Abstract
Background
Obesity is prevalent in PR and has been associated with prostate cancer (PCa) mortality and aggressiveness. Polymorphisms (SNPs) rs9930506 and rs9939609 in the FTO gene have been associated with both obesity and PCa. The aim of this work was to ascertain whether the presence of these SNPs is associated with PCa risk and severity in a cohort of Puerto Rican men.
Methods and findings
The study population consisted of 513 Puerto Rican men age ranging from 40–79 years old who underwent radical prostatectomy (RP) as the first treatment for PCa and 128 healthy Puerto Rican men age ranging from 40–79 years old. Genomic DNA (gDNA) was extracted and SNPs were determined by Real-Time PCR. PCa severity was defined based on RP stage and Gleason Score. The relationship of FTO SNPs with demographic, clinical characteristics, PCa status and PCa severity were assessed. Logistic regression models with a 95% confidence interval (CI) determined SNPs interaction with PCa risk and severity odds ratio (ORs).
Results and discussion
BMI, age and PSA were considered as confounders. Hardy-Weinberg equilibrium was present for both SNPs. The heterozygous forms (A/G; T/A) were the most prevalent genotypes and the frequency of alleles and genotypes for both SNPs agreed with those published in 1000 genomes. Results suggest an inverse association between the mutated rs9939609 and the risk of having PCa (OR: 0.53, 95% CI: 0.31–0.92) and a positive association with overweight (OR: 1.05, 95% CI: 0.68–1.62). Importantly, among the cases that were overweight, those with mutated rs9939609 had a greater chance of high severity PCa (OR: 1.39, 95% CI: 0.84–2.32) although these results were not statistical significant upon adjustment. Limitations of the study were the relatively small cohort and lack of access to the weight history of all our subjects.
Conclusion
Results offer a research line to be followed with an expanded number of subjects that may provide a better statistical significance, to unravel the high mortality rate in this population.
Keywords: Obesity, Prostate cancer, Polymorphisms, SNPs
Introduction
Cancer is the leading cause of death in Puerto Rico (PR), and prostate cancer (PCa) was the most common cancer (17.6%) in PR from 2008 to 2012 according to reports from the Puerto Rico Cancer Registry [1]. The age-adjusted incidence for this period was 141.1 cases per 100,000 men, and the age-adjusted mortality rate was 25.6 per 100,000 men [1]. This mortality rate almost doubles the rate for Hispanics in the US as reported by SEER for the same period [2]. In PR, there are a total of 21 municipalities out of 78 totals in the island, with an age-adjusted death rate of 31.2 or higher [1].
Increased mortality and aggressiveness of PCa have been associated with obesity [3–8]. In 2007, the SNP rs9930506 in the fat mass and obesity-associated (FTO) gene region was associated with BMI and risk of obesity [9]. Located at the 16q12.2, the FTO gene (HGNC: 24678) codes for a 2-oxoglutarate dependent nucleic acid demethylase implicated in DNA repair, post-translational modification and gene modification [10–14]. Genome-wide association studies (GWAS) have associated FTO with several systemic conditions, lipids’ metabolism [9,12–17], body mass index (BMI) [18–22] and particularly, with obesity [12,19,20]. Some meta-analysis studies have found a relationship among FTO and specific cancer types, including PCa [16,21–23]. All these findings are related to a cluster of single-nucleotide SNPs (SNPs) in the FTO gene first intron which is inherited in linkage disequilibrium (LD) [9,15,18,24,25], and includes among others the rs9930506 and rs9939609 SNPs as reported by Loos et al. [26]. In this GWAS study done by Loos’ group with an admixed American population which included participants from Colombia, Mexico and PR, they found that the above mentioned FTO SNPs among others, are related in a statistically significant manner to an increased BMI [26].
In FTO rs9930506, the single nucleotide transition of an adenine nucleotide to a guanine nucleotide has been reported to be significantly associated to BMI, particularly when the rare “G” allele is present in the homozygous form [25,27]. In addition, in FTO rs9939609, the single nucleotide transversion of a thymine nucleotide to an adenine nucleotide has been associated with both BMI and type-2 diabetes, in either the heterozygous or the homozygous forms of the mutated “A” allele [25,28].
Studies made in the Puerto Rican population have found that unhealthy nutritional habits and lifestyle are largely responsible for the 69.9% of overweight and obesity in the population [29–32]. Previous work in our laboratory established that in Puerto Rican overweight men with PCa there was an increased risk for more severe disease and a higher prevalence (2.9 fold) of metastatic disease in those individuals with higher BMI [33]. In addition, in another study, we also found a positive association between PCa severity in Puerto Rican patients and high triglycerides levels [34].
The ancestry profiles for PR show a distribution where the European ancestry (EA) is the more prevalent (64%) followed by West African ancestry (WAA) (21%) and native ancestry (NA) (15%) [35–38]. FTO mutations have been associated with PCa in populations with high EA [39,40]. Considering this, we enquired if FTO SNPs (rs9930506 and rs9939609) could have a similar relationship in Puerto Ricans and could be influencing the PCa phenotype in this population. Here we report the results of an exploratory pilot study that examines the possible relationship between FTO rs9930506 and rs9939609 with PCa risk and PCa severity in this understudied population.
Materials and Methods
Ethics approval
The Institutional Review Board (IRB) of the University of Puerto Rico Medical Sciences Campus approved our study (IRB approved protocol #8860212).
