Summary
MicroRNA biogenesis is known to be modulated by a variety of RNA binding proteins (RBPs), but in most cases, individual RBPs appear to influence the processing of a small subset of target miRNAs. We herein report that the RNA binding NONO/PSF heterodimer binds a large number of expressed pri-miRNAs in HeLa cells to globally enhance pri-miRNA processing by the Drosha/DGCR8 Microprocessor. Because NONO/PSF are key components of paraspeckles organized by the lncRNA NEAT1, we further demonstrate that NEAT1 also has a profound effect on global pri-miRNA processing. Mechanistic dissection reveals that NEAT1 broadly interacts with NONO/PSF as well as many other RBPs, and that multiple RNA segments in NEAT1, including a “pseudo pri-miRNA” near its 3′ end, help attract the Microprocessor. These findings suggest a bird nest model for a large non-coding RNA to orchestrate efficient processing of almost an entire class of small non-coding RNAs in the nucleus.
Introduction
MicroRNAs (miRNAs) are a class of 21–22nt small noncoding RNAs that are extensively involved in post-transcriptional regulation of gene expression in diverse organisms from plant to animal1–3. While a small fraction of miRNAs is encoded by their own genes, ~80% of annotated miRNAs are derived from various large coding and noncoding transcripts4. These initial transcripts, known as pri-miRNAs, are processed to pre-miRNAs by the Microprocessor consisting of DROSHA and DGCR8 in the nucleus, and after nuclear export by Exportin 5, pre-miRNAs are further processed into mature miRNAs by DICER before entering the RNA Induced Silencing Complex (RISC)5.
While the general miRNA biogenesis pathway has been elucidated and the core machineries for each processing step have been identified and well characterized, it is also known that each step during miRNA biogenesis is subjected to modulation, resulting in homeostatic expression of miRNAs in a highly cell type and tissue specific manner. At the level of transcription, a recent study reveals a key histone variant involved in the global regulation of pri-miRNA expression6. Pri-miRNA processing has been suggested to take place co-transcriptionally, but the existing evidence is largely based on the characterization of one or a few pri-miRNAs, but not genome-wide7–10. Therefore, while co-transcriptional pri-miRNA processing has been a popular, and to a large extent, widely accepted concept, we actually do not know to what degree a given pri-miRNA is processed co-transcriptionally or post-transcriptionally, which is an important question for understanding miRNA biogenesis and regulation in specific cells, tissues, and organs under normal physiological conditions or during disease processes.
During transcription and after transcription, a large number of RNA binding proteins (RBPs) and RNA helicases as well as post-translational modifications of these regulators have been documented to modulate miRNA biogenesis in individual processing steps5. To date, however, all characterized RBPs appear to module a single or a small subset of miRNAs via interacting with specific cis-acting elements and/or secondary structures in pri- or pre-miRNAs. Such specific modulation likely contributes to differential expression of miRNAs, even among those expressed from the same pri-miRNA transcripts.
Paraspeckles were discovered in 2002 through the identification of specific RBPs that are localized adjacent to nuclear speckles where most pre-mRNA processing factors are concentrated11,12. Both speckles and paraspeckles are permanent nuclear subdomains in most cell types, but their functions have been a continuous subject for debate and investigation13–18. Interestingly, each of these nuclear subdomains is associated with an abundant large noncoding RNA (lncRNA), known as MALAT1 in speckles and NEAT1 in paraspeckles19,20 with NEAT1, but not MALAT1, being essential for maintaining the structural integrity of the corresponding nuclear subdomain17,21,22. Thus far, the only known or postulated function for paraspeckles is the retention of certain hairpin-containing RNAs, particularly those derived from expressed Alu repeats23,24, and another specific lncRNA besides NEAT123, and sequestration of various RBPs25,26. However, both MALAT1 and NEAT1 have been shown to interact with some actively transcribed genes in the nucleus15,27, thus begging the question of whether these lncRNAs and/or their associated nuclear subdomains are more actively involved in regulated gene expression, rather than simply serving as some sort of storage sites for various RNAs and proteins.
In the present study, we initially pursued differential expression of miRNAs processed from the same pri-miRNAs, which led to the elucidation of both NEAT1 and key paraspeckle components in the global regulation of pri-miRNA processing. Interestingly, NEAT1 harbors an apparently pseudo miRNA, which is poorly processed into mature miRNA, and we found that this pseudo miRNA functions to attract the Microprocessor, while other RNA sequences and/or secondary structures in NEAT1 provide a general binding platform for various RBPs, some of which are engaged in extensive interactions with expressed pri-miRNAs. Our findings suggest a bird nest model for an lncRNA-organized machinery to globally enhance pri-miRNA processing, which also reveals critical insights into the formation and function of paraspeckles in the nucleus.
Results
Identification of key paraspeckle components involved in pri-miRNA processing
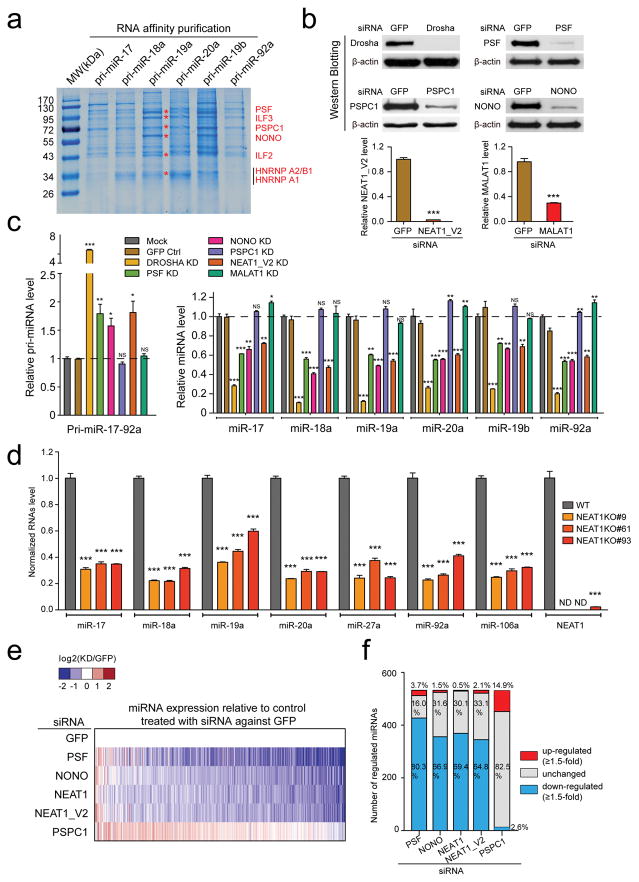
We initially wished to understand how different miRNAs encoded in the same primary transcripts were differentially processed in the cell. For instance, the primary miR-17–92a transcript gave rise to 6 mature miRNAs with dramatic difference in abundance in HeLa cells (Supplementary Fig. 1a, the primers used for quantitative analysis are listed in Supplementary Table 1). Knockdown of either DICER or combined knockdown of AGO 1–4 did not alter the relative abundance of individual miRNAs from the pri-miR-17–92a locus (Supplementary Fig. 1b–e, the antibodies used for Western blotting are listed in Supplementary Table 2), implying differential miRNA processing at the pri-miRNA level, which is known to be modulated by various RBPs5. We therefore prepared individual biotinylated pri-miRNAs from the miR-17–92a locus to compare their relative efficiencies in pulling down specific proteins from HeLa nuclear extracts. Pri-miR-19a and 19b appeared to be more efficient in pulling down several proteins, which we identified by mass spectrometry to correspond to two classes of RBPs (Fig. 1a, the peptides identified by mass spectrometry are listed in Supplementary Table 3). We confirmed binding of these proteins on multiple pri-miRNAs by Western blotting (data not show) and by direct in vivo crosslinking (see below). One class contains NONO (aka P54NRB), PSF (aka SFPQ), PSPC1 (aka PSP1), all of which are key RBP constituents of paraspeckles28, and the other class consists of ILF3 (aka NF90) and ILF2 (aka NF45) previously implicated in nuclear export of a viral dsRNA29. We also identified hnRNP A2/B1 and A1, the latter of which has been previously shown to enhance pri-miR-18a processing30. Because pri-miRNAs are hairpin-containing RNAs, we chose to focus on the RBPs associated with paraspeckles whose sole function elucidated to date is to retain or sequestrate various Alu-derived hairpin RNAs, an lncRNA, and RBPs in the nucleus23–26,31.
