Abstract
Background
Long non-coding RNAs (lncRNAs) have emerged as crucial regulators of tumor progression. However, the effects of lncRNAs in preeclampsia are not entirely clear. The aim of this study was to demonstrate the potential of lncRNAs to serve as biomarkers in preeclampsia.
Material/Methods
The RNA expression levels of lncRNAs (NR_027457, BC030099, AF037219, NR_024178, AF085938, G43016, G36948, NR_029420, NR_024015, AK002210, NR_026643, and AL049277) in the serum of patients with preeclampsia and in the serum of normal controls were measured by qRT-PCR. The area under the curve (AUC), the optimal cut-off values, the specificity, and the sensitivity of NR_027457, AF085938, G36948, and AK002210 were analyzed by receiver operating characteristic (ROC) curve analysis. We designed RNA interference species to suppress NR_027457 and G36948 and identified the roles of NR_027457 and G36948 in the functions of a trophoblast cell line (HTR-8/SVneo).
Results
The qRT-PCR results indicated that NR_027457 and AF085938 were significantly up-regulated, whereas G36948 and AK002210 were significantly down-regulated in preeclampsia. We found that NR_027457, AF085938, G36948, and AK002210 had potential diagnostic value for the detection of preeclampsia. Furthermore, the levels of NR_027457, AF085938, G36948, and AK002210 in the serum of patients were significantly different before vs. after surgery. The silencing of NR_027457 inhibited the proliferation, migration, and invasion abilities of HTR-8/SVneo cells, while the silencing of G36948 promoted the proliferation, migration, and invasion abilities of HTR-8/SVneo cells.
Conclusions
NR_027457, AF085938, G36948, and AK002210 can serve as potential diagnostic biomarkers in preeclampsia, and NR_027457 and G36948 might be involved in the pathogenesis of preeclampsia.
MeSH Keywords: Cell Migration Assays; Pre-Eclampsia; RNA, Long Noncoding
Background
Preeclampsia, which is characterized by high blood pressure and proteinuria, is a pregnancy-specific disease that can cause a series of problems to both mother and fetus [1,2]. Without proper and timely prevention, it may progress to a worse stage known as eclampsia, which is accompanied by seizures and other severe impairments, including the development of hemolysis, elevated liver enzymes, and low platelet (HELLP) syndrome, hemorrhagic or ischemic stroke, liver damage and dysfunction, kidney injury, and acute respiratory distress syndrome (ARDS) [3–5]. Preeclampsia usually occurs 20 weeks after the beginning of pregnancy, and it can be subdivided into 2 types: that which occurs before 34 weeks of gestation is termed early onset preeclampsia, and late onset preeclampsia, which occurs from ≥34 weeks to delivery [6,7]. Preeclampsia involves multiple risk factors, such as hypertension in pregnancy, diabetes, autoimmune disorders, and previous preeclampsia [8, 9]. The identification of pregnant women at risk for preeclampsia provides an opportunity for its prevention [10]. Unfortunately, most proposed primary prevention agents, including oral antioxidants, dietary, aspirin, and activity restrictions, lack proven efficacy [11–15]. In addition, preeclampsia is a multi-system disease that is associated with adverse maternal and fetal outcomes; therefore, to avoid morbidity and mortality, we cannot misdiagnose true patients with preeclampsia according to the clinical diagnostic criteria. Research by the International Society for the Study of Hypertension in Pregnancy (ISSHP) has also found that the lack of a consensus is the major reason for the controversy regarding the counselling, management, and documentation of pregnancy outcomes [16]. Therefore, preeclampsia has been defined as both hypertension and proteinuria (defined as ≥300 mg per 24 h or a spot urine protein/creatinine ratio of ≥30 mg/mmol) [16,17]. Many studies have been published on the functions and mechanisms of preeclampsia [18–20]. A greater understanding of the genetic and molecular pathways involved in the pathogenesis of preeclampsia is essential and may provide future targets for therapeutic action.
Reliable biomarkers for prediction and diagnosis would contribute to maternal health, and several promising markers have been suggested, including PlGF, PP13, endothelin-1, plasma-P-selectin, and PAPP-A [21–23]. Although preeclampsia can be diagnosed through the measurement of blood pressure and proteinuria as well as by other biochemical or biophysical markers, the expression of lncRNAs (long non-coding RNAs) and the roles they play in preeclampsia are unclear. LncRNA is defined as a non-protein coding transcript that is longer than 200 nucleotides and that functions as a regulator of gene expression; lncRNAs have vital effects on imprinting control, cell differentiation, immune responses, human diseases, and other biological processes [24,25]. An advantage to targeting lncRNAs over messenger RNAs (mRNAs) is that targeting a single lncRNA has the potential to affect multiple downstream pathways and therefore can amplify the effect of a single target [26]. Targeting multiple pathways may also lead to unwanted adverse effects, so the functions of lncRNAs should be completely characterized before they are used as therapeutic targets. Many different methods are currently being developed to silence lncRNAs in cancer, including siRNAs, antisense transcripts, and aptamers, which have potentially to be adapted for use in the treatment of preeclampsia [24,27].
