI and rdfs:label) | Definition (iao:definition) | Axiom |
|---|---|---|
| DIDEO_00000056 evidence information from clinical study | An evidence information content entity that is about a clinical drug trial. | ‘evidence information content entity’ and (‘is about’ some (assay and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some (‘chemical entity’ or ‘drug product’)))) |
| DIDEO_00000071 evidence information from drug-drug interaction clinical trial | An evidence information content entity that is about a clinical drug trial that has at least two drugs as its specified input. | ‘evidence information content entity’ and (‘is about’ some (‘scientific observation’ and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘is bearer of’ some ‘object drug role’))) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘is bearer of’ some ‘precipitant drug role’))))) |
| DIDEO_00000072 evidence information from non-randomized drug-drug interaction clinical trial | An evidence information content entity that is about a clinical drug trial that has at least two drugs as its specified input. | ‘evidence information content entity’ and (‘is about’ some (assay and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘bearer of’ some ‘object drug role’))) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘bearer of’ some ‘precipitant drug role’))))) |
| DIDEO_00000073 evidence information from parallel groups drug-drug interaction clinical trial’ | An evidence information content entity that is about a clinical drug trial that has at least two drugs as its specified input, and that does not have group randomization as a part, and that realizes a clinical study design that has parallel group design as a part. | ‘evidence information content entity’ and (‘is about’ some (‘scientific observation’ and (not (has_part value ‘group randomization’)) and (realizes some (concretizes some (‘clinical study design’ and (has_part some ‘independent measure design’)))) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘is bearer of’ some ‘object drug role’))) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘is bearer of’ some ‘precipitant drug role’))))) |
| DIDEO_00000074 evidence information from randomized drug- drug interaction clinical trial | An evidence information content entity that is about a clinical drug trial that has at least two drugs as its specified input and does have group randomization as a part. | ‘evidence information content entity’ and (‘is about’ some (‘scientific observation’ and (has_part some ‘group randomization’) and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘is bearer of’ some ‘object drug role’))) and (has_specified_input some ((‘chemical entity’ or ‘drug product’) and (‘is bearer of’ some ‘precipitant drug role’))))) |
| DIDEO_00000075 evidence information from pharmacokinetic trial | An evidence information content entity that is about a clinical drug trial that focusses on pharmacokinetics. | ‘evidence information content entity’ and (‘is about’ some (‘scientific observation’ and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some (organism and (participates_in some ‘pharmacokinetic process’))) and (has_specified_input some (‘chemical entity’ or ‘drug product’)))) |
| DIDEO_00000076 evidence information from genotyped pharmacokinetic trial | An evidence information content entity that is about a clinical drug trial that focusses on pharmacokinetics and that has organisms as participants that participated in genotyping. | ‘evidence information content entity’ and (‘is about’ some (‘scientific observation’ and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some (organism and (participates_in some ‘pharmacokinetic process’) and (participates_in some genotyping))) and (has_specified_input some (‘chemical entity’ or ‘drug product’)))) |
| DIDEO_00000103 evidence information from phenotyped pharmacokinetic trial | An evidence information content entity that is about a clinical drug trial that focusses on pharmacokinetics and that has organisms as participants that participated in phenotyping. | ‘evidence information content entity’ and (‘is about’ some (‘scientific observation’ and (realizes some (concretizes some ‘clinical study design’)) and (has_specified_input some (organism and (participates_in some ‘pharmacokinetic process’) and (participates_in some ‘phenotype characterization’))) and (has_specified_input some (‘chemical entity’ or ‘drug product’)))) |
Next for Step 2, we created a dataset that would enable automated decision support for people who need to evaluate PDDI evidence items. As Table 1 shows, evidence definitions for more specific kinds of evidence can become fairly complicated. Decision support should simplify the cognitive load of a person evaluating an evidence item.
We developed a proof-of-concept system that was used to test whether an OWL reasoner and formal evidence type definitions can efficiently and consistently infer the specific evidence type from Table 1 based on basic evidence types (e.g., clinical study) and design features (e.g., ‘randomization’). Combining the evidence instance data with the OWL equivalent class axioms from DIDEO should enable an OWL reasoner to infer the specific evidence type of each instance. We tested this in Step 4 shown in Figure 1.
