Abstract
microRNAs (miRs) are a class of small non-coding RNAs that have been demonstrated to have a crucial role in tumorigenesis of human cancers, including gastric cancer (GC). Previous results have established that miR-100 participated in the development of GC; however, the underlying mechanism remains largely unknown. The preesent study utilized reverse transcription-quantitative polymerase chain reaction to analyze the expression of miR-100 in GC tissues and adjacent normal tissues. The present results indicated that the expression of miR-100 was downregulated in GC tissues when compared to the adjacent normal tissues. Furthermore, low miR-100 expression was observed to be associated with lymph node metastasis, tumor diameter and tumor stage. In addition, Kaplan-Meier analysis revealed that patients with low miR-100 expression tended to have a shorter overall survival. The miR-100 was further identified as an independent prognostic factor for overall survival. Notably, the levels of chemokine (CXC motif) receptor 7 (CXCR7) were inversely correlated with miR-100 in GC cell lines. Furthermore, miR-100 overexpression or CXCR7 depletion decreased in vitro GC cell proliferation. Bioinformatics analysis indicated that miR-100 may bind to the 3′-untranslated region of CXCR7 to prevent the initiation of protein translation. Thus, miR-100 may function as a tumor suppressor in GC, partly by regulating the expression of CXCR7, and the regulation of miR-100 expression may be a potential strategy for the treatment of GC patients.
Keywords: microRNA-100, CXCR7, gastric cancer, proliferation, biomarker
Introduction
Gastric cancer (GC) ranks as the second most frequently diagnosed cancer worldwide (1). Although the incidence and mortality rate of GC is decreasing in many developed countries owing to the widely used of the latest diagnostic and therapeutic technologies, yet GC is still the second-leading cause of cancer-related mortalities (2,3). At the same time, the prevalence of GC in Eastern Asia including China, Japan and Korea is still at a relative high level (4). Therefore, indentifying novel biomarkers with high sensitivity and specificity for GC early detection and prognosis prediction are still urgently needed.
MicroRNAs (miRNAs) are a class of small, endogenous, single-strand non-coding RNAs with a length of 17–25 nucleotides (5), which exerted their function through binding to the 3′ untranslated region (3′ UTR) of their target mRNAs (6). Accumulating evidences have revealed that altered expression of miRNAs play an important role in tumorigenesis of many human cancers (7–9). In the mean time, studies have demonstrated that a single cancer can be driven by different miRNAs (7,10,11), while a single miRNA can be found aberrant expressed in different cancers (11,12). As for GC, a number of miRNAs have reported to participate in a variety of physiological processes such as cell proliferation, differentiation, and apoptosis via regulating several target genes (13–15). miR-100, one member of the miR-100 family, is located at chromosome 11 at 11q24.1 (16). Extensive studies have been performed to evaluate the role of miR-100 in tumors but the results seemed to be controversial. For example, miR-100 was found to acting as tumor suppressor by deregulation its target genes in hepatocellular carcinoma (17), epithelial ovarian cancer (18), and breast cancer (19). However, miR-100 was also found to acting as oncogene in renal cell carcinoma (20) and acute myeloid leukemia (16). Therefore, the clinical significance of miR-100 in GC and the underlying mechanism still remained to be elucidated.
Chemokine (CXC motif) receptor 7 (CXCR7), also called as RDC-1, plays a key role in cell survival and tumor development (21). Growing evidences have found CXCR7 was highly expressed in a variety of human cancers, including GC (22–24). In particular, CXCR7 functions as a tumor promoter in esophageal squamous cancer and its expression can be regulated by miR-100 (25). However, whether or not miR-100 can regulate the expression of CXCR7 in GC requires more extensive investigations.
In the present study, the expression of miR-100 in GC tissues and cell lines was evaluated and its association with the clinicopathological features was explored. The effect of miR-100 on the overall survival of patients with GC was investigated. Furthermore, the role and mechanism of miR-100 on cell proliferation in vitro was explored. Understanding the clinical significance of miR-100 in GC may point to a new potential therapeutic target for GC.
Materials and methods
Cell lines and clinical samples
The normal gastric epithelial cell line GES-1 and the gastric cancer cell line BGC823 were obtained from the Chinese Institute of Biochemistry and Cell Biology (Shanghai, China). Cells were incubated in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 U/ml streptomycin in a humidified atmosphere with 5% CO2 at 37°C.
