Abstract
Although ∼98% of the human genomic output is transcribed as non-protein coding RNA, <2% of the protein data bank structures comprise RNA. This huge structural disparity stems from combined difficulties of crystallizing RNA for X-ray crystallography along with extensive chemical shift overlap and broadened linewidths associated with NMR of RNA. While half of the deposited RNA structures in the PDB were solved by NMR methods, the usefulness of NMR is still limited by the high cost of sample preparation and challenges of resonance assignment. Here we propose a novel strategy for resonance assignment that combines new strategic 13C labeling technologies with filter/edit type NOESY experiments to greatly reduce spectral complexity and crowding. This new strategy allowed us to assign important non-exchangeable resonances of proton and carbon (1′, 2′, 2, 5, 6 and 8) nuclei using only one sample and <24 h of NMR instrument time for a 27 nt model RNA. The method was further extended to assigning a 6 nt bulge from a 61 nt viral RNA element justifying its use for a wide range RNA chemical shift resonance assignment problems.
INTRODUCTION
Ribonucleic acids (RNAs) participate in diverse functions ranging from catalysis, gene regulation, protein synthesis, signaling, splicing, to viral infectivity (1–8). This extensive functional diversity derives from an ability to assume complex architectures, and is further enhanced by motions that are intrinsic to these structures (3–6). However, there is a dearth of solved RNA structures, reflected in the inverse 98:2 correlation between the genomic outputs of RNAs and proteins and the number of solved 3D structures of RNA and protein structures (2:95) deposited in the protein databank (PDB) (9–11). The combined difficulties of crystallizing RNA, along with the rapid signal decay and spectral crowding associated with NMR studies, makes structural studies challenging. Furthermore, the average size of the ∼300 deposited RNA NMR structures in the PDB is only 25 nt. While 50% of these structures in the PDB were solved by NMR methods, the usefulness of NMR is still limited by difficulties in resonance assignments.
Resonance assignment of RNA is non-trivial. In an effort to tackle this knowledge gap using NMR, a large suite of multi-nuclear experiments have been developed and various strategies proposed to assign resonances in RNA molecules (12–28). Chemically, RNA has only four bases that look essentially the same, whereas proteins feature 22 chemically distinct amino acids. This profound lack of chemical diversity within RNA leads to severe spectral crowding of the information rich proton resonances. Implementation of through-bond experiments has enabled automated sequential assignment of resonances in proteins, but these experiments are difficult to implement for RNA (25–29). Therefore, sequential assignment with nuclear Overhauser effect spectroscopy (NOESY) has become the main tool for assigning RNA proton resonances.
An unambiguous starting point is a critical step in the chemical shift assignment strategy for RNAs, and traditionally this begins with NOE-based sequential assignment of the exchangeable resonances combined with through-bound multi-dimensional experiments (21, 29). Here, we propose an alternative approach that starts with non-exchangeable resonances, but still relies on through-space NOESY experiments. However, we now combine isotope filtering and editing with our new 13C/12C alternative labeling patterns within the sugar and base moiety of the nucleotide building blocks (30–32) to systematically assign non-exchangeable proton resonances as a powerful alternative to previously established methodologies (21, 24, 29). The combination of an asymmetric labeling pattern and isotopic filtered/edited NOESY experiments (16, 33) allows us to logically distinguish between intra- and inter-nucleotide NOEs for sequential resonance assignment. The filtering/editing NOESY strategy has previously been implemented using 15N/14N labeling strategies for duplex formation between labeled and unlabeled strands of RNA, protein:RNA complexes, and samples containing only uniformly 13C/15N labeled uridine and unlabeled AGC rNTPs (16, 34, 35).
As a model system, we use a 27 nt RNA encompassing a helical fragment from the highly conserved aminoacyl-tRNA anticodon site (A-site) of 16S rRNA on the 30S ribosomal subunit (36). This novel strategy that combines filtering and editing NOESY techniques with our new labeling scheme enables rapid and unambiguous assignment for adenine H2 inter-nucleotide NOESY cross-peaks with sequential and cross-stand H1′ resonances. We successfully assigned all the H1′, H2′, H2, H5, H6 and H8 resonances of the 27 nt A-site RNA from a single NMR sample using only three 2D 1H–1H NOESY experiments. To further demonstrate the usefulness of our new methodology, a 61 nt viral RNA element was transcribed with a similar selective, alternating 13C-labeling pattern and proved equally amenable to our novel assignment strategy. In addition to the traditional NOESY contacts expected in A-form RNA helices, we report a number of noncanonical, readily assigned NOESY cross-peaks that occurred in the 6 nt bulge of this large RNA. Further, using previously reported structures that contained bulges as models, we show that contacts accentuated by our labeling strategy and filter/edit experiments should be a generally observed phenomenon. This improved resonance assignment strategy for RNAs will accelerate access to the growing field of RNA biophysics.
MATERIALS AND METHODS
NTP and oligonucleotide synthesis
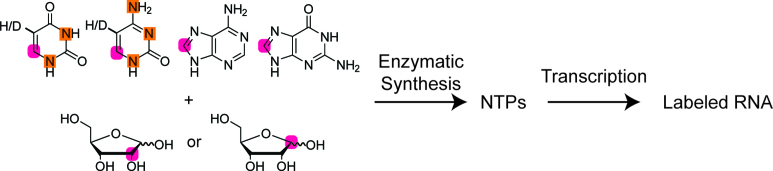
Labeled NTPs (A, C, G and U) were strategically synthesized from selectively 13C-labeled commercially available ribose (Omicron Biochemicals) and chemically synthesized base for an in vitro T7 polymerase based transcription. A schematic of the potential NTP isotopic labeling patterns afforded by our method is shown in Figure 1. Chemo-enzymatic reactions were carried out as previously described with further detail in the Supplementary Material (30–32).
Figure 1.
Following previously published methods, D-ribose-1-13C or D-ribose-2-13C was enzymatically coupled separately to 8-13C-guanine (32), 6-13C-1-15N-cytosine ((30, 31)), 8-13C-adenine, or 6-13C-1-15N-uracil which were then phosphorylated to their respective rNTPs. Atoms enriched with 13C are highlighted with purple circles and atoms enriched with 15N are indicated by orange squares. Purified NTPs were used to synthesize alternatively labeled RNA samples for NMR resonance assignment. Bacterial A-site RNA (27 nt) was transcribed from 2′,8-13C ATP, 1′,8-13C GTP, 2′,6-13C UTP, and 1′,6-13C CTP while the 61 nt viral RNA element was transcribed from 2′,8-13C ATP, 1′,8-13C GTP, 1′,6-13C-5-2H UTP, and 1′,6-13C-5-2H CTP. The common underlying method utilizes 2′,8-13C ATP for both RNAs as explained in the text.
RNA was transcribed from DNA template (Integrated DNA Technologies) with T7 RNA polymerase (in-house produced) and a logically predetermined combination of our site-selectively 13C-labeled NTPs using standard methods (24, 37). Transcribed RNA was purified by denaturing PAGE and electro-eluted from gel slices. Electro-eluted RNA was buffer exchanged in 3K MWCO centrifuge tubes (Millipore) against EDTA, followed by extensive washes with ddH2O. Additional washing of the RNA over SOURCE 15Q resin (GE Healthcare Lifesciences) was occasionally performed before buffer exchange to remove acrylamide contamination. The sample was then lyophilized to dryness after addition of NMR buffer (final concentration of 10 mM sodium phosphate, 0.1 mM EDTA, 0.1% NaN3, and 0.1 mM DSS at pH 6.4) and re-suspended in ∼300 μl of 99.98% D2O to a final concentration of 0.3–1 mM.
NMR experiments
NMR experiments were performed on an Ultrashield Plus 600 MHz Avance III Bruker spectrometer with a room temperature TXI 600SB H-C/N-D-05 Z BTO probe. NMR experiments in D2O were collected at 25°C and exchangeable proton experiments in H2O were collected at 15°C (optimized from several 1D imino experiments at various temperatures from 5 to 35°C). Data were collected and analyzed using TopSpin 3.2, NMRFx Processor (38) and NMRViewJ (39).
