Abstract
Rock salt represents a potential host rock formation for the final disposal of radioactive waste. The interactions between indigenous microorganisms and radionuclides, e.g. uranium, need to be investigated to better predict the influence of microorganisms on the safety assessment of the repository. Hence, the association process of uranium with two microorganisms isolated from rock salt was comparatively studied. Brachybacterium sp. G1, which was isolated from the German salt dome Gorleben, and Halobacterium noricense DSM15987T, were selected as examples of a moderately halophilic bacterium and an extremely halophilic archaeon, respectively. The microorganisms exhibited completely different association behaviors with uranium. While a pure biosorption process took place with Brachybacterium sp. G1 cells, a multistage association process occurred with the archaeon. In addition to batch experiments, in situ attenuated total reflection Fourier-transform infrared spectroscopy was applied to characterize the U(VI) interaction process. Biosorption was identified as the dominating process for Brachybacterium sp. G1 with this method. Carboxylic functionalities are the dominant interacting groups for the bacterium, whereas phosphoryl groups are also involved in U(VI) association by the archaeon H. noricense.
Introduction
Rock salt is a potential host rock for a final repository of radioactive waste [1]. Many microorganisms are indigenous to rock salt and are able to survive under these extreme environmental conditions, namely high salinities and the lack of nutrients [2–4]. These microorganisms can influence the mobility of radionuclides should the radionuclide be released in a worst-case scenario. Possible bioassociation interactions include the following processes [5–7]: bioaccumulation (uptake of the radionuclide by the cell), biotransformation (change to the oxidation state of the radionuclide), biomineralization (formation of an insoluble precipitate containing the radionuclide and a compound released by the microbes) and biosorption (passive binding of the radionuclide to functional groups of living or dead biomass).
Halophilic microorganisms can be classified as moderate halophiles, which require 0.5 – 2.5 M NaCl, and extreme halophiles, which require 2.5 – 5.2 M NaCl [8–10]. Even though Bacteria as well as Archaea have halophilic members, the extreme halophiles belong commonly to Archaea [8]. Nevertheless, it is important to include thermodynamics and interaction mechanisms of all possible microorganisms in the safety assessment of a potential repository. Only a few studies have been reported regarding the investigation of halophilic microorganisms with radionuclides [11–14]. A study by Francis and coworkers [12] compared the interactions of halophilic archaea and bacteria, as well as non-halophilic bacteria, with uranium. Transmission electron microscopy (TEM) images showed that uranium bound to the cell surface of all investigated microorganisms. Additionally, intracellular granules were observed within the bacterium Halomonas (WIPP A1). However, there is still little knowledge available about the molecular interactions of radionuclides with microorganisms under highly saline conditions and as function of time.
The composition of the microbial cell wall is crucial for a passive biosorption process. The cell envelopes of Archaea and Bacteria are entirely different. Archaea lack a universal cell wall polymer and have a different lipid composition than Bacteria, the isoprenoid side chains are ether-linked, whereas bacterial are mostly ester-linked [15]. Instead of the bacterial rigid cell wall component peptidoglycan (murein), a crystalline protein layer on the cell surface is present for most archaea [15]. Early studies with the extremely halophilic archaeon Halobacterium salinarum (formerly named Halobacterium halobium) showed the presence of such a surface layer (S-layer) protein [16]. This was also found for Halobacterium noricense DSM15987T, the strain of our interest [17]. The S-layer proteins of Haloarchaea are glycosylated and are adapted to high salinity, e.g. they have an enriched amount of acidic residues [18]. The moderately halophilic bacterium used in this work, Brachybacterium sp. G1, belongs to Gram-positive bacteria. The outer phase of the cell wall is composed of peptidoglycan layers with anchored molecules, such as sugars and amino acids. For Brachybacterium faecium DSM4810T, the sugars galactose, glucose and glucosamine are present [19, 20].
In this study, we investigated the bioassociation of Brachybacterium sp. G1, an isolate of the German salt dome Gorleben, with uranium as a function of time, uranium concentration and dry biomass (DBM) concentration. Studies were performed at a pH value of 5.5, which is a typical pH for brines analyzed from US and German subterranean salt formations [21, 22]. Due to its high inventory in nuclear waste, e.g. through production of enriched nuclear fuel or as a by-product of spent-fuel, uranium was used as a representative radionuclide in this study [23]. Under oxic conditions U(VI) is the dominant oxidation state, which is highly soluble. Whereas under reducing conditions U(IV) is dominant and immobile, favoring the retardation of this radionuclide [24]. In addition to microbially induced reduction, the formation of insoluble minerals by microorganisms (biomineralization) is another possible retention process. The formation of uranyl phosphate minerals has been commonly observed [25–27].
The localization of uranium was visualized by electron microscopy, and cell viability was verified by fluorescence microscopy. Furthermore, we used in situ attenuated total reflection Fourier-transform infrared spectroscopy (ATR FT-IR) to elucidate the bioassociation process at a molecular level within a timeframe of 2 h. The obtained results regarding the bioassociation of uranium by Brachybacterium sp. G1 were compared with the results from the extremely halophilic archaeon H. noricense [11].
Materials and methods
Isolation of Brachybacterium from a Gorleben rock salt sample
Gorleben halite was retrieved from a depth of 840 m within the Knäuelsalz layer, in an area known to contain a high concentration of total hydrocarbons - up to 400 ppm C1-C40 and aromatic substances [28]. A significant green biofilm was removed from the sampling area prior to the aseptic removal of halite samples from underneath (Fig 1). Samples were triple-sealed in sterile bags and stored in the dark at ambient temperature until processing in the laboratory.
Fig 1. Sampling area from the Gorleben halite in a depth of 840 m within the Knäulsalz layer.
Approximately 50 g of the hydrocarbon-containing halite was inoculated into 500 mL of low-nutrient medium (per L: 0.5 g yeast extract, 0.5 g casamino acids, 20 g MgCl2·6H2O, 2 g KCl, 0.2 g CaCl2·2 H2O, 0.015 g sodium pyruvate, 1.1 mL of 1 M stock Tris-HCl, pH 8.5, 1 mL American Type Culture Collection trace elements) and incubated aerobically at 22 ± 2°C [29]. The halite sample was the only form of sodium chloride added to the medium for this incubation, resulting in approximately 10% (w/v) NaCl in solution. Growth in the inoculated sample was observed within 3 weeks. Control samples remained sterile. The samples were subcultured onto SW-10 agar [9], yielding growth of yellow-pigmented colonies. The 16S ribosomal RNA gene was amplified with an in situ PCR, using the primers 7f and 1513r (initial denaturation at 95°C for 3 min; followed by 5 cycles initial amplification of 95°C denaturation (1.5 min), 59°C annealing (40 s, in each of the 5 cycles a temperature gradient of -1°C), and 72°C elongation (1.5 min); followed by 25 cycles of 94°C denaturation (1 min), 55°C annealing (40 s) and 72°C elongation 1.5 min, and a final 20 min elongation at 72°C). Amplicons were purified with the QuickStepTM2 PCR Purification kit (EdgeBio, MD, USA) prior to submission to GATC Biotech AG (Cologne, Germany) for sequencing. The 16S7f and 16S1513r sequences were aligned using MEGA 6.06 software (ClustalW) [30] and compared to those from GenBank using the Basic Local Alignment Search Tool (BLAST) server at the National Center for Biotechnology Information (NCBI) [31]. A phylogenetic dendrogram was generated with MEGA 6.06 [30] using the neighbor-joining algorithm and was validated by bootstrap analysis using 500 trial replicates.
