Abstract
At the moment, one of the actual trends in medical diagnostics is a development of methods for practical applications such as point-of-care testing, POCT or research tools, for example, whole genome amplification, WGA. All the techniques are based on using of specific DNA polymerases having strand displacement activity, high synthetic processivity, fidelity and, most significantly, tolerance to contaminants, appearing from analysed biological samples or collected under purification procedures. Here, we have designed a set of fusion enzymes based on catalytic domain of DNA polymerase I from Geobacillus sp. 777 with DNA-binding domain of DNA ligase Pyrococcus abyssi and Sto7d protein from Sulfolobus tokodaii, analogue of Sso7d. Designed chimeric DNA polymerases DBD-Gss, Sto-Gss and Gss-Sto exhibited the same level of thermal stability, thermal transferase activity and fidelity as native Gss; however, the processivity was increased up to 3-fold, leading to about 4-fold of DNA product in WGA which is much more exiting. The attachment of DNA-binding proteins enhanced the inhibitor tolerance of chimeric polymerases in loop-mediated isothermal amplification to several of the most common DNA sample contaminants—urea and whole blood, heparin, ethylenediaminetetraacetic acid, NaCl, ethanol. Therefore, chimeric Bst-like Gss-polymerase will be promising tool for both WGA and POCT due to increased processivity and inhibitor tolerance.
INTRODUCTION
DNA polymerases are essential components of the cellular machinery responsible for DNA replication and repair. The superfamily of DNA polymerases is divided into seven families A, B, C, D, X, Y and RT based on amino acid sequence comparisons as well as crystal structure analyses homology (1). Each family plays a specific role in nucleic acid metabolism, whereby the DNA polymerase structure and enzymatic features are accurately suited for a corresponding function (2). Thus, the in vivo family A of DNA polymerases, which includes Taq-polymerase and Bst-polymerase, serves to fill the gaps present after Okazaki fragment maturation during DNA replication or during excision repair processes (3). Family A enzymes are single-chain proteins (4) possessing three enzymatic activities: polymerase, 5′–3′ exonuclease, which is unique among all DNA polymerases and 3′–5′ exonuclease (absent in several enzymes) (5,6).
Various family A DNA polymerases are used in vitro for artificial amplification of DNA in biotechnology and molecular diagnostics. Most practical applications use thermostable enzymes including numerous variants of polymerase chain reaction (PCR) with Taq-polymerase. However, several techniques, such as methods of isothermal amplification, require less enzyme thermostability and focus more on the ability to provide strand displacement activity (7–10). This activity allows the amplification of nucleic acid at a constant temperature without implementing thermal cycling for DNA strand separation. Bst-DNA polymerase is the large fragment of the A family DNA polymerase from Bacillus stearothermophilus, which is now known as Geobacillus stearothermophilus, and comprises all essential enzymatic properties. Bst-like polymerases are widely used in isothermal amplification of DNA due to their temperature optimum around 60°C and inherent strand displacement activity. Nevertheless, the improvement of several properties of natural enzymes, specifically, processivity and inhibitor tolerance can significantly enhance enzyme applicability in a variety of practices. Processivity is the ability of the enzyme to catalyse the reaction without dissociation from the DNA template. This is a crucial characteristic of DNA synthesis on long stretches, defining the amount and quality of the resulting DNA products. In turn, contiguous DNA fragments are preferable, allowing further precise sophisticated analysis. In light of the booming interest in point-of-care testing or in-field diagnostics, where the purification of DNA from specimens is hindered, the ability of DNA polymerase to amplify DNA in the presence of various contaminants is crucial for proper analysis. Strand-displacement ability also has an advantage in point-of care testing (POCT) by simplifying the equipment and reducing the cost of analysis. Taken together, the high inhibitor tolerance and existence of strand-displacement activity gather paramount importance by providing the possibility to develop simple and robust nucleic acid tests.
One of the most effective strategies in enzyme engineering is a construction of chimeric proteins derived from natural enzymes and protein domains with desirable properties. This approach has been successfully applied to improve Pfu-polymerase and Φ29 polymerase (11,12). Conjugation with additional DNA-binding domains or whole proteins has greatly increased the processivity of the enzymes leading to the amplification of much longer DNA fragments than synthesis by intact DNA polymerases. The strategy used for fusion designs was previously successfully developed for improving Pfu-, Taq-, Bst- and Φ29 polymerases (11–14). Specifically, the DNA-binding protein Sso7d from thermophilic archaea Sulfolobus solfataricus was attached to Pfu-polymerase as well as the DNA-binding domain of TopoV of Methanopyrus kandleri to Φ29 polymerase. Bst-polymerase has also been fused with the DNA-binding domain of TopoV; however, the resulting chimeric protein has not been tested in practical applications (13).
Previously, we cloned and characterized the Bst-like polymerase Gss-polymerase, DNA polymerase I from Geobacillus sp. 777 suitable for isothermal amplification of DNA (15). In the present work, we designed a set of chimeric polymerases on the basis of Bst-like Gss-polymerase and DNA-binding domains using the DNA-binding domain of DNA ligase Pyrococcus abyssi (16) and Sto7d protein, a counterpart of Sso7d from Sulfolobus tokodaii.
MATERIALS AND METHODS
Construction of LF Gss pol and Pfu-polymerase fusions
Gss-His
The Gss-pol nucleotide sequence was amplified using Gss-F1 and Gss-R1 primers with NdeI and NotI restriction sites, allowing the in-frame ligation into the pET23a vector (Novagen, USA). PCR was carried out using previously constructed pQE-LF-Gss (15), which contains the nucleotide sequence of the large fragment of DNA polymerase I from Geobacillus sp. 777. The resultant 1.79-kbp DNA fragment and pET23a vector were digested with NdeI and NotI (SibEnzyme Ltd., Russia), ligated, and transformed into Escherichia coli XL1-Blue cells according to the standard protocols (17). The fidelity of the resulting recombinant plasmid named pET-Gss was confirmed by sequence analysis using primers Gss-F1 and Gss-R1 (Table 1).
Table 1. Oligonucleotides.
| Name | 5′-sequence-3′ | Restriction site |
|---|---|---|
| Gss-F1 | TATACATATGGAATCGCCGTCATCAGAAG | NdeI |
| Gss-R1 | GAGTGCGGCCGCTTTCGCGTCATACCATGTC | NotI |
| Gss-F2 | GAAAATCCGCCAAGCGTT | |
| Gss-R2 | GAGTGCGGCCGCTTATTTCGCGTCATACCATGTC | NotI |
| Gss-F3 | GACAGGGATCCGAATCGCCGTCATCAGAAG | BamHI |
| Gss-R3 | AACGCTTGGCGGATTTTC | |
| Gss-R4 | ACTFTCAGGTACCTTTCGCGTCATACCATGTC | KpnI |
| Gss-R5 | ACCGCCACCGCCGGTACCTTTCGCGTCATACCATGTC | |
| DBD-F1 | TCATGCATATGAGGTACATAGAGCTGGCCCA | NdeI |
| DBD-R1 | ATTCGGATCCCTTTATTGGCTTACCAATCTGAATT | BamHI |
| DBD-F2 | TCATGGGTACCATTCACATTGGTAAGCCAATAAAGAGGTACATAGAGCTGGCCCA | KpnI |
| DBD-R2 | GACATGCGGCCGCCTTTATTGGCTTACCAATCTGAATT | NotI |
| Sto-F1 | GTACCGGCGGTGGCGGTGTAACAGTAAAGTTCAAGTATAA | |
| Sto-R1 | ATGTCACTTCCAATTCTTCTCCCTT | |
| Sto-F2 | AAGGGAGAAGAATTGGAAGTAGACAT | |
| Sto-R2 | CTTTGCGGCCGCTTTCTTTCCAGATTTTTCTAACATTTG | |
| Sto-F3 | GTCTCGCTAGCATGGTAACAGTAAAGTTCAAGTATAA | NheI |
| Sto-R3 | GCATGGATCCACCGCCACCGCCTTTCTTTCCAGATTTTTCTAACATTT | BamHI |
| pET-R | CAACTCAGCTTCCTTTCGG | |
| M13-HEX | HEX-CACGACGTTGTAAAACGAC | |
| Rev-Cy3 | Cy3-TGGCTGCTTCTAAGCCAACATCCT | |
| Rev-G | CTAGTGAGGATGTTGGCTTAGAAGCAGCCA | |
| Rev-A | CTAGTAAGGATGTTGGCTTAGAAGCAGCCA | |
| Rev-T | CTAGTCAGGATGTTGGCTTAGAAGCAGCCA | |
| Rev-C | CTAGTTAGGATGTTGGCTTAGAAGCAGCCA | |
| Rev-long | GCTAAGTGGGAAACGATGTGGAGTTGCCCAGACAACCAGGATGTTGGCTTAGAAGCAGCCA | |
| Rev-stop | GCAACTCCACATCGTTTCCCACTTAGC | |
| Rev-blunt | AGGATGTTGGCTTAGAAGCAGCCA | |
| FIP | CAGCATCCCTTTCGGCATACCAGGTGGCAAGGGTAATGAGG | |
| BIP | GGAGGTTGAAGAACTGCGGCAGTCGATGGCGTTCGTACTC | |
| LF | GAATGCCCGTTCTGCGAG | |
| LB | TTCAGTTCCTGTGCGTCG | |
| F3 | GGCGGCAGAGTCATAAAGCA | |
| B3 | GGCAGATCTCCAGCCAGGAACTA | |
| N9 | NNNNNNNNN |
Gss
The partial C-terminus coding sequence of Gss-pol was amplified using Gss-F2/Gss-R2 primers and previously constructed pET-Gss. The resulting 0.74-kbp DNA fragment and pET-Gss plasmid were digested with AsuII and NotI (SibEnzyme Ltd., Russia), ligated, and transformed into E. coli XL1-Blue cells according to the standard protocols. The resulting plasmid was named pET-Gss-native.