Study population
Recruitment of cases and controls was done at five geographical regions in the island: metro, north, south, east and west. These were defined according to Via et al. [38], with some modifications. For specific towns in each region please see supplementary data: Supplementary Table I.
Our cohort included 641 subjects (513 cases and 128 controls). Figure 1 shows the enrollment process. Cases group was a convenience sample of available medical records of previously consented at the time of diagnosis PCa patients. We reviewed a total of 665 records of Hispanic Puerto Rican (living in PR with parents and grandparents born in PR) PCa patients, aged 40–79 years old, diagnosed with PCa between 2005 and 2012, which underwent radical prostatectomy (RP) as the initial treatment for PCa, and with available RP paraffin embedded tissue (FFPE). Exclusion criteria included history of any previous cancer, another therapy scheme received prior to RP, viral co-morbidities such as HIV or HCV, missed demographic data such as anthropometric measures, or impossibility to access FFPE blocks. Five hundred and thirteen (513) individuals complied with the inclusion criteria.
Figure 1.
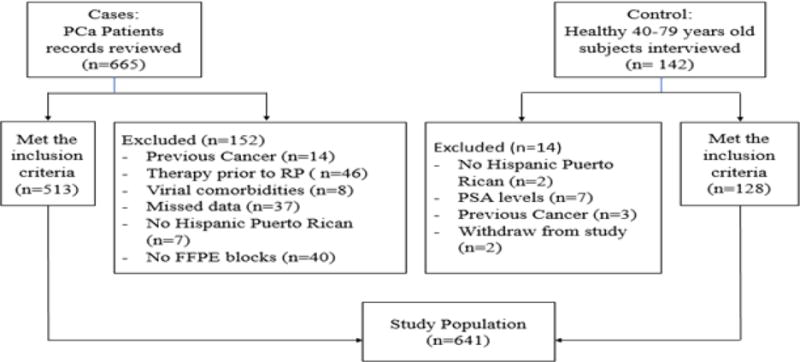
Study population enrollment process: 641 individuals were included in the study after applying the inclusion criteria.
For the control group, we interviewed 142 previously consented healthy Hispanic Puerto Rican men, as defined above, without prior history of viral comorbidities (HIV, HCV) or cancer and that were willing to donate blood. Additional controls’ inclusion criteria were PSA level (≤ 2.5 ng/mL) and normal digital rectal examination (DRE) at the time of the recruitment. These potential participants were recruited through community organizations, referrals from clinical laboratories and volunteers at the University of Puerto Rico Medical Sciences Campus and general hospitals. Twelve (12) subjects were excluded since they did not meet the inclusion criteria and two withdrew from the study. One hundred twenty-eight (128) individuals were recruited as controls.
Definitions
BMI was calculated according to the World Health Organization and the Centers for Disease Control formula (weight [kg] / height2 [m2]) [17] and dichotomized in two categories established for statistical purposes: normal/underweight (<25 kg/m2) and overweight/obese (≥ 25 kg/m2).
Severity was defined using both the prostatectomy’s Gleason Score and the American Joint Committee on Cancer (AJCC) PCa Pathologic Staging (PS) [41]. Low severity was defined as GS ≤ 7 (3 + 4) and PS ≤ pT2c. High severity was defined as GS ≥ 7 (4 + 3) regardless of PS, or GS ≤ 7 (3 + 4) with PS ≥ pT3a.
Data collection procedures
Cases
Clinical and demographic information were collected from medical records at the University District Hospital and at the Urology and Oncology Institute, in San Juan, Puerto Rico. Collected information consisted of: age at the time of PCa diagnosis, height and weight at diagnosis for BMI calculation, pre-RP PSA and prostatectomy’s GS and PS from pathology report.
Controls
once the consent process was completed, clinical and demographic information was collected via an interview prior to blood draw. The data accrued included age, weight, height, PSA value and DRE within the last six months and current physical address.
DNA extraction
Genomic DNA (gDNA) was extracted from 513 FFPE non-tumor seminal vesicles from RP pathology specimens. The procedure was done following Weiss et al. [42], with some modifications. Briefly, cylinders of FFPE tissue were cut using a 3.5 mm puncher. Paraffin was removed using octane and methanol (10:1). Subsequently, DNA was extracted with QIAGEN Gentra® Puregene® Tissue Kit (QIAGEN Inc, Valencia CA, USA). Tissue was incubated for four days at 54°C in 550 μL of Cell Lysis Solution (QIAGEN Gentra® Puregene® Tissue Kit), to which 10 μL of recombinant PCR grade Proteinase K (199 μg/μL) (Roche Applied Science, Indianapolis IN, USA) were added daily. After incubation, samples were cooled to room temperature, heated at 95°C and then cooled in ice to remove proteins via cold precipitation using 200 μL of Protein Precipitation Solution (QIAGEN Gentra® Puregene® Tissue Kit).
gDNA was also extracted from whole blood (10 mL) from healthy Puerto Rican men (n=128) collected in EDTA tubes. DNA was extracted using Gentra® Puregene® Blood Kit (QIAGEN Inc, Valencia CA, USA) following manufacturer’s guidelines. White blood cells (WBC) pellets were obtained through selective red blood cells lysis using RBC Lysis Solution (QIAGEN Gentra® Puregene® Blood Kit). WBC pellets were lysed and RNA was removed with a mix of Cell Lysis Solution (QIAGEN Gentra® Puregene® Blood Kit) and RNAse A Solution (QIAGEN Inc, Valencia CA, USA). Subsequently, proteins were removed by cold precipitation using Protein Precipitation Solution (1 mL) as explained above.