To determine the DBM, cells with a defined optical density were pelleted in previously dried (48 h, 70°C) and weighed tubes, dried for 48 h at 70°C and weighed. This determination was made in triplicate.
Cultivation of the microorganisms
Brachybacterium sp. G1. The moderately halophilic bacterium was cultivated in DifcoTM Marine Broth 2216 (Becton, Dickinson and Company, USA) adding NaCl to a final concentration of 10% (1.7 M). The media were filter-sterilized (pore size 0.22 μm) after autoclaving to remove precipitates. Cell suspensions were shaken at 140 rpm at room temperature. The growth was investigated as a function of different parameters. The pCH+ dependency was studied in a range of 3.2–10.7 at a constant NaCl concentration of 4.5% (0.8 M). Salt-dependent studies were performed at pCH+ 7.6 ± 0.2, whereas the ionic strength was varied from 0 to 5.1 M using different ratios of NaCl and MgCl2.
Halobacterium noricense. The extremely halophilic archaeon H. noricense DSM15987T was purchased from the Leibniz Institute DSMZ - German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). It was cultivated in modified DSM372 medium [11]. During growth, the cultures were shaken at 140 rpm in the dark in a water bath at 30°C.
Cells were harvested during late exponential growth by centrifugation at 10,000 x g and 18°C for 10 min and washed three times with the appropriate NaCl solution (3 M NaCl for H. noricense / 1.7 M NaCl for Brachybacterium) at pCH+ 6 to prepare cells for batch studies (see below).
pH correction at high salinity
Due to the high content of salt, the measured pH (negative logarithm of the hydrogen ion activity) had to be corrected. Therefore, the term pCH+, as hydrogen ion concentration, was introduced. According to the empirical equation of Borkowski et al. [32], the relationship between pCH+ and the measured pH is a linear function of the ionic strength:
| (1) |
K is linear to ionic strength:
| (2) |
For a 3 M NaCl solution K is 0.49, for a 1.7 M NaCl solution the pH has to be corrected by adding a K of 0.24.
Uranium bioassociation in batch systems
For uranium bioassociation studies, washed cells from the late exponential growth phase were used. A defined amount of Brachybacterium sp. G1 cells, expressed as DBM, were suspended in 5 mL of 1.7 M NaCl solution at pCH+ 6 and diluted with 5 mL uranium solution, containing 1.7 M NaCl at pCH+ 6. The uranium stock solution was prepared by dissolving UO2(NO3)2 x 6 H2O (Spectrum Chemical Mfg. Corp., USA) in 1 M HCl and heating it to dryness three times. It was then precipitated with sodium hydroxide and dissolved in 0.2 M HCl to a final concentration of 230 mM U(VI). Depending on the experiment, the uranium concentration ranged from 10 – 120 μM. At the investigated pCH+ value, the dominant species is the 3:5 uranyl hydroxo complex—(UO2)3(OH)5+ [33]. For batch experiments DBM concentrations from 0.075–0.125 g/L were used. They were carried out under aerobic conditions in 50 mL tubes, at 25°C in the dark and shaken at 150 rpm. First, time-dependent bioassociation studies with U(VI) were performed over time frames of 5 min up to 48 h. Later, for uranium as well as DBM concentration dependence experiments, an exposure time of 2 h was used. After the designated exposure time, a 500 μL aliquot of a cell suspension of Brachybacterium sp. G1 was filtered with 100 kD Amicon® 2 mL centrifugal filters (Merck Millipore) for 15 min at 13,000 x g. The concentration of unbound uranium in the filtrate was measured with ICP-MS (Inductively coupled plasma mass spectrometry, Agilent 7500ce; Agilent Technologies, Santa Clara, CA, USA). The experiments with Brachybacterium sp. G1 were performed at CEMRC, NM, USA, whereas studies with H. noricense DSM15987T were done at HZDR.
Therefore, the bioassociation studies with H. noricense DSM15987T were slightly modified. After washing, a defined amount of H. noricense cells was pelleted and resuspended in 10 mL uranium solution, pCH+ 6, containing 3 M NaCl [11]. After exposure, the suspension was centrifuged (18°C, 10,000 x g, 10 min). For analyzing the amount of unbound uranium, a sample from the supernatant was analyzed by ICP-MS (Elan 9000 Perkin Elmer, Waltham, MA, USA). Cells were washed with 3 M NaCl, pCH+ 6 for further experiments.
The amount of bound uranium was normalized to DBM or reported as the percentage of added uranium. Control samples without cells were treated in the same way such that abiotic uranium removal from the solution, due to precipitation and/or chemical sorption to the vials could be excluded. Except when otherwise stated, experiments were performed as triplicates and reported errors are standard deviations.
Verification of cell viability
Cell suspensions (1 mL) treated under different experimental conditions were centrifuged at 18°C and 10,000 x g for 10 min. For Live/Dead staining the Kit LIVE/DEAD® BacLightTM Bacterial Viability Kit L7012, (Molecular ProbesTM Inc., Eugene, OR, USA) was used according to the manufacturer´s instructions. The samples were analyzed with an Olympus BX-61 (Olympus Europa Holding GmbH, Hamburg, Germany) microscope using the imaging software ‘‘cellSense Dimension 1.11”. Fluorescence was excited by light with wavelengths between 420 and 460 nm using a super wideband filter mirror unit. The suitability of this kit for halophilic microorganisms was demonstrated by Leuko et al. [34].
Scanning electron microscopy coupled with energy dispersive X-ray spectroscopy
The localization of U(VI) on cells of Brachybacterium sp. G1 was determined using scanning electron microscopy (SEM) coupled with energy dispersive X-ray (EDX) spectroscopy for elemental mapping. Cells exposed to 20 μM U(VI) for 2 h and 48 h were dried on a silicon wafer at room temperature. No fixation steps were performed to avoid preparation-induced changes. The samples were analyzed at ProVIS, Centre for Chemical Microscopy at the Helmholtz Centre for Environmental Research (Leipzig, Germany) with a scanning electron microscope (Zeiss Merlin VP Compact, Carl Zeiss Microscopy, Germany) coupled with an EDX detector (Quantax X-Flash, Bruker Nano GmbH, Berlin, Germany). The electron-acceleration energy was set to 5.8 kV.
In situ ATR FT-IR spectroscopy
The association processes of U(VI) on cells of Brachybacterium sp. G1 as well as H. noricense were monitored using in situ ATR FT-IR spectroscopy with a sub-minute time resolution [32, 33]. The application of reaction-induced difference spectroscopy allows the detection of very small absorption changes provoked by the bioassociation process despite the presence of a very strong absorbing background, i.e. water and microbial cell film.
The microbial cells were prepared as a thin film directly on the ATR crystal´s surface, which was a horizontal diamond crystal (A = 12.57 mm2, DURASamplIR II, Smiths Inc.) with nine internal reflections on the upper surface and a 45° angle of incidence. An aliquot of 7.5 μL of a freshly harvested and three times washed suspension of H. noricense DSM15987T or Brachybacterium sp. G1 with a concentration of 0.5 g/L DBM was pipetted onto the crystal and dried under a gentle flow of nitrogen. This procedure was repeated.
For the in situ sorption experiment the cell film was at first equilibrated by flushing a blank solution (3 M NaCl for H. noricense, 1.7 M NaCl for Brachybacterium sp. G1, at pCH+ 6) for 60 min using a flow cell (V = 200 μL) at a rate of 200 μL/min (Equilibrium phase). Subsequently, the sorption reactions were induced by rinsing the microbial film for the next 120 min with a 40 μM U(VI) solution, equal in background electrolyte and pCH+ to the blank solution. In order to gain information on the reversibility of the process, the blank solution was flushed for another 60 min in a third step.