Fig. 1. Involvement of paraspeckle-associated proteins and lncRNA in pri-miRNA processing.
(a) Coomassie brilliant blue staining of proteins captured by individual pri-miR-17–92a from HeLa nuclear extracts. Specific proteins identified by mass spectrometry are indicated on the right. (b) Knockdown of three paraspeckle-associated proteins, NEAT1_V2 and MALAT1, respectively, quantified by Western blotting and RT-qPCR. (c) The expression of pri-miR-17–92a (left) and individual mature miRNAs from the pri-miR-17–92a locus (right) in response to knockdown of paraspeckle-associated factors and NEAT1, determined by RT-qPCR. (d) RT-qPCR was performed to confirm NEAT1 knockout (KO) with CRISPR/Cas and their impact on miRNA expression. (e) miRNA profiling in response to specific knockdowns as in c relative to control treated with siRNA against GFP. Color key on top indicates changes in log2 scale. (f) Summary of up-regulated (≥1.5-fold), no change, or down-regulated (≥1.5-fold) numbers of miRNAs based on small RNA-seq in response to specific knockdowns as in e. Uncropped images of Western blots in b are shown in Supplementary Data Set 1. Data in b,c,d are presented as mean ± SEM (n=3, technical replicates). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant, determined by two-tailed Student’s t test. ND, not detectable. Data source for the bar graphs are reported in Source Data for Figure 1.
We first determined by RT-qPCR whether individual paraspeckle-associated RBPs we identified might affect mature miRNA production. By using two independent siRNAs against each RBP, we found that knockdown of NONO or PSF, but not PSPC1, reduced the expression of all miRNAs from the miR-17–92a locus with corresponding increase in their pri-miRNA in HeLa cells (Fig. 1b, c; Supplementary Fig. 2a–c). We further confirmed these results by using individual miRNA sensor reporters (Supplementary Fig. 2d, e). We noted that the effects of NONO and PSF knockdowns were relatively weak compared to DROSHA knockdown, implying a degree of positive influence of NONO and PSF on pri-miRNA processing, rather than being essential for the process. However, we could not rule out the possibility that the residual proteins still provided part of the essential function. This is particularly pertinent to PSPC1, which even caused a minor increase in the expression of multiple miRNAs (Fig. 1c, right). We therefore attempted to use CRISPR/Cas to generate knockout cell lines for each of these RBPs. Knockout of NONO or PSF caused cell lethality, consistent with their involvement of in many critical cellular functions, including transcription and pre-mRNA splicing32–34. In contrast, PSPC1 appeared dispensable for cell viability. Using two independent PSPC1 null cell lines (we confirmed the absence of detectable PSPC1 protein by Western blotting), we found that ablation of PSPC1 significantly increased the expression of multiple miRNAs we examined (Supplementary Fig. 2f, g), an effect also evident from partial knockdown of PSPC1 by siRNA (Fig. 1c). Because NONO and PSF, but not PSPC1, are required for the structural integrity of paraspeckles22,28, which we also confirmed (Supplementary Fig. 3a), we chose to first focus on understanding the mechanism for these two paraspeckle components to stimulate pri-miRNA processing.
Global effect of NONO/PSF as well as NEAT1 in pri-miRNA processing
Given the correlation of paraspeckle disassembly with compromised pri-miRNA processing in NONO and PSF knockdown cells, we were curious about the role of NEAT1, an lncRNA required for the organization and maintenance of paraspeckles21,22,24,35. We observed a similar effect on pri-miRNA processing upon knockdown of NEAT1, and by contrast, knockdown of MALAT1, an lncRNA associated with nuclear speckles36,37, showed no effect (Fig. 1b, c). We obtained similar effects of NEAT1 knockdown by using a “stealth” siRNA in which the sense-strand is modified so that only the antisense strand can enter the RISC to minimize potential off-target effect26 or by generating NEAT1 null cells with CRISPR/Cas (Fig. 1d; Supplementary Fig. 3b). None of the knockdowns had measurable influence on the expression of the Microprocessor DROSHA/DGCR8 or multiple other paraspeckle-associated RBPs we examined (Supplementary Fig. 3c–f). These data revealed key roles of not only specific paraspeckle components but also its organizing lncRNA in enhancing miRNA biogenesis at the pri-miRNA level.
The data presented above were based on analysis of miRNAs from the miR-17–92a locus. To explore potential functional impact genome-wide, we performed small RNA-seq in response to knockdown of NONO, PSF, or PSPC1 in HeLa cells. Because human NEAT1 expresses two isoforms, V2 (23kb full-length NEAT1, 20kb in mice) and V1 (3.7kb 5′ portion of V2, 3.2kb in mice), the latter of which results from an early polyadenylation event18,35,38, we separately knocked down NEAT1_V1 and V2 (note that V1 knockdown would also diminish V2, and thus, we label it as NEAT1). We performed small RNA-seq under each treatment condition in duplicate and included a spike-in RNA during library construction for quantitative analysis (see Methods, sequencing statistics is shown in Supplementary Table 4). The amount of spike-in RNA as well as fragments of other noncoding RNAs, such as tRNAs, snoRNAs, and rRNAs, showed a linear relationship between duplicated experiments despite different sequencing depths (Supplementary Fig. 4a, b), and upon normalization against both external and internal reference RNAs39, all duplicated libraries showed high reproducibility (Supplementary Fig. 4c, d). We thus combined uniquely mapped reads from duplicated libraries, obtaining ~20 million total uniquely mapped reads under each treatment condition. We plotted the miRNA levels from each knockdown against those from control treated with siRNA against GFP. Strikingly, 64–80% from a total of 532 expressed miRNAs with read number >30 in control siRNA-treated HeLa cells were down regulated upon knockdown of NONO, PSF, and NEAT1 (both V1 and V2), and again, PSPC1 knockdown showed the opposite effect on many miRNAs (Fig. 1e, f). We validated the sequencing results by RT-qPCR on a large panel of miRNAs, (Supplementary Fig. 4e, f). These findings revealed a global role of specific paraspeckle-associated RBPs and NEAT1 in pri-miRNA processing.
