Abstract
Endosomal entrapment is a key hurdle for most intracellular protein-based therapeutic strategies. We report a general strategy for efficient delivery of proteins to the cytosol through co-engineering of proteins and nanoparticle vehicles. The proteins feature an oligo(glutamate) sequence (E-tag) that binds arginine-fuctionalized gold nanoparticles, generating hierarchical spherical nanoassemblies. These assemblies fuse with cell membranes, releasing the E-tagged protein directly into the cytosol. Five different proteins with diverse charges, sizes, and functions were effectively delivered into cells, demonstrating the generality of our method. Significantly, the engineered proteins retained activity after cytosolic delivery, as demonstrated through the delivery of active Cre recombinase and Granzyme A to kill cancer cells.
Keywords: Cytosolic protein delivery, membrane fusion, protein engineering, nanoparticles, hierarchical nanoassembly
TOC image
Nanoassemblies delivering protein into cytosol through a membrane fusion-like mechanism.
Intracellular delivery of proteins into cells is crucial for therapeutic development,1 cellular imaging and diagnosis,2 genome engineering,3,4 and synthetic biology applications.5 Native enzymes and transcription factors are particularly attractive ‘biologics’ for intracellular enzyme replacement therapy,6,7 however effective delivery of these potential therapeutics remains elusive.7 Endosomal entrapment is a key hurdle: nanocarrier-based delivery methods result in only a fraction of the entrapped cargo (often ~1%) escaping into the cytosol.8 Protease-mediated degradation and exocytosis of the remaining entrapped cargo make these strategies ultimately highly inefficient.8,9,-10 Delivery through membrane disruption methods can provide efficient cytosolic protein delivery, however, these methods generally require additional osmolytic surfactants,11 hypertonic agents,12 or mechanical distortion techniques13 that are harmful for the cells and have challenges for in vivo enzyme applications.
Recently, our laboratory has developed a nanocapsule-based14 strategy for cytosolic protein delivery.15,16,-17 This method, however, is limited to proteins whose pI values are below ~7. More recently, we developed an engineering strategy to form spherical hierarchical self-assemblies between proteins and nanoparticles.18 In our approach, proteins tagged with recombinantly attached oligo-glutamic acid (E-tagged proteins) self-assemble with positively charged gold nanoparticles (2 nm core diameter) that carry arginine functionality on their surface (ArgNP).14,19 We report here the use of these assemblies in a general strategy for direct cytosolic delivery of proteins into mammalian cells, with versatility demonstrated using five different proteins with spanning a range of charge, size, and functions (Figure 1).
Figure 1.
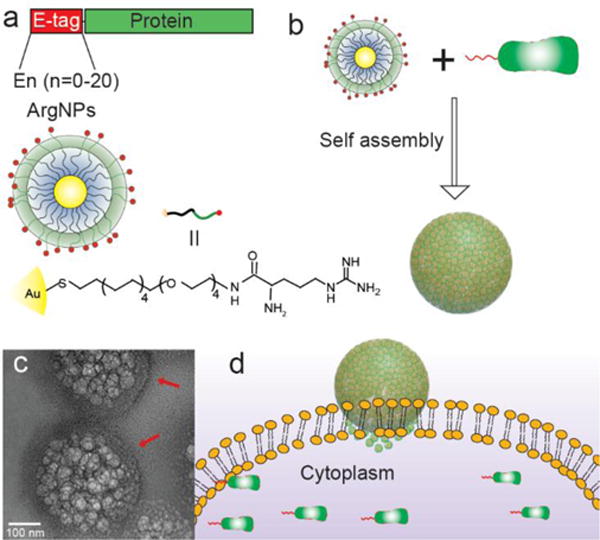
Co-engineering of E-tagged proteins and nanoparticles for direct cytoplasmic protein delivery. a) Strategy for protein engineering, and the chemical structure of arginine functionalized gold nanoparticles (ArgNPs). b) Simple mixing of E-tagged proteins and ArgNPs provides hierarchical nanoassemblies. c) Representative transmission electron micrograph (TEM) of GFP-E10:ArgNPs assemblies. Red arrow indicates nanoparticle coating on the nanoassembly surface. d) Proposed fusion-like mechanism for direct cytosolic protein delivery.
