Abstract
The CST (CTC1–STN1–TEN1) complex mediates critical functions in maintaining telomere DNA and overcoming genome-wide replication stress. A conserved biochemical function of the CST complex is its primase-Pol α (PP) stimulatory activity. In this report, we demonstrate the ability of purified human STN1 alone to promote PP activity in vitro. We show that this regulation is mediated primarily by the N-terminal OB fold of STN1, but does not require the DNA-binding activity of this domain. Rather, we observed a strong correlation between the PP-stimulatory activity of STN1 variants and their abilities to bind POLA2. Remarkably, the main binding target of STN1 in POLA2 is the latter's central OB fold domain. In the substrate-free structure of PP, this domain is positioned so as to block nucleic acid entry to the Pol α active site. Thus the STN1–POLA2 interaction may promote the necessary conformational change for nucleic acid delivery to Pol α and subsequent DNA synthesis. A disease-causing mutation in human STN1 engenders a selective defect in POLA2-binding and PP stimulation, indicating that these activities are critical for the in vivo function of STN1. Our findings have implications for the molecular mechanisms of PP, STN1 and STN1-related molecular pathology.
INTRODUCTION
Telomeres, the nucleoprotein assembly at the ends of eukaryotic chromosomes, are critical for maintaining genome stability; they protect the chromosome ends from degradation, end-to-end fusion and other abnormal reactions (1–3). In most organisms, telomeric DNA consists of short repeats that are rich in G and C residues on the 3′ and 5′-end bearing strands, which are accordingly designated as the G- and C-strand, respectively. Owing to the end replication problem (4), the maintenance of telomere DNA through rounds of cell division requires not only semi-conservative DNA replication, but also extension of the terminal repeat tracts. The extension of the G- and C-strands of telomere tracts are executed sequentially by the telomerase and the primase-Pol α (PP) complex; telomerase lengthens the G-strand through reverse transcription of an integral RNA template subunit (5–8), whereas PP ‘fills in’ the C-strand by copying the extended G-tail (9–11).
A major regulator of telomere DNA synthesis is the CST (CTC1–STN1–TEN1) complex, a conserved G strand-binding complex that modulates the activities of both telomerase and PP (12–14). CST is notable for its structural resemblance to replication protein A (RPA) (15,16), a genome-wide replication and repair complex that coats single stranded DNA promiscuously. However, unlike RPA, CST has more dedicated functions at telomeres. In particular, a deficiency in CST subunits causes prominent defects in both telomere replication and telomere C-strand fill-in synthesis (17–22). Depletion of CST also impaired cellular recovery from replication stress, suggesting an additional, non-telomeric function (18). Indeed, a recent report suggests that CST promotes recovery from replication stress, especially at GC-rich loci, by recruiting RAD51 to stabilize stalled forks and promote fork restart (23). The physiologic importance of CST is underscored by both murine models of CST deficiency and human diseases. For example, CTC1-null mice manifest global cellular proliferative defects and die prematurely from complete bone marrow failure (17). In addition, mutations in CTC1 and STN1 have been shown to cause Coats plus syndrome, which is characterized by neurologic defects and telomeropathy phenotypes such as bone marrow failure and liver fibrosis (24,25).
Biochemical and structural analyses of the CST complex have revealed multiple activities that presumably underpin its cellular functions. These include binding to single-stranded telomeric and non-telomeric DNA (26–28), stimulation of PP (29,30), inhibition of telomerase (31) and interaction with telomeric proteins (31,32). The molecular bases of these activities and their relationships to the cellular functions of CST remain incompletely understood. In this regard, it is worth noting that a recent characterization of a STN1 mutant with reduced DNA-binding activity revealed selective defects in telomere duplex replication and resolution of replication stress, but not other functions of CST, providing an illustration of the intricate relationship between the biochemical activities of CST and its cellular functions (27). Among the unresolved issues, the molecular mechanism by which CST stimulates PP is of particular interest. This stimulatory activity is likely to account for the ability of CST to promote C-strand synthesis and telomere replication. Yet the physical interactions between PP and CST subunits have not been delineated, and whether the DNA binding activity of CST plays a role in this stimulation is also unclear.
Studies of CST mechanisms have been hampered by difficulties in isolating adequate quantities of the complex for detailed biochemical investigations. We recently overcame this obstacle by obtaining high levels of CST from Candida glabrata (26). Using fungal proteins, we demonstrated that CST stimulates DNA synthesis by enhancing both the RNA priming and primase-to-polymerase switch steps of the PP reaction (30). In addition, we found that the stimulatory activity is mediated primarily by the Stn1 subunit, and that this subunit makes multiple, functionally important contacts with the Pol12 subunit of PP. Pol12, one of the four subunits of PP, is often referred to as the regulatory subunit of Pol α, and is named POLA2 or p70 in mammals. In the current study, we extend our analysis to the equivalent human proteins and showed that similar to fungi, human STN1 alone can enhance the activity of human PP. We mapped the stimulatory activity of human STN1 to its N-terminal OB fold domain, but showed that the DNA-binding activity of this domain is not required. Rather, we observed a strong correlation between the PP-stimulatory activity of STN1 variants and their abilities to bind POLA2 (the Pol12 ortholog in mammals). Interestingly, the target of STN1-binding in POLA2 is another OB fold domain, underscoring the versatility of this motif in mediating protein–protein interactions. A disease-causing mutation in human STN1 engenders a selective defect in POLA2-binding and PP stimulation, suggesting that these activities are critical for the physiologic function of STN1. Our findings establish a conserved mechanism for PP stimulation by STN1, and reveal an unexpected similarity between the regulation of telomere G- and C-strand extension.
MATERIALS AND METHODS
Preparations of proteins
The PP complex was obtained by mixing separately purified two-subunit primase and two-subunit polymerase. The two-subunit polymerase was expressed and purified from High Five insect cells by a combination of Q-Sepharose, FLAG affinity chromatography and glycerol gradient (33). The two-subunit primase was produced in Escherichia coli using the pET-Hp48-HisHp58 expression construct and purified via Ni-NTA chromatography and glycerol gradient (34).
STN1F (amino acid 1–368), STN1N (1–190) and STN1C (191–368) with C-terminal FLAG or Strep tag were generated by polymerase chain reaction (PCR) and cloned downstream of the T7 promoter and the His6-SUMO tag in the pSMT3 vector. Each protein was expressed in BL21-CodonPlus E. coli and purified by Ni-NTA chromatography as previously described (30,35). In some cases, the proteins were further processed by ULP1 cleavage (to remove the His-SUMO tag), and finally FLAG or Strep-Tactin affinity chromatography. The FLAG purification step was performed as before (30). For Strep-Tactin chromatography, the ULP1 treated fraction was supplemented with 1/20 vol. of 1M Tris–HCl, pH 8.0 and then applied to a column pre-equilibrated in buffer W (100 mM Tris, pH 8.0, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol). After washing with five column volumes of buffer W, the Strep-tagged protein was eluted with buffer W containing 7 mM desthiobiotin. Peak protein fractions were pooled and used directly in various biochemical assays. The concentrations of the purified proteins were all estimated by comparing their Coomassie staining intensities to a standard curve of specified quantities of bovine serum albumin (BSA). The RPA complex (gift of the Hurwitz lab) was purified as previously described (36). RPA2-Strep was isolated using exactly the same protocol as that for STN1-Strep. To generate the STN1-Strep/GST-TEN1 complex, the TEN1 open reading frame was cloned into pGEX-6P1 and the resulting construct co-transformed with pSMT3-STN1-Strep into BL21-CodonPlus E. coli. Following protein induction and extract preparation, the STN1/TEN1 complex was purified by Ni-NTA chromatography, ULP1 cleavage and glutathione chromatography.