Thus, we attempted to seek potential and valid lncRNAs to serve as biomarkers for the efficient and precise prediction and diagnosis of preeclampsia. Beyond that, we investigated the impact of alterations in candidate lncRNAs on cell function when their expression deviated from the normal level. The experimental results indicated that NR_027457, AF085938, G36948, and AK002210 may be potential predictive and diagnostic biomarkers for preeclampsia. Moreover, the expression changes in these lncRNAs would cause cell functional disorders.
Material and Methods
Patients and samples
Samples from patients who were diagnosed with preeclampsia were collected between 2011 and 2016 from the Lianyungang Maternal and Children’s Hospital; control samples were obtained from normal controls (pregnant women who were completely healthy) and were also provided by Lianyungang Maternal and Children’s Hospital. The patient data were fully recorded, and the samples were processed according to standard protocols. The exclusion criteria were as follows: patients with diabetes, chronic nephritis, chronic hypertension, heart diseases, autoimmune diseases, multiple pregnancies, thrombophilic conditions, vaginal delivery, fetal malformations, and HELLP syndrome. The clinical characteristics assessed at enrollment are shown in Table 1. The blood samples were centrifuged for 15 min at 3000×g at 4°C, and then the serum was collected and stored at −80°C. All protocols used in this study were accepted by the patients and were approved by the Ethics Review Board of Lianyungang Maternal and Children’s Hospital.
Table 1.
The clinical characteristics assessed at enrollment.
| Characteristic | Preeclampsia (n=81) | No Preeclampsia (n=81) | P-value |
|---|---|---|---|
| Age (years) | |||
| ≤35 | 57 (70.37%) | 59 (72.84%) | 0.43 |
| >35 | 24 (29.63%) | 22 (27.16%) | |
| BMI at enrollment | 26.9±1.48 (22.8–31.7) | 24.2±1.12 (21.4–28.6) | <0.001 |
| Blood pressure | |||
| Systolic | 113±4.81 (108–119) | 110±2.07 (101–114) | <0.001 |
| Diastolic | 69±4.74 (62–73) | 66±4.43 (61–70) | <0.001 |
| Proteinuria | 2.7±0.85 (0.3–6.4) | 0.12±0.05 (0–0.3) | <0.001 |
| Gestational age (years) | 33.2±2.69 (27–38) | 33.3±1.88 (26–38) | 0.78 |
| Previous pregnancy | 15 (19.75%) | 19 (23.46%) | 0.28 |
RNA extraction and quantitative real-time PCR (qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cDNA was synthesized using Oligod (T)18 primer, and the synthesis was catalyzed by PrimeScript® RT reagent Kit (Perfect Real Time) (TaKaRa, Japan). The primers used in the qPCR experiments are shown in Table 2. The reaction mixture contained 10 μL of 2×SYBR Premix Ex Taq™ (TaKaRa, Japan), 0.4 μL of 50×ROX Reference Dye II, 0.5 μL of 10 μM forward and reverse primers, 2 μL of template cDNA and ddH2O in a final volume of 20 μL. The qPCR reaction was performed with a denaturation step of 10 min at 95°C; followed by 35 cycles at 95°C (10 s), 60°C (40 s), 72°C (30 s), and a final step at 72°C for 10 min. The relative lncRNA expression levels were calculated using the 2−ΔΔCt method [28].
Table 2.
The primer sequences of the related lncRNAs.
