Abstract
AdeR–AdeS is a two-component regulatory system, which controls expression of the adeABC efflux pump involved in Acinetobacter baumannii multidrug resistance. AdeR is a response regulator consisting of an N-terminal receiver domain and a C-terminal DNA-binding-domain. AdeR binds to a direct-repeat DNA in the intercistronic region between adeR and adeABC. We demonstrate a markedly high affinity binding between unphosphorylated AdeR and DNA with a dissociation constant of 20 nM. In addition, we provide a 2.75 Å crystal structure of AdeR DNA-binding-domain complexed with the intercistronic DNA. This structure shows that the α3 and β hairpin formed by β5–β6 interacts with the major and minor groove of the DNA, which in turn leads to the introduction of a bend. The AdeR receiver domain structure revealed a dimerization motif mediated by a gearwheel-like structure involving the D108F109-R122 motif through cation π stack interaction. The structure of AdeR receiver domain bound with magnesium indicated a conserved Glu19Asp20-Asp63 magnesium-binding motif, and revealed that the potential phosphorylation site Asp63OD1 forms a hydrogen bond with Lys112. We thus dissected the mechanism of how AdeR recognizes the intercistronic DNA, which leads to a diverse mode of response regulation. Unlocking the AdeRS mechanism provides ways to circumvent A. baumannii antibiotic resistance.
INTRODUCTION
Acinetobacter baumannii (A. baumannii) is now recognized as serious nosocomial pathogen and is included as one of the ESKAPE organisms (1). It is the most rapidly emerging multidrug resistant Gram-negative nosocomial pathogen, presenting mainly in pneumonia (particularly ventilator-associated), meningitis, bacteremia and urinary tract infections (2). A. baumannii is resistant to most of the commonly used broad spectrum antimicrobials, presenting a great challenge for the control and eradication of infections (3). Multiple mechanisms have been described to be responsible for multidrug resistance in A. baumannii, and of particular importance are the adaptive and mutational resistance mediated by multidrug efflux pumps (4).
The Resistance-Nodulation-Cell Division (RND) superfamilies of multidrug efflux pumps are mainly identified in Gram-negative bacteria. These efflux pumps actively transport a broad range of substrates, including antimicrobials, out of the cell via a ternary complex that spans the inner membrane, the periplasm and outer membrane (5). In A. baumannii, three RND efflux pumps AdeABC, AdeFGH and AdeIJK have been characterized to be tightly associated with its multidrug resistance phenotype (6–8). Of these, the AdeABC efflux conferred resistance to aminoglycosides, tetracyclines, fluoroquinolones, chloramphenicol, erythromycin, trimethoprim and tigecycline (6,9,10). The AdeABC efflux is genetically linked and tightly controlled by the AdeR–AdeS two-component regulatory system (11). Overproduction of the AdeABC efflux pump resulting in decreased antimicrobial susceptibility was reported to be caused by mutations in the adeRS genes (8,12–14). These genes encode a classical two-component regulatory system (TCS) consisting of a transmembrane sensor kinase and a response regulator. TCS constitute the dominant bacterial signaling system allowing them to adapt to environmental stimuli and display an intrinsic feedback mechanism to survive under stress responses (15–17). Within AdeRS, the histidine kinase AdeS senses environmental stimuli, while the response regulator AdeR mediates the cellular response by receiving a phosphoryl signal from AdeS and further stimulates the expression of its target genes (15,17,18). It is not only the production of the AdeABC efflux pump that is mediated; it is proposed that AdeRS also regulates genes required for biofilm formation and virulence, but in a strain-specific manner (19). Therefore, two-component systems have been highlighted to potentially serve as an effective drug target, especially by targeting the response regulator (16,20).
The response regulator AdeR consists of a CheY-like receiver domain (RD, amino acid residues 1–127) and an OmpR/PhoB type DNA binding domain (DBD, residues 138–247). Unlike most OmpR/PhoB type response regulators which bind to the promoter region of their target genes (21,22), AdeR recognizes a 10bp perfect direct-repeat DNA sequence in a 168bp intercistronic region between adeR and adeABC (23).
We combined biophysical and structural biology approaches to elucidate the mechanism by which AdeR recognizes this intercistronic DNA and how this is regulated through its receiver domain. The structural details of the AdeR receiver domain and the AdeR DNA binding domain in complex with the intercistronic region DNA are provided and complemented with mutagenesis and thermodynamic analysis. We further validated the assembly of full length AdeR in complex with the DNA segment in solution with small angle X-ray scattering. The involvement of AdeR regulation related to its mutations in clinical isolates of the multidrug resistant A. baumannii and its potential in antimicrobial strategy are discussed.
EXPERIMENTAL PROCEDURES
Cloning and site-directed mutagenesis
All strains were grown in Lysogeny Broth (LB, 10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl), unless otherwise indicated. All genes were cloned from A. baumannii ATCC19606, and Escherichia coli DH5α was used for transformations. Constructs were generated to produce a (1) full-length AdeR (AdeR_FL), (2) the receiver domain with linker sequence (AdeR_RD, 1–137) and (3) the DNA binding domain (AdeR_DBD, 138–247). The AdeR_FL, AdeR_RD and AdeR_DBD were cloned into a modified pET-28a vector with an N-terminal His6-tag and overexpressed in E. coli BL21 (DE3). Point mutations were generated through the PCR based site-directed mutagenesis approach using the pET28a-AdeR_FL construct (24). All primers used are listed in Supplementary Table S1. Point mutations were verified by sequencing.
Protein overexpression and purification
The E. coli BL21 (DE3) cells transformed with pET28-AdeR_FL, pET28-AdeR_RD or pET28-AdeR_DBD, were grown to an OD600 of 0.7 at 28°C in LB medium supplemented with 100 μg/ml Kanamycin. After induction with 1 mM IPTG, cultures were incubated for 6 h prior to harvesting by centrifugation at 7000g (Beckman) and the resulting cell pellets frozen at –20°C. The frozen cell pellets were resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl) and lysed by sonication at 4°C. The lysate was further centrifuged at 25 000g to remove cell debris.
Filtered lysate supernatant was loaded onto a Ni-NTA column (Qiagen) pre-equilibrated with loading buffer and washed with 5% elution buffer (25 mM Tris pH 8.0, 300 mM NaCl, 500 mM imidazole) followed by a 5–100% gradient elution with an ÄKTA purifier system. The fractions containing the eluted proteins were concentrated and loaded on Superdex75 16/60 (GE Healthcare) size exclusion chromatography column in 25 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol. The main protein fractions were concentrated for further usage. The protein concentration was measured with a NanoDrop® spectrometer (Thermo Fisher) using the extinction coefficient generated from ExPASy ProtParam program (25). All the mutated AdeR proteins followed the same purification approach as used for the wild type.
Crystallization and data collection
The recombinant AdeR_DBD protein was complexed with the intercistronic DNA by mixing a 2:1 molar ratio of AdeR_DBD protein and the synthesized 25 bp DNA (5′ TAAAGTGTGGAGTAAGTGTGGAGAA 3′), respectively. The complex was further purified and buffer exchanged in 25 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol using a Superdex75 16/60 (GE Healthcare) size-exclusion chromatography column. For crystallization screening, the purified complex was concentrated to 6–8 mg/ml and the AdeR_RD recombinant protein was concentrated to 15–20 mg/ml. Crystallization screening was performed in a 392-well Hampton crystallization plate via sitting-drop vapor diffusion at 20°C against commercially available sparse matrix screens (Molecular Dimensions and Hampton Research). Selected hits were repeated and optimized manually with a crystallization drop containing 0.5 μl of purified protein plus 0.5 μl of reservoir solution. The AdeR_DBD–DNA complex was crystallized in 0.2 M ammonium citrate dibasic, 20% PEG 3350, and the AdeR_RD was crystallized in 0.1 M sodium acetate trihydrate pH 4.6 and 2 M sodium formate. AdeR_RD in the following buffer 5.3 mM BeSO4, 35 mM NaF, 7 mM magnesium chloride, was crystallized under multiple conditions and the reported crystal structure was discovered in 2.8 M sodium acetate trihydrate at pH 7.0. All the obtained crystals were soaked in cryoprotectant solution containing the reservoir solution supplemented with 15–30% (v/v) glycerol and were flash-frozen in liquid nitrogen. X-ray diffraction data were collected on the Beamlines BL18U and BL19U at Shanghai Synchrotron Radiation Facility (SSRF) at 100 K.