Infrared spectra were continuously recorded on a Bruker Vertex 80/v vacuum spectrometer, equipped with a Mercury Cadmium Telluride (MCT) detector with a low frequency cut-off at 600 cm−1. Spectral resolution was 4 cm−1 and spectra were averaged over 256 scans. Müller et al. [35, 36] give further details on the experiments performance and on the calculation of difference spectra.
Results and discussion
Phylogenetic characterization of the isolate
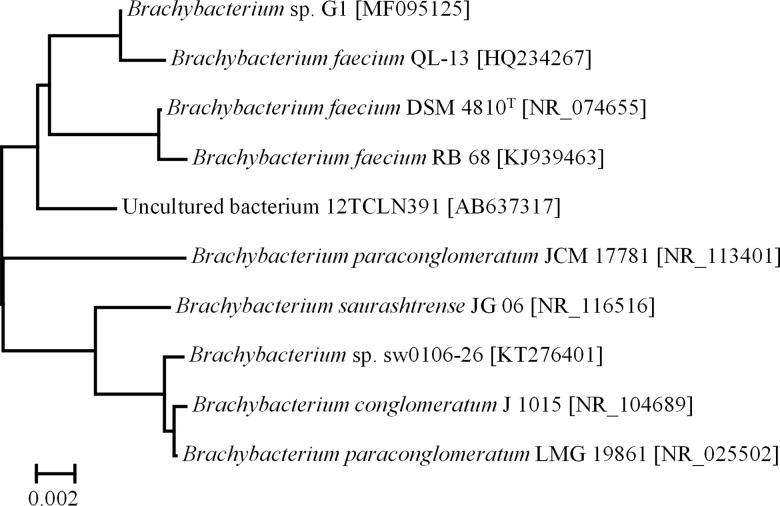
The obtained isolate Brachybacterium sp. G1 (Accession No. MF095125) showed 99% sequence similarity to the 16S rRNA gene sequence of Brachybacterium faecium strain QL-13 (Accession No. HQ234267), Brachybacterium faecium strain DSM4810T (Accession No. NR_074655) and Brachybacterium faecium strain RB 68 (Accession No. AKJ939463) (Fig 2). Brachybacterium faecium strain QL-13 was isolated from a natural Populus euphratica of disused ancient Kiyik River Xinjiang in China and Brachybacterium faecium strain RB 68 was isolated from continental shelf sediments of the Arabian Sea. The type strain Brachybacterium faecium DSM4810T was first isolated from poultry deep litter [37, 38]. Other Brachybacterium species were isolated for instance from Antarctic sea ice brine [39] and the Great Salt Lake, Utah [40].
Fig 2. Phylogenetic dendrogram (neighbor-joining method) of Brachybacterium sp. G1 and its closest phylogenetic relatives based on an alignment of 16S rRNA gene sequences (aligned with ClustalX-MEGA 6.06).
GenBank accession numbers are shown in brackets.
Characterization of the growth of the moderately halophile bacterium
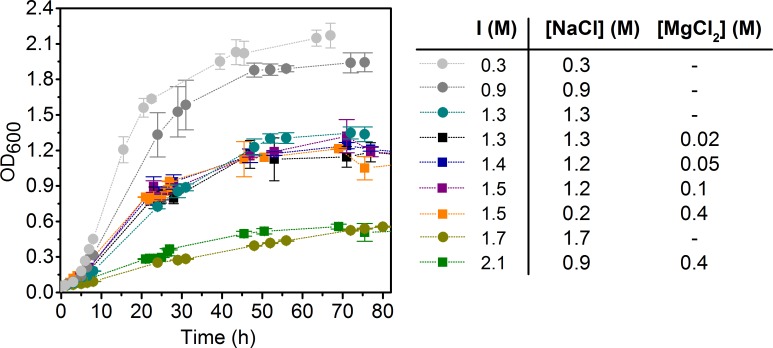
Growth studies on the Brachybacterium sp. G1 isolate showed a tolerance range of 0.3-2.1 M NaCl with an optimum growth between 0.3-1.3 M NaCl. No growth was observed for salt concentrations above 3.4 M NaCl. From variations of the NaCl / MgCl2 ratios it can be concluded that the total ionic strength is more relevant than the composition of the salt fraction. For instance, the same growth behavior was observed for ionic strength of 1.5 M having two different ratios of salt (Fig 3). Brachybacterium sp. G1 grew best with an ionic strength below 0.9 M. No differences occurred in the range of 1.3 M-1.5 M, but elevating the ionic strength to 1.7 M and higher led to an increased inhibition of growth. This suggests that Brachybacterium sp. G1 is a moderately halophilic bacterium [8, 10]. Junge and coworkers isolated a Brachybacterium sp. from Antarctic Sea ice brine and got similar results. The strain was able to grow at relatively high salinities but the presence of salt was not essential, supporting the assumption that they were physiologically adapted to grow at high salt concentrations. This was explained either by a slow adaption from a low salt containing environment or the growth within a various salt containing environment as a selective advantage [39]. The strain is moderately alkaliphilic with a somewhat restricted pCH+ range between pCH+ 7.1–9.1, whereas the optimum is pCH+ 8.1 and minimal growth occurs at pCH+ values 6.1 and 10.1 (S1 Fig). Like other Brachybacterium species [19, 37], Brachybacterium sp. G1 exhibits two morphologies: large cocci occurring in pairs, tetrads, or clumps and short rods in early exponential phase.
Fig 3. Growth of Brachybacterium sp. G1 as a function of ionic strength and salt composition.
Circles indicate NaCl containing media, squares indicate the addition of MgCl2.
