Abstract
The Human antigen R protein (HuR) is an RNA-binding protein that recognizes U/AU-rich elements in diverse RNAs through two RNA-recognition motifs, RRM1 and RRM2, and post-transcriptionally regulates the fate of target RNAs. The natural product dihydrotanshinone-I (DHTS) prevents the association of HuR and target RNAs in vitro and in cultured cells by interfering with the binding of HuR to RNA. Here, we report the structural determinants of the interaction between DHTS and HuR and the impact of DHTS on HuR binding to target mRNAs transcriptome-wide. NMR titration and Molecular Dynamics simulation identified the residues within RRM1 and RRM2 responsible for the interaction between DHTS and HuR. RNA Electromobility Shifts and Alpha Screen Assays showed that DHTS interacts with HuR through the same binding regions as target RNAs, stabilizing HuR in a locked conformation that hampers RNA binding competitively. HuR ribonucleoprotein immunoprecipitation followed by microarray (RIP-chip) analysis showed that DHTS treatment of HeLa cells paradoxically enriched HuR binding to mRNAs with longer 3′UTR and with higher density of U/AU-rich elements, suggesting that DHTS inhibits the association of HuR to weaker target mRNAs. In vivo, DHTS potently inhibited xenograft tumor growth in a HuR-dependent model without systemic toxicity.
INTRODUCTION
The Human antigen R (ELAVL1, HuR) is an ubiquitously expressed RNA-binding protein, belonging to the ELAVL (Embryonic Lethal Abnormal Vision)-like family, that preferentially binds U- and AU-rich elements (AREs) abundant in the 3′ untranslated regions (3′UTRs) of certain mRNAs. It is mainly localized within the nucleus (90%), where it exerts post-transcriptional functions such as splicing (1–4) and alternative polyadenylation (5–7), and is able to shuttle to the cytoplasm, where it mainly regulates the fate of target RNAs (8). HuR regulates cellular responses to differentiation, senescence, inflammatory factors, and immune stimuli by tightly controlling the post-transcriptional fate of specific mRNAs (9–12). Notably, HuR binds to and regulates the half-life of mRNAs and/or the translation of mRNAs encoding key inflammatory cytokines and interleukins, such as tumor necrosis factor-α (TNFα) (13) and interleukin IL-1β, IL-3 (14), IL-6 (15), IL-8, IL-10, IL-4, CXCL1 (16–18), in turn governing the development and maturation of B and T lymphocytes (19,20). HuR is highly expressed in many cancer types, and is believed to promote tumorigenesis by interacting with mRNAs encoding proteins implicated in cell proliferation and survival, angiogenesis, invasion, pharmacoresistance and metastasis (21–27). The role of HuR in inflammation and cancer has prompted the search for inhibitors/modulators to interfere with its biological activity (28–32).
A number of natural and synthetic molecules have been found to interfere with the formation of HuR/mRNA complexes in vitro (29,32–35). The structural basis of the interaction of such molecules with HuR is still poorly characterized. HuR contains three highly conserved RNA recognition motifs (RRMs) among which the first two, RRM1 and RRM2, bind with high affinity to U/AU-rich RNA (36). By contrast, the third domain, RRM3, contributes to the interaction of HuR with poly(A) tails of target mRNA, and is believed to be involved in mRNA-induced cooperative assembly of HuR oligomers (37) (Figure 1A). Each RRM domain adopts a β1–α1–β2–β3–α2–β4 topology with the two α-helices packed in an antiparallel four-stranded β-sheet. Residues at conserved positions located on β-strands 1 and 3 are essential for mRNA binding, and are either involved in stacking interactions with mRNA bases or inserted between two sugar rings (38). At present, two crystal structures of the isolated RRM1 domain (PDB codes 3HI9 and 4FXV (39)) and two of the RRM1–RRM2 domains (PDB codes 4ED5 (40) and 4EGL) are available in the Protein Data Bank (PDB). Conformational changes occurring on the tandem RRM1–RRM2 domains are crucial for mRNA binding (40). As suggested by the crystal structures, the tandem construct adopts an ‘open’ conformation in the free form and a ‘closed’ conformation when the RRM1 and RRM2 domains bind mRNA (Figure 1B and C). This hypothesis is supported by SAXS data that show an equilibrium among ‘closed’ and ‘open’ conformations for HuR in solution, in the absence of mRNA. When a target mRNA sequence is present, the two domains form a stable complex with mRNA and adopt a ‘closed’ globular conformation around the mRNA strand (41).
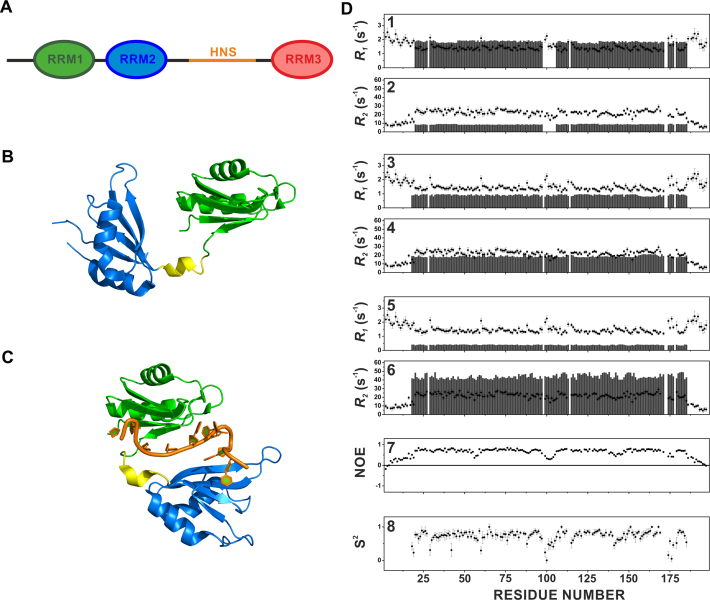
Figure 1.
Multidomain organization of HuR (A). The RRM1–RRM2 tandem domains (RRM1 aminoacids (aa) Thr20-Pro98 and RRM2 aa Ala106-Asn186) are separated by a short linker of 7 residues (aa Ser99-Asp105), while RRM3 (aa Trp244-Asn322) is connected to the other two domains by a long hinge region of about 60 residues (aa Pro187-Gly243), which includes the HuR Nucleocytoplasmic Shuttling (HNS) sequence, responsible for nuclear/cytoplasmic shuttling of HuR. RRM1 is represented in green, RRM2 in blue and RRM3 in red. The HuR Nucleocytoplasmic Shuttling Sequence (HNS) is indicated in orange. Cartoon representation of the ‘open’ structure of the tandem RRM1–RRM2 domains crystallized in the absence of RNA (pdb code 4ED5) (B), and of the ‘closed’ structure of the tandem RRM1–RRM2 domains in complex with RNA (pdb code 4EGL) (C). The two domains and the linker are highlighted with different colors (RRM1 in green, linker in yellow and RRM2 in blue). (D) Comparison of experimental backbone 15NH R1 values for RRM1–RRM2 (data collected at 298 K, black filled circles) with the calculated values (grey bars) for isolated RRM1 and RRM2 domains (1), for monomeric RRM1–RRM2 construct (3) and for rigid dimeric adduct (5). Comparison of experimental backbone 15NH R2 values for RRM1–RRM2 (data collected at 298 K, black filled circles) with the calculated values (grey bars) for isolated RRM1 and RRM2 domains (2), for monomeric RRM1–RRM2 construct (4) and for rigid dimeric adduct (6). Experimental NOE values for RRM1–RRM2 (data collected at 298 K) (7) and S2 order parameter calculated with the program TENSOR2 (8).
Dihydrotanshinone-I (DHTS) is a natural compound present in Salvia miltiorrhiza that interferes with the formation of HuR/RNA complexes (31). However, there is currently no detailed information about the specific interaction of DHTS with HuR or about the perturbations of the RNA-binding abilities of HuR transcriptome-wide. Here, we report the analysis of the interaction between DHTS and HuR by NMR, Molecular Dynamics simulation, and mutagenesis experiments. We have characterized the internal dynamics of the HuR RRM1–RRM2 domains, and have used this information to analyze the role of the two domains in ligand binding. In this respect, the identification of the flexibility of the two domains, RRM1 and RRM2, was particularly interesting. Moreover, ribonucleoprotein immunoprecipitation followed by microarray analysis revealed that DHTS dysregulates HuR by enriching HuR binding towards longer mRNAs highly rich in U/AU-rich 3′UTRs, including the mRNAs that encode apoptotic and cell-cycle regulatory proteins in cells, and inhibits cancer cell growth in vivo.