RESULTS
Engineering E-tagged green fluorescent proteins (GFP) for direct cytoplasmic delivery
We began our studies using GFP to facilitate imaging without complications arising from fluorescent tagging. We expressed a series of GFPs carrying different lengths of E-tags at the C-terminus and self-assembled them with ArgNP particles.18 The assemblies were then screened for delivery efficiency in cultured HeLa cells. After incubating GFP-En:ArgNPs assemblies with HeLa cells for 3 h, the cellular uptake of GFP-En was monitored using confocal laser scanning microscopy (CLSM) and flow cytometry. GFP delivery efficiency increased as the length of E-tag increased, with maximum delivery at GFP-E10 (Figure 2d, Figure S2), consistent with the assembly process.18 The highest GFP delivery intensity observed at a molar ratio of 1: 3 ArgNP (250 nM) to GFP-E10 (750 nM), as determined by flow cytometry analysis (Figure S3).
Figure 2.
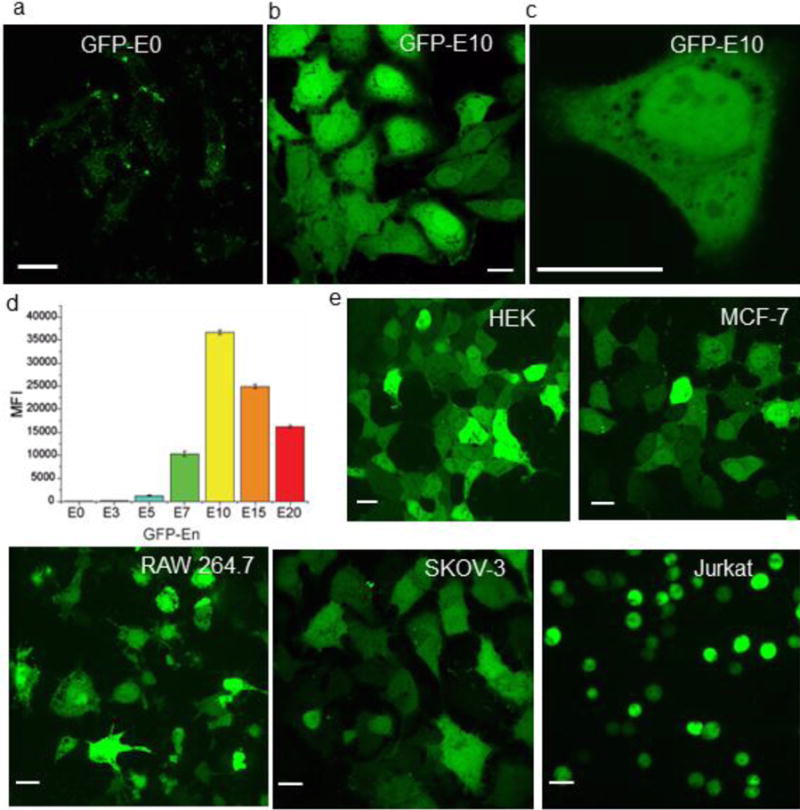
Confocal microscopy images showing nanoparticle-mediated cytoplasmic delivery of E-tagged GFP in Hela cells a) Unmodified GFP (GFP-E0) is not delivered into cell. b) GFP-E10 is delivered efficiently into the cytosol. c) Enlarged image of a cell after delivery of GFP-E10, indicating thorough distribution of delivered protein in the cytoplasm and nucleus. d) Flow cytometry data (mean fluorescence intensity, MFI) showing GFP-En delivery efficiency increases as the E-tag length increases, reaching maximum at GFP-E10. e) GFP-E10 delivery in different cell lines. Scale bar= 20 μm.
Confocal microscopy investigation revealed that the delivered GFP-E10 was evenly distributed throughout the cytoplasm (Figure 2b-c; Figure S4 for z-stacking images), without the punctate fluorescence seen with delivery vehicles with endosomal uptake (Figure S5).20,21 We also observed fluorescence in the nucleus, consistent with the ability of small proteins such as GFP to diffuse through the nuclear pore (Figure 2b-c).22 Direct cytoplasmic delivery of GFP-E10 is also evident from live cell video imaging. As shown in Figure 3b and in Supplementary movie 1, the release of GFP-E10 was observed at ~15 min post-incubation, with complete protein delivery obtained in as little as 40 min. We choose five additional mammalian cell lines to demonstrate the generality of the delivery process: human embryonic kidney cells (HEK), mammary epithelial cells (MCF-7), mouse macrophage (RAW 264.7), human ovarian cancer cells (SKOV-3), and T-lymphocyte cells (Jurkat). As shown in Figure 2e and Figure S6, efficient cytosolic delivery of nanoassembly-mediated GFP-E10 was evident in all of these cell lines, indicating the broad-spectrum capabilities of our system.
Figure 3.