Uranium bioassociation behavior
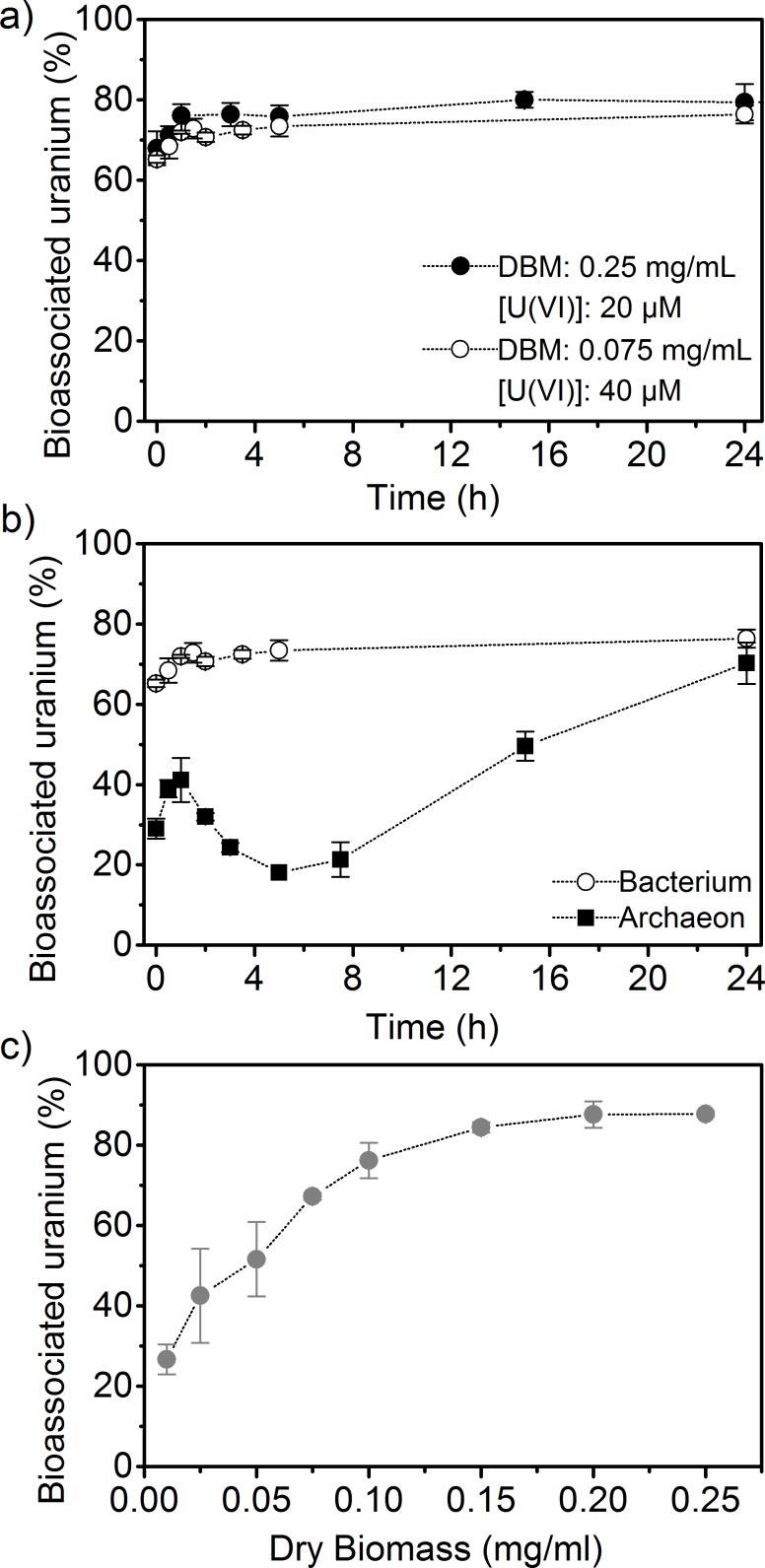
The uranium bioassociation behavior by cells of Brachybacterium sp. G1 was studied as a function of different parameters to gain insights into the interactions of radionuclides with halophilic bacteria. The parameters tested were exposure time, DBM and uranium concentration. Their impacts are summarized in Fig 4A. No differences in bioassociation behavior were apparent when varying either DBM (0.25 mg/mL and 0.075 mg/mL) or uranium concentrations (20 and 40 μM). The high bioassociation values obtained at low DBM (0.075 mg/mL) and increased uranium concentration (40 μM) demonstrate the high sorption capacity of Brachybacterium towards uranyl species. Under these conditions, the amount of U(VI) associated with the bacterium increased from 870 ± 40 mgU/gDBM after 1 h to 971 ± 29 mgU/gDBM after 24 h of exposure. This fast association is characteristic for a biosorption process, which refers to a metabolism-independent association of a sorbate, in our case uranium, to biomass [5, 41] and has been reported for a number of bacteria [42, 43]. Another Actinobacteria, Arthrobacter simplex, adsorbed only 58 mgU/gDBM under slightly different conditions (pH 4.6, 42 μM U(VI), 0.05 mg/mL DBM, 30°C) [43]. In contrast to the simple biosorption process, the bioassociation behavior of uranium (40 μM) with the archaeon H. noricense was found to be different [11] (Fig 4B). H. noricense association with uranium was a rather complex process including multiple steps. Within the first hour of exposure a typical biosorption was observed, followed by a release of uranium for the next four hours and eventually a biomineralization process [11]. Such a multistage uranium bioassociation process was also seen for Microbacterium sp. A9, a bacterium isolated from the Chernobyl exclusion zone [44].
Fig 4.
Bioassociation studies with Brachybacterium sp. G1 and uranium (pCH+ 6, 1.7 M NaCl). a) Time-dependent association with different dry biomass and uranium concentrations, b) comparison to H. noricense (3 M NaCl, 0.5 mg/mL DBM) [11] at 40 μM, c) dry biomass dependent study [U(VI)] = 20 μM).
For a better characterization of the high binding capacity, a DBM dependent study was performed with Brachybacterium sp. G1. Because the association process is relatively fast, an incubation time of 2 h was chosen. No significant increase of associated uranium (20 μM) was detected for DBM higher than 0.1 mg/mL (Fig 4C). Above this level sorption values increased from 76.1 ± 4.4% to 87.8 ± 0.7%. At low DBM (< 0.05 mg/mL) less than 50% of added uranium was associated. For corresponding experiments with H. noricense, an exposure time of 48 h was used, taking the slower and more complex association behavior into account. The obtained results show that DBM below 0.4 mg/mL are not sufficient to associate 50% of initial uranium. Biomass concentrations between 0.5 and 0.7 mg/mL result in the highest sorption values between 89.0 ± 0.2% and 91.0 ± 0.6%. At the higher DBM of 1.8 mg/mL a decrease to 75.3 ± 2.0% was recorded, indicating that the use of a large amount of biomass is not necessary. A reason for this is that cells can interact with each other and decrease the availability of potential binding sites [45, 46].
In conclusion, experiments with Brachybacterium sp. G1 were performed with a DBM of 0.075 mg/mL, but for studies with H. noricense the higher DBM of 0.5 mg/mL was necessary. A possible explanation for the high sorption capacity of Brachybacterium sp. G1 is the high number of carboxyl groups within the peptidoglycan layer of the cell wall [47]. The vast majority of these groups are deprotonated at pCH+ 6.0 and, therefore represent potential binding sites for the predominant positively charged uranyl species. The low DBM also promote a high sorption capacity.
Microscopic investigation of uranium bioassociation
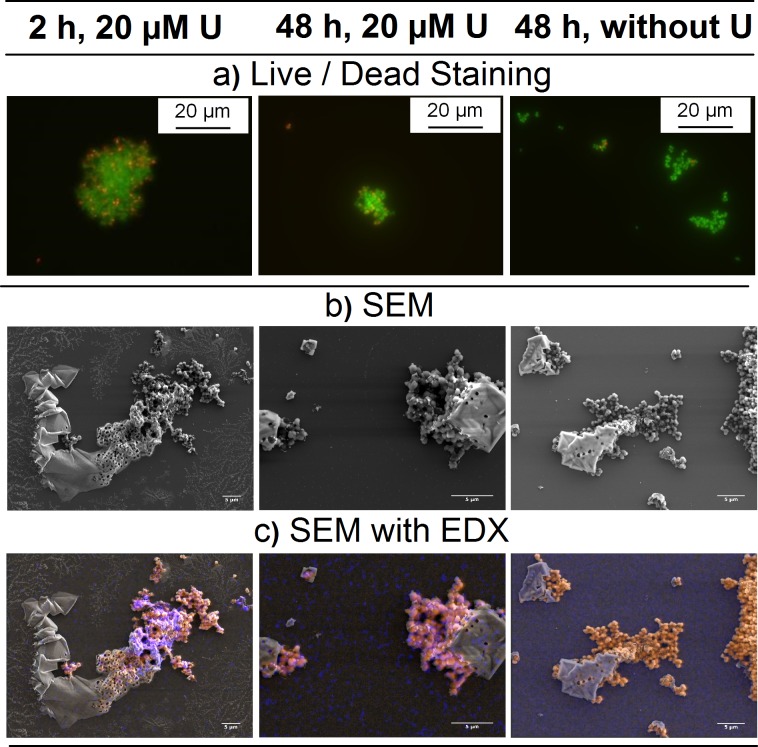
Cells incubated with an initial uranium concentration of 20 μM were investigated with live/dead staining and electron microscopy (Fig 5). From the results, it was determined that cell agglomeration was not induced by the presence of uranium, as the distribution regarding evenly dispersed and agglomerated cells did not change upon exposure to uranium. Rather, a change in distribution occurred due to growth phase. With increasing growth time, the amount of agglomerated cells increased. Cells of Brachybacterium sp. G1 used for uranium interaction studies were harvested in the late exponential phase, a possible explanation for the observed agglomeration. Additionally, agglomeration may have been a response to elevated salt concentration at the ionic strength used for the experiment (1.7 M). Nevertheless, uranium affected neither cell viability nor appearance within the first 48 h. In contrast, a significant change in cell appearance was observed for H. noricense [11]: huge agglomerates formed as a function of exposure time and uranium concentration.