To express FLAG3-tagged POLA2 variants, we first modified the pSMT3 vector by inserting a FLAG3 tag between the NotI and XhoI site of pSMT3 to give pSMT3-FLAG3. Full length POLA2 or truncation variants (including 1–334, 1–414, NTD (1–154) and OB (204–334)) were then generated by PCR and cloned in between the BamHI and NotI site of pMST3-FLAG3. As in the case of STN1 fusion proteins, the His-SUMO-POLA2-FG3 fusion proteins were expressed in BL21-CodonPlus E. coli and purified by Ni-NTA. Following ULP1 cleavage, the samples containing a mixture of His-SUMO and POLA2-FG3 proteins were used directly as inputs for pull down analysis.
Primase-Pol α activity assays and product analysis
Unless otherwise indicated, the coupled primase-polymerase assays were performed in 20 μl volume containing 40 mM Tris–HCl, pH 7.6, 30 mM potassium acetate, 13 mM Mg acetate, 5 mM dithiothreitol, 0.05 mM EDTA, 5% glycerol, 0.1 mg ml−1 BSA, ∼1–2 nM PP, variable concentrations of STN1, 0.15 μM poly-dT (∼300 nt long, from Midland Certified Reagent Company, Inc.), 2 mM adenosine triphosphate (ATP) and 7.5 μM dATP (containing 5 μCi/nmole P32-dATP). After incubation at 32°C for 60 min, reactions were treated with 100 μl STOP solution (20 mM Tris–HCl, pH 8.0, 20 mM EDTA) and 100 μl proteinase K solution (10 mM Tris–HCl, pH 8.0, 0.5% sodium dodecyl sulphate, 0.15 mg/ml proteinase K), and incubated at the same temperature for 30 min. The products from the assays were recovered by ethanol precipitation in the presence of 2.5 M ammonium acetate, 15 μg/ml glycogen and 15 μg/ml tRNA. The samples were dissolved in 90% formamide, 20 mM EDTA, 1 mg/ml xylene cyanol and 1 mg/ml bromophenol blue, boiled for 5 min and then applied to a 13% acrylamide/7M urea/1X Tris/Borate/EDTA (TBE) gel. The gels were dried and exposed to a Phosphor screen, and the signals analyzed using the ImageQuant software. Only discreet RNA-DNA chimeras in the 10–30 nt size range were included in the quantification of activities. One unit of activity is defined as that generated by PP alone in the standard assays. Statistical significance of the difference in activities between wild-type STN1N and mutants was determined using two-tailed t-test (GraphPad Prism).
To test the effect of STN1 on primase activity only, the primase reaction was performed with or without STN1 using 2 mM unlabeled ATP as the only nucleotide. The reactions were extracted with phenol/chloroform/isoamyl alcohol, and the DNA template/RNA primer mix were then recovered and subjected to extension using just the two subunit Pol α (i.e. POLA1 and POLA2) in the presence of P32-dATP. To test the effect of STN1 on just the DNA polymerase activity, we used poly-dT/rA10 (150 nM poly-dT and 450 nM rA10 (purchased from IDT)) as the pre-primed substrate and labeled dATP as the sole nucleotide.
Analysis of STN1–POLA2 interaction
Purified STN1-Strep proteins and POLA2-FG3 proteins were used for interaction analysis using Strep-Tactin beads. Wild-type full length STN1, STN1C, STN1N as well as STN1 mutants were individually bound to equilibrated Strep-Tactin beads (300 μl of 0.1 μg/μl protein per 30 μl beads) at 4°C for 2 h. The samples were spun down at 4000 rpm for 30 sec and the supernatant was discarded. The beads were further washed 3–4 times with Buffer W (100 mM Tris–HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% Glycerol). A total of 180 μg of POLA2 fragments were next added to STN1-bound Strep-Tactin beads at 0.3 μg/μl, and the NaCl concentration of the mix adjusted to 160 mM. The binding was allowed to proceed at 4°C for 2 h, followed by washes using Buffer W containing 160 mM NaCl and supplemented with 0.1% NP-40 and 2.5 mM MgCl2. The proteins were eluted in 3× bed volume buffer W containing 5 mM d-Desthiobiotin. The samples were analyzed by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blot to detect POLA2 (anti-FG antibody) and STN1 (anti-Strep antibody). Relative binding was expressed as the ratio of POLA2 to STN1 signals in the elution samples (normalized to the value for wild-type STN1N). Statistical significance of the difference in relative binding was determined using two-tailed t-test (GraphPad Prism).
STN1-DNA crosslinking assays
Crosslinking reactions (26) were conducted in 13 μl mixtures that include 50 mM Hepes-NaOH, pH 7.5, 10% glycerol, 50 mM NaCl, 2 mM Mg-acetate, 1 mM DTT, 2 nM 5′ P32-labeled oligonucleotide containing a single Iodo-dU substitution (∼50 000 c.p.m.), and indicated concentrations of STN1. The binding was allowed to proceed at 22°C for 10 min, followed by 20 min of UV irradiation (Model UVM-57, UVP Inc.) on ice. The covalent conjugates were separated in SDS-PAGE (10%) and detected by PhosphorImager analysis.