| Gene | Sequence of primers (5′-3′) |
|---|---|
| NR_027457 | F: GTCCTTTGCAGCAACATGGA R: TTCAACCCATGCACCTCTCT |
| BC030099 | F: CAGCTGAGCGGGACATTTAC R: GGTCTGAAAGGATGGAGGCT |
| AF037219 | F: TGAAGCTGTTGAGGGAGGAG R: CCCTCCACCTTCTTTCCCAT |
| NR_024178 | F: ACAGGCTAGGAGAGAAGGGA R: CTTAAGCATCTTCACGGCCC |
| AF085938 | F: GAGCAACACAAGCAGCATCT R: TGAGAGGATATTGTGGCCGA |
| G43016 | F: TCAGTAATGCTCTAGTCCAGGG R: ACAGGTTGCTTGGTCTCAC |
| G36948 | F: AGACCTACCCTCGACCCAG R: TGGGAGTTCTGCTGGATTCA |
| NR_029420 | F: GAGACCTTGGCCAAACACAG R: TTGTCAGCTTTCTTGTGGGC |
| NR_024015 | F: CGCCCATTCTGCTCTTCTTC R: GGAGGTTTCAGAGATGGGCT |
| AK002210 | F: CTACCCCAGCTCGCATCATA R: TGACTTCGATCTGCCCACTT |
| NR_026643 | F: CTGGTACTGGGATTTGGGGT R: TGCTGACCTGGACAAGAGAG |
| AL049277 | F: AGCCTTTCCACTCCCAACTT R: TCTCCCCAGACTTGCCAAAC |
| GAPDH | F: TATGATGATATCAAGAGGGTAGT R: TGTATCCAAACTCATTGTCATAC |
Cell culture and siRNA transfection
The human immortalized first trimester trophoblast cell line HTR8/SVneo was used in the Transwell assay as previously depicted [29]. Briefly, HTR8/SVneo cells were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, USA) with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, USA) and were maintained in a humid atmosphere at 5% CO2 and 37°C. The cells were incubated in serum-free RPMI 1640 medium for different time intervals for serum starvation. Torin1 stimulation was performed by incubating the cells in fresh medium containing different concentrations of Torin1 for 24 h. Transfection of small interfering RNA (siRNA) was performed using Lipofectamine 2000 (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
Cell proliferation assay
An MTT assay was performed to evaluate the effects of si-lncRNA as previously reported [30]. Briefly, the cells were harvested with trypsin and seeded at a concentration of 2×105 cells/well in a 24-well plate. Then, 20 μl of MTT reagent was added to each well, and the cells were incubated at 37°C for 4 h, followed by the addition of 150 μl dimethylsulfoxide (DMSO). The absorbance was determined sequentially at 48 h using a microplate reader at 490 nm (Sunrise™; Tecan Group Ltd., Männedorf, Switzerland).
Cell migration and invasion assay
A Transwell assay was performed to examine the invasion and migration abilities of cells transfected with si-NR_027457, si-AF085938, si-G36948, or si-AK002210. For the invasion assay, 2×105 cells were suspended in 200 μl medium and then plated into the upper chamber of the Transwell plate (8 μm pore size; Corning Inc.; NY, USA), which was coated with Matrigel. Then, 0.5 ml medium supplemented with 10% FBS was added to the lower chamber. After incubation at 37°C for 24 h, a portion of cells remained on the surface of the upper membrane; these cells were removed with a cotton swab, and the cells on the surface of the reverse membrane were stained with crystal violet. The stained cells were examined by confocal microscopy, and the numbers of cells were calculated in at least 5 random images, including the upper, lower, left, right, and middle areas of each image. The general manipulation steps of the migration assay were the same as those described above but without Matrigel.
Statistical analysis
All statistical analysis was performed with SPSS version 19.0 (SPSS Inc, Chicago, IL, USA). The t test was used to detect variations in the data between patients and normal controls. Receiver operating characteristic (ROC) curves were generated to estimate the utility of each parameter as a tool for predicting preeclampsia, and the area under the ROC curves and the related parameters were also calculated in order to analyze the diagnostic points. The log-rank test was used to analyze survival, and the Kaplan-Meier method was used to plot the survival curves. In all analyses, p-value<0.05 was considered statistically significant.
