Abstract
Deposition of replication-independent histone variant H3.3 into chromatin is essential for many biological processes, including development and reproduction. Unlike replication-dependent H3.1/2 isoforms, H3.3 is expressed throughout the cell cycle and becomes enriched in postmitotic cells with age. However, lifelong dynamics of H3 variant replacement and the impact of this process on chromatin organization remain largely undefined. Using quantitative middle-down proteomics we demonstrate that H3.3 accumulates to near saturation levels in the chromatin of various mouse somatic tissues by late adulthood. Accumulation of H3.3 is associated with profound changes in global levels of both individual and combinatorial H3 methyl modifications. A subset of these modifications exhibit distinct relative abundances on H3 variants and remain stably enriched on H3.3 throughout the lifespan, suggesting a causal relationship between H3 variant replacement and age-dependent changes in H3 methylation. Furthermore, the H3.3 level is drastically reduced in human hepatocarcinoma cells as compared to nontumoral hepatocytes, suggesting the potential utility of the H3.3 relative abundance as a biomarker of abnormal cell proliferation activity. Overall, our study provides the first quantitative characterization of dynamic changes in H3 proteoforms throughout lifespan in mammals and suggests a role for H3 variant replacement in modulating H3 methylation landscape with age.
INTRODUCTION
Chromatin is a highly complex and dynamic nucleoprotein structure that controls all DNA-templated processes by modulating the accessibility of DNA to nuclear factors, such as transcription and DNA repair machineries. Dynamic changes in chromatin organization are central for regulating transcriptional programs during development and in response to internal and external stimuli. Dysregulation of chromatin-associated pathways may lead to various pathological conditions such as developmental abnormalities, cancer and age-related diseases (1–3).
Regulation of chromatin architecture and function is largely accomplished by post-translational modifications (PTMs) of histone proteins (4,5). Over 20 distinct types of histone PTMs have been described (6), among which the most abundant ones are acetylation and methylation of lysine residues (7). Histone PTMs can be deposited on and removed from chromatin by different enzymes, known as histone PTM ‘writers’ and ‘erasers’ (8). Histone PTMs exert their regulatory effects via two main mechanisms. First, histone PTMs serve as docking sites for various nuclear proteins—histone PTM ‘readers’—that specifically recognize modified histone residues through their modification-binding domains (9). Recruitment of these proteins at specific genomic loci promotes key chromatin processes, such as transcriptional regulation and DNA damage repair (10,11). Second, some histone PTMs, such as acetylation, directly affect chromatin higher-order structure and compaction (12), thereby controlling chromatin accessibility to protein machineries such as those involved in transcription (13). Chromatin states defined by histones PTMs can be transmitted through cell divisions and are thought to play a role in the inheritance of specific gene expression profiles (14–16).
Another level of chromatin regulation is accomplished by a dynamic exchange of canonical histones with specific histone variants (17). Histone variants are non-allelic isoforms of canonical histones that differ in their primary sequence and functional properties. Unlike canonical histones that are expressed and incorporated into chromatin preferentially, although not exclusively (18,19), during the S phase in DNA synthesis-coupled (DSC) manner, histone variants are expressed throughout the cell cycle and replace their canonical counterparts via DNA synthesis-independent (DSI) nucleosome assembly pathways (20). Substitution of canonical histones with histone variants can influence the structural properties of the nucleosome, thereby affecting different chromatin processes (21). In particular, the evolutionarily conserved histone variant H3.3 was recently implicated in transcriptional regulation, retrotransposon silencing and maintenance of genome integrity (22–25). In metazoans, H3.3 differs from canonical H3 isoforms, i.e. H3.2 and H3.1, by only four and five amino acid residues, respectively. These amino acid substitutions in H3.3 mediate its specific interaction with chaperone complexes HIRA and ATRX/DAXX which deposit H3.3 at distinct chromatin regions, including gene bodies, telomeres and pericentric chromatin (26–28). Depletion of H3.3 as well as a disruption of H3.3 nucleosome assembly pathways results in early embryonic lethality in mice and impairs late gastrulation in Xenopus (22,29).