A total of 39 pairs of human gastric cancer tissues and adjacent noncancerous tissues were obtained from the gastric cancer patients, who underwent gastrectomy at Nantong Cancer Hospital between September 2005 and October 2010. The tissues were immediately frozen in liquid nitrogen and stored at −80°C until further processing. No patients have ever received any adjuvant treatments before surgery. The clinicopathological features were collected from medical records and summarized in Table I. This study was approved by the Ethics Committee of Nantong Cancer Hospital. Informed consent form has been obtained from all the participated patients.
Table I.
Relationship between miR-100 expression and clinicopathological features.
| miR-100 expression level | ||||
|---|---|---|---|---|
| Variable | Cases | High | Low | P-value |
| Age | 0.224 | |||
| ≥50 | 22 | 5 | 17 | |
| <50 | 17 | 4 | 13 | |
| Gender | 0.655 | |||
| Male | 19 | 4 | 15 | |
| Female | 20 | 5 | 15 | |
| Tumor diameter (cm) | 0.034 | |||
| ≥5 | 21 | 3 | 18 | |
| <5 | 18 | 6 | 12 | |
| Differentiation | 0.071 | |||
| Well/moderate | 20 | 6 | 14 | |
| Poor | 19 | 3 | 16 | |
| Tumor stage | 0.008 | |||
| I–II | 13 | 2 | 11 | |
| III | 26 | 7 | 19 | |
| Lymph node metastasis | 0.047 | |||
| Negative | 19 | 6 | 13 | |
| Positive | 20 | 3 | 17 | |
miR-100, microRNA-100.
Cell transfection
The miR-100 mimic, inhibitor, and negative control (NC) miRNA were purchased from RibiBio (Guangzhou, China). The siRNA targeting CXCR7 and negative control siRNA were designed and synthesized by RibiBio. Lipofectamine 2000 (Invitrogen, USA) was used to transfect siRNA, miR-100 inhibitor, miR-100 mimic and negative controls into the GES-1 and BGC823 cell line. All the procedures were performed following the manufacturer's instructions. The resulted cell lines were incubated in the aforementioned conditions. After 48 h incubation, the cell lines were harvested for RT-qPCR, western blot, and cell proliferation analyses.
RNA isolation and quantitative real-time PCR (RT-qPCR)
The expression level of miR-100 and CXCR7 was analyzed using the RT-qPCR method. Total RNA was extracted from the cultured cells using the Trizol reagent (Beyotime, Jiangsu. China) based on the provided protocol. The concentration and purity of RNA were spectrophotometrically determined by measuring the optical density (A260/280>2.0, A260/230>1.8) using a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientifc, Wilmington, DE, USA). To measure the expression of miR-100, 1 µg total RNA was used for reverse transcription to synthesize miRNA cDNA using miRNA cDNA synthesis kit (Invitrogen, USA) according to the instructions. RT-qPCR was performed using the Express SYBR-Green miRNA RT-qPCR kit (Invitrogen, USA). The PCR procedure was as follows: denaturation at 95°C for 2 min (1 cycle) and 95°C for 10 sec and 60°C for 1 min (40 cycles). U6 snRNA was used as an internal control for normalization.
To measure the expression of CXCR7, 1 µg total RNA was reverse transcribed to first-strand cDNA using the BeyoRT II cDNA first-strand synthesis kit (Beyotime, China). RT-qPCR was carried out using the BeyoFast SYBR Green qPCR Mix (Beyotime, China) with the optimized procedure (denaturation at 95°C for 10 min at 1 cycle, 95°C for 30 sec and 58°C for 30 sec (40 cycles)). β-actin was used as an endogenous control. The primers used in this study were as follows: miR-100: forward: 5′-GCTCTGAACCGTAGATCCGAAC-3′, reverse: 5′-GTGCAGGGTCCGAGGT-3′; U6 snRNA: forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse: 5′-AACGCTTCACGAATTTGCGT-3′, CXCR7: forward: 5′-AGCAGCAGGAGGAAGATGGT-3′, reverse: 5′-TCTCATTGTTGGACGCAGAC-3′, β-actin: forward: 5′-AGAAAATCTGGCACCACACC-3′, reverse: 5′-TAGCACAGCCTGGATAGCAA-3′. Fold-change in expression was calculated using the 2−ΔΔCt method (26).
