Abstract
The TREX complex (TREX) plays key roles in nuclear export of mRNAs. However, little is known about its transcriptome-wide binding targets. We used individual cross-linking and immunoprecipitation (iCLIP) to identify the binding sites of ALYREF, an mRNA export adaptor in TREX, in human cells. Consistent with previous in vitro studies, ALYREF binds to a region near the 5′ end of the mRNA in a CBP80-dependent manner. Unexpectedly, we identified PABPN1-dependent ALYREF binding near the 3′ end of the mRNA. Furthermore, the 3′ processing factor CstF64 directly interacts with ALYREF and is required for the overall binding of ALYREF on the mRNA. In addition, we found that numerous middle exons harbor ALYREF binding sites and identified ALYREF-binding motifs that promote nuclear export of intronless mRNAs. Together, our study defines enrichment of ALYREF binding sites at the 5′ and the 3′ regions of the mRNA in vivo, identifies export-promoting ALYREF-binding motifs, and reveals CstF64- and PABPN1-mediated coupling of mRNA nuclear export to 3′ processing.
INTRODUCTION
Eukaryotic gene expression is a complicated multi-step process that begins with transcription. Newly transcribed pre-mRNAs need to undergo several processing steps, including capping at the 5′ end, splicing to remove introns, and polyadenylation at the 3′ end. The mature mRNAs are then transported from the nucleus to the cytoplasm for translation into proteins. Nuclear export of mRNAs is a key step in eukaryotic gene expression that is physically and functionally coupled to transcription and pre-mRNA processing steps (1–9). This coupling on one hand enhances mRNA export efficiency, and on the other hand might ensure that only fully processed mRNAs and packaged into mRNP can be exported to the cytoplasm for translation (10–14).
The TREX complex (TREX) is one of the key players in mRNA export. It mainly contains three parts, including the THO sub-complex, the RNA helicase UAP56, and the RNA-binding protein ALYREF (2,5,15,16). In yeast, TREX is recruited co-transcriptionally (2,3,17), whereas in humans, it is recruited during a late step of splicing (5). Based on this evidence, mRNA export is thought to be coupled to transcription in yeast and is directly coupled to splicing in metazoans. Consistent with this view, splicing has been shown to promote both TREX recruitment and mRNA export in both Xenopus oocyte and in mammalian cells (1,5,18).
In yeast, the binding of the ALYREF homologue, Yra1, to the mRNA is highest at the 5′ end and decreases towards the 3′ end (19). In humans, the whole TREX complex is recruited to a region near the 5′ end of the mRNA (20,21). This recruitment is achieved through the interaction of TREX components ALYREF and THO with the nuclear cap-binding complex (CBC) (16,20,21). This 5′ specific binding was observed with in vitro synthesized pre-mRNAs that was not polyadenylated. Accumulating evidence supports the link between polyadenylation and mRNA export (6,8,22,23). In yeast, the CF1A component, Pcf11, interacts with Yra1 and enhances its recruitment (6). In mammals, the CFI subunit CFI-68 directly interacts with the mRNA export receptor NXF1 and promotes mRNA export (8). In addition, the THO component, THOC5, was shown to associate with several 3′ processing factors and regulate alternative polyadenylation of a subset of genes (22,23). However, whether these interactions affect TREX recruitment in human cells remains to be investigated.
By linking mRNA export to transcription and pre-mRNA processing, TREX exerts vital roles in the gene expression pathway. TREX is essential for embryogenesis, organogenesis and cellular differentiation (22,24), and has also been implicated in viral infections, cancer, neurotoxicity and intellectual disability (25–29). Thus, a greater understanding of TREX binding on RNAs is important for appreciating the potential impact of the complex on gene expression regulation.
In the TREX complex, ALYREF acts as an mRNA export adaptor by mediating the interaction between the mRNA and the mRNA export receptor NXF1 (30,31). ALYREF harbors a canonical RNA-binding motif and has been shown to directly bind RNAs (32,33). Thus, to obtain a global view for TREX-binding on RNAs, we identified the transcriptome-wide binding sites of the human ALYREF. We found that, consistent with its master role in mRNA export, the vast majority of ALYREF binding sites reside in exonic sequences of mature mRNAs. In agreement with previous in vitro studies (20,21), ALYREF shows a clear enrichment at a region near the 5′ end of mRNA in a CBP80-dependent manner. Unexpectedly, in vivo, ALYREF also binds a region near the 3′ end of the mRNA, and PABPN1 is specifically required for this binding. Furthermore, we uncovered the ALYREF-CstF64 interaction as a novel link between mRNA export and the 3′ processing machinery that significantly enhances the overall binding of ALYREF on the mRNA. In addition, we identified specific ALYREF-binding motifs that enhance intronless mRNA export. Together, our data demonstrate how ALYREF associates with the mRNA and show that ALYREF is likely recruited to mRNAs not only by CBC, but also by PABPN1 and the 3′ processing machinery.
MATERIALS AND METHODS
Plasmids and antibodies
The Smad (cS) and β-globin (cG) cDNA constructs were described previously (34). The cS-control, cS-(AGGUA)5 and cS-(CUUCG)5 plasmid were constructed by inserting the annealing oligo containing 5 tandem repeats of the control motif (UAAAA) or ALYREF-binding motif (AGGUA or CUUCG) between XbaI and ApaI sites of the Smad cDNA plasmid. The same strategy was used to make the cG-control, cG-(AGGUA)5 and cG-(CUUCG)5 plasmids. The ALYREF antibody was raised against full-length GST-ALYREF (Abclonal). Antibodies against PABPN1 (Abclonal), CBP80 (Abclonal), CstF64 (Bethyl), CPSF1 (Abcam), CPSF3 (Bethyl) or 6*His (Abclonal) were purchased.
Cell culture, DNA transfection and RNAi
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Biochrom). Plasmid DNAs were transfected using Lipofectamine 2000 (Invitrogen), and siRNAs were transfected with RNAimax (invitrogen). The siRNA sequences are listed in Supplementary Table S4.
iCLIP-seq
The iCLIP assay was carried out as described (35,36). Briefly, HeLa cells were irradiated with UV light at 300 mJ/cm2. After cell lysis, RNAs were partially fragmented using high (5 U/ml) or low (0.5 U/ml) concentrations of RNase I, followed by IPs with or without (no antibody control) 10 μg of the ALYREF antibody immobilized on protein A Dynabeads (Life Technologies). After extensive washing, immunoprecipitated RNAs were ligated at the 3′ ends to a RNA adapter and radioactively labeled by T4 polynucleotide kinase (Fermentas). The protein-RNA complexes were then transferred to a nitrocellulose membrane. For iCLIP cDNA library preparation, fragmented RNAs were purified and reverse-transcribed with a primer containing a barcode. The resulting cDNAs were purified by PAGE, circularized by single-stranded DNA ligase (Epicentre), linearized by restriction enzyme cleavage, and amplified by PCRs. High-throughput sequencing of iCLIP cDNA libraries from three biological replicates was performed on one lane of an Illumina hiseq 2000 flow cell with a 100 nt run length. The ALYREF iCLIP experiments in control, CBP80, and PABPN1 knockdown cells were performed on an Illumina hiseq 2000 flow cell with a 150 nt run length. The ALYREF iCLIP experiments in control and CstF64 knockdown cells were performed on an Illumina hiseq X-10 flow cell with a 150 nt run length. Hiseq sequencing reads of iCLIP experiments and filtered crosslink sites were deposited to NCBI GEO database (GSE99069).
iCLIP-seq data analysis
iCLIP data were analyzed according to a published method (37,38). Briefly, jemultiplexer (version 1.0.0) from the Galaxy environment was used to demultiplex the different experiments by their sample barcodes. The reads containing parts of the 3′ Solexa primer were trimmed from the 3′ end using fastx-toolkit. These iCLIP reads after random barcode evaluation and barcode removal, were mapped to the human genome (hg19) using bowtie2, allowing 2 mismatches. Only uniquely mapped reads were used for further analysis. The first nucleotide in the genome upstream of a mapping cDNA tag was defined as ‘cross-link nucleotide’, and the total of corresponding cDNA sequences was defined as ‘cDNA count’ at this site. The clustered binding sites were identified using the filtering algorithms (37). We performed the evaluation of clustered binding sites with the recommended default parameters (FDR < 0.05, cluster sites within a window of 10 nts). Only the cluster binding sites and the cluster cDNA tag were used in further analyses.