For Step 1 we used classes for the detailed evidence types shown in Table 1 in DIDEO (created in prior work) [11]. For Step 2, we drew on prior work [10] that had manually assigned a study type from Table 1: 30 publications were queried from the Drug Interaction Knowledge Base (http://dikb.org/), representing five publications for each of the six clinical study evidence types defined in DIDEO (see Table 2). Listing 1 shows the SPARQL query used to query a single evidence type from Table 2. The interested reader can modify the dikbEvidence:Evidence_type and PubMed identifier to retrieve any of the other items listed.
Table 2.
Five publications for each of the six clinical study evidence types defined in DIDEO
| Evidence type by DIKB label | Publications (PubMed Identifier) |
|---|---|
| EV_PK_DDI_NR | 10445377,10907965, 15876900,11147928, 8801057 |
| EV_PK_DDI_Par_Grps | 12911366,11910262, 17571477,9855322, 15518608 |
| EV_PK_DDI_RCT | 9542477,16778714, 11563412,9757151, 19242403 |
| EV_CT_Pharmacokinetic | 8911886,15834460, 14747427,1487561, 1438031 |
| EV_CT_PK_Genotype | 11452243,19142106, 16765147,17429316, 8689810 |
| EV_CT_PK_Phenotype | 8513845,7690693, 2007317, 1412613, 8823236 |
Listing 1.
(Step 1) A query used to retrieve one of the 30 evidence types from the Drug Interaction Knowledge Base (https://dbmi-icode-01.dbmi.pitt.edu/dikb/snorql)
SELECT DISTINCT ?ev ?quote
WHERE {
?ev dikbEvidence:Evidence_type
dikbEvidence:EV_PK_DDI_NR.
?ev rdfs:seeAlso
<http://www.ncbi.nlm.nih.gov/pubmed/10445377>.
?ev siocns:content ?quote.
}
|
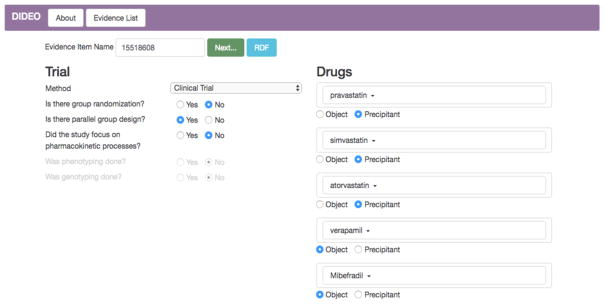
As part of Step 3 a questionnaire tool, previously developed for the CAFE project (https://cafe-trauma.com), was modified and used to manually enter high level type and simple design features for each study in Table 2 (see Fig. 3). We then exported RDF instances created by the tool for each evidence item. We put these into a single file for import into an RDF store. Our RDF store was the community version of the Stardog (http://stardog.com/) RDF store, which comes with a built-in OWL description logic reasoner. We loaded RDF file of evidence instance data along with following OWL files:
Figure 3.
(Step 3) The questionnaire tool used to enter high level type and simple design features for each study in Table 2.
DIDEO: http://purl.obolibrary.org/obo/dideo/release/2016-07-29/dideo.owl
RO core: http://purl.obolibrary.org/obo/ro/releases/2016-09-02/core.owl
RO BFO classes minimal: http://purl.obolibrary.org/obo/ro/releases/2016-09-02/bfo-classes-minimal.owl
BFO (classes only) http://purl.obolibrary.org/obo/bfo/2014-05-03/classes-only.owl
RO annotations: http://purl.obolibrary.org/obo/ro/releases/2016-09-02/annotations.owl
Finally for Step 4, we developed a set of competency questions and associated SPARQL queries (see Table 3) that could be implemented in an application similar to that shown in Figure 3 in order to provide automated support to evidence evaluators for determining an evidence item’s type. We tested that each SPARQL query ran successfully over the inferred RDF store.
Table 3.
