Abstract
Background & Aims
Inhibitors of the epidermal growth factor receptor (EGFR) are the first-line therapy for patients with metastatic colorectal tumors without RAS mutations. However, EGFR inhibitors are ineffective in these patients, and tumor level of EGFR does not associate with response to therapy. We screened human colorectal tumors for EGFR-positive myeloid cells and investigated their association with patient outcome. We also performed studies in mice to evaluate how EGFR expression in tumor cells and myeloid cells contributes to development of colitis-associated cancer and ApcMin-dependent intestinal tumorigenesis.
Methods
We performed immunohistochemical and immunofluorescent analyses of 116 colorectal tumor biopsies to determine levels of EGFR in tumor and stroma; we also collected information on tumor stage and patient features and outcomes. We used the Mann-Whitney U and Kruskal-Wallis tests to correlate tumor levels of EGFR with tumor stage, and the Kaplan-Meier method to estimate patients’ median survival time. We performed experiments in mice lacking EGFR in intestinal epithelial cells (Villin-Cre; Egfrf/f and Villin-CreERT2; Egfrf/f mice) or myeloid cells (LysM-Cre; Egfrf/f mice) on a mixed background. These mice were bred with ApcMin/+ mice; colitis-associated cancer and colitis were induced by administration of dextran sodium sulfate (DSS), with or without azoxymethane (AOM), respectively. Villin-CreERT2 was activated in developed tumors by administration of tamoxifen to mice. Littermates that expressed full-length EGFR were used as controls. Intestinal tissues were collected; severity of colitis, numbers and size of tumors, and intestinal barrier integrity were assessed by histologic, immunohistochemical, quantitative reverse transcription polymerase chain reaction, and flow cytometry analyses.
Results
We detected EGFR in myeloid cells in the stroma of human colorectal tumors; myeloid cell expression of EGFR associated with tumor metastasis and shorter patient survival time. Mice with deletion of EGFR from myeloid cells formed significantly fewer and smaller tumors than the respective EGFR-expressing controls in an ApcMin/+ background as well as after administration of AOM and DSS. Deletion of EGFR from intestinal epithelial cells did not affect tumor growth. Furthermore, tamoxifen-induced deletion of EGFR from epithelial cells of established intestinal tumors in mice given AOM and DSS did not reduce tumor size. EGFR signaling in myeloid cells promoted activation of STAT3 and expression of survivin in intestinal tumor cells. Mice with deletion of EGFR from myeloid cells developed more severe colitis after DSS administration, characterized by increased intestinal inflammation and intestinal barrier disruption, than control mice or mice with deletion of EGFR from intestinal epithelial cells. EGFR-deficient myeloid cells in the colon of DSS-treated LysM-Cre; Egfrf/f mice had reduced expression of interleukin 6 (IL6), and epithelial STAT3 activation was reduced compared with controls. Administration of recombinant IL6 to LysM-Cre; Egfrf/f mice given DSS protected them from weight loss and restored epithelial proliferation and STAT3 activation, compared with administration of DSS alone to these mice.
Conclusions
Increased expression of EGFR in myeloid cells from the colorectal tumor stroma associates with tumor progression and reduced survival time of patients with metastatic colorectal cancer. Deletion of EGFR from myeloid cells, but not intestinal epithelial cells, protects mice from colitis-induced intestinal cancer and ApcMin-dependent intestinal tumorigenesis. Myeloid cell expression of EGFR increases activation of STAT3 and expression of survivin in intestinal epithelial cells and expression of IL6 in colon tissues. These findings indicate that expression of EGFR by myeloid cells of the colorectal tumor stroma, rather than the cancer cells themselves, contributes to tumor development.
Keywords: Tumor Microenvironment, Cytokine, CRC, Colon Cancer
Abbreviations used in this paper: AOM, azoxymethane; BrdU, bromodeoxyuridine; CAC, colitis-associated cancer; CRC, colorectal cancer; DSS, dextran sodium sulfate; EGFR, epidermal growth factor receptor; FITC, fluorescein isothiocyanate; HCC, hepatocellular carcinoma; IEC, intestinal epithelial cell; IEL, intestinal epithelial layer; IHC, immunohistochemistry; IL, interleukin; IP, intraperitoneal; LP, lamina propria; pSTAT3, tyrosine-705-phosphorylated STAT3; rIL6, recombinant IL6; TMA, tissue microarray
Editor's Notes.
Background and Context
EGFR inhibitors are used in combination with chemotherapy in a subset of metastatic colorectal cancer patients. However, therapy response does not correlate to EGFR expression in tumor cells.
New Findings
The researchers show that EGFR expression and activation in myeloid cells promotes colorectal cancer in mice and correlate with bad prognosis in metastatic colorectal cancer patients.
Limitations
The exact myeloid cell population expressing EGFR, thereby influencing tumor growth is yet unknown.
Impact
EGFR expression in myeloid cells is a novel biomarker for the prognosis of colorectal cancer.
Colorectal cancer (CRC) is the third most common cancer in the United States with 5-year survival rates less than 15% for patients with metastasis.1 CRC originates from intestinal epithelial cells (IECs) at the crypt base due to multistage loss of tumor suppressor genes—especially APC—with concomitant accumulation of activating oncogenic mutations, such as in the KRAS gene.2 Besides heritable genetic alterations and environmental factors, one risk factor for tumor development is inflammatory bowel disease, leading to so-called colitis-associated cancer (CAC).3 As first-line treatment of metastatic CRC, combinations of chemotherapies together with targeted therapies like angiogenic (vascular endothelial growth factor) inhibitors and anti–epidermal growth factor receptor (EGFR) antibodies are used.4
The EGFR is a receptor tyrosine kinase that is implicated in a variety of epithelial cancers by controlling cellular proliferation, differentiation, barrier integrity, and survival.5 60%–80% of patients with CRC overexpress EGFR, which is associated with poor prognosis.6 Targeted inhibition of EGFR using monoclonal antibodies like cetuximab and panitumumab, represents one of the standard therapies of metastatic CRC and—combined with chemotherapies—provides survival benefit over chemotherapy alone.7 However, treatment response is limited to patients without activating KRAS mutations.4 Interestingly, treatment response does not correlate with the levels of EGFR expression in tumor cells. There also are a considerable number of nonresponders to anti-EGFR therapies in patients with KRAS wild-type state,8 highlighting the complex and converse roles of EGFR in CRC development.
Several studies indicate a protective role of EGFR in CRC. Using the IL10−/− mouse model of CAC, it was shown that reduced EGFR signaling in the antimorphic Egfrwa5/+ or the hypomorphic Egfrwa2/wa2 background9, 10 augments colitis severity and accelerates and increases tumor development. Furthermore, azoxymethane/dextran sodium sulfate (AOM/DSS)-induced CAC is more invasive in Egfrwa5/+ mice11 and Egfrwa2/wa2 mice exhibit increased severity of DSS- or oxazolone-induced colitis.12, 13 In a clinical trial, localized EGFR stimulation alleviates symptoms of colitis.14 Different studies also support a pro-tumorigenic role of EGFR: diminished EGFR signaling in Egfrwa2/wa2 mice or by treatment with pharmacological EGFR inhibitors reduces tumor formation in the AOM/DSS model of CAC and in the ApcMin model of intestinal tumorigenesis.15, 16, 17 Finally, patient data show that EGFR is required for formation of aberrant crypt foci.18
However, it is unknown how the influence of EGFR on tumorigenesis depends on the cell type from which it is expressed. Interestingly, reduced EGFR signaling in all cells by use of Egfrwa2 mice leads to defective intestinal adaptation after small bowel resection, whereas conditional EGFR deletion in IECs neither affects adaptation19 nor severity of DSS- or oxazolone-induced colitis.13 Conversely, lack of EGFR selectively in myeloid cells reduces severity of DSS-induced colitis.20 These observations collectively point toward a role of EGFR in nonepithelial cells of the intestine, namely myofibroblasts and monocytes/macrophages of the lamina propria (LP), in which EGFR expression has been reported.21, 22
Macrophages are common in CRC and influence tumor behavior at multiple levels.23 Their function seems to be dependent on tissue, location, and microenvironment, as both positive and negative outcomes of CRC have been associated with macrophage infiltration.24, 25, 26, 27 Interleukin 6 (IL6) production by macrophages has been implicated in the prognosis of CAC, as IL6-deficient mice show reduced tumor burden.28 Moreover, elevated serum levels of IL6 directly correlate with poor clinical prognosis in different human cancers.29 Recently we could demonstrate in a mouse model of chemically induced hepatocellular carcinoma (HCC) that EGFR expression in tissue-resident liver macrophages (Kupffer cells) plays a tumor-promoting role by regulating IL6 production in response to tissue injury and thus HCC formation. Further, EGFR expression in Kupffer cells of patients with HCC correlates with poor prognosis.30
In this study, we screened human CRC tumor biopsies for the presence of EGFR-positive myeloid cells to analyze if this impinges on disease outcome. Moreover, by using mouse models, we investigated in which specific cell type EGFR is required for colitis and CAC as well as oncogene-driven CRC development. We demonstrate that EGFR is expressed on myeloid cells within the tumor of patients with CRC which negatively affects overall survival of patients with metastatic CRC. In mice, expression of EGFR in myeloid cells, but not in IECs, promotes AOM/DSS-induced CAC and ApcMin-dependent intestinal tumorigenesis and protects from DSS-induced colitis in an IL6-dependent manner. This study provides mechanistic insight into the complex cell type–specific role of EGFR in CRC and finally marks a step toward the improvement of individualized cancer treatment.
Methods
Clinical Material
Histological blocks of 124 patients diagnosed with CRC, who had undergone surgical resection without any preoperative treatment between 2008 and 2011, were selected to obtain tissue microarrays (TMA) on informed consent as described.31 From the most representative areas of each donor tissue sample, a single core with a diameter of 2 mm was arranged into one recipient paraffin block (3.0 × 2.5 cm) using a semiautomatic tissue arrayer (Galileo TMA, Isenet, LLC, Philadelphia, PA). All cases were diagnosed at National Cancer Institute Fondazione ‘G. Pascale’ of Naples and at Medical Oncology, Seconda Università degli Studi of Naples, and staged according to the TNM classification (version dependent on year of diagnosis). Clinicopathological characteristics including demographics and staging features were evaluated. EGFR expression scores were obtained from immunohistochemistry (IHC) staining by 2 independent persons by intensity. For all analyses, the same TMAs were used. Due to different dropping out of cores, patient numbers varied slightly among the histological analyses.
Colitis and Tumorigenesis
Colitis was induced by administering 2.5% DSS (0216011080; MP Biomedicals, Santa Ana, CA) in autoclaved drinking water for 5 days followed by 5 days of normal water. Colon tumors were induced as described with mild modifications.32 Briefly, mice were injected intraperitoneally (IP) with 7.5 mg/kg AOM (A5486; Sigma-Aldrich, St. Louis, MO) followed by regular diet and water for 5 days. After that, mice received water with 2.5% DSS for 5 days, followed by maintenance on regular water for 16 days and 2 additional DSS cycles (5 days of 2.5% DSS and 4 days of 2% DSS). Mice were sacrificed 9 days after the last treatment. For inducible EGFR deletion after tumor development, 33 days after AOM, mice were injected IP with 1 mg tamoxifen (sunflower seed oil/ethanol mixture, 10:1; Sigma-Aldrich)33 for 5 days, followed by tamoxifen injection every 2 days until analysis. Recombinant murine IL6 (12340065; ImmunoTools, Friesoythe, Germany) or vehicle was administered as described.34, 35 For IL6 depletion, mice were injected with 100 μg anti-mouse IL6 antibody and isotype (12-4301; Thermo Fisher Scientific, Waltham, MA), respectively.
Additional Methods
All further methods can be found in the Supplementary Materials and Methods section.