MATERIALS AND METHODS
Cell lines and reagents
Human cervical adenocarcinoma HeLa cells (ATCC® CCL2™), colon carcinoma cells HCT116 (ATCC; Manassas, VA) were cultured in standard Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (Gibco/Invitrogen), 1% l-glutamine (Gibco/Invitrogen), 1% penicillin–streptomycin (Invitrogen), and growth conditions at 37°C in 5% humidified CO2 incubators. Creation and characterization of CRISPR/Cas9-mediated knockout of the ELAVL1 gene in HCT116 cells was accomplished as described (42). Dihydrotanshinone I (D0947) was purchased from Sigma and dissolved in ultrapure dimethylsulfoxide (DMSO, Amresco, N182) to 10 mM final concentration. Antibodies used recognized HuR (sc-71290; from Santa Cruz Biotechnology), His tag (anti-6x His (ab1187; from Abcam)) and β-actin (Clone C4; MP Biomedicals).
Cell and tumor growth assays
Transient transfection of cells with a HuR expression construct (pcDNA3.1/Zeo/HuR-Flag) or empty vector was accomplished using Lipofectamine Plus (Gibco/Invitrogen) as described (43) for 48 h, following addition of 10 μM DHTS or vehicle. Cell survival was assayed using the MTT-based cell growth determination kit (Sigma-Aldrich) as previously described (30).
Anchorage- and serum-independent growth assays were accomplished by plating cells (20 cells per well) on 96-well ultra-low attachment plates (Corning) in in spheroid growth medium (DMEM supplemented with 10 mM HEPES, N2 supplement (1×; Gibco), B-27 supplement (1×; Gibco), Insulin Transferrin Selenium (1×; Gibco), FGF (10 ng/ml; Gibco) and EGF (20 ng/ml; Gibco)). After 3 days of growth, spheroids were treated with 10 μM DHTS or vehicle for 15 days changing medium every 3 days. Individual spheroids (n = 5–10/time point) were imaged every 3 days and area was measured using ImageJ software.
Six week-old athymic nude (Nu/Nu) mice were purchased from Jackson Laboratories and maintained under sterile conditions in cage micro-isolators according to approved IACUC guidelines. Parental HCT116 and a representative HuR knockout clone (2 × 106 cells) used between passages 14 and 23 were resuspended in PBS containing 50% Matrigel (Corning) and injected into the dorsal subcutaneous tissue (three mice/group with two tumors/mouse). Mice (three per group) received intraperitoneal (IP) injections of DHTS (10 mg/kg) dissolved in PBS/5% N-methyl pyrrolidine (NMP) (Sigma-Aldrich) or vehicle control every 48 h. Tumor volumes and body weight were measured three times per week using a caliper, and tumor volumes were calculated using the formula: volume = length × width2/2. Upon termination of the experiment, mice were euthanized and tumors were harvested.
AlphaScreen and RNA electromobility shift (REMSA) assays
Amplified Luminescent Proximity Homogenous Assay (Alpha) with a 5′-Biotinylated RNA probe (Bi-TNF, 5′-AUUAUUUAUUAUUUAUUUAUUAUUUA) was used with FL-HuR and four studied mutants. 384-well optiplates (PerkinElmer; 6007299) were used with 20 μl final volume in each well. Hooking points of all the respective proteins were determined. Reagents were used in the nanomolar range using the AlphaScreen His detection kit (PerkinElmer) in alpha buffer (25 mM HEPES pH 7.4, 100 mM NaCl, 0.1% BSA). Donor and acceptor beads were used at 10 μg/ml as their final concentration, proteins and constructs were according to their hooking points and incubated for 1 hour at room temperature. Plates were read for fluorescence signals in an Enspire plate reader instrument (PerkinElmer; 2300-001A), and specific binding was calculated by subtracting the background, obtained in the absence of the protein. For REMSA experiments, equimolar concentrations of purified RRMs and FAM-TNF RNA probes were used (32) in REMSA buffer (20 mM HEPES pH 7.5, 50 mM KCl, 0.5 μg BSA, 0.25% glycerol) together with reference doses of DHTS were run on the native Polyacrylamide (6%) gel, in 0.5× TBE buffer at 55 V for 90 min. The gel was analysed by a Typhoon instrument (GE Healthcare; 00-4277-85 AC).
R 1, R2 and NOE measurements
The experiments for the determination of 15N longitudinal and transverse relaxation rates and 15N–1H NOE (44) were recorded at 298 K and 700 MHz on 15N-enriched samples of the RRM1–RRM2 tandem domains of HuR. 15N Longitudinal relaxation rates (R1) were measured using a sequence modified to remove cross-correlation effects during the relaxation delay (45). Inversion recovery times ranging between 2.0 and 2500 ms, with a recycle delay of 3.5 s, were used for the experiments. 15N transverse relaxation rates (R2) were measured using a Carr-Purcell-Meiboom-Gill (CPMG) sequence (45,46), with delays ranging between 8.5 and 237.4 ms and a refocusing delay of 450 μs. Longitudinal and transverse relaxation rates were determined by fitting the cross-peak intensities as a function of the delay to a single-exponential decay using the Origin software. Heteronuclear NOE values were obtained from the ratio of the peak height for 1H-saturated and unsaturated spectra. Theoretical predictions of NH R1 and R2 values for RRM1–RRM2 tandem domains were made using HYDRONMR (47) and the pdb structure 4ED5 (40), considering (i) the isolated domains, (ii) the monomeric and (iii) the dimeric constructs.
Molecular dynamics simulation and analysis
The HuR-DHTS complex, as issuing from docking calculations (see SI for details), was submitted to MD simulation with NAMD (48) using the ff99SBildn Amber force field parameters (49,50) for proteins and the parameters recently developed by Allnér and co-workers for ions (51). Parameters for DHTS were generated in two steps. Initially, charges were computed using the restrained electrostatic potential (RESP) fitting procedure (52). The ESP was first calculated by means of the Gaussian09 package (53) using a 6–31G* basis set at Hartree-Fock level of theory, and then the RESP charges were obtained by a two-stages fitting procedure using the program RED (54,55). Missing bond, angle, torsion and improper torsion angle parameters were then generated using Antechamber (56). The complex was then solvated in a 15 Å layer cubic water box using the TIP3P water model parameters. Neutrality was ensured by adding five further Cl− ions. The final system size was ∼74 Å × 93 Å × 74 Å for a total number of atoms of ∼48 000. A 10 Å cutoff (switched at 8.0 Å) was used for atom pair interactions. The long-range electrostatic interactions were computed by means of the particle mesh Ewald (PME) method using a 1.0 Å grid spacing in periodic boundary conditions. The RATTLE algorithm was applied to constrain bonds involving hydrogen atoms, and thus a 2 fs integration time step could be used. More details in the Supplementary Methods.
RIP-chip protocol
To analyze the influence of DHTS on the interaction of HuR with endogenous mRNAs, immunoprecipitation (IP) of endogenous ribonucleoprotein complexes was performed as described previously (57). Briefly, HeLa cells were lysed in 20 mM Tris–HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.5% NP-40 for 10 min on ice and centrifuged at 15 000 × g for 10 min at 4°C. The supernatants were incubated for 2 h at 4°C with protein G Sepharose beads (GE Healthcare) coated either with anti-HuR or with control IgG antibodies (both from Santa Cruz Biotechnology). The beads were washed with NT2 buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), followed by incubation with 20 units of RNase-free DNase I for 15 min at 37°C to remove the DNA. The samples were then incubated for 15 min at 55°C with 0.1% SDS–0.5 mg/ml proteinase K to digest proteins. Microarray analysis was performed in duplicate (GEO number GSE94360). The RNA from the IP samples was extracted using phenol–chloroform, precipitated, and used for cDNA microarray analysis or RT-qPCR validation.
Analysis of enriched mRNAs
GC content, length and secondary structure density (computed as the fraction of unpaired nucleotides) for the UTRs of DEC and INC genes were obtained from the AURA 2 database (58), and plotted with the R software. The enrichment of post-transcriptional regulatory elements was performed with the Regulatory element enrichment feature of AURA 2. Gene ontology and pathway enrichment analyses were performed with the Enrichr tool (59) on GO (Biological process, Molecular function and Cellular Component) and pathways (KEGG and Reactome) libraries, using a five genes overlap and minimum combined score of 2 as significance threshold. GO terms were clustered by semantic similarity with the GoSemSim R package (60), and the cluster score computed as the average combined score of composing terms.
RESULTS
NMR resonance assignment
The 2D 1H–15N HSQC spectrum of tandem RRM1–RRM2 domains shows well-dispersed signals in agreement with a uniform and folded protein structure (Supplementary Figure S1). All the residues (but Pro172), including those forming the linker region that is crucial for the protein function, have been assigned (Supplementary Table S1, BMRB code: 27002). Our assignment of the tandem RRM1–RRM2 domains was also compared with the ones reported for the isolated RRM1 domain (61) as in the RRM1–RRM2 tandem domains (35). The resonances of residues forming the RRM1 domain are almost the same in the isolated domain (61), and in the RRM1–RRM2 construct. As reported by Wang and coworkers (35), this observation suggests that the RRM1 and RRM2 domains do not interact with each other when they form the tandem construct. Based on TALOS+ predictions, each domain in the RRM1–RRM2 construct is constituted by two α-helices and four β-strands, in agreement with the previously reported crystal structures of the RRM1 and RRM1–RRM2 domains of HuR (39,40,62) (Supplementary Figure S2).