Western blot analysis
Total protein was extracted from the cells using RIPA lysis buffer (Beyotime, China) according to the instructions provided by the manufacturer. The protein concentration was quantified using the Bradford protein concentration determination kit (Beyotime, China) according to the recommended protocol. Equal amount of protein samples were separated using 10% SDS-PAGE gel and then transferred to PVDF membrane. The membrane was then incubated with primary antibody (anti-CXCR7: 1:1,000, bs-4897R, Bioss antibody Inc., Woburn, MA, USA; anti-β-actin: 1:1,000, bs-50545R, Bioss antibody Inc., USA) at 4°C overnight after blocked by 5% fat-free milk in TBST. After that, the membrane was incubated with TBST for three times. Following that, the membrane was incubated with horseradish peroxidase (HRP) conjugated secondary antibody (1:1,000, bs-0296G-HRP, Bioss antibody Inc., USA) for 1 h at room temperature. The band was then developed using the enhanced chemiluminescence kit (Beyotime, USA) according to the supplier's recommendation. The results of protein expression were quantified with Quantity One software (Bio-Rad, Hercules, CA, USA). The expression of β-actin was used as an internal control. Each sample was repeated in triplicate.
Luciferase assay
The CXCR7-3′ UTR (both wild type and mutant type) was obtained from GenePharma Co. Ltd., (Shanghai, China) and inserted into pmirGLO plasmid (Promega, Madison, WI, USA). Co-transfection was performed in BGC-823 cells with Lipofectamine 2000 (Invitrogen, USA). At 48 h after transfection, the relative luciferase activity was analyzed using the Dual-luciferase reporter assay kit (Promega, USA).
Cell proliferation analysis
The cell proliferation rate was analyzed using the widely used MTT assay. In brief, the cells were incubated into 96-well plate at a density of about 1×104 cells/ml. The cell proliferation rate was analyzed at the selected time points (0, 24, 48, and 72 h after seeding). At the selected time points, 20 µl MTT (5 mg/ml, Beyotime, USA) was added into each well and the cells were cultured at the aforementioned condition for additional 4 h. After that, the medium was discarded and 150 µl DMSO was added to each well. The optical density of each well was measured using a multi-wavelength spectrophotometer (Powerwave XS, Bio-Tek, Taipei, Taiwan, R.O.C.) at 490 nm. Each sample was repeated in triplicate.
Statistical analysis
Data analysis was performed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). Data were presented as mean ± SD of at least three independent experiments. The difference between groups was analyzed using Chi-square test. The overall survival was analyzed using the Kaplan-Meier method and log-rank test. Prognostic factors analyses were estimated by multivariate Cox regression. P-value less than 0.05 was considered to be statistically significant.
Results
The expression of miR-100 in GC tissues and cell line
To examine the expression pattern of miR-100 in GC, 39 pairs of GC tissues and the adjacent noncancerous tissues were quantified by RT-qPCR. As shown in Fig. 1A, we found the expression level of miR-100 was down-regulated in 30 of 39 (76.92%) GC tissues. Therefore, these patients were classified into the low miR-100 expression group and the rest of them were classified into the high miR-100 expression group accordingly. Also, the average expression level of miR-100 in tumor tissues was significantly lower in GC tissues than in adjacent noncancerous tissues (P<0.05, Fig. 1B). In addition, the expression level of miR-100 in GC cell line (BGC823) and normal gastric epithelial cell line (GES-1) was measured by the same method. As expected, the expression of miR-100 in BGC823 cell line was significantly lower than in GES-1 cell line (P<0.05, Fig. 1C). Collectively, our results revealed the miR-100 was frequently down-regulated in GC tissues and cell line.
Figure 1.