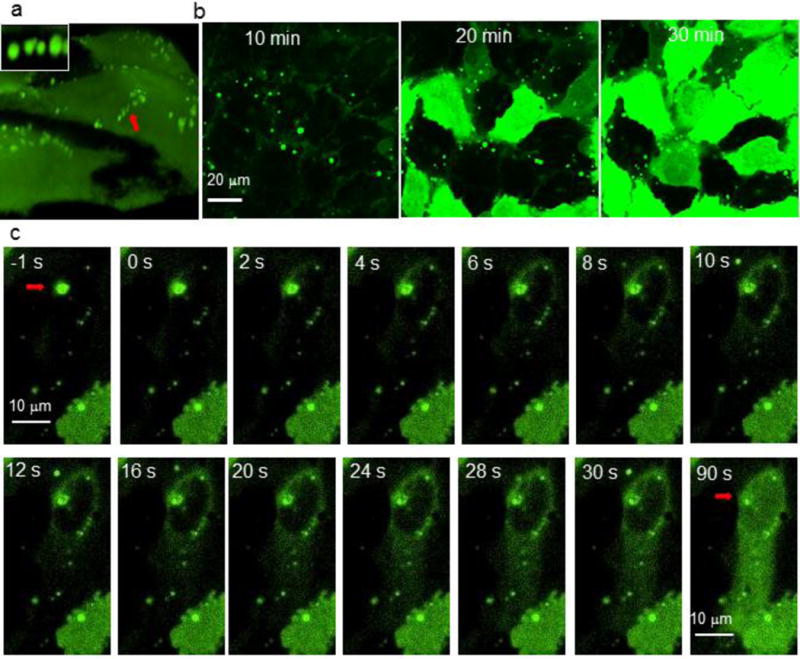
Micrographic evidence for a a fusion-like mechanism for nanoassembly-mediated direct cytosolic protein delivery. a) Confocal microscopy images showing nanoassemblies bound to the cell membrane (indicated by red arrow) in SKOV-3 cells (also see supplementary movie 3). Inset—enlarged image of nanoassemblies. 3D image was reconstructed from z-stacking images. Also, see supplementary movie 3. b) Time-lapse confocal microscopy imaging revealing the direct cytosolic delivery of GFP-E10 in HeLa cells (see supplementary movie 1). Representative still-images showing at 10, 20, and 30 min after nanoassemblies were added to the cell culture dish. c) A single nanoassembly (red arrow) was fused to the cell membrane (at -1s), which then rapidly released encapsulated GFP-E10 into the cytosol. Delivered GFP-E10 was distributed thorough the cytosol (after 30s), and the nucleus (90s) (also see supplementary movie 2).
A fusion-like process facilitates direct cytosolic protein delivery
Insight into the delivery process was obtained using time-lapse confocal microscopy. After contact of a nanoassembly with the cell membrane (Figure 3a), encapsulated GFP-E10 was quickly released into the cytosol (Figure 3c), reaching the opposite end of the cell in less than 30s and visible in the nucleus after 90s (Figure 3c, Supplementary movie 2). Significantly, free mCherry protein in the media did not enter the cell during nanoassembly mediated GFP-E10 delivery (Figure S7), indicating that delivery does not occur through a “hole punching” process.23 Mechanistic insight was obtained through cholesterol depletion assays, where complete inhibition of protein delivery (Figure S8) was observed. This shut-down suggests that the delivery process occurs through a lipid raft mediated process.24
Generality of protein delivery
We selected five different proteins with a range of charge (pI), size, and function: Prothymosin-α (PTMA) (pI= 3.71, MW= 11.8 kDa, chromatin remodeling protein);25 GFP (pI= 5.9, MW= 27 kDa, imaging protein);26 Granzyme A (GzmA) (pI= 9.14, MW= 29.0 kDa, cytolytic protein secreted by T-cells);27 Cre recombinase (Cre) (pI= 9.60, MW= 38.5 kDa, DNA recombinase);28 Histone 2A (H2A) (pI= 10.60, MW= 13.5 kDa, DNA packaging protein in the nucleosome) (Figure 4a).29
Figure 4.