Reproducibility among three independent biological replicates was examined by pearson correlation analysis. Only iCLIP binding sites with high confidence, existing in two of three replicates, were used in further analysis (allow 5 nt deviation). Genomic annotations of mRNA exon, mRNA intron and intergenic region were based on the Ensembl Gene Track form UCSC database. For genes with different isoforms, the longest transcripts were used as representative mRNAs.
The length of mature mRNA was evenly divided into 2000 bins to examine the distribution of ALYREF binding sites. Permutation sample of binding sites distribution was generated by randomizing all binding sites with 99% confidence.
The base line in grey indicated the occurrence of binding site at each bin in permutation sample. The regions above the base line are defined as enrichment peaks of ALYREF bindings. All the source codes for scripts used for analyzing the data are provided as supplementary material.
Motif analysis
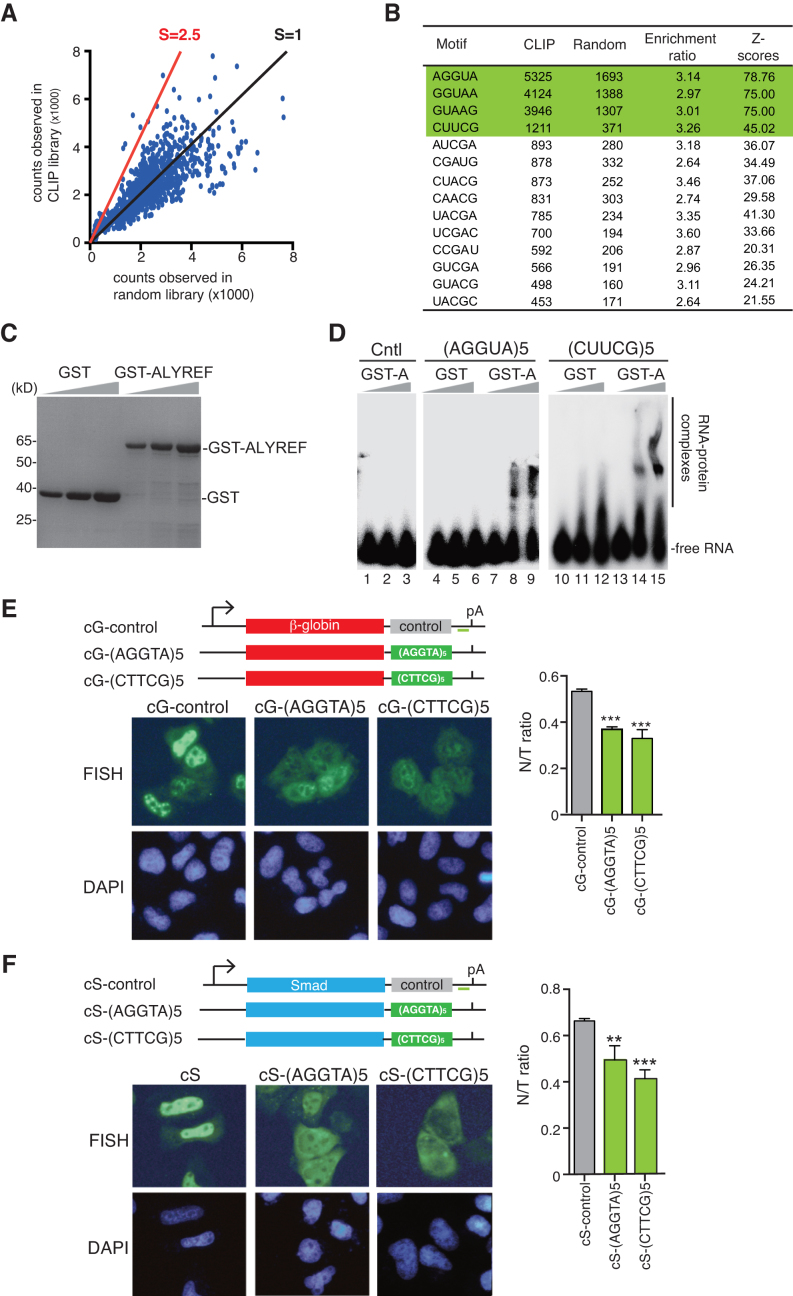
To identify ALYREF-binding motifs in the iCLIP dataset, we evaluated the occurrence of all pentamers within a –30 nt to +30 nt window around the binding sites excluding the binding sites in the 5′ and the 3′ regions. The positions of cross-linking sites were randomized within the same regions of the gene to establish random library. The enrichment of each pentamer was determined by comparing the occurrence of each pentamer around the binding sites (occurrence in iCLIP library) with at randomized cross-linking positions. Pentamers were then ranked by their enrichment, and the top 14 pentamers are shown in Figure 5B (enrichment rate > 2.5).
Figure 5.
Identification of functional ALYREF-binding motifs. (A) Analysis of ALYREF binding motif. The enrichment ratio of the occurrences in the ALYREF binding sites (occurrence in iCLIP library, vertical ordinate) to those in randomized binding site (occurrence in random library, horizontal ordinate) of each pentamer is shown. The red line indicates the position with the enrichment ratio of 2.5. Z-scores are used to evaluate statistical significance of enrichment of each motif. (B) The sequences of motifs with enrichment ratio bigger than 2.5. (C) Purified GST and GST-ALYREF used in EMSA were separated by SDS-PAGE, followed by Coomassie staining. (D) 0.1 pmol of 32P-labeled RNAs containing 5 tandems repeats of AGGUA or CUUCG were incubated with 0, 3 and 10 pmol of purified GST-ALYREF or GST. The protein–RNA complexes were fractionated on a 5% native-PAGE gel and visualized using autoradiography. The position of free RNAs and protein-bound RNAs are indicated. As another control, same amount of the RNA containing five tandem repeats of UAAAA was incubated with GST-ALYREF. (E) Schematic of the β-globin reporter constructs. The CMV promoter, BGH polyA sites and the location of the FISH probe (vector probe) that detects a region of pcDNA3 vector (indicated by a short green line) are shown. Tηεσε constructs were transfected into HeLa cells. 12 h after transfection, FISH was preformed to detect the indicated mRNAs. DAPI staining was used to indicate the nuclei. N/T ratios were determined for 30 cells per construct in each experiment. The graph shows the average N/T ratios from three independent experiments, and error bars indicate the standard deviations. Statistical analysis was performed using Student's t test. **P < 0.01; ***P < 0.001. (F) Same as (E), except that Smad DNA constructs were used.
Reverse transcription and PCRs
Total RNAs were extracted using TRI Reagent (Sigma) and treated with the RNase-free RQ DNase I (Promega) for 2 h at 37°C. After phenol/chloroform extraction and ethanol precipitation, RNAs were reverse transcribed with M-MLV reverse transcriptase (Promega). For qPCRs, cDNAs were amplified using GoTaq qPCR Master Mix (Promega) according to the manufacturer's protocol. RT-qPCRs were carried out following the MIQE guideline, and MIQE checklist is provided in supplementary material. For PCRs, cDNAs were amplified using KOD-plus DNA polymerase (Toyobo). Primers sequences are listed in Supplementary Table S5.
Electrophoretic mobility shift assay (EMSA)
The 32P-labeled RNAs containing five tandems repeats of motif UAAAA (as control), AGGUA or CUUCG were generated by in vitro transcription using the T7 RNA polymerase. 0.1 pmol of 32P-labeled RNA (∼2000 cpm) were mixed with 4 μl of binding buffer (8 mM MgCl2, 0.25 μg/μl tRNA, 4 U RNasin), 1.2 μl of native loading buffer (60% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol), and 6 μl of 0 pmol, 3 or 10 pmol of GST or GST-ALYREF. After incubation at 37°C for 15 min, 10 μl of the mixture was loaded into a 5% native gel. Radioactivity was detected by PhosphorImager.