DBD-Gss fusion
Pab-DBD and partial N-terminus Gss-pol nucleotide sequences were amplified using DBD-F1/R1 and Gss–F3/Gss-R3 primer sets (Table 1), respectively. PCR was carried out using previously constructed pET-DBD (16), containing the nucleotide sequence of the DBD of ATP-dependent DNA ligase from P. abyssi, and pET-Gss. The resultant PCR fragments were digested with BamHI, followed by ligation according to the standard protocols. The obtained fusion DNA fragment was eluted from the agarose gel, digested with NdeI/SalI, and ligated with pET-Gss (NdeI/SalI). The resultant plasmid was named pET-DBD-Gss.
Gss-DBD fusion
The Pab-DBD and partial C-terminus LF Gss-pol nucleotide sequences were amplified using DBD-F2/R2 and Gss–F2/Gss-R4 primer sets (Table 1), respectively. PCR was carried out using previously constructed pET-DBD, and pET-Gss. The resultant PCR fragments were digested with KpnI, followed by ligation according to the standard protocols. The fusion DNA fragment was eluted from the agarose gel, digested with AsuII/NotI, and ligated with pET-Gss (AsuII/NotI). The resultant plasmid was named pET-Gss-DBD.
Sto-Gss fusion
Parts of the Sto7d sequence were amplified using Sto-F3/R1 and Sto-F2/R3 (Table 1) primers, and the resulting DNA fragments were fused via PCR with Sto-F3/R3 primers. Genomic DNA isolated from S. tokodaii and pET-Gss were used as templates for PCR. Gss nucleotide sequence was amplified using Gss-F3/pET-R primers set. The resultant PCR fragments were digested with BamHI, followed by ligation according to the standard protocols. The obtained fusion DNA fragment was eluted from the agarose gel, digested with NheI/SalI and ligated with pET23a vector (NheI/SalI). The resultant plasmid was named pET-Sto-Gss.
Gss-Sto fusion
Parts of the Sto7d sequence were amplified using Sto-F1/R1 and Sto-F2/R2 (Table 1) primers, and the resulting DNA fragments were fused via PCR with Sto-F1/R2 primers. Genomic DNA isolated from S. tokodaii and pET-Gss were used as templates for PCR. Sto7d and partial C-terminus Gss nucleotide sequences (amplified with Gss-F2/R5 primers) were used for amplification of the fusion DNA fragment, with primers Gss-F2/Sto-R2 (Table 1). The fusion DNA fragment was eluted from the agarose gel, digested with AsuII/NotI and ligated with pET-Gss (AsuII/NotI) according to the standard protocols. The resultant plasmid was named pET-Gss-Sto.
Expression and purification of polymerases
The BL21 (DE3) pLysS (Promega, USA) strain of E. coli cells harbouring the encoding polymerase plasmid were grown to OD600 = 0.3 in LB medium at 37°C. Four litres of LB in an LiFlus GX fermenter (Biotron Inc., South Korea) were inoculated with 40 ml of the previously obtained culture, and the cells were grown to OD600 = 0.6 at 37°C. Expression was induced by the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG) up to 1 mM concentration. After induction for 3 h at 37°C, the cells were harvested by centrifugation at 4000 × g and stored at −70°C. The cell pellets were resuspended in the lysis buffer (50 mМ Tris–HCl pH 8.0, 0.3 М NaCl, 2.5 mM MgCl2, 0.1 mM CaCl2, 1 mM PMSF), treated for 30 min at 37°C with DNAse I (1 μg/ml) for DNA digestion followed by centrifugation at 14 000 × g. The resulting cell supernatant was incubated for 30 min at 60°C and centrifuged a second time at 14 000 × g to obtain a clarified lysate.
His-Gss, Gss-His, DBD-Gss, Gss-DBD, Sto-Gss and Gss-Sto were loaded onto a 5-ml IMAC (Bio-Rad, USA) column pre-equilibrated with buffer A (50 mМ Tris–HCl pH 8.0, 0.3 М NaCl), followed by washing the column with 25 ml of buffer A. Bound proteins were eluted using buffer B (Buffer A with 0.3 mM imidazole).
Gss polymerase was loaded onto a 4-ml Affi-Gel (Bio-Rad, USA) column pre-equilibrated with buffer C (50 mМ Tris–HCl pH 8.0, 0.1 М NaCl), followed by washing the column with 20 ml of buffer C. Bound proteins were eluted by a 0–50% linear gradient of buffer D (50 mМ Tris–HCl, pH 8.0, 1.5 М NaCl); Gss polymerase was eluted by 50–150 mM NaCl.
Enzyme fractions were pooled and loaded onto a 2-ml Macro-Prep Ceramic Hydroxyapatite Type I (Bio-Rad, USA) column pre-equilibrated with buffer E (25 mМ K2HPO4, pH 8.0). The column was washed with 15 ml of buffer E, and bound proteins were eluted by a 0–30% linear gradient of buffer F (1 М K2HPO4, pH 8.0). The fractions containing enzymes were pooled, and dialysed against storage buffer (10 mМ Tris–HCl, pH 7.5, 50 mМ KCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 0.5% Tween 20), followed by storage at +4°C. All fractions from each step were analysed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. The concentrations of purified proteins were measured using the Bradford assay.
Thermal shift assay
The thermal stability of the enzymes was analysed by heating in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, USA). Reactions were conducted in 20 μl containing 1 × enzyme storage buffer, 1 × SYPRO Orange (Invitrogen, USA) and 2 μM of enzymes. The temperature was increased from 55 to 85°C, an increment of 1°C per 50 s with collecting the fluorescent signal in fluorescence resonance energy transfer (FRET) mode.
Electrophoretic mobility shift assay
Oligonucleotides Rev-Cy3 (24-mer) and Rev-G (30-mer) (Table 1) were hybridized in 1 × loop-mediated isothermal amplification (LAMP) buffer at a 1:2 molar ratio; the resulting DNA fragment with 5′-overhang and Rev-Cy3 were used as DNA substrates for electrophoretic mobility shift assay. The reactions were carried out in 10 μl containing 1 × LAMP buffer, 0.5 pmol of DNA substrate and the indicated amounts of enzymes. After incubation for 15 min at 55°C, the samples were immediately loaded into 5% native polyacrylamide gels with 0.5 × Tris/Borate/EDTA (TBE) buffer. The gels were subjected to electrophoresis for 45 min at 10 V/cm and 4°C on 24 × 10 cm plates. After electrophoresis the gels were scanned using a Pharos FX molecular imager (Bio-Rad, USA). The resulting autoradiograms were analysed using Quantity One software (Bio-Rad, USA).
Processivity assay
Processivity was measured using primer extension. The HEX-labelled primer M13-HEX (Table 1) was hybridized to ssDNA of M13mp8 phage at a 1:2 molar ratio (50 and 100 nM, respectively) in 1 × LAMP buffer (20 mM Tris–HCl pH 8.9, 10 mM (NH4)2SO4, 10 mM KCl, 0.5% Tween-20 and 4 mM MgSO4). After annealing, 5 μl of primed template was mixed with 5 μl of enzyme in 1 × LAMP buffer and 0.2 mM dNTP on ice. The reactions were incubated for 10 min at 55°C and quenched by the addition of 10 μl of 0.125 M EDTA. The reaction products were analysed on an ABI PRISM 3130 instrument using Peak Scanner 1.0 software (Applied Biosystems, CA, USA). Processivity was calculated as described in Ricchetti et al. (18) using the following equation: P = [[(1 × I(1)]+[(2 × (I(2)]+…+[(n) × (I(n))])/[I(1)+I(2)+…+I(n)]], where P: processivity, I: area of each peak, n: number of nt added.