gDNA was recovered from supernatants by precipitation with isopropanol and glycogen (Sigma-PCR grade from Mussels, Sigma-Aldrich, St. Louis MO, USA). After washing with 70% ethanol in nuclease-free water (QIAGEN Inc, Valencia CA, USA), DNA was dissolved in DNA Hydration Solution (QIAGEN Gentra® Puregene® Tissue Kit). Concentrations and purity were assessed by means of a Thermo Fisher NanoDrop 2000. Absorbance 260/280 ratio of 1.8 – 2.0 was considered as acceptable DNA purity. DNA was diluted to 20 ng/μL in nuclease-free water (QIAGEN Inc., Valencia CA, USA). DNA quality was evaluated by PCR amplification of the G3PDH housekeeping gene and subsequent electrophoresis on 1.5% agarose gels. Samples that failed QC were re-purified according to Gentra® Puregene® Kits manufacturer’s guidelines or new aliquots were made.
Allelic discrimination
An Applied Biosystems Real-Time PCR StepOne™ 2011 system was used for quantitative detection of rs9930506 (A/G) and rs9939609 (T/A) SNPs. Genotyping assays were performed following the manufacturer’s specifications for the Applied Biosystems TaqMan® Universal Master Mix II NO UNG and custom Applied Biosystems TaqMan® SNP Human Small-Scale (40X Concentration) with fluorescent-based (VIC®and FAM™) probes. The probe linked to VIC® dye detected rs9930506 wild type “A” allele and the rs9939609 wild type “T” allele was detected by the probe linked to FAM™ dye. The final reaction volume was set to 12.5 μL.
Basic QC procedures for SNPs genotyping were performed. Following the manufacturer’s recommendations, nuclease-free water was used as negative control in triplicate in each run (No Template Control=NTC). As an inside laboratory QC procedure and since there are no commercial controls available, we randomly selected 3 samples per SNP and treated them as blind samples running them twice. In addition, in the same run we included triplicates of samples with known results as positive controls. The concordance was of the 99%. Some runs results were also analyzed with the “TaqMan® Genotyper Software”. This Life Technologies software contains the call for improved automated genotyping and includes quality control tools for troubleshooting of experiments algorithms, with 100% match. Six (6) samples for rs9930506 and seven (7) samples for rs9939609 failed genotyping. These samples were repeated after double-checking for aliquots’ purity, concentration and integrity.
Hardy-Weinberg equilibrium and haplotype-based association test
We performed alleles’ frequency calculation using the Hardy-Weinberg equation [43]. Genotypes and allele’s frequencies were compared to the PR population genetics composition reported by 1000 Genomes [44]. Hardy-Weinberg Equilibrium (HWE) was tested using the Chi Square Goodness of fit test with one degree of freedom for each SNP (rs9930506 and rs9939609) for both cases and controls. The analysis was performed using the CRAN “The Hardy Weinberg Package” for R software environment for statistical computing and graphics, following procedures reported by Graffelman [45]. Haplotype-based association test including frequencies and D′ [46] was performed using the 3BY3 program included in the Statistical Genetics Utility Programs (UtilWin) by Jurg Ott.
Statistical Analysis
Univariate analysis was performed to characterize the sample (n=641), according to demographic and clinical characteristics. Fisher exact test of independence was used to assess the relationship between geographical regions of PR and age, BMI, PSA and SNPs. Contingency tables were generated to assess the relationship of demographic and clinical characteristics with PCa, using Chi-square or Fisher’s exact test for categorical variables. To test the difference in proportion we used the Two-proportion Z-test. Continuous variables were analyzed using t-test or Wilcoxon Mann-Whitney test if continuous variables did not have a normal distribution. Normality of the continuous variables by groups (cases vs controls) was assessed by means of the Shapiro-Wilk test for normality [47]. Logistic regression models were conducted to estimate the odds ratio (ORs) with 95% confidence interval (CI) of the SNPs rs9930506 and rs9939609, both of which have a three-level categorical variable (homozygous wild type, heterozygous and homozygous mutated), in relation with PCa in general, PCa low severity and PCa high severity, respectively. We first estimated the ORs of rs9930506 genotypes A/A vs. A/G and A/A vs G/G or rs9939609 T/T vs. T/A and T/T vs. A/A for the presence or absence of PCa. We then estimated the ORs of rs9930506 genotypes A/A vs. A/G and A/A vs. G/G or rs9939609 T/T vs. T/A and T/T vs A/A in relation to PCa severity (low or high). All models were adjusted for BMI, age, and PSA and were tested to evaluate first level of interaction using the Likelihood Ratio Test (LRT). Variables statistically associated with PCa severity (p<0.05) in the crude and adjusted logistic regression models were included in the multivariate logistic regression models. All analyses were performed using Stata for Mac release 13.0 (Stata Corporation, College Station, Texas).
Results
Cases and controls were distributed among the five geographical areas previously described. The enrollment process did not target any specific area.