For sequential assignment of non-exchangeable proton resonances (1′, 2′, 2, 5, 6, 8), we implemented three different filter/edit type NOESY experiments as well as a 13C-HSQC. Filter/edit type NOESY experiments were classified by the dimension (F1 = indirect detected dimension and F2 = direct detected dimension) as well as the proton chemical shift observed (a = all protons, e = edited, only protons attached to 13C, and f = filtered, only protons attached to 12C). The F1eF2e NOESY experiment was a standard NOESY-HSQC with an initial double purge module to dephase 1H–12C magnetization adapted from previously published methods (16). F1eF2f and F1fF2e NOESY experiments were standard Bruker pulse sequences (hsqcgpnowgx33d and noesyhsqcgpwgx13d, respectively). F1eF2e, F1eF2f, F1fF2e NOESY and a standard 1H–13C HSQC were collected on the alternatively labeled A-site RNA sample in <24 h of total instrument time. The F1eF2f NOESY experiment on the 61 nt viral RNA element was collected in 14 h of instrument time.
In the absence of published or deposited peak lists available for the 27 nt bacterial A-site RNA, we performed imino NOESY and through-bond heteronuclear correlation experiments on a uniformly 13C/15N- labeled A-site RNA to confirm our resonance assignment. The following experiments were run in H2O for a total of 59 h of NMR instrument time: 1D 1H imino with Watergate, imino NOESY with jump-return or Watergate, 1H–15N-HSQC, 2-bond 1H–15N HSQC, and HNN-COSY (40–46). Imino resonance assignment was confirmed with the automated assignment method, RNA-Pairs (25). Heteronuclear correlation data was also collected for the same sample in D2O with the following NMR experiments for a total of 27 h of NMR instrument time: 13C-HSQC, HCN and HCCH-COSY (47–51). Parameters for all NMR experiments are provided in Supplementary Table S1.
RESULTS AND DISCUSSION
General selective labeling strategy
Based on previously published chemo-enzymatic synthesis methods from our laboratory, we devised a new isotopic labeling scheme using isolated 13C labels within ribose and base moieties to eliminate 13C–13C spin–spin coupling and enable us to edit or filter observable chemical shift signals within NOESY experiments (30, 31). Our approach uses strategic 13C-labeling of the ribose C1′ or C2′ positions (coupled to purine/pyridimine base 13C-labeled at the C8/C6 positions via enzymatic synthesis) guided by the nucleotide distribution within our RNA of interest. Our chemo-enzymatic synthesis also affords the option of deuterating the pyrimidine C5 position, which is convenient for larger RNAs. We selected two RNAs to demonstrate the assignment strategy that arises from our labeling scheme.
In order to demonstrate the sequential assignment of helical regions of RNA we present an exhaustive example on the 27 nt A-site RNA. We also demonstrate the usefulness of our labeling scheme for assigning more difficult unstructured regions of RNA with a 6 nt bulge within a 61 nt viral RNA element. For A-Site RNA, 13C-labeling of the A and U ribose C2′ position allowed convenient separation of all peaks in the typically crowded H2′ and C2′ regions of a 13C-HSQC spectrum since only A and U resonances were observable in this region. The remaining G and C residues were 13C-labeled at the C1′ position, significantly reducing chemical shift overlap in the H1′ dimension. For base residues, pyrimidine C6 and purine C8 were 13C-labeled, and A C2 and pyrimidine C5 were unlabeled, minimizing the usual crowding in a 2D 1H–1H NOESY spectrum when implementing filtering and editing techniques.
We choose this particular labeling scheme for three reasons: (i) based on BMRB statistics, pyrimidine and purine resonances have well separated chemical shifts, thus we elected to 13C-label one purine type nucleotide and one pyrimidine type nucleotide for each C1′ and C2′ position (52). (ii) For A-site RNA, there were only four A and five U residues compared to nine each of G and C residues, therefore by 13C-labeling A and U at C2′ we were less likely to contend with chemical shift overlap for H2′ resonances. (iii) Labeling A at the C2′ position rather than C1′ avoids intra-nucleotide NOESY cross-peaks arising from A-H2 to H1′ in a NOESY F1eF2f spectrum and thus only detects sequential intra-strand and cross-strand A-H2 to 13C-labeled (G/C) H1′ cross-peaks. As a general approach, choosing which nucleotides to label at the C1′ or C2′ position can be accomplished by predicting chemical shift dispersion from solved structures using programs like RAMSEY or RNAShifts2 and minimizing predicted chemical shift overlap (53, 54), logically selecting your labeling pattern based on specific nucleotide distribution within the candidate RNA (as performed here with A-site RNA), or even synthesizing four separate 13C single nucleotide specifically labeled RNAs (e.g. 13C-ATP only) and empirically determining the optimum 13C-C1′/C2′ labeling scheme.
The cost of input NTPs was also significantly reduced using our chemo-enzymatic 13C-labeling methods. To obtain NMR quantities of RNA (100+ nmol), we typically use 20 μmol of each NTP in a 10 ml transcription reaction. The site-specifically 13C-enriched NTPs used in this study are not commercially available, thus the closest comparison is 13C/15N-uniformly labeled NTPs (Sigma-Aldrich or Cambridge Isotopes) with listed prices of 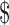 250/20 μmol NTP. At these prices, a typical NMR sample would cost
250/20 μmol NTP. At these prices, a typical NMR sample would cost 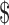 1000. The cost of the chemo-enzymatic synthesis of site-specifically 13C-enriched NTPs used in this study provides an approximate savings of 10–50-fold, but does not include set-up costs. Nonetheless even after accounting for such issues, these nucleotides can still be prepared at a significantly-reduced cost.
1000. The cost of the chemo-enzymatic synthesis of site-specifically 13C-enriched NTPs used in this study provides an approximate savings of 10–50-fold, but does not include set-up costs. Nonetheless even after accounting for such issues, these nucleotides can still be prepared at a significantly-reduced cost.
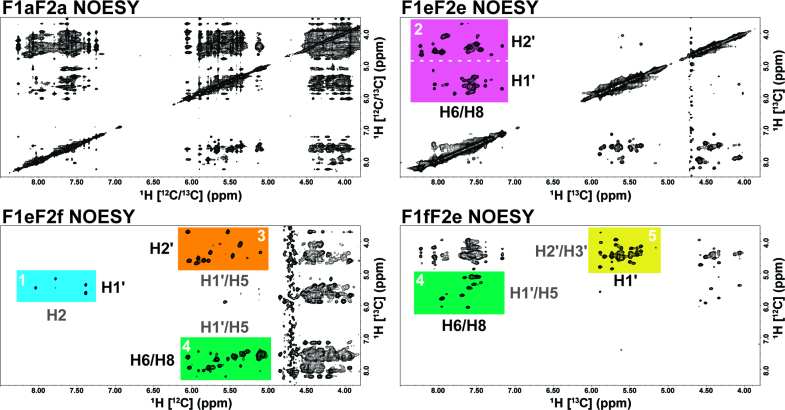
In addition to reduced cost, the power of filtering/editing the signal of the NOESY spectra was immediately evident for our alternatively 13C-labeled RNA sample when compared with a standard 2D 1H–1H NOESY, F1aF2a NOESY (Figure 2). By separating half of the nucleotides into the H1′ or H2′ region, spectral crowding of the ‘fingerprint’ region is essentially reduced to at most half for the F1eF2e NOESY. Spectral crowding was significantly reduced through isotopic filtering/editing in all NOESY spectra to the point that full resonance assignment could be accomplished from a combination of only three 2D 1H–1H NOESY spectra. Specific regions of the various NOESY spectra contain valuable structural information and our labeling strategy allowed us to develop a logical resonance assignment scheme based on the unique information content of each region. In Figure 2, we have highlighted these regions in a color coded manner which is discussed in further detail below.
Figure 2.
Four separate NOESY spectra from the alternatively labeled bacterial A-site RNA with color coded regions labeled by expected proton cross-peaks (13C-label in black and 12C-label in gray) on the edge of each color coded region. Each spectrum is labeled by the dimension (F1 = indirect and F2 = direct) followed by the type of proton observed (a = all [13C-label and 12C-label], e = edited [13C-label], and f = filtered [12C-label]). The axis of each spectrum is also labeled by the type of observed proton resonance with the attached carbon isotope given in brackets. For example, the F1eF2e NOESY in the top right panel contains only cross-peaks from the 13C-labeled non-exchangeable protons. The color coded regions are as follows (white number): 1 = sky blue, 2 = reddish purple, 3 = orange, 4 = bluish green and 5 = yellow. The utility of these regions are described in detail within the text and the color and numbering of the highlighted regions correspond to the steps for logical resonance assignment.