Polymerase activity assay
Incorporation of radiolabelled nucleotides was used to assay the polymerase activity of the enzymes. The reaction mix (10 μl) contained 1 μg of activated calf thymus DNA, 0.2 mM of each dNTP, 4 μBq of α-[32P]-dATP, 20 mM Tris–HCl pH 9.0, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgCl2 and 1 mg/ml bovine serum albumin. The reactions were initiated by adding the enzyme on ice; samples were immediately transferred to a preheated thermal cycler for incubation at 60°C for 30 min followed by quenching by the addition of 5 μl of 0.125 M EDTA. The reaction products were collected on DE81 paper (Sigma–Aldrich Corp.), washed twice with 0.5 М Na2HPO4 and counted in a Pharos PX (Bio-Rad Laboratories, Inc., USA). One unit of enzyme specific activity was defined as the amount of enzyme that incorporated 10 nmol of dNTP into acid insoluble material in 30 min at 60°C.
Thermal stability, optimal temperature and ion concentration
To determine thermal stability, the aliquots of polymerase activity reaction buffer (described above) containing an identical amount (0.2 U) of enzyme were incubated at 50, 60°C or 70°C for 0.5–5 h. The reactions were chilled on ice, and the polymerase activity was measured as mentioned above at 60°C.
The temperature optimum of the enzymes was investigated by measuring polymerase activity at a range of 40–80°C by 5°C per step, whilst other conditions were identical to those described above for the polymerase activity assay.
The optimal ion concentrations were examined similarly as in the polymerase activity assay, using concentrations of K2SO4, NH4Cl, KCl, (NH4)2SO4 in the range of 50–500 mM.
Terminal transferase assay
DNA substrates were prepared by hybridizing Rev-Cy3 (24-mer) with Rev-blunt (24-mer) in 1 × LAMP buffer (Table 1). Reactions were carried out in 5 μl containing 1 × LAMP buffer, 0.5 pmol of DNA substrate, 0.4 mM dNTPs and 100 nM of enzyme. After incubation for 20 min at 55°C, the reactions were quenched by the addition of 5 μl of formamide and denaturated at 95°C for 10 min. The heated samples were immediately loaded onto 15% polyacrylamide denaturing gels containing 8 M Urea with 0.5 × TBE buffer. Gels were run at 50°C and scanned using a Pharos FX molecular imager (Bio-Rad, USA). Resulting autoradiograms were analysed using Quantity One software (Bio-Rad, USA).
Strand displacement assay
Oligonucleotides Rev-Cy3 (24-mer), Rev-long (61-mer) and Rev-stop (27-mer) (Table 1) or Rev-Cy3 and Rev-long were hybridized at molar ratios of 1:10:10 and 1:10, respectively, in 1 × LAMP buffer. Reactions were carried out in 10 μl containing 1 × LAMP buffer, 0.2 mM dNTPs, 0.5 pmol of DNA substrate (calculated at Rev-Cy3) and 50 nM of enzyme. After incubation for an indicated time at 55°C, the reactions were quenched by the addition of 10 μl of formamide, followed by denaturation at 95°C for 10 min. The heated samples were immediately loaded onto 15% polyacrylamide denaturing gels containing 8 M urea with 0.5 × TBE buffer. The gels were run at 50°C and scanned using a Pharos FX molecular imager (Bio-Rad, USA). Resulting images were analysed using Quantity One software (Bio-Rad, USA).
Fidelity assay
Enzyme fidelity was verified according to Sharma et al. (19). Four oligonucleotides [Rev-G, Rev-A, Rev-T and Rev-C (Table 1)] were designed to contain different nucleobases (G, A, T, C, respectively) at the +1 position of the template with respect to the 3′-end of the primer. DNA substrates were prepared by hybridizing Rev-Cy3 (24-mer) with either of Rev-G, Rev-A, Rev-T and Rev-C (30-mer) in 1 × LAMP buffer at a 1:2 molar ratio. The reactions were carried out in 5 μl containing 1 × LAMP buffer, 0.5 pmol of DNA substrate, 0.4 mM dNTPs and 100 nM of enzyme. After incubation for 1 min at 55°C, the reactions were quenched by the addition of 5 μl of formamide and denaturated at 95°C for 10 min. Heated samples were immediately loaded onto 15% polyacrylamide denaturing gels containing 8 M Urea with 0.5 × TBE buffer. Gels were run at 50°C and scanned using a Pharos FX molecular imager (Bio-Rad, USA). Resulting autoradiograms were analysed using Quantity One software (Bio-Rad, USA).
Whole genome amplification
Whole genome amplification (WGA) was performed as described in (8) with slight changes. The WGA reactions were performed in a 50 μl volume containing 1 × LAMP buffer, 0.4 mM dNTP, 20 μM primers N9 (Table 1), 5 ng of human genomic DNA and 100 nM enzyme. Reactions were incubated at 50°C for 5 h followed by enzyme inactivation at 95°C for 5 min. The resulting WGA products were purified by phenol–chloroform extraction and analysed on 1.5% agarose gels stained with ethidium bromide.
Quantitative PCR analysis
Quantitative analysis of WGA products was performed using TaqMan probes. Twenty-one loci were used for this analysis (Supplementary Table S1). Each WGA sample was analysed in triplicate. A standard curve was generated to determine the loci copy number in the amplified DNA relative to genomic DNA. The standard curve was generated from 50, 25, 12.5, 6.25, 3.12 and 1.56 ng of unamplified genomic DNA and 10 ng of WGA product. The amplification fold was calculated as a ratio of DNA quantity after WGA to initial DNA quantity for each locus.
The PCR reactions were performed in 20 μl containing 65 mM Tris–HCl, pH 8.9, 24 mM (NH4)2SO4, 0.05% Tween-20, 3 mM MgSO4, 0.2 mM dNTP, 300 nM primers, 100 nM TaqMan probe (Table 1) and 1 U of Taq-polymerase. Amplification was carried out in the CFX96 Real-Time PCR Detection System (Bio-Rad, USA) according to the following program: 95°C for 3 min followed by 40 cycles of 95°C for 10 s, and 60°C for 30 s with a collection of fluorescent signals at each respective channel.
Droplet digital PCR
ddPCR was performed using the QX100 system (Bio-Rad, USA) according to the manufacturer's recommendations. Reactions in a 20 μl volume contained 1 × ddPCR™ Supermix for Probes (No dUTP), 900 nM primers and 250 nM probes (Supplementary Table S1), 5 U of HindIII and 30 ng (around 10 000 genome equivalent) of WGA product or genomic DNA. The reactions were incubated at room temperature for 20 min for template digestion followed by droplet generation in DG8™ Cartridges. Prepared droplets were placed in PCR plates, and PCR was processed according to the following program: 95°C for a 10 min, then 45 cycles of 95°C for 30 s and 57°C for 1 min with a 2°C ramp rate. Droplets were read in a Droplet Reader, and the results were analysed using Quanta Soft software.
Real-time loop-mediated amplification
LAMP reactions were performed in a 16 μl volume containing 1 × LAMP buffer, 0.4 mM dNTP, 0.2 μM F3/B3, 0.4 μM loopF/loopB, 0.8 μM FIP/BIP (Table 1) (20), 1 ng of Lambda DNA (SibEnzyme, Russia), 1 μM SYTO 82 and 50 nM of enzymes.
The enzyme’s tolerance was analysed by adding different inhibitors including heparin, EDTA, NaCl, ethanol, urea and whole blood (collected from five healthy individuals) to the LAMP reactions at various concentrations. Amplification was carried out with the CFX96 Real-Time PCR Detection System (Bio-Rad, USA) according to the following program: 1.5 h at 59°C, collecting fluorescent signals (channel HEX) at 1-min intervals.
RESULTS
Cloning, overexpression and purification of Gss and Pfu fusions
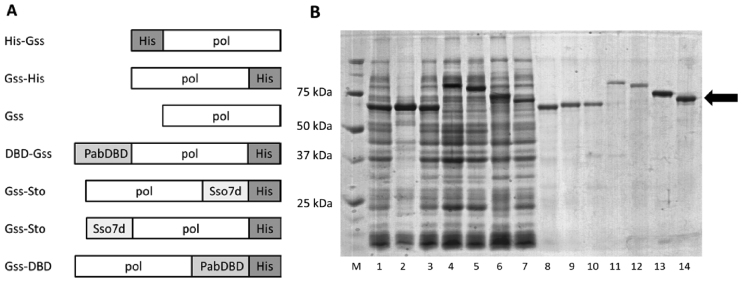
Four fusions were designed of Gss polymerase with Sto7d protein and the DNA-binding domain of P. abyssi DNA ligase (DBD): Gss-Sto with Sto7d at the N-terminus, Gss-Sto with Sto7d at the C-terminus, DBD-Gss with DBD at the N-terminus and Gss-DBD with DBD at the C-terminus. Additional three variants of the Gss polymerase were designed with a His-tag at the N-terminus (His-Gss), at the C-terminus (Gss-His) and without a His-tag (Gss). The compositions of all chimeric proteins are presented in Figure 1A.
Figure 1.