Results
EGFR Expression in Myeloid Cells Is a Prognostic Factor for Metastatic CRC
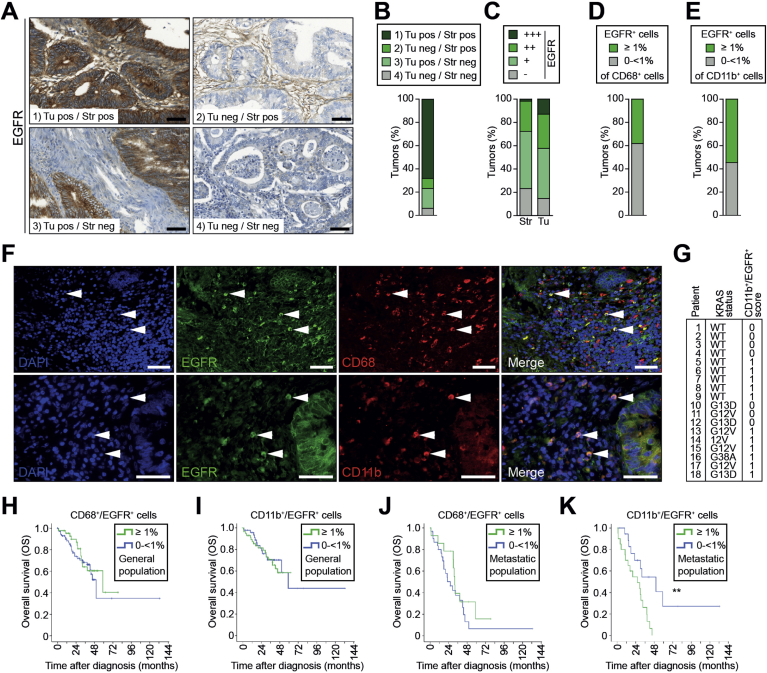
We have previously shown that presence of EGFR in tumor-associated macrophages (EGFR+ TAMs) in patients with HCC correlated with decreased disease-free and overall survival.30 Therefore, we investigated 116 human TMA samples of patients with surgically resected CRC for EGFR expression in both stromal and tumor cells by IHC (Figure 1A). EGFR expression was highly variable in intensity and location and patients could be subdivided into 4 groups: (1) positive in tumor and stroma; (2) negative in tumor and positive in stroma; (3) positive in tumor and negative in stroma; or (4) negative in tumor and stroma, with more than 60% of patients belonging to group 1 (Figure 1A and B). To visualize differences in EGFR expression in stroma and tumor, samples were further classified as IHC score +++ (n = 15), ++ (n = 34), + (n = 50), and − (n = 17) for tumor cells and IHC score +++ (n = 2), score ++ (n = 30), + (n = 57), and − (n = 27) for stromal cells according to the respective staining intensity (Figure 1C, Supplementary Figure 1A). With respect to group classification according with EGFR expression in stroma and tumor, correlation analyses showed no association with clinicopathological features, with exception of metastatic status at diagnosis (P = .03) (Supplementary Table 1A). No correlation between any clinical parameter and EGFR intensity in tumor or stromal cells was found (Supplementary Table 1B,C). Next, we investigated by immunofluorescence if specific stromal cell types showed EGFR expression. We performed double staining for EGFR and the macrophage marker CD68 as well as for the pan myeloid marker CD11b. Most of the analyzed samples showed presence of CD68+ (n = 114/118) or CD11b+ (n = 106/108) cells in the stromal compartment (Figure 1F). The presence of CD68+/EGFR+ and CD11b+/EGFR+ double positive cells varied highly, with 60% and 36% being the maximum of the total CD68+ and CD11b+ cell population, respectively (Supplementary Figure 1B). Staining for tyrosine-1068-phosphorylated EGFR (pEGFR) showed a strong correlation between EGFR expression and its phosphorylation, indicating that EGFR in myeloid cells is indeed activated (Supplementary Figure 1B–D). To stratify patients for the absence or presence of EGFR in myeloid cells or TAMs, samples were scored as negative (−) if they contained less than 1% and as positive (+) if they contained ≥1% of double positive cells out of the respective total number of CD68+ TAMs or CD11b+ myeloid cells. According to this classification, 73 samples were negative and 45 were double positive for CD68 and EGFR (Figure 1D), and 49 samples were negative and 59 were double positive for CD11b and EGFR (Figure 1E). When correlating these results with patient data, CD68+/EGFR+ cells did not show any association with clinicopathological features. However, moderately differentiated tumors (G2) displayed a higher percentage of double positive CD11b+/EGFR+ cells compared with less differentiated tumors (G3) (P = .03) (Supplementary Table 1D,E). The distribution of double positive myeloid cells was comparable among the 18 patients with known RAS status. From the patients with wild-type RAS, 5 of 9 scored positive (>1%) for CD11b+/EGFR+ cells and similarly, 6 of 9 patients were positive among the patients with mutated RAS (Figure 1G). In the total population, overall survival was not influenced by the presence of CD68+/EGFR+ or CD11b+/EGFR+ cells (Figure 1H and I). In patients with metastatic disease (at diagnosis or during follow-up), no correlation was found between survival and presence of CD68+/EGFR+ double positive TAMs (Figure 1J). However, metastatic patients showed significantly reduced overall survival with CD11b+/EGFR+ double positive cells in their tumors when compared with those who were negative (Figure 1K), suggesting that the presence of EGFR+ myeloid cells and not of just TAMs is negatively affecting overall survival of patients with metastatic CRC.
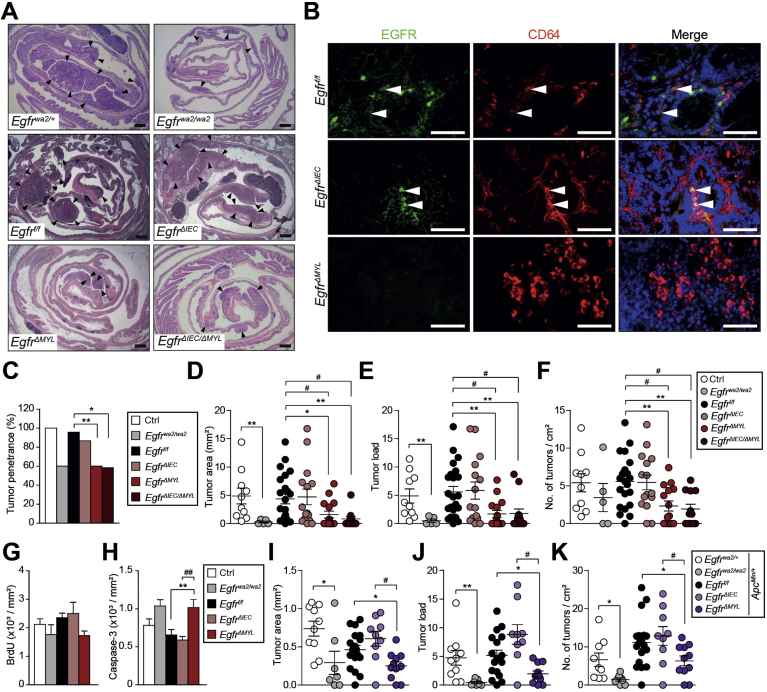
Figure 1.
EGFR expression in CD11b+ myeloid cells reduces overall survival of patients with metastatic CRC. (A) IHC showing EGFR expression in human CRC. Four different EGFR staining patterns: (1) tumor/stroma EGFR positive, (2) tumor negative/stroma positive, (3) tumor positive/stroma negative, (4) tumor and stroma negative. Scale bars 50 μm. (B) Patient stratification according to staining patterns shown in (A) (n = 116). (C) Patient stratification according to immunohistochemical EGFR expression intensity in tumor (Tu) and stromal (Str) cells (n = 116). (D, E) Patients were divided into 2 groups: low (0 to <1%) and high (≥1%), according to percentage of (D) CD68+/EGFR+ in the total CD68+ stromal population (n = 118) and (E) CD11b+/EGFR+ cells in the total CD11b+ stromal population (n = 108), determined by immunofluorescence shown in (F). (F) Immunofluorescence double staining on CRC samples shows presence of both CD68+/EGFR+ (upper panel) and CD11b+/EGFR+ (lower panel) cells in the stroma. Scale bars 50 μm. (G) Distribution of CD11b+/EGFR+ (0: 0 to <1%; 1: ≥1%) among Patients with CRC with wild-type or mutant RAS. (H–K) Overall survival (OS) of all (H and I) and metastatic (J and K) patients with CRC with low (0 to <1%) or high (≥1%) numbers of (H and J) CD68+/EGFR+ cells among the total CD68+ stromal population (general population: 118 patients; n = 73 with low and n = 45 with high counts, metastatic population: 45 patients; n = 31 with low and n = 14 with high counts) and (I and K) CD11b+/EGFR+ cells among the total CD11b+ stromal population (general population: 108 patients; n = 49 with low and n = 59 with high counts, metastatic population: 39 patients; n = 19 with low and n = 20 with high counts). Metastatic patients with high numbers of CD11b+/EGFR+ cells had a median OS of 26.2 months; 95% confidence interval, 9.1–43.4. Those with low CD11b+/EGFR+ numbers had a median OS of 50.2 months; 95% confidence interval, 22.0–78.2). **P = .005, log-rank test. neg, negative; pos, positive.
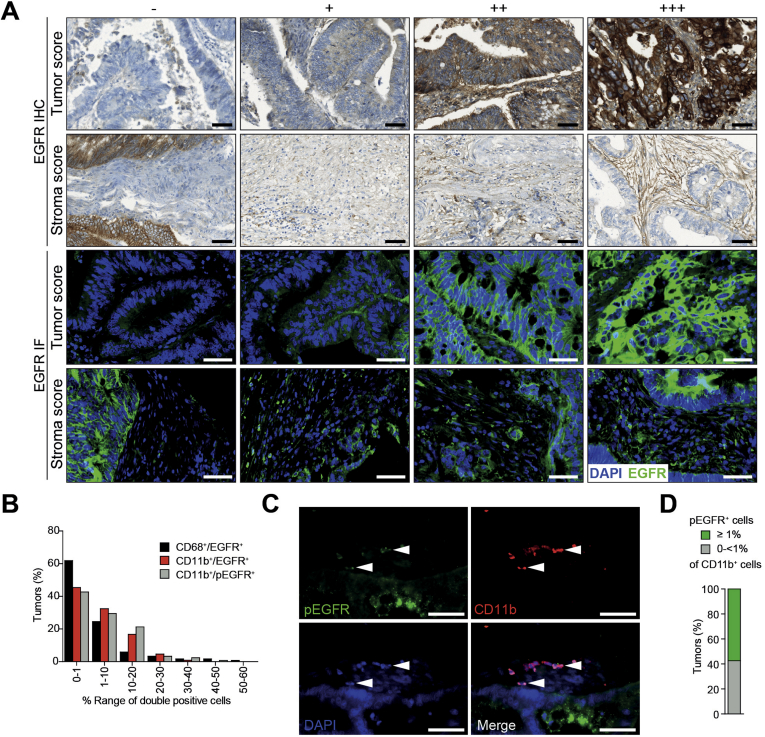
Supplementary Figure 1.
EGFR expression and phosphorylation in tumor and stromal cells of patients with CRF. (A) Representative images used for IHC scoring of EGFR expression (−, +, ++, +++) in tumor and stromal cells (upper panels). Intensities of EGFR determined by IF (lower panels) were comparable with IHC. (B) Distribution of tumors according to the percentage of CD68+/EGFR+ (n = 118), CD11b+/EGFR+ (n = 108), and CD11b+/pEGFR+ (n = 113) cells in the total CD68+ or CD11b+ stromal population, respectively, determined by IF double staining shown in (C) and Figure 1F. (C) IF double staining on CRC samples shows presence of CD11b+/pEGFR+ cells in the stroma. Scale bars 50 μm. (D) Patients were divided into 2 groups: low (0 to <1%) and high (≥1%), according to percentage of CD11b+/pEGFR+ cells in the total CD11b+ stromal population (n = 113), determined by IF shown in (C).