Table 1 shows the distribution of cases and controls in each region. The higher percentage of both, cases and controls came from the metro and north areas. The lower percentage came from the West.
Table 1.
Study population distribution across Puerto Rico by participant status.
| Region | Control (n=128) n (%) | Case (n=513) n (%) |
|---|---|---|
| Metro | 49 (38.28) | 227 (44.25) |
| North | 25 (19.53) | 120 (23.39) |
| East | 48 (37.50) | 98 (19.10) |
| South | 6 (4.69) | 39 (7.60) |
| West | 0 (0.00) | 29 (5.65) |
We found that there was a statistically significant association between the geographical regions of PR and age among cases (p<0.001), but not among controls (p=0.46). In addition, no statistically significant association was found between the geographical regions of PR and BMI among cases (p=0.89) or controls (p=0.90). Moreover, there was no association between geographical regions and neither of the SNPs (rs9930506 among cases (p=0.85) and controls (p=0.94); rs9939609 among cases (p=0.92) and controls (p=0.42)).
Table 2 shows the demographic features and FTO genotype of our cohort. Overall, 63% of individuals were younger than 60 years, 82 % of the subjects were classified as overweight/obese, and 45% of the subjects had levels of PSA ≥ 4 ng/mL.
Table 2.
Demographic characteristics and FTO genotype of study population (n=641).
| Characteristic | n (%) |
|---|---|
| Age (years) | |
| <60 | 404 (63.03) |
| ≥ 60 | 237 (36.97) |
| Mean ± SD | 56.87 ± 7.70 |
| BMI (kg/m2) | |
| <25 | 116 (18.10) |
| ≥ 25 | 525 (81.90) |
| Mean ± SD | 28.65 ± 4.72 |
| PSA (ng/mL) | |
| <4.0 | 350 (54.60) |
| ≥ 4.0 | 291 (45.40) |
| Mean ± SD | 5.22 ± 5.43 |
| rs9930506 | |
| A/A | 266 (41.50) |
| A/G | 311 (48.52) |
| G/G | 64 (9.98) |
| rs9939609 | |
| T/T | 237 (36.97) |
| T/A | 309 (48.21) |
| A/A | 95 (14.82) |
Frequency of the heterozygous and the wild type genotypes as well as the allele frequencies concurred with what was reported by 1000 Genomes (99 ± 0.08%) for the Puerto Rican population. For rs9930506 the allele frequencies were A: 0.66 and G: 0.34, while for rs9939609 the allele frequencies were T: 0.61 and A: 0.39. Using these frequencies, we performed a haplotype test in our cohort and found that four possible haplotypes are present in our cohort (D′ = 0.77) (A/T = 0.56; G/T = 0.05; A/A = 0.09; G/A = 0.29). Allele distributions for rs9930506 and rs9939609 followed HWE among both cases (rs9930506: ϰ2= 1.58, p-value = 0.21; rs9939609: ϰ2 = 0.04, p-value = 0.37) and controls (rs9930506: ϰ2 = 3.56, p-value = 0.38; rs9939609: ϰ2 = 2.14, p-value = 0.47).
Table 3 shows the clinical and demographic characteristics of the study sample by participant status. The cases (58.16 ± 6.60) were older than the controls (51.68 ± 9.47), in a statistically significant manner (p ≤ 0.05). Cases had significant higher proportion of participants older than 60 years compared to controls (15.62% vs. 42.30%, respectively; (p=0.02). There was a higher proportion of overweight/obese participants (BMI ≥ 25 kg/m2) among controls compared to cases (88.20% vs. 80.31%, respectively; (p=0.05)). FTO rs9930506 was not significantly associated to PCa in the Chi square test (p=0.255). In contrast FTO rs9939609 (p=0.01), age (p<0.05), BMI (p=0.04) and PSA (p<0.05) were associated to PCa.
Table 3.
Description of clinical and demographic characteristics and FTO genotype by participant status.
| Control (n=128) n (%) | Case (n=513) n (%) | p-value | |
|---|---|---|---|
| Age (mean ± SD) | <0.05a | ||
| <60 | 108 (84.38) | 296 (57.70) | |
| ≥ 60 | 20 (15.62) | 217 (42.30) | 0.02c |
| Mean ± SD | 51.68 ± 9.47 | 58.16 ± 6.60 | <0.05b |
| BMI kg/m2 | 0.04a | ||
| Underweight/normal | 15 (11.72) | 101 (19.62) | |
| Overweight/Obese | 113 (88.28) | 412 (80.31) | 0.051c |
| Mean ± SD | 30.40 ± 6.21 | 28.22 ± 4.17 | <0.05b |
| PSA (ng/ml) | <0.05a | ||
| <4.0 | 128.0 (100.00) | 163 (31.77) | |
| ≥ 4.0 | 0 (0%) | 350 (68.23) | |
| Mean ± SD | 0.78 ± 0.52 | 6.36 ± 5.56 | <0.05b |
| rs9930506 | 0.245a | ||
| A/A | 45 (35.16) | 221(43.08) | |
| A/G | 70 (54.99) | 241(46.98) | |
| G/G | 13 (10.16) | 51(9.94) | |
| rs9939609 | 0.007a | ||
| T/T | 32 (25.00) | 205 (39.96) | |
| T/A | 72 (56.25) | 237 (46.20) | |
| A/A | 24 (18.75) | 71 (13.84) |
p-values using chi-square or Fisher exact test for categorical variables
Wilcoxon Mann-Whitney test for continuous variables and
Two-proportion Z-test.