Schematic representation of Gss fusions (A), expression (lanes 1–7) and purification (lanes 8–14) of chimeric proteins (B). Enzymes were expressed in either Escherichia coli strains BL-21 (DE3) pLysS (Gss, Gss-His, DBD-Gss, Gss-DBD, Sto-Gss, Gss-Sto) or XL10-Gold (His-Gss), and purified using affinity and ion-exchange chromatography. M—Precision Plus Protein standards (Bio-Rad, USA), 1, 8—Gss, 2, 9—His-Gss, 3, 10—Gss-His, 4, 11—DBD-Gss, 5, 12—Gss-DBD, 6, 13—Sto-Gss, 7, 14—Gss-Sto. Chimeric proteins are marked by black arrows.
DBD, Sto7d and His-tag were fused with the N- or C-terminus of Gss polymerase in order to identify any location effects of the His tag on polymerase properties. Chimeric proteins were expressed in E. coli cells and purified with affinity and ion-exchange chromatography (Figure 1B).
Thermal stability of chimeric proteins depends on additional domain and its localization
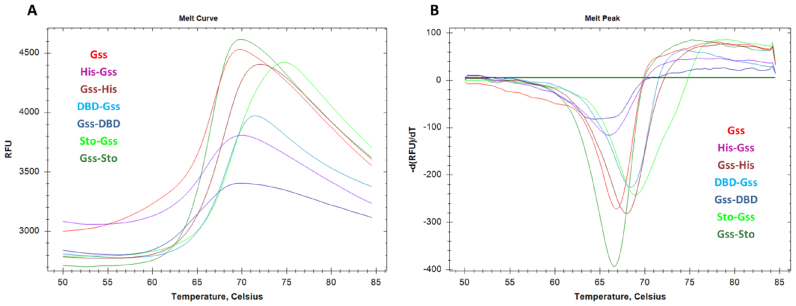
The thermal stability of an enzyme is a crucial characteristic in practical applications of Bst-like polymerases; these enzymes should remain stable and active for several hours at 50–60°C for optimal performance in WGA. The stability of the chimeric enzymes at various temperatures was investigated using differential scanning fluorimetry (DSF). DSF is based on the ability of certain fluorescent dyes to bind with hydrophobic regions of proteins. Generally during thermal denaturation soluble globular proteins undergo structural rearrangement, which leads to the exposure of inner hydrophobic regions out of the protein globule. Emerging hydrophobic surfaces can be bound by fluorescent dyes followed by a detectable increase in fluorescent signal. A change of protein stability is indicated by a shift of the thermal profile, and the magnitude of the shift indicates the degree of folding alteration. It should be noted that the absolute magnitude of the shift can differ between native and fusion proteins. In this case, a change in the melting temperature will be a relevant comparative parameter.
As expected, the presence of any additional amino-acid sequences (His-tag, DBD or Sto7d) influenced the thermal stability of the respective chimeric enzymes (Figure 2 and Table 2). Inverse results were observed for His-tagged and DBD- or Sto-modified polymerase: the presence of a His-tag at the N-terminus slightly decreased the thermal stability of Gss, whereas the presence of a DBD or Sto at the N-terminus magnified the temperature of Gss’s unfolding. The basal fluorescence level of Gss-DBD significantly exceeded the level of the other enzymes confirming a high disturbance of Gss-DBD folding, although the thermal stability of Gss-DBD is in the common range for studied chimeric enzymes. The latter observation is consistent with a complete loss of Gss-DBD activity, which is implied to be a result of misfolding (see below).
Figure 2.
Thermal shift assay of chimeric polymerases. Thermal denaturation profiles (A) and first derivatives of fluorescent curves (B) are presented; curve color indicates the particular polymerase. Each experiment was triplicated, typical curves are presented.
Table 2. Specific activity, thermostability and processivity of Gss-polymerase derivatives.
| Enzyme | Specific activity, 105U/mg | Melting temperature, °C | Processivity, bp |
|---|---|---|---|
| Gss native | 0.87 | 66.8 ± 0.2 | 98 |
| His-Gss | 1.04 | 65.9 ± 0.2 | 105 |
| Gss-His | 0.32 | 68.0 ± 0.2 | 110 |
| DBD-Gss | 0.10 | 68.6 ± 0.2 | 362 |
| Gss-DBD | no activity | 64.7 ± 0.2 | |
| Sto-Gss | 0.89 | 68.9 ± 0.2 | 298 |
| Gss-Sto | 0.94 | 66.8 ± 0.2 | 322 |
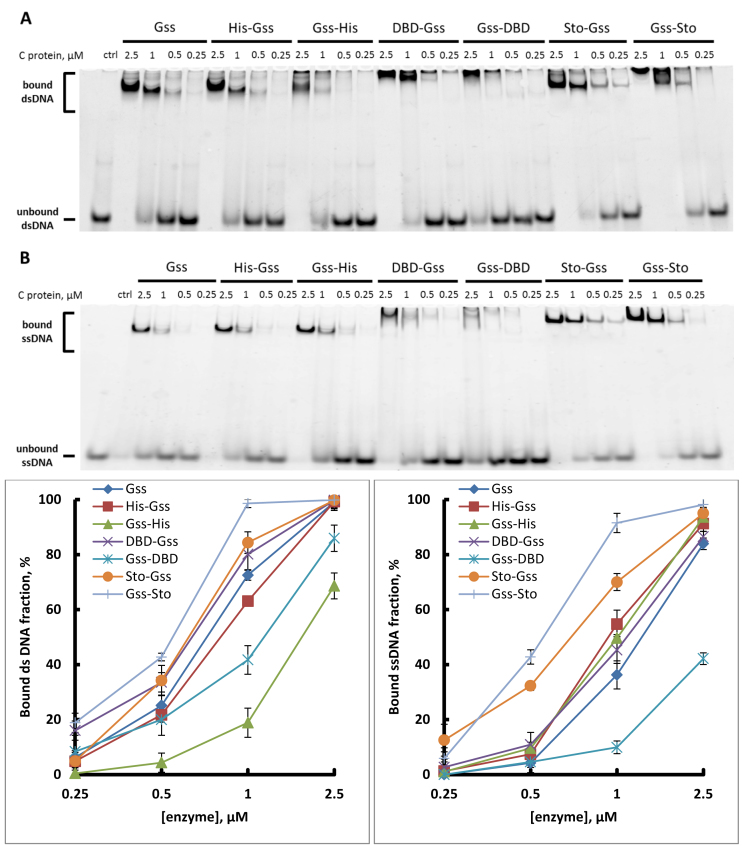
Addition of Sto7d or N-terminus DBD improves binding of enzyme with DNA
DNA-binding affinity is a key parameter responsible for the processivity of DNA polymerases. For processive synthesis of long DNA stretches during WGA analysis, the tight binding of the DNA template is indispensable. The fusion of Gss with Sto7d increased the ability of the enzyme to bind both double- and single-stranded DNA (Figure 3). Surprising, that the addition of DBD-domain at N- or C-end differently affected the enzyme ability to bind DNA molecule. DBD-Gss showed a similar affinity as Gss without additional DNA-binding domain; however, the Gss-DBD binding ability was expectedly low. Moreover, this experiment demonstrated that the joining of Sto to N-terminal part of Gss extremely increased the ss/ds DNA-binding ability of the chimeric enzyme.
Figure 3.
DNA-binding capacity of chimeric enzymes. Indicated amounts of enzymes were incubated with either 5′-overhang dsDNA (A) or ssDNA (B) at 55°C. Graphs below present the quantitative analysis of electropherograms.
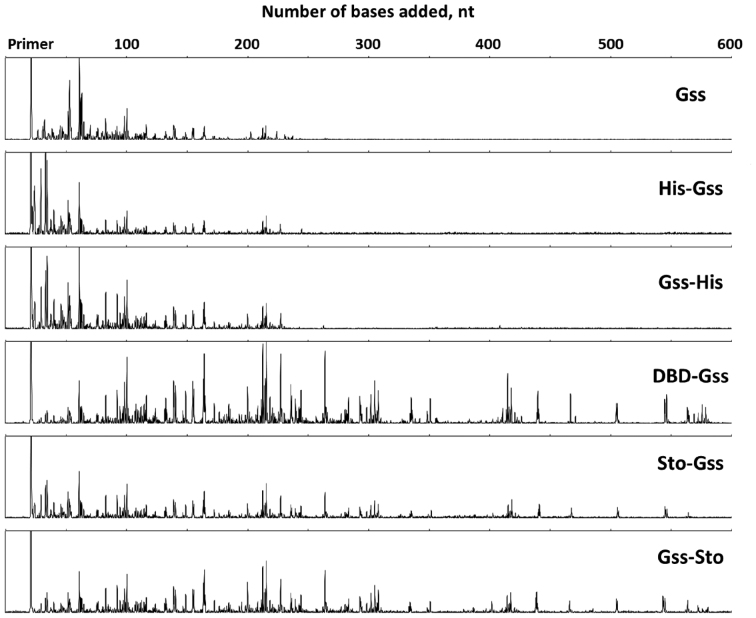
Addition of Sto7d or N-terminus DBD increases processivity of enzyme
Sophisticated analysis after WGA often require producing long DNA fragments. This could be reached by increasing the processivity of used enzyme. Thus, the processivity, which is the ability of DNA polymerase to catalyse DNA synthesis without dissociation from DNA, is one of the main enzyme characteristics determining the applicability of the enzyme for WGA. We tested this capacity of all fused proteins using long phage DNA as a template. It should be noted that the Gss-DBD mutant completely lost its own polymerase activity (see below), thus it was excluded from subsequent experiments. Obtained results demonstrated that the presence of the His-tag did not affect the ability of the enzymes to catalyse polymerization, whereas the attachment of either N-DBD or Sto7d drastically increased the processivity of the corresponding chimeric enzymes (Table 2 and Figure 4). Moreover, DBD-Gss displayed the highest level of the reaction yield.