Data processing and structure determination
X-ray diffraction data were integrated and scaled using XDS suite (26). The structure of AdeR_RD was determined with the maximum-likelihood molecular replacement implemented in the Phaser program suite (27), using the truncated structure of the Bacillus subtilis YycF/WalR receiver domain (PDB: 2ZWM) as a search model. This model had a sequence identity of 37%. The structure of the AdeR_RD in complex with Mg2+ was solved using the obtained AdeR_RD structure as the molecular replacement search model. The structure of AdeR_DBD in complex with the intercistronic DNA was solved using the BALBES molecular replacement platform. As a template model, the sequence of the AdeR_DBD. B. subtilis YycF/WalR DNA binding domain (PDB: 2DIV) was selected (28,29). The program COOT and PHENIX suite were used for further manual model rebuilding and refinement (30,31). The crystal structure of AdeR_RD was finally solved at 1.4 Å in spacegroup P3221 with the following cell dimensions a = b = 65.3 Å, c = 50.3 Å and α = β = 90°, γ = 120°. The structure was refined to a final Rwork/Rfree of 0.1844/0.2045. The AdeR_RD structure contains one molecule of AdeR_RD in the asymmetric unit, including the residues 10–129, and 105 water molecules. The structure of AdeR_RD in complex with Mg2+ was solved at 1.6 Å resolution using similar cell parameters as AdeR_RD, and refined to a final Rwork/Rfree of 0.1758/0.2074. The structure of the AdeR_DBD, bound to the intercistronic DNA complex was solved at 2.75 Å in spacegroup P1 with a = 55.5 Å, b = 71.4 Å, c = 78.4 Å and α = γ = 102°, β = 105°. Global NCS and TLS parameters were applied in the final refinement. The final structure was refined to a Rwork/Rfree of 0.2462/0.2846. In total, there are six AdeR_DBD molecules in the asymmetric unit with four of them interacting with two 25bp DNA molecules, respectively, and 77 water molecules were observed in the asymmetric unit. All the data collection and refinement statistics are summarized in Table 1.
Table 1. X-ray data collection and refinement statistics.
| Crystal | AdeR_RD | AdeR_RD Mg | AdeR_DBD&DNA complex |
|---|---|---|---|
| Data collection | |||
| Spacegroup | P3221 | P3221 | P1 |
| a, b, c (Å) | 65.3, 65.3, 50.3 | 64.8, 64.8, 50.0 | 55.5, 71.4, 78.4 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 102, 105, 102 |
| Resolution (Å) | 37.58–1.40 (1.45–1.40) | 28.04–1.60 (1.65–1.60) | 43.82–2.75 (2.85–2.75) |
| R merge | 0.115 (1.06) | 0.045 (0.58) | 0.106 (0.981) |
| R meas | 0.118 | 0.046 | 0.124 |
| Multiplicity | 19.1 (15.5) | 18.8 (14.0) | 3.6 (3.6) |
| CC (1/2) | 0.998 (0.831) | 1 (0.915) | 0.996 (0.601) |
| CC* | 1 (0.953) | 1 (0.978) | 0.999 (0.867) |
| I/σ(I) | 15.20 (1.92) | 36.84 (3.31) | 9.97 (1.29) |
| Completeness (%) | 99.83 (98.37) | 99.09 (96.05) | 97.95 (97.83) |
| Wilson B-factor (Å2) | 16.60 | 23.54 | 58.79 |
| Refinement | |||
| Total reflections | 470 483 (37 348) | 305 468 (21 852) | 99 115 (10 127) |
| Unique reflections | 24 660 (2412) | 16 262 (1560) | 27 914 (2791) |
| R work/Rfree | 0.1884/0.2045 | 0.1758/0.2074 | 0.2462/0.2846 |
| Number of atoms: | |||
| Macromolecules | 930 | 958 | 6459 |
| Water | 105 | 76 | 77 |
| Ligand | 1 | ||
| Average B-factor (Å2) | 29.00 | 36.09 | 49.05 |
| Protein (Å2) | 28.20 | 35.84 | 49.28 |
| Water (Å2) | 36.30 | 39.21 | 29.73 |
| Ramachandran plot: | |||
| Favored/allowed (%) | 97/3 | 96/4 | 93/7 |
| Root-mean-square-deviation: | |||
| Bond lengths (Å) | 0.010 | 0.008 | 0.003 |
| Bond angle (°) | 1.40 | 1.11 | 0.84 |
Statistics for the highest resolution shell are shown in parentheses.
Isothermal titration calorimetery
All the ITC experiments were performed with a Microcal ITC200 calorimeter (GE Healthcare) at 25°C. The protein and DNA samples were buffer exchanged to 25 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, using size-exclusion chromatography prior to the experiments. The experiments were conducted by injecting AdeR_FL and AdeR_FLR231A protein (100–120 μM), AdeR_DBD or AdeR_FLR122A recombinant protein (200–250 μM), into 10–14 μM of the 25 bp intercistronic region DNA. The injections contained 0.4 μl for the first injection and 2 μl for the second to 19th injection, with 120 s intervals. All the ITC data were analyzed with the supplemented Microcal ITC data analysis package under the one binding site mode.
Electrophoretic mobility shift assay
The electrophoretic mobility shift assay (EMSA) experiments were carried out by applying the protocol of the LightShift Chemiluminescent EMSA Kit (Thermo Scientific) using biotin to detect protein–DNA interaction. The nucleotides used for the EMSA had the same sequence as those used for ITC but were labeled with biotin at their 5′ primer. The binding reactions were incubated in 10 μl volume containing 1 pmol DNA and a protein concentration gradient in the binding buffer (10 mM Tris, 50 mM KCl, 1 mM DTT, 5% glycerol, pH 7.5) for 30 min. The reaction mixtures were loaded onto a 5% native polyacrylamide gel and separated in 0.5 × TBE at 100 V on ice, followed by western blotting for biotin.
Small angle X-ray scattering
SAXS data were collected at the National Center for Protein Science, Shanghai, BL19U2 beamline in Shanghai Synchrotron Radiation Facility (32). Measurements were carried out at 293K within a momentum transfer range from 0.04 to 0.45 Å−1. Protein or Protein-DNA complex samples were measured under batch mode at solute concentrations in 25 mM Tris pH 8.0, 150 mM NaCl, 5% glycerol buffer. Twenty frames were collected for each sample and no measurable radiation damage was detected. Data processing was performed by following the procedure described previously (33). Briefly, buffer subtraction and extrapolation were performed with the program PRIMUS; the Guinier region was evaluated with the Guinier approximation. The Rg (radius of gyration), I0 (forward scattering), Dmax (maximum particle dimension) and the distance distribution function P(r) were determined with the program GNOM (34). The rigid body and the ab initio modeling were performed with the SASREF, DAMMIF and DAMAVER program respectively under the ATSAS package (35). CRYSOL from ATSAS package (35,36) was used to resolve the respective discrepancy of crystal structure or models to the experimental data. The molecular weight determination from SAXS data was performed using the MoW2 online resource (37). SAXS data collection, analysis and statistical details are summarized in Supplementary Table S2.