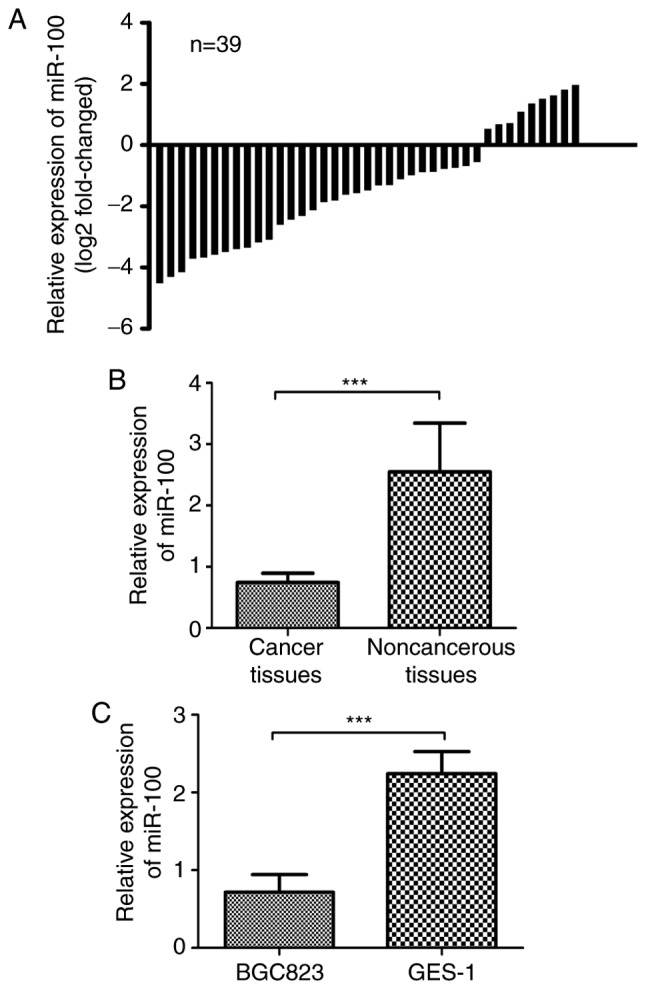
miR-100 is downregulated in gastric cancer. (A) RT-qPCR analysis of the expression of miR-100 in 39 pairs of gastric cancer tissues and surrounding noncancerous tissues. The data were represented as log2-fold change (cancer/normal). (B) The expression of miR-100 in gastric cancer tissues was significant lower than in the surrounding noncancerous tissues. (C) RT-qPCR analysis the expression of miR-100 in gastric cancer cell line (BGC823) and normal gastric epithelial cell line (GES-1) (NS: Not significance, ***P<0.001). miR-100, microRNA-100.
The correlation between miR-100 expression and clinicopathological features of GC
We then analyzed the correlation between miR-100 expression and the clinicopathological features in GC patients. As summarized in Table I, we found the miR-100 expression was closely associated with lymph node metastasis (P=0.047), tumor diameter (P=0.034) and tumor stage (P=0.008). However, no significant association was observed between miR-100 expression and age, gender, and differentiation (all P>0.05).
Clinical significance of miR-100 in GC
To explore the prognostic significance of miR-100 in GC patients, we measured the correlation between miR-100 expression and the overall survival of 39 GC patients using Kaplan-Meier analysis and log-rank test. We found the miR-100 expression was significantly correlated with the GC patients' overall survival (P=0.041, Fig. 2). In other words, the patients with low miR-100 expression had a poorer overall survival compared with the patients with high miR-100 expression. Furthermore, we found the miR-100 expression showed unfavorable prognosis in GC patients (P=0.024), along with lymph node metastasis (P=0.042), tumor diameter (P=0.031) and tumor stage (P=0.014) (Table II). Besides that, the miR-100 expression was proven to be a poor independent predictor for GC patients through multivariate analysis (P=0.019, Table II).
Figure 2.
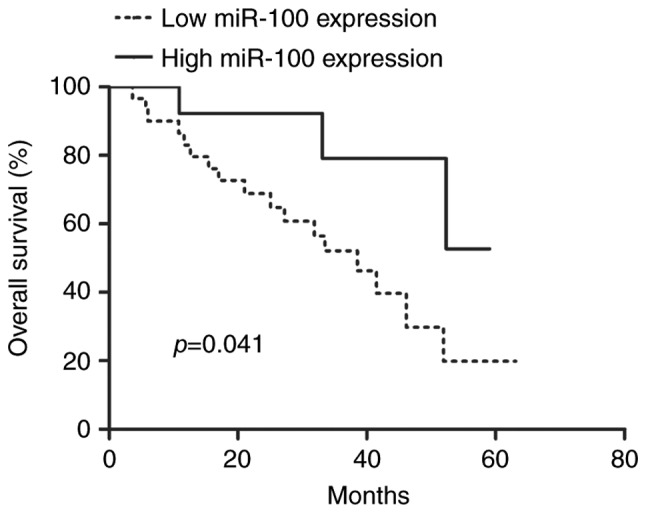
Kaplan-Meier overall survival curve for the high miR-100 expression group (n=9) and the low miR-100 expression group (n=30). The difference in overall survival rate between the two groups was significant. miR-100, microRNA-100.