A growing body of evidence indicates that H3.3 plays an essential role in maintaining and regulating chromatin organization in postmitotic cells. In oocytes, DSI deposition of H3.3 into the chromatin is required for efficient de novo DNA methylation and transcriptional transitions associated with oocyte maturation (30). H3.3 controls gene expression programs and plasticity in mammalian neurons, where it progressively accumulates and replaces canonical H3.1/2 isoforms with age (31). H3.3 accumulation was also observed in non-proliferating chicken tissues and senescent human fibroblasts (32,33). In line with these findings, the progressive replacement of H3.1/2 by the H3.3 was hypothesized to constitute a common feature of chromatin organization in postmitotic cells (34). This hypothesis is particularly intriguing in light of emerging evidence indicating that H3.3 is functionally distinct from its canonical counterparts (35,36). In particular, recent studies have demonstrated that the conserved amino acid variations between canonical H3.1/2 isoforms and H3.3 variant can directly affect the deposition and functional readout of PTMs of these proteins (35,37,38). However, lifelong dynamics of H3 variant replacement in mammals and the effect of this process on the H3 modification landscape remain largely undefined.
Here, we provide quantitative mass spectrometry (MS)-based middle-down proteomic characterization of dynamic changes in histone H3 proteoforms at five time points across the lifespan in mice, covering ages 3–24 months. We demonstrate that the histone variant H3.3 progressively accumulates in various mouse somatic tissues with age, resulting in near complete replacement of the canonical H3.1/2 isoforms by the age of 18 months. H3 variant replacement is associated with profound changes in global levels of a number of individual and combinatorial H3 methyl PTMs, among which some PTMs, such as K36me2 and K27me2K36me2, exhibit stable lifelong enrichment on H3.3. Bioinformatic analysis of 128 quantitative MS datasets obtained in this study reveals several unexpected features of H3 modification landscape organization, including the strong correlation between global levels of H3 methylation at lysines 9(K9), 27(K27) and 36(K36). Finally, we show that H3.3 accumulation profile is not restricted to mouse cells, but is also observed in non-tumor adult human hepatocytes, while the chromatin of hepatocarcinoma cells is preferentially marked by the H3.1/2. All proteomic MS datasets obtained in the study were deposited into the PRIDE database (dataset identifier PXD005300) and the CrosstalkDB resource (http://crosstalkdb.bmb.sdu.dk/).
MATERIALS AND METHODS
Animals and tissue collection
Male C57BL/6J mice were obtained from a study approved by the Danish Animal Ethics Inspectorate (J. no. 2011/561-1950). Wild-type mice were bred in the Biomedical Laboratory, University of Southern Denmark under a 12 h:12 h light:dark cycle (lights on at 6:30 a.m.). Food and water were available ad libitum. Mice were sacrificed by cervical dislocation at ages of 3, 5, 10, 18 and 24 months. Liver, kidney, brain and heart were excised, rinsed in ice-cold phosphate-buffered saline (PBS) and were immediately snap-frozen. Tissues were stored at –80°C until further processing.
Primary human hepatocytes
Primary adult human hepatocytes were purchased from BioreclamationIVT (Brussels, Belgium).
Human tissue samples
Protein lysates of human hepatocarcinoma tissues and matched normal liver tissues were purchased from Acris Antibodies GmbH and from Origene Inc.
Sample preparation for proteomic analysis
Mouse tissues were homogenized on ice in nuclei isolation buffer (NIB; 15 mM Tris–HCl (pH 7.5), 60 mM KCl, 11 mM CaCl2, 5 mM NaCl, 5 mM MgCl2, 250 mM sucrose, 1 mM dithiothreitol, 10 mM sodium butyrate and 0.3% NP-40) supplemented with protease (Complete; Roche) and phosphatase inhibitors (PhosSTOP, Roche) using a pestle tissue grinder (Wheaton Science Products). Nuclei were pelleted by centrifugation (1200 × g for 10 min) and washed twice with ice-cold NIB (without NP-40). Histones were acid extracted from the resulting nuclei pellet as previously described (39).
Immunoblot analysis
Immunoblot analysis was performed as described previously (40). The following antibodies were used: anti-H3.3 (1:1000; ab176840, Abcam) or anti-H3 (1:5000; ab1791, Abcam) primary antibodies, followed by secondary horseradish peroxidase (HRP)-conjugated antibodies (1:5000; ab6721, Abcam).