Results
The expression levels of 12 lncRNAs in the serum of patients with preeclampsia
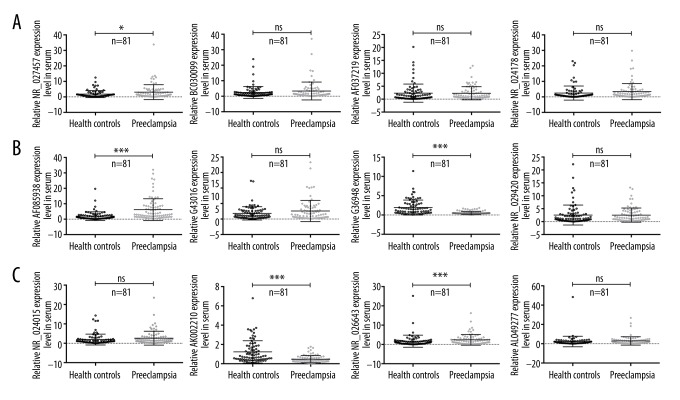
This study first determined the genome-wide lncRNA expression patterns in placentas derived from patients with preeclampsia using a Human LncRNA Microarray [27]. The results showed that clusters of lncRNAs were aberrantly expressed in placentas from patients with preeclampsia. Simultaneously, this study also demonstrated that LOC391533, LOC284100, and CEACAMP8 might contribute to the underlying mechanism of preeclampsia. In our study, we randomly selected relatively distinct lncRNAs, such as NR_027457, BC030099, AF037219, NR_024178, AF085938, G43016, G36948, NR_029420, NR_024015, AK002210, NR_026643, and AL049277. In this research, serum samples from all 81 patients with preeclampsia were obtained, and the expression levels of 12 various lncRNAs were detected by qRT-PCR according to the method described in a previous study. Compared with the normal controls, 4 lncRNAs were found to be differentially expressed in the serum samples of patients with preeclampsia; these lncRNAs were NR_027457, AF085938, G36948, and AK002210. The qRT-PCR results indicated that NR_027457 and AF085938 were significantly up-regulated, while G36948 and AK002210 were significantly down-regulated, and no significant difference was observed in the remainder of the lncRNAs that were tested (Figure 1A–1C).
Figure 1.
The expression levels of 12 different lncRNAs in the serum samples of patients with preeclampsia were examined by qRT-PCR. (A–C) In all, 81 serum samples were collected from patients with preeclampsia and from normal controls. The statistical results for the expression levels of lncRNA were demonstrated in healthy controls and in patients with preeclampsia, and each lncRNA is presented in an individual plot. The X axis shows the categories, the Y axis represents the relative expression levels of the lncRNAs (2−ΔΔCt), and ns indicates no significance (* P<0.05, ** P<0.01, *** P<0.001).
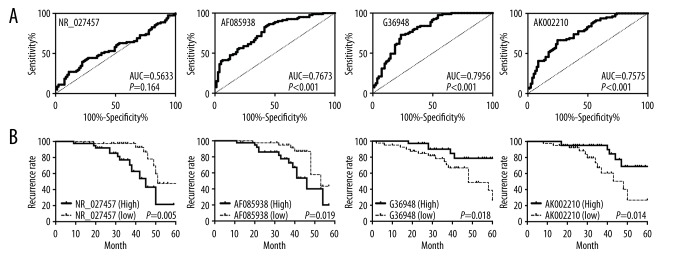
The correlation between the expression levels of 4 lncRNAs and preeclampsia
The differential expression of the lncRNAs NR_027457, AF085938, G36948, and AK002210 was verified by qRT-PCR, suggesting that they may have potential value as diagnostic markers of preeclampsia; moreover, the results clearly confirmed this via the ROC curves and the area under the ROC curves (AUC) of NR_027457, AF085938, G36948, and AK002210 (Figure 2A). Among these, the AUCs of AF085938 (AUC=0.7673, P<0.001), G36948 (AUC=0.7956, P<0.001), and AK002210 (AUC=0.7575, P<0.001) were larger than 0.75, which indicated that the performance of these 3 lncRNAs can be considered “fair” in the prediction of preeclampsia (Figure 2A). However, the results of NR_027457 (AUC=0.5633, P=0.164) were not as good as those for the previously mentioned 3 lncRNAs (Figure 2A). In addition, the Kaplan-Meier analysis showed that patients with higher expression of NR_027457 and AF085938 would be more likely to experience preeclampsia at an earlier time than patients who expressed lower levels; patients with lower expression of G36948 and AK002210 would also be more likely to experience preeclampsia at an earlier time than patients with higher levels. In summary, patients with high expression levels of NR_027457 and AF085938 are more likely to relapse than patients with low expression levels of NR_027457 and AF085938; patients with low expression levels of G36948 and AK002210 are more likely to relapse than those patients with high expression levels of G36948 and AK002210. Therefore, NR_027457, AF085938, G36948 and AK002210 indicate a good prognosis and have the potential to be valid markers for the prediction of preeclampsia (Figure 2B).
Figure 2.
The good prognostic effects of NR_027457, AF085938, G36948, and AK002210 in patients with preeclampsia. (A) The ROC curve for the 4 lncRNAs in their ability to predict preeclampsia. (B) Kaplan-Meier curve for the same lncRNAs in patients with a high or a low expression level of a particular lncRNA.