Logical assignment of non-exchangeable proton resonances
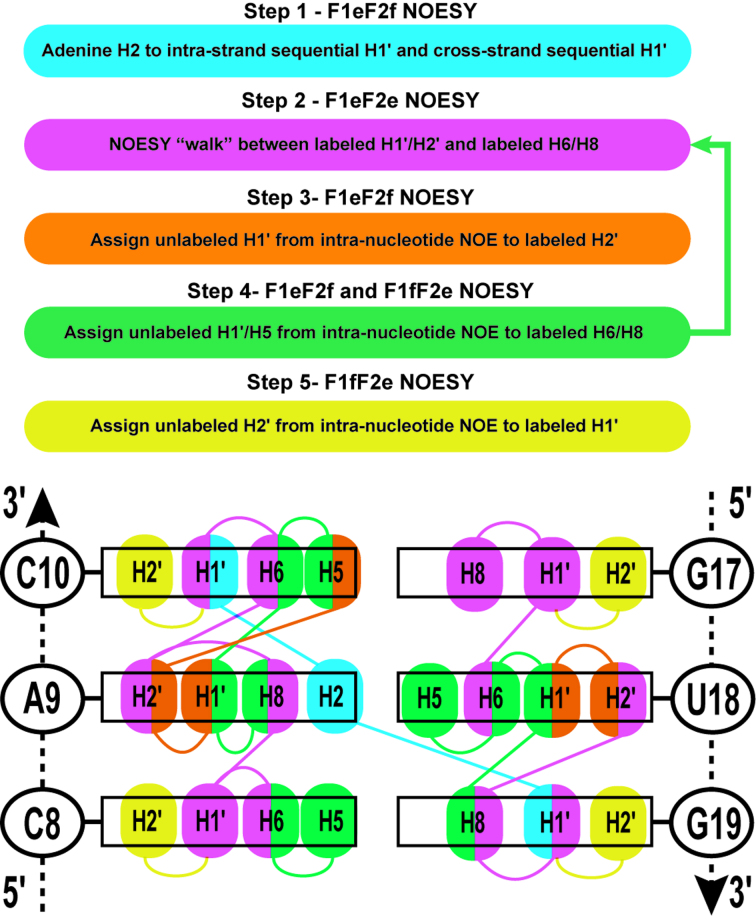
A logical sequential assignment strategy arising from the alternative labeling of RNA is depicted in Figure 3 alongside a schematic with the NOE cross-peaks expected for three stacked base-pairs in the upper-stem of the 27 nt bacterial A-site RNA based on our 13C-labeling scheme. The stepwise assignment scheme works with any given 13C-C1′/C2′ alternating pattern, assuming that A is 13C-C2′ labeled and not 13C-C1′-labeled. The various edited/filtered NOESY spectra (F1eF2e, F1eF2f and F1fF2e) then provide useful separation of cross-peaks into identifiable regions, enabling a logical assignment as well as simplification of spectral analysis. Colors and numbers depicted in Figure 3 correspond to the regions of the spectra shown in Figure 2. Step 1 is essential to the assignment process by identifying the starting points for the NOESY ‘walk’ and our unique labeling simplifies the assignment of the H1′ inter-nucleotide cross-peaks to adenine H2 resonances. When A-H2 resonances are difficult to assign, a 1H–1H ‘imino’ 2D NOESY provides clear A-H2 cross-peaks with imino 1H in base-paired regions. Step 2 is the typical NOESY ‘walk’ of the fingerprint region, but with our labeling scheme H2′ cross-peaks are easily assigned by removing crowding from H3′, H4′, H5′ and H5′' resonances. Step 3 is useful for assigning the unlabeled (12C) H1′ (e.g. of A/U for our selectively labeled A-site RNA sample) which is further used in step 4 to confirm intra- and inter-nucleotide NOEs with labeled (13C) resonances (e.g. H6/H8). Step 5 is the final process of assigning the remaining unlabeled (12C) H2′ resonances (e.g. of C/G). We demonstrate this assignment process using the A9-C10 stacked nucleotides within the same upper-stem of A-site RNA.
Figure 3.
The sequential NOESY ‘walk’ strategy is given as a step-by-step process color coded to match the regions of the NOESY spectra in Figure 2. Step 4 allows cross-validation of the assignments from the sequential NOESY ‘walk’ in step 2. A 3 bp region of the upper-stem of the 27 nt bacterial A-site RNA demonstrates the NOE cross-peaks expected from each of the NOESY regions in Figure 2. To simplify NOE cross-peaks, only non-exchangeable H1′, H2′, H2, H5, H6 and H8 atoms are labeled within this schematic figure and are color-coded by the spectral regions from which they were assigned.
Step 1. An unambiguous starting point for the NOESY ‘walk’ is a critical step for resonance assignment. For this step, we analyzed the sky blue region from the F1eF2f NOESY experiment (Figure 4). The base region of the F2 filtered dimension (6.5–8.5 ppm) reveals only cross-peaks arising from A-H2 resonances since the remaining base proton (H6/H8) resonances in this region are filtered by the removal of proton signals attached to 13C-labeled nuclei. The crowded ‘fingerprint’ NOESY region is reduced to five cross-peaks between H2 (12C-labeled) resonances and 13C-labeled H1′ resonances (Figure 4A and C). By editing the F1 dimension for 13C-labeled protons, only H1′ resonances from 13C-C1′ labeled nucleotides (e.g. C/G) are observed and both unlabeled H5 and H1′ resonances are rejected. Thus, no intra-nucleotide NOEs are observed for A-H2 to H1′ since only the C2΄ position of A is labeled with 13C, thus we do not see any cross-peaks for A20-H2. Even though an F1fF2f NOESY experiment is sufficient to identify these cross-peaks, it is unnecessary. The F1eF2f NOESY provided adequate starting points to begin our NOESY ‘walk’. In more complex samples, a standard 2D ‘imino’ 1H–1H NOESY can be used to assign A-H2 resonances before performing Step 1.
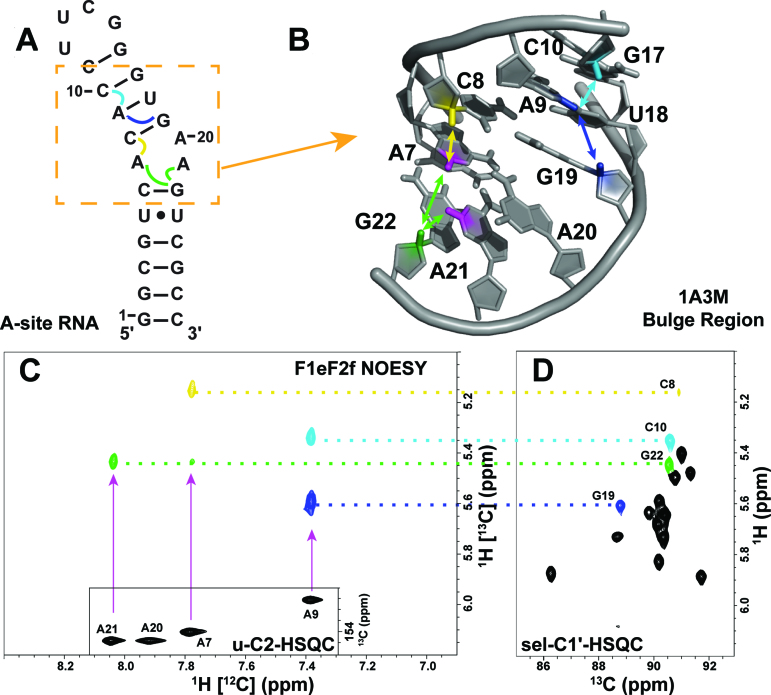
Figure 4.
(A) The secondary structure of A-site RNA with inter-nucleotide NOEs expected in the F1eF2f NOESY indicated with curly colored lines. The bulge region is highlighted with an orange dashed box. (B) The 3D structure of the bulge region with the A7-H2 (magenta), A9-H2 (dark blue), A21-H2 atoms (magenta) and their NOEs with C8-H1′ (yellow), C10-H1′ (light blue), G19-H1′ (dark blue) and G22-H1′ (green) marked with colored lines and arrows. (C) The sky blue region of the F1eF2f NOESY spectrum is depicted with an inset of the C2-H2 HSQC region of the uniformly labeled A-site RNA. Cross-peaks are colored to correlate with their C1′-H1′ partner in the C1′-H1′ HSQC of the alternatively labeled A-site sample (sel-C1′-HSQC). (D) Labeled peak assignments are connected by dotted color-coded lines to the corresponding ones in C.