Internal dynamics of RRM1–RRM2 tandem domains
To characterize protein dynamics, measurements of longitudinal (R1) and transverse (R2) relaxation rates of backbone amide nitrogens at 700 MHz, 1H Larmor frequency, and 298 K were performed on 15N-enriched samples of the RRM1–RRM2 construct. Theoretical estimates of R1 and R2 values were calculated for the following model structures: (i) the isolated RRM1 (T20-P98) and RRM2 (A106-N186) domains (Figure 1D, panels 1, 2), (ii) the monomeric RRM1–RRM2 construct (Figure 1D, panels 3, 4) and, (iii) the dimeric adduct (Figure 1D, panels 5, 6) (PDB 4ED5). Figure 1D shows the experimental R1 and R2 values (black circles) as well as the theoretical ones (grey bars). The comparison of experimental R1 and R2 data with theoretical values calculated for the isolated RRM1 and RRM2 domains shows that experimental R1 values were smaller and R2 values are larger than their theoretical counterparts (Figure 1D, panels 1, 2). At the same time, experimental R1 values were higher than theoretical estimates calculated for the monomeric construct in solution (Figure 1D, panel 3), indicating that the RRM1–RRM2 construct did not behave as a rigid body but instead displayed inter-domain flexibility, simulating a protein of lower molecular weight (63–65). Experimental R2 values were instead slightly higher than their theoretical counterparts, indicating the occurrence of possible aggregation phenomena (Figure 1D, panel 4). On the other hand, experimental R1 values were dramatically higher, and R2 dramatically lower than theoretical values calculated for the rigid dimer (Figure 1D, panel 5, 6), suggesting that the RRM1–RRM2 dimer was not present in solution as a stable complex. Further indication of the presence of inter-domain flexibility was provided by the 15N–1H NOE values for the linker residues Ser99 (0.46), Ser100 (0.34), Glu101 (0.31), Val102 (0.30), Ile103 (0.32) and Lys104 (0.40) (Figure 1D, panel 7). The small NOE values of the residues in the linker between the two domains are evidence of fast motions on ps-ns timescale (faster than the overall protein-tumbling rate). Very small NOE values are found also for the N- and C-terminal tails, and for some residues in the loops of RRM1 (Val56, Ala57, Gly58) and RRM2 (Gln141, Thr142, Leu145) domains. The order parameter S2 calculated with the program TENSOR2 (66) starting from experimental R1, R2 and NOE values is also reported in Figure 1D, panel 8. The S2 values confirm the presence of high flexibility between the two domains, and for some loops within each domain.
DHTS stabilizes HuR in a closed conformation and competes with mRNA for binding
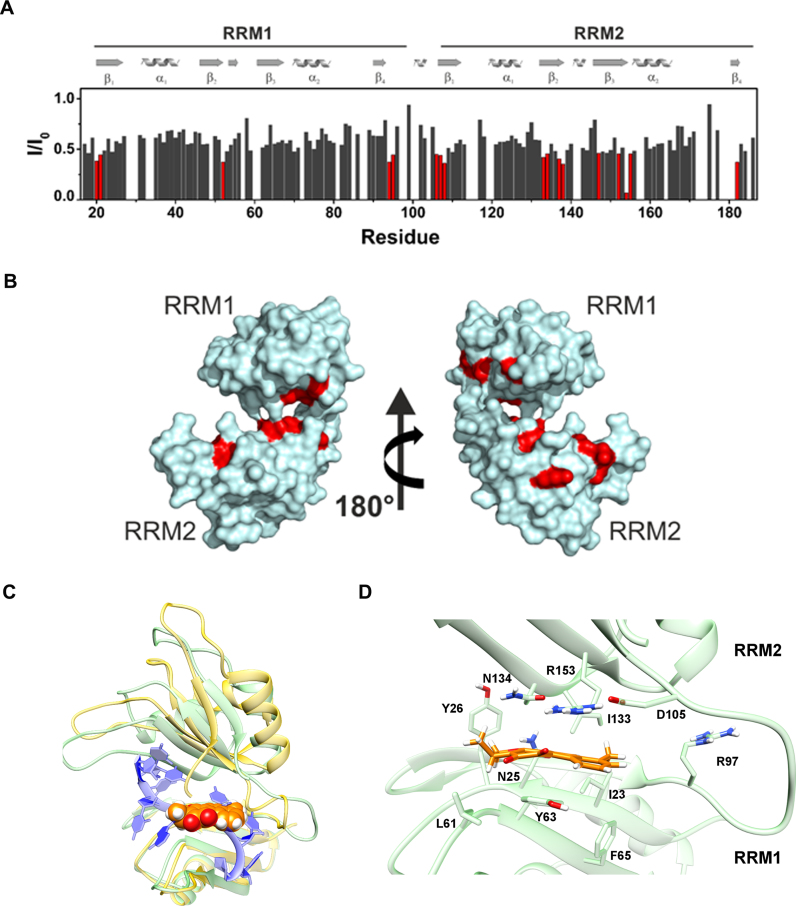
The interaction of HuR with DHTS was investigated through solution NMR. The significant precipitation of the ligand in the solution, occurring at the high concentrations required by the NMR analysis, prevented the estimation of the affinity constant. Nevertheless, after the addition of increasing amounts of DHTS to the protein, we observed a generalized decrease in signal intensity, with few residues (Thr20, Asn21, Ile52, Ser94, Tyr95, Ala106, Asn107, Leu108, Ile133, Asn134, Val137, Leu138, Arg147, Ile152, Phe154, Asp155, Lys182) experiencing a larger effect. These residues were located in the β-platform of both RRM domains (Figure 2A and B). The generalized decrease of signal intensity, and the distribution of affected residues over the large surfaces of the β-platform in each domain suggested an alteration of the equilibrium among ‘closed’ and ‘open’ conformations upon ligand binding. Specifically, the decrease of signal intensity was consistent with a mechanism where the small molecule stabilizes a ‘closed’ conformation of HuR. After the addition of DHTS to the protein, only negligible chemical shift changes occur. The residues experiencing the largest chemical shift perturbations are located close to the β-platforms (Supplementary Figure S3).
Figure 2.
Graphical representation of the intensity changes per residues of RRM1–RRM2 HuR protein (50 μM in solution) in the presence of DHTS (200 μM) (A) and surface representation of the closed conformation of HuR (pdb: 4ED5) (B) with the residues exhibiting the highest decreases in signal intensities highlighted in red. (C) Global view of the HuR (green cartoons)-DHTS (orange spheres) complex. Note how the insertion of DHTS into the mRNA binding cleft and the further closure of the latter, as compared to the mRNA-bound conformation (yellow), prevents the accommodation of the mRNA strand (blue ribbons). (D) Theoretical DHTS binding mode, as suggested by our MD simulation. DHTS and HuR residues involved in binding interactions are displayed as sticks.
With the dual aim of bringing some insights into DHTS binding mode and mechanism of action, we carried out a molecular modeling study. We employed a ‘tandem’ approach of docking calculations and molecular dynamics (MD) simulations in the attempt to follow the protein/ligand reciprocal conformational mutations. We first performed a docking calculation to the whole mRNA binding surface of HuR by means of AutoDock 4.2, which converged to a single binding pose. As a result, DHTS was found at the center of the mRNA binding cleft, which is shaped by the RRM1 and RRM2 domains (residues 18–95 and 107–185, respectively), and in proximity of the inter-domain linker. To allow both the ligand and the protein to fully adapt to each other, we performed an extended (0.5-μs) MD simulation. During the first 100 ns of simulation, DHTS was displaced from its starting position laterally along the surface of the RRM domain β-sheets, though always remaining bound to the HuR surface (Supplementary Figure S4A and B). This displacement was accompanied by a HuR conformational shift towards a ‘closed’ form featuring the two RRM domains even closer to each other (Supplementary Figure S4C), and establishing further inter-residues contacts (Supplementary Figure S4D) lacking in the mRNA-bound conformation, e.g. a salt bridge between Asp-105 and Arg-153, a backbone hydrogen bond between Ile-133 and Asn-25 or a hydrophobic contact between Ile-133 and Ile-23. For the sake of comparison, a further 500 ns MD was performed, starting from the X-ray HuR-RNA complex. The simulation resulted in an overall greater structural stability as compared to the HuR–DHTS complex (Supplementary Figure S5A and B). Furthermore, we observed neither a decrease in the distance between the two RRM domains nor a significant increase in the number of inter-domain residue contacts (Supplementary Figure S5C and D).