EGFR Signaling in Myeloid Cells Promotes Formation of CAC and ApcMin/+-driven Intestinal Tumors
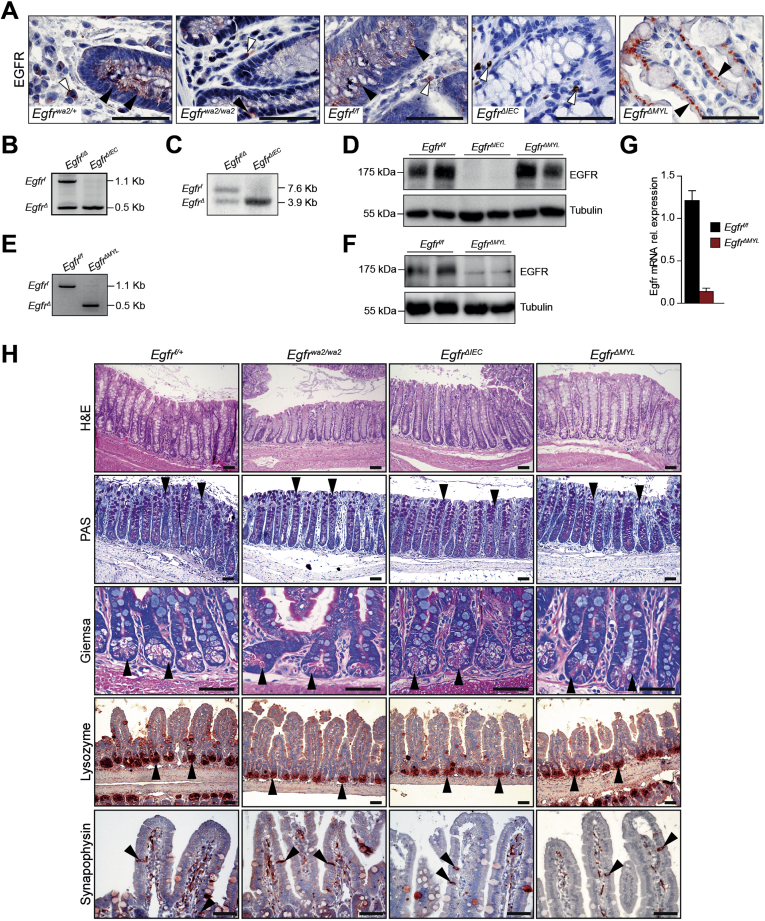
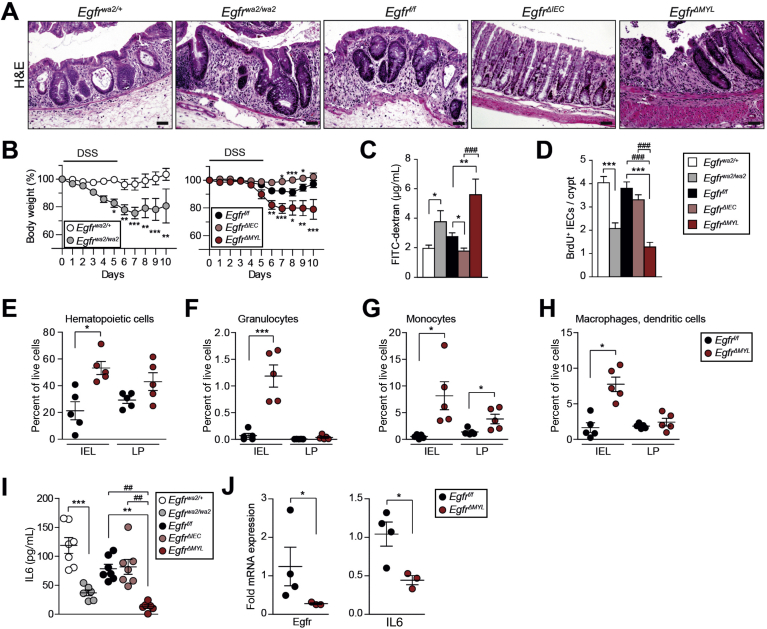
Because we observed that EGFR expression in myeloid cells correlated with patient overall survival, whereas expression in epithelial cells did not, we further investigated EGFR signaling in different intestinal cell populations during development of CAC in vivo. We used mouse models with conditional EGFR deletion in either IECs (EgfrΔIEC) or myeloid cells (EgfrΔMYL). EgfrΔIEC mice were generated by breeding Egfrf/f mice with Villin-Cre mice that start to express Cre recombinase at approximately embryonic day 10.36 EgfrΔMYL animals were obtained by crossing Egfrf/f animals with LysMCre/+ mice, which express Cre recombinase in myeloid cells.37 As a comparison model for overall reduced EGFR signaling, we used hypomorphic Egfrwa2/wa2 mice.10 Successful EGFR deletion in IECs of EgfrΔIEC mice was shown by IHC, also demonstrating abundant EGFR expression in nonepithelial cells of the LP in both Egfrf/f and EgfrΔIEC mice. Accordingly, IHC analysis did not show any EGFR expression in LP cells of EgfrΔMYL mice, whereas expression was maintained in IECs (Supplementary Figure 2A). IEC- and myeloid cell–specific EGFR deletion in EgfrΔIEC and EgfrΔMYL mice was confirmed by polymerase chain reaction, Southern blot, Western blot, and quantitative reverse-transcriptase polymerase chain reaction analysis in isolated IECs and cultured bone marrow–derived macrophages, respectively (Supplementary Figure 2B–G). All mice were viable and displayed no obvious gut abnormalities. Histological examination of adult Egfrwa2/wa2, EgfrΔIEC, and EgfrΔMYL mice showed proper cellular composition and differentiation in both colon and small intestine (Supplementary Figure 2H), as shown previously.17, 19, 20 These data demonstrate that neither lack of EGFR in IECs or myeloid cells nor overall EGFR depletion affects intestinal structure or cell differentiation.
Supplementary Figure 2.
EGFR expression and intestinal development. (A) EGFR IHC staining on DSS-treated colons of indicated mice, black arrowheads depict IECs, white arrowheads depict LP cells. (B) PCR and (C) Southern blot analysis of genomic DNA from purified colonocytes showing successful Cre-mediated recombination of the floxed Egfr allele in EgfrΔIEC mice. (D) Western blot confirming absence of EGFR protein in purified colonocytes from EgfrΔIEC mice. (E) PCR analysis of purified bone marrow–derived macrophages showing successful Cre-mediated recombination of the floxed Egfr allele in EgfrΔMYL mice. (F) Western blot confirming loss of EGFR protein in bone marrow–derived macrophages from EgfrΔMYL mice. (G) Quantitative reverse transcriptase PCR analysis of EGFR mRNA expression of bone marrow–derived macrophages showing absence of EGFR in EgfrΔMYL mice. mRNA expression levels were normalized to TBP (Egfrf/f n = 2, EgfrΔMYL n = 5). (H) Hematoxylin-eosin (H&E) and periodic acid-Schiff (PAS) staining on colon sections, arrowheads depict goblet cells. Giemsa staining on small intestine, arrowheads point to refractive eosinophilic granules of paneth cells. IHC staining for lysozyme and synaptophysin, arrowheads demonstrate presence of paneth cells and enteroendocrine cells, respectively. Scale bars 50 μm.
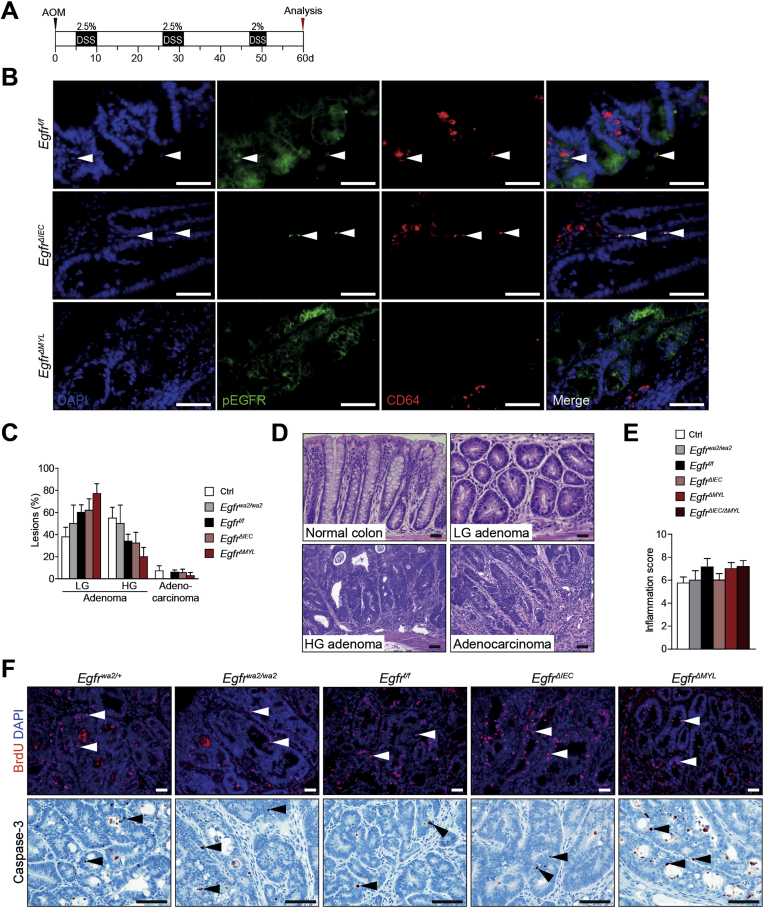
As chronic colitis is a significant risk factor for development of CRC, we applied the AOM/DSS model for CAC investigation (Supplementary Figure 3A).38 CAC developed in animals of all genotypes (Figure 2A) and CD64+/EGFR+ monocytes/macrophages were detected in all tumors except those derived from EgfrΔMYL mice (Figure 2B). Moreover, EGFR was activated in CD64+ cells of Egfrf/f and EgfrΔIEC mice, as evidenced by the presence of CD64+/pEGFR+ double positive cells. No pEGFR was detectable in EgfrΔMYL mice (Supplementary Figure 3B). Tumor penetrance, area, load, and multiplicity were similar in EgfrΔIEC and Egfrf/f mice (Figure 2C–F), suggesting that EGFR signaling in IECs is not required for CAC formation. In contrast, EgfrΔMYL mice with EGFR ablation in myeloid cells had a significantly lower tumor penetrance than controls (Figure 2C). Further, EgfrΔMYL animals exhibited significant reduction in tumor area, load, and multiplicity, with most tumors localized in the distal colon (Figure 2A and D–F). Tumor area and load of Egfrwa2/wa2 mice with hypomorphic EGFR alleles in all body cells were also significantly smaller than those of controls (Figure 2D and E), confirming previous studies15 and suggesting that myeloid cell–specific deficiency of EGFR signaling impairs CRC development. EgfrΔIEC/ΔMYL compound conditional knockout mice lacking EGFR in both IECs and myeloid cells had significantly reduced tumor number and size and significantly reduced overall tumor burden compared with Egfrf/f mice (Figure 2A and C–F). As tumor development in EgfrΔIEC/ΔMYL mice was analogous to EgfrΔMYL mice, we concluded that EGFR signaling in myeloid cells is the major contributor to CAC oncogenesis. Tumors of all genotypes were predominantly low-grade adenomatous lesions, with EgfrΔMYL mice having the highest proportion. Only a minor percentage of tumors progressed to high-grade adenomas or adenocarcinomas (Supplementary Figure 3C and D). There was no difference in the histological inflammation score among genotypes (Supplementary Figure 3E). All genotypes showed comparable levels of proliferation (Figure 2G, Supplementary Figure 3F). However, the rate of apoptosis was significantly increased in tumors of EgfrΔMYL mice (Figure 2H, Supplementary Figure 3E). These data indicate that EGFR signaling in myeloid cells supports the survival of CRC cells, thereby promoting tumorigenesis.
Supplementary Figure 3.