In addition to a sequential inter-nucleotide NOE transfer between the A-H2 (i) proton and the H1′ proton of the next nucleotide (i+1) in the 5′→3′ direction, A-H2 protons involved in base-pairing, or stacking in A-helical regions, will also ‘talk’ with H1′ protons on the nucleotide proceeding (j+1) its cross-strand base-pair partner (j) in the 5′→3′. Examples of these cross-strand NOEs are shown in Figure 4 for A7-H2 to G22-H1′ and A9-H2 to G19- H1′. Both types of inter-nucleotide NOE interactions provide excellent starting points for resonance assignment as they are the only cross-peaks observed in the sky blue highlighted region of the F1eF2f NOESY spectrum in Figure 2. A7-H2 and A21-H2 share a cross-strand or same-strand NOE contact, respectively, with G22-H1′ as predicted from the 2D and 3D model and easily identified with a complimentary 13C HSQC (Figure 4D). While this is the only NOE observed in this region for A21-H2, as expected, A7-H2 has an additional same-strand NOE cross-peak with C8-H1′, making assignment of these resonances straightforward. The two cross-peaks to A9-H2 are ambiguously assigned as either G19-H1′ or C10-H1′ in the F1eF2f NOESY spectrum—its cross-strand and same-strand partners. However, this ambiguity is removed when we compared the F1eF2e and F1eF2f NOESY spectral regions using the logical assignment process illustrated for representative cases in steps 2–4 (Figure 3). G19-C8 and C10-C6 are both labeled with 13C, and because G19 is followed by A20 (no 13C label at C1′) whereas C11, that follows C10, has a 13C label at C1′ label we can readily distinguish the residues. We can also use NMRViewJ prediction to help remove ambiguity in this step as we did with the 61 nt viral RNA element. The assignment of A-H2 inter-nucleotide NOE cross-peaks with sequential and cross-strand H1′ resonances provides a powerful tool for RNA resonance assignment and our labeling scheme provides a means of simplifying the NOESY spectrum, allowing rapid identification of these important cross-peaks.
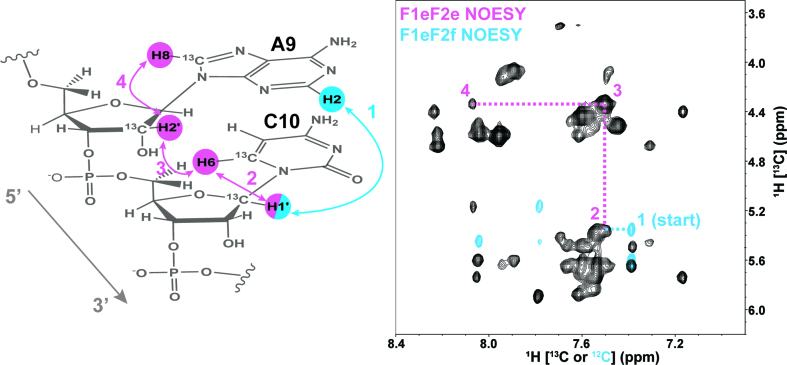
Step 2. We demonstrate our strategic NOESY ‘walk’ with the sequentially stacked base pairs A9-C10. We chose residue C10 as a starting point since we had identified its H1΄ chemical shift using Step 1 discussed above. The following Steps 2–5 are color coded to match regions of the various NOESY spectra with corresponding information content. An overlay of the essential F1eF2f NOESY region (sky blue), which identifies the A9-H2 to sequential C10-H1′ resonance, with the expanded ‘fingerprint’ region of the F1eF2e NOESY region (reddish purple) illustrates this sequential walk (Figure 5). The starting point residue C10-H1′ is identified as NOESY cross-peaks 2, 3 and 4 (reddish purple) and the strong cross-peak between A9-H2′ and C10-H6 as peak 3 (Figure 5). These strong inter-nucleotide H2′ to H6/H8 NOEs form distinctive and diagnostic signatures for resonance assignment because of the short (∼2 Å) distance between the atoms in stacked A-form helical regions of RNA (21, 29).
Figure 5.
The sequential NOESY ‘walk’ strategy for steps 1 and 2 is highlighted for the A9 and C10 stacked residues. All carbons are 12C labeled unless otherwise identified. The F1eF2f NOESY spectrum (sky blue peaks) is overlaid with the reddish purple F1eF2e NOESY region (black peaks) and includes the peaks labeled for each NOE connectivity (color coded and numbered to match NOESY spectrum) expected in the sequential A9-C10 example.
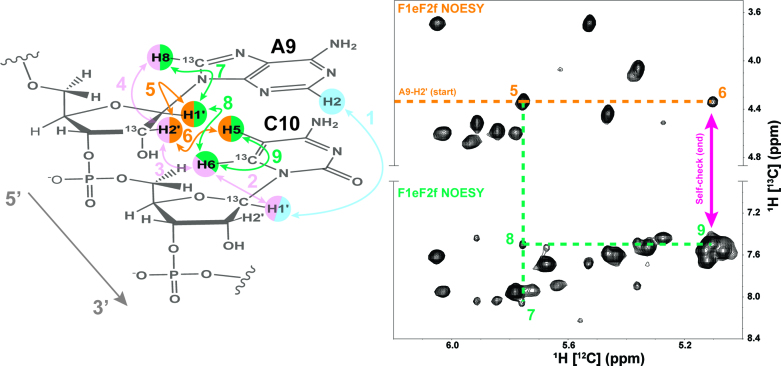
Steps 3 and 4. To cross-validate assignment of the 13C-labeled protons from Steps 1 and 2, we used filtering techniques to include cross-peaks from 12C-labeled protons for Steps 3 and 4 of the sequential assignment process (Figure 6). Again starting with the previously assigned A9-H2′, we could immediately identify the resonances of A9-H1′ (peak 5) and C10-H5 (peak 6), Step 3. In turn, assignment of the 12C-labeled A9-H1′ resonance helped identify the inter-nucleotide NOE involving C10-H6 (peak 7) and intra-nucleotide NOE with A9-H8 (peak 8), Step 4. The strong intra-nucleotide NOE cross-peak between 13C-labeled C10-H6 and 12C-labeled C10-H5 (peak 9) served as an internal consistency check (magenta arrow) to confirm the resonance assignments from all previous steps and the correlation between peaks 6 and 9. Peak 7 also correlated with the previously assigned peak 4 from Figure 5 and, therefore served as an internal check. Thus, the different NOESY spectra contain two self-check points that allowed confirmation of resonance assignment of the stacked residues A9 and C10.
Figure 6.
The sequential NOESY ‘walk’ for Steps 3 and 4 are color coded and numbered to match the peaks in the F1eF2f NOESY spectrum regions. Previous assignment steps are faded within the A9-C10 example for visual reference. The A9-H2′ starting point is labeled and follows the dotted orange line to peaks 5 and 6. Extension of the A9-H1′ assignment is marked by the dotted bluish-green line to peaks 7 (A9-H8) and 8 (C10-H6). Peak 9 from the C10-H6 to C10-H5 serves as a self-check with peak 6 (magenta arrow).
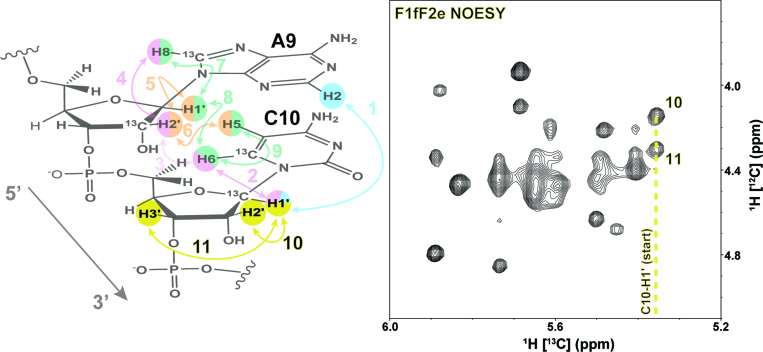
Step 5. In the final step of the assignment process (Figure 7), the unlabeled (12C) H2′ resonance is unambiguously identified by an NOE contact to 13C-labeled H1′ cross-peaks in the yellow region of the F1fF2e NOESY spectrum. Starting from assignment of the 13C-labeled C10-H1′ resonance, two strong intra-nucleotide cross-peaks are labeled as peak 10 and peak 11. The stronger NOESY cross-peak (peak 10) is identified as the C10-H2′ resonance and peak 11 is assigned to C10-H3′. The yellow region of the F1fF2e NOESY contains cross-peaks from intra-nucleotide H3′ and H2′ resonances. Therefore, supplementing this experiment with HCCH-COSY correlations from a uniformly 13C-labeled sample confirm the H2′ cross-peaks (Supplementary Figure S4).
Figure 7.