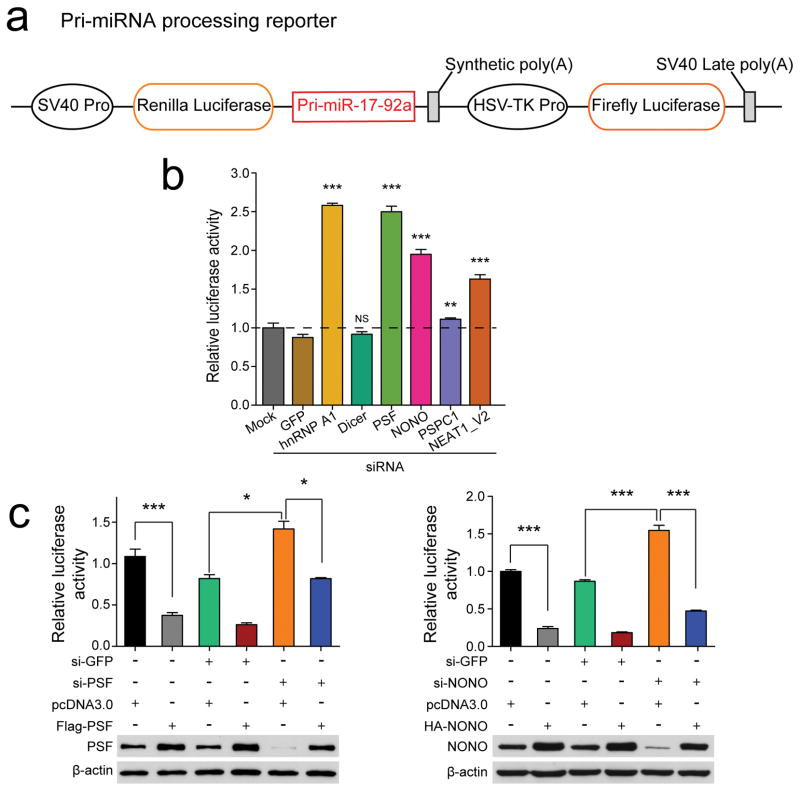
To further demonstrate compromised pri-miRNA processing, we constructed a pri-miRNA processing reporter by inserting the pri-miR-17–92a sequence in the 3′UTR of the Renilla reporter (illustrated in Fig. 2a; compromised pri-miRNA processing would lead to increased Renilla activity). Knockdown of NONO, PSF, or NEAT1 all caused elevated Renilla activities, similar to knockdown of hnRNP A1 as previously shown30, while knockdown of either DICER or PSPC1 had no effect (Fig. 2b). These observations imply a more direct role of NONO/PSF and NEAT1, but not PSPC1, in pri-miRNA processing. We next performed overexpression/rescue experiments and demonstrated that overexpression of NONO and PSF each stimulated pri-miRNA processing and their siRNA-resistant cDNAs rescued the defects in specific siRNA-treated cells (Fig. 2c). These data provide further evidence for the involvement of NONO/PSF in enhancing pri-miRNA processing by the Microprocessor.
Fig. 2. Function of NONO-PSF and NEAT1 analyzed with the pri-miRNA processing reporter.
(a) Top: The pri-miRNA processing reporter containing pri-miR-17–92a cloned into the 3′UTR of the Renilla luciferase in psiCHECK2 vector. (b) Relative luciferase activities of the pri-miRNA processing reporter in response to knockdown of individual RBPs as indicated or NEAT1_V2. (c) Relative luciferase activities of the pri-miRNA processing reporter in response to overexpression of siRNA-resistant PSF (left) or NONO (right). Bar graphs in b and c are presented as mean ± SEM (n=3, cell culture). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant, determined by two-tailed Student’s t test. Data source for the bar graphs are reported in Source Data for Figure 2. Uncropped images of Western blots in c are shown in Supplementary Data Set 1.
Prevalent binding of NONO/PSF on expressed pri-miRNAs in HeLa cells
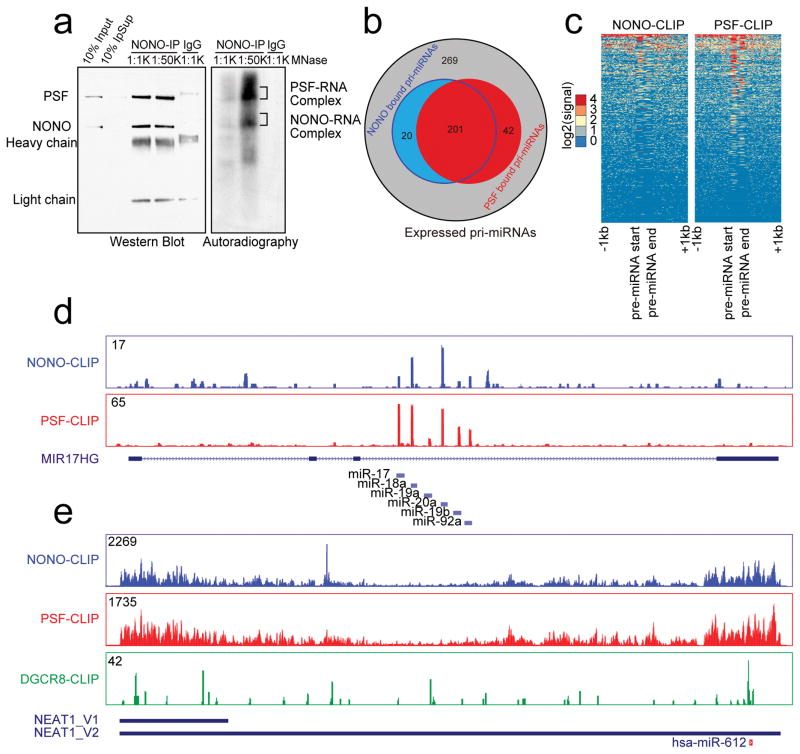
To investigate how NONO/PSF might facilitate global pri-miRNA processing, we performed UV CrossLinking ImmunoPrecipitation coupled with deep sequencing (CLIP-seq) to identify their direct RNA targets. Both anti-NONO and anti-PSF antibodies efficiently brought down the NONO/PSF heterodimer, as reported earlier40,41, each of which was crosslinked to RNA, as detected by 32p-labeling with T4 polynucleotide kinase (Fig. 3a; Supplementary Fig. 5a). We separately isolated protein-RNA adducts after trimming RNA with micrococcal nuclease (MNase) for CLIP-seq library construction. Libraries from reciprocal immunoprecipitation showed high reproducibility among all NONO/PSF CLIP-seq experiments (Supplementary Fig. 5b). We thus combined uniquely mapped, PCR duplicate-removed reads, obtaining ~14M reads for NONO and ~18M reads for PSF (sequencing statistics is listed in Supplementary Table 4). The deduced NONO and PSF binding peaks were similarly distributed in the human genome with a large fraction on intronic and 3′UTR regions (Supplementary Fig. 5c), consistent with their established roles in pre-mRNA processing32,34,42.
Fig. 3. Genome-wide analysis of NONO-PSF-RNA interactions.