Middle-down mass spectrometry analysis
Acid-precipitated bulk histones were resuspended in 100 mM NH4HCO3 (pH 4) to a final protein concentration of ∼1 mg/ml and subjected to endoproteinase GluC digestion (Calbiochem, 1:10 enzyme to substrate ratio) performed for 6 h at room temperature. The resulting peptide mixtures were analyzed by the on-line nanoflow weak cation exchange–hydrophilic interaction liquid chromatography–tandem mass spectrometry (WCX-HILIC-MS/MS) method adapted from our previous studies (41,42). About 4 μg of peptide mixture were separated using Dionex Ultimate 3000 high-performance liquid chromatography (HPLC) system (Thermo Scientific) equipped with a two-column setup, consisting of a reversed-phase trap column (3 cm, 100 μm i.d., 360 μm o.d., packed with ReproSil, Pur C18AQ 3 μm; Dr Maisch) and a WCX-HILIC analytical column (15 cm, 75 μm i.d. 360 μm o.d., packed with PolycatA 1000 Å, 1.9 μm, PolyLC Inc). Loading buffer was 0.1% formic acid (Merck Millipore) in water. Buffer A and B were prepared as described (43). Peptides were eluted directly into an ESI tandem mass spectrometer (Orbitrap Fusion ETD, Thermo Fisher Scientific) using a 130-min linear gradient of 65–85% buffer B at a flow rate of 230 nl/min. The MS instrument was controlled by Xcalibur software (Thermo Fisher Scientific). The nanoelectrospray ion source (Thermo Fisher Scientific) was used with a spray voltage of 2.2 kV. The ion transfer tube temperature was 275°C. Data acquisition was performed in the Orbitrap for both precursor ions and product ions. MS survey scans were obtained for the m/z range of 400–750 in the Orbitrap with maximum ion injection time of 50 ms, auto gain control target 5 × 105, mass resolution of 60 000 and three microscans per each full MS event. The four most intense ions with MS1 signal higher than 20 000 counts were selected for fragmentation using electron transfer dissociation (ETD). In order to fragment only histone H3 N-terminal peptide proteoforms the following criteria for the precursor ion selection were applied: m/z range of 667–703 and charge +8; m/z range of 593–625 and charge +9; m/z range of 533–562 and charge +10; m/z range of 485–512 and charge +11. MS/MS spectra were measured in the Orbitrap with maximum ion injection time 100 ms, auto gain control target 2 × 105, mass resolution of 30 000 and three microscans per MS/MS.
Middle-down MS data processing and analysis
Database search
Raw MS files were processed and searched with Mascot (version 2.3.2, Matrix Science, London, UK) using the Proteome Discoverer (version 1.4, Thermo Fischer Scientific) data analysis package. Spectra were deconvoluted using the Xtract tool (Thermo Fisher Scientific) with the following parameters: S/N threshold was set to 1, mass resolution at 400 m/z was 30 000. The following parameters were used in the Mascot search: MS mass tolerance: 1.05 Da, to include possible errors in isotopic recognition; MS/MS mass tolerance: 0.02 Da; enzyme: GluC with no missed cleavage allowed; mono- and dimethylation (KR), trimethylation (K) and acetylation (K) were set as variable modifications. MS/MS spectra were searched against mouse histone database consisting of histone H3.1/H3.2 and H3.3 N-terminal sequences covering amino acid (aa) residues 1–50.
Validation of PTM site assignments
Database search result files from Mascot were exported into CSV file format and processed by isoScale slim (http://middle-down.github.io/Software, (41)) with a mass tolerance of 0.015 Da and only c/z fragment ions allowed. Only PTMs with at least one site determining ion before and after the assigned PTM site were accepted.
Peptide quantification
H3 N-terminal peptides were quantified based on the total ion current of their MS/MS spectra (MS2-TIC) using the isoScale slim software. In a case of mixed MS/MS spectra generated as a result of co-fragmentation of co-selected isobaric peptides, i.e. peptides which share the same sequence and have the same masses but have distinct localizations of PTMs, isoScale slim calculated the fragment ion relative ratio (FIRR) (44) of such peptides and divided the MS2-TIC for this ratio.