A schematic of the final assignment steps for unlabeled H2′ and H3′ from Step 5 of the logical assignment process with yellow highlighted arrows and numbers. The previous assignment steps and protons are faded for visual reference. The F1fF2e NOESY (yellow region) with starting point (C10-H1′) and cross-peaks to C10-H2′ and C10-H3′ are labeled 10 and 11, respectively.
Overall, the separation of peaks between 13C-labeled (edited) and 12C-labeled (filtered) regions of the NOESY spectra greatly reduced crowding, allowed logical assignment strategies, and prevented breaks in the sequential NOESY ‘walk’ that are usually observed for selective labeling strategies. Working through Steps 1–5 of our proposed logical resonance assignment process, we could fully assign the non-exchangeable H1′, H2′, H2, H5, H6 and H8 resonances (Supplementary Table S2).
Confirmation of resonance assignment
Imino NOESY resonances
As is good practice with any RNA system studied by NMR, we confirmed the secondary structure of our construct by analyzing imino proton resonances. For our RNA model system, there was no overlap in imino resonances once an ideal temperature was selected from 1D experiments. All expected resonances could be assigned easily from a 2D 1H–1H NOESY experiment (Supplementary Figure S5). Our model RNA system did not require the use of 15N-edited NOESY experiments to assign the imino resonances. However for more complex systems, automated tools such as RNA-PAIRS can aid imino assignment (25). Indeed, RNA-PAIRS analysis of the 1H–1H NOESY and 1H–15N HSQC yielded a 100% probability for assignment of the upper stem, and our imino assignment results agree with previously published assignments of bacterial A-site RNA (55, 56).
With imino resonances assigned, we confirmed all our previous non-exchangeable resonance assignments through the spin-diffusion patterns observed within G–C base-pairs. In A-form helical regions of RNA, magnetization is easily transferred from G-H1 imino protons to C-H5 and to H1′ of the sequential residue (5′→3′) on same strand, as well as the cross-strand H1′ of the sequential residue to the base-paired C (5′→3′) (23, 29). All expected cross-peaks from this spin-diffusion pattern were observed for base-paired G-H1 resonances (Supplementary Figure S6) confirming the validity of our strategic non-exchangeable resonance assignment process.
Adenine H2
To further confirm these assignments, several heteronuclear correlation experiments were recorded using a uniformly 13C/15N-labeled bacterial A-site RNA sample. The H2 resonances for three of the four adenines were identified in a two-bond 15N HSQC experiment, but the A20-H2 resonance was not visible due to line-broadening from chemical exchange as previously reported (56). The two-bond 15N HSQC experiment was combined with HNN-COSY and 1H–1H NOESY (Supplementary Figure S7) experiments to confirm the resonance assignment of A9-H2. As expected, we observed a strong cross-peak between U18-H3 and A9-H2 in the 1H-1H NOESY spectrum. With the through-bond HNN-COSY experiment, we could correlate the H3 of U18 with the hydrogen bond donor, U18-N3, and acceptor, A9-N1. With the 2-bond 15N HSQC, we could correlate the N1 and N3 chemical shifts of A9 with its H2 resonance, further confirming the assignment of A9-H2. Cases of overlap could be resolved by separating either H2 and C2 resonances or 1H–1H NOESY cross-peaks into a third dimension (16,17,57). After assigning adenine H2 resonances, through-bond TROSY relayed HCCH-COSY experiments can also be used to correlate H2 with H8 resonances for medium to small RNAs (58). A9-H2 was further confirmed with the 2-bond 15N HSQC identifying the N1 and N3 chemical shifts of A9. However this step was not necessary with our strategy for assignment.
Pyrimidine intra-nucleotide NOEs
To further verify our assignment, we used the JCC coupling splitting of the pyrimidine C6 resonances to identify these resonances in the HSQC spectra of the uniformly 13C/15N-labeled bacterial A-site RNA sample. We performed a HCN correlation experiment on the alternatively labeled bacterial A-site RNA sample to further confirm previous assignment of the H6 resonances as belonging to either U or C (Supplementary Figure S8). Based on previously published data from the 14 nt UUCG tetraloop RNA, we could confirm the resonance of C14 (12, 59). The resonance for C24 was absent in the HCN correlation spectra, most likely due to line-broadening from chemical exchange which matches previous reports (56). U-C6 resonances were confirmed from the lack of corresponding resonances at C1′ in the HCN spectrum and from the chemical shift of U-N1, which is upfield of the C-N1 resonances. The H1′ to H6 correlation of C was used to confirm intra-nucleotide C-H1′ to H6 cross-peaks in Step 2 of the logical resonance assignment process.
Unlabeled H2′ and H5 resonances
Finally, the unlabeled protons for H5 and H1′ are easily confirmed with through-bond HCCH-COSY experiments on the uniformly 13C/15N-labeled bacterial A-site RNA sample. The HCCH-COSY is a fairly sensitive experiment applicable to larger RNAs and can facilitate assignment of more complex systems. By comparing the F1fF2e NOESY spectral (yellow) region with the HCCH-COSY spectra of the uniformly labeled sample, we could determine that all strong cross-peaks observed in this region were from intra-nucleotide H1′ to H2′ NOEs (data not shown).
Expansion to larger RNA and noncanonical bulge assignment
To further test the utility of our method for larger RNA systems, we incorporated a similar labelling scheme into a 61 nt hepadnaviral RNA element. Here, we chose to 13C-label the U residues at the C1′ position rather than the C2′ position to observe even more inter-nucleotide cross-peaks from the F1eF2f NOESY experiment performed in Step 1 of our novel assignment strategy. Additionally, for U and C the H5 position of the base was deuterated. Intended as a strategy for increasing the signal intensity of the H6 position, deuteration had unforeseen benefits for the assignment of the bulge discussed below. The RNA tested here contains 8 A-U base-pairs with two sets of sequentially stacked A-U pairs. The resulting F1eF2f NOESY spectrum captured all expected inter-nucleotide H2 (12C-labeled) to H1′ (13C-labeled) cross-strand and intra-strand NOEs (Figure 8).
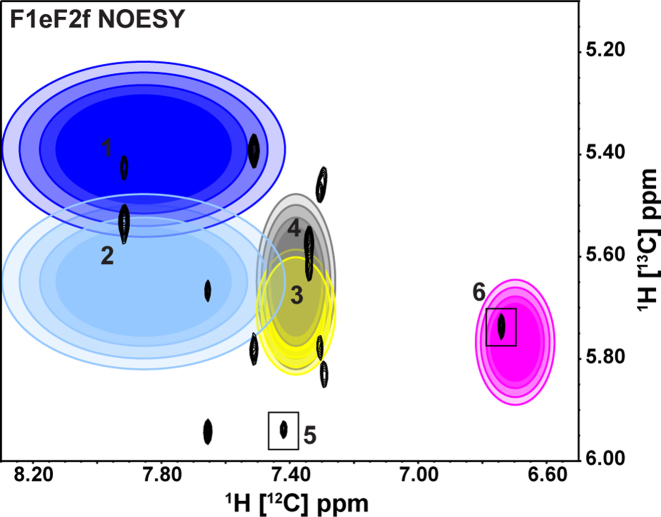
Figure 8.
The F1eF2f NOESY spectrum of a 61 nt hepadnaviral RNA element transcribed from 2′,8-13C ATP, 1′,8-13C GTP, 1′,6-13C-5-2H UTP and 1′,6-13C-5-2H CTP. All inter-nucleotide NOEs are observed for the A-H2 → H1′ (14 total peaks) arising from the eight A–U base-pairs found in the 61-nt viral RNA element. Two 5′-A form sets of stacked A-U base-pairs resulting in one observable inter-nucleotide NOE since the sequential 3′-A of the stacked A-U base-pair does not contain a 13C-labeled H1′. Peaks (5 and 6) from these unique 5′-A within A–U stacked base-pairs are highlighted with black boxes. Additional starting points (peaks 1–4 and 6) are highlighted with colored ellipses with increasing radii representing 68, 80, 90 and 95% probabilities for predicted NOE cross-peak using NMRViewJ chemical shift prediction resulting in a total of six unambiguous starting points (39).