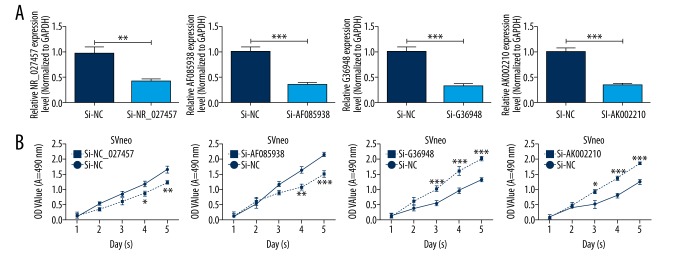
The effects of lncRNA NR_027457, AF085938, G36948, and AK002210 knockdown on cell proliferation
The patient-derived serum samples were tested, and the results showed that the lncRNAs NR_027457, AF085938, G36948, and AK002210 had high potential to be valid markers for the prediction of preeclampsia. For further research and for the validation of the availability of selecting these lncRNAs as markers, in vitro experiments were conducted. SVneo cells were transfected with the synthesized interfering oligonucleotide sequences si-NR_027457, si-AF085938, si-G36948, and si-AK002210 or with the corresponding control siRNA. As the results showed, the expression of all 4 lncRNAs in SVneo cells transfected with siRNAs was dramatically decreased compared with the control groups transfected with the negative control (si-NC) (Figure 3A). The results of the MTT assay showed that the proliferation rate of si-NR_027457- and si-AF085938-transfected cells was significantly inhibited, which suggests that the silencing of NR_027457 and AF085938 suppressed the proliferation of SVneo cells. In contrast, the proliferation rate of si-G36948- and si-AK002210-transfected cells was significantly promoted, which suggests that the silencing of G36948 and AK002210 promotes the proliferation of SVneo cells (Figure 3B). In summary, NR_027457, AF085938, G36948, and AK002210 play roles in cell proliferation.
Figure 3.
The silencing of NR_027457, AF085938, G36948, and AK002210 affected the proliferation abilities of SVneo cells. SVneo cells were transfected with negative control siRNAs (si-NC), NR_027457 siRNAs (si-NR_027457), AF085938 siRNAs (si-AF085938), G36948 siRNAs (si-G36948), or AK002210 siRNAs (si-AK002210). (A) The expression levels of NR_027457, AF085938, G36948 and AK002210 were detected by qRT-PCR assays (** P<0.01, *** P<0.001). (B) The proliferation abilities were measured by MTT assay in treated SVneo cells (* P<0.05, ** P<0.01, *** P<0.001).
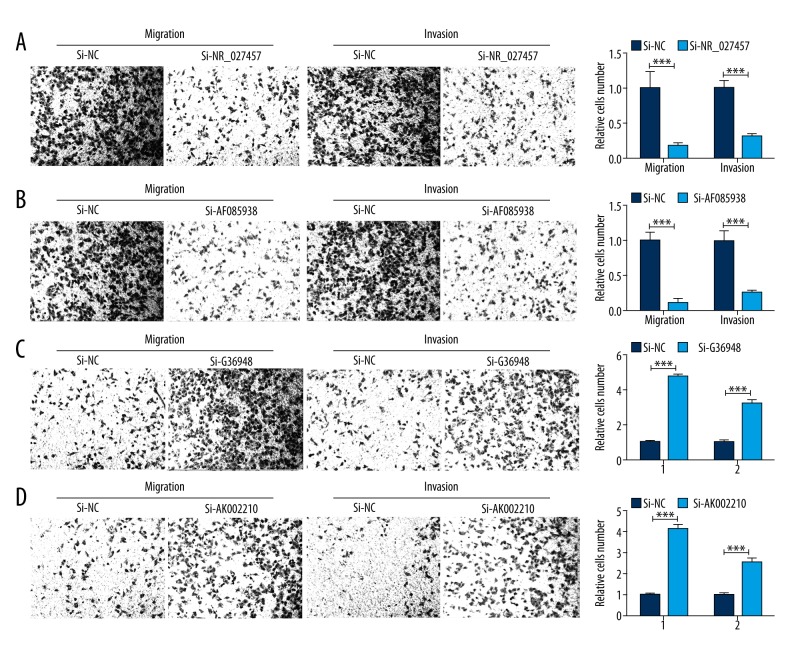
The effects of lncRNA NR_027457, AF085938, G36948, and AK002210 knockdown on cell invasion and migration
To explore the effects of NR_027457, AF085938, G36948, and AK002210 on the invasion and migration abilities of the cells, a Transwell assay was performed using SVneo cells transfected with si-NR_027457, si-AF085938, si-G36948, and si-AK002210 or with the negative control siRNA. The results showed that the knockdown of NR_027457 and AF085938 significantly suppressed the cell migration and invasion abilities in vitro (Figure 4A, 4B), while the silencing of G36948 and AK002210 in SVneo cells significantly promoted the cell migration and invasion abilities (Figure 4C, 4D). These results suggest that NR_027457, AF085938, G36948, and AK002210 act as important regulators in the SVneo cell line.