Table 4 summarizes the association between the rs9939609 and PCa risk and PCa severity. We found that having at least one “A” allele decreases by 51% the possibility of PCa risk and for low severity PCa. This significance was maintained when adjusted by age, BMI and PSA, except for high severity PCa, that has no statistical significance in the adjusted model (p=0.41).
Table 4.
Association between rs9939609 genotype and prostate cancer risk and severity.
| Variables | AT vs TT | AA vs TT | ||
|---|---|---|---|---|
| Odds ratio (95%CI) | p | Odds ratio (95%CI) | p | |
| Unadjusted | ||||
| Cases (n=513) vs. Controls (n=128) | 0.51 (0.32–0.81) | <0.01 | 0.46 (0.25–0.84) | 0.01 |
| Low severity cases (n=370) vs. controls (n=128) | 0.50 (0.31–0.81) | <0.01 | 0.46 (0.25–0.87) | 0.02 |
| High severity cases (n=143) vs. controls (n=128) | 0.54 (0.31–0.93) | 0.03 | 0.45 (0.21–0.95) | 0.04 |
| Adjusted by age, PSA, and BMI | ||||
| Cases (n=513) vs. controls (n=128) | 0.53 (0.31–0.92) | 0.03 | 0.44 (0.21–0.92) | 0.03 |
| Low severity cases (n=370) vs. controls (n=128) | 0.51 (0.29–0.90) | 0.02 | 0.42 (0.19–0.92) | 0.03 |
| High severity cases (n=143) vs. controls (n=128) | 0.65 (0.25–1.73) | 0.41 | 0.58 (0.15–2.22) | 0.43 |
OR’s adjusted by all variables in the model simultaneously. No significant first level interactions in the multivariate model using the LRT.
For all the study population, subjects with the “A” allele had a higher possibility of being overweight (OR: 1.05, 95% CI: 0.68 – 1.62). Among the cases, those with mutated FTO rs9939609 tend to have a greater chance of having high severity PCa compared with the wild type subjects (OR: 1.07, 95% CI: 0.71 – 1.62). In addition, also among the cases those that were overweight showed a higher probability of having high severity PCa (OR: 1.39, 95% CI; 0.84 – 2.32). None of the above results achieved statistical significance.
Discussion
Both rs9930506 and rs9939609 have been associated to obesity in the European population [24–27,28], and obesity has been associated to higher risk of PCa in other populations [40,47–49]. Given the high percentage of the European heritage in the Puerto Rican population, in this pilot study we hypothesized that either or both of these SNPs could influence the risk of PCa and its phenotype in a cohort of Puerto Rican men. To our knowledge, this is the first study that attempts to associate the obesity related gene FTO with the risk of developing PCa or its severity in the Puerto Rican population.
For genetic association studies conducted in admixed populations such as Puerto Rican, sample size is crucial. Fesinmeyer et al. reported in 2008 the effect of FTO SNPs rs9930506 and rs9939609 in BMI and obesity in some mixed populations, in a sample of 7,346 Hispanics with 80% power [50]. In that study, the researchers reported that there is correlation between linkage disequilibrium and FTO SNPs in Hispanics (r2: 0.63–0.91). Despite the fact that our cohort is comparatively small, we found that the LD was similar (r2=0.64) as in the Feismeyer study. In addition, frequency of both alleles and genotypes for rs9930506 and rs9939609 for the Puerto Rican population in our analysis concurs with 1000 genomes and as well the expected Hardy-Weinberg (HWE) distribution.
The associations of FTO SNPs and PCa risk and severity have been analyzed in other populations with European heritage. Li et al. [23] performed a meta-analysis that included a study by Meyer et al. [51] which found no association between FTO rs8050136 and PCa risk and also a study by Lewis et al. [40] which found an inverse association between PCa risk and FTO rs9939609 in a white European cohort [40]. In this study, we also report association between the FTO rs9939609 mutant allele “A” and PCa risk. Of note, establishing comparisons among different studies is hindered by the fact that studies have different inclusion criteria for controls and different definitions of high and low severity. Lewis’ group set PCa low severity cut off to GS ≤ 6 and ours was set as ≤ 7 (3 + 4). They used two different groups of controls in terms of PSA levels; we set up a PSA cut off of ≤ 2.5 ng/mL. Acknowledging these differences, our results concur with Lewis et al. in terms of the inverse association of the FTO rs9939609 mutant allele “A” and PCa risk.
The average age for PCa diagnosis is 66 years old [2]. In the Puerto Rican population being older than 60 years old is a risk factor for PCa as reported by Soto-Salgado et al. [52]. Besides that, there have been reports (Hussein et al) that show that PCa may appear early in life (≤ 55 years old) [53], and that this early PCa onset may be associated and promoted by several genes [54]. Furthermore, these researchers state that PCa may remain undetectable in its early stages for many years until different factors like obesity, propitiate its development to a more aggressive stage [49,53,54]. In our cohort, the mean age at PCa diagnosis was 58 years old, particularly in the overweight group, which could suggest an agreement with Hussein [53]. We cannot conclude that FTO is responsible for PCa development at early age in PR, but the poor dietary patterns in this population [29,34,55] may influence the FTO effect on obesity and hence in PCa. Studies in other populations with a high European heritage had suggested that consumption of animal protein, milk and dairy protein might increase the PCa risk by hormonal changes [17,56].