Figure 4.
Processivity of the chimeric enzymes. Each trace represents one lane from a sequencing gel and each peak represents a single primer extension product. Reaction time was 10 min for each enzyme. The x-axis indicates the primer extension product length, which is determined based on size markers run on the same gel (trace not shown).
One of the main characteristics is the molar activity of DNA polymerase. We analysed this activity by the incorporation of 10 nmol of dAMP into the acid insoluble fraction in 30 min at 60°C using α-[32P]-dATP and activated calf thymus DNA as a template. Specific characteristics of the chimeric proteins are presented in Table 2. Fusion with DBD was associated with a significant decrease or complete loss of the specific activity of Gss.
The resultant data revealed Sto-Gss and Gss-Sto chimeras as the most promising enzymes because of the combination of its high processivity and molar activity.
DBD and Sto7d improve salt tolerance of the fused proteins
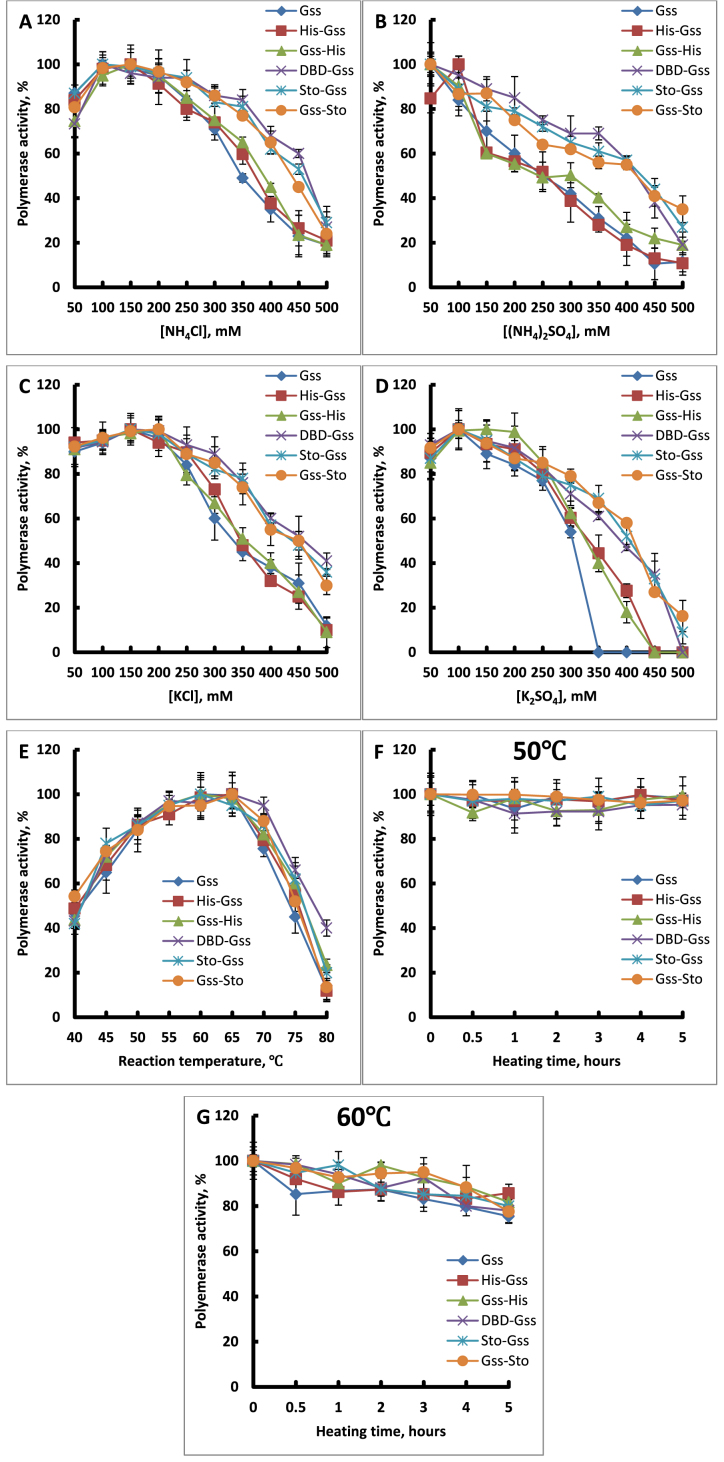
Binding of protein with DNA involves electrostatic interactions related to the ionic strength of the solution. High salt concentrations hinder protein–DNA interactions acting as a screen of charged surfaces. From this point of view, the attachment of an additional DNA-binding domain should increase the ability of the enzyme to preserve binding with DNA in a solution with a high salt concentration. We tested the influence of the most common ions present in the all biological samples such as NH4+, K+, Cl− and SO42− (Figure 5A–D). As expected, the chimeric enzymes demonstrated a higher tolerance to mono- and divalent ions in comparison to the native enzyme.
Figure 5.
Ion concentrations (A–D), temperature (E) and thermostability (F and G) assays of the chimeric polymerases. Enzyme characteristics were determined via incorporation of α-[32P] dAMP in activated calf thymus DNA. Each experiment was triplicated. Optimal temperature and thermal stability was determined in standard polymerase activity assays.
As it was mentioned above, the application of Bst-like DNA polymerases is associated with the use of high temperatures; therefore, ‘good tools’ for amplification should be resistant to prolonged exposure of these conditions. In this field, we examined our chimeras in two ways: the activity of the enzyme in the temperature range from 40 to 80°C and thermal stability at 50 and 60°C for 5.5 h. The results demonstrated that optimal temperature for DNA synthesis was also left unaltered despite the presence of the DBD or Sto7d domains (Figure 5E). The thermal stability of the chimeric enzymes was also unchanged—all enzymes lost their activity only after 0.5 h at 70°C (Figure 5F, G and unpresented data). Thus, we can conclude that the introduction of N-DBD or Sto7d into the whole enzyme leads to higher processivity, less anion/cation sensitivity and no change in thermal stability of the chimeras in comparison with the native enzyme.
Additional domains do not influence on terminal transferase activity
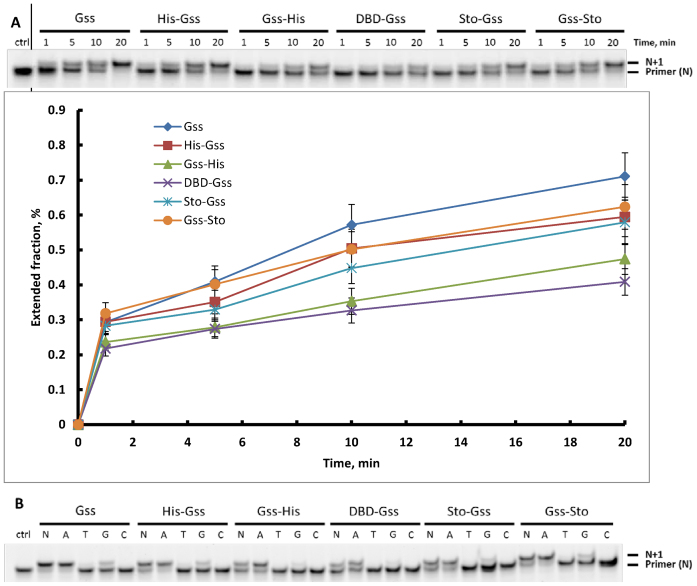
Several practical applications of DNA polymerases (e.g. tailing of DNA fragments during NGS library preparation) require terminal transferase activity, during which the enzyme adds nucleotides at the 3′-terminus of a DNA fragment in a template-independent manner. In this sense, the reaction rates as well as a number of incorporated nucleotides and nucleotide specificity are significant characteristics defining the enzyme’s potential for NGS library preparation.
In the present study, we tested the capacity of the constructed chimeric proteins to catalyse these reactions on the qualitative and quantitative levels. It was found that the fusion of additional domains with native Gss does not alter the preferences during the nucleotide transferase reaction (Figure 6). All chimeric enzymes and native Gss utilized purines, and adenosine was incorporated at a higher rate than guanosine.