Figure 4.
The results of the Transwell assays revealed that the silencing of NR_027457, AF085938, G36948, and AK002210 suppressed the cell migration and invasion capabilities in vitro. Images obtained from the Transwell assay illustrate the migratory and invasive abilities of treated cells compared with those of cells treated with si-control; the statistical results are shown alongside the images. (A) The result of si-NR_027457 and the corresponding control. (B) The result of si-AF085938 and the corresponding control. (C) The result of si-G36948 and the corresponding control. (D) The result of si-AK002210 and the corresponding control (* P<0.05, ** P<0.01, *** P<0.001).
Discussion
Previous studies on the prediction, diagnosis, and treatment of preeclampsia have mainly concentrated on physiological and biochemical indexes [24,31–33]. However, no single biomarker can be regarded as a standard and valid diagnostic marker [34,35]. Some new predictive markers and therapeutics have been explored, and angiogenic factors, including soluble fms-like tyrosine kinase 1, an endogenous vascular endothelial growth factor inhibitor, have been implicated [36–39]. We attempted and managed to solve this problem by other approaches using lncRNAs as new diagnostic targets. LncRNAs have been demonstrated to be reliable biomarkers and are commonly used as biomarkers for the prediction and diagnosis of cancers [24,40,41]. To investigate the roles of lncRNAs in preeclampsia, above all, we explored numerous prior studies of lncRNA and preeclampsia to find some clues. The research by X He et al. provided a series of lncRNAs that were up-regulated or down-regulated in patients who were clinically diagnosed with preeclampsia by microarray [27]. From those lncRNAs, we selected NR_027457, BC030099, AF037219, NR_024178, AF085938, G43016, G36948, NR_029420, NR_024015, AK002210, NR_026643, and AL049277 as our research objects.
To confirm the different expression levels of these lncRNAs, serum samples were collected from patients with preeclampsia and from healthy controls and were tested by qRT-PCR. The results showed that the expression levels of NR_027457, AF085938, G36948, and AK002210 in the serum derived from patients with preeclampsia deviated from their normal expression levels, in accordance with a previous study [27]. Then, the data were further analyzed using the ROC curve and Kaplan-Meier analysis. As we expected, the AUC of these 4 lncRNAs were all larger than 0.5, and the AUC of AF085938, G36948, and AK002210 were even larger than 0.75 according to the ROC curves. Moreover, the significant potential of these lncRNAs to be diagnostic markers of preeclampsia was also shown by the Kaplan-Meier analysis. Since most diseases involve the dysfunction of cells, and preeclampsia is no exception, we sought to determine the roles of these 4 lncRNAs in cells that may be affected in preeclamptic patients by performing in vitro cell experiments (MTT analysis and Transwell assay). The expression of these 4 lncRNAs was knocked down in vitro by transfection of si-lncRNAs. The results showed that in the siRNA-transfected cells, the capacity of the cells to proliferate, migrate, and invade was remarkably affected. A large number of studies have drawn similar conclusions, and the lncRNA SPRY4-IT1 was demonstrated to modulate proliferation, migration, apoptosis, and network formation in the trophoblast cell line HTR-8SV/neo [42]. The lncRNA MEG3 was found to promote apoptosis and suppress the migration of trophoblast cells [43]. The lncRNA CCAT1 promotes colon cancer cell proliferation and invasion [44]. All these results indicate that NR_027457, AF085938, G36948, and AK002210 can be selected as biomarkers for the diagnosis of preeclampsia. Although the results were promising, some issues still need to be resolved. In vitro experiments have demonstrated that these 4 lncRNAs play roles in cell proliferation, migration, and invasion, but no in vivo data were obtained; therefore, the mechanism is still unclear. In addition, the number of included patients was too small, and more data are required to validate the conclusion of this study. Still, this research may provide a new perspective and a useful reference in the diagnosis of preeclampsia.
Conclusions
We demonstrated that NR_027457, AF085938, G36948, and AK002210 may serve as potential diagnostic biomarkers for preeclampsia. However, further studies are needed to validate this observation on the prognostic and therapeutic effects of NR_027457, AF085938, G36948, and AK002210 in preeclampsia, and in vivo studies are also needed. Moreover, it will be necessary to further explore the functions and mechanisms of NR_027457, AF085938, G36948, and AK002210 in preeclampsia.
Footnotes
Conflict of interests
None.