In the final HuR–DHTS complex, which remained stable for the rest of the simulation, DHTS is accommodated in a narrow, elongated and mostly hydrophobic pocket (Figure 2C) shaped by residues of the two RRM domain β-sheets (RRM1:Ile-23, Asn-25, Tyr-26, Leu-61, Tyr-63, Phe-65; RRM2: Asp-105, Ile-133, Asn-134, Arg-153) and of inter-domain linker (Ile-103 side chain and Lys-104, Arg-97 and Ala-96). The aromatic rings of DHTS establish several π-interactions, among which a cation-π interaction with Arg-153 and a NH–π interaction with the Asn-134 side chain. However, although DHTS is gripped between the two domains, it does not bind HuR rigidly, but rather gently sways along the binding surface (Figure 2D). Taken together, our structural data indicate that DHTS competes with RNA for the binding to HuR, interacting with the β-platform of both RRM domains in the proximity of the interdomain linker, and thus stabilizing HuR in a closed conformation.
The β-platforms of RRM domains are involved in DHTS inhibitory activity
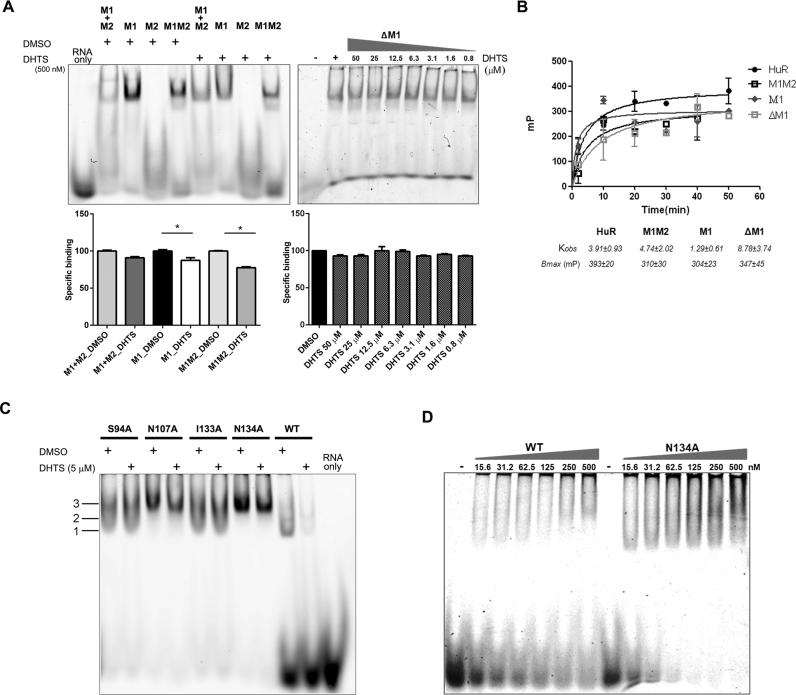
To support NMR and molecular modeling that addressed a specific interacting region on HuR, we produced HuR protein domains made of the first RRM domain (RRM1), of the second RRM domain (RRM2), of the RRM1–RRM2 wild-type RRM domains, and of RRM1 lacking 14 aminoacids at the C-terminus (ΔRRM1, missing residues from Ser94 to Asn107) and performed in vitro RNA binding experiments. Some of these residues belong to the inter-domain linker (Ser99–Asp105) and the others to the β-platform regions of the two domains, where some aminoacids experienced decreased intensity in the presence of DHTS (Ser94–Tyr95 in RRM1 and Ala106–Asn107 in RRM2). HuR domains were produced in Escherichia coli using the pET42 plasmid (31). We obtained the same purity for all the protein isoforms (Supplementary Figure S6). As single domains lose RNA-binding activity very quickly, the in vitro activity of the single protein domains and of their combination was evaluated by REMSA, mixing equimolar concentration of each, freshly prepared, with 0.2 μM of the FAM-ARE ssRNA probe (Figure 3A). The RRM1–RRM2 isoform was used as positive control because it displayed a similar Kd (2.62 ± 0.6 nM (31)) to the FL HuR protein, and because it was used in the NMR experiments (35). RRM1 retained the capability to recognize mRNA substrates (40), however with a lower affinity compared to the RRM1–RRM2 construct, probably indicating a change in the stoichiometry of cooperative protein binding (Figure 3A, Supplementary Figure S7) (40,62).
Figure 3.
The inter-domain region between RRM1 and RRM2 is crucial for RNA and DHTS binding. (A) On left, representative REMSAs of at least three independent protein preparations of recombinant RRM1 + RRM2 (M1 + M2) domains, RRM1 (M1), RRM2 (M2), RRM1–RRM2 (M1M2) HuR proteins. REMSAs were performed with 0.2 μM of protein, 0.2 μM of Cy-3 RNA probe and DMSO or DHTS at indicated doses. On right, representative REMSA performed with 2.5 μM of ΔRRM1 and 75 fM of probe RNA titrated with DHTS (concentration as shown in the legend). Densitometric quantification plotted below represents specific HuR–RNA binding challenged by DHTS. Mean ± SD refers to three independent experiments (n = 3, * indicates t-test P-value < 0.05). (B) Kinetic saturation binding experiment by fluorescence polarization. 200 nM wild-type protein or mutants were incubated with FAM-ARE RNA probe (100 nM). Full-length HuR and RRM1–RRM2 tandem domains (M1M2) have similar Kobs. RRM1 (M1) is binding faster (Kobs of ∼1 min), ) but deletion of the inter-domain region abolishes the binding properties of RRM1 (ΔM1) (Kobs of ∼8 min). (C) RNA- and DHTS-interacting amino acids are crucial for DHTS and RNA binding, and for protein dimerization. Representative REMSAs of at least three independent protein preparations of recombinant full-length HuR and indicated muteins. REMSAs were performed with 0.2 μM of protein, 0.2 μM of Cy-3 RNA probe, and DMSO or 5 μM DHTS. Muteins are insentive to DHTS and show different binding patterns to the RNA probe. (D) Representative REMSAs of at least three independent protein purification performed with increasing concentration of WT and HuR single point mutant N134A with 75 fM of probe RNA.
Importantly, the RRM1–RNA complex was still sensitive to DHTS. After removing 14 aminoacids, the binding capacity of ΔRRM1 to RNA was slightly reduced (∼20%) in comparison with the RRM1 domain (Figure 3A). Conversely, ΔRRM1 became resistant to DHTS, suggesting that this region is important for DHTS inhibitory activity (Figure 3A). REMSA performed with RRM2 and in vitro complementation of the two domains (RRM1 + M2 and ΔRRM1 + M2) did not provide information about the contact region of DHTS (Supplementary Figure S7B–D). By using fluorescence polarization, we analyzed the binding kinetics of proteins (200 nM) toward the FAM–ARE RNA probe (100 nM). We confirmed that full-length HuR and RRM1–RRM2 tandem domains behave almost similarly (average Kobs ∼4 min), reaching equilibrium after 10 min. On the other hand, the RRM1 domain rapidly recognized the substrate (Kobs ∼1 min), but this affinity was significantly impaired in the ΔRRM1 construct (Kobs of ∼8 min) (Figure 3B). Circular dichroism (CD) experiments performed at 10 μM concentration of both reagents, and NMR measurements ruled out a putative interaction between DHTS and RNA (Supplementary Figure S8A and B). Collectively, these findings show that the residues forming the β-platform and placed at the C-terminal in RRM1 domain are critical for RNA binding and the inhibitory activity of DHTS.
Single point mutations in the HuR–DHTS interacting region abolishes DHTS efficacy
Based on NMR data and an initial raw model of DHTS binding as derived by docking calculations, residues Ser94, Asn107, Ile133 and Asn134 of the RRM1–RRM2 inter-domain region were mutated to probe their relevance for DHTS binding. We produced residue-to-Alanine mutations in the full-length HuR sequence by site-directed mutagenesis (67) (Supplementary Figure S9). The preservation of the folding of the mutants was assessed by 1H 1D NMR spectra (Supplementary Figure S10). By reacting equimolar amount of wild-type HuR or muteins with 0.2 μM FAM–ssRNA, we observed an inhibitory effect of DHTS only with wild-type HuR, while any functional single point mutation led to resistance to DHTS (Figure 3C). Interestingly, we observed a qualitatively distinct binding profile among muteins: at least three discrete supershifts, numbered according to the molecular weight (1 being the lightest, 2, and 3), were detectable for wild-type HuR. Supershift 3 was markedly enriched in N134A and N107A, and to a lesser extent in I133A muteins (Figure 3C). This behavior could result from a higher efficiency in recognition and dimerization along the mRNA substrate, or alternatively from an aggregation-prone tendency of muteins that therefore aggregate on the same molecule of RNA probe. Indeed, mutants showed significantly lower Kd values, i.e. an increased affinity in saturation binding experiments with respect to wild-type HuR (Figure 3D, Supplementary Figures S11 and S12). Additionally, the raw signals at the hook point values for protein and RNA probes were significantly reduced in intensity compared to wild-type HuR, indicating a suboptimal assay environment, compatible with ligand self-aggregation (68).