Figure 6.
Terminal transferase activity was defined under the incorporation of dNMP on the blunt-ended dsDNA fragment. (A) Kinetics of terminal transferase reaction. Enzymes were incubated for indicated times with DNA substrate and dNTPs at 55°C. The graph below reflects the percent of elongated DNA. (B) Nucleotide preferences under the incorporation of dNMP into blunt-ended DNA.
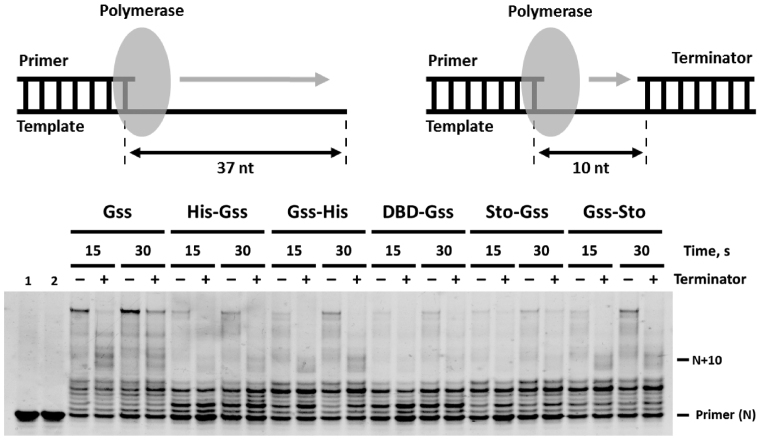
Additional domains do not dramatically influence on the strand displacement activity
The unique feature of Bst-like polymerases, which makes them useful for practical applications, is an ability to displace the upstream DNA chain during amplification. Since we are exploring chimeric enzymes in biotechnology methods, the occurrence of strand displacement activity is a significant point. As follows from the experimental data obtained by using partial DNA duplexes, the presence of the His-tag, DBD or Sto7d did not substantially influence the strand displacement activity of the corresponding enzymes (Figure 7). Interestingly, that the kinetics of product accumulation using gapped DNA substrates varied for different enzymes, which is probably a result of differences in molar activity. Stopping of DNA synthesis at the length of product exceeded size of a gap (10 nt) can be explained by the absence of a 5′-phosphate group on the downstream primer, which is an important recognition component for DNA polymerases (21).
Figure 7.
Strand displacement activity of the chimeric enzymes. Enzymes were incubated with either primed template or primed template with terminator for indicated times at 55°C. Controls: 1—primed template, 2—primed template with terminator.
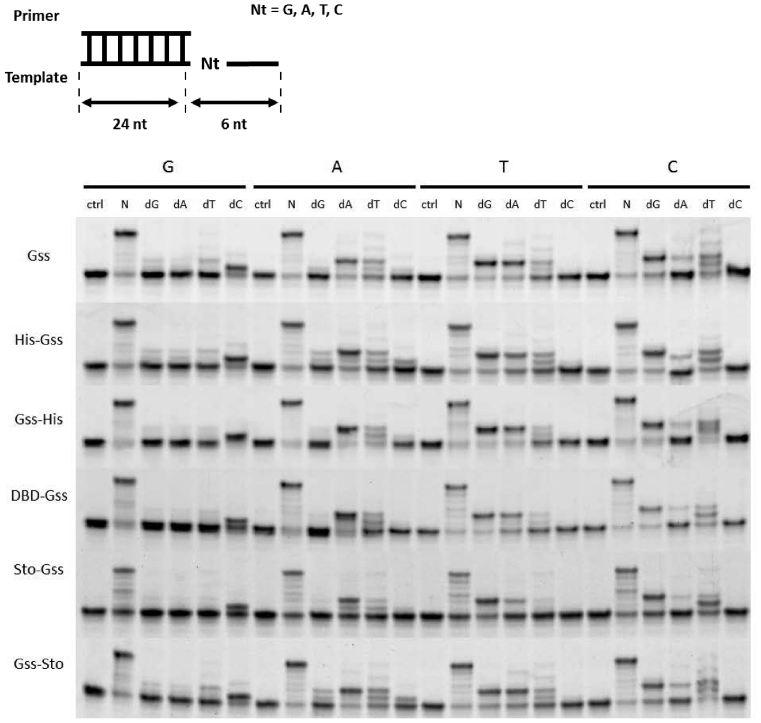
Additional domains do not influence on the fidelity of DNA synthesis
The fidelity of DNA synthesis is an important criterion for maintenance of genome integrity. In terms of WGA, a high fidelity of DNA polymerase used is a critical point at WGA product’s formation during sophisticated analyses such as somatic mutation identification in tumour specimens (e.g. FFPE-derived DNA). Our data suggest that attaching either a His-tag, DBD or Sto7d did not interfere with the fidelity of the respective enzymes (Figure 8). It should be noted that the excessive amounts of a single nucleotide in combination with the short oligonucleotide templates in the reaction mixes can lead to supposed error rate of the studied enzymes.
Figure 8.
Fidelity of DNA synthesis catalysed by the chimeric enzymes. Template oligonucleotides contained different nucleobases at the +1 position with respect to the 3′-end of the primer. Reactions were performed in the presence of either mixture or one of the dNTPs.
Gss fusions in WGA
The amount of DNA produced and the amplification bias for each locus indicate the feasibility of the WGA product for further analysis. Twenty loci were chosen for analysis with a different GC-content in the range of 42–58% with a length of 83–202 bp (Table 3). Thus, we took into account the influence of GC-content on the efficacy of WGA. Different length of quantitative PCR (qPCR) amplicons afforded the possibility to investigate the length of actual WGA products suitable for further PCR analysis.
Table 3. Loci for WGA quantification.
| Locus | Localization | Length, bp | GC-content |
|---|---|---|---|
| MET | 7q31 | 100 | 42 |
| F II | 11p11 | 110 | 42.7 |
| BDNF | 11p13 | 135 | 43 |
| KRAS | 12p12.1 | 95 | 43.2 |
| IL10RA | 11q23 | 83 | 43.4 |
| PPARG C1A | 4p15.1 | 167 | 43.7 |
| ALB | 4q13.3 | 94 | 45.7 |
| CKMM | 19q13.32 | 115 | 46.1 |
| BCO1 | 16q23.2 | 196 | 46.4 |
| APOA2 | 1q23.3 | 171 | 48.0 |
| ADIPOQ | 3q27 | 161 | 48.4 |
| CYP1B1 | 2p22.2 | 199 | 48.7 |
| NBPF3 | 1p36.12 | 147 | 49.6 |
| EGFR | 7p12 | 84 | 50 |
| BHMT | 5q14.1 | 149 | 51.7 |
| MTHFR | 1p36.3 | 120 | 51.7 |
| F XI | 4q35 | 116 | 53.4 |
| ALDH1L1 | 3q21.3 | 142 | 54.2 |
| ERBB2 | 17q12 | 100 | 57 |
| PER2 | 2q37.3 | 202 | 57.9 |
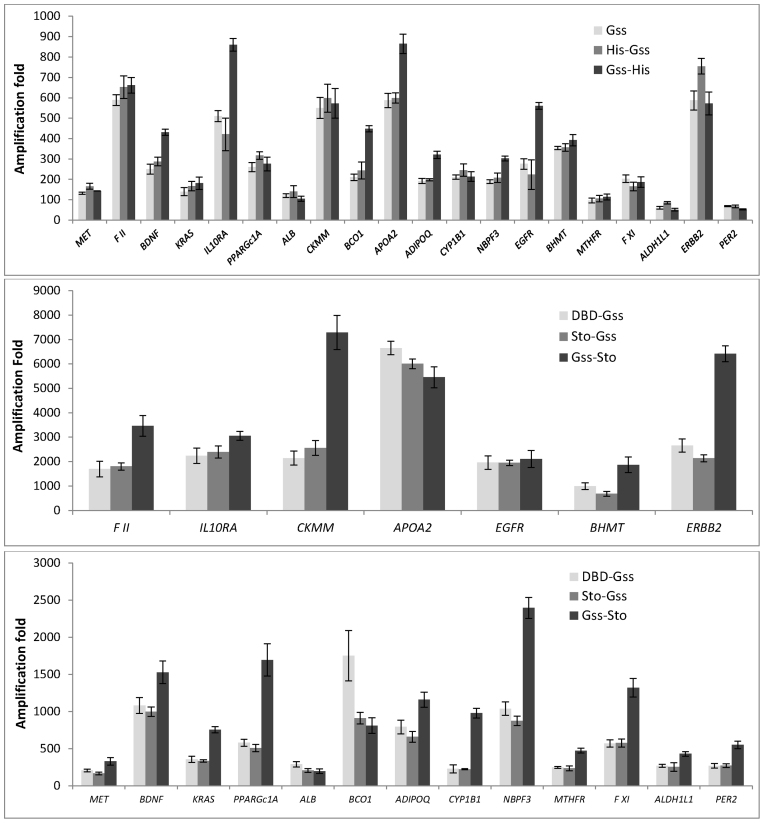
qPCR analysis of WGA products revealed that active chimeric enzymes with additional DNA-binding domains DBD-Gss, Sto-Gss and Gss-Sto outperformed Gss and its His-tag derivatives. The amount of WGA product generated by Gss-Sto of 16 out of 20 loci exceeded the amount of product of DBD-Gss or Sto-Gss (Figure 9). However, the amplification bias for each locus by Gss-Sto during WGA was also high.