(a) Immunoprecipitated NONO-PSF crosslinked to RNA. The complex was treated with two different concentrations of MNase (1:1,000 or 1:50,000 dilution); RNA in the complex was 32p-labeled with T4 polynucleotide kinase. Proteins and RNA were visualized by Western blotting (left) and autoradiography (right). Indicated bands (right) were individually isolated for CLIP-seq library construction. (b) Venn Diagram showing overlapped pri-miRNAs bound by NONO and PSF in HeLa cells. (c) Footprint of NONO and PSF on pri-miRNAs. (d) Representative NONO and PSF binding tracks on the pri-miR-17–92a transcript. (e) The binding profiles of NONO and PSF on NEAT1 in comparison with the published DGCR8 CLIP-seq signals44. Y-axis in d and e shows CLIP-seq read density in each case. The region encoding for miR-612 is indicated at bottom. Uncropped images of Western blots and autoradiography in a are shown in Supplementary Data Set 1.
Importantly, we found that both NONO and PSF bound 263 transcribed pri-miRNAs, about 2/3 of expressed pri-miRNAs in HeLa cells (Fig. 3b, c), as illustrated on their highly discrete binding on all 6 pri-miRNAs encoded in the pri-miR-17–92a locus (Fig. 3d), as well as on many other representative pri-miRNAs (Supplementary Fig. 5d). We noted that such PSF binding on expressed pri-miRNA was not as prevalent in HepG2 as we observed in HeLa cells based on similar analysis of the existing PSF eCLIP data from the ENCODE consortium (data not shown, see further in Discussion), and in addition, the number of down-regulated miRNAs was clearly larger than that of NONO/PSF-bound pri-miRNAs. These observations imply potential cell type specificity with respect of NONO/PSF binding on expressed pri-miRNAs and suggest roles of other paraspeckle-associated RBPs in pri-miRNA processing, which were likely affected by induced paraspeckle disassembly28.
Interestingly, we also detected prevalent binding of the NONO/PSF heterodimer on NEAT1, but with a dramatically distinct binding pattern compared to their discrete binding on pri-miRNAs (Fig. 3e). Such continuous binding on both ends of NEAT1 is consistent with the proposed structure of paraspeckles with both of its ends exposed at the periphery of this subnuclear domain43. For comparison, we also displayed the published DGCR8 CLIP-seq data on NEAT144. Although the DGCR8 CLIP-seq read density is relatively low on the lncRNA, the reads showed two binding clusters, one at the 5′ end and the other on pri-miR-612 at the 3′ end of NEAT1 (marked at the bottom of Fig. 3e). Interestingly, although NEAT1 is extremely abundant in the cell, we found that mature miR-612 was nearly undetectable from our small RNA-seq experiments or by RT-qPCR (data not shown). We further confirmed its poor processing by using a pri-miR-612 processing reporter in comparison with the reporter derived from pri-miR-17–92a in response to Microprocessor knockdown (Supplementary Fig. 6a). This finding suggested that miR-612 might be a “pseudo” miRNA and its primary function might serve as an anchor for attracting the Microprocessor to NEAT1. These data therefore begin to paint a general picture in which NEAT1 might function as a scaffold, not only for a large number of RBPs, but also for the Microprocessor, thereby facilitating their kinetic interactions that lead to more efficient pri-miRNA processing in the nucleus.
NEAT1 mediates the interaction of NONO/PSF with the Microprocessor
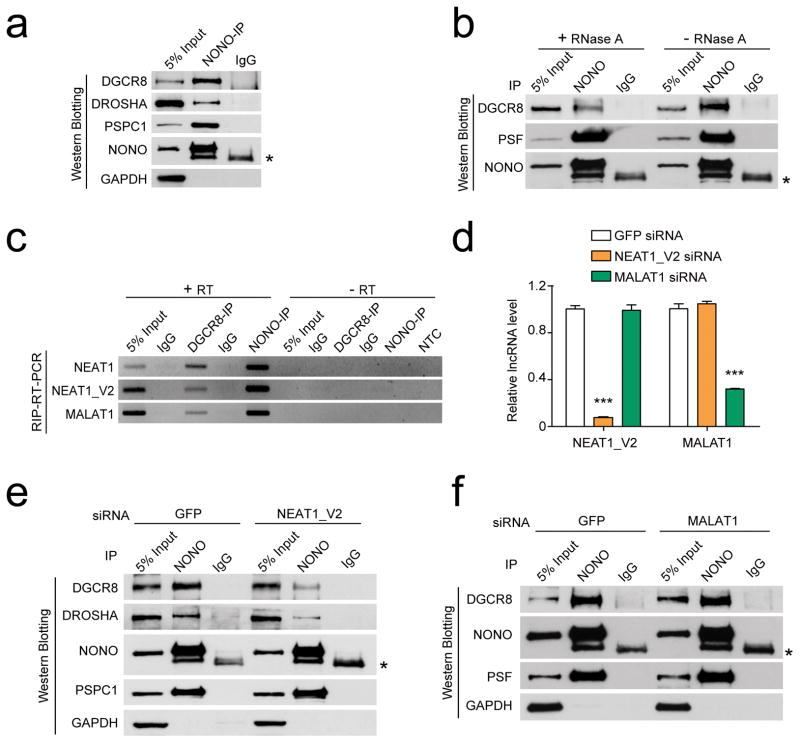
To provide evidence for NEAT1-mediated interactions between NONO/PSF and the Microprocessor, we performed reciprocal immunoprecipitation with NONO and DGCR8, finding that NONO was indeed able to bring down both endogenous and exogenous FLAG-tagged DGCR8 in HeLa cells (Fig. 4a; Supplementary Fig. 6b, c). Importantly, RNase A treatment greatly reduced the interactions of NONO with DGCR8, but not with PSF, indicating direct protein-protein between NONO and PSF and RNA-mediated interactions between NONO/PSF and the Microprocessor (Fig. 4b). As predicated, specific antibodies against DGCR8 and NONO also brought down both isoforms of NEAT1 (Fig. 4c). Interestingly, both also pulled down MALAT1, which is known to interact with numerous RBPs involved in pre-mRNA splicing37. To determine whether any of these lncRNAs mediated the interactions between NONO/PSF and the Microprocessor, we performed siRNA knockdown (Fig. 4d), finding that NEAT1 knockdown largely abolished the in vivo interactions of NONO with the Microprocessor without affecting its interaction with PSPC1 (Fig. 4e), and by contrast, MALAT1 knockdown showed no impact on any of these interactions (Fig. 4f). These data strongly suggested that NEAT1 specifically bridged the interactions between paraspeckle components and the Microprocessor in the cell.
Fig. 4. NEAT1 bridges NONO-PSF and the Microprocessor.
(a) Co-IP of endogenous NONO with PSPC1 and the Microprocessor DROSHA/DGCR8. (b) RNA-dependent interactions between NONO-PSF and the Microprocessor. (c) RT-PCR analysis of NEAT1_V1, NEAT1_V2, and MALAT1 in NONO and DGCR8 immunoprecipitants. (d) Knockdown efficiency of NEAT1_V2 and MALAT1, quantified by RT-qPCR and normalized against GAPDH mRNA. (e, f) Western blotting analysis of NONO and the Microprocessor interactions in response to knockdown of NEAT1_V2 (e) or MALAT1 (f). * indicates IgG heavy chain. Uncropped images of western blots in a,b,c,e, and f are shown in Supplementary Data Set 1. Bar graphs in d are presented as mean ± SEM (n=3, technical replicates). ***P < 0.001, determined by two-tailed Student’s t test. Data source for the bar graphs are reported in Source Data for Figure 4.