Fluorescence in situ hybridization (FISH)
After fixation with 4% PFA in 1× PBS, cells were permeabilized with 1× PBS/0.1% Triton for 15 min. Following two washes with 1× SSC/50% Formamide, the cells were incubated with the 70 nt vector probe at 37°C for 12 h which were labeled at the 5′ end with Alexa Fluor 488 NHS Ester and high-performance liquid chromatography purified. The probe sequence is AAGGCACGGGGGAGGGGCAAACAACAGATGGCTGGCAACTAGAAGGCACAGTCGAGGCTGATCAGCGGGT. Images were captured with a DP72-CCD camera (Olympus) on an inverted microscope using DP-BSW software (Olympus). FISH quantitation was carried out using ImageJ 1.33u software (National Institutes of Health). N/T ratios were calculated as described (18).
Immunoprecipitations
For each assay, 106 cells were suspended with 250 μl of IP buffer (1 × PBS, 0.1% Triton, 50 U/ml RNase A, 0.2 mM PMSF and protease inhibitor). The lysates was centrifuged at 13 000 g for 15 min. The supernatant was mixed with 20 μl of antibodies cross-linked to protein A beads. The mixtures were rotated overnight at 4°C, followed by four washes with 1.5 ml of IP buffer. Proteins were eluted with sodium dodecyl sulfate (SDS) loading buffer. Samples were separated by SDS-PAGE, followed by western blotting.
GST pull-downs
All the recombined fusion proteins (GST tag or His tag) were expressed in Escherichia coli and purified. For each pull down reaction, 8 μg of purified GST-eIF4A3 or GST-ALYREF bound to 20 ml of Glutathione Sepharose 4B was mixed with 8 μg of His-CstF64 or His-Cntl (His-KIT) protein in pull-down buffer (1 × PBS with 0.1% Triton, 0.2 mM PMSF, 50 U/ml RNase A and protease inhibitor). The mixtures were rotated overnight at 4°C and beads were washed for five times. Proteins were eluted with SDS loading buffer, separated by SDS-PAGE, followed by staining with Coomassie blue and western blotting.
Nuclear and cytoplasmic RNA preparation
Nuclear and cytoplasmic fractionation was carried out as described (39). Briefly, 4 × 105 of HeLa cells were suspended in 1 ml of lysis buffer (10 mM Tris at pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% Igepal, 2 mM VRC) and incubated for 5 min on ice. The lysate was centrifuged at 1000 g for 3 min at 4°C, and the pellet and the supernatant were kept as the nuclei and the cytoplasmic fraction, respectively. To obtain pure cytoplasmic RNA, the supernatant fraction was further centrifuged at 13 000 rpm for 10 min at 4°C and then collected carefully to a new tube. To obtain pure nuclear RNA, the nuclear pellets were subjected to one additional wash with 1 ml of lysis buffer containing 0.5% deoxycholic acid and one additional wash with the lysis buffer. Finally, the purified nuclei were re-suspended in 100 μl of lysis buffer. Nuclear or cytoplasmic RNAs were extracted using TRI Reagent (Sigma).
RNA-seq
PolyA RNAs were isolated using NEBNext ploy(A) mRNA magnetic isolation module (NEB), and the libraries were generated using NEBNext Ultra directional RNA library prep kit (NEB). RNA-seqs were performed on an Illumina hiseq 2000 flow cell with a 150 nt run length. Reads were trimmed with fastx-toolkit, and mapped against human genome (hg19) with Tophat2, and gene expression was quantified by RPM using HTseq (40). Input of iCLIP samples were sequences similarly except that instead of polyA RNA enrichment, rRNAs were depleted using the NEBNext rRNA depletion kit (NEB). RNA-seq data are available at NCBI GEO database (GSE99069).
Northern blotting
10 μg of nuclear or cytoplasmic RNAs was fractionated on 13% urea-PAGE gel and transferred to positively charged nylon membrane (GE). After cross-linking by UV irradiation, the membrane was hybridized with PerfectHyb plus hybridization buffer (Toyobo) containing 10 ng/μl of 32P-end-labeled DNA oligonucleotide probe to U6 snRNA or tRNA−lys at 42°C overnight. The probe sequences are shown in Supplementary Table S6.
RESULTS
ALYREF iCLIP reveals its specific RNA-bindings in vivo
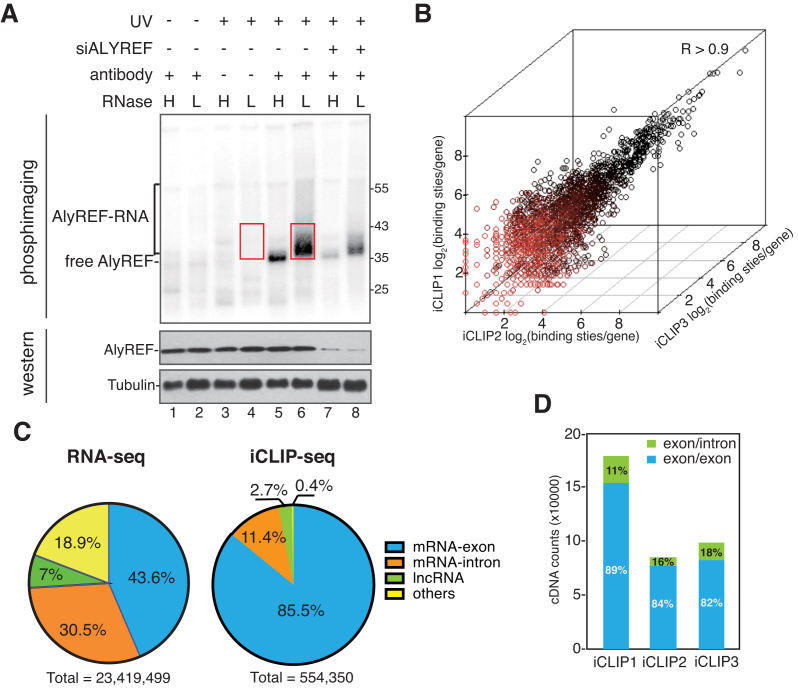
To analyze transcriptome-wide binding of the key mRNA export adaptor ALYREF, we performed iCLIP-seq in HeLa cells using an ALYREF antibody. ALYREF-RNA complexes shifting upwards from the size of ALYREF were apparently observed (Figure 1A, lane 6), and the shift was less pronounced in the presence of high concentration of RNase I (Figure 1A, lane 5). The radioactive signal significantly decreased when ALYREF was knocked down (Figure 1A, lanes 7–8), and disappeared when cells were not cross-linked (Figure 1A, lanes 1 and 2) or no antibody was used in immunoprecipitation (IP) (Figure 1A, lanes 3 and 4). We purified, reverse-transcribed and PCR-amplified cross-linked RNAs present in the ALYREF immunoprecipitates, using those present in no antibody samples as a quality controls (Figure 1, boxed regions; Supplementary Figure S1A). These RNAs as well as RNAs extracted from the iCLIP inputs were sequenced. To access experimental variation, this experiment was biologically repeated in triplicate.
Figure 1.
ALYREF binding sites are enriched on exons of mature mRNAs. (A) Detection of ALYREF-RNA complexes. RNase I treated (H: 5 U/ml; L: 0.5 U/ml) and 32P-labeled RNP complexes were immunoprecipitated with or without the ALYREF antibody from normal and ALYREF knockdown cells. After size-separation using denaturing gel electrophoresis, ALYREF-RNA complexes were transferred to a nitrocellulose membrane. The upper panel shows the autoradiograph of the nitrocellulose membrane. The lower panel shows the western blotting result using the indicated antibodies for input of IP. Red box indicates the region that was extracted for subsequent analyses. (B) Correlation of ALYREF iCLIP-seq replicates. Clustered ALYREF binding sites in each gene are plotted for three independent biological replicates (Spearman correlation coefficient, R > 0.90 for all comparisons). (C) ALYREF binding sites are enriched at mRNA exon. Left pie chart shows the percentage of RNA-seq reads that uniquely mapped to four human genome regions: mRNA exons, mRNA-introns, lncRNAs or others. Right pie chart shows the percentage of clustered ALYREF binding sites mapped to different human genome regions. (D) ALYREF mainly binds mature mRNAs. Percentages of the iCLIP tag in ALYREF binding sites specifically mapped to the exon-exon junction (blue) or exon-intron junction (green).