Fig 5.
Micrographs of Brachybacterium sp. G1 incubated with and without uranium (20 μM U, pCH+ 6, 1.7 M NaCl) a) Fluorescence microscopy images of live/dead stained cells (green fluorescence–alive, red fluorescence– dead), b) Electron microscopy images (secondary electrons), c) Mapping of organic elements (orange, C, N, O) and uranium (blue). Scale bar on SEM images is 5 μm. SEM images are enlarged in SI (S2 Fig).
For SEM experiments, no further sample preparations steps were performed and 1.7 M NaCl was used as background electrolyte. Hence, salt crystals are visible on SEM images. Elemental mapping by EDX showed the association of uranium with cells of Brachybacterium sp. G1. The strong EDX signal after 2 h confirmed the fast association process for this microorganism.
Spectroscopic investigation of uranium bioassociation
ATR FT-IR spectroscopy has been proven to be very sensitive to reactions occurring at interfaces between water and solids of mineral or biological origin [11, 48, 49]. It provides in situ, but also time-resolved molecular information [35]. In a flow-through experiment, the absorption properties of an immobile phase, e.g. a microbial film, are directly monitored throughout the surface processes. Surface reactions can be identified in real time with a resolution in the sub-minute range from the pure bacteria up to complete bioassociation with the metal, e.g. U(VI) including all intermediate steps. The kinetic spectral information obtained might be complementary to results of other spectroscopic techniques mainly investigating batch samples.
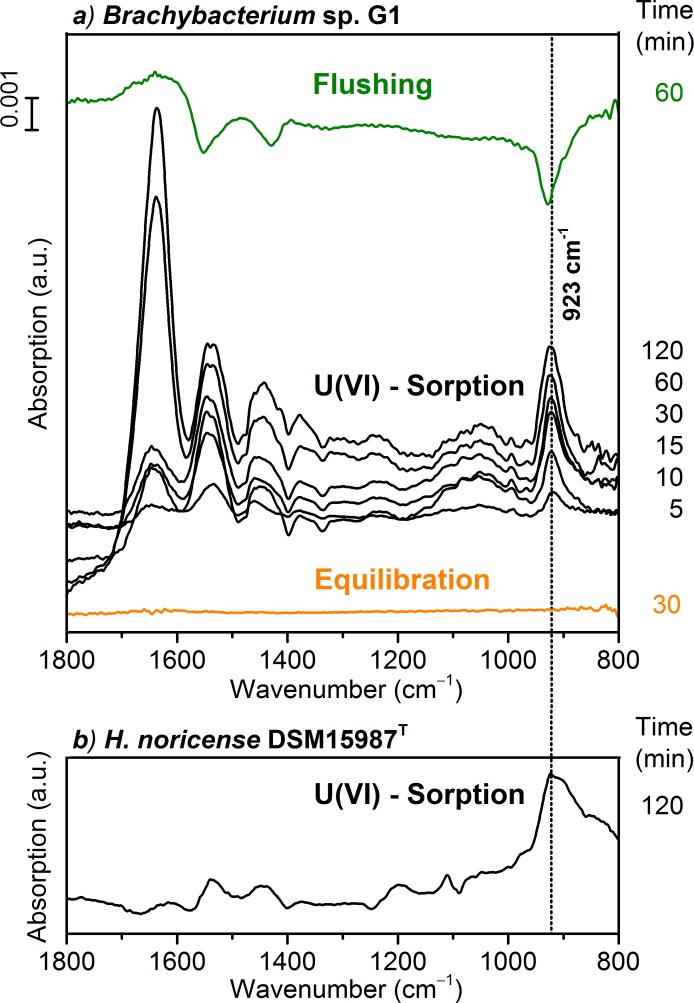
Prior to studying the bioassociation on Brachybacterium sp. G1 cells with in situ ATR FT-IR, the stability of the stationary microbial film on the ATR crystal´s surface was verified. The film was flushed with a blank solution for a prolonged time as an equilibration step. The difference spectrum obtained (Fig 6, red trace) showed no significant absorption changes in the spectral region from 1800 to 800 cm-1 within a time period of 30 min and therefore, indicates a sufficiently stable microbial film under the prevailing continuous flow conditions.
Fig 6.
a) In situ ATR FT-IR difference spectra of U(VI) sorption on Brachybacterium sp. G1 cells ([U(VI)] = 40 μM, pCH+ 6, 1.7 M NaCl). The “Equilibration” spectrum confirms a stable bacterial film on the ATR crystal. “U(VI)—sorption” spectra were recorded at different times after induction of U(VI) association. “Flushing” shows the reversibility. b) For comparison the spectrum of U(VI) bioassociation on H. noricense cells after 120 min ([U(VI)] = 40 μM, pCH+ 6, 3 M NaCl) [11].
In general, we observed seven modes in the two investigated microorganisms, whereby six of them were occurring for the extremely halophilic archaeon and four for the moderately halophilic bacterium (Table 1). The IR spectra of the bioassociation process of Brachybacterium sp. G1 calculated from the spectra recorded before and after distinct times of U(VI) exposure are shown in Fig 6A (middle black traces). These spectra exhibit absorption bands with maxima at 1540, 1430 and 923 cm-1. The intensities of the bands increased during the two hours indicating U(VI) association with the bacterial film. After 120 min, no further intensity increase was observed, showing steady state conditions. For comparison, a corresponding spectrum of the archaeon H. noricense obtained at 120 min after U(VI) exposure is given in Fig 6B. A tentative assignment of the spectral features to functional groups of the microbial system is given in Table 1.
Table 1. Tentative assignment of infrared bands observed in difference spectra of in situ ATR FT-IR with H. noricense DSM15987T ([U(VI)] = 40 μM, [NaCl] = 3 M, pCH+ 6) [11] and Brachybacterium sp. G1 ([U(VI) = 40 μM, [NaCl] = 1.7 M, pCH+ 6).
| Bands with local maxima (cm-1) | Bands with local maxima (cm-1) | Tentative assignment to vibrational mode [50–53] |
|---|---|---|
| H. noricense [11] | Brachybacterium sp. G1 | |
| - | 1638 | Amide-I |
| 1535 | 1540 | νas (COO-)/Amide-II |
| 1435 | 1430 | νs (COO-) |
| 1197 | - | νas (P-O) |
| 1111 | - | νas (P-O) |
| 925 | 923 | ν3 (UO2) coordinated to R–COO− |
| 901 | - | ν3 (UO2) coordinated to phosphoryl residues |
Vibrational modes above 1200 cm-1 are observed for both microorganisms. In the spectra of Brachybacterium sp. G1, several bands are observed which can be attributed to general vibration modes of proteins such as amide-I and amide-II. These modes are usually observed around 1650 and 1550 cm-1 [50, 52].