Table II.
Univariate and multivariate analyses of overall survival rate.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| miR-100 | 2.704 | 1.140–6.417 | 0.024 | 2.764 | 1.186–6.441 | 0.019 |
| Age | 2.309 | 0.867–6.150 | 0.094 | – | – | – |
| Gender | 2.044 | 0.701–5.960 | 0.191 | – | – | – |
| Tumor diameter | 2.643 | 1.093–6.395 | 0.031 | 2.637 | 1.090–6.383 | 0.032 |
| Differentiation | 2.489 | 0.985–6.293 | 0.054 | – | – | – |
| Tumor stage | 2.832 | 1.237–6.483 | 0.014 | 3.232 | 1.335–7.823 | 0.009 |
| Lymph node metastasis | 2.560 | 1.036–6.329 | 0.042 | 2.578 | 1.044–6.367 | 0.040 |
HR, hazard ratio; CI, confidence interval; miR-100, microRNA-100.
Upregulation of miR-100 inhibits cell proliferation in vitro
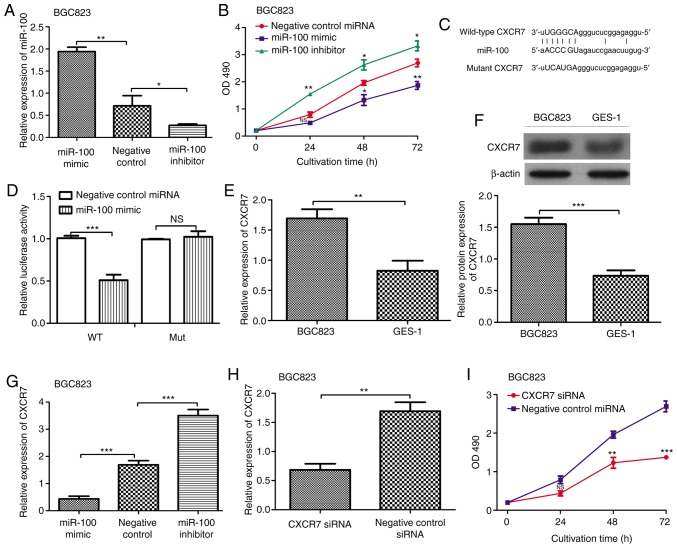
To understand the role of miR-100 expression in tumor progression of GC, we analyzed the effect of miR-100 expression on GC cell proliferation through miR-100 mimic, miR-100 inhibitor, negative control miRNA transfection. It is noteworthy the BGC823 cell line with miR-100 mimic transfection had the highest miR-100 expression compared with the cell line with miR-100 inhibitor or negative control miRNA transfection (Fig. 3A). Then, the cell proliferation rate of the above-mentioned cell lines was analyzed. Conversely, we found the BGC823 cell line with miR-100 mimic had the lowest cell proliferation rate compared with the cell line with miR-100 inhibitor or negative control miRNA transfection (Fig. 3B). Taken together, our results suggested that the miR-100 expression could inversely regulate cell proliferation rate and thus we deduct the miR-100 might play a tumor suppressor role in GC.
Figure 3.