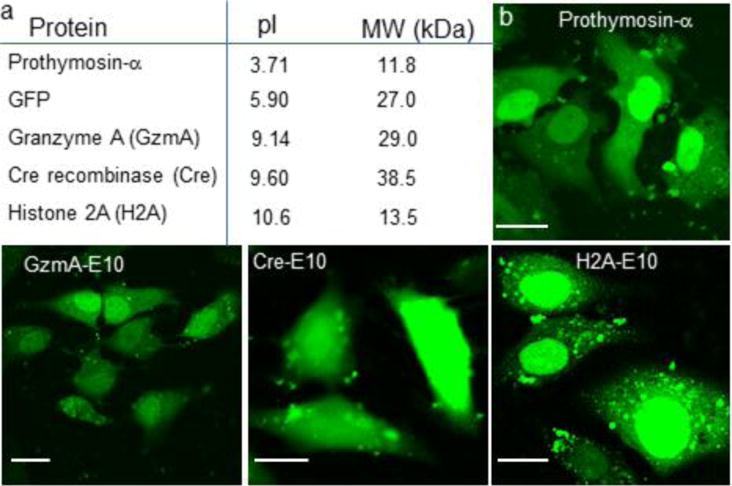
Direct cytoplasmic delivery of multiple E-tagged proteins with widely varying size and charge. a) List of proteins delivered here with their respective charge (pI) and size (MW). b) Nanoassembly-mediated delivery of these E-tagged proteins (FITC-labelled) indicated the even distribution in the cytoplasm and nucleus as shown through confocal microscopy images. Note that, except Cre recombinase, these proteins have an inherent nuclear signal, which is also reflected by their preferential accumulation in the nucleus. Scale bar= 20 μm.
We attached an E10-tag to these proteins similar to GFP, at either the N- or C-terminus (Methods). For imaging studies these proteins (except GFP) were labeled with fluorescein isothiocyanate (FITC) and assembled with ArgNPs at the appropriate molar ratios (Methods). These nanoassemblies were incubated with HeLa cells at 37°C and 5% CO2 for 3 h. As shown in Figure 4b, all E10-tagged proteins were evenly distributed in the cytosol as well as the nucleus, establishing the generality of the protocol.
Functional delivery of Cre recombinase provides efficient gene recombination
Cre recombinase delivery provides an attractive tool for cellular engineering and synthetic biology applications. After achieving successful delivery of FITC-tagged Cre-E10, we tested its activity and function in the delivered cells. Cre excises out (delete) genes flanked by a recognition sequence ‘loxP’.30 To generate a simple readout of Cre activity, we generated a plasmid (loxP-dsRedSTOP-loxP-GFP) and delivered it into human embryonic kidney cells (HEK) for transient expression of the cassette. These cells exhibit red fluorescence, but will turn green after delivery of active Cre. When Cre-E10 was delivered into these reporter cells, red-to-green fluorescence conversion was observed using confocal microscopy and flow cytometry (Figure 5). Prominent expression of GFP was observed after 48 h of delivery, indicating that Cre-E10 was delivered in active form.
Figure 5.
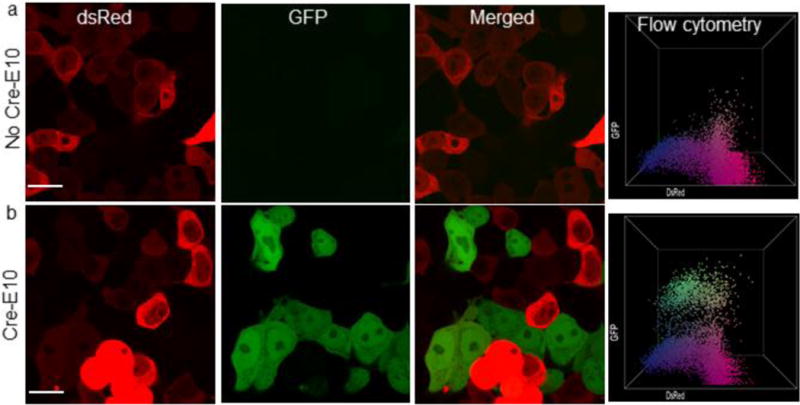
Nanoassembly mediated Cre-E10 delivery provided efficient gene recombination. Confocal micrograph and flow cytometry data before (a), and after (b) Cre-E10 delivery in HEK cells. Delivery of Cre-E10 efficiently floxed dsRed gene, thus turning on GFP expression (b). Scale bar= 20 μm.