EGFR signaling in myeloid cells in the AOM/DSS-dependent model of CAC. (A) Scheme of AOM/DSS-administration to induce CAC; 7.5 mg/kg AOM was injected IP; DSS was given in drinking water (black areas) followed by regular water (white areas). (B) IF double staining for CD64 and pEGFR. Arrowheads depict presence of CD64+/pEGFR+ cells. (C) Grading of neoplastic lesions. Percentage of low-grade (LG) and high-grade (HG) adenomas as well as adenocarcinomas in indicated genotypes (n ≥4). (D) Hematoxylin-eosin stainings of different lesion grades as indicated. (E) Histological assessment of inflammation scores on colon sections of AOM/DSS-treated mice (n ≥4). (F) BrdU IF and cleaved Caspase-3 IHC stainings, arrowheads depict BrdU+ and cleaved Caspase-3+ nuclei in tumor cells, respectively. Scale bars 50 μm.
Figure 2.
EGFR signaling in myeloid cells promotes formation of colitis-associated and ApcMin-driven intestinal tumorigenesis. (A) Hematoxylin-eosin colon staining of AOM/DSS-treated mice. Arrowheads depict tumors. Scale bars 1 mm. (B) Immunofluorescence double staining of EGFR/CD64 on colorectal tumors. Arrowheads depict presence of EGFR+/CD64+ cells. Scale bars 50 μm. (C) Tumor penetrance in AOM/DSS-treated mice (ctrl [control], n = 10; Egfrwa2/wa2, n = 5; Egfrf/f, n = 22; EgfrΔIEC and EgfrΔMYL, n = 15; EgfrΔIEC/ΔMYL, n = 12). *P < .05, **P < .01, Fisher’s exact test. (D–F) Analysis of tumor formation in AOM/DSS-treated mice (see [C] for number of mice). (D) Tumor area, (E) load (tumor area (×10–2)/colon area), and (F) multiplicity (number of tumors/cm2 colon area). (G) Proliferation index (BrdU-incorporated tumor cells/area, 9 tumors from 3 different mice were counted). Data are mean ± SEM. (H) Apoptotic index (cleaved Caspase-3+ tumor cells/area, 14–28 tumors from 3–6 different mice were counted). (I–K) Analysis of tumor formation in the ApcMin background (ApcMin/+;Egfrwa2/+, n = 9; ApcMin/+;Egfrwa2/wa2, n = 8; ApcMin/+;Egfrf/f, n = 18; ApcMin/+;EgfrΔIEC, n = 9; ApcMin/+;EgfrΔMYL, n = 11). (I) Tumor area, (J) load, and (K) multiplicity. Data from (D–K) are mean ± SEM, *P < .05, **P < .01, t test. # P < .05, ## P < .01, 1-way analysis of variance with Dunn’s posttest.
To investigate the role of EGFR signaling in a colitis-independent intestinal tumor background, we used the ApcMin/+ model, in which we confirmed that reduced EGFR in all cells leads to tumor reduction.17 Similar to the AOM/DSS model, no change in tumor size, load, and multiplicity was observed in ApcMin/+;EgfrΔIEC mice when compared with ApcMin/+;Egfrf/f controls. In contrast, deletion of EGFR in myeloid cells led to markedly reduced tumor size, load, and number in ApcMin/+;EgfrΔMYL animals (Figure 2I–K). Therefore, EGFR signaling in myeloid cells promotes intestinal tumor formation also in an oncogene-driven model of intestinal tumorigenesis.
Targeting EGFR in IECs of Established Tumors Does Not Lead to Tumor Shrinkage
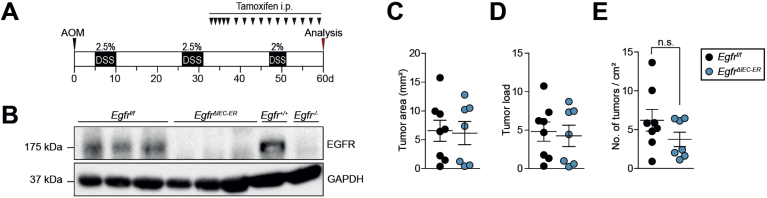
As targeted EGFR inhibition is one of the first-line treatments in metastatic CRC, we depleted EGFR in IECs after AOM/DSS-dependent tumor formation using the tamoxifen-inducible Villin-CreERT2 line (EgfrΔIEC-ER) (Figure 3A). Successful EGFR depletion in IECs was confirmed by Western blot (Figure 3B). The EgfrΔIEC-ER mice developed tumors comparable to EgfrΔIEC mice, and EGFR deletion had no effect on tumor number, size, and load (Figure 3C–E). These results demonstrate that EGFR inhibition in tumor cells of CRC does not have any therapeutic benefit, suggesting that the positive outcomes following EGFR inhibition in patients with metastatic CRC might be due to EGFR blockade in myeloid cells. Additionally, these results rule out the possibility of any influence of genetic adaptation and compensatory mechanism to the constitutive absence of EGFR in the EgfrΔIEC mice.
Figure 3.
Targeting EGFR in IECs of established tumors does not lead to tumor shrinkage. (A) Scheme of inducible EGFR deletion in the AOM/DSS model. Villin-CreERT2 activation was mediated by tamoxifen injection after tumors had developed. (B) Western blot confirming loss of EGFR expression in purified colonocytes from EgfrΔIEC-ER mice on Cre induction. (C–E) Analysis of tumor growth in EgfrΔIEC-ER mice and controls (n = 7–8) treated with tamoxifen showing (C) tumor area, (D) load, and (E) multiplicity.
EGFR Signaling in Myeloid Cells Promotes STAT3 Activation in Colorectal Tumor Cells
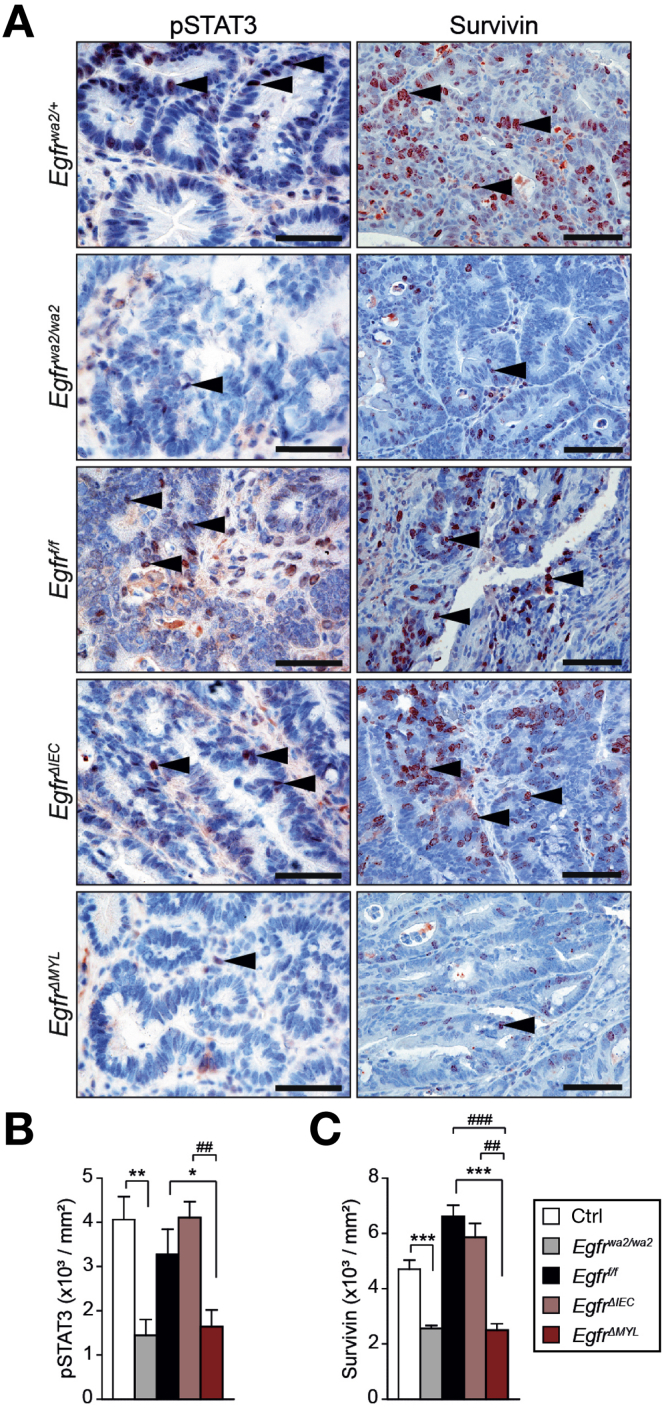
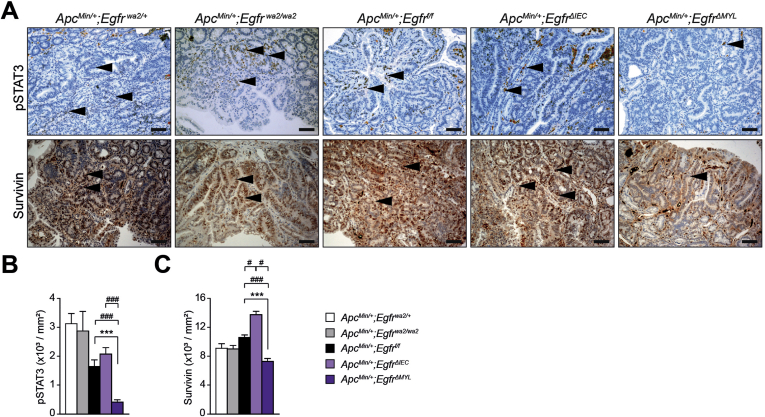
Independent studies have established that activation of the transcription factor STAT3 is an important factor governing CRC formation.28, 39 Therefore, we assessed STAT3 activation in tumor cells by IHC for tyrosine-705-phosphorylated STAT3 (pSTAT3). In EgfrΔIEC mice and controls, we observed comparable numbers of pSTAT3-positive nuclei. In contrast, tumors from Egfrwa2/wa2 and EgfrΔMYL mice showed significant reduction of nuclear pSTAT3 when compared with control mice (Figure 4A and B). Moreover, Egfrwa2/wa2 and EgfrΔMYL tumors displayed significantly decreased expression of the antiapoptotic protein survivin, which is a downstream target of STAT3 (Figure 4A and C). Also in the ApcMin/+ model, ApcMin/+;EgfrΔMYL tumors showed significant reductions of both pSTAT3+ and survivin+ nuclei compared with controls (Supplementary Figure 4A–C), further corroborating these results. These data suggest that myeloid EGFR signaling promotes STAT3 activation and STAT3-dependent expression of survivin in intestinal tumor cells.
Figure 4.
EGFR signaling in myeloid cells promotes colitis-associated CRC development via STAT3 activation in tumor cells. (A) IHC for pSTAT3 and survivin in the AOM/DSS model; arrowheads depict pSTAT3+ and survivin+ nuclei in tumor cells, respectively. Scale bars 50 μm. (B) pSTAT3 index (pSTAT3+ tumor cells/tumor area, 9–12 tumors from 3–4 different mice were counted). (C) Survivin index (survivin+ tumor cells/tumor area, 11–28 tumors from 3 different mice were counted). Data are mean ± SEM. *P < .05, **P < .01, ***P < .001, t test. ##P < .01, ###P < .001, 1-way analysis of variance with Dunn’s posttest.
Supplementary Figure 4.
EGFR signaling in myeloid cells promotes ApcMin-driven CRC development via STAT3 activation in tumor cells. (A) IHC staining for pSTAT3 and survivin in the ApcMin/+ model, arrowheads depict pSTAT3+ and survivin+ nuclei in tumor cells, respectively. Scale bars 50 μm. (B) pSTAT3 index was defined by dividing the number of pSTAT3+ tumor cells by the tumor area in ApcMin/+ mice (13–25 tumors from 3–4 different mice were counted). (C) Survivin index was defined by dividing the number of survivin+ tumor cells by area of tumors in ApcMin/+ mice (9–30 tumors from 3–5 different mice were counted). Data are mean ± SEM. ***P < .001, t test. #P < .05, ###P < .001, 1-way analysis of variance with Dunn’s posttest.