Relative quantification of single and combinatorial H3 PTMs
The relative abundance of each unique H3 N-terminal peptide proteoform, i.e. unmodified or modified H3.1/2 or H3.3 N-terminal peptide bearing single or combinatorial PTM, hereinafter referred to as H3 proteoform, was calculated by summing MS2-TIC for all peptide-spectrum matches (PSMs) corresponding to the given proteoform and dividing the sum intensity by the total intensity of all quantified H3 N-terminal proteoforms. For example, the relative abundance of H3.3K27me2K36me2 proteoform was calculated as the sum of MS2-TICs for all H3.3K27me2K36me2 PSMs divided by the sum of MS2-TICs for all H3.1/2 and H3.3 PSMs.
The relative abundance of each single or combinatorial PTM (for definitions see Table 1) on total H3 (also referred to as global level) was calculated by summing the relative abundances of both H3.1/2 and H3.3 proteoforms carrying the given PTM. For example, the relative abundance of combinatorial PTM H3K27me2K36me2 was calculated as the sum of relative abundances of H3.1/2K27me2K36me2 and H3.3K27me2K36me2.
Table 1. Definitions for distinct modification types and quantitative MS measurements used in the study.
| Modification type | Meaning | Examples |
| Single PTM | Specified aa residue within H3 N-terminal tail is modified by a given PTM; other aa residues are unmodified | H3K9me2: H3K9 is dimethylated; other H3 N-terminal tail aa residues are unmodified |
| Individual PTM | Specified aa residue within H3 N-terminal tail is modified by a given PTM; other aa residues can either be modified or unmodified | H3K9me2: H3K9 is dimethylated; other H3 N-terminal tail aa residues can either be modified or unmodified |
| Combinatorial PTM | Specified aa residues within H3 N-terminal tail are modified by a given PTM(s); other aa residues are unmodified | H3K9me2K27me3: H3K9 and H3K27 are di- and trimethylated, respectively; other H3 N-terminal tail aa residues are unmodified |
| Binary PTM | Two specified aa residues within H3 N-terminal tail are modified by given a PTM(s); other aa residues can either be modified or unmodified | H3K9me2K27me3: H3K9 and H3K27 are di- and trimethylated, respectively; other H3 N-terminal tail aa residues can either be modified or unmodified |
| Quantitative MS measurement | Meaning | Examples |
| Relative abundance (RA) of PTM on total H3 | Proportion (%) of all H3 N-terminal tails carrying a given PTM | It is important to distinguish the difference between the RAs of single and individual PTMs: |
| RA of PTM on H3.3 (H3.1/2) | Proportion (%) of H3.3 (H3.1/2) N-terminal tails carrying a given PTM | RA of a single PTM H3K9me2 is 1% means that 1% of all H3 N-terminal tails is modified by K9me2 with NO other modifications present on them. |
| RA of an individual PTM H3K9me2 is 1% means that 1% of all H3 N-terminal tails is modified by K9me2 alone or in combination with any other H3 PTM(s). |
The relative abundance of each single or combinatorial PTM on H3.3 (H3.1) was calculated by normalizing the relative abundance of H3.3 (H3.1/2) proteoforms carrying the given PTM by the relative abundance of H3.3 (H3.1/2) calculated as a sum of relative abundances of all H3.3 (H3.1/2) proteoforms.
Relative quantification of individual H3 PTMs
The relative abundance of each individual modification (for definition see Table 1) on total H3 (also referred to as global level) was calculated as the sum of relative abundances of all H3 proteoforms containing the given modification. For example, the relative abundance of H3K9me2 was calculated by summing the relative abundances of all H3 proteoforms containing K9me2 alone or in combination with any other H3 PTM(s).
The relative abundance of each individual modification on H3.3 (H3.1/2) was calculated as the sum of relative abundances of all H3.3 (H3.1/2) proteoforms containing the given modification normalized by the relative abundance of H3.3 (H3.1/2).