NEAT1_V1 enhances pri-miRNA processing in a NEAT1_V2 dependent manner
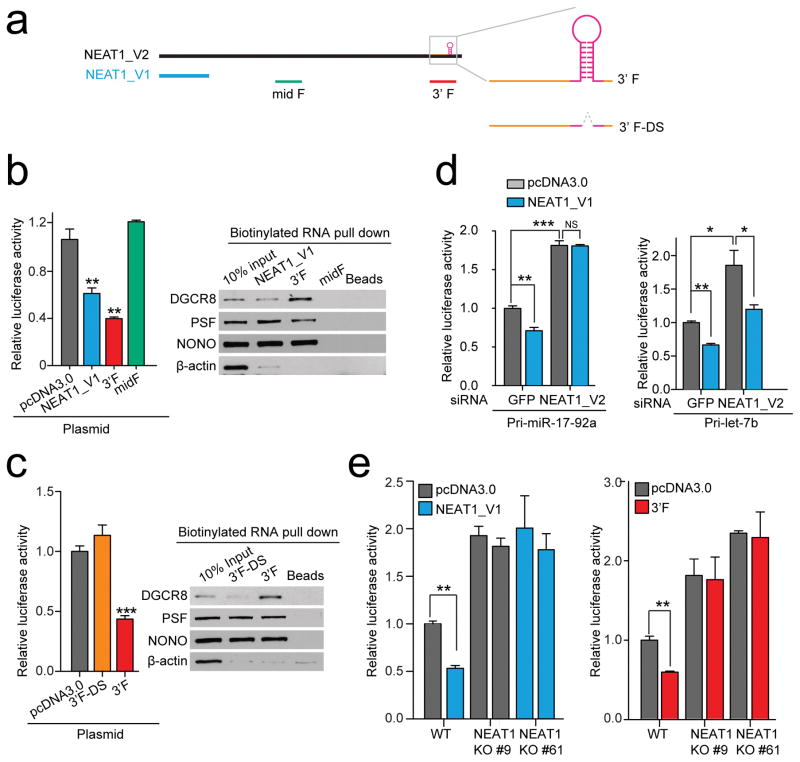
Because the NONO/PSF heterodimer interacts with numerous regions in NEAT1 and the smaller V1 isoform of NEAT1 has been shown to enhance paraspeckle formation28,45, we next determined whether V1 and some representative fragments from full-length NEAT1 (Fig. 5a) were able to enhance pri-miRNA processing. Using the pri-miR-17–92a processing reporter (Fig. 2a), we found that transfected V1 was indeed stimulatory to pri-miR-17–92a processing, so was a V2 fragment from the 3′ end (3′F), but not a middle fragment (midF) (Fig. 5b, left). When the stem-loop of pri-miR-612 in the 3′F was deleted (3′F-DS), the enhancement effect was lost (Fig. 5c, left). To further explore the molecular basis for enhanced pri-miR-17–92a processing, we incubated nuclear extracts with various in vitro transcribed RNAs to determine their abilities to bridge the interactions between NONO/PSF and the Microprocessor. We found that both V1 and the 3′F, but not its pri-miR-612 deleted version (3′F-DS), were able to efficiently bring down NONO/PSF and DGCR8 (Fig. 5b, c, right).
Fig. 5. NEAT1-mediated interaction networks for enhancing pri-miRNA processing.
(a) Illustration of NEAT1_V1, NEAT1_V2 and derived RNA fragments. Highlighted on the right are pri-miR-612 near the 3′ end of NEAT1_V2 and the 3′ fragments before (3′F) or after deletion of the pre-miR-612 stem-loop (3′F-DS). (b, c) Enhanced processing of the pri-miR-17–92a reporter by NEAT_V1 and 3′F, but not a middle fragment from NEAT1_V2 (midF) or 3′F-DS (left panels). The right panels show RNA pulldown results from HeLa nuclear extracts, analyzed by Western blotting for NONO-PSF and DGCR8. (d) Knockdown of NEAT1_V2 diminished the enhancement of pri-miRNA processing by overexpressed NEAT_V1 on the pri-miR-17–92a (left) or pri-Let-7b (right) processing reporter. (e) Knockout of NEAT1_V2 prevented the enhancement of pri-miRNA processing by overexpressed NEAT_V1 and the 3′F on the pri-miR-17–92a processing reporter. Bar graphs in b,c,d, and e are presented as mean ± SEM (n=3, cell culture). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant, determined by two-tailed Student’s t test. Uncropped images of Western blots in b and c are shown in Supplementary Data Set 1 and data source for the bar graphs are reported in Source Data for Figure 5.
Because an earlier observation indicated that V1 was able to enhance the appearance of paraspeckles, but only in the presence of full-length NEAT128,45, we next tested whether full-length NEAT1 was required for enhanced pri-miR-17–92a processing by V1. We observed that the enhancement was lost in NEAT1 knockdown cells (Fig. 5d, left). We made a similar observation with another Let-7b based pri-miRNA processing reporter although V1 continued to show some effect on this reporter in NEAT1 depleted cells (Fig. 5d, right). These observations suggest a broad effect of full-length NEAT1. To further confirm this finding, we took advantage of NEAT1 null cell lines we generated (Fig. 1d ) and tested the requirement of full-length NEAT1 for stimulated pri-miRNA processing by the small V1 isoform or the 3′F of V2. Consistent with the NEAT1 knockdown experiments, we observed that both overexpressed V1 and the 3′F were able to enhance pri-miRNA processing in wild-type, but not NEAT1 null cells (Fig. 5e). Combined, these data suggest the function of full-length NEAT1 in providing a platform for enhanced pri-miRNA processing in the nucleus.
Evidence for the involvement of paraspeckles in pri-miRNA processing
A recent study suggested two populations of NEAT1-containing ribonucleoprotein particles (RNPs) in mammalian cells, one in the form of numerous microscopic structures throughout the nucleus and the other as paraspeckles46, the latter of which may be the aggregated form of the former. In literature, the data on the involvement of paraspeckles have been controversy at this point. On one hand, it has been demonstrated that at least a fraction of pri-miRNAs are processed co-transcriptionally9 and retarded release of pri-miRNAs from chromatin appears to be important for their efficient processing7,47. Accordingly, DGCR8 has been localized in a largely diffused pattern in the nucleoplasm7. These data suggest that paraspeckles visible under microscope may not correspond to cellular locations for pri-miRNA processing. On the other hand, one report indicated that the FLAG-tagged exogenous DGCR8 was localized adjacent to nuclear speckles48 and certain induced pri-miRNAs (e.g. pri-miR-155) also became localized near nuclear speckles7, although none of these studies verified adjacent nuclear speckles as paraspeckles by co-staining with a paraspeckle marker. Therefore, although inconclusive, these existing data suggest that a subset of pri-miRNAs, especially those highly induced ones, as well as a fraction of the Microprocessor were detectable in a localized fashion under certain experimental conditions.