Functional delivery of Granzyme A efficiently kills cancer cells
GzmA delivery provides a direct therapeutic application of intracellular protein delivery. Granzymes are cytolytic enzymes that are produced by cytotoxic T-cells and released into target cells to kill them,31 a process used in adoptive cancer immunotherapy.32 The killing efficiency of delivered GzmA-E10 in HeLa cells was evaluated by incubating nanoassemblies containing GzmA-E10 were incubated with HeLa cells at 37°C and 5% CO2 for 3 h followed by washing. Cell death was assessed immediately or 24 h after, using confocal microscopy, phosphatidylserine staining, Alamar blue cell viability test, and caspase 3/7 staining. As shown in Figure 6, GzmA-E10 mediated cell death was observed 24 h after the delivery. Staining of phosphatidylserine, which is expressed on granzyme-mediated dead cells, confirmed that the slow cell death was indeed caused by delivered GzmA-E10 activity (Figure 6).33 On the other hand, GFP-E10 delivery did not cause cell death, indicating the specificity of delivered GzmA-E10 (Figure 6, bottom panel). Further, delivered GzmA-E10 killed the cells through a caspase 3/7 independent pathway (Figure S9), another hallmark of granzyme-A mediated cell death.34 Cell viability test confirmed the GzmA-mediated cell death (Figure S10). Taken together, nanoassembly-mediated GzmA-E10 delivery may provide an efficient means for intracellular protein therapy for cancer treatment.
Figure 6.
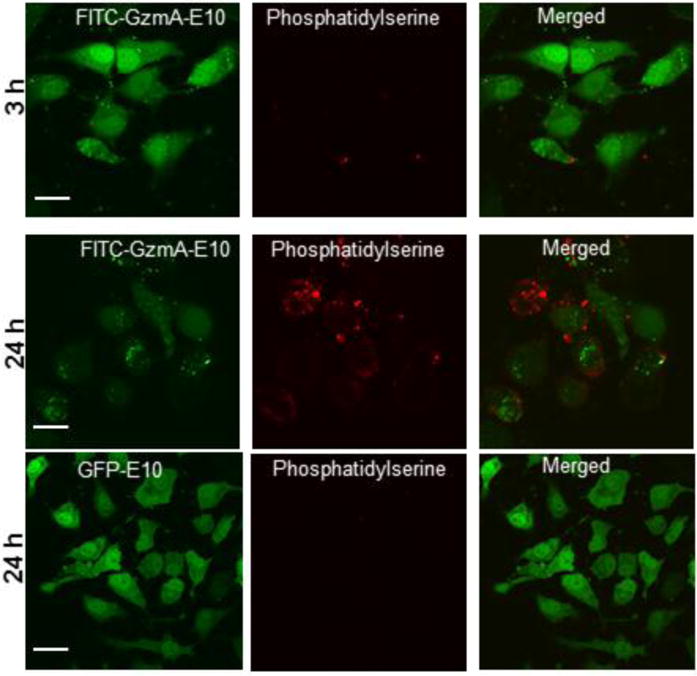
Nanoassembly-mediated delivery of functional Granzyme A-E10 (GzmA-E10) effectively killed HeLa cells. Confocal microscopy images showing FITC-GzmA-E10 or GFP-E10 delivered cells along with phosphatidylserine staining. a) 3, and 24 h after GzmA-E10 delivery. b) 24 h after GFP-E10 delivery. Note that 24 h after GzmA-E10 delivery cells died, which also showed phosphatidylserine staining, confirming the GzmA-E10 mediated cell death. Scale bar= 20 μm.
CONCLUSIONS
In summary, we present here a general method for direct cytoplasmic protein delivery through co-engineering of proteins (E-tagged) and functionalized nanoparticles. The versatility of our method was established by delivering five proteins with diverse sizes, charges, and functions, as well as through delivery to multiple cell types. This system immediately provides a useful tool for cell biology applications in vitro, and offer ways for imaging intracellular protein trafficking and dynamics. In longer-term, the protein-particle coengineering strategy presented here offers a direction for the creation of enzyme/protein replacement therapeutics.
METHODS
Engineering E-tagged protein
A series of glutamic acid tags (E-tag) with different length was inserted to the C-terminus of GFP according to our previous method.18 Similarly, a E10 tag was inserted to Cre recombinase (N-term) (Addgene plasmid id= 36915),35 Granzyme A (N-term) (Addgene plasmid id= 8823),36 Histone 2A (C-term) (Addgene plasmid id= 36207),37 through restriction cloning.
Recombinant proteins were expressed in E. coli BL21 Rosetta strain using standard protein expression protocol. Briefly, protein expression was carried out in 2xYT media with an induction condition of 1 mM IPTG and 18/25 °C for 16 hours. At this point, the cells were harvested and the pellets were lysed by using 1% Triton-X-100 (30 min, 37 °C) /DNase-I treatment (10 min). Proteins were purified using HisPur cobalt columns. Note that except GFP-En, proteins were eluted using high salt concentration buffer (2 M NaCl, 300 mM Imidazole) due to the high positive charge of the proteins. Proteins were finally preserved in PBS buffer containing 300 mM salt, (Histone 2A, 750 mM salt). The purity of native proteins was determined using 12% SDS-PAGE gel.