RESULTS
Purification and thermodynamics of AdeR and its interaction with the intercistronic region DNA
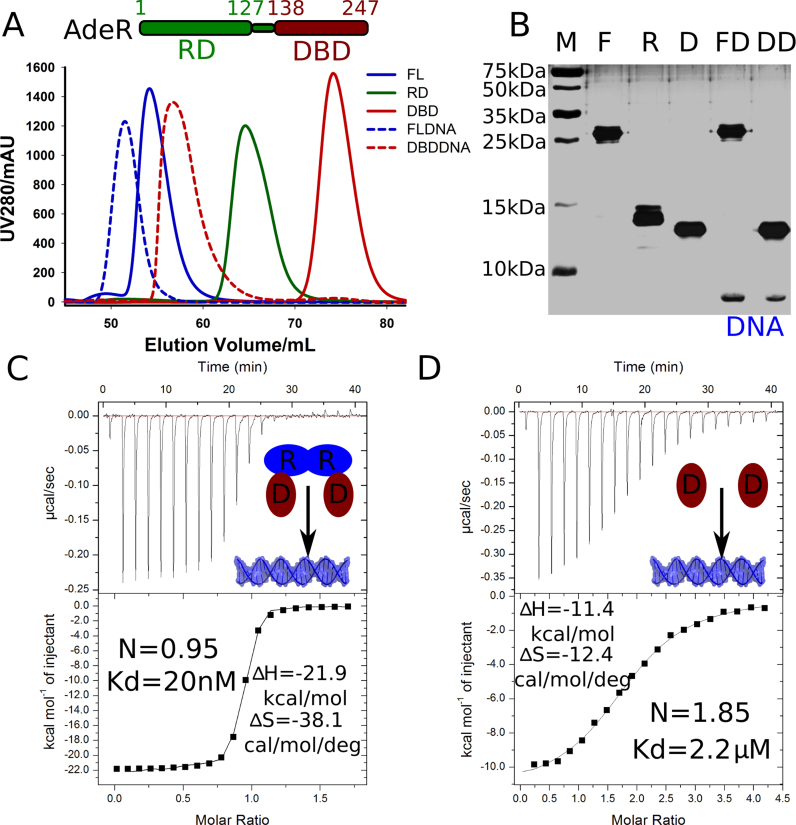
The full length AdeR protein consists of a receiver domain (RD) and an OmpR/PhoB-like DNA binding domain (DBD). To investigate the AdeR interaction with the adeR-adeABC intercistronic DNA, we constructed and purified the recombinant full length AdeR (AdeR_FL), as well as the AdeR receiver domain (AdeR_RD), and AdeR DNA binding domain (AdeR_DBD), separately (Figure 1A). The elution profile of size-exclusion chromatography, calibrated with a standard protein mixture, revealed that both AdeR_FL and AdeR_RD eluted as a dimer, while AdeR_DBD eluted as a monomer. The molecular weight determination was cross-validated using small angle X-ray scattering (Supplementary Table S2). A DNA fragment consisting of the direct repeat sequence in the intercistronic region between adeR and the RND efflux pump adeABC was previously identified to interact with AdeR (23). Here, we show that both AdeR_FL and AdeR_DBD alone can bind a 25 bp DNA fragment containing this direct repeat sequence, forming a complex with a distinct chromatographic behavior (Figure 1A). All the size-exclusion chromatography purified samples were further analyzed by SDS-PAGE followed with silver staining, to visualize both protein and protein–DNA complex content (Figure 1B).
Figure 1.
Purification and thermodynamics of AdeR and its interaction with the intercistronic DNA. (A) Schematic view of AdeR and size exclusion chromatography profile of AdeR full length (FL), receiver domain (RD), DNA binding domain (DBD). FLDNA and DBDDNA are the complex of AdeR FL and DBD with the intercistronic DNA region, respectively. (B) Silver staining SDS PAGE of SEC purified AdeR samples from panel A, the DNA band is highlighted. (C and D) The isothermal titration calorimetery of AdeR_FL and AdeR_DBD with the intercistronic DNA, the stoichiometry (N), dissociation constant; enthalpy (ΔH) and entropy (ΔS) are denoted.
The interactions between AdeR_FL or AdeR_DBD and DNA were quantified by isothermal titration calorimetery (ITC) (Figure 1C and D). The dimer AdeR_FL binds to the intercistronic DNA in a markedly high-affinity interaction with an equilibrium dissociation constant (Kd) of 20 nM, a molar reaction enthalpy (ΔH) of –21.9 kcal/mol, and entropy of –38.1 cal/mol/deg (Figure 1C). The AdeR_DBD monomer was also able to interact with DNA, but with an almost 100 times weaker interaction as indicated by the Kd of 2.2 μM, a molar reaction enthalpy ΔH of –11.4 kcal/mol, and entropy of –12.4 cal/mol/deg (Figure 1D).
Crystal structure of the AdeR receiver domain
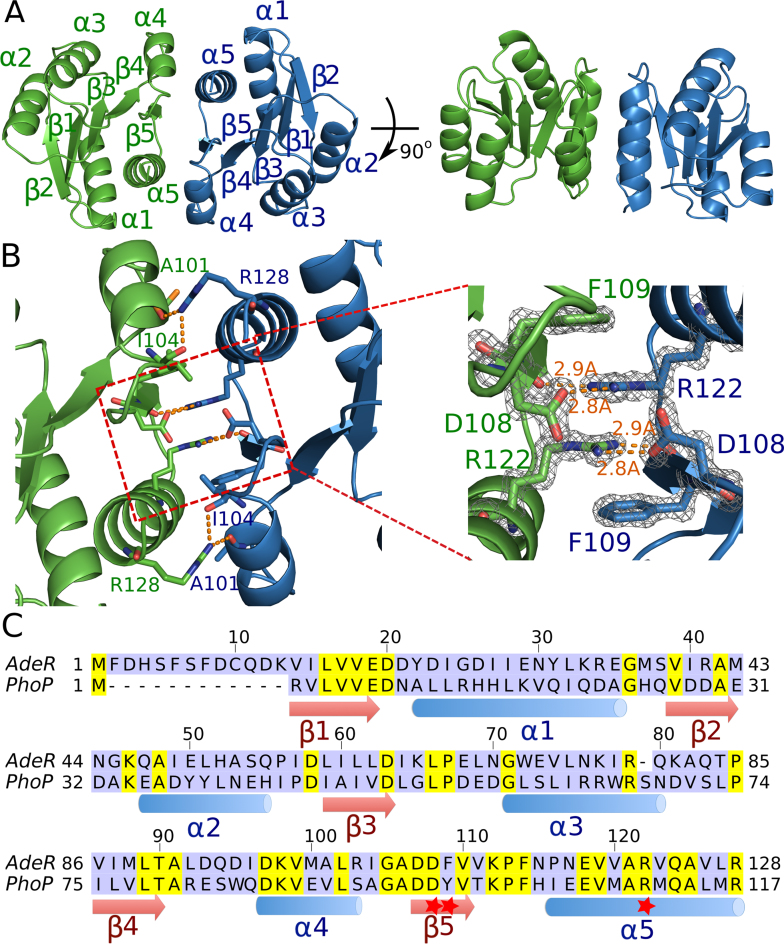
To explore why the binding affinity drops after removing the receiver domain, we first solved the crystal structure of AdeR_RD at 1.4 Å. The crystals presented one AdeR_RD molecule in the asymmetric unit (Table 1). AdeR_RD has the canonical (βα)5 repeat motif of the RR regulatory domain of two component systems. It has a helix-sheet-helix sandwich with the five β strands forming a parallel β sheet in the center surrounded by α1 and α5 on one side, and α2, α3, α4 on the other side (Figure 2A). AdeR_RD assembles as a compact dimer, generating a 2-fold symmetry. The dimerization interface is formed by α4–β5–α5 with a buried surface of 792.5 Å2, determined using the PDBPISA server (Figure 2A). Arg128 interacts via hydrogen bonds with both Ala101 and Ile104 at both sides of the dimerization interface (Figure 2B). In the core region of the dimerization interface, Arg122 also forms strong hydrogen bonds with the main chain at residue Phe109 and with the side chain of Asp108, with bond lengths of 2.9 and 2.8 Å, respectively. Noteworthy, the benzene ring of Phe109 and the side chain of Arg122 from one monomer form a gear wheel-like structure with the other monomer by forming a stack cation π interaction. This structure is further stabilized by a hydrogen bond between Arg122 and Asp108 at both sides (Figure 2B). The key residues involved in this dimerization interface, D108-F109-R122 are relatively conserved in A. baumannii AdeR and are also found in other response regulators such as the well-studied PhoP, where F109 is replaced by Y (Figure 2C).