In total, we observed 14 NOE peaks within the previously labelled sky blue region of the F1eF2f NOESY spectrum instead of the typical 16 inter-nucleotide NOEs expected for eight A–U base-pairs. We did not observe the remaining intra-strand NOEs because there are two sets of stacked A–U base-pairs in the RNA and the 5′-A does not have an observable cross-peak with the 3′-A in the stacked base-pairs since the sequential 3′-A-H1′ is 12C-labeled. We used RNA chemical shift predictions from secondary structure as implemented in NMRViewJ to identify six unambiguous starting points (peak 1–6) as indicated by ellipses (predicted chemical shift) and boxes (logically assigned as 5′-A in AA stacked base-pairs) in Figure 8 (39, 60). Each color-coded ellipse represents the predicted region of an expected cross-peak between A-H2 and its respective inter-nucleotide sequential H1′ or cross-strand H1′. For each expected cross-peak, a total of four concentric ellipses with increasing radii and greater transparency are drawn to represent the predicted probability (68, 80, 90 and 95%, respectively) of finding the expected cross-peak. Peak 1 and peak 2 represent the same A-H2 resonance and the chemical shift predictor distinguishes the inter-nucleotide sequential H1′ (peak 1) from the cross-strand H1′ (peak 2) since only peak 2 falls within the 95% region of the light blue ellipse. The yellow and grey ellipses indicate that peak 3 (intra-strand sequential NOE) is expected downfield of peak 4 since peak 4 is outside of the 95% probability of the yellow ellipse. The two 5′-A in the AA base-pair stacks are easily distinguished (peak 5 and 6) with the cross-peak of peak 6 contained within the predicted magenta ellipse. Chemical shift predictions provide similar separation of the remaining intra-strand sequential and cross-strand NOEs with some ambiguity. Our initial guesses were confirmed through our stepwise assignment process in which all H1′, H2′, H2, H6 and H8 resonances in helical regions were assigned for the 61 nt RNA following the general approach outlined in Steps 1–5. For longer stretches of A–U stacked base-pairs, an additional F1fF2f NOESY experiment would identify sequential inter-nucleotide A-H2 to A-H1′ (12C-labeled) NOEs. All A-H2 resonances are easily confirmed with an imino NOESY ‘walk’.
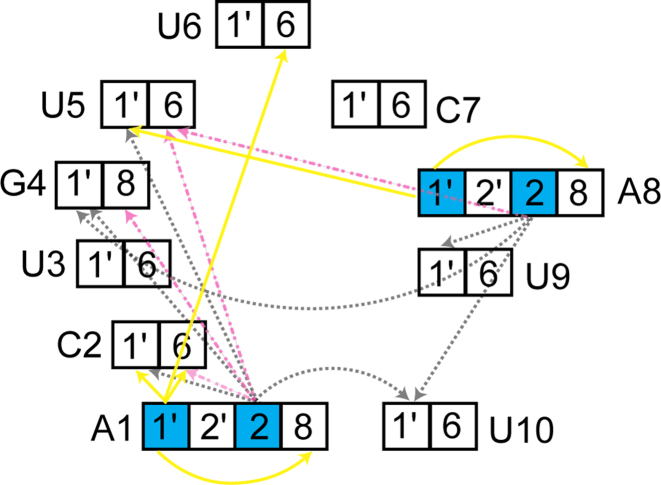
An unexpected application of our alternate labeling scheme was uncovered when working with this RNA (Figure 9). The element contains a 6 nt bulge that would prove difficult to assign using traditional assignment techniques. Bulge regions do not have the canonical contacts (imino/sugar to base) that are intrinsic to A-form helices. Further, due to the large size of the RNA, through-bond phosphorous experiments were unfeasible (61–63). When examining the F1eF2f NOESY, a number of unidentified peaks from the H2s (12C-labeled) of A1 and A8 were noted. In the F1eF2f NOESY, cross-peaks arise between a proton attached to a 12C nucleus within <5 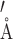 of proton attached to a 13C nucleus. With our labeling pattern, this occurs in four discrete uncrowded regions of the spectrum due to distinct chemical shifts of the nuclei involved. H2 to H8/H6 contacts, H2 to H1′, H1′ to H1′ and H1′ to H8/H6 contacts were observed where the first nuclei is the filtered nucleus, and the second the edited nucleus. Contacts of A-H1′ to G/C/U-H1′ were observable due to the lack of H5 protons afforded by deuterating C5. Using these contacts, four of the six base residues were immediately assigned and the remainder could be inferred.
of proton attached to a 13C nucleus. With our labeling pattern, this occurs in four discrete uncrowded regions of the spectrum due to distinct chemical shifts of the nuclei involved. H2 to H8/H6 contacts, H2 to H1′, H1′ to H1′ and H1′ to H8/H6 contacts were observed where the first nuclei is the filtered nucleus, and the second the edited nucleus. Contacts of A-H1′ to G/C/U-H1′ were observable due to the lack of H5 protons afforded by deuterating C5. Using these contacts, four of the six base residues were immediately assigned and the remainder could be inferred.
Figure 9.
A 6 nt bulge from the 61 nt hepadnaviral RNA element was assigned using a F1eF2f NOESY experiment. In helical regions, well defined contacts are seen from the H2 of adenine to a number of H1′ residues. However, in the bulge, the versatility of our labeling scheme is realized. The RNA was labeled with 1′,8-13C GTP, 2′,8-13C ATP, 1′,6-13C-5-2H UTP, and 1′,6-13C-5-2H CTP. Protons attached to 12C nuclei are highlighted in cyan. During the F1eF2f NOESY experiment three classes of cross-peaks were defined. H2 to G/U/C H1′ contacts are depicted in grey dotted lines. H2 to base (H6/H8) contacts are shown in dot/dash magenta lines. Finally, adenine H1′ to both base and sugar are shown as solid yellow.
While this pattern of NOE contacts was readily assigned in the 61 nt RNA, it was unclear that these are a general phenomenon. To address this concern, we examined RNA structures with bulges already deposited in the PDB (10). As a first example we used the Domain IIa of the IRES from HCV (64). The five nucleotide asymmetric central bulge contains three adenines (Supplementary Figure S9). A number of unique contacts are seen when analyzing the same contacts discussed above (Supplementary Table S3). No intra-residue contacts were counted for H1′ to H8 contacts, although they were always observed for Ade. Of great note, the 11 conformers of the structure when only including bulge residues are relatively well defined with a RMSD of 0.28 ± 0.09 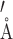 . If the HCV RNA was labeled with the patterns described above, we predict a number of contacts throughout the bulge would appear in a F1eF2f spectrum. We also analyzed the bulge of A-Site and HIV-TAR, two well-characterized RNAs. Both RNAs had significantly higher RMSDs of 1.1 ± 0.2
. If the HCV RNA was labeled with the patterns described above, we predict a number of contacts throughout the bulge would appear in a F1eF2f spectrum. We also analyzed the bulge of A-Site and HIV-TAR, two well-characterized RNAs. Both RNAs had significantly higher RMSDs of 1.1 ± 0.2 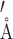 and 2.9 ± 0.9. This made a similar analysis of possible NOESY contacts difficult because different reported minimal energy structures had different numbers of possible contacts. Bulges are most likely highly dynamic in solution, thus differences in structure may be less of an artifact and more of realistic phenomenon. However, it is possible that the lowest energy conformations of the bulge were not captured because of an inability to observe the contacts that we observe here, which could undoubtedly provide constraints in hard-to-define bulge regions. Regardless, a similar analysis as above for the lowest energy structure is shown in Supplementary Tables S4 and S5. Assuming these structures are representative, a number of contacts to the H2 and H1′ of Ade would again be observed.
and 2.9 ± 0.9. This made a similar analysis of possible NOESY contacts difficult because different reported minimal energy structures had different numbers of possible contacts. Bulges are most likely highly dynamic in solution, thus differences in structure may be less of an artifact and more of realistic phenomenon. However, it is possible that the lowest energy conformations of the bulge were not captured because of an inability to observe the contacts that we observe here, which could undoubtedly provide constraints in hard-to-define bulge regions. Regardless, a similar analysis as above for the lowest energy structure is shown in Supplementary Tables S4 and S5. Assuming these structures are representative, a number of contacts to the H2 and H1′ of Ade would again be observed.
CONCLUSIONS
Identifying A-H2 resonances is of great importance when assigning NOESY peaks as they provide unique same-strand and cross-strand inter-nucleotide connectivities to H1′ (29). A similar strategy has been adopted for large RNAs in which multiple samples with and without deuteration at the A-H2 position are used to determine cross-peaks arising from H2 to H1′ through-space transfers (14, 65). Through deuteration, this strategy provides tools for assigning large RNAs by eliminating extraneous NOESY transfer pathways, improving signal/noise, and reducing spectral crowding and complexity. Drawbacks of this approach include cost of deuterated NTPs and the need for six or more deuterated samples. For large RNAs (>25 kDa), this deuteration strategy may be the only feasible method for NMR resonance assignment. For small to medium RNAs (<25 kDa), we recommend our new isotopic labelling strategy as a means of reducing overall resonance assignment time and material cost. We have shown a full analysis of a 27 nt RNA as well as the expansion of our method to a 61 nt viral RNA element including the assignment of often difficult bulge regions. As we have begun to explore with our H5 deuterated sample, combining site-specific deuteration with our selective 13C-labeling might prove useful for RNAs >70 nt.