Nanoparticle synthesis and characterization
Arginine-functionalized gold nanoparticles (ArgNPs) were synthesized according to our previous protocol.14 Briefly, after synthesizing the arginine-functionalized thiol ligand, ArgNPs were prepared by conventional place-exchange reaction of 2-nm sized 1-pentanethiol-protected gold nanoparticles (Au-C5) with HS-C11-TEG-NH-Arginine. The resultant ArgNPs products were dissolved in distilled water, and purified by dialysis in 5 mM phosphate buffer (PB).
Nanoassembly fabrication
Nanoassemblies of ArgNPs with various E-tagged proteins were fabricated through a simple mixing method, according to our previous method.18 ArgNPs (50 μM stock in 5 mM PB, pH 7.4) were first added to 100 μL of 1×PBS in a vial, followed by adding the E-tagged protein at appropriate molar ratio: GFP-En (1:3; 250:750 nM ArgNPs/GFP-En); Cre-E10 (2:1; 250:125 nM ArgNPs/Cre-E10); GzmA-E10 (1:2 250:500 nM ArgNPs/GzmA-E10); H2A-E10 (1:3; 250:750 nM ArgNPs/H2A-E10), Prothymosin-α (1:3; 250:750 nM ArgNPs/PTMA). The assemblies were incubated at room temperature for another 10 min. Following the incubation, DMEM (without FBS and antibiotics) was added to the assemblies to make the final volume up to 1000 μL. The nanoassemblies were then immediately added to cells grown for overnight in cell culture dish (round bottom confocal dish, 35 mm, MatTek) plates for delivery experiments.
Transmission electron microscopy (TEM)
Fabricated nanoassemblies from the above step was directly used for TEM imaging. Briefly, 10 μL of the assembly solution was drop-cast on to a TEM grid (carbon film- 400 mesh copper, electron microscopy sciences) and allowed to dry at room temperature (overnight). Samples were then imaged by using JEOL 2000FX TEM.
Cell culture
80-100k cells were grown in a 24-well plate in following media: low glucose DMEM (HeLa, HEK, RAW 264.7, and MCF-7); RPMI-1640 (Jurkat); and McCoy’s 5a media (SKOV-3). All the media contained 10% FBS and 1% antibiotics (Antibiotic-Antimycotic, Corning). Cells were grown for overnight at 37 °C under 5% CO2, then washed with 1×PBS (twice) before incubation with nanoassemblies. For confocal studies, ~240,000 cells were seeded per dish.
Delivery
Assembled E-tagged protein:ArgNPs nanoassemblies (preassembled in 100 μL PBS for 10 min, plus 400 μL media for 24 well plate or 900 μL media for confocal dish as mentioned above) were immediately transferred to confluently grown cells. Cells were then incubated with nanoassemblies at 37 °C and 5% CO2 for 3 h. Delivery efficiency was determined by Confocal microscopy or flow cytometry when necessary.
Flow cytometry
Flow cytometry experiments were performed using LSR-II flow system. Briefly, after the completion of nanoassembly-mediated protein delivery or Cre recombinase activity (mentioned in the delivery and Cre activity assay section), cells were briefly washed, then trypsinized. Cells were then collected in 1% BSA solution (~100,000 cells). Flow cytometry experiments were performed immediately after that.
Confocal microscopy and Time-lapse imaging
Confocal microscopy imaging was performed using either Zeiss 510 Meta laser scanning microscope or Nikon spinning disc microscope. Time-lapse real-time video imaging was performed in the Nikon spinning disc microscope.
Cre-lox system generation
Briefly, the plasmid was constructed by placing a dsRed-STOP fluorescent protein sequence between loxP recognition sites, and a GFP sequences downstream of it [loxP-dsRed(STOP)-loxP-GFP]. Active Cre can excise out the loxP genes which will turn the color to GFP.
Assessing Cre recombinase activity
Human embryonic kidney (HEK) cells were transiently transfected with Retroviral loxP-DsRed-STOP-loxP-GFP system to create a testbed for Cre recombinase activity. 48 h after the transfection, Cre-E10 was delivered into these cells using nanoassemblies. Cells were left to grow for another 72 h, then used confocal microscopy or flow cytometry to assess the Cre activity by checking GFP positive cells.