Figure 2.
Crystal structure of the AdeR receiver domain. (A) Crystal structure of the AdeR receiver domain dimer in different orientations. The AdeR receiver domain consist of five α helices and five β stands forming a parallel β in between, the α4–β5–α5 motif is involved in the dimerization. (B) Detailed interaction of residues involved in the AdeR receiver domain dimerization, the zoom-in view highlights D108, F109 and R122, forming a stable gear wheel like structure. The density map of D108, F109 and R122 are shown with 2Fo – Fc = 3σ. (C) Sequence alignment of AdeR and the response regulator PhoP receiver domain, secondary structures are denoted and the red star indicates the key residues involved in AdeR dimerization.
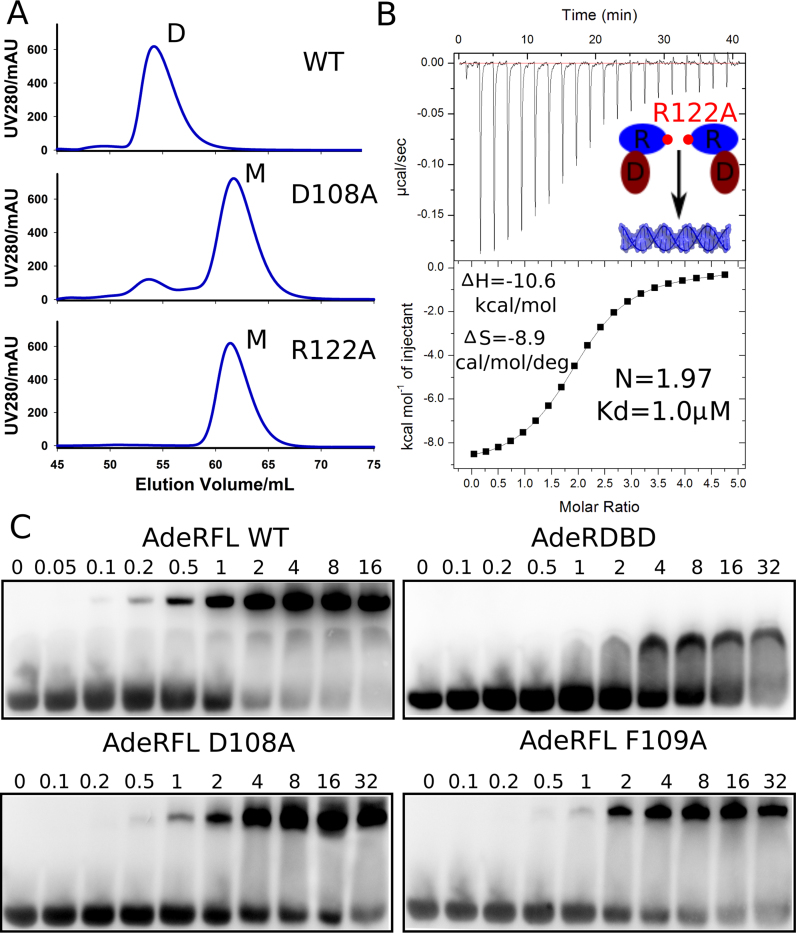
To investigate the necessity of the receiver domain dimerization for binding to the intercistronic DNA, we constructed multiple point mutations of residues involved in AdeR dimer interactions. In size-exclusion chromatography, the wild type AdeR_FL was purified as a homogenous dimer, while the D108A mutant presented predominantly as a monomer. Mutation to alanine of the key residue R122, involved in the cation π interaction and in hydrogen bonds with D108 and F109, completely disrupted the dimerization of AdeR (Figure 3A). In addition, ITC analysis of DNA binding of this AdeR R122A monomer demonstrated a Kd of 1 μM and a 2:1 stoichiometry with a molar reaction enthalpy ΔH of –10.6 kcal/mol and entropy of –8.9 cal/mol/deg (Figure 3B). Notably the binding affinity of AdeR_FL R122A to the intercistronic DNA was consistent with the AdeR DNA-binding domain on its own. We further cross-validated the mutagenesis study by an EMSA experiment. The mobility shift indicated that binding of AdeR_FL to the intercistronic DNA was saturated at around 2 μM, while AdeR_DBD, AdeR_FL D108A and AdeR_FL F109A all exhibited similar binding but could not be saturated even at 32 μM (Figure 3C).
Figure 3.
Mutagenesis studies of AdeR receiver domain. (A) Size-exclusion chromatography analysis shows that the residues D108A and R122A could disrupt the full length dimerization of AdeR. (B) Isothermal titration calorimetry experiment indicates the thermodynamics of AdeR full length R122A mutation exhibits a similar profile as the AdeR DNA binding domain alone. (C) Electrophoretic mobility shift assay validates that D108A and F109A interact with the intercistronic DNA similar to the AdeR DNA binding domain alone in a much lower affinity compared to wildtype AdeR full length.
The interaction between the AdeR DNA binding domain and the intercistronic DNA
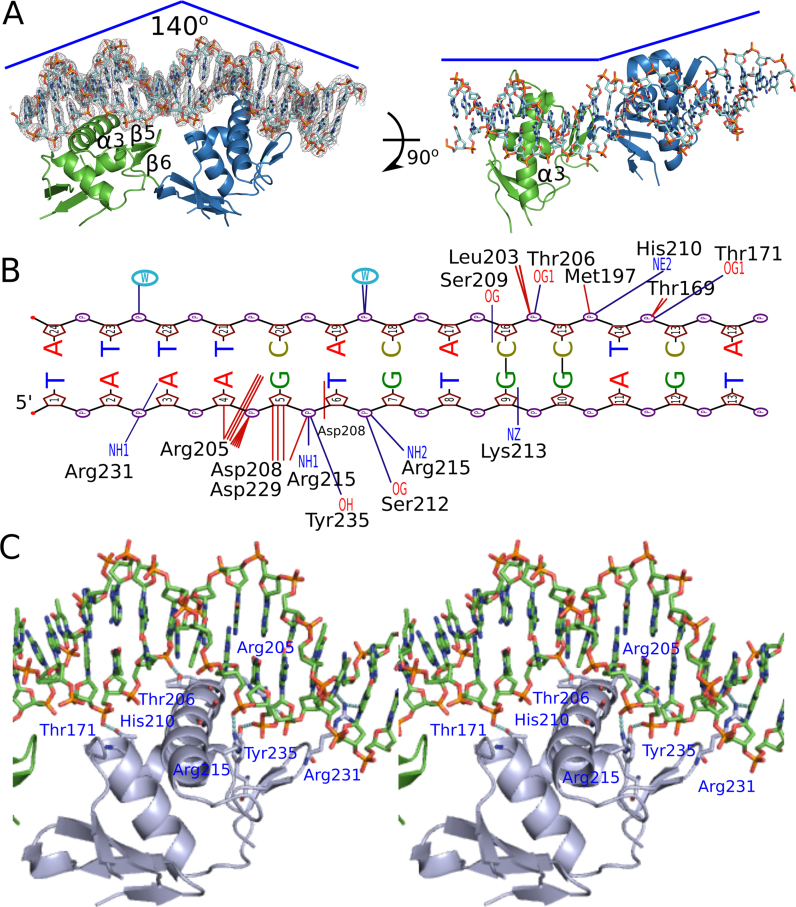
AdeR binds to a 10 bp direct repeat DNA sequence containing only a single random nucleotide in between (AAGTGTGGAGNAAGTGTGGAG), present in the intercistronic region between the genes encoding AdeR and the efflux pump AdeABC. The crystal structure of recombinant AdeR_DBD protein in complex with a 25 bp DNA segment containing this repeat sequence was solved at 2.75 Å in the P1 spacegroup. There were six monomers of AdeR_DBD, with four of them binding to two DNA molecules in the asymmetric unit. The fifth AdeR_DBD molecule was trapped between two DNA molecules and the last AdeR_DBD molecule barely interacted with the DNA (Supplementary Figure S1). The AdeR_DBD forms a traditional OmpR/PhoB-like DNA binding domain with three α helices and five β strands. Each AdeR_DBD molecule binds to the DNA with a contact surface of about 485 Å2, mainly through the α3 and β hairpin formed by β5 and β6 (Figure 4A). This binding of the AdeR_DBD to the DNA resulted in a 140 degrees bend towards the protein that is similar to that seen with the response regulator PhoP (Figure 4A, left panel). The two AdeR_DBD monomers are not perfectly aligned with the DNA strand direction (from 5′ to 3′), and hence they introduce a curvature of the DNA strand (Figure 4A, right panel). In addition, the alignment of AdeR_DBD bound and unbound to DNA indicated a conformational change of the β5 and β6 hairpin to adapt to the minor groove, leading to and more residues tracing of were traced of the flexible loop linked to the α3 helix resulting from the DNA binding (Supplementary Figure S1). There are at least 10 hydrogen bonds and multiple van der Waals interactions involved in the formation of the AdeR_DBD/DNA complex. The detailed interaction was plotted with NUCPLOT as shown in Figure 4B (38). The NH2 groups from positive residues such as Arg205, Arg215, Arg231 and the OH group from Tyr235, the OG group from Ser209, Ser212 and Thr171 form hydrogen bonds with the backbone of the DNA helix. Specific interactions of AdeR with the DNA including Arg231–3A’ (the primer indicates the template strand), Arg205–4A’-5G’, Asp208–6T’, Lys213–9G’ and Ser209–16C (Figure 4B), establish the basis for the recognition sequence specificity. The main α3 helix interacts with the major groove of the DNA, while Arg231 from the β hairpin formed by β5 and β6 inserts into the minor groove of the DNA (Figure 4C).