Source of support: The study was supported by the Social Development Project of Lianyungang Science and Technology Bureau (Grant Number SH1126)
References
- 1.Kornacki J, Skrzypczak J. [Results of Doppler examinations in fetuses of mothers with early- and late-onset preeclampsia]. Ginekol Pol. 2014;85:504–8. doi: 10.17772/gp/1761. [in Polish] [DOI] [PubMed] [Google Scholar]
- 2.Mol BWJ, Roberts CT, Thangaratinam S, et al. Pre-eclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 3.Jido TA, Yakasai IA. Preeclampsia: A review of the evidence. Ann Afr Med. 2013;12:75–85. doi: 10.4103/1596-3519.112395. [DOI] [PubMed] [Google Scholar]
- 4.Haram K, Mortensen JH, Nagy B. Genetic aspects of preeclampsia and the HELLP syndrome. J Pregnancy. 2014;2014:910751. doi: 10.1155/2014/910751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duarte AG. ARDS in pregnancy. Clin Obstet Gynecol. 2014;57:862–70. doi: 10.1097/GRF.0000000000000067. [DOI] [PubMed] [Google Scholar]
- 6.Lee G, Tubby J. Preeclampsia and the risk of cardiovascular disease later in life – A review of the evidence. Midwifery. 2015;31:1127–34. doi: 10.1016/j.midw.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Lisonkova S, Sabr Y, Mayer C, et al. Maternal morbidity associated with early-onset and late-onset preeclampsia. Obstet Gynecol. 2014;124:771–81. doi: 10.1097/AOG.0000000000000472. [DOI] [PubMed] [Google Scholar]
- 8.Pare E, Parry S, McElrath TF, et al. Clinical risk factors for preeclampsia in the 21st century. Obstet Gynecol. 2014;124:763–70. doi: 10.1097/AOG.0000000000000451. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed R, Dunford J, Mehran R, et al. Pre-eclampsia and future cardiovascular risk among women: a review. J Am Coll Cardiol. 2014;63:1815–22. doi: 10.1016/j.jacc.2014.02.529. [DOI] [PubMed] [Google Scholar]
- 10.Leffert LR. What’s new in obstetric anesthesia? Focus on preeclampsia. Int J Obstet Anesth. 2015;24:264–71. doi: 10.1016/j.ijoa.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Inversetti A, Smid M, Candiani M, et al. Predictive biomarkers of pre-eclampsia and effectiveness of preventative interventions for the disease. Expert Opin Biol Ther. 2014;14:1161–73. doi: 10.1517/14712598.2014.912271. [DOI] [PubMed] [Google Scholar]
- 12.Atallah A, Lecarpentier E, Goffinet F, et al. Aspirin for prevention of preeclampsia. Drugs. 2017;77(17):1819–31. doi: 10.1007/s40265-017-0823-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slomski A. Prophylactic low-dose aspirin reduces risk of preterm preeclampsia. JAMA. 2017;318:1099. doi: 10.1001/jama.2017.13073. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg K. Aspirin lowers the risk of preeclampsia. Am J Nurs. 2017;117:62. doi: 10.1097/01.NAJ.0000525880.12596.d7. [DOI] [PubMed] [Google Scholar]
- 15.Greene MF, Solomon CG. Aspirin to prevent preeclampsia. N Engl J Med. 2017;377:690–91. doi: 10.1056/NEJMe1708920. [DOI] [PubMed] [Google Scholar]
- 16.Tranquilli AL, Dekker G, Magee L, et al. The classification, diagnosis and management of the hypertensive disorders of pregnancy: A revised statement from the ISSHP. Pregnancy Hypertens. 2014;4:97–104. doi: 10.1016/j.preghy.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Hossain N, Khan N, Shah N, et al. Spot urine protein-creatinine ratio and 24-h urine protein excretion: Diagnostic accuracy in women with pre-eclampsia. Pregnancy Hypertens. 2014;4:87–90. doi: 10.1016/j.preghy.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Xu C, Li X, Guo P, Wang J. Hypoxia-induced activation of JAK/STAT3 signaling pathway promotes trophoblast cell viability and angiogenesis in preeclampsia. Med Sci Monit. 2018;24:4909–17. doi: 10.12659/MSM.905418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, Zhao J, Yi J, et al. Association between gene polymorphisms on Chromosome 1 and susceptibility to pre-eclampsia: An updated meta-analysis. Med Sci Monit. 2016;22:2202–14. doi: 10.12659/MSM.896552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakacak M, Kilinc M, Serin S, et al. Changes in Copper, Zinc, and Malondialdehyde levels and superoxide dismutase activities in pre-eclamptic pregnancies. Med Sci Monit. 2015;21:2414–20. doi: 10.12659/MSM.895002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawfield A, Freedman BI. Pre-eclampsia: The pivotal role of the placenta in its pathophysiology and markers for early detection. Ther Adv Cardiovasc Dis. 2009;3:65–73. doi: 10.1177/1753944708097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saleh L, Samantar R, Garrelds IM, et al. Low soluble fms-like tyrosine Kinase-1, Endoglin, and Endothelin-1 levels in women with confirmed or suspected preeclampsia using proton pump inhibitors. Hypertension. 2017;70:594–600. doi: 10.1161/HYPERTENSIONAHA.117.09741. [DOI] [PubMed] [Google Scholar]