(Step 4) A set of competency questions and associated SPARQL queries that could be used by an application similar to that shown in Figure 3 to implement automated support to evidence evaluators for determining an evidence item’s type.
| Competency Question | SPARQL Query |
|---|---|
| Which evidence items stem from non-randomized clinical studies on drug-drug interactions without parallel group design? |
select * {
?evidence a obo:DIDEO_00000072 .
filter not exists {
?evidence a obo:DIDEO_00000073 .
}
}
|
| Which evidence items stem from non-randomized clinical studies on drug-drug interactions with a parallel group design? |
select * {
?evidence a obo:DIDEO_00000073 .
}
|
| Which evidence items stem from randomized clinical studies on drug-drug interactions? |
select * {
?evidence a obo:DIDEO_00000074 .
}
|
| Which evidence items stem from clinical studies targeting pharmacokinetics without genotyping or phenotyping? |
select * {
?evidence a obo:DIDEO_00000075 .
filter not exists {
?evidence a obo:DIDEO_00000076 .
} .
filter not exists {
?evidence a obo:DIDEO_00000103 .
} .
}
|
| Which evidence items stem from clinical studies targeting pharmacokinetics and using genotyping? |
select * {
?evidence a obo:DIDEO_00000076 .
}
|
| Which evidence items stem from clinical studies targeting pharmacokinetics and using phenotyping? |
select * {
?evidence a obo:DIDEO_00000103 .
}
|
Results
Table 4 shows the results of running the competency question SPARQL queries (see Table 3) over the inferred RDF store. Each evidence type classification corresponds exactly to the manual classification from Table 2, thereby validating the data model and OWL ontology while showing proof of concept that complex evidence type classification can be obtained from the answers to simple questions.
Table 4.
(Step 4) Retrieval results from running the SPARQL queries (Table 3, Column B) on the Stardog triplestore.
| DIDEO annotation | PMID of Publications |
|---|---|
| Evidence information from non-randomized DDI clinical trial (DIDEO_00000072) | 10445377, 10907965, 15876900, 11147928, 8801057 |
| Evidence information from parallel groups DDI (DIDEO_00000073) | 12911366, 11910262, 17571477, 9855322, 15518608 |
| Randomized drug-drug interaction clinical trial (DIDEO_00000074) | 9542477,16778714,11563412, 9757151, 19242403 |
| Pharmocokinetic trial (DIDEO_00000075) | 8911886, 15834460, 14747427, 1487561, 1438031 |
| Genotyped pharmacokinetic trial (DIDEO_00000076) | 11452243, 19142106, 16765147, 17429316, 8689810 |
| Phenotyped pharmacokinetic trial (DIDEO_000000103) | 8513845, 7690693, 2007317, 1412613, 8823236 |
Discussion
DIDEO now provides OWL classes for many of the evidence types within the PDDI domain of scientific discourse. The current work shows the feasibility of using these formal definitions to build decision support that helps evidence evaluators to determine the specific types of studies or experiments they review. Instead of having to master complex written evidence type definitions, such as those shown in Table 1, evaluators will only need to answer a small number of simple questions about the study, such as those shown Figure 3. With that information, simple SPARQL queries can be run over an inferred RDF dataset similar to the one used in this study to return the specific evidence type as a URI from DIDEO. By simplifying the task of evidence type assignment, a team of curators should be able to produce more correct and consistent work over the many hundreds of evidence items they need to manage.
Conclusions
In this work we showed proof-of-concept for the technical infrastructure, showing that PDDI evidence item data we added into a triple store together with DIDEO evidence types enable an OWL reasoner to infer an evidence item’s specific type. In future work we plan to conduct a user study to test the hypothesis that this approach improves the correctness and consistency of evidence type assignment by evidence base curators. We also plan to extend the decision support to include questions that can be used to infer the quality of a study. We then plan to conduct a user study that compares interrater agreement between two groups of PDDI experts – one provided with evidence evaluation decision support, the other using their usual procedures. Based on our hypothesis, the group of partipants given decision support should show much greater interrater agreement than the comparison group. We will also compare speed, ease, and user preference. If our hypothesis is shown to be true, it would be a promising step forward toward reducing the large differences in content found in PDDI knowledge bases that are designed to help clinicians make management decisions [6,7].