Milk and dairy products high consumption are a key feature in Puerto Rican nutritional habits [29,55]. Milk contains high concentrations of miRNA-29s rich exosomes, which attack DNA methyltransferases, reducing the methylation of FTO CpG islands, stimulating FTO expression [17]. Moreover, in animal model’s overexpression of FTO increases the expression of the RUNX1T1 transcription factor in embryonic fibroblasts stimulating the number of adipocytes [57]. Knowing that FTO expression can modulate obesity onset [19,58] due to its stronger expression during childhood, it is feasible to think that throughout adulthood the high consumption of foods that stimulate FTO overexpression would contribute to the high incidence of obesity, and hence PCa risk. Unfortunately, we did not have access to the lifetime health records of our participants, so we are unable to establish the timeline for obesity onset in our cohort.
The heterozygous form (T/A) of rs9939609 was the most prevalent in our cohort. This would suggest that rs9939609 “A” allele may be implicated in the phenotype of PCa and the male population in PR could have a more severe PCa if they have the “A” genotype, thus contributing to the high mortality rate from the disease. Our results prompt us to inquire if patients with high grade PCa at an early age and the FTO mutated rs9939609 will also have high triglycerides levels, given that in a previous work our group found that age and high triglycerides levels are related to high grade PCa [34]. This premise remains unclear in the literature. Authors like De Luis et al. have related high triglycerides levels to the FTO rs9939609 homozygous form (T/T) in Spanish obese men [59], while others like Grunnet et al. report an association to the FTO rs9939609 heterozygous form (A/T) [60]. We faced the limitation that we did not have access to the serum lipid levels of all our patients at the diagnosis.
We propose that FTO could be another gene that may be implicated in the early onset of PCa in this Hispanic population and that this gene is impacting PCa by its effect on overweight.
Conclusion
Notwithstanding our limitations, we found a definite association between the presence of rs9939609 with the risk of having PCa in this cohort of Puerto Rican men and the early onset of disease in a relatively young group of Puerto Rican men. To our better knowledge, no other study has characterized the Puerto Rican population with respect to the FTO rs9930506 or rs9939609 SNPs and their association with PCa. The alleles and genotypes frequency for both SNPs agreed with those published in 1000 genomes and these findings contribute to advance in the direction of having a genomic database of the Puerto Rican population for PCa. Further studies are needed to understand the mechanisms that may be involved in this early onset of PCa in this population. These results provide additional information to the complex nature of PCa in this population.
Supplementary Material
Acknowledgments
We want to acknowledge Lorena González, MS and Naydi Pérez, MS for all the help received in the statistical analysis of the study results. We thank the personnel at the University District Hospital, San Pablo Pathology Group and the Urology and Oncology Institute for the assistance regarding recruitment and samples. We also recognize the contribution of Luis E. Vázquez Quiñones, Ph. D. in editing this manuscript.
Funding
Support was received from Award number 8U54MD 007587-03, G12 MD007600 both from the National Institute on Minority Health and Health Disparities, and by Award Grant Number #CA096297/CA096300 from the National Cancer Institute of the National Institute of Health.
Footnotes
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Minority Health and Health Disparities or the National Institutes of Health.
References
- 1.Zavala ZD, Tortolero LG, Torres CCR, Alvarado OM, Traverso OM, et al. Cancer in Puerto Rico, 2008–2012. Puerto Rico Central Cancer Registry; San Juan, PR: 2015. pp. 125–126. [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, Miller D, Bishop K, et al. SEER Cancer Statistics Review, 1975–2013. National Cancer Institute; Bethesda, MD: 2016. [Google Scholar]
- 3.Allott EH, Masko EM, Freedland SJ. Obesity and prostate cancer: Weighing the evidence. Eur Urol. 2013;63:800–809. doi: 10.1016/j.eururo.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassett JK, Severi G, Baglietto L, MacInnis RJ, Hoang HN, et al. Weight change and prostate cancer incidence and mortality. Int J Cancer. 2012;131:1711–1719. doi: 10.1002/ijc.27414. [DOI] [PubMed] [Google Scholar]
- 5.Bhaskaran K, Douglas I, Forbes H, Dos-Santos-Silva I, Leon DA, et al. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5.24 million UK adults. The Lancet. 2014;384:755–765. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamie K, Oberfoell S, Kwan L, Labo J, Wei JT, et al. Body mass index and prostate cancer severity: Do obese men harbor more aggressive disease on prostate biopsy? Urology. 2013;81:949–955. doi: 10.1016/j.urology.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Häggström C, Stocks T, Ulmert D, Bjørge T, Ulmer H, et al. Prospective study on metabolic factors and risk of prostate cancer. Cancer. 2012;118:6199–6206. doi: 10.1002/cncr.27677. [DOI] [PubMed] [Google Scholar]