Analysis of interplay between histone PTMs
To assess which PTMs tend to attract or repel each other within H3 N-terminal tail we used the interplay score that we recently introduced (41,45). This score was calculated as Ixy = log2(Fxy/FxFy), where Ixy is the interplay score between the modifications X and Y, Fxy is the co-frequency and Fx or Fy are the relative abundances of the individual modifications in the dataset. The co-frequency of the two modifications X and Y (also referred to as a relative abundance of binary PTM) was calculated by summing the relative abundances of all H3 proteoforms simultaneously decorated by both modifications. Positive interplay score values indicate a tendency to co-exist more often than if the two modifications were completely independent of each other, while negative values indicate the opposite.
Targeted LC–MS/MS analysis of human tissue lysates
Tumor and non-tumor human liver tissue lysates from patients 1–4 were separately pooled by equal protein mass. A 60 μg protein aliquot from each pool was run on 4–12% SDS-PAGE gel followed by staining with Coomassie Blue. The protein bands corresponding to histone H3 (and its truncated proteoform (42,46)) were cut out from SDS-PAGE gels. Histones were in-gel propionylated and digested according to (47). Resulting peptide mixtures were analyzed in duplicates using targeted bottom-up LC–MS method adapted from (48). Relative abundance of H3.3 (H3.1/2) variant was calculated as the sum of integrated MS1 intensities for all H3.3 (H3.1/2) peptide proteoforms covering aa residues 27–40, including all differentially modified H3.3 (H3.1/2) peptides, normalized by the total intensity of all H3 peptides covering residues 27–40.
Statistical analysis
Tissues derived from 3-, 5- and 10-month-old mice were analyzed in four biological replicates and tissues derived from 18- and 24-month-old mice were analyzed in two biological replicates. Each biological sample was analyzed by LC-MS/MS in two technical replicates. Differences in H3 PTM abundances between age groups were determined using the Limma package in R/Bioconductor. Differences in H3 PTM abundances between H3 variants were determined using paired t-test. All p-values were corrected for multiple testing using the Benjamini–Hochberg method. Statistically significant differences are indicated by asterisks (P < 0.05 [*], P < 0.01 [**] and P < 0.001 [***]), n.s. indicates nonsignificant.
RESULTS
Histone variant H3.3 replaces canonical H3.1/2 isoforms in mouse somatic tissues with age
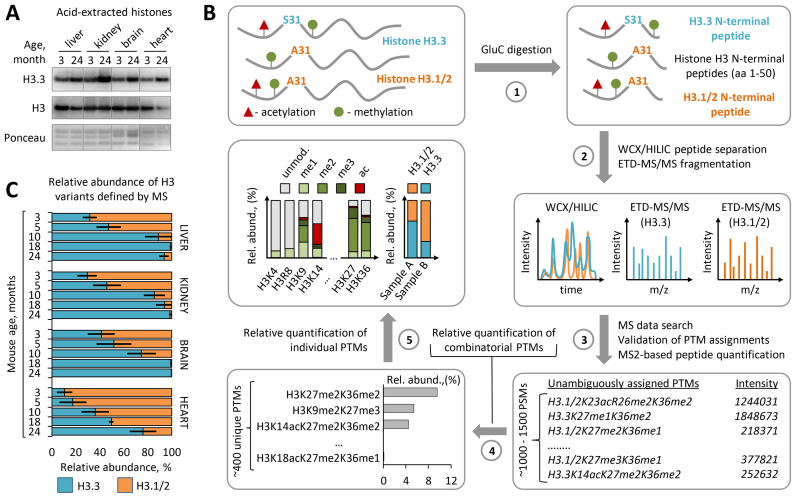
Histone variant H3.3 was previously shown to accumulate in the mouse brain with age (31,49). We first investigated whether H3.3 also accumulates in other mouse tissues. To address this, we analyzed the relative abundance of H3 variants in chromatin isolated from liver, kidney, heart and brain tissues, derived from 3- to 24-month-old C57BL/6J mice. Immunoblot analysis with histone variant–specific antibodies demonstrated pronounced enrichment of the H3.3 in all analyzed tissues harvested from old mice as compared to those tissues obtained from young mice (Figure 1A). To quantitatively assess dynamics of H3.1/2 replacement by the H3.3 we determined the relative abundances of histone H3 variants in the chromatin at five time points across mouse lifespan using middle-down MS. Middle-down MS is based on the analysis of long (≥5 kDa) N-terminal histone peptides and enables detection, characterization and quantitation of a distinct H3 species, including canonical H3.1/2 isoforms and H3.3 variant as well as their differentially modified proteoforms bearing single or combinatorial PTMs (Figure 1B, Supplementary Figure S1) (41,42). In agreement with the report by Maze et al. (31), we found that H3.3 progressively accumulates in the mouse brain during adulthood resulting in near complete replacement of canonical H3.1/2 isoforms by the age of 18 months (Figure 