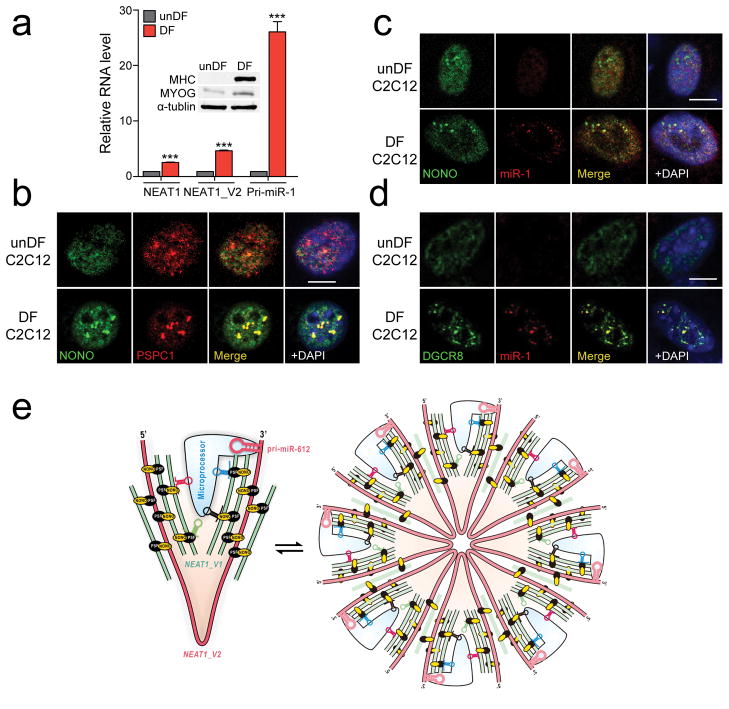
Given such controversy, we sought to localize DGCR8 by immunochemistry and pri-miRNAs by fluorescence in situ hybridization (FISH) under various conditions, but we rarely detected signals in paraspeckles. Reasoning that such localization might become detectable with only highly expressed pri-miRNAs, we took advantage of the dramatic induction of the pri-miR-1 gene in C2C12 cells upon differentiation49, which we confirmed, and interestingly, we also detected an increase in NEAT1 expression in differentiated C2C12 cells (Fig. 6a). By immunostaining with PSPC1, we found that undifferentiated (unDF) C2C12 myoblasts exhibited detectable paraspeckles, but the appearance of paraspeckles became much stronger after C2C12 differentiation (DF) into myotubes, consistent with induced NEAT1 (Fig. 6b). We next performed FISH for pri-miR-1 and found that induced pri-miR-1 was indeed detectable in multiple foci, colocalizing with NONO only on differentiated C2C12 cells (Fig. 6c). Under these conditions, we further observed that induced pri-miR-1 also colocalized with DGCR8 (Fig. 6d). It is important to disclose that we observed such DGCR8 foci under FISH conditions, but rarely saw such foci under standard immunostaining conditions even on differentiated C2C12 cells (data not shown). These observations imply that DGCR8 may be engaged in other RNA metabolism pathways in the nucleoplasm, as suggested by its broad RNA binding profile44, which may mask its localization in paraspeckles, but under FISH conditions, some of DGCR8 interactions were weakened in the nucleoplasm despite on fixed cells while those in paraspeckles were preserved, thus providing a plausible explanation to the controversy in the field. Importantly, our data now suggest the involvement of NEAT1-containing RNPs in global modulation of pri-miRNA processing, either in the form of microscopic structures throughout the nucleus or in a more aggregated form in nuclear paraspeckles, similar to co-transcriptional and post-transcriptional pre-mRNA splicing with respect to nuclear speckles.
Fig. 6. Localization of induced pri-miR-1 in paraspeckles in differentiated C2C12 cells and the proposed bird nest model.
(a) The expression levels of NEAT1 and pri-miR-1 quantified by RT-qPCR in undifferentiated (unDF) and differentiated (DF) C2C12 cells. Inset shows the induction of the differentiation marker MHC and MYOG by Western blotting. (b) Enhanced paraspeckles after C2C12 differentiation, detected by NONO immunostaining. (c) FISH analysis of inducible pri-miR-1. No pri-miR-1 signal was detectable in C2C12 cells before differentiation and colocalization of induced pri-miR-1 with NONO on paraspeckles in differentiated C2C12 cells. (d) Colocalization of induced pri-miR-1 with DGCR8 in differentiated C2C12 cells. Scale bars in b,c,d: 10 μm. (e) The proposed bird nest model for NEAT1-orchestrated enhancement of pri-miRNA processing by the Microprocessor. Multiple RBPs, including the NONO-PSF heterodimer, extensively interact with NEAT1_V2, on which additional NEAT1_V1 and NEAT1_V2 may be added to build a bird nest-like structure. Various RBPs may also bring pri-miRNAs to the nest and various RNA secondary structures in NEAT1, including a poorly processed pri-miR-612 near the 3′ end of NEAT1_V2, may help recruit the Microprocessor. These NEAT1-containing RNPs may exist in both the microscopic form (left) and become “aggregated” to generate larger structures visible as paraspeckles (right). In both forms, such RNA-orchestrated structures may create the proximity between pri-miRNAs and the Microprocessor to enhance the kinetics of pri-miRNA processing. Bar graphs in a are presented as mean ± SEM (n=3, technical replicates). ***P < 0.001; NS, not significant, determined by two-tailed Student’s t test. Uncropped images of Western blots are shown in Supplementary Data Set 1 and data source for the bar graphs are reported in Source Data for Figure 6.
Discussion
Considering all data presented in this study, we propose a bird nest model for NEAT1-mediated enhancement of pri-miRNA processing (Fig. 6e, left). The lncRNA NEAT1 may provide a scaffold for the NONO/PSF heterodimer and many other RBPs28. These RBPs may bind additional NEAT1_V1 and V2 isoforms as well as pri-miRNAs to form a bird nest-like structure in the nucleus. As NEAT1 likely contains various hairpin structures that resemble pri-miRNAs, as exemplified by pri-miR-612, those secondary structures may also help attract the Microprocessor, thus facilitating kinetic interactions between pri-miRNAs and their processing machinery. These microscopic NEAT1-containing RBPs may become further aggregated to give rise to the appearance of paraspeckles (Fig. 6e, right), especially after cell differentiation during which some specific components of paraspeckles are induced35.
Although a role(s) of paraspeckles visible under a microscope in certain aspects of regulated gene expression will continue to be a subject of debate, our current data provide evidence for its active participation in post-transcriptional pri-miRNA processing and perhaps other RNA metabolism activities. It is well known in the field that DGCR8 is largely distributed in a diffused pattern in the nucleus, which likely reflects its involvement in multiple RNA metabolism pathways, consistent with its limited interaction with expressed pri-miRNAs and NEAT1 (see the bottom panel in Fig. 3e) based on the published DGCR8 RNA binding profile44. We now found that, under certain conditions, such as on cells treated for FISH, a fraction of DGCR8 became detectable on paraspeckles where it colocalized with highly induced pri-miR-1 in differentiated C2C12 cells. This observation implies that the localization of DGCR8 in paraspeckles might be largely masked in most cell types. Because ~80% of pri-miRNAs reside in introns of pre-mRNAs4,8 and the structural integrity of paraspeckle depends on on-going transcription12,45, it is attempting to speculate that its spatial relationship with splicing factor-enriched speckles might result from co-processing of certain pri-miRNAs and pre-mRNAs in the nucleus.