In total, high-throughput sequencing generated 15.6, 10.8 and 20.4 million raw reads from ALYREF iCLIP1, iCLIP2 and iCLIP3, respectively (Supplementary Table S1). After the elimination of PCR amplification artifacts and mapping to the human genome, in total 930 479 cross-linking nucleotides were identified in the three replicates of ALYREF iCLIP, each representing a uniquely cross-linked RNA molecule. To reduce false positive hits and to increase the resolution of the data, we applied filtering algorithms to identify clustered binding sites (36,37). In the these ALYREF iCLIP samples, we identified 554,350 clustered binding sites (FDR < 0.05) in total. In contrast, in no antibody controls, only 302,241 cDNAs from all biological triplicates were mapped to the human genome, and 17,643 clustered binding sites were identified. We used cDNAs or the clustered cross-linking nucleotides located at clustered binding sites for further analysis.
To validate the reproducibility of our data, we examined the correlation of ALYREF binding sites per gene between biological replicates and found a high level of reproducibility between experiments (Spearman correlation coefficient; R > 0.90 for all comparisons) (Figure 1B). This consistency between replicates allowed us to merge these biological replicates. Note that only iCLIP binding sites with high confidence, existing in two of three replicates, were used in further analysis.
ALYREF mainly binds mature mRNAs in vivo
To obtain a first overview of the ALYREF binding, we analyzed the distribution of ALYREF clustered binding sites in the human genome by defining four regions (mRNA-exons, mRNA-introns, lncRNAs, etc.). 85.5% of ALYREF binding sites were located in mRNA-exons, and 11.4% were distributed to mRNA-introns (Figure 1C, right pie). The rest 3.1% were located in long noncoding RNAs (lncRNAs) and other RNAs. To establish an accurate reference for ALYREF binding specificity, we analyzed RNA-seq data for the iCLIP inputs. In total, we obtained ∼23 million uniquely mapped reads (Figure 1C, left pie), among which 43.6% were overlapped with mRNA-exons and 30.5% were mapped to mRNA-introns (Figure 1C, left pie). Relative to the abundance in RNA-seq data, ALYREF binding on exons were apparently enriched, indicating that ALYREF mainly binds to of mRNA-exons in vivo.
To examine whether ALYREF mostly binds exons of pre-mRNAs or spliced mRNAs, we selectively computed iCLIP cDNAs mapping to exon/exon junctions or exon/intron junctions. As shown in Figure 1D, in all three replicates, >80% of iCLIP cDNAs were mapped to exon/exon junctions, indicating that ALYREF mainly binds mature mRNAs in vivo. This result is consistent with previous in vitro studies on TREX recruitment (5,16,20). Together, we conclude that ALYREF mainly binds mature mRNAs globally.
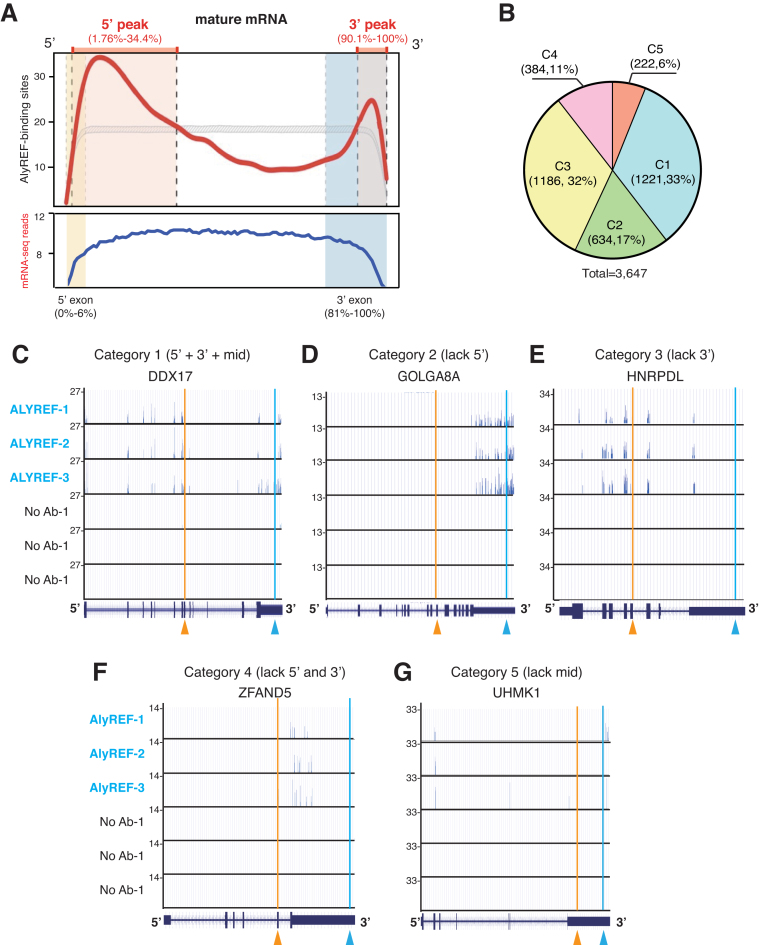
ALYREF is enriched at the 5′ and the 3′ regions of the mRNA
Previously, using in vitro transcribed RNAs lacking a polyA tail, we found that ALYREF mainly binds a region near the 5′ end of the mRNA (20). Similarly, PAR-CLIP of Yra1, the yeast counterpart of ALYREF, revealed that it binds to about the 5′ half of the mRNA (19). To examine where ALYREF binds on the mRNA globally, mature mRNAs were binned into 2000 discrete units, and ALYREF binding sites across each bin were calculated. Consistent with the previous studies in HeLa nuclear extract and in yeast (20) (19), we found a clear enrichment of ALYREF binding sites at the 5′ region of the mRNA (Figure 2A). This enrichment forms a specific ALYREF binding peak in the 5′ one third of the mature mRNA (Figure 2A). Note that when input sequencing reads were mapped across these 200 units, no such enrichment was detected (Figure 2A). Different from previous observation that ALYREF mainly binds the 5′ most exon (19), we found that the 5′ ALYREF-binding peak did not mostly fit into the first exon (Figure 2A; see Discussion).
Figure 2.
ALYREF binds different regions on mature mRNAs. (A) Genome-wide distribution profile of ALYREF binding sites along mature mRNAs. The mature mRNA is divided into 2000 bins. The y values show the total occurrence of binding sites in three repeats at each bin. The median length of 5′ most and 3′ most exons are shown in yellow and blue block, respectively. (B) Five categories of mRNAs according to ALYREF binding at different regions. The pie chart shows the percentage of mRNAs in each category. C1, ALYREF binds at all three regions; C2, lacks ALYREF binding at the 5′ region; C3, lacks ALYREF binding at the 3′ region; C4, lacks ALYREF binding at the 5′ and the 3′ regions; C5, lacks ALYREF binding at the middle region (C5). The number and percentage of mRNAs in each category are shown. (C–G) Screenshots of the UCSC genome browser for representative mRNAs in each category. The y-values represent the cDNA counts of each binding site. The orange arrowheads indicate the boundary of 5′/middle regions and the blue ones indicate the boundary of middle/3′ regions.
In addition to the 5′ peak, unexpectedly, we found an apparent ALYREF binding peak at the 3′ region (Figure 2A). By comparison of this peak with the median size of the 3′ most exon, we found that this 3′ peak (90.1–100%) was located in the 3′ most exon (81–100%). This result indicates that ALYREF is also recruited to the 3′ end of the mRNA in vivo. In addition, compared to the controls, ALYREF binding sites were also apparent in the middle region of the mRNA (Supplementary Figure S1B), indicating that ALYREF might also bind middle exons in vivo.