EGFR Signaling in Myeloid Cells Protects From Colitis
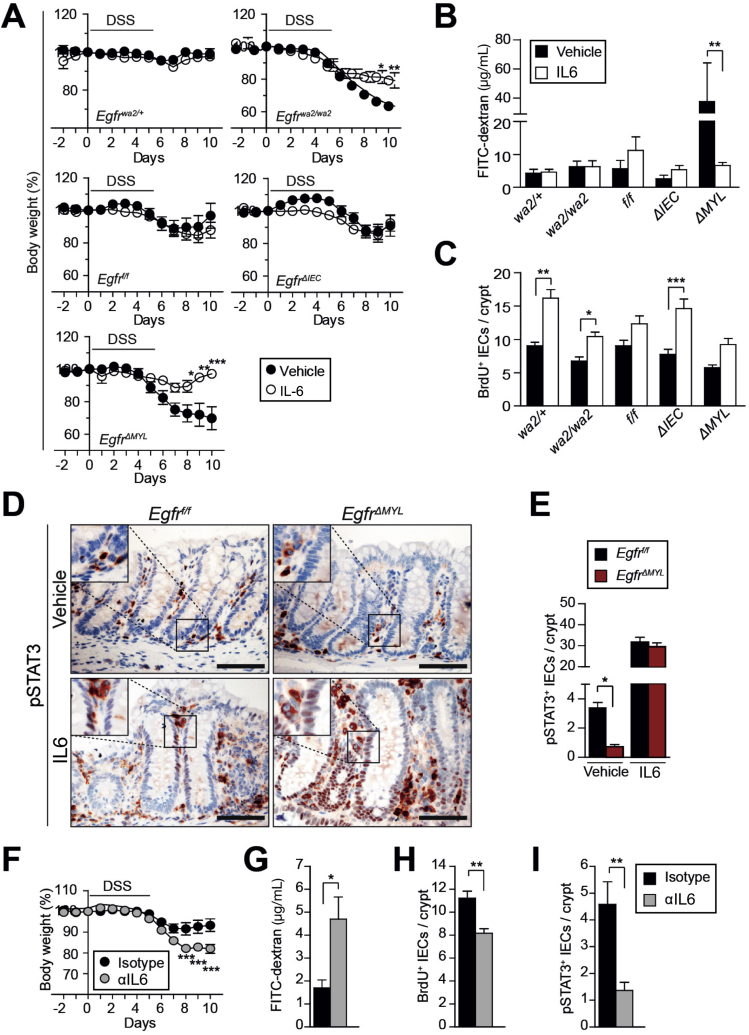
To investigate the mechanisms responsible for the observed phenotypes, we analyzed early phases of CAC development when chronic inflammation acts as a major driver. Therefore, we applied the DSS-dependent model of colitis (Supplementary Figure 5A) in mice lacking the EGFR in various cell types. Histological analysis showed that the colons of EgfrΔIEC mice were less inflamed than in Egfrf/f mice (Figure 5A). In fact, EgfrΔIEC mice showed significant gain of body weight during the entire period of DSS administration when compared with the respective controls (Figure 5B). In contrast to EgfrΔIEC mice, DSS administration to Egfrwa2/wa2 and EgfrΔMYL mice resulted in extensive damage of the colonic mucosa, large ulcerated regions, and severe inflammation (Figure 5A), leading to significant loss of body weight when compared with controls (Figure 5B), which was accompanied by diarrhea. These observations confirm previous studies describing increased severity of DSS-induced colitis in Egfrwa2/wa2 mice.12 Our data show that EGFR signaling in myeloid cells and not in IECs protects from DSS-induced colitis.
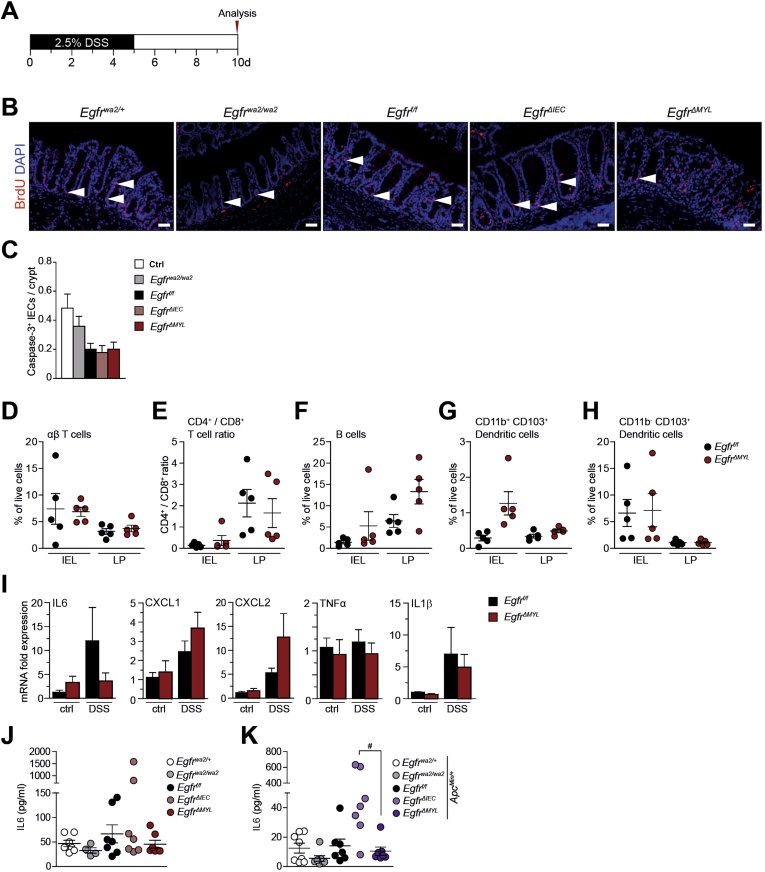
Supplementary Figure 5.
EGFR signaling in myeloid cells protects from colitis. (A) Scheme of the DSS-dependent colitis model. On day 10, mice were starved for 4 hours before oral administration of FITC-dextran. (B) BrdU IF staining, arrowheads depict BrdU+ IECs. Scale bars 50 μm. (C) Quantification of cleaved Caspase-3+ IECs per crypt as readout for apoptosis (30 crypts per mouse were counted, n = 3–4). (D–H) Flow cytometric analysis of the IEL and LP immune cell fraction of colons from DSS-treated mice shows percentage of live cells of (D) TCRβ+ αβ T cells, (E) ratio of CD4+/CD8+ T cells, (F) CD19+ B cells, (G) CD11c+/CD11b+/CD103+, and (H) CD11c+/CD11b-/CD103+ dendritic cell subsets. (I) Quantitative reserve transcriptase PCR analysis of IL6, CXCL1, CXCL2, tumor necrosis factor-α, and IL1β mRNA expression. Relative mRNA expression levels were determined in whole colon tissues on day 10 (n = 3) and normalized to Cyclophilin. Data are mean ± SEM. (J–K) IL6 serum levels in endpoint mice with (J) AOM/DSS treatment (n = 4 for Egfrwa/wa, otherwise n = 7) and (K) the ApcMin background (n ≥7). Data are mean ± SEM. #P < .5, 1-way analysis of variance with Dunn’s posttest.
Figure 5.
EGFR signaling in myeloid cells protects from colitis. (A) Hematoxylin-eosin staining of colons of DSS-treated mice at day 10. Scale bars 50 μm. (B) Body weight during DSS treatment (n ≥6). Data are mean ± SEM. *P < .05, **P < .01, ***P < .001, 2-way analysis of variance. (C) FITC-dextran concentration in serum as readout for intestinal permeability (n ≥5). (D) Number of BrdU+ IECs per crypt (20–30 crypts per mouse were counted, Egfrwa2/wa2, n = 2 mice; otherwise n = 3). (E–H) Flow cytometric analysis of the IEL and LP immune cell fraction of colons from DSS-treated mice showing percentage of live cells of (E) CD45+ hematopoietic cells, (F) CD11b+/Gr1+/Ly6Clo granulocytes, (G) CD11b+/Gr1int/Ly6Chi monocytes, and (H) CD11c+/CD11b+/CD103− macrophages and dendritic cells. (I) IL6 serum levels in DSS-treated mice at day 7 (n ≥6). (J) EGFR and IL6 messenger RNA expression levels of MACS-sorted CD11b+ colonic myeloid cells of DSS-treated mice at day 5. Data from (C–J) are mean ± SEM. *P < .05, **P < .01, ***P < .001, t test. ##P < .01, ###P < .001, 1-way analysis of variance with Dunn’s posttest.
DSS has been described to target the crypt epithelial cells, thereby altering intestinal barrier permeability. Therefore, we investigated epithelial barrier function by oral administration of fluorescein isothiocyanate (FITC)-dextran.38 EgfrΔMYL and Egfrwa2/wa2 mice displayed significantly higher serum levels of FITC-dextran during colitis, demonstrating loss of barrier integrity. In contrast, EgfrΔIEC mice showed reduced levels of FITC-dextran (Figure 5C), confirming a previous study, which demonstrated that inhibition of EGFR prevents loss of barrier permeability.40
Crypt cell proliferation is an important factor contributing to intestinal homeostasis and epithelial regeneration during mucosal injury. Analysis of bromodeoxyuridine (BrdU) incorporation showed significantly reduced BrdU+ IECs in the crypts of Egfrwa2/wa2 and EgfrΔMYL mice when compared with respective control mice. In contrast, the number of proliferating nuclei in EgfrΔIEC animals was comparable with controls (Figure 5D, Supplementary Figure 5B). Moreover, cleaved Caspase-3 staining in IECs did not reveal any major differences in apoptosis between EgfrΔMYL and EgfrΔIEC mice (Supplementary Figure 5C).
To investigate if EGFR loss on myeloid cells alters the immune cell composition during DSS-induced colitis, we separated the intestinal epithelial layer (IEL) from the LP of EgfrΔMYL and Egfrf/f control mice and performed flow cytometric analysis to characterize the immune cells in the respective layers. Because DSS-induced ulcers frequently reach down to the LP (Figure 5A), the number of immune cells in the IEL fraction was higher than in the LP. Total CD45+ immune cells were overall significantly enriched in the IEL fraction of EgfrΔMYL mice (Figure 5E). The populations of TCRβ+ and CD4+/CD8+ T cells as well as CD19+ B cells and CD103+ dendritic cells were similar among genotypes (Supplementary Figure 5D–H). Consistent with the higher inflammation and barrier breakdown, granulocytes and monocytes, as well as macrophages/CD103− dendritic cells were markedly increased in the IEL of EgfrΔMYL mice (Figure 5F–H) and monocyte numbers were also higher in the LP fraction (Figure 5G). Our data reveal that EGFR expression in myeloid cells is protective against DSS-induced colitis by maintaining mucosal integrity and proliferation of epithelial cells.
EGFR Signaling in Myeloid Cells Promotes STAT3 Activation Via Regulation of IL6 Production
LP cells secrete various prosurvival factors during inflammation that support epithelial regeneration. One important factor is IL6, a potent inducer of STAT3 activation.28, 41 It was demonstrated that IL6/STAT3 can protect mice from DSS-induced colitis.28 Further, we recently described the importance of EGFR signaling in liver macrophages for production of IL6 in a hepatic injury model of liver cancer.30 Colons of DSS-treated EgfrΔMYL mice showed a noticeable decrease in IL6 messenger RNA expression, which is consistent with a previous study.20 In addition, these colons contained higher messenger RNA levels of chemoattractants, such as CXCL1 and CXCL2. However, the expression levels of tumor necrosis factor-α and IL1β were unchanged (Supplementary Figure 5I). IL6 levels were also significantly reduced in the serum of DSS-treated Egfrwa2/wa2 and EgfrΔMYL mice but not in EgfrΔIEC animals (Figure 5I). In tumor-bearing AOM/DSS-treated and ApcMin/+ mice, there was a nonsignificant trend of reduced IL6 in serum of EgfrΔMYL mice (Supplementary Figure 5J–K). Importantly, magnetic activated cell sorted CD11b+ colonic myeloid cells of DSS-treated EgfrΔMYL mice, which lacked EGFR, expressed significantly less IL6 than the respective controls (Figure 5J).
These data suggest that the observed phenotypes of Egfrwa2/wa2 and EgfrΔMYL mice in colitis and CAC formation are due to reduced IL6 production by EGFR-deficient myeloid cells.