miR-100 regulates the expression of CXCR7 in vitro. (A) RT-qPCR analysis of the expression of miR-100 in gastric cancer cell line (BGC823) after miR-100 mimic, inhibitor, and negative control miRNA transfection. (B) MTT assay to determine the level of cell proliferation in gastric cancer cell line (BGC823) after miR-100 mimic, inhibitor, and negative control miRNA transfection. (C) Putative miR-100-binding sequence within the 3′-UTR of CXCR7 mRNA. (D) Luciferase gene reporter gene assays were performed to verify the relationship between miR-100 and CXCR7. (E) RT-qPCR and (F) Western blot analysis of the expression of CXCR7 in gastric cancer cell line (BGC823) and normal gastric epithelial cell line (GES-1). (G) RT-qPCR analysis of the expression of CXCR7 in gastric cancer cell line (BGC823) after miR-100 mimic, inhibitor, and negative control miRNA transfection. (H) RT-qPCR analysis of the expression of CXCR7 in gastric cancer cell line (BGC823) after CXCR7-sepcific siRNA and negative control siRNA transfection. (I) MTT assay to determine the level of cell proliferation in gastric cancer cell line (BGC823) after CXCR7-sepcific siRNA and negative control siRNA transfection (NS: Not significance, *P<0.05, **P<0.01, ***P<0.001). miR-100, microRNA-100; CXCR7, Chemokine (CXC motif) receptor 7; siRNA, small-interfering RNA; OD490, optical density at 490 nm.
CXCR7 is a target gene of miR-100
To further explore the mechanisms underlying the inhibitory role of miR-100 in the proliferation of GC cells, the computational prediction website TargetScan was used to identify functionally targets of miR-100. Interestingly, we found the CXCR7 contains a putative target sequence of miR-100 in 3′-UTR (Fig. 3C). Meanwhile, the luciferase reporter assay demonstrated that the luciferase activity in BGC-823 cells with wild-type 3′-UTR CXCR7 and miR-100 mimic transfection was significantly lower than those with mutant 3′-UTR CXCR7 and miR-100 mimic transfection (Fig. 3D). Then, the expression of CXCR7 in GC cell line BGC823 and normal gastric epithelial cell line GES-1 was examined by RT-qPCR and western blot. The data showed that the expression of CXCR7 in the GES-1 cell line was significantly decreased compared to that in the BGC823 cell line (Fig. 3E and F). Furthermore, the expression of CXCR7 in the miR-100 mimic, miR-100 inhibitor, and negative control miRNA transfection BGC823 cell line was examined. The results presented in Fig. 3G illustrated that the transfection of miR-100 mimic could decrease CXCR7 mRNA expression level, which means the expression of CXCR7 could be inversely regulated by miR-100. In addition, the effect of CXCR7 expression on BGC823 cell proliferation was analyzed through CXCR7-specific siRNA and negative control siRNA transfection. The results demonstrated that the CXCR7-specific siRNA could impair both the expression of CXCR7 and the cell proliferation of BGC823 cell line (Fig. 3H and I). These results demonstrated that up-regulation of miR-100 expression could suppress the cell proliferation of GC cell line BGC823, at least partly through down-regulating the expression of CXCR7.
Discussion
Dysregulation of miRNAs in diseases especially in tumor has attracted extensive attentions in recent years (7–12). Mounting evidences have demonstrated that aberrant expression of several miRNAs is associated with survival outcome of cancer patients (27,28) and therefore could be regarded as potential therapeutic targets for these cancers. Take GC as an example, numerous miRNAs have been identified involved in GC development, but their underlying molecular mechanism in GC development is still poorly understood (7,14). Therefore, in this present study, we aimed to identify new miRNA biomarker that could be used as predictor for the overall survival of GC patients and also with the hope to reveal the underlying mechanism of how miRNA participated in the development of GC.
miR-100 has been found aberrant expressed in several human cancers but its role in tumorigenesis is still controversial (16–20). The reason for that might be it can regulate different downstream target genes and therefore it can have different and even opposite functions in tumors. Therefore, in this study, we first examined the expression pattern of miR-100 in GC tissues. Our results showed that the expression of miR-100 was clearly downregulated in GC tissues compared with the surrounding noncancerous tissues (P<0.05), which implies the miR-100 might play an important role in the progression of GC. Following, we analyzed the expression of miR-100 in GC cell line. Not surprisingly, the same trend was found when compared with the expression of miR-100 in GC cell line and the normal gastric epithelial cell line.
Since miR-100 is frequently down-regulated in GC tissues, we then analyzed the association between miR-100 expression and clinicopathological features. We found the miR-100 expression was closely associated with lymph node metastasis (P=0.047), tumor diameter (P=0.034) and tumor stage (P=0.008). The survival analysis demonstrated that the low miR-100 expression predicts poor prognosis in GC patients, which highlighted the importance of miR-100 in GC. Finally, we identified the miR-100 could be used as an independent predictor for the overall survival of GC patients through the univariate and multivariate analyses. Taken together, we explored the expression pattern of miR-100 in GC and revealed the clinical importance of miR-100 in GC. However, the biological function of miR-100 in GC is still unknown.