RESULTS
Human STN1 alone stimulates the synthesis of RNA-DNA chimeras by human primase-Pol α (PP)
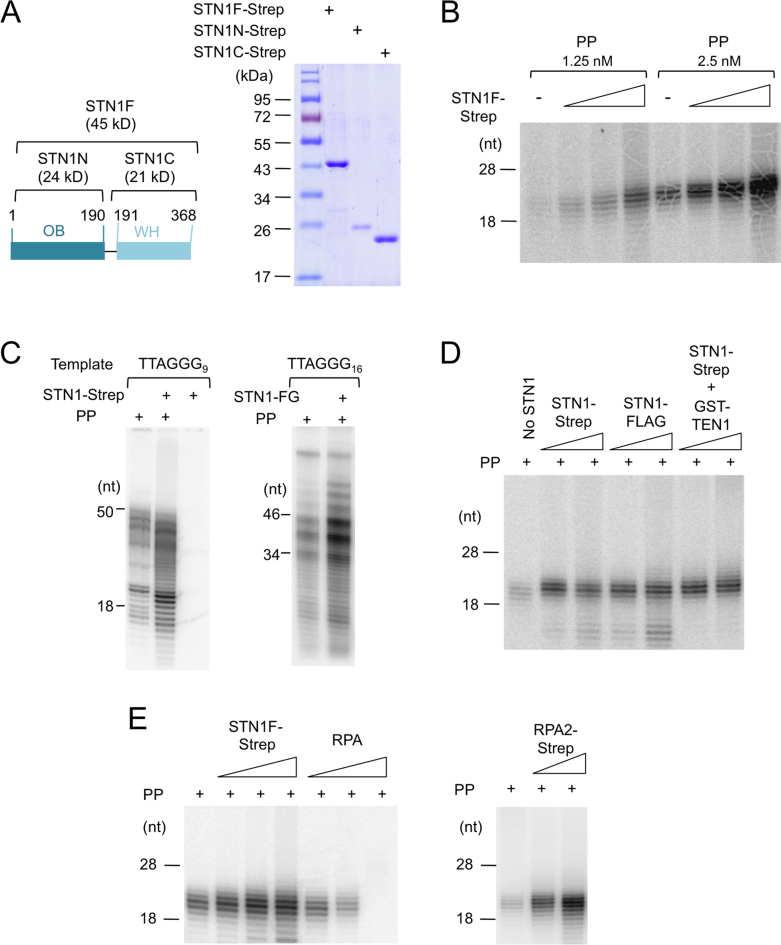
Because C. glabrata Stn1 alone was sufficient to stimulate the cognate PP activity in vitro, we tested human STN1 for a comparable activity, and found that it, too, can enhance the synthesis of RNA–DNA chimera by human PP (Figure 1A and B). The magnitude of stimulation is positively correlated with STN1 concentration, and the stimulation can be observed on both the poly-dT and telomere G-strand templates (Figure 1B and C). Notably, in the previous C. glabrata study, we utilized a His-SUMO-Stn1 fusion protein (30). In the current work, we were able to remove the His-SUMO tag by ULP1 cleavage, and further purify human STN1 through either a C-terminal FLAG or C-terminal Strep-tag II tag. The different variants of STN1 fusion protein behave similarly in the PP stimulation assays, indicating that the stimulatory activity was not dependent on the tags (Figure 1D).
Figure 1.
Human STN1 stimulates the synthesis of RNA–DNA chimera by Primase-Pol α; the effects of STN1, STN1–TEN1 and RPA on PP activity (A) Human STN1s (FL, N and C) purified by two-step affinity chromatography (Ni-NTA and Strep-Tactin) were analyzed by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). (B) Human PP (1.25 and 2.5 nM) was tested in the coupled primase-polymerase assay using poly-dT as template in the presence of 0.17, 0.5 and 1.5 μM of full length STN1-Strep (STN1F-Strep). (C) Human PP (2.5 nM) was assayed for the synthesis of RNA–DNA chimera using the indicated G-strand templates in the absence or presence of Strep-tagged or FLAG-tagged STN1 (1 μM). (D) Human PP (1 nM) was assayed for the synthesis of RNA–DNA chimera using Poly-dT template in the presence of two different concentrations (0.5 and 1.5 μM) of STN1-Strep, STN1-FLAG or STN1-Strep/GST-TEN1 complex. (E) Human PP (2 nM) was assayed for the synthesis of RNA–DNA chimera using Poly-dT template in the presence of increasing concentrations of STN1-Strep (0.16, 0.5 and 1.5 μM) or RPA (0.053, 0.16 and 0.5 μM). In the right panel, 0.5 and 1.5 μM RPA2-Strep was added to the PP reaction.
Previous analysis of C. glabrata CST indicates that while the complex can significantly enhance the synthesis of RNA–DNA chimera in a coupled primase-polymerase assay, it has only a modest effect on the isolated primase activity and no effect on the uncoupled, pre-primed DNA polymerase activity (30). We interpret the results to mean that the complex primarily affects the primase-to-polymerase switch. Notably, all of these earlier assays were performed using the four-subunit C. glabrata PP holoenzyme. Because in the current study, we purified the two-subunit human primase and two-subunit human polymerase separately, we are able to analyze the effects of STN1 on the isolated sub-assemblies. Similar to the findings in C. glabrata, we observed only small effects of STN1 on these isolated enzyme activities (i.e. ∼30% increase in primase activity and no increase in DNA polymerase activity), suggesting that STN1 affects primarily the coupling between primase and polymerase (Supplementary Figure S1A and B).
STN1 is part of the CST complex in mammalian cells. We sought to compare the stimulatory activity of STN1 to that of the full CST complex, but were unable to express and purify adequate amounts of the latter for this analysis. However, we were successful at preparing high levels of STN1–TEN1, and found that this subcomplex stimulated PP to the same extent as STN1 alone (Figure 1D). Thus, TEN1 evidently does not affect the stimulatory activity of STN1 on PP in vitro.
CST exhibits structural similarity to RPA, especially in regard to the STN1–TEN1 and RPA2–RPA3 subcomplex. RPA has also been reported to modulate Pol α activity in a variety of assays. Interestingly, we found that in our standard assay, while RPA inhibited the synthesis of RNA–DNA chimera, RPA2 alone has a stimulatory effect (Figure 1E). RPA2 may thus have a STN1-like stimulatory activity that is normally masked by the rest of the RPA complex.
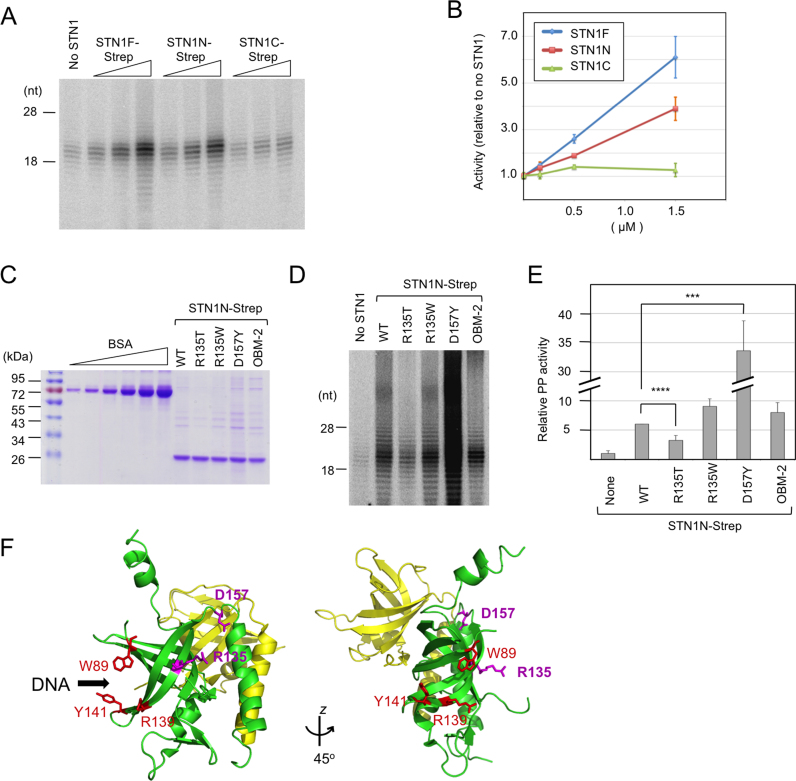
The PP-stimulatory activity of STN1 resides mostly in its N-terminal OB fold domain, and is impaired by selective point mutations in this domain
Human STN1, like most other STN1 family members, consists of an N-terminal OB fold domain and a C-terminal winged helix domain. We purified and tested each domain (named STN1N and STN1C) in PP stimulation assays, and detected a strong activity in the N-terminal domain only. Like the effects of full length protein on PP, those of the N-terminus OB fold are concentration dependent (Figure 2A and B), and can be observed on both poly-dT and G-strand templates (data not shown). Because we have demonstrated earlier that full length human STN1 can stimulate the C. glabrata PP complex (30), we also tested the N and C-terminus of human STN1 in this ‘heterologous’ assay (Supplementary Figure S2). Consistent with findings in the ‘purely human’ assay system, the N-terminus exhibited a weak stimulatory activity, whereas the C-terminus alone is largely inactive. Thus, the OB fold of STN1 is likely to enhance PP activity by employing a conserved structural feature.