IL6 Protects EgfrΔMYL Mice Against DSS-induced Damage
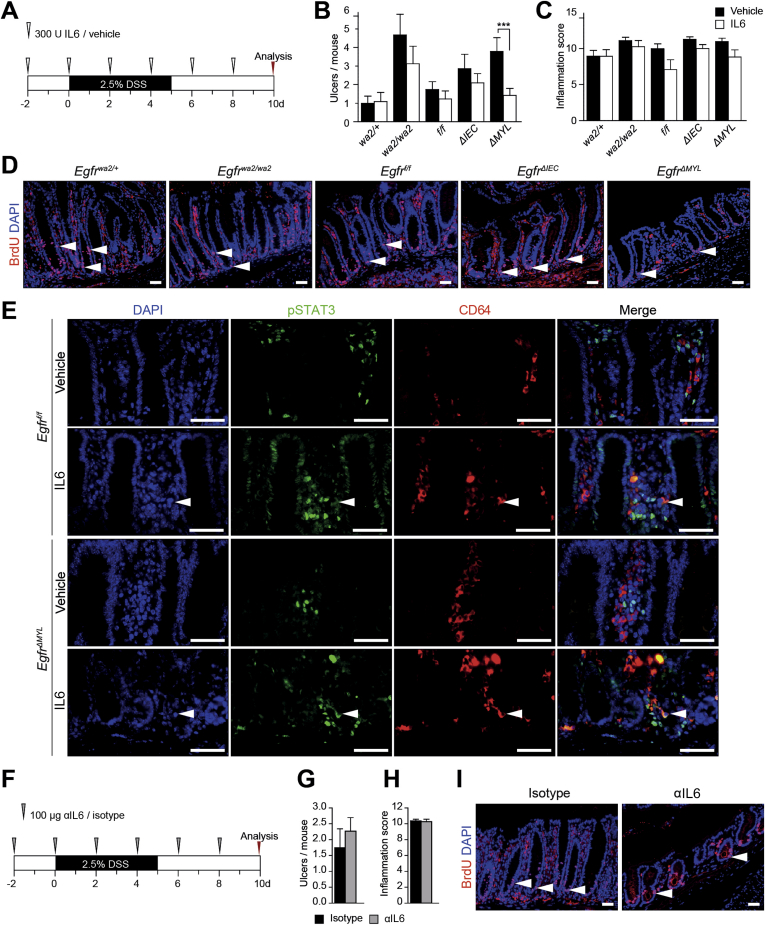
To assess the role of IL6 during colitis in EgfrΔMYL mice, we administered recombinant IL6 (rIL6) during DSS treatment (Supplementary Figure 6A). Indeed, exogenous IL6 protected EgfrΔMYL mice from DSS-induced weight loss, whereas Egfrwa2/wa2 mice were partially rescued (Figure 6A). Although the severity of inflammation was similar in all genotypes, administration of rIL6 significantly decreased the number of ulcers in EgfrΔMYL mice (Supplementary Figure 6B and C) and prevented from loss of epithelial barrier function (Figure 6B). rIL6 treatment was also able to restore epithelial proliferation during colitis (Figure 6C, Supplementary Figure 6D). STAT3 activation (pSTAT3+ nuclei in IECs), which was significantly reduced during DSS treatment of EgfrΔMYL mice, was fully restored on rIL6 administration (Figure 6D and E). pSTAT3 was present in CD64+ myeloid cells of both Egfrf/f and EgfrΔMYL mice after IL6 induction (Supplementary Figure 6E). Taken together, we were able to rescue EgfrΔMYL mice from DSS-induced damage by administering rIL6, suggesting that myeloid EGFR signaling is responsible for IL6 release and protection of the intestinal epithelium during colitis.
Supplementary Figure 6.
IL6 protects from DSS-induced colitis. (A) Scheme of IL6 or vehicle administration during colitis induction, on day 10 mice were starved for 4 hours before oral FITC-dextran administration. (B and C) Histological assessment of (B) number of ulcers and (C) inflammation scores on colon sections of mice supplemented with IL6 during DSS treatment. Data are mean ± SEM. ***P < .001, t test. (D) BrdU IF stainings, arrowheads depict BrdU+ IECs. (E) IF double staining for pSTAT3 and CD64. Arrowheads depict presence of pSTAT3+/CD64+ cells. (F) Scheme of anti-IL6 or isotype control antibody administration during colitis induction, on day 10 mice were starved for 4 hours before oral FITC-dextran administration. (G and H) Histological assessment of number of ulcers (G) and inflammation scores (H) on colon sections of IL6-depleted mice (n ≥6). Data are mean ± SEM. (I) BrdU IF staining, arrowheads depict BrdU+ IECs. Scale bars 50 μm.
Figure 6.
IL6 protects from DSS-induced colitis. (A–E) Administration of vehicle or recombinant IL6 to DSS-treated mice. (A) Body weight following IL6 or vehicle administration during DSS treatment (n ≥3). (B) FITC-dextran concentration in serum as readout for intestinal permeability (n ≥3). (C) Number of BrdU+ IECs per crypt (30–40 crypts per mouse were counted, n = 3 mice). (D) pSTAT3 IHC of DSS-treated colons. Scale bars 50 μm. (E) Quantification of pSTAT3+ IECs per crypt (30 crypts per mouse were counted, n ≥4). (F–I) Antibody-mediated IL6 depletion during DSS-induced colitis in wild-type mice. (F) Body weight of IL6-depleted mice and controls during DSS treatment (n ≥3). (G) FITC-dextran concentration in serum as readout for intestinal permeability (n ≥7). Quantification of (H) BrdU+ IECs per crypt (30–80 crypts per mouse were counted, n ≥4) and (I) pSTAT3+ IECs per crypt (30–80 crypts per mouse were counted, n ≥3). All data are mean ± SEM. *P < .05, **P < .01, ***P < .001, t test except (A and F), 2-way analysis of variance.
To confirm the importance of IL6 for colitis induction, we depleted IL6 from Egfrf/f mice during DSS treatment to replicate the phenotype of EgfrΔMYL mice (Supplementary Figure 6F). Although IL6 depletion did not influence the overall colitis severity, a mild increase in ulcers was observed (Supplementary Figure 6G and H). However, IL6-depleted mice showed significant weight loss compared with isotype controls during DSS treatment (Figure 6F). They further displayed a significant increase of intestinal barrier permeability (Figure 6G). This defect in gut barrier integrity was further substantiated by a strong reduction in proliferation of crypt cells (Figure 6H, Supplementary Figure 6I). Additionally, we observed a significant decrease in nuclear pSTAT3 signaling in IL6-depleted mice (Figure 6I). In summary, these observations suggest that IL6 is required to prevent DSS-induced damage and that its depletion establishes a phenotype similar to EgfrΔMYL mice.
Discussion
To date, the absence of KRAS and NRAS (exons 2, 3, and 4) mutations serves as the only clinically approved predictive biomarker for EGFR-targeted therapies of metastatic CRC. Besides RAS, mutations in genes like BRAF and PIK3CA have been suspected to predict unresponsiveness of anti-EGFR therapy. However, several clinical studies delivered partly conflicting results and, as these mutations occur in a rather low percentage of patients, this highlights the need for a more reliable strategy for patient stratification.42
We show for the first time that EGFR expression in myeloid cells is a negative prognostic factor for overall survival of patients with metastatic CRC and further demonstrate a pro-tumorigenic role of myeloid EGFR in mouse models of AOM/DSS-induced and ApcMin-dependent intestinal tumorigenesis, mimicking early stages of human disease. In a previous study of HCC, we made a similar discovery. Most importantly, we demonstrated a correlation between EGFR expression in Kupffer cells/liver macrophages and poor prognosis for patients with HCC,30 suggesting that the ability of EGFR to support tumor formation via myeloid cells might extend to other inflammation-associated cancers. Intriguingly, whereas the presence of CD68+/EGFR+ macrophages in patients with CRC did not affect overall survival, the number of CD11b+/EGFR+ myeloid cells was a bad prognostic factor for overall survival of patients with metastatic disease. This highlights that cells other than macrophages, such as granulocytes or myeloid suppressor cells, might be responsible for the observed phenotypes. These myeloid cells might need EGFR signaling for the secretion of immune-modulatory cytokines, such as IL6, to promote tumor growth. Consistent with this, ApcMin mice have fewer polyps in an IL6-deficient background.43 Our findings offer an explanation for the nonresponsiveness of more than 50% of patients with CRC with wild-type RAS to EGFR-targeted treatment.42 Based on our results, we predict that patients with EGFR+ myeloid cells respond to anti-EGFR treatment (55% in our patient cohort), whereas patients in whom EGFR is expressed only in tumor cells (45% in our patient cohort), might not benefit. Interestingly, the distribution of CD11b+/EGFR+ myeloid cells was similar among the 18 patients with known RAS status, suggesting that the presence of oncogenic RAS in tumor cells does not influence EGFR expression on myeloid cells. Because patients with mutant RAS in tumor cells do not respond to anti-EGFR treatment, it is possible that EGFR inhibitors are effective only if they can act on both tumor and myeloid cells. This situation would occur only in patients with CRC with wild-type RAS in whom EGFR is expressed on tumor as well as on myeloid cells. Clinical follow-up studies are necessary to evaluate a patient stratification strategy based on myeloid EGFR expression, ultimately providing better personalized application of EGFR-directed therapy.
The role of EGFR during early and late stages of CRC is highly complex and likely depends on time and cell type of expression. In previous studies based on the Egfrwa2/wa2 model, it was postulated that EGFR expression in IECs is necessary for colon cancer formation.15, 16, 17, 18 However, our results demonstrate that EGFR signaling is required in myeloid cells for CRC development, as EGFR deletion in IECs does not affect tumor growth. Thus, impaired CAC and CRC formation in Egfrwa2/wa2 mice is likely due to attenuated EGFR signaling in the myeloid compartment. Dubé et al,11 however, show that EGFR inhibition accelerates CAC development in Egfrwa5/+ mice. These results are contradictory to our results and the results of Dougherty et al,15 which were obtained with Egfrwa2/wa2 mice, pointing toward a broader and even dominant-negative effect of the Wa5 variant of EGFR. Moreover, factors like mouse strain, intestinal microbiome, diet, and experimental setup also can affect the outcome.
We further deleted EGFR exclusively on IECs, clearly and unequivocally showing that it is dispensable for tumor formation in both AOM/DSS-induced and ApcMin-dependent intestinal tumorigenesis. This is supported by previous investigations, in which EGFR expression on IECs is dispensable during colitis and after surgical resection.13, 19 Last, lack of EGFR in IECs of EgfrΔIEC mice also may contribute to greater barrier integrity, as EGFR inhibition has been shown to prevent increase in barrier permeability.40, 44 Besides showing that EGFR is dispensable in IEC during tumor initiation, we also demonstrate that it is not essential for tumor progression, as EGFR ablation on IECs had no effect on preexisting tumors, when we recapitulated the therapeutic situation. These results clearly demonstrate that EGFR deletion in tumor cells did not affect tumor growth.
Colitis, which can ultimately lead to tumor formation, is a complicated process involving a variety of immune cells and a network of cytokines and chemokines. We demonstrate that EGFR ablation in myeloid cells aggravates DSS-induced colitis, which can be rescued by IL6 administration. Results of Lu et al20 describe that EGFR deletion in myeloid cells improves colitis via increased IL10 production. The variation in observations might be explained by differences in concentration and duration of DSS treatment, as well as by distinct time points of analysis and different Egfrf/f lines; however, they reported impaired IL6 transcription during colitis after myeloid-specific EGFR deletion, thus confirming our results. IL6 is known to be primarily produced by cells of the innate immune system, such as myeloid cells, during an immune response.3 In chemically induced HCC, we previously showed that EGFR expression in Kupffer cells induces IL6 production.30 In CAC, IL6 stimulates survival and proliferation of IECs.28 Supporting this, we here observe that selective deletion of EGFR in myeloid cells, but not in IECs, critically impairs systemic IL6 levels during colitis, which is coupled with a proliferation defect of IECs. This is in line with observations in IL6−/− mice.28 Thus, our results suggest that the protective function exerted by EGFR on IECs might occur indirectly via activation of EGFR signaling in myeloid cells with subsequent IL6 secretion.