Furthermore, contributions from carboxylate groups representing preferred binding sites of U(VI) species have to be considered at frequencies around 1550 and 1420 cm-1. Thus, in comparison to spectral data obtained from H. noricense, stronger contributions from protein modes are derived during the coordination of U(VI) to Brachybacterium sp. G1, especially after a sorption time above 60 min. The spectra clearly revealed an enormous increase in band intensity of the amide-I band after 60 min of U(VI) exposure, indicating a change in association behavior. It is conceivable that the spectra represent an uptake of U(VI) by the bacterium and subsequent enhanced interaction with intracellular proteins. This hypothesis needs to be investigated in more detail.
In the spectral range from 1200 to 1000 cm-1, bands mainly representing modes of phosphoryl functionalities but also of sugars residues, can be observed. In case of the archaeon H. noricense, distinct bands at 1197 and 1111 cm-1 correspond to the U(VI) interaction with phosphoryl groups [11, 50]. In contrast, for Brachybacterium sp. G1 no specific bands are observed, indicating no involvement of phosphoryl groups in U(VI) association. Thus, the broad band observed in the range from 1150 to 1000 cm-1 is more likely to indicate vibrational modes of sugar residues mainly of the cell wall. The peptidoglycan layers of Brachybacterium show a variety of anchored sugar molecules; for example galactose, glucose and glucosamine [19, 20]. Vibrational modes observed in the spectral region around 1000 cm-1 correspond to ν(C-O) stretching of the C-O-C glycosidic bond [53], which is present in high numbers in peptidoglycan. We assume that the polysaccharides, or more precisely the glycosidic bonds, do not serve as primary binding sites for U(VI) ions, but they are affected by the biosorption process of uranium leading to a signal in the respective IR spectra.
The absorption band between 950 and 900 cm-1 is assigned to the asymmetric stretching mode of the uranyl moiety ν3(UO2). In the spectrum of Brachybacterium sp. G1, this band shows a constant frequency maximum and bandwidth throughout the time interval of U(VI) exposure indicating the presence of a predominant species. The frequency of the band maximum at 923 cm-1 can be attributed to the coordination of uranyl ions to carboxylate functional groups [51, 52], and/or to aqueous species of U(VI), which also reveal a similar frequency in IR spectra under the prevailing conditions [54]. The spectrum of the subsequent flushing step showed negative bands at 1552, 1429 and 928 cm-1, representing the release of U(VI) species. These bands can be assigned to the antisymmetric and symmetric stretching modes of carboxylate groups and to the ν3(UO2) mode, respectively. The appearance of these bands during the release of uranyl species strongly suggests a coordination of uranyl species predominantly to carboxylate residues. This assumption is supported by the spectrum of H. noricense. The band of the ν3(UO2) mode shows an asymmetric shape and the 2nd derivative indicates maxima at 925 and 905 cm-1, indicating an additional coordination of the uranyl species to phosphoryl groups as was found previously [11, 50, 52].
Conclusions
A fundamental understanding of interactions of halophilic microorganisms with radionuclides and heavy metals is of high importance for the final storage of nuclear waste in salt rock. Therefore, the U(VI) interactions with a moderately halophilic bacterium as well as an extremely halophilic archaeon [11] were studied in detail. It was shown that the two investigated microorganisms interact with uranium in different ways. Uranium binds to Brachybacterium sp. G1 in the beginning in a pure biosorption process, meaning a fast removal from solution. In situ ATR FT-IR hinted a bioaccumulation after 1 h. In contrast the bioassociation with H. noricense is a multistage association process underlying different mechanisms [11]. The sorption capacity of both microorganisms differs significantly. For Brachybacterium sp. G1 maximal sorption capacities up to 971 ± 29 mgU/gDBM were reached after 24 h with 40 μM U(VI), whereas H. noricense achieved merely 9.3 ± 0.4 mgU/gDBM [11] after 14 d. Infrared spectroscopic studies showed the involvement of carboxylate groups in uranium association for both microorganisms, however phosphoryl interactions were only visible for H. noricense.
The bioassociation of uranium with cells of the two halophilic microorganisms is not only different with regard to the kinetics and the related mechanisms but also to the functional groups, which are involved within the first two hours, as studied by in situ ATR FT-IR spectroscopy. Because cells were harvested during late exponential phase and placed into a nutrient-free solution that did not promote growth, the possibility that cells were actively metabolizing was minimized. In this case, an important factor for biosorption is the cell wall structure, which differs significantly between the two investigated strains. The cell wall of the Gram-positive bacterium Brachybacterium sp. G1 consists of peptidoglycan layers with anchored teichoic acids and amino acids, like glutamic acid, glycine and alanine. The presence of glucosamine, galactose and glucose has also been shown [19, 20]. Next to potential sites for metal interactions, e.g. amino and hydroxyl functional groups, distinctly negatively charged sites like glutamic carboxyl and teichoic phosphodiester groups are crucial for metal sorption processes. Beveridge et al. [55] modified amine and carboxyl groups of the cell wall of the Gram-positive bacterium Bacillus subtilis to determine the dominant functional group in metal binding. From the modification of the carboxyl group it was concluded that carboxyl groups of glutamic acid represent the most potent binding sites. The predominant interaction of U(VI) with carboxylate and the absence of significant interactions of the actinide with phosphoryl groups demonstrated by in situ ATR FT-IR spectroscopy as well as the high content of glutamic acids in the cell wall of Brachybacterium faecium DSM4810T [19] support this thesis.
In the case of bioassociation of uranium with H. noricense, a more complex mechanism was considered. Due to the multistage process, it was hypothesized that several different processes are involved. The cell membrane of the extremely halophilic archaeon H. noricense consists of derivatives of phosphatidylglycerol (PG), such as PG sulfate and PG methylphosphate, and S-layer proteins [17]. As described in the literature, the S-layer proteins of Haloarchaea are highly glycosylated, enriched with acidic residues and contain sulfated acid residues [18, 56]. Consequently, the infrared spectroscopic studies from the first two hours of bioassociation showed that in addition to carboxylate functional groups, phosphoryl groups are involved in the bioassociation process.
Both microorganisms are efficiently capable of immobilizing aqueous U(VI) in the lower micro molar range by association of about 80% within 48 h. However, both microorganisms interact in different ways with the actinide. Brachybacterium sp. G1 sorbs uranium within a short time frame on the cell surface and form a biocolloid, which can be further migrated. On the contrary H. noricense interact with uranium in multiple steps, which result in a potential formation of a biomineral [11]. The biomineralization of uranium implies the immobilization of uranium. Therefore, our study provides a detailed understanding of different interaction processes of halophilic bacteria and archaea indigenous to rock salt, which influences the migration behavior of uranium in a rock salt environment.
Supporting information
Growth media was filtered DifcoTM Marine Broth 2216 (Becton, Dickinson and Company, United Sates) with a final NaCl concentration of 4.5% (0.8 M).
(PDF)
Electron microscopy images of Brachybacterium sp. G1 incubated with uranium (20 μM U, pCH+ 6, 1.7 M NaCl) for a) 2 h, b) 48 h and c) without uranium for 48 h. Mapping of organic elements (orange; C, N, O) and uranium (blue).