According to these data, when Ser94, Asn107, Ile133 and Asn134 are mutated into Alanine, DHTS does not bind to any mutein. The four residues are thus crucial in providing the required flexibility to HuR for its mRNA binding function.
DHTS prevents HuR binding to low AU-rich density mRNA, but enriches it to high AU-rich density species
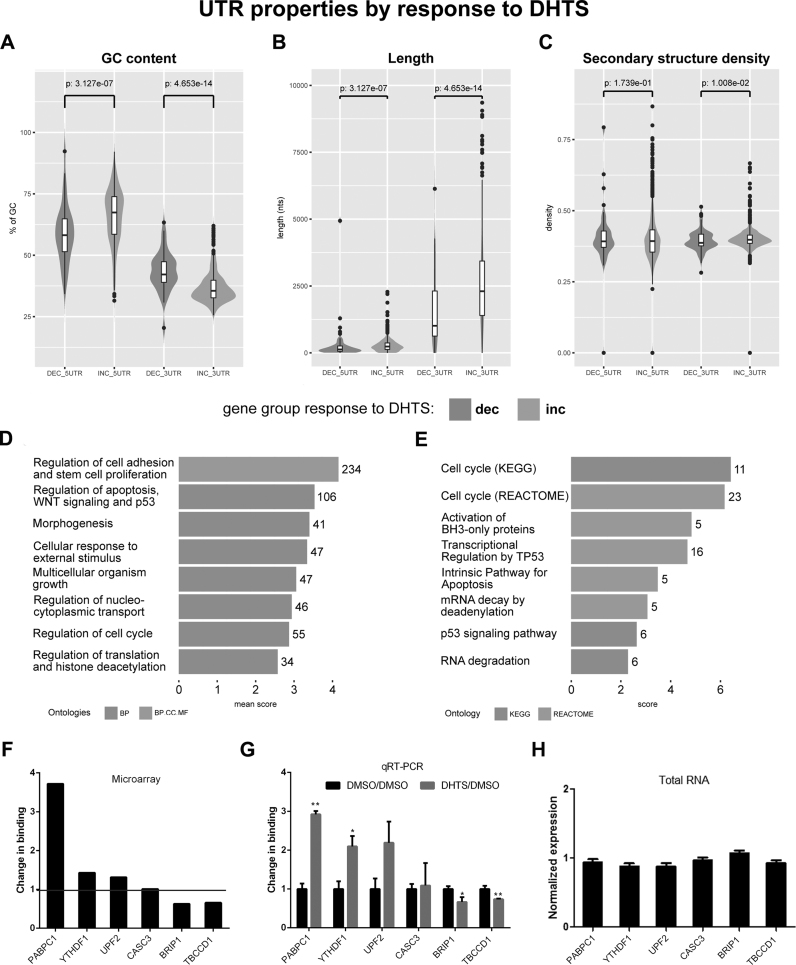
We evaluated the ability of DHTS to disrupt the interaction of HuR with its target mRNAs by employing a transcriptome-wide approach. We performed a RIP-chip (ribonucleoprotein immunoprecipitation followed by microarray) experiment in HeLa cells, and observed that, out of the 2306 mRNAs bound to HuR, DHTS only reduced binding of 79 transcripts. Conversely, 558 mRNAs were enriched after treatment with DHTS. Therefore, contrary to our expectations, we observed an overall enrichment of mRNAs bound to HuR after treatment with DHTS. We reasoned that DHTS could displace mRNAs that had lower affinity to HuR than DHTS itself had, and, paradoxically, provide the opportunity for mRNAs with higher affinity for HuR than DHTS to bind in higher copy numbers to HuR. We observed that HuR-bound mRNAs had relatively higher frequency of U/AU-rich segments compared to the frequency transcriptome-wide, as expected (69). However, the U/AU density in UTRs in enriched mRNAs was significantly higher than that in downregulated mRNAs (Supplementary Table S2, median of 1.37 ARE nts/100 nts for enriched genes, 0.97 for depleted genes; mean of 2.24 ARE nts/100 nts for enriched genes, 1.49 for depleted genes; maximum of 81.97 ARE nts/100 nts for enriched genes, 7.92 for depleted ones). Moreover, the highest differences were observed in the 3′UTR (Figure 4A), where the percentage of AU bases increased from 58% (depleted) to 65% (enriched). Additionally, 3′UTRs, but not 5′UTRs, were significantly longer for the enriched mRNAs (Figure 4B), while no differences in the density of secondary structure elements were observed (Figure 4C).
Figure 4.
Enriched and depleted mRNAs and their UTRs have distinct properties. (A) Distribution of GC content for depleted (dec) and enriched (inc) UTRs, with Wilcoxon test-P-values of the differences. (B) Length distribution for depleted (dec) and enriched (inc) UTRs, with Wilcoxon test-P-values of the differences. (C) Secondary structure density (computed as the fraction of unpaired nucleotides) of depleted (dec) and enriched (inc) UTRs, with Wilcoxon test-P-values of the differences. (D) Gene Ontology enrichment analysis for the enriched gene set. Number of genes belonging to each terms cluster is shown at the end of the corresponding bar. Mean score represents the mean of the Enrichr combined score for all belonging terms. GO classes found in each cluster are represented by the bars color. (E) KEGG and REACTOME pathway enrichment analysis for the enriched gene set. Number of genes belonging to each pathway is shown at the end of the corresponding bar. The score is the Enrichr combined score for the pathway. Pathway database of each entry is represented by the bars color. RIP of HuR in DHTS-treated HeLa cells. (F) Microarray data for selected targets. Fold enrichment of each mRNA during DHTS treatment is reported, black bars represent microarray values obtained. (G) Validation of microarray data by RT-qPCR. Gray bars represent fold enrichment of each mRNA during DHTS treatment compared with the control condition (DMSO). Comparison between microarray (F) and RT-qPCR data (G) shows similar results for enriched transcripts (PABPC1, YTHDF1 and UPF2 mRNAs), unchanged transcripts (CASC3 mRNA) and depleted transcripts (BRIP1 and TBCCD1 mRNAs). In RT-qPCR validation experiments, RPLP0 mRNA was used as an endogenous control mRNA that did not bind to HuR. Error bars represent SD. P-value is *<0.05. **<0.001. Mirocrarray experiments were done in duplicate (n = 2), qRT-PCR in triplicate (n = 3). (H) RT-qPCR analysis of mRNAs bound to of HuR showing no changes in total expression levels after DHTS treatment. RPLP0 mRNA was used as an endogenous control. Error bars represent SD. Experiments were done in triplicate (n = 3).
Functional analyses of depleted genes provided no considerable enrichment, due to the small size of the set and because it was partially composed of unannotated mRNAs. Enriched mRNAs (Figure 4D and E, Supplementary Table S3) showed that, during DHTS treatment, HuR bound preferentially to mRNAs encoding proteins with functions in the regulation of gene expression, cell cycle progression, and apoptosis. Data validation also suggested that the changes in HuR binding were independent of changes in total mRNA levels, as mRNA abundance was generally unchanged (Figure 4F and G). In summary, the ability of DHTS to displace HuR-bound RNAs was specifically limited to mRNAs with a low affinity for HuR, which generally displayed lower AU content in their 3′UTRs.