Figure 4.
Crystal structure of AdeR DNA binding domain in complex with the intercistronic DNA. (A) Overall structure of AdeR_DBD with DNA complex. Two monomers of AdeR_DBD binds with the two direct repeat intercistronic regions of DNA. The α3 and β hairpin formed by the β5 and β6 are involved in the binding of major and minor groove respectively. The density of the DNA in the left panel is shown in 2Fo – Fc = 1.5σ. (B) NUCPLOT of the detailed interaction between one AdeR_DBD and a single repeat of the intercistronic DNA. The dark blue represents the hydrogen bonds. (C) Detailed main interaction between the AdeR DNA binding domain and the intercistronic DNA.
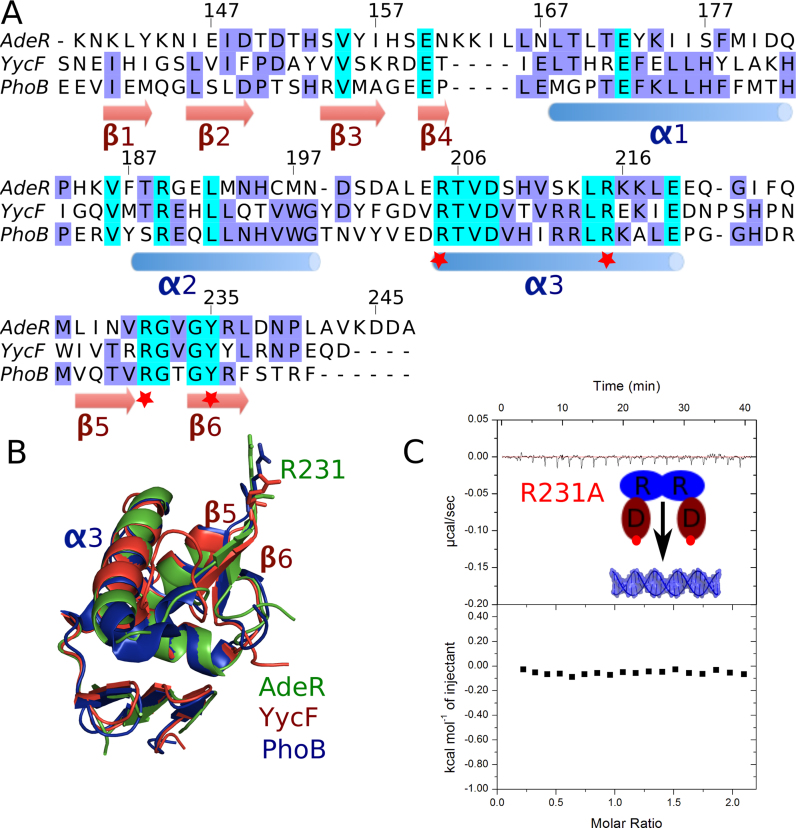
Overall, the AdeR DNA binding domain shows a relatively low sequence identity with other response regulators such as YycF and PhoB (15–20%) (Figure 5A). Interestingly, the recognition sequence of AdeR is approximately two times longer (10bp) than the one of YycF and PhoB (5–6 bp). In addition, this sequence has a much shorter gap between the two binding boxes, i.e. 1 bp with AdeR, and 5 bp with both YycF and PhoB (29). Despite this low sequence identity and the difference in the recognition DNA sequence, the DNA binding motif α3 and the β hairpin formed by β5 and β6 display high similarity and the key residues involved in the DNA backbone interaction are still conserved.
Figure 5.
Sequence alignment of AdeR DNA binding domain and mutagenesis study. (A) Sequence alignment of AdeR with the OmpR/PhoB like response regulator YycF and PhoB DNA binding domains. The red star indicates the key positive residues involved in the protein and DNA interaction. (B) Superposition of AdeR (Green) with YycF (Red) and PhoB (Blue) DNA binding domains with RMSD of 0.8 and 1.0 Å respectively; R231 is shown in stick. (C) The R231A mutation in AdeR full length could abolish its interaction with the intercistronic DNA as shown by isothermal titration calorimetery.
The AdeR DNA binding domain presents a structure highly similar to YycF and PhoB with a RMS of 0.8 and 1 Å, respectively, between matching Cα positions, while the Arg231 residue from the β hairpin pitch is oriented in a highly similar conformation allowing it to insert in the minor groove of the DNA (Figure 5B). Mutation of this key residue to Ala completely abolishes the DNA binding as indicated by ITC (Figure 5C) and EMSA experiments (Supplementary Figure S2). Furthermore, a double mutation of Arg205 and Arg215 from the α3 helix also results in the loss of interaction between AdeR and its recognition target (Supplementary Figure S2). Indeed, the mutation of Arg205 to alanine was also demonstrated in the B. subtilis PhoP causing the failure of transcription activation (39).