ALYREF binds different regions on individual mRNAs
With the ALYREF general binding rule in mind, we next looked into details for ALYREF binding on individual mRNAs. ALYREF clustered binding sites were apparently and reproducibly detected on 3647 mRNAs in total. According to ALYREF binding at the 5′, the 3′, and the middle regions, mRNAs fall into 5 categories (Figure 2B). In the first category (1221, 33%), ALYREF binds at all of these three regions. The DDX17 mRNA shows a typical example for this category (Figure 2C). The second category (634, 17%) represents mRNAs lacking the 5′ ALYREF binding, with the GOLGA8A mRNA as a typical example (Figure 2D). In the third category, ALYREF binding at the 3′ is specifically missing (1186, 32%), with the HNRPDL mRNA as an example (Figure 2E). The fourth category represents mRNAs (384, 11%) harboring ALYREF bindings in the middle region, but not in the 5′ and the 3′ region, with the ZFAND5 mRNA as a typical example (Figure 2F). In the last category (222, 6%), ALYREF binds mRNAs at both the 5′ and the 3′ regions, but not at the middle region, exemplified by the UHMK1 mRNA (Figure 2G). Together, these data demonstrate that ALYREF binding on mRNAs are heterogeneous.
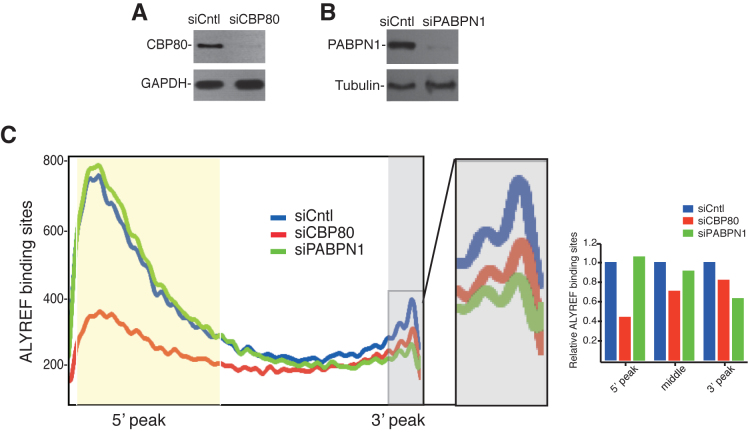
CBP80 mainly facilitates ALYREF binding at the 5′ region of the mRNA
Previous studies in HeLa nuclear extracts revealed that the 5′ cap facilitates TREX recruitment through the interaction between ALYREF and CBP80, which is the large subunit of CBC (20). To investigate how the 5′ cap/CBP80 affects ALYREF binding globally, we carried out ALYREF iCLIP in cells depleted of control or CBP80 (Supplementary Table S2). CBP80 was efficiently depleted as revealed by western blotting analysis (Figure 3A). Depletion of CBP80 significantly reduced ALYREF binding at the 5′ region (Figure 3C), indicating that CBC plays a key role in recruiting ALYREF to the 5′ region of the mRNA in the cells. Notably, ALYREF binding at the middle and the 3′ regions was also reduced to some extent (Figure 3C), suggesting that CBP80 might also moderately affect ALYREF binding at the these regions of the mRNA.
Figure 3.
The effect of CBP80 and PABPN1 on ALYREF bindings on the mRNA. (A) Western blotting to examine the knockdown efficiency of CBP80. Tubulin was used as a loading control. (B) Western blotting to show the knockdown efficiency of PABPN1. GAPDH was used as a loading control. (C) The effect of CBP80 and PABPN1 knockdown on distribution of ALYREF binding sites. The horizontal ordinate represents the 2000 bin of mature mRNA. The vertical ordinate show the number of ALYREF binding sites in all mRNA in each bin. Lines in blue, red and green show distribution of ALYREF binding sites in control, CBP80, or PABPN1 knockdown. Part of the 3′ region was enlarged to clearly visualize the change of ALYREF binding. Quantification data of ALYREF-binding sites in the 5′, the middle, and the 3′ region of the mRNA in CBP80, or PABPN1 knockdown cells relative to control cells are shown in the graph.
PABPN1 down-regulation specifically affects the 3′ ALYREF binding
We next sought to investigate how ALYREF is recruited to the 3′ end of the mRNA. In the previous study, no ALYREF binding was observed at the 3′ end region of in vitro transcribed mRNAs that neither were polyadenylated nor contained a polyA tail (20). We speculated that the nuclear polyA-binding protein (PABPN1) might be involved in ALYREF recruitment to the 3′ end of the mRNA. To examine this possibility, we carried out ALYREF iCLIP in PABPN1 knockdown cells (Figure 3B, Supplementary Table S2), and compared ALYREF binding sites with those in the control cells. As shown in Figure 3C, PABPN1 knockdown moderately, but specifically, attenuated ALYREF binding at the 3′ region (Figure 3B and Supplementary Figure S3). These results suggested that PABPN1 is specifically involved in ALYREF recruitment to the 3′ region of the mRNA.
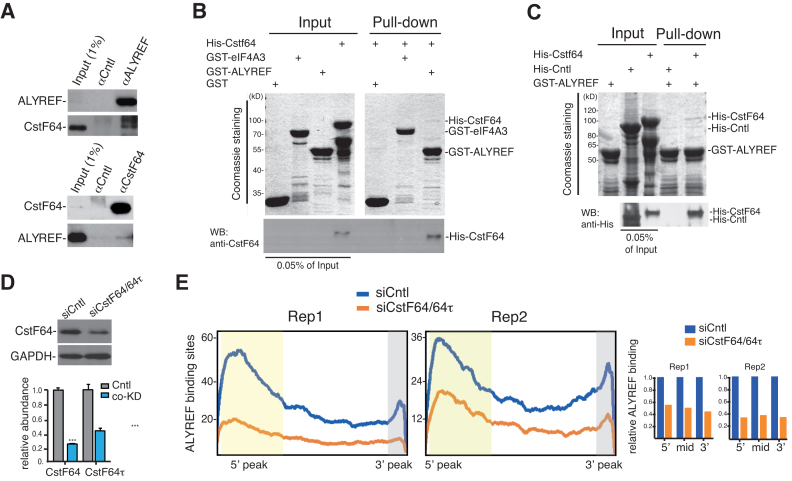
CstF64 directly interacts with ALYREF and is required for overall ALYREF binding
Considering that PABPN1 knockdown only led to moderately reduced ALYREF binding at the 3′ end region, we speculated that the 3′ processing factors might also facilitate this ALYREF binding. To identify possible associations of ALYREF with the 3′ processing factors, we carried out IP with the ALYREF antibody from RNase-treated HeLa cell lysate, followed by western blotting with antibodies to 3′ processing factors (Supplementary Figure S4). We found that CstF64, a component of the CstF complex, but not components of the CPSF complex, including CPSF1 and CPSF3, was co-immunoprecipitated with ALYREF (Figure 4A, upper panel; Supplementary Figure S4). Reciprocal IP with an antibody to CstF64 showed that ALYREF was specifically co-precipitated by the CstF64 antibody, but not the IgG (Figure 4A, lower panel), indicating that ALYREF associates with CstF64 in the cells. Note that this association is supported by a previous study that identified ALYREF as a Flag-CstF64-associating protein (41). To examine whether the interaction between ALYREF and CstF64 was direct, we carried out GST pull-down using purified recombinant proteins. Coomassie stained gels showed that GST-ALYREF, but not the control GST-eIF4A3 or GST, reproducibly pulled down His-CstF64 (Figure 4B). As an additional control, a His tagged control protein that has similar molecular weight to CstF64, was not pulled down by GST-ALYREF (Figure 4C). These data demonstrate a direct interaction between ALYREF and CstF64.
Figure 4.