DHTS is effective on HuR-positive models in vitro and in vivo
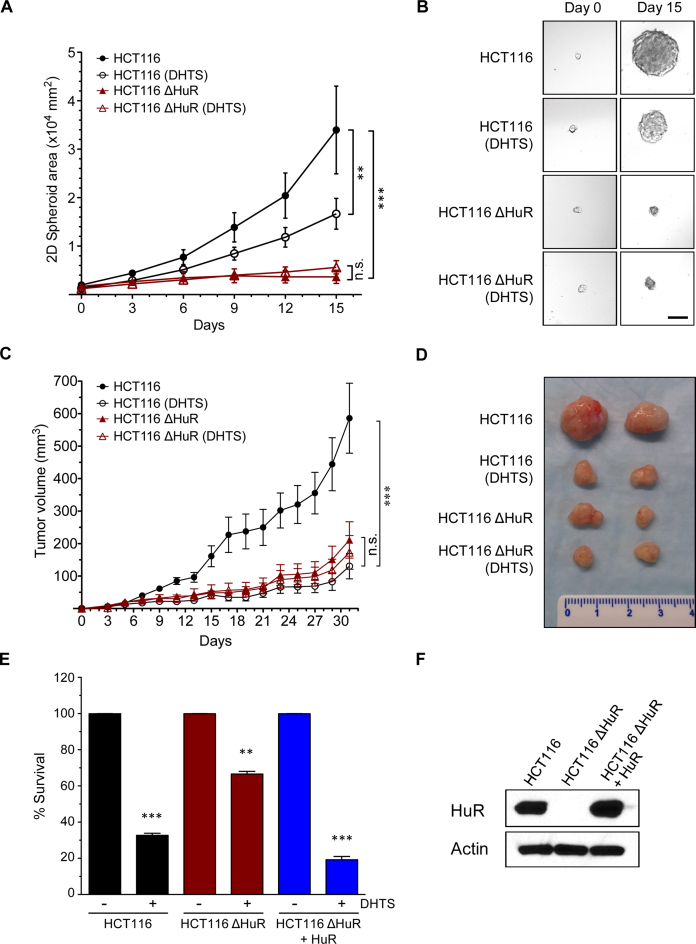
To evaluate this mechanism of action, in which a limited displacement of RNAs from HuR may be effective in HuR-dependent tumor growth, we studied tumor growth in vitro and in vivo. HCT116 colon cancer cells were used as a model, based on high endogenous levels of HuR and their sensitivity to HuR inhibition (30). Creation and characterization of CRISPR/Cas9-mediated knockout of the ELAVL1 gene in HCT116 cells was accomplished as described (42). HCT116 and HuR-knockout cells (HCT116ΔHuR) were grown under anchorage- and serum-independent conditions to facilitate tumorspheroid growth, and HuR was observed to be needed for tumorsphere growth (Figure 5A and B). In the presence of DHTS, growth of HCT116 spheroids was attenuated 2-fold, whereas DHTS did not impact HCT116ΔHuR sphere growth (Figure 5A and B). To test the effects of DHTS on tumor growth in vivo, mice bearing HCT116 cell xenografts received IP injections of DHTS (10 mg/kg body weight) or vehicle every 48 h. Over the course of the experiment, DHTS was well tolerated and mice did not display any signs of acute toxicity and maintained similar weights. Significant anti-tumor effects of DHTS were observed, with approximately a 4-fold reduction in tumor size (Figure 5C and D). Additionally, the efficacy of DHTS was strictly dependent on the presence of HuR. HCT116ΔHuR cells grew significantly more slowly and formed smaller tumors, but were completely insensitive to DHTS. Furthermore, this decreased growth sensitivity of DHTS in HuR-deficient cells could be restored with expression of HuR in HCT116ΔHuR cells (Figure 5E and F). These results indicate that DHTS has significant antitumor activity in vitro and in vivo without major systemic toxicity, along with demonstrating specificity of HuR inhibition.
Figure 5.
DHTS efficacy relies on HuR presence in vivo. (A) Spheroid growth of parental HCT116 (HCT116) and HuR-knockout cells (HCT116ΔHuR) treated with DHTS (10 μM) or vehicle. DHTS was added after 3 days of culture in spheroid growth medium on ultra-low adherence substrate and spheroid growth was tracked by imaging for 15 days. P-value is **<0.01, ***<0.001, n.s. = not significant. (B) Representative tumorsphere images from day 0 and day 15 of DHTS treatment. Scale bar = 100 μm. (C) Tumor growth of parental HCT116 and HuR-knockout cells (HCT116ΔHuR) xenografts in nude mice treated with 10 mg/kg DHTS or vehicle control every 48 h. P-value is ***<0.001, n.s. = not significant. (D) Representative tumors excised at day 31 are shown. (E) HuR-knockout cells (HCT116ΔHuR) were transfected a HuR-expression construct (+HuR), along with empty vector transfected parental HCT116 and HCT116ΔHuR cells. Cells were treated with DHTS (10 μM) or vehicle, and cell growth was assessed 6 days after the treatment using MTT assay. Cell survival was normalized to the respective control and are the average of three experiments. P-value is **<0.01, ***<0.001. (F) Western blot showing HuR complementation in HCT116ΔHuR+HuR cells and absence of HuR in HCT116ΔHuR cells. Actin was used as a loading control.
DISCUSSION
Previous efforts targeted towards HuR disruptors (28–30,32,70) identified small molecules that can inhibit the HuR–RNA interaction in the nanomolar range. Here, we have characterized from a structural and functional perspective the mechanism of action of DHTS, a disruptor of the HuR–RNA interaction, identifying the protein regions that promote the interaction, and providing hints for the rational design of more potent HuR inhibitors. Additionally, we showed the cellular effects of DHTS treatment on HuR ability to bind mRNAs, and we described a paradoxical enrichment of mRNAs containing longer 3′UTRs with increased AU content. Such effects result in dysregulation of HuR function, specific to cells that are strictly dependent on HuR function. Starting from the relaxation measurements of RNA-free protein in solution, we described the events preceding RNA binding. The analysis of NMR relaxation data indicates that the free RRM1–RRM2 construct is largely monomeric in solution. However, the observed R2 values are slightly higher than those expected for a monomeric protein, suggesting the presence of an equilibrium with a fraction of protein experiencing a larger rotational correlation time. On the other hand, the discrepancy of observedR1 data with respect to the theoretical values calculated for a rigid RRM1–RRM2 monomer are explained by the presence of inter-domain flexibility. The low NOE values suggest a significant conformational plasticity of the protein that is needed for mRNA binding. Therefore, even if it is reported that the RRM1–RRM2 tandem construct forms a separated functional unit from the RRM3 domain (71), RRM1 and RRM2 domains are not rigidly held together but undergo independent motions that can facilitate the recognition of the RNA partner. This confirms observations from the crystal structure of non-complexed HuR, where no contacts between the two domains were detected (40,62) and supports earlier results that phosphorylation at the linker (e.g. Ser100) had a profound impact on HuR binding to target mRNAs (72). Moreover, the large R1 values and small NOE values seen for the linker indicate that this region highly influences the conformational change of the protein from an open/flexible free conformation towards a closed one bound to mRNA.
As previously reported (40), binding of mRNA to HuR can occur starting from the open/flexible state of the protein, where HuR first binds the mRNA strand via its RRM1 domain. As a result of the subsequent conformational change, the linker and the RRM2 domain bind the mRNA filament to form a stable complex (40). Therefore, the linker takes part in the binding allowing the reciprocal reorientation of the two domains and establishing interactions with the RNA strand. Addition of DHTS to unbound HuR results in a generalized decrease of the signal intensity of the NMR resonances, with some residues experiencing larger effects. It is likely that this behavior is due, at least in part, to a change in the conformational dynamics of the protein. Accordingly, upon complex formation with DHTS, in our MD simulation we observed a rather limited inter-domain flexibility, as a result of the shift in the conformational equilibrium between HuR forms in favor of a closed conformation. This result was not anticipated since, at least in principle, a change in the conformational dynamics of HuR could also have been achieved by stabilizing an open form of HuR. Indeed, residues located on the β-platforms of RRM1 and RRM2 domains, in the same regions involved in the binding with the mRNA strand, experience a deep quenching.
In our MD simulation, we observed a further closure of the RNA-binding groove, as compared to the RNA-bound conformation of HuR, and an ensuing increase in the number of inter-domain contacts, which could explain why the largest decreases in signal intensity were observed in residues belonging to these β-platforms. Deletion of the C-terminus region of RRM1 reduces the binding affinity of RRM1 to the mRNA probe but, importantly, abolishes the interaction between RRM1 and DHTS. This experimental validation of the NMR data and molecular modeling points to the residues next to the linker as being key structural elements responsible of the interaction between DHTS and HuR. Single aminoacid mutagenesis of Ser94, Asn107, Ile133, and Asn134 into alanine highlighted the importance of these residues in maintaining the equilibrium between the free protein in the monomer/dimer form and the closed-bound-to-RNA protein dimer, avoiding its aggregation on the RNA target. Interestingly, muteins are fully resistant to DHTS, further supporting that this small molecule competes for the same protein regions interacting with the target RNA. Collectively, these results suggest that DHTS stabilizes an unproductive ‘closed’ conformation of HuR and prevents the physiological re-orientation of the two domains needed to bind the target mRNA. Stabilization of such ‘closed’ conformation alters the protein dynamics, producing the observed generalized decrease of signal intensity of the resonances observed upon the addition of DHTS to the RRM1–RRM2 tandem domains. Unfortunately, the non-optimal solubility of DHTS hampered the quantitative assessment of its Kd.