- 23.Panaitescu AM, Akolekar R, Kametas N, et al. Impaired placentation in women with chronic hypertension that develop preeclampsia. Ultrasound Obstet Gynecol. 2017;50(4):496–500. doi: 10.1002/uog.17517. [DOI] [PubMed] [Google Scholar]
- 24.Aguilo F, Di Cecilia S, Walsh MJ. Long non-coding RNA ANRIL and polycomb in human cancers and cardiovascular disease. Curr Top Microbiol Immunol. 2016;394:29–39. doi: 10.1007/82_2015_455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Wu Z, Fu X, Han W. lncRNAs: Insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Balgkouranidou I, Matthaios D, Karayiannakis A, et al. Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res. 2015;778:46–51. doi: 10.1016/j.mrfmmm.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 27.He X, He Y, Xi B, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One. 2013;8:e81437. doi: 10.1371/journal.pone.0081437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Arimura G, Ozawa R, Maffei ME. Recent advances in plant early signaling in response to herbivory. Int J Mol Sci. 2011;12:3723–39. doi: 10.3390/ijms12063723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Ye Y, Yang X, et al. Expression-based in silico screening of candidate therapeutic compounds for lung adenocarcinoma. PLoS One. 2011;6:e14573. doi: 10.1371/journal.pone.0014573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sohlberg S, Stephansson O, Cnattingius S, Wikstrom AK. Maternal body mass index, height, and risks of preeclampsia. Am J Hypertens. 2012;25:120–25. doi: 10.1038/ajh.2011.175. [DOI] [PubMed] [Google Scholar]
- 32.Lucovnik M, Blickstein I, Verdenik I, et al. Impact of pre-gravid body mass index and body mass index change on preeclampsia and gestational diabetes in singleton and twin pregnancies. J Matern Fetal Neonatal Med. 2014;27:1901–4. doi: 10.3109/14767058.2014.892069. [DOI] [PubMed] [Google Scholar]
- 33.Karumanchi SA, Bdolah Y. Hypoxia and sFlt-1 in preeclampsia: The “chicken-and-egg” question. Endocrinology. 2004;145:4835–37. doi: 10.1210/en.2004-1028. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed H. Promoter methylation in prostate cancer and its application for the early detection of prostate cancer using serum and urine samples. Biomark Cancer. 2010;2:17–33. doi: 10.4137/BIC.S3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastek JA, Elovitz MA. The role and challenges of biomarkers in spontaneous preterm birth and preeclampsia. Fertil Steril. 2013;99:1117–23. doi: 10.1016/j.fertnstert.2013.01.104. [DOI] [PubMed] [Google Scholar]
- 36.Savaj S, Vaziri N. An overview of recent advances in pathogenesis and diagnosis of preeclampsia. Iran J Kidney Dis. 2012;6:334–38. [PubMed] [Google Scholar]
- 37.Uddin MN, Allen SR, Jones RO, et al. Pathogenesis of pre-eclampsia: Marinobufagenin and angiogenic imbalance as biomarkers of the syndrome. Transl Res. 2012;160:99–113. doi: 10.1016/j.trsl.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Park HJ, Shim SS, Cha DH. Combined screening for early detection of pre-eclampsia. Int J Mol Sci. 2015;16:17952–74. doi: 10.3390/ijms160817952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith TA, Kirkpatrick DR, Kovilam O, Agrawal DK. Immunomodulatory role of vitamin D in the pathogenesis of preeclampsia. Expert Rev Clin Immunol. 2015;11:1055–63. doi: 10.1586/1744666X.2015.1056780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 41.Martens-Uzunova ES, Bottcher R, Croce CM, et al. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–51. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Zou Y, Jiang Z, Yu X, et al. Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo. PLoS One. 2013;8:e79598. doi: 10.1371/journal.pone.0079598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Zou Y, Wang W, et al. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J Cell Biochem. 2015;116:542–50. doi: 10.1002/jcb.25004. [DOI] [PubMed] [Google Scholar]
- 44.He X, Tan X, Wang X, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–88. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]