Figure 2.
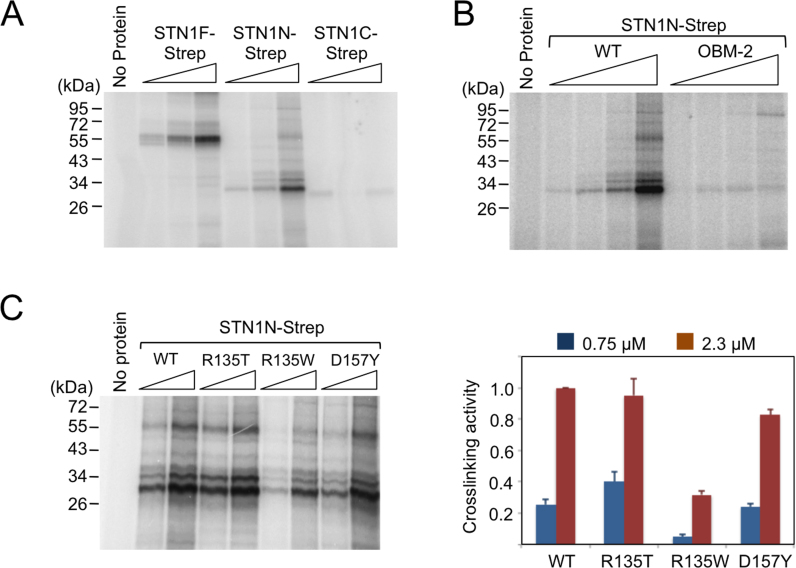
PP stimulation by STN1 variants. (A) Human PP (2 nM) was assayed using the poly-dT template in the presence of increasing concentrations (0.17, 0.5 and 1.5 μM) of STN1F, STN1N and STN1C. (B) Total signal for each sample in the 10–30 nt product size range as determined by assays shown in A were quantified from PhosphorImger scans, normalized against the signal for the ‘no STN1’ sample and plotted. Data (averages ± S.D.) are from three independent experiments. (C) Purified STN1N and the R135T, R135W, D157Y and OBM mutants were analyzed on 10% SDS-PAGE along with bovine serum albumin (BSA) standards (0.25, 0.5, 1.0, 2.0, 4.0 and 8.0 μg). (D) Human PP (1 nM) was assayed using the poly-dT template in the presence of wild-type or mutant STN1N (1.5 μM) as indicated. (E) The assays in D were repeated three times. The signals in each series of assays were normalized against the ‘STN1N WT’ sample and plotted (average ± S.E.M.). One unit was defined as the average activity of the ‘no STN1’ sample. Note that the bar for D157Y is compressed so that the differences between other STN1N variants are better visualized. P-values as determined by two-tailed t-test are designated by asterisks as follows: ***: 0.05 > P > 0.01; ****: 0.01 > P. (F) Using the crystal structure of STN1N–TEN1 complex as template (PDB ID: 4JOI), the STN1 residues mutated and analyzed in this study were mapped onto the STN1N OB fold using Pymol. Residues altered in the OBM-2 mutant (shown in red) fall on the presumed DNA binding surface of STN1 based on analogy with RPA2. In contrast, the residues mutated in Coats plus (R135 and D157Y) fall on a different face of STN1.
Several point mutations in the OB fold of STN1 have been previously reported to affect its cellular function or biochemical activities. Specifically, the R135T and D157Y mutations are found in Coats plus patients, and a W89A/R139L/Y141A triple mutation (OBM) impairs the DNA-binding activity of the CST complex (25,27). We therefore purified STN1N-Strep bearing the disease mutations and a slight variant of the OBM mutations (W89A/R139A/Y141A, named OBM-2), and analyzed their ability to stimulate PP in vitro (Figure 2C and D). For comparative purposes, we also tested a R135W mutant in which the basic R135 was substituted by the aromatic Trp rather than the hydrophilic Thr. Interestingly, the R135T mutation reduced the stimulatory activity by 2-fold; the R135W and OBM mutations had no effect, whereas D157Y enhanced the stimulatory activity (Figure 2D and E). The different effects of mutations, which map to different surfaces of STN1 (Figure 2F), suggest that PP stimulation may require an interaction mediated by just one face of the protein.
The OB fold domain of STN1 binds directly to the POLA2, the regulatory subunit of human Pol α
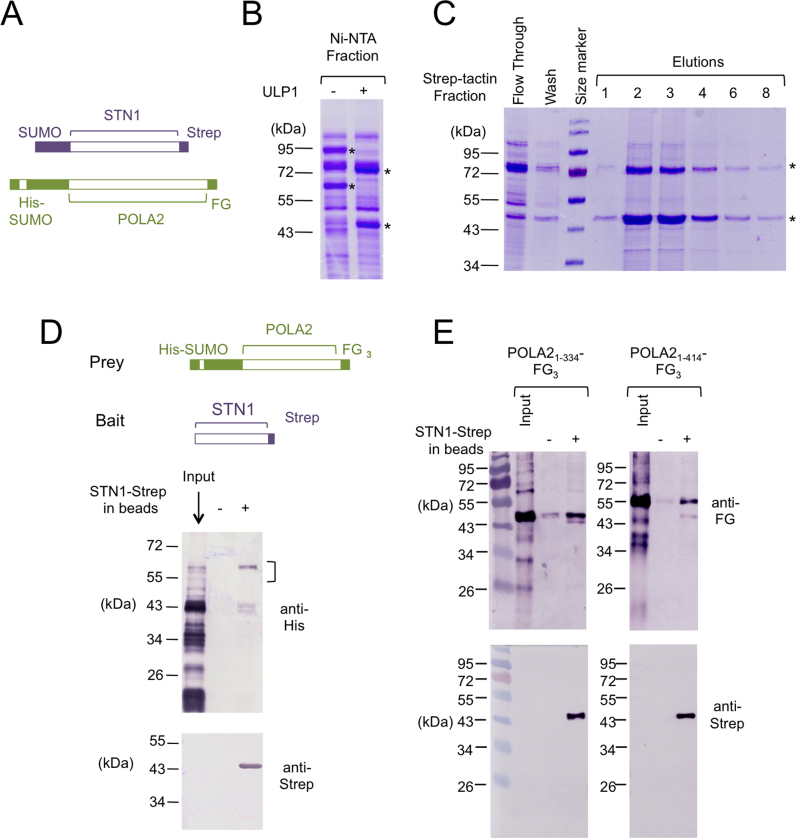
In our previous analysis of C. glabrata Stn1, we showed that this protein forms a stable complex with the Pol12 subunit of PP. To determine if this is true for the human homologs, we first co-expressed SUMO-STN1-Strep and His-SUMO-POLA2-FLAG fusion proteins in E. coli, and then used sequential affinity chromatography to assess complex formation. Notably, following Ni-NTA purification and ULP1 cleavage, we observed co-elution of POLA2-FLAG with STN1-Strep in the subsequent Strep-Tactin chromatography step, indicating the two proteins bind to each other (Figure 3A–C).