In other experiments, we characterized the ability of DHTS to inhibit HuR activity in vitro, to modulate its post-transcriptional function in cell models, and its specificity towards other RNA-binding proteins. Additionally, we observed that DHTS inhibits the association step of HuR to its target RNAs, and that its cytotoxicity against cancer cells was HuR-dependent (31). The stable ‘closed’ form of the protein blocks the access to HuR for low affinity target RNAs. Indeed, paradoxically, the mRNAs with longer 3′UTR and higher U/AU content were more abundantly loaded on HuR after DHTS treatment, as they likely bind more avidly to HuR than DHTS itself. The levels of HuR-target RNAs were not changed during DHTS treatment, so a ‘post-binding’ mechanism of regulation can be inferred. Nevertheless, DHTS-dependent HuR dysregulation has a strong anti-cancer activity in vivo, as observed using a xenograft model of colon cancer (HCT116 cells). These findings are consistent with other results using colon, leukemia, cervical, and breast cancer cells, and indicate that DHTS can penetrate tumors effectively (73–76). The absence of systemic toxicity in treated animals supports the idea that general inhibition of HuR by small molecules can be a therapeutic avenue for future efforts, although the effects on the immune system should be evaluated in a non-immuno-compromised mouse model. Notably, HCT116ΔHuR knockout cells in vivo showed a limited ability to develop tumors, but the extent of DHTS growth inhibition on these tumors did not match the effects on WT tumors. While we cannot discount that loss of HuR may impact cancer cell drug uptake, these results indicate in vivo specificity of DHTS and support the view that DHTS requires HuR for its antitumor influence. Finally, experimental and theoretical studies here reported suggest that the mechanism of action of DHTS is that of a competitive inhibitor of mRNA binding to HuR. These observations will be the ground for a rational design and synthesis of more potent small-molecule HuR disruptors.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro (AIRC) [17153 to A.P.]; University of Trento [Intramural funding 40201031 to A.P.]; Fondazione Cariplo [40102636 to A.P.]; AXonomIX research project (to A.Q.) financed by the Provincia Autonoma di Trento, Italy; National Institute of Aging Intramural Research Program of the National Institutes of Health [to M.G., K.A., R.M.]; National Institutes of Health [R01 CA134609 to D.D.]; NIH/NCI Cancer Center Support Grant [P30 CA168524 to D.D.]; Prevent Cancer Foundation and an Institutional Development Award (IDeA) from NIH/ NIGMS [P30 GM103495 to R.P.]. Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro (AIRC).
Conflict of interest statement. None declared.
REFERENCES
- 1. Akaike Y., Masuda K., Kuwano Y., Nishida K., Kajita K., Kurokawa K., Satake Y., Shoda K., Imoto I., Rokutan K.. HuR regulates alternative splicing of the TRA2β gene in human colon cancer cells under oxidative stress. Mol. Cell. Biol. 2014; 34:2857–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Izquierdo J.M. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J. Biol. Chem. 2008; 283:19077–19084. [DOI] [PubMed] [Google Scholar]
- 3. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M., Tuschl T., Ohler U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N.. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011; 43:340–352. [DOI] [PubMed] [Google Scholar]
- 5. Al-Ahmadi W., Al-Ghamdi M., Al-Haj L., Al-Saif M., Khabar K.S.A.. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation. Nucleic Acids Res. 2009; 37:3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai W., Zhang G., Makeyev E. V. RNA-binding protein HuR autoregulates its expression by promoting alternative polyadenylation site usage. Nucleic Acids Res. 2012; 40:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dutertre M., Chakrama F.Z., Combe E., Desmet F.-O., Mortada H., Polay Espinoza M., Gratadou L., Auboeuf D.. A recently evolved class of alternative 3′-terminal exons involved in cell cycle regulation by topoisomerase inhibitors. Nat. Commun. 2014; 5:3395. [DOI] [PubMed] [Google Scholar]
- 8. Brennan C.M., Steitz J.A. HuR and mRNA stability. Cell. Mol. Life Sci. 2001; 58:266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Srikantan S., Gorospe M.. HuR function in disease. Front. Biosci. 2012; 17:189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papadaki O., Milatos S., Grammenoudi S., Mukherjee N., Keene J.D., Kontoyiannis D.L.. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J. Immunol. 2009; 182:6779–6788. [DOI] [PubMed] [Google Scholar]
- 11. Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I.. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 2014; 53:506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang W., Yang X., Cristofalo V.J., Holbrook N.J., Gorospe M.. Loss of HuR is linked to reduced expression of proliferative genes during replicative senescence. Mol. Cell. Biol. 2001; 21:5889–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sakai K., Kitagawa Y., Hirose G.. Binding of neuronal ELAV-like proteins to the uridine-rich sequence in the 3′-untranslated region of tumor necrosis factor-alpha messenger RNA. FEBS Lett. 1999; 446:157–162. [DOI] [PubMed] [Google Scholar]
- 14. González-Feliciano J.A., Hernández-Pérez M., Estrella L.A., Colón-López D.D., López A., Martínez M., Maurás-Rivera K.R., Lasalde C., Martínez D., Araujo-Pérez F. et al. The role of HuR in the post-transcriptional regulation of interleukin-3 in T cells. PLoS One. 2014; 9:e92457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zha W., Wang G., Pecora B.S., Studer E., Hylemon P.B., Pandak W.M., Zhou H.. Role of RNA-binding protein HuR and CUGBP1 in LPS-induced interleukin-6 expression in macrophages. FASEB J. 2010; 24:494.7–494.7. [Google Scholar]
- 16. Kim Y., Noren Hooten N., Dluzen D.F., Martindale J.L., Gorospe M., Evans M.K.. Posttranscriptional regulation of the inflammatory marker C-reactive protein by the RNA-binding protein HuR and MicroRNA 637. Mol. Cell. Biol. 2015; 35:4212–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gubin M.M., Techasintana P., Magee J.D., Dahm G.M., Calaluce R., Martindale J.L., Whitney M.S., Franklin C.L., Besch-Williford C., Hollingsworth J.W. et al. Conditional knockout of the RNA-binding protein HuR in CD4+ T cells reveals a gene dosage effect on cytokine production. Mol. Med. 2014; 20:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan J., Ishmael F.T., Fang X., Myers A., Cheadle C., Huang S.-K., Atasoy U., Gorospe M., Stellato C.. Chemokine transcripts as targets of the RNA-binding protein HuR in human airway epithelium. J. Immunol. 2011; 186:2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Diaz-Muñoz M.D., Bell S.E., Fairfax K., Monzon-Casanova E., Cunningham A.F., Gonzalez-Porta M., Andrews S.R., Bunik V.I., Zarnack K., Curk T. et al. The RNA-binding protein HuR is essential for the B cell antibody response. Nat. Immunol. 2015; 16:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeMicco A., Naradikian M.S., Sindhava V.J., Yoon J.-H., Gorospe M., Wertheim G.B., Cancro M.P., Bassing C.H.. B cell-intrinsic expression of the HuR RNA-binding protein is required for the T cell-dependent immune response in vivo. J. Immunol. 2015; 195:3449–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazan-Mamczarz K., Hagner P.R., Corl S., Srikantan S., Wood W.H., Becker K.G., Gorospe M., Keene J.D., Levenson A.S., Gartenhaus R.B.. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene. 2008; 27:6151–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang W., Furneaux H., Cheng H., Caldwell M.C., Hutter D., Liu Y., Holbrook N., Gorospe M.. b. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 2000; 20:760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Latorre E., Carelli S., Raimondi I., D’Agostino V., Castiglioni I., Zucal C., Moro G., Luciani A., Ghilardi G., Monti E. et al. The ribonucleic complex HuR-MALAT1 represses CD133 expression and suppresses epithelial-mesenchymal transition in breast cancer. Cancer Res. 2016; 76:2626–2636. [DOI] [PubMed] [Google Scholar]
- 24. Latorre E., Tebaldi T., Viero G., Spartà A.M., Quattrone A., Provenzani A.. Downregulation of HuR as a new mechanism of doxorubicin resistance in breast cancer cells. Mol. Cancer. 2012; 11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Latorre E., Castiglioni I., Gatto P., Carelli S., Quattrone A., Provenzani A.. Loss of protein kinase Cδ/HuR interaction is necessary to doxorubicin resistance in breast cancer cell lines. J. Pharmacol. Exp. Ther. 2014; 349:99–106. [DOI] [PubMed] [Google Scholar]
- 26. Abdelmohsen K., Gorospe M.. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA. 2010; 1:214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levy N.S., Chung S., Furneaux H., Levy A.P.. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 1998; 273:6417–6423. [DOI] [PubMed] [Google Scholar]
- 28. Meisner N.-C., Hintersteiner M., Mueller K., Bauer R., Seifert J.-M., Naegeli H.-U., Ottl J., Oberer L., Guenat C., Moss S. et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat. Chem. Biol. 2007; 3:508–515. [DOI] [PubMed] [Google Scholar]