Assessing GzmA activity
After GzmA-E10 delivery, cells were incubated for another 3, 12, 24, or 48 h. Phosphatidylserine (PS) staining was performed in these cells according to standard manufacturer’s protocol. Further, caspase 3/7 activity assay was performed to assess GzmA-E10 activity in the delivered cells according to manufacturer’s protocol. Additionally, Alamar blue cell viability test was performed using standard protocol. Briefly, Alamar blue test was carried out after treating the cells with appropriate nanoassemblies for 3 h, and then washed and left the cells to grow in DMEM media (10% FBS, but without antibiotics) for another 21 h. At this point cells were washed carefully, and treated with Alamar blue.
Cholesterol depletion
To investigate the delivery mechanism, endocytic and membrane fusion inhibitors were used. Cells were pretreated with endocytic inhibitor [wortmannin (100ng/mL), chlorpromazine (1μg/mL)], and membrane fusion inhibitor [methyl-β-cyclodextrin (MBCD, 5mg/mL)] for 1 h, as reported previously.38 At the same time nanoassemblies were prepared and kept ready. Inhibitor-treated cells were washed with 1×PBS twice, then the nanoassembly solutions were immediately applied to the cells for protein delivery. Confocal microscopy or flow cytometry experiments were performed after 3 h of nanoassembly incubation.
Supplementary Material
Acknowledgments
This research was supported by the NIH (GM077173 and EB022641) and the NSF (CHE-1506725). We thank James Chambers, the director of the light microscopy facility (LMF) at the Institute for Applied Life Sciences, UMass Amherst for his assistance in confocal microscopy measurements.
Footnotes
Author contributions
R.M. conceived the idea and designed the experiments; G.Y.T. synthesized the particle; R.M. performed all the experiments and data analysis with help from M.R., T. T. and K. S. on protein expression and purification; V.M.R. supervised the project; R.M. and V.M.R. wrote the manuscript.
Notes
The authors declare the following competing financial interest(s): V.M.R and R.M. submitted a non-provisional patent to USPTO (Application number PCT/US2016/015711) on the invention.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at http://pubs.acs.org.)
Figure S1-10, and description of supplementary movies (PDF)
Supplementary movie 1 (AVI)
Supplementary movie 2 (AVI)
Supplementary movie 3 (AVI)
References
- 1.Mitragotri S, Burke PA, Langer R. Overcoming the Challenges in Administering Biopharmaceuticals: Formulation and Delivery Strategies. Nat Rev Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu Z, Biswas A, Zhao M, Tang Y. Tailoring Nanocarriers for Intracellular Protein Delivery. Chem Soc Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 3.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mout R, Ray M, Lee YW, Scaletti F, Rotello VM. In Vivo Delivery of CRISPR/Cas9 for Therapeutic Gene Editing: Progress and Challenges. Bioconjug Chem. 2017;28:880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil AS, Collins JJ. Synthetic Biology: Applications Come of Age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leader B, Baca QJ, Golan DE. Protein Therapeutics: A Summary and Pharmacological Classification. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 7.Walsh G. Biopharmaceutical Benchmarks 2014. Nat Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 8.Stewart MP, Sharei A, Ding X, Sahay G, Langer R, Jensen KF. In Vitro and Ex Vivo Strategies for Intracellular Delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 9.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, Manygoats K, Seifert S, Andree C, Stöter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-Based Analysis of Lipid Nanoparticle-Mediated siRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 10.Fu A, Tang R, Hardie J, Farkas ME, Rotello VM. Promises and Pitfalls of Intracellular Delivery of Proteins. Bioconjug Chem. 2014;25:1602–1608. doi: 10.1021/bc500320j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erazo-Oliveras A, Najjar K, Dayani L, Wang TY, Johnson GA, Pellois JP. Protein Delivery into Live Cells by Incubation with an Endosomolytic Agent. Nat Methods. 2014;11:861–867. doi: 10.1038/nmeth.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Astolfo DS, Pagliero RJ, Pras A, Karthaus WR, Clevers H, Prasad V, Lebbink RJ, Rehmann H, Geijsen N. Efficient Intracellular Delivery of Native Proteins. Cell. 2015;161:674–690. doi: 10.1016/j.cell.