Solution assembly of AdeR and its complex with the intercistronic DNA
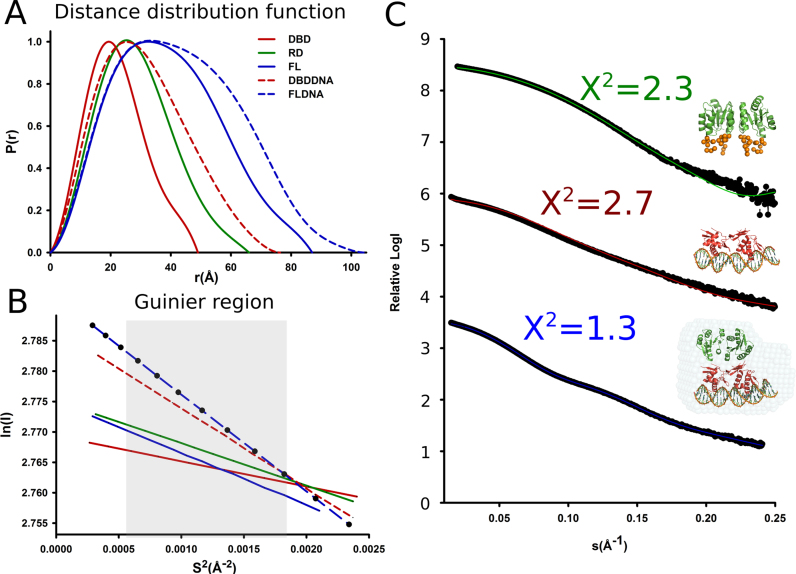
To elucidate the structure of AdeR and its complex with the intercistronic DNA in solution, we continued its structural characterization with small angle X-ray scattering supplemented with rigid body modeling. Highly monodisperse preparations of AdeR_FL, AdeR_RD, AdeR_DBD and AdeR_FL, AdeR_DBD complexed with the intercistronic DNA at different concentrations were used for SAXS measurements (Figure 1A, Supplementary Table S2). Subsequently, the molecular weight of the measured samples could be judged using the SAXS MoW2 package (37). This indicated that the oligomerization assembly is consistent with the SEC calculations as well as the binding studies via ITC (Supplementary Table S2). The distance distribution function and the Guinier region of the measured samples were generated, and described the quality of the SAXS measurements (Figure 6A and B). The SAXS experimental scattering data of the AdeR_RD and AdeR_DBD binary complexes with DNA revealed a compatible fit with the crystal structures after adding the missing terminal residues or simple SASREF modeling (Figure 6C). The AdeR_FL and DNA complex model were generated using the DAMMIF and DAMAVER program in P1 symmetry; the AdeR_RD is in a head-to-head dimer while the AdeR_DBD in complex DNA is in a head-to-tail dimer form. Furthermore, the crystal structures of the AdeR_RD and AdeR_DBD-DNA complexes were successfully superpositioned onto the Ab Initio model and agreed very well with a X2 value of 1.3 (Figure 6C) lending support that the crystal structure reflects the solution assembly.
Figure 6.
Solution assembly of AdeR and its complex with the intercistronic region DNA. (A) Distance distribution function. (B) Guinier region analysis of SAXS measure AdeR protein and its complex with DNA. C. The SAXS experiment scattering curve and their fit to the crystal structure of the AdeR receiver domain (green top), AdeR DNA binding domain with DNA complex (red middle), the DAMMIF model of AdeR full length with DNA complex (blue bottom). The AdeR receiver domain and the AdeR DNA binding domain complexed with DNA are superimposed onto the AdeR full length DNA complex DAMMIF model.
Phosphorylation mimic and magnesium binding study of AdeR
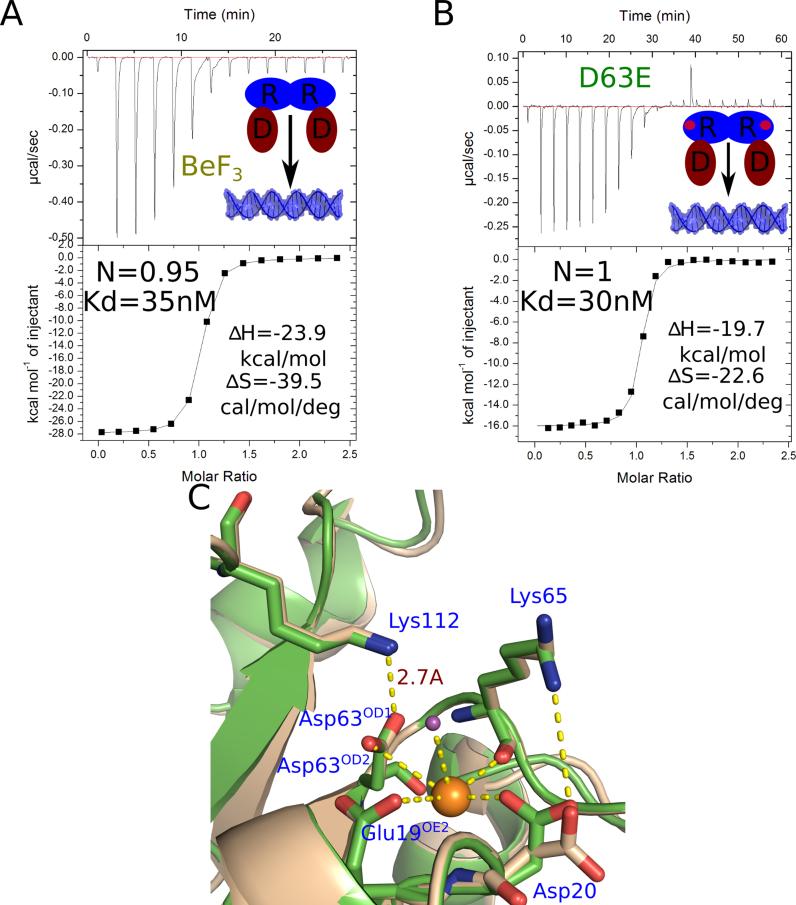
To investigate the activation mechanism of AdeR, we carried out an aspartate phosphorylation mimic study of AdeR. BeF3 is widely used as a phosphate analog in protein aspartate phosphorylation especially with response regulators such as the well characterized PhoP families (21,40). In the case of response regulator VarR and RstA, supplementing with BeF3 can increase the binding affinity of the response regulator to their target DNA fragments (21, 41). However, the isothermal titration calorimetery experiment of AdeR with the intercistronic DNA fragment in the presence of BeF3 and magnesium had no influence on the AdeR binding affinity to its target DNA (Figure 7A). This finding corresponds with previous response regulator phosphorylation mimic studies (42) where the mutation of aspartate into asparagine showed no interference, the mutation of D63 to E in AdeR also showed no affect in its interaction with the target DNA as indicated by ITC (Figure 7B).
Figure 7.
Phosphorylation mimic and magnesium binding study of AdeR. (A) Isothermal titration calorimetry of AdeR_FL interaction with the intercistronic DNA in the presence of BeF3 and magnesium. (B) Isothermal titration calorimtery of AdeR_FL D63E with the intercistronic DNA, the stoichiometry (N), dissociation constant; enthalpy (ΔH) and entropy (ΔS) are denoted. (C) Alignment of AdeR bound (green) and unbound (wheat) with magnesium ion (orange sphere). The phosphorylation site D63OD2 atom together with the Glu19, Asp20 side chain, Lys65 backbone and one water molecule are involved in the binding of magnesium. The binding of magnesium causes a conformational change of Asp20. The D63OD1 atom forms a strong hydrogen bond with the Lys112 amino group with a distance of 2.7 Å.
To validate whether BeF3 is not able to effectively mimic the AdeR D63 phosphorylation or that phosphorylation indeed has no effect on AdeR interaction with its target DNA, we crystallized the AdeR receiver domain in the presence of BeF3 and Mg2+. Although we could not observe any density for BeF3 in the structure, we solved the crystal structure of the AdeR receiver domain in complex with one magnesium ion to 1.6 Å (Table 1). The phosphorylation site D63 is directly involved in the binding with the magnesium ion through its OD2 atom together with the Glu19, Asp20 side chain, 1 water molecule and Lys65 backbone (Figure 7C). The alignment of AdeR with or without magnesium binding indicated a conformational switch of the Asp20 residue (Figure 7C). The Glu, Asp and phosphorylation site D-forming pocket for the binding of magnesium is highly conserved in the PhoP response regulator family (Figure 2C). Surprisingly, the D63 OD1 atom that was expected to be phosphorylated forms a strong hydrogen bond with Lys112 amino group with a distance of 2.7 Å (Figure 7C). This Lys112 is highly conserved between PhoP families and AdeR (Figure 2C), however, the strong hydrogen bond between Lys112 and the phosphorylation site was not observed in other PhoP response regulator family members.