Figure 9.
Fold of WGA with chimeric enzymes. Each WGA sample was analysed in triplicate. A standard curve was generated to determine the locus copy number in amplified DNA relative to genomic DNA at each locus. Fold amplification values were calculated as a ratio of DNA quantity after WGA to initial DNA quantity for each locus.
Surprisingly, there was no correlation between the efficacy of WGA and GC-content of the amplicon (Supplementary Figure S1). Additionally, there was no dependence between qPCR efficacy and amplicon length, implying uniform amplification of DNA in WGA for the length range.
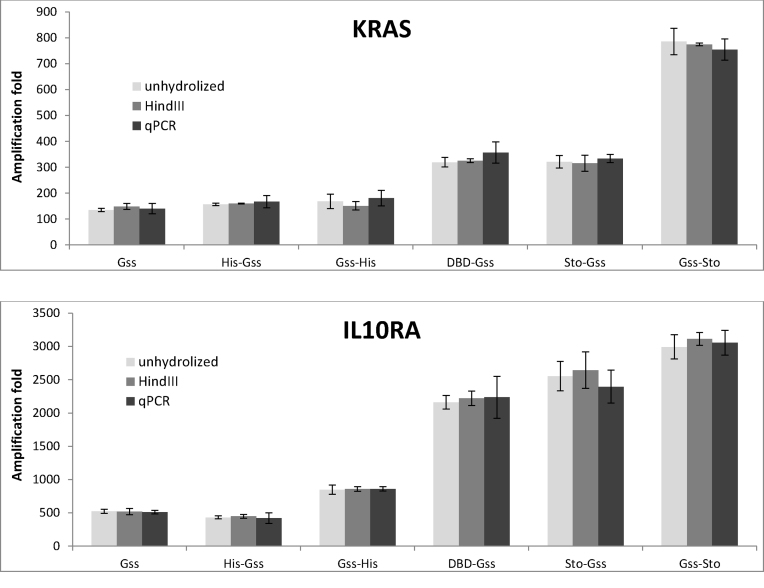
DNA structure is known to influence the efficacy of amplification. Supercoiled DNA hinders amplification and disables DNA quantification via qPCR, leading to an underestimation of the WGA product. To determine the influence of DNA structure on quantitative yield of WGA, the amount of DNA was measured by droplet digital PCR (Figure 10). For this purpose, we chose the IL10RA and KRAS genes, which are comparatively similar in their GC-content and amplicon length. In this case, no significant effect of DNA state was detected. It can be concluded that the data obtained by ddPCR and qPCR are congruent.
Figure 10.
Efficacy of ddPCR performed using chimeric DNA polymerases. Each WGA sample was analysed in triplicate. Amplification fold for each locus was calculated as a ratio of DNA quantity after WGA to initial DNA quantity for each locus.
Sto7d and DBD fusions have higher inhibitor tolerance in LAMP
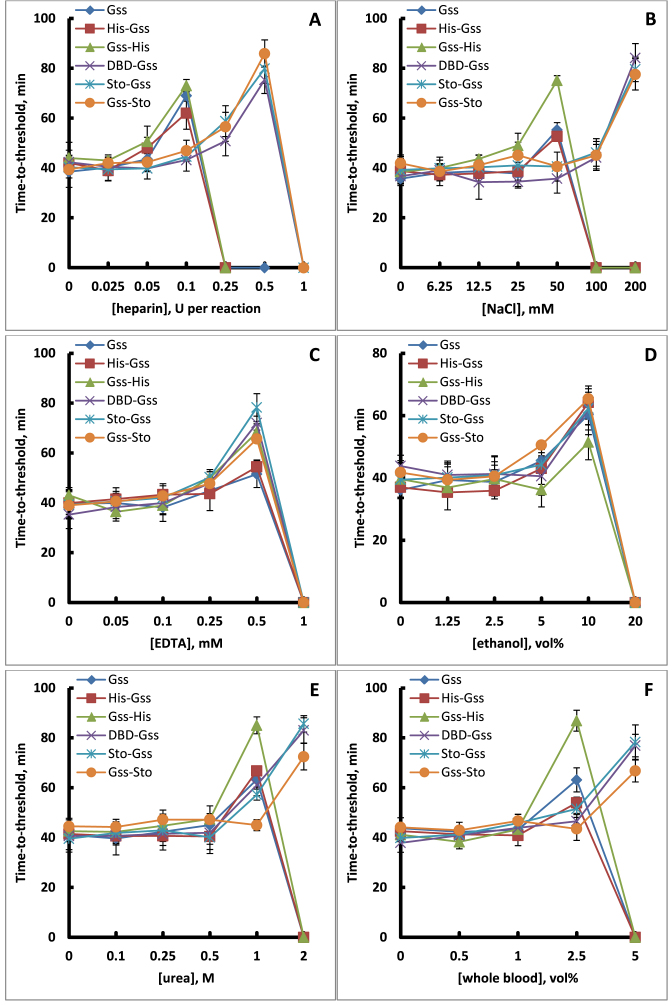
One perspective development in point-of-care or in-field diagnostics is quantitative loop-mediated isothermal amplification, qLAMP. qLAMP is associated with a high tolerance of DNA polymerase to different inhibitors remaining after sample purification. Therefore, we examined the influence of heparin, NaCl, EDTA, ethanol, urea and whole blood on the ability of fused proteins to act as DNA polymerase in qLAMP (Figure 11).
Figure 11.
Robustness of qLAMP with Gss derivatives. qLAMP reactions were conducted using a model system with a Lambda DNA as template and SYTO-82. Resulting time-to-threshold (Tt) values were plotted against concentration of different inhibitor (A-F).
The results showed that DBD and Sto7d did not affect the resistance of the chimeras to high concentrations of EDTA and ethanol. However, additional DNA-binding domains provided increased reaction yields in spite of the presence of heparin, urea and up to 200 mM NaCl. In parallel, we checked the ability of fused proteins to perform qLAMP in the presence of whole blood components. It was found that DBD-Gss, Sto-Gss and Gss-Sto mutants successfully catalysed this reaction in whole blood up to 5 vol%.
DISCUSSION
The attachment of additional domains to a protein is a prominent strategy to vary enzyme properties including solubility, thermostability and contaminant resistance. Successful examples in this field include Pfu, Taq and Φ29 polymerases, which have been fused with various DNA-binding domains in order to increase their processivity and salt tolerance (11–14). However, analogous fusions of Bst-polymerase, which is widely used for diagnostic and research goals, with additional thermostable DNA-binding domains demonstrated only improved thermostability, whilst the other enzymatic characteristics remained unknown (13). The present study was aimed to fill this gap and create enzymes with improved characteristics for practical applications, specifically for WGA and LAMP.
Here, we constructed chimeric Bst-like polymerases based on catalytic domain of DNA polymerase I from Geobacillus sp. 777 with DNA-binding domain of DNA ligase P. abyssi either at N- or C-terminus or with DNA-binding protein Sto7d from thermophilic archaea S. tokodaii at N- or C-end. In contrast to data obtained by Pavlov et al. (13) fusions of Gss with DBD or Sto7d did not increase the thermostability of chimeric proteins compared to native Gss. Using DSF and functional analysis, we demonstrated the same temperature optimum and half-life time of the enzymes at different temperatures (Figure 1). This contradiction can be resulted from the difference between methods of thermostability analysis used on. In any case, thermal stability of Bst-like polymerases including Gss and its derivatives is sufficient for practical applications, so unaltered stability of DBD-Gss, Sto-Gss and Gss-Sto does not diminish their value for WGA and LAMP.
The polymerase activity of the chimeric enzymes, which was evaluated as the ability of enzyme to incorporate dNMPs in activated DNA, was disturbed by additional domains (Table 2). Adjunction of the DNA-binding domain to the N-terminus of Gss caused a tenfold decrease in the DBD-Gss activity compared to that of the native Gss, whereas joining the DBD to the C-terminus of Gss led to a complete loss in its ability to amplify DNA. Such drastic changes can be explained by conformational (folding) disturbances that occur after fusion with the DBD. The same effect was described for chimeric Taq-polymerase with thioredoxin-binding domain, which retained only ∼15% of the wild-type Taq-polymerase activity (22). In our case, a complete loss of the activity may have resulted from the increased size of the enzyme and location of the DBD at the C-terminus. Despite such a dramatic reduction in polymerase activity, enzymes fused with the DNA-binding domains showed extremely high processivity (Table 2 and Figure 4). Simultaneously, these enzymes demonstrated the same fidelity as native Gss and have advantages in higher levels of terminal transferase activity and processivity (Figures 5–7). Thus, we constructed chimeric enzymes, which can be potentially useful to WGA applications.