Evidence annotation is already a significant part of the biomedical enterprise, with computer-supported manual annotation used by an entire community and additional efforts to synthesize clinical research (e.g. Cochrane Reviews). We believe that approaches to simplify annotation processes, using incremental formalization and granular information, will be essential for increasing the availability of searchable (and in some cases algorithmically-synthesized) information. Generalizations of the work presented in this paper have the potential to greatly increase the impact of the curation enterprise.
Acknowledgments
This project was supported by a grant from the National Library of Medicine: “Addressing gaps in clinically useful evidence on drug-drug interactions” (R01LM011838). JS was partially supported by training grant T15LM007059 from the National Library of Medicine/National Institute of Dental and Craniofacial Research.
RDF descriptions for the 30 clinical studies generated in Step 3 are freely available from https://goo.gl/Wk84gq.
References
- 1.Bouvy JC, Bruin MLD, Koopmanschap MA. Epidemiology of adverse drug reactions in Europe: A review of recent observational studies. Drug Saf. 2015;38:437–453. doi: 10.1007/s40264-015-0281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; US Department of Health and Human Services. National Action Plan for Adverse Drug Event Prevention [Internet] 2014 Available from: https://health.gov/hcq/pdfs/ade-action-plan-508c.pdf.
- 2.Magro L, Moretti U, Leone R. Epidemiology and characteristics of adverse drug reactions caused by drug-drug interactions. Expert Opin Drug Saf. 2012;11:83–94. doi: 10.1517/14740338.2012.631910. [DOI] [PubMed] [Google Scholar]
- 3.Hennessy S, Leonard CE, Gagne JJ, Flory JH, Han X, Brensinger CM, Bilker WB. Pharmacoepidemiologic methods for studying the health effects of drug-drug interactions. Clin Pharmacol Ther. 2016;99:92–100. doi: 10.1002/cpt.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Percha B, Altman RB. Informatics confronts drug-drug interactions. Trends Pharmacol Sci. 2013;34:178–184. doi: 10.1016/j.tips.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang LM, Wong M, Lightwood JM, Cheng CM. Black box warning contraindicated comedications: concordance among three major drug interaction screening programs. Ann Pharmacother. 2010;44:28–34. doi: 10.1345/aph.1M475. [DOI] [PubMed] [Google Scholar]
- 6.Saverno KR, Hines LE, Warholak TL, Grizzle AJ, Babits L, Clark C, Taylor AM, Malone DC. Ability of pharmacy clinical decision-support software to alert users about clinically important drug–drug interactions. J Am Med Inform Assoc. 2011;18:32–37. doi: 10.1136/jamia.2010.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheife RT, Hines LE, Boyce RD, Chung SP, Momper JD, Sommer CD, Abernethy DR, Horn JR, Sklar SJ, Wong SK, Jones G, Brown ML, Grizzle AJ, Comes S, Wilkins TL, Borst C, Wittie MA, Malone DC. Consensus recommendations for systematic evaluation of drug-drug interaction evidence for clinical decision support. Drug Saf. 2015;38:197–206. doi: 10.1007/s40264-014-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brochhausen M, Schneider J, Malone D, Empey P, Hogan WR, Boyce RD. Towards a foundational representation of potential drug-drug interaction knowledge. Proc. of DIKR 2014/IWOOD 2014/OBIB 2014 – Workshops of ICBO; Houston TX, USA. October 6–7, 2014; 2014. http://ceur-ws.org/Vol-1309/paper2.pdf. [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce R, Collins C, Horn J, Kalet I. Computing with evidence: Part I: A drug-mechanism evidence taxonomy oriented toward confidence assignment. J Biomed Inform. 42:979–989. doi: 10.1016/j.jbi.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brochhausen M, Empey PE, Schneider J, Hogan WR, Boyce RD. Adding Evidence Type Representation to DIDEO. Proc. Of the Joint International Conference on Biological Ontology and BioCreative; Corvallis OR, USA. August 1–4, 2016; 2016. http://ceur-ws.org/Vol-1747/IP02_ICBO2016.pdf. [Google Scholar]