DISCUSSION
We provided here systematical structural insights into AdeR, the key regulator of the AdeABC efflux pump involved in multidrug resistance of A. baumannii. AdeR recognizes an intercistronic region direct repeat DNA sequence located between the adeR and efflux pump adeABC operon. The full length AdeR interacts with the intercistronic region with a dissociation constant of 20 nM, which is almost 100 times higher than the DNA binding domain alone. The conserved OmpR/PhoB-like DNA binding domain of AdeR uses the long α3 helix and the β hairpin formed by β5 and β6 to bind the major and minor groove of the intercistronic region, respectively. AdeR binding to the DNA introduced a bend of ∼140° toward AdeR and a light curvature along the DNA strand. Mutation from the pitch residue R231A in the β hairpin, or R205A and R215A from α3 could completely abolish this recognition. The receiver domain of AdeR dimerizes through the α4–β5–α5 motif D108F109 and R122, forming a stable gear wheel-like structure by stack cation π interaction. Mutation of these key residues disrupts the dimerization of AdeR and further weakens its interaction with the intercistronic DNA. The SAXS model indicated that the full length AdeR assembles with the intercistronic DNA in a head-to-head arrangement of the receiver domain, and head-to-tail positioning of the DNA binding domain. This assembly may play a role in the sequential binding of the response regulator to the two-half target DNA sites, and affects the downstream gene activation through recruitment of the RNA polymerase σ subunit by the transaction loop (21,43).
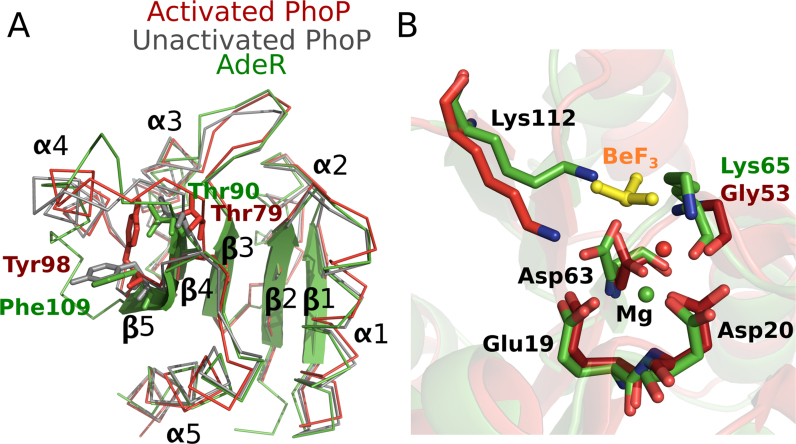
The activation of most response regulators is mediated by phosphoryl group transfer from its cognate histidine kinase, however some of the atypical orphan response regulators were also reported to use a phosphorylation independent regulatory mechanism (18,22,44,45). In the canonical response regulator such as PhoP, phosphorylation could stabilize its dimerization to further increase the binding affinity to DNA (18,46). AdeR D63 is aligned to the phosphorylation site of PhoP D52, and is thus predicted to be the target phosphorylation residue (Figure 2C). A switch involving two key residues, Thr79 in β4 and Tyr98 in β5, is proposed as the major conformational change during activation of PhoP, reported as the common mechanism for all response regulators (21,47,48). However, structural alignment of the AdeR receiver domain to activated and unactivated PhoP indicates a dramatically different conformation of AdeR α4 and its loop connection with β4&β5 (Figure 8A). Nevertheless, the unphosphorylated and magnesium bound AdeR switch residues Thr90 (PhoP_Thr79) and Phe109 (PhoP_Tyr98) are consistent with the unactivated conformation state (Figure 8A). More importantly, Phe109 is directly involved in the dimerization of the AdeR receiver domain (Figure 2B). A conformational change of Phe109 to the activated state as in PhoP would disrupt the cation π interaction between Phe109 and Arg122, and would weaken the gear wheel-like structure, hence destabilizing the AdeR dimer. The interaction of unphosphorylated AdeR to the intercistronic region DNA exhibits a dissociation constant of 20nM, which is much higher than other PhoP-like response regulators such as RstA in an activated state (21). The high affinity binding of AdeR to its target DNA sequence without phosphorylation may maintain a basal level of transcription of the efflux pump in non-stimulated environment and establish a rapid-reaction mode against antibiotic stress. Furthermore, structural analysis of AdeR in complex with magnesium indicated that the magnesium-binding motif was conserved between the AdeR and PhoP family. However, the AdeR potential phosphorylation atom Asp63OD1 can form a strong hydrogen bond with the Lys112. The alignment of AdeR-Mg and the activated PhoP-Mg-BeF3 structure revealed a severe clash between the Lys112 and the BeF3 ion (Figure 8B). Therefore, the strong hydrogen bond interaction between Lys112 and Asp63OD1 may affect the phosphorylation by using an autoinhibition mechanism, as in the ubiquitin ligase CBL which uses both an autoinhibition and a phosphorylation dependent activation mechanism (49). The hydrogen bond between Lys112 and Asp63OD1 could prevent non-specific phosphorylation and, hence, activation of AdeR from low-molecular-weight phosphate donors. Activation of AdeR may specifically be initiated by its interaction with AdeS as this induces the conformational change of Lys112 to release the phosphorylation site Asp63OD1. The further structural investigation of AdeR and AdeS complexes will definitely shine light on the activation mechanism.
Figure 8.
Structural alignment of the AdeR receiver domain (Green) with activated (Red) and unactivated PhoP (Grey). (A) PhoP Thr79 (AdeR Thr90) and Tyr 98 (AdeR Phe109) are the switch residues involved in the conformational change during PhoP activation. The α4 and the loops between α4 and β4&β5 are distinct with neither activated nor unactivated PhoP. (B) Alignment of the magnesium-binding motif of activated PhoP and AdeR in complex with magnesium. The BeF3 (yellow stick) of PhoP has a severe clash with the Lys112 in AdeR, which forms a hydrogen bond with Asp63. Magnesium ions are shown in spheres.
Besides D63, there are multiple potential phosphorylation D sites in AdeR that correlate with the expression of the efflux pump AdeABC (14,50). These residues may play a direct or indirect role in the phosphorylation of AdeR. The residue D20, for example, is occupied in the same pocket as D63 and its mutation to E was found in a clinical isogenic A. baumannii strain and was demonstrated to enhance the efflux pump activity (13,50). We have now shown that D20 is directly involved in the magnesium binding of AdeR. Mutation to E may stabilize the interaction, and hence enhance the AdeS phosphoryl transfer. Nonetheless, additional phospho-proteomic effort will definitely help to identify the AdeR aspartate phosphorylation profile and enlighten the regulation mechanism of the AdeRS two-component system. Besides occurring near the phosphorylation site, the mutational hot spots of AdeR from clinical multidrug resistant isolates could also be located in the receiver domain dimerization interface such as A91L and P116L, or the DNA binding interface i.e. E219A (11,14,51).
A simple BLAST analysis of the AdeR 10bp intercistronic region recognition sequence resulted in more than 250 identical results in Acinetobacter spp. including genomic DNA, plasmids, and resistance islands, with many of them containing multiple copies of AdeRS–AdeABC systems. The tight correlation of the AdeRS two-component system with expression of the efflux pump AdeABC, and its specificity in A. baumannii, makes it a potential therapeutic target against multidrug resistant A. baumannii. The mechanistic insight of the AdeR recognition of its target DNA with the atomic resolution structure paves the way for the structural based putative ligand binding site discovery, and aid the in silico drug target screening for A. baumannii.
ACCESSION NUMBERS
PDB access codes: 5X5J, 5X5L and 5XJP.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Savvas Savvides, Prof. Joanne Lemieux and M.Sc. Stijn De Waele for their critical reading of the manuscript. We thank the staff from BL18U1/BL19U1 beamline and Dr Na Li from BL19U2 beamline for technical support during data collection in National Center for Protein Sciences Shanghai (NCPSS) at Shanghai Synchrotron Radiation Facility.