- 29. Wu X., Lan L., Wilson D.M., Marquez R.T., Tsao W., Gao P., Roy A., Turner B.A., McDonald P., Tunge J. a et al. Identification and validation of novel small molecule disruptors of HuR-mRNA interaction. ACS Chem. Biol. 2015; doi:10.1021/cb500851u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blanco F.F., Preet R., Aguado A., Vishwakarma V., Stevens L.E., Vyas A., Padhye S., Xu L., Weir S.J., Anant S. et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016; doi:10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. D’Agostino V.G., Lal P., Mantelli B., Tiedje C., Zucal C., Thongon N., Gaestel M., Latorre E., Marinelli L., Seneci P. et al. Dihydrotanshinone-I interferes with the RNA-binding activity of HuR affecting its post-transcriptional function. Sci. Rep. 2015; 5:16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D’Agostino V.G., Adami V., Provenzani A.. A novel high throughput biochemical assay to evaluate the HuR protein-RNA complex formation. PLoS One. 2013; 8:e72426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chae M.-J., Sung H.Y., Kim E.-H., Lee M., Kwak H., Chae C.H., Kim S., Park W.-Y.. Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-α mRNA. Exp. Mol. Med. 2009; 41:824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zucal C., D’Agostino V., Loffredo R., Mantelli B., Thongon N., Lal P., Latorre E., Provenzani A.. Targeting the multifaceted HuR protein, benefits and caveats. Curr. Drug Targets. 2015; 16:499–515. [DOI] [PubMed] [Google Scholar]
- 35. Wang Z., Bhattacharya A., Ivanov D.N.. Identification of small-molecule inhibitors of the HuR/RNA interaction using a fluorescence polarization screening assay followed by NMR validation. PLoS One. 2015; 10:e0138780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan X.C., Steitz J.A.. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheiba R.M., de Opakua A.I., Díaz-Quintana A., Cruz-Gallardo I., Martínez-Cruz L.A., Martínez-Chantar M.L., Blanco F.J., Díaz-Moreno I.. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol. 2014; 11:1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maris C., Dominguez C., Allain F.H.-T.. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005; 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- 39. Benoit R.M., Meisner N.-C., Kallen J., Graff P., Hemmig R., Cèbe R., Ostermeier C., Widmer H., Auer M.. The x-ray crystal structure of the first RNA recognition motif and site-directed mutagenesis suggest a possible HuR redox sensing mechanism. J. Mol. Biol. 2010; 397:1231–1244. [DOI] [PubMed] [Google Scholar]
- 40. Wang H., Zeng F., Liu Q., Liu H., Liu Z., Niu L., Teng M., Li X.. The structure of the ARE-binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallogr. D. Biol. Crystallogr. 2013; 69:373–380. [DOI] [PubMed] [Google Scholar]
- 41. Kim H.S., Wilce M.C.J., Yoga Y.M.K., Pendini N.R., Gunzburg M.J., Cowieson N.P., Wilson G.M., Williams B.R.G., Gorospe M., Wilce J.A.. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011; 39:1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lal S., Cheung E.C., Zarei M., Preet R., Chand S.N., Mambelli-Lisboa N.C., Romeo C., Stout M.C., Londin E., Goetz A. et al. CRISPR knockout of the HuR gene causes a xenograft lethal phenotype. Mol. Cancer Res. 2017; doi:10.1158/1541-7786.MCR-16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Young L.E., Moore A.E., Sokol L., Meisner-Kober N., Dixon D.A.. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012; 10:167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barbato G., Ikura M., Kay L.E., Pastor R.W., Bax A.. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992; 31:5269–5278. [DOI] [PubMed] [Google Scholar]
- 45. Kay L.E., Nicholson L.K., Delaglio F., Bax A., Torchia D.. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 1992; 97:359–375. [Google Scholar]
- 46. Peng J.W., Wagner G.. Investigation of protein motions via relaxation measurements. Methods Enzymol. 1994; 239:563–596. [DOI] [PubMed] [Google Scholar]
- 47. García de la Torre J, Huertas M.L., Carrasco B.. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000; 147:138–146. [DOI] [PubMed] [Google Scholar]
- 48. Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K.. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005; 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A.. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995; 117:5179–5197. [Google Scholar]
- 50. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E.. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010; 78:1950–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Allnér O., Nilsson L., Villa A.. Magnesium ion–water coordination and exchange in biomolecular simulations. J. Chem. Theory Comput. 2012; 8:1493–1502. [DOI] [PubMed] [Google Scholar]
- 52. Bayly C.I., Cieplak P., Cornell W., Kollman P.A.. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993; 97:10269–10280. [Google Scholar]
- 53. Gaussian 09, Revision A.02 Gaussian 09 Citation. 2009; http://gaussian.com/g09citation/. [Google Scholar]
- 54. Dupradeau F.-Y., Pigache A., Zaffran T., Savineau C., Lelong R., Grivel N., Lelong D., Rosanski W., Cieplak P.. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. PCCP. 2010; 12:7821–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vanquelef E., Simon S., Marquant G., Garcia E., Klimerak G., Delepine J.C., Cieplak P., Dupradeau F.-Y.. R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011; 39:W511–W517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang J., Wang W., Kollman P.A., Case D.A.. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006; 25:247–260. [DOI] [PubMed] [Google Scholar]
- 57. Abdelmohsen K., Srikantan S., Yang X., Lal A., Kim H.H., Kuwano Y., Galban S., Becker K.G., Kamara D., de Cabo R. et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009; 28:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dassi E., Re A., Leo S., Tebaldi T., Pasini L., Peroni D., Quattrone A.. AURA 2: Empowering discovery of post-transcriptional networks. Transl. (Austin, Tex.). 2014; 2:e27738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A.. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013; 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yu G., Li F., Qin Y., Bo X., Wu Y., Wang S.. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010; 26:976–978. [DOI] [PubMed] [Google Scholar]
- 61. Mujo A., Lixa C., Carneiro L.A.M., Anobom C.D., Almeida F.C., Pinheiro A.S.. 1H, 15N and 13C resonance assignments of the RRM1 domain of the key post-transcriptional regulator HuR. Biomol. NMR Assign. 2014; doi:10.1007/s12104-014-9592-9. [DOI] [PubMed] [Google Scholar]
- 62. Wang H., Zeng F., Liu H., Teng M., Li X.. Crystal structure of two tandem RNA recognition motifs of Human antigen R. 2012; https://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=4EGL.
- 63. Bertini I., Fragai M., Luchinat C., Melikian M., Mylonas E., Sarti N., Svergun D.I.. Interdomain flexibility in full-length matrix metalloproteinase-1 (MMP-1). J. Biol. Chem. 2009; 284:12821–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bertini I., Calderone V., Fragai M., Jaiswal R., Luchinat C., Melikian M., Mylonas E., Svergun D.I.. Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12. J. Am. Chem. Soc. 2008; 130:7011–7021. [DOI] [PubMed] [Google Scholar]
- 65. Cerofolini L., Fields G.B., Fragai M., Geraldes C.F.G.C., Luchinat C., Parigi G., Ravera E., Svergun D.I., Teixeira J.M.C.. Examination of matrix metalloproteinase-1 in solution: a preference for the pre-collagenolysis state. J. Biol. Chem. 2013; 288:30659–30671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dosset P., Hus J.-C., Blackledge M., Marion D.. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000; 16:23–28. [DOI] [PubMed] [Google Scholar]
- 67. Morrison K.L., Weiss G.A.. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001; 5:302–307. [DOI] [PubMed] [Google Scholar]
- 68. Arkin M.R., Glicksman M.A., Fu H., Havel J.J., Du Y.. Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Iversen PW. Inhibition of protein-protein interactions: non-cellular assay formats. Assay Guidance Manual. 2004; Bethesda: Eli Lilly & Company and the National Center for Advancing Translational Sciences. [PubMed] [Google Scholar]
- 69. de Silanes I.L., Zhan M., Lal A., Yang X., Gorospe M.. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2987–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu X., Lan L., Smith A., Marquez R., Wilson D., Rogers S., Gao P., Lovell S., Karanicolas J., Dixon D. et al. Abstract 2449: Targeting an ‘undruggable’ RNA-binding protein: Discovery of small molecule inhibitors of HuR for novel breast cancer therapy. Cancer Res. 2015; 75:2449–2449. [Google Scholar]
- 71. Scheiba R.M., Aroca Á., Díaz-Moreno I.. HuR thermal stability is dependent on domain binding and upon phosphorylation. Eur. Biophys. J. 2012; 41:597–605. [DOI] [PubMed] [Google Scholar]
- 72. Abdelmohsen K., Pullmann R., Lal A., Kim H.H., Galban S., Yang X., Blethrow J.D., Walker M., Shubert J., Gillespie D. a et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007; 25:543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang L., Hu T., Shen J., Zhang L., Chan R.L.-Y., Lu L., Li M., Cho C.H., Wu W.K.K.. Dihydrotanshinone I induced apoptosis and autophagy through caspase dependent pathway in colon cancer. Phytomedicine. 2015; 22:1079–1087. [DOI] [PubMed] [Google Scholar]
- 74. Liu J.-J., Wu H.-H., Chen T.-H., Leung W., Liang Y.-C.. 15,16-Dihydrotanshinone I from the functional food Salvia miltiorrhiza exhibits anticancer activity in human HL-60 leukemia cells: in vitro and in vivo studies. Int. J. Mol. Sci. 2015; 16:19387–19400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ye Y., Xu W., Zhong W., Li Y., Wang C.. Combination treatment with dihydrotanshinone I and irradiation enhances apoptotic effects in human cervical cancer by HPV E6 down-regulation and caspases activation. Mol. Cell. Biochem. 2012; 363:191–202. [DOI] [PubMed] [Google Scholar]
- 76. Tsai S.-L., Suk F.-M., Wang C.-I., Liu D.-Z., Hou W.-C., Lin P.-J., Hung L.-F., Liang Y.-C.. Anti-tumor potential of 15, 16-dihydrotanshinone I against breast adenocarcinoma through inducing G1 arrest and apoptosis. Biochem. Pharmacol. 2007; 74:1575–1586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.