Figure 3.
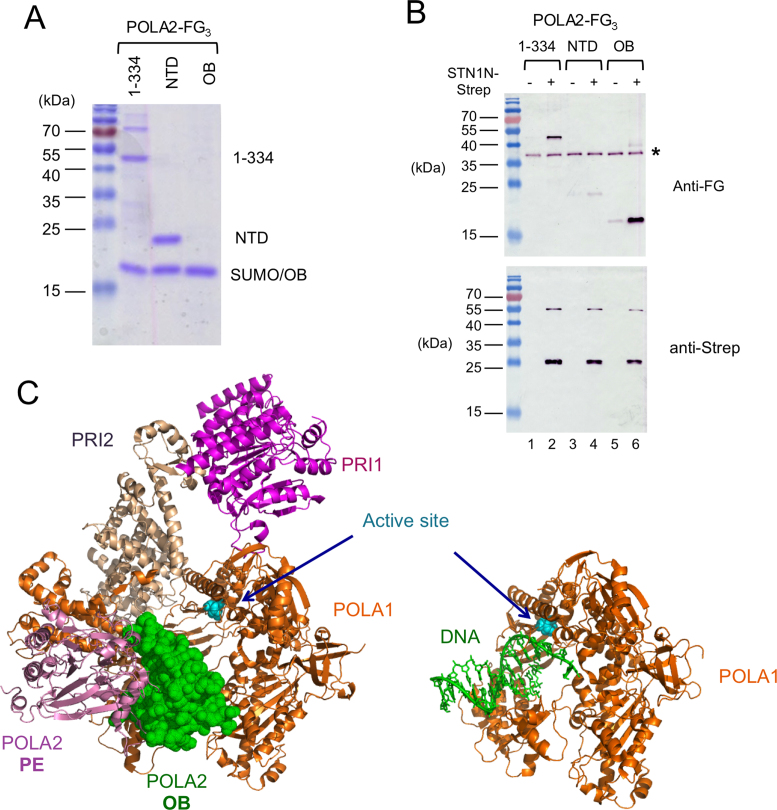
Co-purification of STN1 and POLA2 following co-expression and affinity chromatography; Mapping of the region of POLA2 that binds to STN1 (A) The locations and identities of the tags in the co-expressed fusion proteins are schematically illustrated. (B) The Ni-NTA elution fraction containing SUMO-tagged STN1 and POLA2 fusion proteins were digested with ULP1 to remove the SUMO tag. The fractions before and after ULP1 treatment were analyzed by SDS-PAGE and Coomassie staining. (C) The ULP1 treated Ni-NTA fraction was applied to a Strep-Tactin column. The fractions obtained at different chromatography steps as indicated were analyzed by SDS-PAGE and Coomassie staining. (D) The bait and prey proteins used for the pull down assays are schematically illustrated at the top, and the results shown at the bottom. Note that even though the prey expressed in Escherichia coli was a His-SUMO fusion protein containing full length POLA2, the Ni-NTA fraction used for the binding assay (input) contained mostly proteolyzed POLA2. Only a large fragment of ∼58 kD (marked by a vertical bracket) was efficiently pulled down by STN1. (E) The binding of STN1-Strep to two N-terminal fragments of POLA2 (1–334 and 1–414) was analyzed.
To further analyze the interaction between STN1 and POLA2, we sought to develop an in vitro pull down assay using purified proteins. However, while STN1 can be expressed and purified as an individual polypeptide, full length POLA2 is severely proteolyzed when expressed in the absence of STN1. Nevertheless, using the pool of truncated POLA2 in pull down assays, we were able to detect specific binding of STN1-Strep to N-terminal POLA2 fragments that are more than ∼320 amino acid long (Figure 3D). (Note that the minimal His-SUMO-tagged POLA2 protein that can bind STN1-Strep is about 58 kDa, implying that the minimal POLA2 fragment is about 42 kDa, given the apparent His-SUMO mobility in SDS-PAGE of 16 kDa). The absence of shorter fragments in the pull down samples suggests that the OB fold domain of POLA2 (spanning amino acids 210–320) is required for binding STN1 (see (37) for the domain organization of POLA2). Based on these initial assays, we experimented with several truncated variants of POLA2, and identified two variants (i.e. 1–334 and 1–414) that can be expressed in intact forms as His-SUMO-POLA2-FG3 fusion proteins. We then partially purified these deletion mutants by Ni-NTA chromatography, and subjected them to ULP1 cleavage followed by pull down using STN1-Strep as the bait. Notably, the 1–334 and 1–414 fragments of POLA2 can each be specifically retained by and eluted from STN1-Strep-coated resin but not control resin, indicating that they bind specifically to STN1 (Figure 3E). Likewise, STN1-Strep can be specifically retained on FLAG beads coated with POLA21–334-FG3 (Supplementary Figure S3A). Because both truncated forms of POLA2 lack the C-terminal PDE (phosphoesterase) domain, this domain is evidently not required for STN1 interaction.
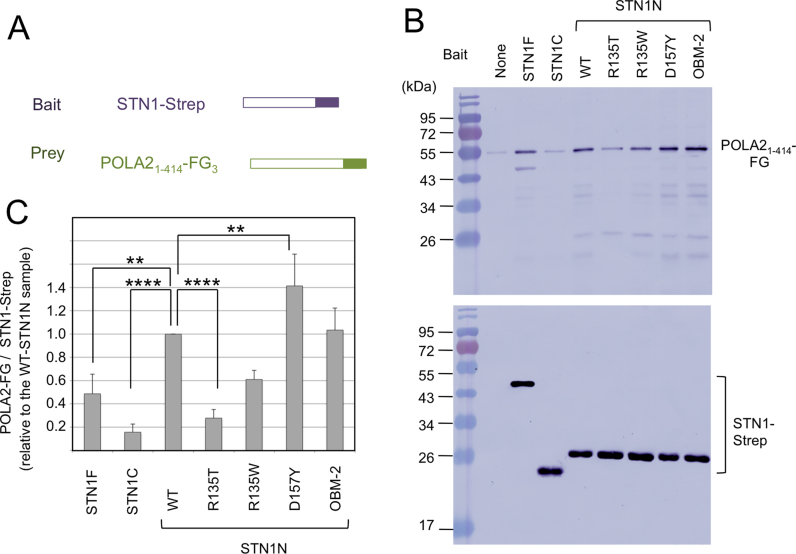
The PP stimulatory activities of STN1 variants correlate with their POLA2-binding, but not DNA-binding activities
Next we tested the binding of POLA21–414 to individual domains of STN1. Interestingly, the binding of POLA21–414 to the OB fold domain is consistently stronger than that to the full length protein, whereas the binding to the winged helix domain is just slightly above background (Figure 4A–C). Thus, both the PP stimulatory and POLA2-binding activities of STN1 appear to reside primarily in the N-terminal OB fold structure. Moreover, the effects of OB fold point mutations on its binding to POLA21–414 are quite similar to those on PP stimulation, with the R135T mutant showing a significant reduction in binding and the D157Y mutant showing an increase in binding (Figure 4B and C). These results suggest that the PP stimulatory activity of STN1 is likely to be mediated at least in part through STN1–POLA2 interaction. In support of this idea, we found that both STN1–TEN1 and RPA2, which we showed to be competent in PP stimulation, were also able to bind specifically to POLA2 (Figure 1D and E; Supplementary Figure S3B).