Author contribution: Y.W. and F.Z. designed and conducted the project, Z.O., Y.Y., X.Z. and Y.P. carried out the protein overexpression and purification experiments under the supervision of Y.W. Y.W. and Z.O. performed the crystallography and physiological studies; Y.W. and Z.O. performed the ITC and SAXS experiments. B.D. and P.G.H. provided strains and new reagents. Y.W. and F.Z. wrote the paper with contribution from all the authors, B.D. and P.G.H. made manuscript revision.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [NO.31500051 to Y.W., NO.81501527 to F.Z.]; GOA grant from Ghent University and by BELSPO through IAP 7/44 project iPROS (to B.D.); German Research Council (DFG) [FOR2251 to P.G.H.]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pendleton J.N., Gorman S.P., Gilmore B.F., Pendleton J.N., Gilmore B.F.. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013; 11:297–308. [DOI] [PubMed] [Google Scholar]
- 2. Maragakis L.L., Perl T.M.. Antimicrobial resistance: Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008; 46:1254–1263. [DOI] [PubMed] [Google Scholar]
- 3. Potron A., Poirel L., Nordmann P.. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int. J. Antimicrob. Agents. 2015; 45:568–585. [DOI] [PubMed] [Google Scholar]
- 4. Lucia Fernandez R.E.W.H. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012; 25:661–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li X.Z., Plesiat P., Nikaido H.. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015; 28:337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magnet S., Courvalin P., Lambert T.. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance. Antimicrob. Agents Chemother. 2001; 45:3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damier-Piolle L., Magnet S., Brémont S., Lambert T., Courvalin P.. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008; 52:557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coyne S., Rosenfeld N., Lambert T., Courvalin P., Périchon B.. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010; 54:4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Q., Li X., Zhou H., Jiang Y., Chen Y., Hua X., Yu Y.. Decreased susceptibility to tigecycline in acinetobacter baumannii mediated by a mutation in trm encoding SAM-dependent methyltransferase. J. Antimicrob. Chemother. 2014; 69:72–76. [DOI] [PubMed] [Google Scholar]
- 10. Montaña S., Vilacoba E., Traglia G.M., Almuzara M., Pennini M., Fernández A., Sucari A., Centrón D., Ramírez M.S.. Genetic variability of AdeRS two-component system associated with tigecycline resistance in XDR-acinetobacter baumannii isolates. Curr. Microbiol. 2015; 71:76–82. [DOI] [PubMed] [Google Scholar]
- 11. Marchand I., Marchand I., Damier-piolle L., Damier-piolle L., Courvalin P., Courvalin P., Lambert T., Lambert T.. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 2004; 48:3298–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kröger C., Kary S., Schauer K., Cameron A.. Genetic regulation of virulence and antibiotic resistance in Acinetobacter baumannii. Genes (Basel). 2016; 8:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nowak J., Schneiders T., Seifert H., Higgins P.G.. The Asp20-to-Asn substitution in the response regulator AdeR leads to enhanced efflux activity of AdeB in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016; 60:1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoon E.J., Courvalin P., Grillot-Courvalin C.. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and aders mutations. Antimicrob. Agents Chemother. 2013; 57:2989–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Capra E.J., Michael, Laub T.. The evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012; 66:325–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gotoh Y., Eguchi Y., Watanabe T., Okamoto S., Doi A., Utsumi R.. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 2010; 13:232–239. [DOI] [PubMed] [Google Scholar]
- 17. Groisman E.A. Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 2016; 70:103–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao R., Mack T.R., Stock A.M.. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem. Sci. 2007; 32:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richmond G.E., Evans L.P., Anderson M.J., Wand M.E., Bonney L.C., Ivens A., Chua K.L., Webber M.A., Mark Sutton J., Peterson M.L. et al. . The Acinetobacter baumannii two-component system aders regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio. 2016; 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watanabe T., Okada A., Gotoh Y., Utsumi R.. Inhibitors targeting two-component signal transduction. Adv Exp Med Biol. 2008; 631:229–236. [DOI] [PubMed] [Google Scholar]
- 21. Li Y.C., Chang C.K., Chang C.F., Cheng Y.H., Fang P.J., Yu T., Chen S.C., Li Y.C., Hsiao C.D., Huang T.H.. Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element. Nucleic Acids Res. 2014; 42:8777–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zschiedrich C.P., Keidel V., Szurmant H.. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016; 428:3752–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang T.Y., Huang B.J., Sun J.R., Perng C.L., Chan M.C., Yu C.P., Chiueh T.S.. AdeR protein regulates adeABC expression by binding to a direct-repeat motif in the intercistronic spacer. Microbiol. Res. 2016; 183:60–67. [DOI] [PubMed] [Google Scholar]
- 24. Zheng L., Baumann U., Reymond J.-L.. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A., Servet R.M.. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003; 31:3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kabsch W. Xds. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006; 63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Long F., Vagin A.A., Young P., Murshudov G.N.. BALBES: a molecular-replacement pipeline. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008; 64:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okajima T., Doi A., Okada A., Gotoh Y., Tanizawa K., Utsumi R.. Response regulator YycF essential for bacterial growth: X-ray crystal structure of the DNA-binding domain and its PhoB-like DNA recognition motif. FEBS Lett. 2008; 582:3434–3438. [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 31. Adams P.D., Afonine P. V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li N., Li X., Wang Y., Liu G., Zhou P., Wu H., Hong C., Bian F., Zhang R.. The new NCPSS BL19U2 beamline at the SSRF for small-Angle X-ray scattering from biological macromolecules in solution. J. Appl. Crystallogr. 2016; 49:1428–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wen Y., Behiels E., Felix J., Elegheert J., Vergauwen B., Devreese B., Savvides S.N.. The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence. Nucleic Acids Res. 2014; 42:10134–10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992; 25:495–503. [Google Scholar]
- 35. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P. V., Svergun D.I.. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012; 45:342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Svergun D., Barberato C., Koch M.H.J.. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995; 28:768–773. [Google Scholar]
- 37. Fischer H., De Oliveira Neto M., Napolitano H.B., Polikarpov I., Craievich A.F.. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2010; 43:101–109. [Google Scholar]
- 38. Luscombe N.M., Laskowski R.A., Thornton J.M.. NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997; 25:4940–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y., Abdel-fattah W.R., Hulett F.M.. Residues required for Bacillus subtilis PhoP DNA binding or RNA polymerase interaction: alanine scanning of PhoP effector domain transactivation loop and Helix 3. J. Bacteriol. 2004; 186:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cho H., Wang W., Kim R., Yokota H., Damo S., Kim S.H., Wemmer D., Kustu S., Yan D.. BeF (3) (-) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF (3) (-) complex with phosphoserine phosphatase. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leonard P.G., Golemi-Kotra D., Stock A.M.. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8525–8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krasteva P. V, Fong J.C.N., Shikuma N.J., Beyhan S., Navarro M.V.A.S., Yildiz F.H., Sondermann H.. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010; 327:866–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blanco A.G., Canals A., Bernués J., Solà M., Coll M.. The structure of a transcription activation subcomplex reveals how σ 70 is recruited to PhoB promoters. EMBO J. 2011; 30:3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hong E., Lee H.M., Ko H., Kim D., Jeon B., Jung J., Shin J., Lee S., Kim Y., Jeon Y.H. et al. . Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J. Biol. Chem. 2007; 282:20667–20675. [DOI] [PubMed] [Google Scholar]
- 45. Bourret R.B. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 2010; 13:142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Casino P., Rubio V., Marina A.. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009; 139:325–336. [DOI] [PubMed] [Google Scholar]
- 47. Bachhawat P., Stock A.M.. Crystal structures of the receiver domain of the response regulator PhoP from Escherichia coli in the absence and presence of the phosphoryl analog beryllofiuoride. J. Bacteriol. 2007; 189:5987–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S. Bacterial two-component systems: structures and signaling mechanisms, protein phosphorylation in human health Cai, H. (ed) InTech. 2012. [Google Scholar]
- 49. Dou H., Buetow L., Hock A., Sibbet G.J., Vousden K.H., Huang D.T.. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 2012; 19:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Higgins P.G., Schneiders T., Hamprecht A., Seifert H.. In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob. Agents Chemother. 2010; 54:5021–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hornsey M., Loman N., Wareham D.W., Ellington M.J., Pallen M.J., Turton J.F., Underwood A., Gaulton T., Thomas C.P., Doumith M. et al. . Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J. Antimicrob. Chemother. 2011; 66:1499–1503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.