CstF64 affects the overall binding of ALYREF on the mRNA. (A) CstF64 interacts with ALYREF in vivo. Immunoprecipitations were carried out from RNased HeLa nuclear extract using the ALYREF antibody (upper panels) or the CstF64 antibody (lower panels). IgG serves as a negative control for immunoprecipitation. Western blots using ALYREF and CstF64 antibodies are shown. Note that to clearly show the co-IP of CstF64 and ALYREF, relatively shorter exposure for IPs and longer exposure for co-IPs western blot are shown here. (B) CstF64 directly interacts with ALYREF in vitro. GST-ALYREF was used for pulling down purified His-tagged CstF64, followed by Coomassie staining and western blotting with a CstF64 antibody. GST-eIF4A3 and GST were used as negative controls. Note that for western blot, 0.05% of recombinant proteins shown on Coomassie gels are loaded. (C) GST-ALYREF specifically interacts with CstF64. GST-ALYREF was used for pulling down purified His-tagged CstF64 or a control protein (His-KIT), followed by Coomassie staining and Western blotting with a 6His antibody. Note that for western blot, 0.05% of recombinant proteins shown on Coomassie gels are loaded. (D) CstF64 was efficiently knocked down. Upper panel shows western blotting to examine the protein level of CstF64 in control and CstF64/CstF64τ co-knockdown cells. Lower panel shows RT-qPCRs to examine the relative mRNA levels of CstF64 and CstF64τ to GAPDH in CstF64/CstF64τ co-knockdown cells. Statistical analysis was performed using Student's t test. ***P < 0.001. (E) The effect of CstF64 knockdown on distribution of ALYREF binding sites. The x-axis displays the 2000 bin of mature mRNAs. The y-axis displays the number of ALYREF binding sites in each bin. Lines in blue and orange display enrichment of ALYREF binding sites in control or CstF64 knockdown cells, respectively. Quantification data of ALYREF-binding sites in the 5′, the middle, and the 3′ region of the mRNA in CstF64/64τ knockdown cells relative to control cells are shown in the graph.
To investigate whether CstF64 affects ALYREF binding on the mRNA, we carried out ALYREF iCLIP in CstF64 knockdown cells. Note that CstF64τ, the CstF64 paralog with overlapping functions, was co-knocked down (Figure 4D). In these co-knockdown cells, ALYREF bindings at the 5′, the 3′, as well as the middle regions were all dramatically reduced in two biological replicates (Figure 4E; Supplementary Table S3). Interestingly, similar reductions in ALYREF bindings were observed in these different mRNA regions. These results indicate that CstF64 plays an important role in the overall binding of ALYREF on the mRNA. Together, we conclude that CstF64 interacts with ALYREF and functions in ALYREF recruitment to the mRNA.
Identification of ALYREF-binding motifs that promote mRNA export
We next sought to investigate how ALYREF is recruited to the middle region of the mRNA. ALYREF bindings on exons in the middle regions were highly heterogeneous, even on the same mRNA (i.e. Figure 2C). We reasoned that specific sequences might be important for ALYREF binding to these exons. To investigate this possibility, we selectively analyzed ALYREF binding sites in the middle region of the mRNA. We compared the occurrences of all possible 5-nt motifs (1024) in ALYREF iCLIP library with those in randomized library of exon sequences in the same genomic regions. This analysis let us identify motifs whose occurrences in iCLIP library were significantly enriched (>2.5-fold) (Figure 5A). Sequences of these motifs are shown in Figure 5B.
To examine whether ALYREF binds to these motifs, 5 tandem repeats of the 4 most frequently appeared ones in ALYREF iCLIP library, AGGUA, GGUAA, GUAAG and CUUCG, were used for RNA electrophoretic mobility shift assays (EMSAs). As tandem repeating of AGGUA, GGUAA, and GUAAG resulted in the same sequence, we used AGGUA to represent these three motifs. Incubation of 32P-labeled, in vitro transcribed AGGUA or CUUCG repeats with GST-ALYREF, but not equal amount of GST itself, led to their lower mobility (Figure 5C and D, lanes 4–15). In contrast, equal amount of GST-ALYREF did not cause a detectable shift of a control RNA (Figure 5D, lanes 1–3). These results indicate that ALYREF indeed binds to the RNA motifs identified by iCLIP.
To investigate whether these ALYREF binding motifs could promote mRNA export, we inserted the AGGUA or CUUCG repeat into the 3′ of β-globin cDNA transcript (cG) that otherwise cannot be efficiently exported (18,27,42). As expected, the cG mRNA containing the control motif was largely retained in the nucleus (Figure 5E, cG-control). In contrast, the cG mRNA containing either AGGUA or CUUCG was apparently accumulated in the cytoplasm (Figure 5E). When these motifs were inserted into the Smad cDNA transcripts, similar results were obtained (Figure 5F). Note that insertion of these ALYREF-binding motifs did not introduce cryptic splicing of cG or cS mRNAs (Supplementary Figure S2A and S2B). These data indicate that ALYREF-binding motifs can promote mRNA export.
A correlation between ALYREF-binding motif occurrence and nuclear export of intronless mRNAs
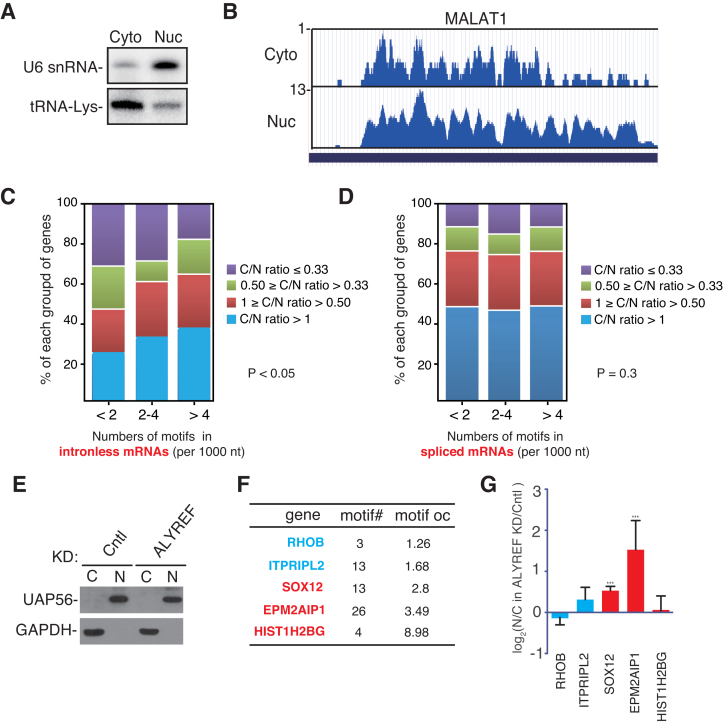
To examine the potential global effect of ALYREF-binding motifs on mRNA export, we purified nuclear and cytoplasm RNAs, followed by RNA-seq. Northern blots with the U6 snRNA and the tRNA probes were used to confirm the purity of nuclear and cytoplasmic RNAs (Figure 6A). Note that the nucleocytoplamic RPM ratio (N/C ratio) of the nuclear lncRNA MALAT1 was more than 13 (Figure 6B), indicating that our RNA-seq data were reliable. We analyzed the correlation of the occurrences of the ALYREF binding motifs, including AGGUA, GGUAA, GUAAG and CUUCG, with cytoplasmic accumulation of mRNAs. According to the occurrences of these motifs, intronless mRNAs were grouped into three categories (<2, 2–4, and >4 per 1000 nt). In each category, according to the C/N RPM ratio, mRNAs were further separated into four groups (<0.33, 0.33–0.5, 0.5–1 or >1). Significantly, the percentage of mRNAs with C/N ratio greater than 1 increased, whereras that of mRNAs with C/N ratio smaller than 0.33 decreased, along with the increasing motif occurrence (P < 0.05) (Figure 6C). These data show a correlation of C/N ratio with the occurrence of ALYREF-binding motifs and support the effect of these ALYREF-binding motifs on genome-wide intronless mRNA nucleocytoplasmic distribution. In contrast to intronless mRNAs, for spliced mRNAs, no similar correlation was observed (P = 0.3) (Figure 6D). To experimentally study how ALYREF functions in nuclear export of intronless mRNAs containing ALYREF-binding motifs, we picked five intronless mRNAs with different motif occurrence, ranging from 1.26 to 8.98, and examined how their nucleocytoplasmic distributions were affected by ALYREF depletion (Figure 6F and G). Interestingly, cytoplasmic accumulations of the intronless mRNAs with motif occurrence greater than 2, including SOX12 and EPM2AIP1, were apparently inhibited by ALYREF knockdown (Figure 6G). In contrast, no significant effect was observed with mRNAs with motif occurrence smaller than 2, including RHOB and ITPRIPL2. Although the HIST1H2BG mRNA contains more than 8 motifs/1000 nt, its nuclear export was not significantly affected by ALYREF knockdown. This is consistent with the finding that the mRNA export receptor NXF1 is recruited to histone mRNAs via SR proteins (4). Together, these data support the possibility that ALYREF-binding motifs promote nuclear export of intronless mRNAs by recruiting ALYREF.