Activation of STAT3 signaling by IL6 family cytokines is required for CRC formation in AOM/DSS-dependent mouse models and protects from colitis.28, 39 Here, we prevent DSS-induced colitis by restoring systemic IL6 levels in EgfrΔMYL mice, thereby reestablishing STAT3 signaling in IECs, which restores IEC proliferation. Our observations describe the requirement of myeloid EGFR signaling for protective IL6 production during intestinal inflammation. In the ApcMin/+ model, STAT3 signaling is essential for promoting tumorigenesis during the initial stages.45 We could observe a reduction of the overall tumor burden in ApcMin/+;EgfrΔMYL mice going along with pSTAT3 reduction, which further supports our model of EGFR-dependent IL6 signaling as tumor promoter. In line with our findings in colitis, the reduced nuclear pSTAT3 levels in the AOM/DSS-induced tumors of EgfrΔMYL mice point to a systemic IL6 deficiency. This is linked with a decreased expression of the STAT3 target gene survivin and increased apoptosis, which further leads to reduced tumor size.
In conclusion, our results imply that EGFR-expressing myeloid cells might be a novel prognostic marker for CRC, providing the possibility to stratify patients who can really benefit from EGFR-directed therapy and to spare others from ineffective treatment and its painful side effects.
Acknowledgments
The authors thank Martina Hammer for maintaining mouse colonies; Sarah Bardakji, Elisabeth Born, and Malgorzata Tryniecki for genotyping; and Temenuschka Baykuscheva-Gentscheva, Elisabeth Glitzner, Sofia Grau, Verónica Moreno-Viedma, and Silke Sohn for technical assistance. The authors thank Marc B. Bissonnette for help and discussions during the initial phases of the project and Stephanie Rost for pilot experiments in GEMMs.
Footnotes
Conflicts of interest There is a patent application PCT/EP2015/061220 covering some aspects of this publication (M.S.). There are no other conflicts to declare.
Funding This work was supported by Austrian Science Fund grants DK W1212 and P25925 (M.S., R.E), P26908 and SFB-F28 (R.E.), and the Austrian Federal Government’s GEN-AU program “Austromouse” 820966 and the European Research Council Advanced Grant 694883 (M.S.). The work of S.R.-J. was supported by grants from the Deutsche Forschungsgemeinschaft (SFB-877, Project A1 and the Cluster of Excellence “Inflammation at Interfaces”).
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2017.03.053.
Supplementary Materials and Methods
Mice
All animal experiments conducted were compliant with federal laws and guidelines of the Medical University of Vienna. Egfrf/f,1 Villin-Cre and Villin-CreERT2,2 LysMCre/+,3 and ApcMin/+4 mice and their genotyping by polymerase chain reaction (PCR) were previously described. Egfr+, Egfrf, and EgfrΔ alleles were detected by PCR using primers R4: GCCTGTGTCCGGGTCTCGTCG, R6: CAACCAGTGCACCTAGCCTGG, Egfr-fwd1: AAGTTTAAGAAACCCCGCTCTACT. Villin-Cre mice were obtained on a DBA/2;C57BL/6J background and bred to a mixed 129/Sv;C57BL/6 genetic background. Littermates with EGFR expression (Egfrf/f, Villin-Cre;Egfrf/+, LysMCre/+;Egfrf/+, Egfrf/+, and Egfrwa2/+ mice) were used as controls to the respective EGFR-deleted animals. Mice from the ApcMin/+ model were analyzed at the age of 3 months. All further experiments were performed in mice between 6 and 8 weeks of age.
Southern Blot Analysis
Intestinal epithelial cells (IECs) were isolated as described.5 Genomic DNA was extracted from IECs by salt and ethanol precipitation; 10 μg DNA was digested with HindIII (New England BioLabs, Ipswich, MA) and probed using an XbaI/HindIII digested fragment from intron 1 of Egfr as described.1
Western Blot, Enzyme-Linked Immunosorbent Assay, and Quantitative Reverse Transcriptase PCR Analysis
IECs were isolated as described previously and lysed in radioimmunoprecipitation assay buffer. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA). Western blot analysis was performed as described.6 Antibodies used were EGFR (06-847, EMD; Millipore) and glyceraldehyde-3-phosphate dehydrogenase (5174; Cell Signaling, Danvers, MA). Interleukin (IL)6 enzyme-linked immunosorbent assay was carried out using the Mouse IL6 enzyme-linked immunosorbent assay Ready-SET-Go.
Bone marrow–derived macrophages were isolated as previously described.7 Total RNA isolation from the distal colon and bone marrow–derived macrophages was performed as described.8 Complementary DNA (cDNA) synthesis was performed using ProtoScript II Reverse Transcriptase (NEB, Ipswich, MA). Total RNA isolation from CD11b+ colonic macrophages was performed using the miRNeasy Micro Kit (Qiagen, Hilden, Germany). cDNA synthesis was done using SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). Quantitative reverse transcriptase PCR reactions and analysis were done as described.8 Primer sequences for IL6, IL1β and cyclophilin have been previously reported.9 Further primers used were tumor necrosis factor-α-F: ATGAGAAGTTCCCAAATGGCC, tumor necrosis factor-α-R: TCCACTTGGTGGTTTGCTACG, CXCL1-F: GCCAATGAGCTGCGCTGT, CXCL1-R: CCTTCAAGCTCTGGATGTTCTTG, CXCL2-F: ATCCAGAGCTTGAGTGTGACGC, CXCL2-R: AAGGCAAACTTTTTGACCGCC, EGFR-F: TTGGAATCAATTTTACACCGAAT, EGFR-R: GTTCCCACACAGTGACACCA, TBP-F: GGGGAGCTGTGATGTGAAGT, TBP-R: CCAGGAAATAATTCTGGCTCAT.
Histology
Mice were injected with 10 mg/kg bromodeoxyuridine (BrdU) 2.5 hours before they were euthanized. The colons were extracted, flushed with ice-cold phosphate-buffered saline (PBS), fixed overnight at 4°C in neutral buffered 4% formaldehyde as “swiss rolls,” which were further dehydrated, embedded in paraffin, and sectioned for subsequent histological analysis. Colitis severity was scored as described.10 Mouse tumor stage was scored and graded by a pathologist in a blinded manner according to described criteria.5 tyrosine-705-phosphorylated STAT3+ epithelial cell counting in the colitis model was performed as described.5 Starting from the distal part of the colon, for each mouse, tyrosine-705-phosphorylated STAT3+ cells were counted in more than 30 whole crypts.
Immunohistochemistry and Immunofluorescence
Sections were dewaxed, rehydrated and subjected to antigen recovery using either citrate buffer pH 6.1 or pH 9 (Dako, Glostrup, Denmark). For immunohistochemistry, endogenous peroxidase was quenched by incubation with 3% hydrogen peroxide in Tris-buffered saline, followed by blocking (10% goat serum, 2% bovine serum albumin, 0.1% Tween in Tris-buffered saline). Sections were then incubated overnight with antibodies against pTyr705-STAT3 (9145), Survivin (2808), cleaved Caspase-3 (9661; all Cell Signaling), Lysozyme (A0099; Dako), EGFR (sc-03; Santa Cruz, Santa Cruz, CA) or Synaptophysin (GTX100865; GeneTex, Irvine, CA). All sections were further treated with SignalStain Boost (Cell Signaling) and AEC Chromogen/Substrate Bulk Kit (ID Labs, London, UK) and counterstained with hematoxylin.
For immunofluorescence (IF) BrdU staining, sections were treated with 2N hydrochloric acid after antigen recovery, blocked as described previously, incubated with BrdU antibody (347580; BD Biosciences, San Jose, CA) and secondary antibody (A-11018; Invitrogen, Carlsbad, CA) and counterstained with 4′,6-diamidino-2-phenylindole.
For CD11b/EGFR, CD11b/pEGFR, or CD68/EGFR IF staining of human paraffin slides, sections were incubated briefly with 3% H2O2 in methanol after antigen recovery followed by biotin block (E-21390; Thermo Fisher Scientific). After blocking, (5% horse serum, 1% bovine serum albumin, 0.1% Triton in PBS), sections were incubated overnight with antibodies against EGFR (4267; Cell Signaling), pTyr1068-EGFR (API 300 AA; Biocare Medical, Pacheco, CA), CD68 (M0814; Dako) and CD11b (IM0190; Beckman Coulter, Brea, CA).
After 1 hour of secondary antibody incubation, each tissue microarray core was completely photographed using a Nikon (Vienna, Austria) Eclipse 80i fluorescence microscope at ×200 magnification or scanned using the Panoramic Digital Slide Scanner or Tissuegnostics (Vienna, Austria) TissueFAXS at the Core Facility Imaging of the Medical University of Vienna. Images were assembled with ImageJ (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD). Counting of CD11b+/EGFR+ and CD11b+/pEGFR+ cells was performed manually for each core using Adobe Photoshop (Adobe Systems, Inc, San Jose, CA). Analysis of CD68+/EGFR+ cells was carried out using Definiens image analysis software (Munich, Germany).
For IF on mouse paraffin slides, the same conditions as for human paraffin slides were applied. Sections were incubated with antibodies against CD64 (139303; Biolegend, San Diego, CA), EGFR (4267), and pTyr1068-EGFR (3777, both Cell Signaling).
Flow Cytometric Analysis and Magnetic Cell Sorting
For subsequent flow cytometric analysis, colitis was induced in EgfrΔMYL and Egfrf/f control mice by dextran sodium sulfate administration. Analysis was done at day 10 using 5 mice per genotype. The fraction of intraepithelial lymphocytes and lamina propria cells were isolated for flow cytometric analysis as previously described.11 Magnetic cell sorting of CD11b+ myeloid cells from the colon of dextran sodium sulfate–treated EgfrΔMYL and Egfrf/f control mice was performed at day 5 using ≥5 mice per genotype. The colon was cut into pieces and digested in RPMI medium with 10% fetal calf serum (FCS), 1% penicillin/streptomycin, 1 mg/mL Collagenase VIII (Sigma-Aldrich, St. Louis, MO) and 1 mg/mL DNase I (Sigma-Aldrich) for 1 hour at 37°C shaking at 300 rpm. Samples were filtered through a 70-μm cell strainer and washed once with Hank’s balanced salt solution containing 10% FCS, 1% penicillin/streptomycin, and 5 mM EDTA and twice with PBS containing 5% FCS, 5 mM EDTA, and 20 μg/mL DNase I. Myeloid cell fractions were enriched by density gradient centrifugation as described.12 Magnetic cell sorting was carried out using a biotinylated CD11b antibody (M1/70; Biolegend) and IMag Streptavidin Particles Plus-DM (BD Biosciences). Sorting quality was tested by flow cytometric analysis, revealing that more than 95% of sorted live cells were CD45+/CD11b+.
Cell suspensions were stained using fluorescently labeled antibodies for 30 minutes at 4°C after blocking with Fc-block (CD16/CD32, 93; Biolegend). The following antibodies were used: TCRβ (H57–597), CD4 (GK1.5), CD8a (53.6–7), CD19 (6D5), CD45 (30-F11, APC-Cy7), Gr1 (RB6–8C5), Ly6G (1A8), F4/80 (BM8), CD11b (M1/70), CD11c (N418; all Biolegend), CD45 (30-F11, PECF594), TCRδ (GL-3), CD44 (IM7), CD103 (2E7), CD335 (29A1.4; all BD Biosciences), Ly6C (HK1.4), I-A/I-E (MHCII, M5/114; all Thermo Fisher Scientific). Data were acquired on LSRFortessa flow cytometer (BD Biosciences, Schwechat, Austria) and analyzed with FloJo software (FlowJo, LLC, Ashland, OR).
Intestinal Barrier Assessment
Intestinal barrier permeability was assessed as described.13 Briefly, food and water were withdrawn 4 hours before oral gavage with 60 mg/kg fluorescein isothiocyanate (FITC)-dextran (Sigma-Aldrich). FITC levels were determined in the serum 4 hours after gavage by absorption at 488 nm using standard dilutions of FITC-dextran as reference.