Complete assignment of the H1′, H2′, H2, H5, H6 and H8 resonances was achieved from a single NMR sample; however, since chemical shift assignments were not publicly available, we found it prudent to confirm these with heteronuclear through-bond experiments on a uniformly 13C/15N-labeled sample. In our work, we used imino 1H–1H NOESY, HNN-COSY, one- and two-bond 15N HSQC, 13C HSQC, HCN and HCCH-COSY experiments to confirm all the non-exchangeable resonance assignments. For more complex RNAs, an orthogonal sample may prove beneficial, if the labelling patterns of the ribose are switched such that GC is 13C-labeled at C2′ and AU is 13C-labeled at C1′. Using an orthogonal labeling scheme one could still follow the general protocol outlined in Steps 2–5 of our logical assignment scheme. Combining this selective labeling strategy with segmental labeling (66–70) and deuteration (8, 71–73) will extend the usefulness of this technique to very large RNA systems. In future work, we aim to synthesize deoxyribonucleotides from our ribonucleotides using ribonucleotide reductase (74) in order to apply a similar resonance assignment strategy to DNA. We also aim to automate the assignment process using our generalized assignment approach.
The benefit of complete resonance assignment from a minimal set of isotopically enriched RNA samples is highly desirable, reducing both the cost and time associated with sample preparation and analysis. Our unique labeling scheme also provides isolated 13C–1H spin systems probes, and these are ideal for measuring relaxation dispersion, RDC and RCSA by removing 13C–13C coupling and reducing spectral crowding (75–78). Although we limit the number of probe sites available, the utility of this labeling scheme for assignment, dynamics and structural refinement by NMR will allow accelerated access to atomic resolution studies of RNA by greatly reducing the bottleneck of RNA chemical shift resonance assignment.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of General Medical Sciences [P50GM103297 to T.K.D. and B.A.J.] (in part); National Science Foundation for NMR Instrumentation [DBI1040158 to T.K.D.]; Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Human Services (to S.L.G.). Funding for open access charge: National Institute of General Medical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Breaker R.R. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011; 43:867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mattick J.S. A new paradigm for developmental biology. J. Exp. Biol. 2007; 210:1526–1547. [DOI] [PubMed] [Google Scholar]
- 3. Serganov A., Nudler E.. A decade of riboswitches. Cell. 2013; 152:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steitz T.A. A structural understanding of the dynamic ribosome machine. Nat. Rev. Mol. Cell Biol. 2008; 9:242–253. [DOI] [PubMed] [Google Scholar]
- 5. Ramakrishnan V. The ribosome emerges from a black box. Cell. 2014; 159:979–984. [DOI] [PubMed] [Google Scholar]
- 6. Hang J., Wan R., Yan C., Shi Y.. Structural basis of pre-mRNA splicing. Science. 2015; 349:1191–1198. [DOI] [PubMed] [Google Scholar]
- 7. Newman A.J., Nagai K.. Structural studies of the spliceosome: blind men and an elephant. Curr. Opin. Struct. Biol. 2010; 20:82–89. [DOI] [PubMed] [Google Scholar]
- 8. Keane S.C., Heng X., Lu K., Kharytonchyk S., Ramakrishnan V., Carter G., Barton S., Hosic A., Florwick A., Santos J. et al. . RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015; 348:917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dayie K.T. Key labeling technologies to tackle sizeable problems in RNA structural biology. Int. J. Mol. Sci. 2008; 9:1214–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dutta S., Burkhardt K., Young J., Swaminathan G.J., Matsuura T., Henrick K., Nakamura H., Berman H.M.. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009; 42:1–13. [DOI] [PubMed] [Google Scholar]
- 12. Nozinovic S., Fürtig B., Jonker H.R.A., Richter C., Schwalbe H.. High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res. 2010; 38:683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu K., Miyazaki Y., Summers M.F.. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2010; 46:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. D’Souza V., Dey A., Habib D., Summers M.F.. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J. Mol. Biol. 2004; 337:427–442. [DOI] [PubMed] [Google Scholar]
- 15. Butcher S.E., Dieckmann T., Feigon J.. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997; 16:7490–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peterson R.D., Theimer C.A., Wu H., Feigon J.. New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA-protein complexes. J. Biomol. NMR. 2004; 28:59–67. [DOI] [PubMed] [Google Scholar]
- 17. Dieckmann T., Feigon J.. Assignment methodology for larger RNA oligonucleotides: application to an ATP-binding RNA aptamer. J. Biomol. NMR. 1997; 9:259–272. [DOI] [PubMed] [Google Scholar]
- 18. Pardi A. Multidimensional heteronuclear NMR experiments for structure determination of isotopically labeled RNA. Methods Enzymol. 1995; 261:350–380. [DOI] [PubMed] [Google Scholar]
- 19. Cromsigt J., van Buuren B., Schleucher J., Wijmenga S.. Resonance assignment and structure determination for RNA. Methods Enzymol. 2001; 338:371–399. [DOI] [PubMed] [Google Scholar]
- 20. Lukavsky P.J., Puglisi J.D.. RNAPack: an integrated NMR approach to RNA structure determination. Methods. 2001; 25:316–332. [DOI] [PubMed] [Google Scholar]
- 21. Wijmenga S.S., van Buuren B.N.M.. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998; 32:287–387. [Google Scholar]
- 22. Varani G., Aboul-Ela F., Allain F.H.-T.. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spectrosc. 1996; 29:51–127. [Google Scholar]
- 23. Fürtig B., Richter C., Wöhnert J., Schwalbe H.. NMR spectroscopy of RNA. ChemBioChem. 2003; 4:936–962. [DOI] [PubMed] [Google Scholar]
- 24. Dayie K.T. Resolution enhanced homonuclear carbon decoupled triple resonance experiments for unambiguous RNA structural characterization. J. Biomol. NMR. 2005; 32:129–139. [DOI] [PubMed] [Google Scholar]
- 25. Bahrami A., Clos L.J., Markley J.L., Butcher S.E., Eghbalnia H.R.. RNA-PAIRS: RNA probabilistic assignment of imino resonance shifts. J. Biomol. NMR. 2012; 52:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aeschbacher T., Schmidt E., Blatter M., Maris C., Duss O., Allain F.H.-T., Güntert P., Schubert M.. Automated and assisted RNA resonance assignment using NMR chemical shift statistics. Nucleic Acids Res. 2013; 41:e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krähenbühl B., Lukavsky P., Wider G.. Strategy for automated NMR resonance assignment of RNA: application to 48-nucleotide K10. J. Biomol. NMR. 2014; 59:231–240. [DOI] [PubMed] [Google Scholar]
- 28. Krähenbühl B., El Bakkali I., Schmidt E., Güntert P., Wider G.. Automated NMR resonance assignment strategy for RNA via the phosphodiester backbone based on high-dimensional through-bond APSY experiments. J. Biomol. NMR. 2014; 59:87–93. [DOI] [PubMed] [Google Scholar]
- 29. Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley Interscience. 1986; NY. [Google Scholar]
- 30. Alvarado L.J., LeBlanc R.M., Longhini A.P., Keane S.C., Jain N., Yildiz Z.F., Tolbert B.S., D’Souza V.M., Summers M.F., Kreutz C. et al. . Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. ChemBioChem. 2014; 15:1573–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alvarado L.J., Longhini A.P., LeBlanc R.M., Chen B., Kreutz C., Dayie T.K.. Chemo-enzymatic synthesis of selectively 13C/15N-labeled RNA for NMR structural and dynamics studies. Methods Enzymol. 2014; 549:133–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Longhini A.P., LeBlanc R.M., Becette O., Salguero C., Wunderlich C.H., Johnson B.A., D’Souza V.M., Kreutz C., Dayie T.K.. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2015; 44:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Breeze A.L. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc. 2000; 36:323–372. [Google Scholar]
- 34. Duszczyk M.M., Zanier K., Sattler M.. A NMR strategy to unambiguously distinguish nucleic acid hairpin and duplex conformations applied to a Xist RNA A-repeat. Nucleic Acids Res. 2008; 36:7068–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Webba da Silva M. NMR methods for studying quadruplex nucleic acids. Methods. 2007; 43:264–277. [DOI] [PubMed] [Google Scholar]