Figure 6.
Genome-wide effect of ALYREF-binding motifs on nucleocytoplasmic mRNA distribution. (A) Northern blots to examine the purities of nuclear and cytoplasmic fractions. The U6 snRNA and tRNA-Lys were used as nuclear and cytoplasmic maker, respectively. (B) Screenshots of the UCSC genome browser for RNA-seq of MALAT1. The y-axis displays the RPM. (C) Analysis of the correlation of ALYREF binding motifs number and the C/N ratios of intronless mRNAs. Intronless mRNAs were divided into three groups based on occurrences of ALYREF binding motifs per 1000 nt. Based on C/N (RPM) ratio in RNA-seq, intronless mRNAs were further divided into fourtwo subgroups. The percentage of mRNA in each subgroup is shown in colored block. Chi-square tests were used to determine the association between occurrences of ALYREF binding motif and C/N ratio, P < 0.05. (D) Same as (C), except that statistical analysis on spliced mRNAs, Chi-square test, P = 0.3. (E) Western blotting to examine the purities of nuclear and cytoplasmic fractions from Cntl and ALYREF knockdown cells. HeLa cells were transfected with control and ALYREF siRNA for 72 hrs before nuclear and cytoplasmic fractions were prepared. UAP56 and GAPDH were used as nuclear and cytoplasmic makers, respectively. (F) The motif numbers (motif#) and occurrence (motif OC, motif numbers per kilobase of mature mRNA) of the five intronless mRNAs are shown. The mRNAs containing less than 2 motifs per kilobase are colored in blue, and those containing more than two motifs per kilobase are colored in red. (G) N/C ratios of intronless mRNAs in ALYREF knockdown cells relative to those in control cells are shown. HeLa cells were transfected with control and ALYREF siRNA for 72 h before nuclear and cytoplasmic fractions were prepared. Nuclear and cytoplasmic RNAs were extracted followed by RT-qPCRs. Statistical analysis was performed using Student's t test. ***P < 0.001, *P < 0.05.
DISCUSSION
In this study, our iCLIP data provide the first global view of the binding of ALYREF, and probably the TREX complex, in mammalian cells. We not only confirmed its binding at the 5′ region of the mRNA as previously identified in vitro, but also identified novel bindings at the 3′ end as well as at the middle region. Furthermore, our study reveals distinct mechanisms for ALYREF bindings at different regions.
Multiple binding regions of ALYREF/TREX on the mRNA in vivo
In the current model for TREX recruitment, CBP80 interacts with ALYREF and facilitates TREX recruitment to the 5′ region of the mRNA (20). This model mostly bases on studies using in vitro transcribed mRNAs without a polyA tail. Consistent with these in vitro studies, our iCLIP data show that ALYREF binding is indeed enriched at the 5′ region in vivo, and depletion of CBP80 significantly reduced this enrichment. Different from in vitro studies demonstrating that ALYREF mostly binds to the first exon (20), we found that in vivo the 5′ ALYREF peak was beyond the first exon. Three possible reasons might have caused this inconsistency. First, in the previous study, lack of ALYREF binding at the middle and the 3′ region of in vitro transcribed RNAs might have caused the 5′ shift of the 5′ ALYREF binding peak. Second, the technique of iCLIP might result in the seemingly attenuated ALYREF binding at the very end of the 5′, as the 5′ cap could interfere with the circulation of the RNA fragment. Third, it is also possible that in vitro conditions did not loyally reflect the ALYREF binding in vivo. Further studies are needed to investigate these possibilities.
Our study identified a novel ALYREF-binding region very close to the 3′ end of the mRNA. This binding peak was not identified in the previous study possibly because the reporter mRNAs used for mapping TREX binding sites neither were polyadenylated nor contain a polyA tail. Consistent with this possibility, we found that knockdown of either PABPN1 or the 3′ processing factor CstF64 clearly influences ALYREF binding at the 3′ region.
ALYREF bindings on individual mRNAs are highly variable. Only one third of mRNAs we analyzed harbors ALYREF binding sites on all three regions. Analysis of the nucleocytoplasmic distribution of mRNAs harboring ALYREF binding site at different regions did not show apparent difference (data not shown). Thus, it seems that as long as ALYREF binds to any of these three regions, the mRNA can be exported. The multiple position binding pattern of ALYREF/TREX might be important for cells to ensure that most mRNAs are exported even they have some specific features unfavorable for TREX binding at a specific region. If this is true, how do the cells ensure that only fully processed mRNAs can efficiently recruit TREX and are exported? Based on our iCLIP data, knockdown of the 3′ processing factor CstF64 not only dramatically weakened ALYREF binding at the 3′ end, but also significantly reduced its bindings at the 5′ and the middle regions. Similarly, CBP80 not only influences ALYREF binding at the 5′ region, but also moderately affects its 3′ binding. Thus, it is possible that in most cases, only when all processing steps are completed, ALYREF/TREX could be efficiently recruited to different regions of the mRNA. It is puzzling how proteins bound at the 3′ end affect ALYREF binding at the 5′ end, or versus. It was reported that mature mRNPs form a looped structure (43). In this way, the 5′ and the 3′ ends of the mRNP might be able to crosstalk with each other and facilitate ALYREF/TREX binding on each other.
The Linkage between 3′ processing and mRNA export
Up to date, multiple linkage have been identified between machines in 3′ processing and mRNA export (6,22,23). Whether these linkages are important for TREX recruitment globally remains unclear. Here, we found that CstF64 directly interacts with ALYREF and plays critical roles in transcriptome-wide ALYREF/TREX recruitment to the mRNA. Among the three factors we examined, CstF64 had the most significant effect on ALYREF binding. What could be the physiological relevance for 3′ processing factors affecting overall bindings of mRNA export factors? Polyadenylation can be considered as the final pre-mRNA processing step, whereas CBC probably binds pre-mRNAs as soon as the mRNA is capped. Upon maturation, ALYREF is recruited via its interaction between CBC. If this cap/CBC-mediated ALYREF recruitment was not affected by other pre-mRNA processing steps, the mRNA could be exported no matter whether it is appropriately spliced and polyadenylated or not. However, this is not the case. Previously, it was shown that both TREX recruitment and mRNA export depends on efficient splicing (5,20). Here, our data showed that knockdown of the 3′ processing factor CstF64 significantly weakens the overall binding of ALYREF, including the 5′ region. In yeast, Yra1 interacts with Pcf11, a component of the cleavage and polyadenylation factor IA (CF IA), and is involved in pre-mRNA 3′ end processing and in transcription termination (6). However, Yra1 mainly binds to the 5′ half of the mRNA (19). It remains unclear why the Yra1-Pcf11 interaction did not result in 3′ Yra1 binding in yeast. Nevertheless, this splicing and polyadenylation-dependent ALYREF/TREX binding would ensure that only fully mature mRNAs can be efficiently exported. In addition, polyadenylation-dependent ALYREF/TREX binding could also make contribution to nuclear export of intronless mRNAs as well as cDNA transcripts.
ALYREF-binding motifs promote mRNA export
We identified multiple ALYREF binding motifs in the middle region of mature mRNAs. Reporter mRNA export assay indicate that these motifs promote export of intronless mRNA, but not spliced mRNA. A recent study reported that β-globin cDNA transcript contains nuclear retention elements and splicing probably overcomes this retention elements (44). Thus, it is possible that the ALYREF-binding motifs we identified also facilitate in overriding the nuclear retention signals. The role of ALYREF-binding motif in promoting mRNA export was also supported by genome-wide sequencing data as well as qRT-PCR results of endogenous intronless mRNAs. Previously, it was reported that intronless mRNAs recruit mRNA export factors via cis-acting elements, including CAR-E (45–50). However, ALYREF-binding motifs we identified here are not similar to CAR-E (Supplementary Figure S5). Furthermore, these studies were mostly carried out with individual mRNAs and might not reflect a general rule for intronless mRNA recruiting mRNA export factors. In addition, most of these motifs were not directly bound by mRNA export adaptors. In this study, we only picked several motifs for examination of ALYREF-binding and export functions, it is highly possible that more, and even stronger ALYREF-binding motifs exist in human genomes. Further studies are needed to explore these possibilities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jing Fan and Ke Wang and other Cheng Lab members for useful discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by grants from National key R&D Program of China [2017YFA0504400]; Ministry of Science and Technology of China [2013CB910402]; National Natural Science Foundation of China [21625302, 31570822, 91540104, 31400676]. Funding for open access charge: National Natural Science Foundation of China [21625302].