Statistics
Human material
For human samples, differences between categorical data were measured by χ2 test. Differences between continuous variables were investigated by Mann-Whitney U test and Kruskal-Wallis test, when appropriate. Kaplan-Meier method was used to estimate median overall survival time, and P values were calculated using log-rank test. All tests were 2-sided, with P < .05 considered to indicate statistical significance. All statistical analyses were carried out using the SPSS package (IBM Corporation, Chicago, IL).
Mouse experiments
The mouse experiments were not randomized and the investigators were not blinded during experiments and outcome assessment. Sample size calculation: For tumor studies, we considered 10 mice per group to detect a relevant difference in means of 1.5 within-group standard deviations at a 2-sided significance level of .05 and a power of 90%, which ensures 80% power in case of a 20% drop-out rate. Mouse experiments were performed as indicated in the figure legends. Quantifications on blinded histological samples were performed by counting/measuring microscopic fields (high-powered fields where indicated) as indicated in the legends. Data are presented as mean ± SEM. Statistical significance was calculated by unpaired Student t test (comparison of 2 experimental groups) or 1-way analysis of variance followed by multiple comparison Dunn’s posttest (comparison of 3 or more experimental groups) for parametric observations, whereas for discontinuous observations, such as scores for colitis and tumors, significance was evaluated by Mann-Whitney U or Kruskal-Wallis test. Significance of tumor penetrance was calculated by Fisher’s exact test. Each tumor measurement contributed by 1 animal is the mean value over several gastrointestinal sections. All calculations were performed using the GraphPad Prism software (San Diego, CA). A 2-sided P < .05 was considered statistically significant.
Author names in bold designate shared co-first authorship.
Supplementary Material
References
- 1.Siegel R., Ma J., Zou Z. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K.W., Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Terzic J., Grivennikov S., Karin E. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 4.Tobin N.P., Foukakis T., De Petris L. The importance of molecular markers for diagnosis and selection of targeted treatments in patients with cancer. J Intern Med. 2015;278:545–570. doi: 10.1111/joim.12429. [DOI] [PubMed] [Google Scholar]
- 5.Sibilia M., Kroismayr R., Lichtenberger B.M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- 6.Haraldsdottir S., Bekaii-Saab T. Integrating anti-EGFR therapies in metastatic colorectal cancer. J Gastrointest Oncol. 2013;4:285–298. doi: 10.3978/j.issn.2078-6891.2013.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azizi E., Kittai A., Kozuch P. Antiepidermal growth factor receptor monoclonal antibodies: applications in colorectal cancer. Chemother Res Pract. 2012;2012:198197. doi: 10.1155/2012/198197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linardou H., Dahabreh I.J., Kanaloupiti D. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 9.Lee D., Cross S.H., Strunk K.E. Wa5 is a novel ENU-induced antimorphic allele of the epidermal growth factor receptor. Mamm Genome. 2004;15:525–536. doi: 10.1007/s00335-004-2384-2. [DOI] [PubMed] [Google Scholar]
- 10.Luetteke N.C., Phillips H.K., Qiu T.H. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- 11.Dube P.E., Yan F., Punit S. Epidermal growth factor receptor inhibits colitis-associated cancer in mice. J Clin Invest. 2012;122:2780–2792. doi: 10.1172/JCI62888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egger B., Buchler M.W., Lakshmanan J. Mice harboring a defective epidermal growth factor receptor (waved-2) have an increased susceptibility to acute dextran sulfate-induced colitis. Scand J Gastroenterol. 2000;35:1181–1187. doi: 10.1080/003655200750056664. [DOI] [PubMed] [Google Scholar]
- 13.Yan F., Cao H., Cover T.L. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121:2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha A., Nightingale J., West K.P. Epidermal growth factor enemas with oral mesalamine for mild-to-moderate left-sided ulcerative colitis or proctitis. N Engl J Med. 2003;349:350–357. doi: 10.1056/NEJMoa013136. [DOI] [PubMed] [Google Scholar]
- 15.Dougherty U., Cerasi D., Taylor I. Epidermal growth factor receptor is required for colonic tumor promotion by dietary fat in the azoxymethane/dextran sulfate sodium model: roles of transforming growth factor-{alpha} and PTGS2. Clin Cancer Res. 2009;15:6780–6789. doi: 10.1158/1078-0432.CCR-09-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fichera A., Little N., Jagadeeswaran S. Epidermal growth factor receptor signaling is required for microadenoma formation in the mouse azoxymethane model of colonic carcinogenesis. Cancer Res. 2007;67:827–835. doi: 10.1158/0008-5472.CAN-05-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts R.B., Min L., Washington M.K. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2002;99:1521–1526. doi: 10.1073/pnas.032678499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen G., Mustafi R., Chumsangsri A. Epidermal growth factor receptor signaling is up-regulated in human colonic aberrant crypt foci. Cancer Res. 2006;66:5656–5664. doi: 10.1158/0008-5472.CAN-05-0308. [DOI] [PubMed] [Google Scholar]
- 19.Rowland K.J., McMellen M.E., Wakeman D. Enterocyte expression of epidermal growth factor receptor is not required for intestinal adaptation in response to massive small bowel resection. J Pediatr Surg. 2012;47:1748–1753. doi: 10.1016/j.jpedsurg.2012.03.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu N., Wang L., Cao H. Activation of the epidermal growth factor receptor in macrophages regulates cytokine production and experimental colitis. J Immunol. 2014;192:1013–1023. doi: 10.4049/jimmunol.1300133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo J., Rodriguez Perez C.E., Nie W. TNF-alpha induces upregulation of EGFR expression and signaling in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2012;302:G805–G814. doi: 10.1152/ajpgi.00522.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eales-Reynolds L.J., Laver H., Modjtahedi H. Evidence for the expression of the EGF receptor on human monocytic cells. Cytokine. 2001;16:169–172. doi: 10.1006/cyto.2001.0966. [DOI] [PubMed] [Google Scholar]
- 23.Biswas S.K., Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 24.Forssell J., Oberg A., Henriksson M.L. High macrophage infiltration along the tumor front correlates with improved survival in colon cancer. Clin Cancer Res. 2007;13:1472–1479. doi: 10.1158/1078-0432.CCR-06-2073. [DOI] [PubMed] [Google Scholar]
- 25.Khorana A.A., Ryan C.K., Cox C. Vascular endothelial growth factor, CD68, and epidermal growth factor receptor expression and survival in patients with Stage II and Stage III colon carcinoma: a role for the host response in prognosis. Cancer. 2003;97:960–968. doi: 10.1002/cncr.11152. [DOI] [PubMed] [Google Scholar]
- 26.Lackner C., Jukic Z., Tsybrovskyy O. Prognostic relevance of tumour-associated macrophages and von Willebrand factor-positive microvessels in colorectal cancer. Virchows Arch. 2004;445:160–167. doi: 10.1007/s00428-004-1051-z. [DOI] [PubMed] [Google Scholar]
- 27.Bailey C., Negus R., Morris A. Chemokine expression is associated with the accumulation of tumour associated macrophages (TAMs) and progression in human colorectal cancer. Clin Exp Metastasis. 2007;24:121–130. doi: 10.1007/s10585-007-9060-3. [DOI] [PubMed] [Google Scholar]
- 28.Grivennikov S., Karin E., Terzic J. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S., Keku T.O., Martin C. Circulating levels of inflammatory cytokines and risk of colorectal adenomas. Cancer Res. 2008;68:323–328. doi: 10.1158/0008-5472.CAN-07-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanaya H., Natarajan A., Komposch K. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. 2014;16:972–981. doi: 10.1038/ncb3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinelli E., Martini G., Cardone C. AXL is an oncotarget in human colorectal cancer. Oncotarget. 2015;6:23281–23296. doi: 10.18632/oncotarget.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greten F.R., Eckmann L., Greten T.F. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Lichtenberger B.M., Tan P.K., Niederleithner H. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140:268–279. doi: 10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 34.Rollwagen F.M., Yu Z.Y., Li Y.Y. IL-6 rescues enterocytes from hemorrhage induced apoptosis in vivo and in vitro by a bcl-2 mediated mechanism. Clin Immunol Immunopathol. 1998;89:205–213. doi: 10.1006/clin.1998.4600. [DOI] [PubMed] [Google Scholar]
- 35.Peters M., Blinn G., Jostock T. Combined interleukin 6 and soluble interleukin 6 receptor accelerates murine liver regeneration. Gastroenterology. 2000;119:1663–1671. doi: 10.1053/gast.2000.20236. [DOI] [PubMed] [Google Scholar]
- 36.el Marjou F., Janssen K.P., Chang B.H. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 37.Clausen B.E., Burkhardt C., Reith W. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 38.Wirtz S., Neufert C., Weigmann B. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 39.Bollrath J., Phesse T.J., von Burstin V.A. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Tripathi A., Lammers K.M., Goldblum S. Identification of human zonulin, a physiological modulator of tight junctions, as prehaptoglobin-2. Proc Natl Acad Sci U S A. 2009;106:16799–16804. doi: 10.1073/pnas.0906773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee M.J., Lee J.K., Choi J.W. Interleukin-6 induces S100A9 expression in colonic epithelial cells through STAT3 activation in experimental ulcerative colitis. PLoS One. 2012;7:e38801. doi: 10.1371/journal.pone.0038801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bronte G., Silvestris N., Castiglia M. New findings on primary and acquired resistance to anti-EGFR therapy in metastatic colorectal cancer: do all roads lead to RAS? Oncotarget. 2015;6:24780–24796. doi: 10.18632/oncotarget.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baltgalvis K.A., Berger F.G., Pena M.M. Interleukin-6 and cachexia in ApcMin/+ mice. Am J Physiol Regul Integr Comp Physiol. 2008;294:R393–R401. doi: 10.1152/ajpregu.00716.2007. [DOI] [PubMed] [Google Scholar]
- 44.Fasano A. Zonulin, regulation of tight junctions, and autoimmune diseases. Ann N Y Acad Sci. 2012;1258:25–33. doi: 10.1111/j.1749-6632.2012.06538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musteanu M., Blaas L., Mair M. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 2010;138:1003–1011. doi: 10.1053/j.gastro.2009.11.049. e1–5. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 1.Natarajan A., Wagner B., Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci U S A. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.el Marjou F., Janssen K.P., Chang B.H. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 3.Clausen B.E., Burkhardt C., Reith W. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 4.Pathria P., Gotthardt D., Prchal-Murphy M. Myeloid promotes formation of colitis-associated colorectal cancer in mice. Oncoimmunology. 2015;4:e998529. doi: 10.1080/2162402X.2014.998529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musteanu M., Blaas L., Mair M. Stat3 is a negative regulator of intestinal tumor progression in Apc(Min) mice. Gastroenterology. 2010;138:1003–1011. doi: 10.1053/j.gastro.2009.11.049. e1–e5. [DOI] [PubMed] [Google Scholar]
- 6.Sibilia M., Fleischmann A., Behrens A. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell. 2000;102:211–220. doi: 10.1016/s0092-8674(00)00026-x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;14:14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanaya H., Natarajan A., Komposch K. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. 2014;16:972–981. doi: 10.1038/ncb3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bollrath J., Phesse T.J., von Burstin V.A. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Greten F.R., Eckmann L., Greten T.F. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Geem D., Medina-Contreras O., Kim W. Isolation and characterization of dendritic cells and macrophages from the mouse intestine. J Vis Exp. 2012;63:e4040. doi: 10.3791/4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vremec D., Shortman K. Dendritic cell subtypes in mouse lymphoid organs: cross-correlation of surface markers, changes with incubation, and differences among thymus, spleen, and lymph nodes. J Immunol. 1997;159:565–573. [PubMed] [Google Scholar]
- 13.Chalaris A., Adam N., Sina C. Critical role of the disintegrin metalloprotease ADAM17 for intestinal inflammation and regeneration in mice. J Exp Med. 2010;207:1617–1624. doi: 10.1084/jem.20092366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.