Enzymes suitable for WGA analysis should display similar efficacies in reactions using DNA templates with different GC-contents, lengths or secondary structures. The presence of GC-rich tracts and the existence of secondary structures are by nature an additional obstacle for WGA, leading to decreasing amplification rates and limiting the size of resulting products. High processivity of an enzyme affords the possibility to preclude delay or halt of synthesis thus increasing both the amount and quality of DNA after WGA. As anticipated, fusion of either DBD or Sto7d with Gss was associated with an increased processivity leading to a better performance in WGA due to higher mean length of synthesized DNA stretches. In the present study, the efficacy of WGA was independent of the GC-content and uniform between amplicons with different lengths using DBD-Gss, Sto-Gss and Gss-Sto. The branched structure of WGA products also did not influence the amplification efficacy obtained by ddPCR.
These data are very promising to test the chimeric enzymes in in-field or point-of-care diagnostic methods such as qLAMP. Point-of-care (POC) diagnostics uses tests at the bedside; therefore, the DNA purification procedure from biological samples should be simplified resulting in the accumulation of different cellular and chemical contaminations in the probe. Different types of glycosaminoglycans, such as a native heparin, are widely presented at the cell surface and in the extracellular matrix. Moreover, its chemical analogues are an extra addition to biological samples to achieve high anticoagulant potency. One of the frightful outcomes of the presence of heparin in the reaction mixtures is its reversible chaotropic properties. Additionally, it can interact with the protein as a non-specific polyanion. Thus, the presence of the heparin fractions in the analysed probe can potentially inhibit the reactions under diagnostic applications. Several blood components (hem, lactoferrin, bile salts, bilirubin and IgG) are known to directly inhibit DNA polymerases (23); however, the exact mechanism of the inhibition remains unknown. Traces of the mentioned inhibitors can lead to false-negative results even after the purification procedure; rates of inhibition can occur at 0.34–2.4% in the case of HIV (24) or hepatitis C virus (25) testing. However, the problem is not only in the cellular components. Through the DNA purification procedure, the analysed sample can be contaminated by other chaotropic agents such as urea, ethanol and monovalent cations, or by chelating agents such as EDTA. All of these components suppress DNA polymerase activity and prevent precise accumulation of DNA in the sample. Thus, inhibitor-tolerable enzymes are urgently required for rapidly developing POC tests. Since LAMP was shown to be more robust than PCR (26), Bst-polymerase can permit the use of complex biological substances for direct analysis. Here, we demonstrated that the additional DNA-binding domain increased not only the processivity of the enzyme, but provided inhibitor tolerance to common DNA sample contaminants. An exact mechanism of inhibition by components of blood is unclear; however, increasing tolerance of the chimeric enzymes implies the role of DNA binding at this process. Further studies should be conducted to investigate the mechanism of inhibition of amplification by whole blood. Increased indifference of chimeric DNA polymerases opens new possibilities for the construction of enzymes that are less susceptible to inhibitors.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
State funded budget project [VI.62.1.5, 0309–2016-0003]; ‘Synthetic biology: development of tools for manipulating with genetic material and perspective agents for therapy and diagnostics’; Russian Ministry of Science and Education [5–100 Excellence Programme].
Conflict of interest statement. None declared.
REFERENCES
- 1. Burgers P.M., Koonin E.V., Bruford E., Blanco L., Burtis K.C., Christman M.F., Copeland W.C., Friedberg E.C., Hanaoka F., Hinkle D.C. et al. . Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001; 276:43487–43490. [DOI] [PubMed] [Google Scholar]
- 2. Hübscher U., Spadari S., Villani G., Maga G.. DNA Polymerases Discovery, Characterization and Functions in Cellular DNA Transactions. 2010; Singapore: World Scientific Publishing Co. Pte. Ltd. [Google Scholar]
- 3. Patel P.H., Suzuki M., Adman E., Shinkai a, Loeb L.A.. Prokaryotic DNA polymerase I: evolution, structure, and ‘base flipping’ mechanism for nucleotide selection. J. Mol. Biol. 2001; 308:823–837. [DOI] [PubMed] [Google Scholar]
- 4. Joyce C.M., Kelley W.S., Grindley N.D.. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J. Biol. Chem. 1982; 257:1958–1964. [PubMed] [Google Scholar]
- 5. Sellmann E., Schröder K.L., Knoblich I.M., Westermann P.. Purification and characterization of DNA polymerases from Bacillus species. J. Bacteriol. 1992; 174:4350–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang Y.P., Downie J.A., Ito J.. Primary structure of the DNA polymerase I gene of an alpha-proteobacterium, Rhizobium leguminosarum, and comparison with other family A DNA polymerases. Curr. Microbiol. 1999; 38:355–359. [DOI] [PubMed] [Google Scholar]
- 7. Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T.. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000; 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aviel-Ronen S., Qi Zhu C., Coe B.P., Liu N., Watson S.K., Lam W.L., Tsao M.S.. Large fragment Bst DNA polymerase for whole genome amplification of DNA from formalin-fixed paraffin-embedded tissues. BMC Genomics. 2006; 7:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fire A, Xu S.Q.. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:4641–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J. et al. . Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y., Prosen D.E., Mei L., Sullivan J.C., Finney M., Vander Horn P.B.. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 2004; 32:1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Vega M., Lázaro J.M., Mencía M., Blanco L., Salas M.. Improvement of φ29 DNA polymerase amplification performance by fusion of DNA binding motifs. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:16506–16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pavlov A.R., Pavlova N. V, Kozyavkin S.A., Slesarev A.I.. Cooperation between catalytic and DNA binding domains enhances thermostability and supports DNA synthesis at higher temperatures by thermostable DNA polymerases. Biochemistry. 2012; 51:2032–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pavlov A.R., Belova G.I., Kozyavkin S.a, Slesarev A.I.. Helix-hairpin-helix motifs confer salt resistance and processivity on chimeric DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:13510–13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oscorbin I.P., Boyarskikh U.A., Filipenko M.L.. Large fragment of DNA polymerase I from Geobacillus sp. 777: cloning and comparison with DNA polymerases I in practical applications. Mol. Biotechnol. 2015; 57:947–959. [DOI] [PubMed] [Google Scholar]
- 16. Oscorbin I.P., Boyarskikh U.A., Zakabunin A.I., Khrapov E.A., Filipenko M.L.. DNA-binding domain of DNA ligase from the Thermophilic Archaeon Pyrococcus abyssi: improving long-range PCR and neutralization of Heparin's inhibitory effect. Appl. Biochem. Biotechnol. 2015; 176:1859–1869. [DOI] [PubMed] [Google Scholar]
- 17. Maniatis T., Fritsch E.F., Sambrook J.. Molecular cloning: a laboratory manual. 1990; 2nd edn, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 18. Ricchetti M., Buc H.. E. coli DNA polymerase I as a reverse transcriptase. EMBO J. 1993; 12:387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sharma A., Nair D.T.. MsDpo4-a DinB homolog from Mycobacterium smegmatis-is an error-prone DNA polymerase that can promote G:T and T:G mismatches. J. Nucleic Acids. 2012; 2012:25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goto M., Honda E., Ogura A., Nomoto A., Hanaki K.-I.. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques. 2009; 46:167–172. [DOI] [PubMed] [Google Scholar]
- 21. Duym W.W., Fiala K.A., Bhatt N., Suo Z.. Kinetic effect of a downstream strand and its 5′-terminal moieties on single nucleotide gap-filling synthesis catalyzed by human DNA polymerase lambda. J. Biol. Chem. 2006; 281:35649–35655. [DOI] [PubMed] [Google Scholar]
- 22. Davidson J.F., Fox R., Harris D.D., Lyons-Abbott S., Loeb L.A.. Insertion of the T3 DNA polymerase thioredoxin binding domain enhances the processivity and fidelity of Taq DNA polymerase. Nucleic Acids Res. 2003; 31:4702–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-soud W.A., Rådström P.. Purification and characterization of PCR-inhibitory components in blood cells purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001; 39:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drosten C., Seifried E., Roth W.K.. TaqMan 5′-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. J. Clin. Microbiol. 2001; 39:4302–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nolte F.S., Fried M.W., Shiffman M.L., Ferreira-Gonzalez A., Garrett C.T., Schiff E.R., Polyak S.J., Gretch D.R.. Prospective multicenter clinical evaluation of AMPLICOR and COBAS AMPLICOR hepatitis C virus tests. J. Clin. Microbiol. 2001; 39:4005–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Francois P., Tangomo M., Hibbs J., Bonetti E.-J., Boehme C.C., Notomi T., Perkins M.D., Schrenzel J.. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011; 62:41–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.