Previous studies demonstrated that miR-100 could inhibit cell proliferation, migration, and differentiation in human cancers (19,29–30). Therefore, the effect of miR-100 on cell proliferation was evaluated in vitro. We found the level of cell proliferation in GC cell line was obviously higher than in normal gastric epithelial cell line (P<0.05). By transferring the miR-100 mimic and inhibitor into GC cell line, we found forced the expression of miR-100 could inhibit GC cell proliferation. However, the transfection of miR-100 inhibitor could promote GC cell proliferation. Collectively, we deducted that miR-100 expression could suppress the cell proliferation in GC.
Numerous miR-100 target genes have been identified in previous literatures (17,19,25,29,30). Using the bioinformatical tools, we found the 3′-UTR of CXCR7 has a putative binding site for miR-100 and the luciferase reporter assay confirmed CXCR7 is a direct target of miR-100. Importantly, the CXCR7 was found overexpressed in GC in previous study (24). Therefore, we first examined the expression of CXCR7 in GC cell line and normal gastric epithelial cell line. In accordance with the previous finding, we found the CXCR7 was overexpressed in GC cell line. Besides that, the expression of CXCR7 was found inversely correlated with the expression of miR-100. Following, the forced downregulate expression of CXCR7 in GC cell line could impair the cell proliferation. Taken together, we deducted the CXCR7 might be a downstream target for miR-100 in GC and the miR-100 could exerts its oncogenic role be targeting CXCR7.
In summary, we found the miR-100 was downregulated in GC and was correlated with the poor prognosis of GC. Another important finding in this study was we found CXCR7 was the downstream target gene of miR-100. These results indicate that miR-100 deregulation may play important roles in tumor growth and that miR-100 may be a potential therapeutic target for the treatment of GC.
References
- 1.Tinico A, Gottardi LF, Boechat ED. Gastric cancer in the excluded stomach 10 years after gastric bypass. Case Rep Surg. 2015;2015:468293. doi: 10.1155/2015/468293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Malvezzi M, Bonifazi M, Bertuccio P, Levi F, La Vecchia C, Decarli A, Negri E. An age-period-cohort analysis of gastric cancer mortality from 1950 to 2007 in Europe. Ann Epidemiol. 2010;20:898–905. doi: 10.1016/j.annepidem.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Sionov RV. MicroRNAs and glucocorticoid-induced apoptosis in lymphoid maligancies. ISRN hematol. 2013;2013:348212. doi: 10.1155/2013/348212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santpere G, Lopez-Valenzuela M, Petit-Marty N, Navarro A, Espinosa-Parrila Y. Differences in molecular evolutionary rates among microRNAs in the human and chimpanzee genomes. BMC Genomics. 2016;17:528. doi: 10.1186/s12864-016-2863-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen Y, Gong JM, Zhou LL, Sheng JH. Mir-451 as a new tumor marker for gastric cancer. Oncotarget. 2017;8:56542, 56545. doi: 10.18632/oncotarget.17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. doi: 10.1186/1476-4598-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donohoe OH, Henshilwood K, Way K, Hakimjavadi R, Stone DM, Walls D. Identification and characterization of cyprinid herpesvirus-3 (CyHV-3) encoded MicroRNAs. PLoS One. 2015;10:e0125434. doi: 10.1371/journal.pone.0125434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Yu JH. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2017 May 8; doi: 10.1139/bcb-2016-0254. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 11.Xie M, Dart DA, Guo T, Xing XF, Cheng XJ, Du H, Jiang WG, Wen XZ, Ji JF. MicroRNA-1 acts as a tumor suppressor microRNA by inhibiting angiogenesis-related growth factors in human gastric cancer. Gastric Cancer. 2017 May 10; doi: 10.1007/s10120-017-0721-x. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Zang Y, Wang T, Pan J, Gao F. miR-215 promotes cell migration and invasion of gastric cancer cell lines by targeting FOXO1. Neoplasma. 2017;64:579–587. doi: 10.4149/neo_2017_412. [DOI] [PubMed] [Google Scholar]