- 8.Tewari R, Rajender S, Natu SM, Dalela D, Goel A, et al. Diet, obesity, and prostate health: are we missing the link? J Androl. 2012;33:763–776. doi: 10.2164/jandrol.111.015578. [DOI] [PubMed] [Google Scholar]
- 9.Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26:266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco S, Frye M. Role of RNA methyltransferases in tissue renewal and pathology. Curr Opin Cell Biol. 2014;31:1–7. doi: 10.1016/j.ceb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gulati P, Avezov E, Ma M, Antrobus R, Lehner P, et al. Fat mass and obesity-related (FTO) shuttles between the nucleus and cytoplasm. Biosci Rep. 2014;34:e00144. doi: 10.1042/BSR20140111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulati P, Yeo GS. The biology of FTO: From nucleic acid demethylase to amino acid sensor. Diabetologia. 2013;56:2113–2121. doi: 10.1007/s00125-013-2999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klungland A, Dahl JA. Dynamic RNA modifications in disease. Curr Opin Genet Dev. 2014;26:47–52. doi: 10.1016/j.gde.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Larder R, Cheung MM, Tung YL, Yeo GS, Coll AP. Where to go with FTO? Trends in Endocrinology & Metabolism. 2011;22:53–59. doi: 10.1016/j.tem.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Hess ME, Brüning JC. The fat mass and obesity-associated (FTO) gene: Obesity and beyond? Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2014;1842:2039–2047. doi: 10.1016/j.bbadis.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Tang H, Dong X, Hassan MM, Abbruzzese JL, Li D. Body mass index and obesity-and diabetes-associated genes and risk for pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:779–792. doi: 10.1158/1055-9965.EPI-10-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnik BC. Milk: An epigenetic amplifier of FTO-mediated transcription? Implications for Western diseases. J Transl Med. 2015;13:385. doi: 10.1186/s12967-015-0746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung YL, Yeo GS, O’Rahilly S, Coll AP. Obesity and FTO: Changing focus at a complex locus. Cell Metab. 2014;20:710–718. doi: 10.1016/j.cmet.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Ursu RI, Badiu C, Cucu N, Ursu GF, Craciunescu I, et al. The study of the rs9939609 FTO gene polymorphism in association with obesity and the management of obesity in a Romanian cohort. J Med Life. 2015;8:232. [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Q, Grant SF. The genetics of human obesity. Ann NY Acad Sci. 2013;1281:178–190. doi: 10.1111/nyas.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Loos RJ, Powell JE, Medland SE, Speliotes EK, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490:267. doi: 10.1038/nature11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo GS. The role of the FTO (Fat Mass and Obesity Related) locus in regulating body size and composition. Mol Cell Endocrinol. 2014;397:34–41. doi: 10.1016/j.mce.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Li G, Chen Q, Wang L, Ke D, Yuan Z. Association between FTO gene polymorphism and cancer risk: evidence from 16,277 cases and 31,153 controls. Tumor Biol. 2012;33:1237–1243. doi: 10.1007/s13277-012-0372-9. [DOI] [PubMed] [Google Scholar]
- 24.Hinney A, Nguyen TT, Scherag A, Friedel S, Brönner G, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS one. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sentinelli F, Incani M, Coccia F, Capoccia D, Cambuli VM, et al. Association of FTO polymorphisms with early age of obesity in obese Italian subjects. Exp Diabetes Res. 2012;872176 doi: 10.1155/2012/872176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loos RJ, Yeo GS. The bigger picture of FTO [mdash] the first GWAS-identified obesity gene. Nat Rev Endocrinol. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colón-López V, Banerjee G, Gertz AM, Ortiz AP, Calo P, et al. Behavioral correlates of fruit and vegetables intake in Puerto Rico: results from the Health Information National Trends Survey. P R Health Sci J. 2013;32:194. [PMC free article] [PubMed] [Google Scholar]
- 30.Ho GY, Figueroa-Vallés NR, La Torre-Feliciano D, Tucker KL, Tortolero-Luna G, et al. Cancer disparities between mainland and island Puerto Ricans. Rev Panam Salud Publica. 2009;25:394–400. doi: 10.1590/s1020-49892009000500003. [DOI] [PubMed] [Google Scholar]
- 31.http://www.lexjuris.com/lexlex/leyes2003/lexl2003083.html
- 32.Sanchez H, Sanchez Aleman AS, Morales GJ, Rivera RJ, Torres CK, et al. Resumen General de la Salud en Puerto Rico 2004–2013. San Juan, Puerto Rico. Departamento de Salud; San Juan, Puerto Rico: 2014. p. 6. [Google Scholar]
- 33.Negrón R, Vásquez A, Nieves M, Guerrios L, Irizarry-Ramírez M. Body mass index affects the diagnosis and progression of prostate cancer in Hispanics. Ethn Dis. 2010;20(1 Suppl 1):S1-162–172. [PMC free article] [PubMed] [Google Scholar]
- 34.Salgado-Montilla J, Salgado MS, Trautmann BS, Sánchez-Ortiz R, Irizarry-Ramírez M. Association of serum lipid levels and prostate cancer severity among Hispanic Puerto Rican men. Lipids Health Dis. 2015;14:111. doi: 10.1186/s12944-015-0096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martínez-Cruzado JC, Toro-Labrador G, Viera-Vera J, Rivera-Vega MY, Startek J, et al. Reconstructing the population history of Puerto Rico by means of mtDNA phylogeographic analysis. Am J Phys Anthropol. 2005;128:131–155. doi: 10.1002/ajpa.20108. [DOI] [PubMed] [Google Scholar]