With respect to paraspeckle-associated RBPs, we noted that NONO/PSF binding on pri-miRNAs were more prevalent in HeLa cells compared to HepG2 cells, implying a degree of cell-type specificity in terms of divided labors of different paraspeckle-associated RBPs in pri-miRNA processing. It is also curious that the paraspeckle-associated protein PSPC1 appears to suppress pri-miRNA processing. This might be related to the observation that PSPC1 belongs to a distinct of paraspeckle-associated RBPs that are not essential for paraspeckle formation nor maintenance, which requires further investigation to under its regulatory mechanism.
Last but not least, we also need to consider the fact that Neat1 null mouse does not have gross phenotype18, suggesting that NEAT1-enhanced pri-miRNA processing may not be essential for cell survival; however, such process may still critically contribute to specific gene expression programs under certain developmental and/or pathological conditions, as seen in Neat1 null animals26,35,50–54. In any case, the data presented in this report suggest a potential new role of NEAT1-containng RNPs, either in its microscopic form or as part of paraspeckles, which provide a new angle to envision and investigate the biological function of this intriguing nuclear subdomain in diverse developmental and disease processes.
Methods
Methods, including statements of data availability and associated accession code and references, are available in the online version of the paper.
Methods
Cell culture, plasmids, transfection, RIP-PCR and RT-qPCR
HeLa and C2C12 cells from ATCC were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) plus 100 U penicillin/streptomycin (Gibico) at 37°C in a humidified incubator with 5% CO2. Both cultured HeLa and C2C12 cells were determined to be free from mycoplasma contamination.
All expression plasmids were cloned in pCDNA3.0 and luciferase reporter plasmids were cloned in pSicheck2 between Xhol I and Not I sites. The siRNA-resistant FLAG-PSF and HA-NONO expression plasmids were generated by a PCR-based method (KOD Plus from TOYOBO) with specific primers containing site-specific mutations, isted in Supplementary Table 1.
Plasmids and siRNAs were transfected into cells with Lipo2000 and RNAi Max, respectively, according to the manufacturer’s protocols. Cells were harvested 48 to 72 hrs post-transfection for subsequent analysis. For double transfection, cells were first transfected with siRNA for 12 hrs followed by plasmid transfection for another 24 hrs. Individual siRNA sequences are listed in Supplementary Table 1. Dual luciferase assays were performed 48 hrs post-transfection.
For RIP-PCR, RNA from immunoprecipitant was extracted with Trizol (ThermoFisher) followed by reverse transcription with M-MLV (Promega) and random hexamers at 37°C for 1 hr. Quantitative analysis of miRNAs was performed with the Qiagen miScript II RT Kit and real-time PCR was conducted by using a SYBR green master mix and gene-specific primers listed in Supplementary Table 1.
Immunoprecipitation, RNA pulldown, and Western blotting
For immunoprecipitation, 2 μg specific antibody was coupled to Dynabeads at 4°C for 2 hrs in 200 μl lysis buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 1 mM PMSF and proteinase inhibitor cocktail), and after wash 3 times with lysis buffer, 300 μl of whole cell extracts was added. The mix was incubated with rotation at 4°C for 2 hrs. For RNase A treatment, immunoprecipitant was incubated in lysis buffer containing 200 μg/ml RNase A (Fermentas) at 37°C for 10 min. The beads were washed 4 times with lysis buffer, re-suspended in 1XSDS loading buffer, and boiled for analysis by SDS-PAGE. Western blotting was performed with standard protocol using specific antibodies listed in Supplementary Table 2.
For preparation of nuclear extracts, 1×107 HeLa cells grown in 10 cm dishes were washed twice with 10 ml ice-cold PBS and collected by centrifugation at 2000 rpm for 5 min at 4°C. Pelleted cells were re-suspended in 1 ml CE-I buffer (10 mM HEPES pH7.4, 60 mM KCl, 1 mM EDTA, 0.075% NP-40, 1 mM DTT) and incubated on ice for 5 min. Nuclei were isolated by centrifugation at 1500 rpm at 4°C for 5 min, washed twice with 0.5 ml CE-II buffer (10 mM HEPES, pH7.4, 60 mM KCl, 1 mM EDTA, 1 mM DTT), and then resuspended in 0.5 ml NE buffer (20 mM HEPES, pH7.4, 420 mM KCl, 4 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 15% glycerol) with brief vortex followed by incubation with rotation at 4°C for 60 min.
For RNA pulldown, biotin-labeled RNAs were prepared with the Biotin RNA labeling mix (Sigma) and T7 RNA polymerase (ThermoFisher). The reaction was carried out at 37°C for 2 hrs, treated with 2 μl DNase I (Promega) at 37°C for 15 min, and then stopped by the addition of 2 μl 0.5 M EDTA (pH8.0). The resulting RNA was purified on Micro Bio-Spin 30 Column (Bio-Rad) and stored in TE buffer (10 mM Tris-HCl, pH7.4). ~3 μg of biotin-labeled RNA was heated to 90°C for 2 min, chilled on ice for 2 min, and incubated in 50 μl RNA structure buffer (10 mM Tris-HCl pH 7.0, 0.1 M KCl, 10 mM MgCl2) at room temperature for 20 min to allow RNA folding. Folded RNA was mixed with ~1 mg of nuclear extracts diluted in RIP buffer (25 mM Tris-HCl pH 7.0, 150 mM KCl, 0.5 mM DTT, 0.5% NP-40, 1 mM PMSF and protease inhibitor cocktail) and incubated at 4°C for 1 hr followed by the addition of 30 μl of streptavidin beads pre-washed twice with RIP buffer. The reaction was further incubated for another 1hr, washed 3 times with cold RIP buffer, and boiled in 30 μl 1XSDS loading buffer for analysis by Western blotting.
Global miRNA profiling and CLIP-seq
Isolated total RNA (5–8 μg) was mixed with 10 μM pre-adenylated 3′ linker in 1× RNA ligase buffer containing 50% (w/v) PEG 8000, 1 μl RNase inhibitor, and 1 μl T4 RNA ligase 2 (NEB), and incubated at room temperature for 1 hrs, then at 16°C for 3 hrs. After 3′ linker ligation, the reaction was size fractionated on 15% polyacrylamide gel and RNAs between 42 to 54 nt were recovered by incubating the corresponding gel slice at 4°C in 600 μl of 0.3 M NaCl with constant agitation. 5′ linker ligation was next performed in 1× RNA ligase buffer containing 50% (w/v) PEG 8000, 1 mM ATP, 10 μM 5′ linker, 1 μl RNase inhibitor and 1 μl T4 RNA ligase 1 (NEB) at 37 °C for 4 hrs. Linker-ligated RNAs were reverse transcribed with Superscript III (Life Technology) and PCR-amplified with Phusion polymerase (ThermoFisher). The product was fractionated on 4% agarose gel and recovered DNA was quantified for deep sequencing.