- 36. Fourmy D., Recht M.I., Blanchard S.C., Puglisi J.D.. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996; 274:1367–1371. [DOI] [PubMed] [Google Scholar]
- 37. Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C.. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Norris M., Fetler B., Marchant J., Johnson B.A.. NMRFx Processor: a cross-platform NMR data processing program. J. Biomol. NMR. 2016; 65:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson B.A., Blevins R.A.. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994; 4:603–614. [DOI] [PubMed] [Google Scholar]
- 40. Hwang T.L., Shaka A.J.. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A. 1995; 112:275–279. [Google Scholar]
- 41. Palmer A.G., Cavanagh J., Wright P.E., Rance M.. Sensitivity improvement in proton-detected two-dimensional heteronuclear correlation NMR spectroscopy. J. Magn. Reson. 1991; 93:151–170. [Google Scholar]
- 42. Kay L., Keifer P., Saarinen T.. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992; 114:10663–10665. [Google Scholar]
- 43. Schleucher J., Schwendinger M., Sattler M., Schmidt P., Schedletzky O., Glaser S.J.J., S⊘rensen O.W., Griesinger C., Srensen O.W., Griesinger C.. A general enhancement scheme in heteronuclear multidimensional NMR employing pulsed field gradients. J. Biomol. NMR. 1994; 4:301–306.8019138 [Google Scholar]
- 44. Dingley A.J., Grzesiek S.. Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2 J NN couplings. J. Am. Chem. Soc. 1998; 120:8293–8297. [Google Scholar]
- 45. Grzesiek S., Bax A.. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993; 115:12593–12594. [Google Scholar]
- 46. Piotto M., Saudek V., Sklenář V.. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992; 2:661–665. [DOI] [PubMed] [Google Scholar]
- 47. Brutscher B., Simorre J.P.. Transverse relaxation optimized HCN experiment for nucleic acids: combining the advantages of TROSY and MQ spin evolution. J. Biomol. NMR. 2001; 21:367–372. [DOI] [PubMed] [Google Scholar]
- 48. Sklenár V., Peterson R.D., Rejante M.R., Feigon J.. Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N,13C-labeled RNA oligonucleotides. J. Biomol. NMR. 1993; 3:721–727. [DOI] [PubMed] [Google Scholar]
- 49. Kay L.E., Xu G.Y., Singer A.U., Muhandiram D.R., Formankay J.D.. A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. Ser. B. 1993; 101:333–337. [Google Scholar]
- 50. Pardi A., Nikonowicz E.P.. Simple procedure for resonance assignment of the sugar protons in carbon-13 labeled RNAs. J. Am. Chem. Soc. 1992; 114:9202–9203. [Google Scholar]
- 51. Nikonowicz E.P., Pardi A.. An efficient procedure for assignment of the proton, carbon and nitrogen resonances in 13C/15N labeled nucleic acids. J. Mol. Biol. 1993; 232:1141–1156. [DOI] [PubMed] [Google Scholar]
- 52. Xue Y., Kellogg D., Kimsey I.J., Sathyamoorthy B., Stein Z.W., McBrairty M., Al-Hashimi H.M.. Characterizing RNA excited states using NMR relaxation dispersion. Methods Enzymol. 2015; 558:39–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brown J.D., Summers M.F., Johnson B.A.. Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. J. Biomol. NMR. 2015; 63:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Frank A.T., Law S.M., Brooks C.L.. A simple and fast approach for predicting (1)H and (13)C chemical shifts: toward chemical shift-guided simulations of RNA. J. Phys. Chem. B. 2014; 118:12168–12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lynch S.R., Puglisi J.D.. Structure of a eukaryotic decoding region A-site RNA. J. Mol. Biol. 2001; 306:1023–1035. [DOI] [PubMed] [Google Scholar]
- 56. Dethoff E.A., Petzold K., Chugh J., Casiano-Negroni A., Al-hashimi H.M.. Visualizing transient low-populated structures of RNA. Nature. 2012; 491:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoffman D.W. Resolution of the 1H-1H NOE spectrum of RNA into three dimensions using 15N-1H two-bond couplings. J. Biomol. NMR. 2000; 16:165–169. [DOI] [PubMed] [Google Scholar]
- 58. Simon B., Zanier K., Sattler M.. A TROSY relayed HCCH-COSY experiment for correlating adenine H2/H8 resonances in uniformly 13C-labeled RNA molecules. J. Biomol. NMR. 2001; 20:173–176. [DOI] [PubMed] [Google Scholar]
- 59. Fürtig B., Richter C., Bermel W., Schwalbe H.. New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J. Biomol. NMR. 2004; 28:69–79. [DOI] [PubMed] [Google Scholar]
- 60. Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004; 278:313–352. [DOI] [PubMed] [Google Scholar]
- 61. Kellogg G.W., Szewczak A.A., Moore P.B.. Two-dimensional hetero-TOCSY-NOESY. Correlation of phosphorus-31 resonances with anomeric and aromatic proton resonances in RNA. J. Am. Chem. Soc. 1992; 114:2727–2728. [Google Scholar]
- 62. Marino J.P., Schwalbe H., Anklin C., Bermel W., Crothers D.M., Griesinger C.. Three-dimensional triple-resonance 1H, 13C, 31P experiment: sequential through-bond correlation of ribose protons and intervening phosphorus along the RNA oligonucleotide backbone. J. Am. Chem. Soc. 1994; 116:6472–6473. [Google Scholar]
- 63. Richter C., Kovacs H., Buck J., Wacker A., Fürtig B., Bermel W., Schwalbe H.. 13C-direct detected NMR experiments for the sequential J-based resonance assignment of RNA oligonucleotides. J. Biomol. NMR. 2010; 47:259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Locker N., Easton L.E., Lukavsky P.J.. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007; 26:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. D’Souza V., Summers M.F.. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004; 431:586–590. [DOI] [PubMed] [Google Scholar]
- 66. Duss O., Diarra Dit Konté N., Allain F.H.-T.. Cut and paste RNA for nuclear magnetic resonance, paramagnetic resonance enhancement, and electron paramagnetic resonance structural studies. Methods Enzymol. 2015; 565:537–562. [DOI] [PubMed] [Google Scholar]
- 67. Nelissen F.H.T., van Gammeren A.J., Tessari M., Girard F.C., Heus H.A., Wijmenga S.S.. Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res. 2008; 36:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Duss O., Maris C., von Schroetter C., Allain F.H.-T.. A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res. 2010; 38:e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tzakos A.G., Easton L.E., Lukavsky P.J.. Preparation of large RNA oligonucleotides with complementary isotope-labeled segments for NMR structural studies. Nat. Protoc. 2007; 2:2139–2147. [DOI] [PubMed] [Google Scholar]
- 70. Kim I., Lukavsky P.J., Puglisi J.D.. NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J. Am. Chem. Soc. 2002; 124:9338–9339. [DOI] [PubMed] [Google Scholar]
- 71. Duss O., Lukavsky P.J., Allain F.H.-T.H.-T.. Isotope labeling and segmental labeling of larger RNAs for NMR structural studies. Adv. Exp. Med. Biol. 2012; 992:121–144. [DOI] [PubMed] [Google Scholar]
- 72. Scott L.G., Tolbert T.J., Williamson J.R.. Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol. 2000; 317:18–38. [DOI] [PubMed] [Google Scholar]
- 73. Miyazaki Y., Irobalieva R.N., Tolbert B.S., Smalls-Mantey A., Iyalla K., Loeliger K., D’Souza V., Khant H., Schmid M.F., Garcia E.L. et al. . Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 2010; 404:751–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kolberg M., Strand K.R., Graff P., Andersson K.K.. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta. 2004; 1699:1–34. [DOI] [PubMed] [Google Scholar]
- 75. Wunderlich C.H., Juen M.A., LeBlanc R.M., Longhini A.P., Dayie T.K., Kreutz C.. Stable isotope-labeled RNA phosphoramidites to facilitate dynamics by NMR. Methods Enzymol. 2015; 565:461–494. [DOI] [PubMed] [Google Scholar]
- 76. Ying J., Grishaev A., Bryce D.L., Bax A.. Chemical shift tensors of protonated base carbons in helical RNA and DNA from NMR relaxation and liquid crystal measurements. J. Am. Chem. Soc. 2006; 128:11443–11454. [DOI] [PubMed] [Google Scholar]
- 77. Hansen A.L., Al-hashimi H.M.. Dynamics of large elongated RNA by NMR carbon relaxation. J. Am. Chem. Soc. 2007; 129:16072–16082. [DOI] [PubMed] [Google Scholar]
- 78. Hansen A.L., Al-Hashimi H.M.. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: towards new long-range orientational constraints. J. Magn. Reson. 2006; 179:299–307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.