(PDF)
Acknowledgments
The authors thank Hannes Junghans and Jessica Paff for their support during an internship, Karsten Heim for acquisition of IR-spectra, Jean-Francois Lucchini for the preparation of uranium stock solution, Sabrina Gurlit, Stefanie Schubert and Michael K. Richmann for various ICP-MS measurements. We also thank Gloria Stolzenberg (Bundesamt für Strahlenschutz), Marcus Altmaier and Volker Metz (Karlsruhe Institute of Technology/Institute for Nuclear Waste Disposal (KIT/INE)) for supporting and facilitating the sampling effort at Gorleben and Xavi Gaona (KIT/INE) for obtaining the salt samples.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Hunsche U, Hampel A. Rock salt—the mechanical properties of the host rock material for a radioactive waste repository. Eng Geol. 1999;52:271–291. [Google Scholar]
- 2.Vreeland RH, Piselli AF Jr., McDonnough S, Meyers SS. Distribution and diversity of halophilic bacteria in a subsurface salt formation. Extremophiles. 1998;2:321–331. [DOI] [PubMed] [Google Scholar]
- 3.Fendrihan S, Legat A, Pfaffenhuemer M, Gruber C, Weidler G, Gerbl F, et al. Extremely halophilic archaea and the issue of long-term microbial survival. Rev Environ Sci Biotechnol. 2006;5(2–3):203–218. doi: 10.1007/s11157-006-0007-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGenity TJ, Gemmell RT, Grant WD, Stan-Lotter H. Origins of halophilic microorganisms in ancient salt deposits. Environ Microbiol. 2000;2(3):243–250. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd JR, Macaskie LE. Biochemical basis of microbe-radionuclide interactions In: Keith-Roach MJ, Livens FR, editors. Interactions of microorganisms with radionuclides. Radioactivity in the environment. 2: Elsevier; 2002. p. 313–342. [Google Scholar]
- 6.Selenska-Pobell S, Merroun ML. Accumulation of heavy metals by microorganisms: Biomineralization and nanocluster formation In: König H, Claus H, Varma A, editors. Prokaryotic cell wall compounds—Structure and Biochemistry. Berlin Heidelberg: Springer-Verlag; 2010. p. 483–500. [Google Scholar]
- 7.Jones DL, Andrews MB, Swinburne AN, Botchway SW, Ward AD, Lloyd JR, et al. Fluorescence spectroscopy and microscopy as tools for monitoring redox transformations of uranium in biological systems. Chem Sci. 2015;6(9):5133–5138. doi: 10.1039/c5sc00661a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrei AS, Banciu HL, Oren A. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol Lett. 2012;330(1):1–9. doi: 10.1111/j.1574-6968.2012.02526.x [DOI] [PubMed] [Google Scholar]
- 9.Ventosa A, Nieto JJ, Oren A. Biology of moderatly halophilic aerobic bacteria. Microbiol Mol Biol R. 1998;62(2):504–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oren A. Life at high salt concentrations. Prokaryotes. 2006;2:263–282. [Google Scholar]
- 11.Bader M, Müller K, Foerstendorf H, Drobot B, Schmidt M, Musat N, et al. Multistage bioassociation of uranium onto an extremely halophilic archaeon revealed by a unique combination of spectroscopic and microscopic techniques. J Hazard Mater. 2017;327:225–232. doi: 10.1016/j.jhazmat.2016.12.053 [DOI] [PubMed] [Google Scholar]
- 12.Francis AJ, Gillow JB, Dodge CJ, Harris R, Beveridge TJ, Papenguth HW. Uranium association with halophilic and non-halophilic bacteria and archaea. Radiochim Acta. 2004;92:481–488. [Google Scholar]
- 13.Ozaki T, Gillow JB, Kimura T, Ohnuki T, Yoshida Z, Francis AJ. Sorption behavior of europium(III) and curium(III) on the cell surfaces of microorganisms. Radiochim Acta. 2004;92:741–748. [Google Scholar]
- 14.Ams DA, Swanson JS, Szymanowski JES, Fein JB, Richmann M, Reed DT. The effect of high ionic strength on neptunium (V) adsorption to a halophilic bacterium. Geochim Cosmochim Ac. 2013;110:45–57. [Google Scholar]
- 15.Albers S-V, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. doi: 10.1038/nrmicro2576 [DOI] [PubMed] [Google Scholar]
- 16.Houwink AL. Flagella, gas vacuoles and cell-wall structure in Halobacterium halobium; an electron microscope study. J Gen Micobiol. 1956;15:146–150. [DOI] [PubMed] [Google Scholar]
- 17.Gruber C, Legat A, Pfaffenhuemer M, Radax C, Weidler G, Busse HJ, et al. Halobacterium noricense sp. nov., an archaeal isolate from a bore core of an alpine Permian salt deposit, classification of Halobacterium sp. NRC-1 as a strain of H. salinarum and emended description of H. salinarum. Extremophiles. 2004;8(6):431–439. doi: 10.1007/s00792-004-0403-6 [DOI] [PubMed] [Google Scholar]
- 18.Eichler J. Facing extremes: archaeal surface-layer (glyco)proteins. Microbiology. 2003;149(12):3347–3351. [DOI] [PubMed] [Google Scholar]
- 19.Schubert K, Ludwig W, Springer N, Kroppenstedt RM, Accolas J-P, Fiedler F. Two coryneform bacteria isolated from the surface of French Gruyère and Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov.. Int J Syst Evol Micr. 1996;46(1):81–87. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi M, Yokota A. Cell-wall polysaccharides in coryneform bacteria. J Gen Appl Microbiol. 1989;35:233–252. [Google Scholar]
- 21.Kloppmann W, Négrel P, Casanova J, Klinge H, Schelkes K, Guerrot C. Halite dissolution derived brines in the vicinity of a Permian salt dome (N German Basin). Evidence from boron, strontium, oxygen, and hydrogen isotopes. Geochim Cosmochim Ac. 2001;65(22):4087–4101. [Google Scholar]
- 22.Swanson JS, Cherkouk A, Arnold T, Meleshyn A, Reed DT. The microbiology of subsurface, salt-based nuclear waste repositories: Using microbiol egology, bioenergetics, and project conditions to help predict mirobial effects on repository performance. 2016;LA-UR-16-28895.
- 23.Lindley BA, Fiorina C, Gregg R, Franceschini F, Parks GT. The effectiveness of full actinide recycle as a nuclear waste management strategy when implemented over a limited timeframe–Part I: Uranium fuel cycle. Prog Nucl Energ. 2015;85:498–510. [Google Scholar]
- 24.Suzuki Y, Suko T. Geomicrobiological factors that control uranium mobility in the environment: Update on recent advances in the bioremediation of uranium-contaminated sites. J Miner Petrol Sci. 2006;101(6):299–307. [Google Scholar]
- 25.Mehta VS, Maillot F, Wang Z, Catalano JG, Giammar DE. Effect of reaction pathway on the extent and mechanism of uranium(VI) immobilization with calcium and phosphate. Environ Sci Technol. 2016;50(6):3128–3136. doi: 10.1021/acs.est.5b06212 [DOI] [PubMed] [Google Scholar]
- 26.Lloyd JR, Renshaw JC. Bioremediation of radioactive waste: radionuclide-microbe interactions in laboratory and field-scale studies. Curr Opin Biotechnol. 2005;16(3):254–260. doi: 10.1016/j.copbio.2005.04.012 [DOI] [PubMed] [Google Scholar]
- 27.Choudhary S, Sar P. Uranium biomineralization by a metal resistant Pseudomonas aeruginosa strain isolated from contaminated mine waste. J Hazard Mater. 2011;186(1):336–343. doi: 10.1016/j.jhazmat.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 28.Pusch M, Hammer J, Kus J, Klosa D, Thiemeyer N, Mingerzahn G. Macro- and microscale distribution of hydrocarbons in the Staßfurt Hauptsalz of the Gorleben salt dome. Zeitschrift der Deutschen Gesellschaft für Geowissenschaften. 2014;165(1):3–14. [Google Scholar]
- 29.Swanson JS, Simmons KA, Norden D, Reed DT. Microbial characterization of halite and groundwater samples from the WIPP. Status Report. 2013;LA-UR-13-26280.