- 36.Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruaño G, Duconge J, Windemuth A, Cadilla CL, Kocherla M, et al. Physiogenomic analysis of the Puerto Rican population. Pharmacogenomics. 2009;10:565–577. doi: 10.2217/pgs.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Via M, Gignoux CR, Roth LA, Fejerman L, Galanter J, et al. History shaped the geographic distribution of genomic admixture on the island of Puerto Rico. PLoS One. 2011;6:e16513. doi: 10.1371/journal.pone.0016513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies NM, Gaunt TR, Lewis SJ, Holly J, Donovan JL, et al. The effects of height and BMI on prostate cancer incidence and mortality: a Mendelian randomization study in 20,848 cases and 20,214 controls from the PRACTICAL consortium. Cancer Causes & Control. 2015;26:1603–1616. doi: 10.1007/s10552-015-0654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis SJ, Murad A, Chen L, Smith GD, Donovan J, et al. Associations between an obesity related genetic variant (FTO rs9939609) and prostate cancer risk. PLoS One. 2010;5:e13485. doi: 10.1371/journal.pone.0013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.AJCC Cancer Staging Atlas. A companion to the seventh editions of the AJCC Cancer Staging Manual and Handbook. 2nd. Springer; Berlin: 2012. [Google Scholar]
- 42.Weiss AT, Delcour NM, Meyer A, Klopfleisch R. Efficient and cost-effective extraction of genomic DNA from formalin-fixed and paraffin-embedded tissues. Vet Pathol. 2011;48:834–838. doi: 10.1177/0300985810380399. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auton ALD, Brooks RM, Durbin EP, Garrison HM, Kang H, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graffelman J. Exploring diallelic genetic markers: the hardy weinberg package. Journal of Statistical Software. 2015;64:1–23. [Google Scholar]
- 46.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet. 2003;4:587. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 47.Razali NM, Wah YB. Power comparisons of shapiro-wilk, kolmogorov-smirnov, lilliefors and anderson-darling tests. Journal of Statistical Modeling and Analytics. 2011;2:21–33. [Google Scholar]
- 48.Chu DI, Freedland SJ. Metabolic risk factors in prostate cancer. Cancer. 2011;117:2020–2023. doi: 10.1002/cncr.25749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leitzmann MF, Rohrmann S. Risk factors for the onset of prostatic cancer: Age, location, and behavioral correlates. Clin Epidemiol. 2012;4:1. doi: 10.2147/CLEP.S16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fesinmeyer MD, North KE, Ritchie MD, Lim U, Franceschini N, et al. Genetic risk factors for BMI and obesity in an ethnically diverse population: Results from the population architecture using genomics and epidemiology (PAGE) study. Obesity. 2013;21:835–846. doi: 10.1002/oby.20268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer TE, Boerwinkle E, Morrison AC, Volcik KA, Sanderson M, et al. Diabetes genes and prostate cancer in the Atherosclerosis Risk in Communities study. Cancer Epidemiol Biomarkers Prev. 2010;19:558–565. doi: 10.1158/1055-9965.EPI-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soto-Salgado M, Suárez E, Torres-Cintrón M, Pettaway CA, Colón V, et al. Prostate cancer incidence and mortality among Puerto Ricans: an updated analysis comparing men in Puerto Rico with US racial/ethnic groups. PR Health Sci J. 2012;31:107–113. [PubMed] [Google Scholar]
- 53.Hussein S, Satturwar S, Van der Kwast T. Young-age prostate cancer. J Clin Pathol. 2015;68:511–515. doi: 10.1136/jclinpath-2015-202993. [DOI] [PubMed] [Google Scholar]
- 54.Lange EM, Johnson AM, Wang Y, Zuhlke KA, Lu Y, et al. Genome-wide association scan for variants associated with early-onset prostate cancer. PloS one. 2014;9:e93436. doi: 10.1371/journal.pone.0093436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soltero SM, Palacios C. Association between dietary patterns and body composition in a group or Puerto Rican obese adults: a pilot study. P R Health Sci J. 2011;30:22–27. [PMC free article] [PubMed] [Google Scholar]
- 56.Allen NE, Key TJ, Appleby PN, Travis RC, Roddam AW, et al. Animal foods, protein, calcium and prostate cancer risk: the European Prospective Investigation into Cancer and Nutrition. Br J Cancer. 2008;98:1574–1581. doi: 10.1038/sj.bjc.6604331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, et al. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun. 2015;6:6792. doi: 10.1038/ncomms7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebert S, Salonurmi T, Keinänen-Kiukaanniemi S, Savolainen M, Herzig KH, et al. Programming effects of FTO in the development of obesity. Acta Physiol. 2014;210:58–69. doi: 10.1111/apha.12196. [DOI] [PubMed] [Google Scholar]
- 59.De Luis DA, Aller R, Izaola O, Primo D, Romero E. Association of the rs9939609 gene variant in FTO with insulin resistance, cardiovascular risk factor and serum adipokine levels in obese patients. Nutr Hosp. 2016;33:573. doi: 10.20960/nh.573. [DOI] [PubMed] [Google Scholar]
- 60.Grunnet LG, Nilsson E, Ling C, Hansen T, Pedersen O, et al. Regulation and function of FTO mRNA expression in human skeletal muscle and subcutaneous adipose tissue. Diabetes. 2009;58:2402–2408. doi: 10.2337/db09-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