Conflict of interest statement. None declared.
REFERENCES
- 1. Luo M.J., Reed R.. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strasser K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondon A.G., Aguilera A., Struhl K., Reed R. et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002; 417:304–308. [DOI] [PubMed] [Google Scholar]
- 3. Zenklusen D., Vinciguerra P., Wyss J.C., Stutz F.. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002; 22:8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang Y., Gattoni R., Stevenin J., Steitz J.A.. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003; 11:837–843. [DOI] [PubMed] [Google Scholar]
- 5. Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R.. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005; 19:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson S.A., Cubberley G., Bentley D.L.. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell. 2009; 33:215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schneider M., Hellerschmied D., Schubert T., Amlacher S., Vinayachandran V., Reja R., Pugh B.F., Clausen T., Kohler A.. The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell. 2015; 162:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruepp M.D., Aringhieri C., Vivarelli S., Cardinale S., Paro S., Schumperli D., Barabino S.M.. Mammalian pre-mRNA 3′ end processing factor CF I m 68 functions in mRNA export. Mol. Biol. Cell. 2009; 20:5211–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meinel D.M., Burkert-Kautzsch C., Kieser A., O’Duibhir E., Siebert M., Mayer A., Cramer P., Soding J., Holstege F.C.P., Strasser K.. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet. 2013; 9:e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maniatis T., Reed R.. An extensive network of coupling among gene expression machines. Nature. 2002; 416:499–506. [DOI] [PubMed] [Google Scholar]
- 11. Mitchell S.F., Parker R.. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014; 54:547–558. [DOI] [PubMed] [Google Scholar]
- 12. Katahira J. [Formation and nuclear export of messenger ribonucleoprotein complexes]. Seikagaku. 2015; 87:75–81. [PubMed] [Google Scholar]
- 13. Heath C.G., Viphakone N., Wilson S.A.. The role of TREX in gene expression and disease. Biochem. J. 2016; 473:2911–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bjork P., Wieslander L.. Integration of mRNP formation and export. Cell. Mol. Life Sci. 2017; 74:2875–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rehwinkel J., Herold A., Gari K., Kocher T., Rode M., Ciccarelli F.L., Wilm M., Izaurralde E.. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 2004; 11:558–566. [DOI] [PubMed] [Google Scholar]
- 16. Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., Cheng H.. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2013; 41:1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abruzzi K.C., Lacadie S., Rosbash M.. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004; 23:2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Valencia P., Dias A.P., Reed R.. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:3386–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baejen C., Torkler P., Gressel S., Essig K., Soding J., Cramer P.. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell. 2014; 55:745–757. [DOI] [PubMed] [Google Scholar]
- 20. Cheng H., Dufu K., Lee C.S., Hsu J.L., Dias A., Reed R.. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006; 127:1389–1400. [DOI] [PubMed] [Google Scholar]
- 21. Nojima T., Hirose T., Kimura H., Hagiwara M.. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007; 282:15645–15651. [DOI] [PubMed] [Google Scholar]
- 22. Tran D.D., Saran S., Williamson A.J., Pierce A., Dittrich-Breiholz O., Wiehlmann L., Koch A., Whetton A.D., Tamura T.. THOC5 controls 3′end-processing of immediate early genes via interaction with polyadenylation specific factor 100 (CPSF100). Nucleic Acids Res. 2014; 42:12249–12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katahira J., Okuzaki D., Inoue H., Yoneda Y., Maehara K., Ohkawa Y.. Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res. 2013; 41:7060–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X., Chang Y., Li Y., Zhang X., Goodrich D.W.. Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol. Cell. Biol. 2006; 26:4362–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dominguez-Sanchez M.S., Saez C., Japon M.A., Aguilera A., Luna R.. Differential expression of THOC1 and ALY mRNP biogenesis/export factors in human cancers. BMC Cancer. 2011; 11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L., Miao Y.L., Zheng X., Lackford B., Zhou B., Han L., Yao C., Ward J.M., Burkholder A., Lipchina I. et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013; 13:676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chi B., Wang K., Du Y., Gui B., Chang X., Wang L., Fan J., Chen S., Wu X., Li G. et al. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res. 2014; 42:7305–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Freibaum B.D., Lu Y., Lopez-Gonzalez R., Kim N.C., Almeida S., Lee K.H., Badders N., Valentine M., Miller B.L., Wong P.C. et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015; 525:129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar R., Corbett M.A., van Bon B.W., Woenig J.A., Weir L., Douglas E., Friend K.L., Gardner A., Shaw M., Jolly L.A. et al. THOC2 mutations implicate mRNA-export pathway in X-linked intellectual disability. Am. J. Hum. Genet. 2015; 97:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Viphakone N., Hautbergue G.M., Walsh M., Chang C.T., Holland A., Folco E.G., Reed R., Wilson S.A.. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012; 3:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hung M.L., Hautbergue G.M., Snijders A.P., Dickman M.J., Wilson S.A.. Arginine methylation of REF/ALY promotes efficient handover of mRNA to TAP/NXF1. Nucleic Acids Res. 2010; 38:3351–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Golovanov A.P., Hautbergue G.M., Tintaru A.M., Lian L.Y., Wilson S.A.. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA. 2006; 12:1933–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gromadzka A.M., Steckelberg A.L., Singh K.K., Hofmann K., Gehring N.H.. A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 2016; 44:2348–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi M., Zhang H., Wang L., Zhu C., Sheng K., Du Y., Wang K., Dias A., Chen S., Whitman M. et al. Premature termination codons are recognized in the nucleus in A reading-frame dependent manner. Cell Discov. 2015; 1:15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP–transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J. Vis. Exp. 2011; 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rossbach O., Hung L.H., Khrameeva E., Schreiner S., Konig J., Curk T., Zupan B., Ule J., Gelfand M.S., Bindereif A.. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol. 2014; 11:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zarnack K., Konig J., Tajnik M., Martincorena I., Eustermann S., Stevant I., Reyes A., Anders S., Luscombe N.M., Ule J.. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013; 152:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hwang H.W., Wentzel E.A., Mendell J.T.. A hexanucleotide element directs microRNA nuclear import. Science. 2007; 315:97–100. [DOI] [PubMed] [Google Scholar]
- 40. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao C., Choi E.A., Weng L., Xie X., Wan J., Xing Y., Moresco J.J., Tu P.G., Yates J.R. 3rd, Shi Y.. Overlapping and distinct functions of CstF64 and CstF64tau in mammalian mRNA 3′ processing. RNA. 2013; 19:1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee E.S., Akef A., Mahadevan K., Palazzo A.F.. The consensus 5′ splice site motif inhibits mRNA nuclear export. PLoS One. 2015; 10:e0122743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Flaherty S.M., Fortes P., Izaurralde E., Mattaj I.W., Gilmartin G.M.. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akef A., Lee E.S., Palazzo A.F.. Splicing promotes the nuclear export of beta-globin mRNA by overcoming nuclear retention elements. RNA. 2015; 21:1908–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gruter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B.K., Izaurralde E.. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998; 1:649–659. [DOI] [PubMed] [Google Scholar]
- 46. Braun I.C., Rohrbach E., Schmitt C., Izaurralde E.. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999; 18:1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lindtner S., Zolotukhin A.S., Uranishi H., Bear J., Kulkarni V., Smulevitch S., Samiotaki M., Panayotou G., Felber B.K., Pavlakis G.N.. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 2006; 281:36915–36928. [DOI] [PubMed] [Google Scholar]
- 48. Palazzo A.F., Springer M., Shibata Y., Lee C.S., Dias A.P., Rapoport T.A.. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007; 5:e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lei H., Dias A.P., Reed R.. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:17985–17990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lei H., Zhai B., Yin S., Gygi S., Reed R.. Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export. Nucleic Acids Res. 2013; 41:2517–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.