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madden T. The BLAST Sequence Analysis Tool. 2013. In: The NCBI Handbook, 2nd edition [Internet]. [Google Scholar]
- 32.Borkowski M, Lucchini J-F, Richmann MK, Reed DT. Actinide (III) solubility in WIPP brine: data summary and recommendations. Los Alamos Report 2009;LA-14360.
- 33.Moulin C, Laszak I, Moulin V, Tondre C. Time-resolved laser-induced fluorescence as a unique tool for low-level uranium speciation. Appl Spectrosc. 1998;52(4):528–535. [Google Scholar]
- 34.Leuko S, Legat A, Fendrihan S, Stan-Lotter H. Evaluation of the LIVE/DEAD BacLight kit for detection of extremophilic archaea and visualization of microorganisms in environmental hypersaline samples. Appl Environ Microbiol. 2004;70(11):6884–6886. doi: 10.1128/AEM.70.11.6884-6886.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller K, Foerstendorf H, Meusel T, Brendler V, Lefèvre G, Comarmond MJ, et al. Sorption of U(VI) at the TiO2–water interface: An in situ vibrational spectroscopic study. Geochim Cosmochim Ac. 2012;76:191–205. [Google Scholar]
- 36.Müller K, Foerstendorf H, Brendler V, Rossberg A, Stolze K, Groschel A. The surface reactions of U(VI) on gamma-Al2O3—In situ spectroscopic evaluation of the transition from sorption complexation to surface precipitation. Chem Geol. 2013;357:75–84. [Google Scholar]
- 37.Collins MD, Brown J, Jones D. Brachybacterium faecium gen. nov., sp. nov., a coryneform bacterium from poultry deep litter. Int J Syst Evol Micr. 1988;38(1):45–48. [Google Scholar]
- 38.Schefferle HE. Coryneform Bacteria in poultry deep litter. J Appl Bact. 1966;29(1):147–160. [Google Scholar]
- 39.Junge K, Gosink JJ, Hoppe HG, Staley JT. Arthrobacter, Brachybacterium and Planococcus isolates identified from Antarctic Sea Ice brine. Description of Planococcus mcmeekinii, sp. nov. Syst Appl Microbiol. 1998;21(2):306–314. doi: 10.1016/S0723-2020(98)80038-6 [DOI] [PubMed] [Google Scholar]
- 40.Bowcutt KM, Burton B, Cox DR. Isolation and characterization of chitin-utilizing halophiles from the Great Salt Lake, Utah. Weber State University ERGO Undergraduate Research Journal. 2008;2:45–56. [Google Scholar]
- 41.Gadd GM. Biosorption: critical review of scientific rationale, environmental importance and significance for pollution treatment. J Chem Technol Biot. 2009;84(1):13–28. [Google Scholar]
- 42.Volesky B, Holan ZR. Biosorption of heavy metals. Biotechnol Prog. 1995;11:235–250. doi: 10.1021/bp00033a001 [DOI] [PubMed] [Google Scholar]
- 43.Veglio F, Beolchini F. Removal of metals by biosorption: a review. Hydrometallurgy. 1997;44:301–316. [Google Scholar]
- 44.Theodorakopoulos N, Chapon V, Coppin F, Floriani M, Vercouter T, Sergeant C, et al. Use of combined microscopic and spectroscopic techniques to reveal interactions between uranium and Microbacterium sp. A9, a strain isolated from the Chernobyl exclusion zone. J Hazard Mater. 2015;285:285–293. doi: 10.1016/j.jhazmat.2014.12.018 [DOI] [PubMed] [Google Scholar]
- 45.Rani MJ, Hemambika B, Hemapriya J, Kannan VR. Comparative assessment of heavy metals removal by immobilized and dead bacterial cells: a biosorption approach. AJEST. 2010;4(2):77–83. [Google Scholar]
- 46.Selatnia A, Boukazoula A, Kechid N, Bakhti MZ, Chergui A, Kerchich Y. Biosorption of lead (II) from aqueous solution by a bacterial dead Streptomyces rimosus biomass. Biochem Eng J. 2004;19(2):127–135. [Google Scholar]
- 47.Bäuerlein E. Biomineralization of unicellular organisms: An unusual membrane biochemistry for production of inorganic nano- and microstructures. Angew Chem Int Ed. 2003;42:614–641. [DOI] [PubMed] [Google Scholar]
- 48.Lefèvre G. In situ Fourier-transform infrared spectroscopy studies of inorganic ions adsorption on metal oxides and hydroxides. Adv Colloid Interface Sci. 2004;107(2):109–123. [DOI] [PubMed] [Google Scholar]
- 49.Jiang W, Saxena A, Song B, Ward BB, Beveridge TJ, Myneni SCB. Elucidation of gunctional groups on Gram-positive and Gram-negative bacterial surfaces using infrared spectroscopy. Langmuir. 2004;20(26):11433–11442. doi: 10.1021/la049043+ [DOI] [PubMed] [Google Scholar]
- 50.Li B, Raff J, Barkleit A, Bernhard G, Foerstendorf H. Complexation of U(VI) with highly phosphorylated protein, phosvitin—A vibrational spectroscopic approach. J Inorg Biochem. 2010;104(7):718–725. doi: 10.1016/j.jinorgbio.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 51.Barkleit A, Foerstendorf H, Heim K, Sachs S, Bernhard G. Complex formation of uranium (VI) with L-phenylalanine and 3-phenylpropionic acid studied by attenuated total reflection fourier transform infrared spectroscopy. Appl Spectrosc. 2008;62(7):798–802. doi: 10.1366/000370208784909607 [DOI] [PubMed] [Google Scholar]
- 52.Barkleit A, Foerstendorf H, Li B, Rossberg A, Moll H, Bernhard G. Coordination of uranium(VI) with functional groups of bacterial lipopolysaccharide studied by EXAFS and FT-IR spectroscopy. Dalton Trans. 2011;40(38):9868–9876. doi: 10.1039/c1dt10546a [DOI] [PubMed] [Google Scholar]
- 53.Kacuráková M, Mathlouthi M. FTIR and laser-Raman spectra of oligosaccharides in water: characterization of the glycosidic bond. Carbohyd Res. 1996;284:145–157. [DOI] [PubMed] [Google Scholar]
- 54.Müller K, Brendler V, Foerstendorf H. Aqueous uranium(VI) hydrolysis species characterized by attenuated total reflextion Fourier-transform infrared spectroscopy Inorg Chem. 2008;47:10127–10134. doi: 10.1021/ic8005103 [DOI] [PubMed] [Google Scholar]
- 55.Beveridge TJ, Murray RGE. Sites of metal deposistion in the cell wall of Bacillus subtilis. J Bacteriol. 1980;141(2):876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.König H, Rachel R, Claus H. Proteinaceous surface layers of Archaea: Ultrastructure and Biochemistry In: Cavicchioli R, editor. Archaea, Molecular and cellular biology. Washington, DC: ASM Press; p. 315–340. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth media was filtered DifcoTM Marine Broth 2216 (Becton, Dickinson and Company, United Sates) with a final NaCl concentration of 4.5% (0.8 M).
(PDF)
Electron microscopy images of Brachybacterium sp. G1 incubated with uranium (20 μM U, pCH+ 6, 1.7 M NaCl) for a) 2 h, b) 48 h and c) without uranium for 48 h. Mapping of organic elements (orange; C, N, O) and uranium (blue).
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.