Duplicated experiments were performed for treatment condition and multiplexed sequenced in one Illumina Hi-seq 4000 lane and the sequencing statistics is listed in Supplementary Table 4. Reads were decoded by index sequences without mismatch, and 1–40nt target sequences were saved for downstream analysis. The 3′ adaptor of reads (NNCTCGTATGCCGTCTTCTGCTTG) were firstly trimmed by using cutadapt program55 with parameters ‘-O 3 -e 0.25 -q 20’. The 5′ random index (NNNTC) was then removed and only reads of ≥16nt were kept for mapping. The annotated miRNAs in miRBase version 20 (ref56) were used as reference. The reads were mapped to the human genome (hg19) with Bowtie program57, and the expression of mature miRNAs were quantified with program miRDeep2 (ref58). The counts for mature miRNAs from two replicates were combined, and the counts from different samples were normalized according to the published procedure39, using the number of reads mapping to rRNAs, snoRNAs, tRNAs, and spike-in RNA (CTCAGGATGGCGGAGCGGTCT) as internal controls. 1.5-fold change or larger of reads from knockdown samples relative to siGFP sample were computed to identify differentially expressed miRNAs as summarized in Supplementary Table 4.
CLIP-seq for NONO and PSF as well as associated data analysis were as previously described59,60, and the peak calling was done with CLIPper59. The distribution of binding was computed with program DeepTools2 (ref61). The sequencing statistics was listed in Supplementary Table 4.
Genomic engineering by CRISPR/Cas
sgRNAs (listed in Supplementary Table 1) were designed using the CRISPR tool at http://crispr.mit.edu. To knockout PSPC1, annealed DNA oligonucleotides were cloned into p×459 at the Bbs I restriction site. To knockout NEAT1, four pairs of sgRNA sequences (#1 and #3; #1 and #4; #2 and #3; #2 and #4) were cloned into the PiggyBac plasmid (PBC2), each under a separate U6 promoter. The PBC2 plasmids containing individual pair of sgRNAs and the plasmid expressing Cas9 were co-transfected in HeLa cells using lipo 2000. After 24 hrs post-transfection, PSPC1 KO cells were selected with puromycin (sigma) for 4 days, and NEAT1 KO cells were selected with hygromycin (Roche) for 4 days. Live cells were cultured in fresh DMEM containing 10% FBS without antibiotics to allow recovery for 1 to 2 days before isolating single clones. For PSPC1 KO cell lines, out of 48 clones obtained two homozygous cloned were picked for functional analysis. For NEAT1 KO, out of 148 clones obtained, four homozygous clones (one from sgRNA pair #1 and #3, one for sgRNA pair # 2 and # 3, two for sgRNA # 2 and #4) were picked for verification by direct sequencing and subsequent functional studies.
RNA FISH on C2C12 cells
C2C12 cells grown to 80% confluence were induced to differentiate into myotubes by incubation in DMEM supplemented with 2% horse serum (Gibico) for two days. FISH was performed as previously described22. Fixed cells on coverslip were dehydrated by 70%, 95%, 100% ethanol for 5 min each and incubated with pre-hybridization buffer (2×SSC, 1XDenhardt’s solution, 50% formamide, 10 mM EDTA, 100 μg/ml yeast tRNA, and 0.01% Tween 20) at 37°C for 1 h. Pre-hybridized coverslips were incubated in hybridization buffer (5% dextran sulfate in pre-hybridization buffer plug Dig-labeled RNA Probe) for 16–18h at 37°C. Coverslips were washed twice with buffer A (2× SSC, 50% formamide, and 0.01% Tween 20) at 37°C for 20 min, twice with buffer B (2× SSC, and 0.01% Tween 20), and once with buffer C (0.1× SSC and 0.01% Tween 20). Coverslips were then blocked with blocking buffer (50mM Tris pH8.0, 100 mM NaCl, 0.1%TritonX-100, 3%NGS, 0.1%BSA) for 1 hr and incubated with anti-DIG antibody (Thermo Fisher). For co-localization with protein markers, cells were blocked with blocking buffer at 37°C for 20 min and incubated with specific antibodies diluted in the blocking buffer for 1 hr. After wash three times with 1×TBST, secondary antibodies fluorescence labeled (Alexa 594 anti-sheep or Alexa 488 anti-rabbit) were applied and incubated for another hr followed by wash three times with 1×TBST and counterstained with DAPI before mounting onto glass slides for microscopy. Images were taken with a Leica SP8 microscope with a 63× objective lens. The primers used for probe preparation are listed in Supplementary Table 1 and the antibodies for immunostaining are listed in Supplementary Table 2.
Statistical analysis
Each experiment presented was based on 3 or 4 technical replicates as indicated in individual figure legends. Data are presented as means ± SEM, and p-values were determined by two-tailed Student’s t test. All experiments were further confirmed by biological repeats.
Data availability
All the deep sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) under series accession number GSE90650. All the source data are available with the paper online. Other data from this study are available upon request.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2017YFA0506400), the 111 Program of China (B06018), NIH (HG004659, GM049369, GM052872), the National Natural Science Foundation of China (31000573), and the Postdoctoral Science Foundation of China (20090451074).
Footnotes
Author contributions
QW, LJ, CS, and XDF designed the experiments; LJ, QW, and CS performed most experiments; JZ, BY, and YuZ analyzed the data; GC, HL, LG, YiZ, YW and GY contributed to sequencing, cell lines, and data interpretation; LJ, CS, YuZ and XDF wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, et al. HP1BP3, a Chromatin Retention Factor for Co-transcriptional MicroRNA Processing. Mol Cell. 2016;63:420–432. doi: 10.1016/j.molcel.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morlando M, et al. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballarino M, et al. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox AH, et al. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 12.Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- 14.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki T, Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci (Elite Ed) 2015;7:1–41. doi: 10.2741/715. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. Rna-a Publication of the Rna Society. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. The Journal of cell biology. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 24.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose T, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imamura K, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 27.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naganuma T, et al. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urcuqui-Inchima S, Castano ME, Hernandez-Verdun D, St-Laurent G, 3rd, Kumar A. Nuclear Factor 90, a cellular dsRNA binding protein inhibits the HIV Rev-export function. Retrovirology. 2006;3:83. doi: 10.1186/1742-4690-3-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 31.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 34.Liang S, Lutz CS. p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA. 2006;12:111–121. doi: 10.1261/rna.2213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & development. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto K, Sakurai H, Sugiura T. Proteomic identification of a PSF/p54nrb heterodimer as RNF43 oncoprotein-interacting proteins. Proteomics. 2008;8:2907–2910. doi: 10.1002/pmic.200800083. [DOI] [PubMed] [Google Scholar]
- 41.Passon DM, et al. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes & development. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 43.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21:4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macias S, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li R, Harvey AR, Hodgetts SI, Fox AH. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA. 2017;23:872–881. doi: 10.1261/rna.059477.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pawlicki JM, Steitz JA. Subnuclear compartmentalization of transiently expressed polyadenylated pri-microRNAs: processing at transcription sites or accumulation in SC35 foci. Cell Cycle. 2009;8:345–356. doi: 10.4161/cc.8.3.7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng C, et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choudhry H, Mole DR. Hypoxic regulation of the noncoding genome and NEAT1. Brief Funct Genomics. 2016;15:174–185. doi: 10.1093/bfgp/elv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choudhry H, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa S, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141:4618–4627. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adriaens C, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 55.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nature structural & molecular biology. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao C, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nature structural & molecular biology. 2014;21:997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the deep sequencing data from this study have been deposited in the Gene Expression Omnibus (GEO) under series accession number GSE90650. All the source data are available with the paper online. Other data from this study are available upon request.