Figure 4.
POLA2-binding by STN1 variants. (A) A schematic depiction of the tagged STN1 and POLA2 proteins used for the pull down assays is shown. (B) Purified, Strep-tagged STN1F, STN1N, STN1C and STN1N mutants were bound to Strep-Tactin beads. Purified POLA21–414-FG3 was used as the prey in the Strep-Tactin pull down/elution experiments. The elutions were analyzed by western blot to assess the binding of POLA2 to STN1 variants and mutants. The level of POLA2 was measured by probing the blot with anti-FG antibody, while that of STN1 by anti-Strep antibody. (C) The ratio of POLA2 to STN1 in the elution samples was calculated, normalized against the value for wild-type STN1N and plotted. Data (averages ± S.E.M) are from three or more independent experiments. P-values as determined by two-tailed t-test are designated by asterisks as follows: **: 0.1 > P > 0.05; ***: 0.05 > P > 0.01; ****: 0.01 > P.
Another important function of the OB fold of STN1 is binding to ssDNA. To examine the potential relationship between DNA-binding and PP stimulation, we analyzed the DNA-binding activities of STN1 variants using a photocrosslinking assay. As predicted, we found that both full length STN1 and the OB fold domain (STN1N) can be efficiently crosslinked to a single stranded oligonucleotide bearing an Iodo-dU residue following UV irradiation (Figure 5A). In contrast, little crosslinking can be detected for the winged helix domain (STN1C). Consistent with previous findings, we found that the OBM mutations, which map to the putative DNA-binding surface of STN1, drastically reduced the crosslinking efficiency of this domain (Figure 5B). In contrast, the disease related mutations either had no effect (R135T and D157Y), or moderately reduced DNA-binding (R135W) (Figure 5C). Importantly, the effects of the mutations on DNA-binding do not parallel the effects on PP stimulation or POLA2 binding, indicating that these activities of STN1 are separable. This conclusion is also consistent with the physical locations of the disease and DNA-binding mutations, which map to different faces of STN1 (Figure 2F) (38).
Figure 5.
STN1-DNA crosslinking. (A) Increasing concentrations of STN1F, STN1N and STN1C (0.25, 0.75, 2.3 μM) were mixed with 2 nM 5′-P32-labeled oligonucleotide containing a single Iodo-dU substitution, irradiated with UV and then analyzed by SDS-PAGE and PhosphorImager scanning. (B) Increasing concentrations (0.083, 0.25, 0.75 and 2.3 μM) of wild-type STN1N and the OBM mutant were subjected to the same DNA crosslinking assay. (C) Two different concentrations of wild-type STN1N and the indicated mutants (0.75 and 2.3 μM) were subjected to the same DNA crosslinking assay. A representative set of assays is shown on the left, and the data (averages ± S.D.) from three independent experiments are plotted on the right. One unit of activity is defined as that generated by 2.3 μM of the wild-type STN1N.
An OB–OB interface underlies the physical interaction between STN1 and POLA2
To define the minimal domain in POLA2 capable of binding STN1 OB fold, we further expressed POLA2NTD (amino acids 1–154) and POLA2OB (amino acids 204–334) based on available structural information, and tested these fragments for binding to STN1N. Consistent with earlier results (see Figure 3D), we found that POLA2NTD is inactive in the pull down assay (Figure 6A and B). In contrast, POLA2OB exhibited slightly greater binding to STN1 than the larger 1–334 fragment (Figure 6A and B). Thus, the interaction between STN1 and POLA2 is mainly mediated by the two OB fold domains within the respective proteins.
Figure 6.
OB–OB interaction between STN1 and POLA2, and the steric clash between DNA and POLA2 OB fold domain near the Pol α active site. (A) Truncated POLA2 variants were expressed as His-SUMO-POLA2-FG3 fusions, purified by Ni-NTA and then subjected to ULP1 cleavage. The resulting preparations that were used as inputs for STN1N-Strep pull down assays were subjected to SDS-PAGE and Coomassie staining. About 1/30 of the input samples were analyzed. Note that the mobility of OB domain is identical to the SUMO fragment. (B) POLA2 fragments were subjected to pull down by STN1N, and the samples eluted from Strep-Tactin beads were subjected to western blot analysis using both anti-FLAG and anti-Strep antibodies. A contaminant that cross-reacts with the FLAG antibody is indicated by *. (C) The crystal structure of the human primase-pol α complex without substrates (PDB ID: 5EXR) is shown on the left and that of a polymerase–DNA complex (PDB ID: 5IUD) shown on the right. The different polypeptides and the DNA are displayed in different colors as matched by the text labels. The active site aspartates of the DNA polymerase are shown in cyan and in sphere representations. The POLA1 polypeptide in the two structures are displayed in the same orientation to illustrate the point that the OB fold of POLA2 in the apo complex occupies the same position as the DNA substrate in the polymerization complex.
DISCUSSION
In the current study, we generalized our previous conclusions regarding the mechanisms of PP stimulation by CST from the fungal proteins to the human homologs. We showed that like its fungal ortholog, the isolated human STN1 subunit alone is sufficient for stimulation, and this stimulation is due primarily to an enhancement of the primase-to-polymerase switch reaction. In addition, like fungal Stn1, the stimulatory activity of human STN1 maps mostly to its N-terminal OB fold domain. Moreover, this stimulation is unrelated to STN1’s DNA-binding activity, and instead mediated through its binding to the OB fold of POLA2. The implications of these findings, as well as their relationship to previous observations, are discussed below.
The initial biochemical characterization of CST purified directly from mouse cells revealed complex effects of the native complex on priming and polymerization (29,39). Following the cloning of CST subunits, the activities of recombinant factors on PP were analyzed in two additional studies. In one study, co-expression of CTC1 and STN1 followed by affinity purification resulted in a preparation capable of stimulating RNA priming by the PP complex (28). Notably, this stimulatory activity was reported to depend on both CTC1 and STN1. In a second study, recombinant CST was shown to stimulate DNA synthesis in Xenopus extract on unprimed ssDNA template, but not on primed template (40). Our results are generally in agreement with these two studies, and the few potential discrepancies can all be readily reconciled. For example, while our experiments highlight the ability of STN1 alone to stimulate PP, they do not rule out a role for CTC1. In the budding yeast, besides the Stn1–Pol12 interaction, another physical contact between CST and PP has been identified between Pol1 and Cdc13 (equivalent to CTC1) (41). It is possible that a similar interaction exists between the mammalian factors, allowing for more efficient stimulation of PP by CST. Consistent with this idea, we found that relatively high concentrations of STN1 (∼0.5–1.5 μM) are needed to observe an appreciable stimulatory effect. Second, in regard to the reaction step simulated by CST, while both our C. glabrata and human studies emphasize an effect of CST on the primase-to-polymerase switch ((30) and the current study), we have also shown a positive effect of the fungal CST on the priming step. Importantly, this positive effect was observed on the full PP complex, whereas the smaller effect of human STN1 on priming was observed on the primase subcomplex alone, which is known to adopt a different conformation in the absence of the two polymerase subunits (42–44).