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Liu Z, Jo MC, Zhang K, Li Y, Zeng Z, Li N, Zu Y, Qin L. CRISPR-Cas9 Delivery to Hard-to-Transfect Cells via Membrane Deformation. Sci Adv. 2015;1:e1500454. doi: 10.1126/sciadv.1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XC, Samanta B, Agasti SS, Jeong Y, Zhu ZJ, Rana S, Miranda OR, Rotello VM. Drug Delivery Using Nanoparticle-Stabilized Nanocapsules. Angew Chem Int Ed. 2011;50:477–481. doi: 10.1002/anie.201005662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang R, Kim CS, Solfiell DJ, Rana S, Mout R, Velázquez-Delgado EM, Chompoosor A, Jeong Y, Yan B, Zhu ZJ, Kim C, Hardy JA, Rotello VM. Direct Delivery of Functional Proteins and Enzymes to the Cytosol using Nanoparticle-Stabilized Nanocapsules. ACS Nano. 2013;7:6667–6673. doi: 10.1021/nn402753y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ray M, Tang R, Jiang Z, Rotello VM. Quantitative Tracking of Protein Trafficking to the Nucleus using Cytosolic Protein Delivery by Nanoparticle-Stabilized Nanocapsules. Bioconjug Chem. 2015;26:1004–1007. doi: 10.1021/acs.bioconjchem.5b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang R, Jiang Z, Ray M, Hou S, Rotello VM. Cytosolic Delivery of Large Proteins using Nanoparticle-Stabilized Nanocapsules. Nanoscale. 2016;8:18038–18041. doi: 10.1039/c6nr07162g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mout R, Tonga GY, Wang L-S, Ray M, Roy T, Rotello VM. Programmed Self-Assembly of Hierarchical Nanostructures through Protein-Nanoparticle Co-Engineering. ACS Nano. 2017;11:3456–3462. doi: 10.1021/acsnano.6b07258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mout R, Ray M, Tonga GY, Lee Y-W, Tay T, Sasaki K, Rotello VM. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. Potent Delivery of Functional Proteins into Mammalian Cells In Vitro and In Vivo using a Supercharged Protein. ACS Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Toro DC, Ryu JH, Chacko RT, Zhuang J, Thayumanavan S. Concurrent Binding and Delivery of Proteins and Lipophilic Small Molecules Using Polymeric Nanogels. J Am Chem Soc. 2012;134:6964–6967. doi: 10.1021/ja3019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dingwall C, Laskey RA. Protein Import into the Cell Nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- 23.Jamur MC, Oliver C. Permeabilization of Cell Membranes. Methods Mol Biol. 2010;588:63–66. doi: 10.1007/978-1-59745-324-0_9. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y, Tang R, Duncan B, Jiang Z, Yan B, Mout R, Rotello VM. Direct Cytosolic Delivery of siRNA Using Nanoparticle-Stabilized Nanocapsules. Angew Chem Int Ed Engl. 2015;54:506–510. doi: 10.1002/anie.201409161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Marquez J, Rodríguez P. Prothymosin Alpha is a Chromatin-Remodelling Protein in Mammalian Cells. Biochem J. 1998;333:1–3. doi: 10.1042/bj3330001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsien RY. The Green Fluorescent Protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman J. The ABCs of Granule-Mediated Cytotoxicity: New Weapons in the Arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 28.Nagy A. Cre Recombinase: The Universal Reagent for Genome Tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 29.Bönisch C, Hake SB. Histone H2A Variants in Nucleosomes and Chromatin: More or Less Stable. Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH, Clevers H. Controlled Gene Expression in Primary Lgr5 Organoid Cultures. Nat Methods. 2011;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 31.Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A Cleaves a Mitochondrial Complex I Protein to Initiate Caspase-Independent Cell Death. Cell. 2008;133:681–692. doi: 10.1016/j.cell.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Restifo NP, Dudley ME, Rosenberg SA. Adoptive Immunotherapy for Cancer: Harnessing the T Cell Response. Nat Rev Immunol. 2012;12:269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman J. Granzyme A Activates Another Way to Die. Immunol Rev. 2010;235:93–104. doi: 10.1111/j.0105-2896.2010.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chowdhury D, Lieberman J. Death by a Thousand Cuts: Granzyme Pathways of Programmed Cell Death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakota L, Brandt R, Heinisch JJ. Triple Mammalian/Yeast/Bacterial Shuttle Vectors for Single and Combined Lentivirus- and Sindbis Virus-Mediated Infections of Neurons. Mol Genet Genomics. 2012;287:313–324. doi: 10.1007/s00438-012-0680-1. [DOI] [PubMed] [Google Scholar]
- 36.Beresford PJ, Kam CM, Powers JC, Lieberman J. Recombinant Human Granzyme A Binds to two Putative HLA-Associated Proteins and Cleaves one of Them. Proc Natl Acad Sci U S A. 1997;94:9285–9290. doi: 10.1073/pnas.94.17.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW, Jr, Royant A. Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93% Nat Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha K, Kim ST, Yan B, Miranda OR, Alfonso FS, Shlosman D, Rotello VM. Surface Functionality of Nanoparticles Determines Cellular Uptake Mechanisms in Mammalian Cells. Small. 2013;9:300–305. doi: 10.1002/smll.201201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