- 14.Jafarzadeh-Samani Z, Sohrabi S, Shirmohammadi K, Effatpanah H, Yadegarazari R, Saidijam M. Evaluation of miR-22 and miR-20a as diagnostic biomarkers for gastric cancer. Chin Clin Oncol. 2017;6:16. doi: 10.21037/cco.2017.03.01. [DOI] [PubMed] [Google Scholar]
- 15.Hu CE, Du PZ, Zhang HD, Huang GJ. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR-145. Cell Physiol Biochem. 2017;42:13–21. doi: 10.1159/000477107. [DOI] [PubMed] [Google Scholar]
- 16.Qin C, Huang RY, Wang ZX. Potential role of miR-100 in cancer diagnosis, prognosis, and therapy. Tumor Biol. 2015;36:1403–1409. doi: 10.1007/s13277-015-3267-8. [DOI] [PubMed] [Google Scholar]
- 17.Zhou HC, Fang JH, Luo X, Zhang L, Yang J, Zhang C, Zhuang SM. Downregulation of microRNA-100 enhances the ICMT-Rac1 signaling and promotes metastasis of hepatocellular carcinoma cells. Oncotarget. 2014;5:12177–12188. doi: 10.18632/oncotarget.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azizmohammadi S, Azizmohammadi S, Safari A, Kosari N, Kaghazian M, Yahaghi E, Seifoleslami M. The role and expression of miR-100 and miR-203 profile as prognostic markers in epithelial ovarian cancer. Am J Tansl Res. 2016;8:2403–2410. [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Q, He M, Guan S, Ma M, Wu H, Yu Z, Jiang L, Wang Y, Zong X, Jin F, Wei M. MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/β-catenin signaling pathway. Tumor Biol. 2016;37:5001–5011. doi: 10.1007/s13277-015-4342-x. [DOI] [PubMed] [Google Scholar]
- 20.Gu L, Li H, Chen L, Ma X, Gao Y, Li X, Zhang Y, Fan Y, Zhang X. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: A systematic review and meta-analysis. Oncotarget. 2015;6:32545–32560. doi: 10.18632/oncotarget.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Teng XY, Meng XP, Wang BS. Expression of stromal cell-dervied factor 1 and CXCR7 ligand receptor system in pancreatic adenocarcinoma. World J Surg Oncol. 2014;12:348. doi: 10.1186/1477-7819-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattermann K, Held-Feindt J, Lucius R, Müerköster SS, Penfold ME, Schall TJ, Mentlein R. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptoic effects. Cancer Res. 2010;70:3299–3308. doi: 10.1158/0008-5472.CAN-09-3642. [DOI] [PubMed] [Google Scholar]
- 24.Nambara S, Iguchi T, Oki E, Tan P, Maehara Y, Mimori K. Overexpression of CXCR7 is a novel prognostic indicator in gastric cancer. Dig Surg. 2017;34:312–318. doi: 10.1159/000452977. [DOI] [PubMed] [Google Scholar]
- 25.Zhou SM, Zhang F, Chen XB, Jun CM, Jing X, Wei DX, Xia Y, Zhou YB, Xiao XQ, Jia RQ, et al. miR-100 suppresses the proliferation and tumor growth of esophageal squamous cancer cells via targeting CXCR7. Oncol Rep. 2016;35:3453–3459. doi: 10.3892/or.2016.4701. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.White NM, Yousef GM. MicroRNAs: Exploring a new dimension in the pathogenesis of kidney cancer. BMC Med. 2010;8:65. doi: 10.1186/1741-7015-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamali Z, Asl Aminabadi N, Attaran R, Pournagiazar F, Ghertasi Oskouei S, Ahmadpour F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2015;51:321–331. doi: 10.1016/j.oraloncology.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Fu HL, Pan HX, Zhao B, Dong BC, Shao L, Fu GS, Wang Q, Li M. MicroRNA-100 inhibits bone morphogenetic protein-induced osteoblast differentiation by targeting Smad1. Eur Rev Med Pharmacol Sci. 2016;20:3911–3919. [PubMed] [Google Scholar]
- 30.Luan YX, Zhang S, Zuo L, Zhou L. Overexpression of miR-100 inhibits cell proliferation, migration, and chemosensitivity in human glioblastoma through FGFR3. Onco Targets Ther. 2015;8:3391–3400. doi: 10.2147/OTT.S85677. [DOI] [PMC free article] [PubMed] [Google Scholar]