The binding of STN1 OB fold to POLA2 is likely to be conserved in evolution. Even though a previous two-hybrid analysis failed to detect an interaction between Saccharomyces cerevisiae Pol12 and the N-terminus of Stn1 (45), such an interaction is supported by other studies (46). In addition, we have shown that In C. glabrata, the N- and C-terminus of Stn1 can each bind Pol12 and stimulate PP, with the N-terminus exhibiting greater activities in both assays (30). Thus, both the fungal and human studies highlight the importance of the STN1 OB fold in regulating PP. Moreover, our observation that RPA2 alone can stimulate PP activity in vitro and bind POLA2 suggest that the RPA complex may also be capable of regulating PP through this mechanism. However, the effects of RPA are likely to be complex, and more studies will be necessary to assess the physiologic relevance of RPA2–POLA2 interaction.
Our findings concerning the mechanisms of STN1 in PP stimulation may be interpreted in light of the recently proposed model of concerted RNA–DNA primer synthesis by the human PP complex (42). This model, based on the crystal structure of the full complex without substrates, as well as those of several subcomplexes with nucleic acids, envisions a switch reaction mediated by a dramatic rotation of PRI2 C-terminus relative to POLA1, which delivers the bound template-RNA primer to the polymerase active site. Importantly, in the substrate-free structure of the PP complex, the nucleic acid entry site for the DNA polymerase is blocked by the OB fold of POLA2 (Figure 6C) (42,47,48). Thus, in order for the template-RNA primer to be delivered by the PRI2 C-terminus, this OB fold must be moved away from the nucleic acid entry site. According to our interaction study, the primary target of STN1 is the OB fold of POLA2, providing a plausible explanation for how the STN1–POLA2 interaction may facilitate the required movement. While the molecular determinants of STN1–POLA2 binding remain largely undefined, the effects of the R135T mutation suggest that this residue, which maps away from the DNA-binding and TEN1-binding surface, may be part of the POLA2-binding surface (Figure 2F). A high resolution view of the STN1–POLA2 interface should help to advance understanding of the molecular basis of the primase-to-polymerase switch reaction.
Our results have implications for STN1 evolution and for STN1-associated telomeropathy. At the sequence level, STN1 is the most highly conserved subunit of the CST complex, and the N-terminal OB fold the more highly conserved domain of this protein. The attribution of the PP stimulatory, POLA2-binding, DNA-binding and TEN1-binding activities of STN1 all to this domain is thus not surprising and rationalizes the finding that the STN1 orthologs in Arabidopsis and Drosophila lack the C-terminal winged helix domain (49,50). Because the winged helix domain is present in the great majority of STN1 orthologs (51), it was likely to be part of the ancestral protein, and to be lost in selected lineages due to its lesser functional importance. In terms of direct disease relevance, it is worth noting that the initial set of telomeropathy patients bearing CST-related mutations all have compound heterozygous mutations in CTC1 (24,52), raising the possibility of a special role for this subunit. However, the recent discovery of two Coats plus patients with homozygous mutations in STN1 rules out a special status for CTC1 in triggering disease (25). Instead, the paucity of disease-associated STN1 mutations may be due to its greater functional importance, and the lethal consequences of most STN1 mutations. We have shown that R135T causes a clear defect in PP stimulation and POLA2-binding in vitro, providing good explanations for the associated cellular pathology such as G-strand accumulation and impaired recovery from replication stress (25). In contrast, the D157Y mutant is more active than wild-type STN1 in PP stimulation, and appears to be largely normal with respect to all the biochemical activities we tested, yet still causes the Coats plus syndrome. In this regard, we note that the D157 residue is partially buried in the interior of STN1, and the expression level of the D157Y mutant in E. coli was substantially lower than that of the wild-type protein (data not shown). This mutant may thus have a subtle conformational defect that impairs an aspect of STN1 function not detected by current assays. Alternatively, hyper-activation of PP may also cause cellular pathology.
Finally, it is worth noting that our study has revealed a surprising similarity between the regulatory mechanisms that control telomere G- and C-strand extension. In G-strand extension, there is compelling evidence that an OB fold domain in the shelterin component TPP1 is responsible both for recruiting telomerase to telomere ends and for stimulating the processivity of telomerase (53,54). Likewise in C-strand extension, we have now shown that the OB fold of STN1 in the CST complex plays a key role in controlling the activity of PP. Thus, telomere-tethered OB folds are central regulators of the telomere maintenance machinery.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Weihang Chai (Washington State University) for human STN1 cDNA, and Jerry Hurwitz (Memorial Sloan-Kettering Cancer Center) for purified human primase and Pol α.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM107287 to N.F.L.]. Funding for open access charge: NIH [GM107287].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jain D., Cooper J.P.. Telomeric strategies: means to an end. Annu. Rev. Genet. 2011; 44:243–269. [DOI] [PubMed] [Google Scholar]
- 2. de Lange T. How telomeres solve the end-protection problem. Science. 2009; 326:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Sullivan R.J., Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010; 11:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olovnikov A. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973; 41:181–190. [DOI] [PubMed] [Google Scholar]
- 5. Lue N.F., Hsu M.. A web of interactions at the ends. Mol. Cell. 2011; 42:269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blackburn E.H., Collins K.. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2011; 3:a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nandakumar J., Cech T.R.. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013; 14:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Autexier C., Lue N.F.. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006; 75:493–517. [DOI] [PubMed] [Google Scholar]
- 9. Adams Martin A., Dionne I., Wellinger R.J., Holm C.. The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol. 2000; 20:786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qi H., Zakian V.A.. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000; 14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 11. Reveal P.M., Henkels K.M., Turchi J.J.. Synthesis of the mammalian telomere lagging strand in vitro. J. Biol. Chem. 1997; 272:11678–11681. [DOI] [PubMed] [Google Scholar]
- 12. Giraud-Panis M.J., Teixeira M.T., Geli V., Gilson E.. CST meets shelterin to keep telomeres in check. Mol. Cell. 2010; 39:665–676. [DOI] [PubMed] [Google Scholar]
- 13. Chen L.Y., Lingner J.. CST for the grand finale of telomere replication. Nucleus. 2013; 4:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price C.M., Boltz K.A., Chaiken M.F., Stewart J.A., Beilstein M.A., Shippen D.E.. Evolution of CST function in telomere maintenance. Cell Cycle. 2010; 9:3157–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gelinas A.D., Paschini M., Reyes F.E., Heroux A., Batey R.T., Lundblad V., Wuttke D.S.. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19298–19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun J., Yu E.Y., Yang Y., Confer L.A., Sun S.H., Wan K., Lue N.F., Lei M.. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009; 23:2900–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gu P., Min J.N., Wang Y., Huang C., Peng T., Chai W., Chang S.. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012; 31:2309–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stewart J.A., Wang F., Chaiken M.F., Kasbek C., Chastain P.D. 2nd, Wright W.E., Price C.M.. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 2012; 31:3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kasbek C., Wang F., Price C.M.. Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J. Biol. Chem. 2013; 288:30139–30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang C., Dai X., Chai W.. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012; 22:1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng X., Hsu S.J., Kasbek C., Chaiken M., Price C.M.. CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res. 2017; 45:4281–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Derboven E., Ekker H., Kusenda B., Bulankova P., Riha K.. Role of STN1 and DNA polymerase alpha in telomere stability and genome-wide replication in Arabidopsis. PLoS Genet. 2014; 10:e1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chastain M., Zhou Q., Shiva O., Whitmore L., Jia P., Dai X., Huang C., Fadri-Moskwik M., Ye P., Chai W.. Human CST facilitates genome-wide RAD51 recruitment to GC-rich repetitive sequences in response to replication stress. Cell Rep. 2016; 16:1300–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anderson B.H., Kasher P.R., Mayer J., Szynkiewicz M., Jenkinson E.M., Bhaskar S.S., Urquhart J.E., Daly S.B., Dickerson J.E., O'Sullivan J. et al. . Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 2012; 44:338–342. [DOI] [PubMed] [Google Scholar]
- 25. Simon A.J., Lev A., Zhang Y., Weiss B., Rylova A., Eyal E., Kol N., Barel O., Cesarkas K., Soudack M. et al. . Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects. J. Exp. Med. 2016; 213:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lue N.F., Zhou R., Chico L., Mao N., Steinberg-Neifach O., Ha T.. The telomere capping complex CST has an unusual stoichiometry, makes multipartite interaction with G-Tails, and unfolds higher-order G-tail structures. PLoS Genet. 2013; 9:e1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhattacharjee A., Stewart J., Chaiken M., Price C.M.. STN1 OB fold mutation alters DNA binding and affects selective aspects of CST function. PLoS Genet. 2016; 12:e1006342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Casteel D.E., Zhuang S., Zeng Y., Perrino F.W., Boss G.R., Goulian M., Pilz R.B.. A DNA polymerase-α⋅primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 2009; 284:5807–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goulian M., Heard C.J.. The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J. Biol. Chem. 1990; 265:13231–13239. [PubMed] [Google Scholar]
- 30. Lue N.F., Chan J., Wright W.E., Hurwitz J.. The CDC13-STN1-TEN1 complex stimulates Pol alpha activity by promoting RNA priming and primase-to-polymerase switch. Nat. Commun. 2014; 5:5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen L.Y., Redon S., Lingner J.. The human CST complex is a terminator of telomerase activity. Nature. 2012; 488:540–544. [DOI] [PubMed] [Google Scholar]
- 32. Wan M., Qin J., Songyang Z., Liu D.. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem. 2009; 284:26725–26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bermudez V.P., Farina A., Tappin I., Hurwitz J.. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J. Biol. Chem. 2010; 285:9493–9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schneider A., Smith R.W., Kautz A.R., Weisshart K., Grosse F., Nasheuer H.P.. Primase activity of human DNA polymerase alpha-primase. Divalent cations stabilize the enzyme activity of the p48 subunit. J. Biol. Chem. 1998; 273:21608–21615. [DOI] [PubMed] [Google Scholar]
- 35. Steinberg-Neifach O., Wellington K., Vazquez L., Lue N.F.. Combinatorial recognition of a complex telomere repeat sequence by the Candida parapsilosis Cdc13AB heterodimer. Nucleic Acids Res. 2015; 43:2164–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Henricksen L.A., Umbricht C.B., Wold M.S.. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994; 269:11121–11132. [PubMed] [Google Scholar]
- 37. Baranovskiy A.G., Tahirov T.H.. Elaborated action of the human primosome. Genes (Basel). 2017; 8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bryan C., Rice C., Harkisheimer M., Schultz D.C., Skordalakes E.. Structure of the human telomeric Stn1-Ten1 capping complex. PLoS One. 2013; 8:e66756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goulian M., Heard C.J., Grimm S.L.. Purification and properties of an accessory protein for DNA polymerase alpha/primase. J. Biol. Chem. 1990; 265:13221–13230. [PubMed] [Google Scholar]
- 40. Nakaoka H., Nishiyama A., Saito M., Ishikawa F.. Xenopus laevis Ctc1-Stn1-Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem. 2011; 287:619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun J., Yang Y., Wan K., Mao N., Yu T.Y., Lin Y.C., Dezwaan D.C., Freeman B.C., Lin J.J., Lue N.F. et al. . Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha. Cell Res. 2011; 21:258–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baranovskiy A.G., Babayeva N.D., Zhang Y., Gu J., Suwa Y., Pavlov Y.I., Tahirov T.H.. Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J. Biol. Chem. 2016; 291:10006–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baranovskiy A.G., Zhang Y., Suwa Y., Babayeva N.D., Gu J., Pavlov Y.I., Tahirov T.H.. Crystal structure of the human primase. J. Biol. Chem. 2015; 290:5635–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baranovskiy A.G., Zhang Y., Suwa Y., Gu J., Babayeva N.D., Pavlov Y.I., Tahirov T.H.. Insight into the human DNA primase interaction with template-primer. J. Biol. Chem. 2016; 291:4793–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Puglisi A., Bianchi A., Lemmens L., Damay P., Shore D.. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008; 27:2328–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petreaca R.C., Chiu H.C., Eckelhoefer H.A., Chuang C., Xu L., Nugent C.I.. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 2006; 8:748–755. [DOI] [PubMed] [Google Scholar]
- 47. Coloma J., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Human DNA polymerase alpha in binary complex with a DNA:DNA template-primer. Sci. Rep. 2016; 6:23784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Klinge S., Nunez-Ramirez R., Llorca O., Pellegrini L.. 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. EMBO J. 2009; 28:1978–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Song X., Leehy K., Warrington R.T., Lamb J.C., Surovtseva Y.V., Shippen D.E.. STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19815–19820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Raffa G.D., Raimondo D., Sorino C., Cugusi S., Cenci G., Cacchione S., Gatti M., Ciapponi L.. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010; 24:1596–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao H., Cervantes R.B., Mandell E.K., Otero J.H., Lundblad V.. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007; 14:208–214. [DOI] [PubMed] [Google Scholar]
- 52. Polvi A., Linnankivi T., Kivela T., Herva R., Keating J.P., Makitie O., Pareyson D., Vainionpaa L., Lahtinen J., Hovatta I. et al. . Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet. 2012; 90:540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhong F.L., Batista L.F., Freund A., Pech M.F., Venteicher A.S., Artandi S.E.. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012; 150:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nandakumar J., Bell C.F., Weidenfeld I., Zaug A.J., Leinwand L.A., Cech T.R.. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012; 492:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.