1C). Interestingly, a nearly identical accumulation pattern for H3.3 was observed in liver and kidney tissues, where H3.3 levels exceeded 99% and 94% of the total H3 pool, respectively, by the age of 18 months. In the heart, the accumulation rate of H3.3 was slower with 76% of canonical H3.1/2 isoforms been replaced by the H3.3 at 24 months of age (Figure 1C). Although the molecular mechanisms modulating histone variant exchange rate remain undefined, previous studies demonstrated that H3 proteins bearing active PTMs, such as acetylation at distinct lysine residues, exhibit faster turnover than H3 bearing repressive PTMs, such as methylation at K9 and K27 (50). Hence, we next compared the relative abundances of these PTMs between the heart and other somatic tissues. We found that heart chromatin is significantly enriched in repressive H3 PTMs, including H3K9me2 and H3K27me3, and is depleted in active H3 PTMs, including H3K14ac, H3K18ac and H3K23ac, as compared to liver, kidney and brain chromatin (Supplementary Figure S2). In line with the slower H3.3 accumulation rate observed in heart, these data link the global rate of H3 variant replacement with age to cell type-specific H3 PTM landscape. On the one hand, the low abundance of active H3 PTMs that are associated with fast histone turnover might slow down the H3 variant replacement rate with age. Alternatively, considering the fact that H3.3 variant is enriched in active PTMs (51), the lower abundance of these PTMs in the heart as compared to other tissues might be a consequence of the lower level of H3.3 in this tissue. Taken together, our results identify progressive age-dependent replacement of canonical H3.1/2 isoforms by the H3.3 variant as a common feature of chromatin organization in somatic mouse cells and indicate that H3.3 accumulates to near saturation levels by the late adulthood, except for the heart where H3.3 accumulation proceed throughout life.
Figure 1.
Histone H3.3 accumulates and replaces canonical H3 isoforms with age. (A) Immunoblot analysis of H3.3 levels in histone extracts from somatic tissues of 3-month-old and 24-month-old C57BL/6J mice. Total histone H3 and Ponceau S staining were used as loading controls. (B) Overview of the middle-down proteomics workflow used in the study. Histones were acid extracted from mouse tissues and digested with endopeptidase GluС (1). The resulting peptide mixtures containing 50 aa residues long H3 N-terminal peptides were analyzed by on-line nano-LC–MS/MS method using weak cation exchange–hydrophilic interaction liquid chromatography (WCX-HILIC) for peptide separation and electron transfer dissociation (ETD) for their fragmentation (2). Acquired MS/MS data were searched against a histone protein database to obtain a set of peptide identifications followed by filtering out of hits with ambiguous PTM site assignments (3). Peptides with unambiguous PTM localizations were quantified based on the total ion current of their MS/MS spectra (MS2-TIC). Obtained quantitative results were used to calculate the relative abundances of distinct H3 proteoforms barring single or combinatorial PTMs, where the sum of all MS2-TIC intensities was considered as 100% (4). The relative abundance of each individual H3 PTM and each H3 variant was calculated by summing up the relative abundances of all H3 proteoforms containing the given PTM or assigned to the given H3 variant, respectively (5). The relative abundance of individual PTMs on H3.3 was calculated as the sum of the relative abundances of H3.3 proteoforms containing the given PTM normalized by the relative abundance of the H3.3 variant. For more details, see the ‘Materials and Methods’ section. (C) Dynamics of H3.3 accumulation in chromatin of various mouse somatic tissues with age as revealed by middle-down MS analysis. Error bars indicate SD (n = 2–4 biological replicates).
H3.3 is depleted in tumor tissues as compared to adjacent non-tumor tissues
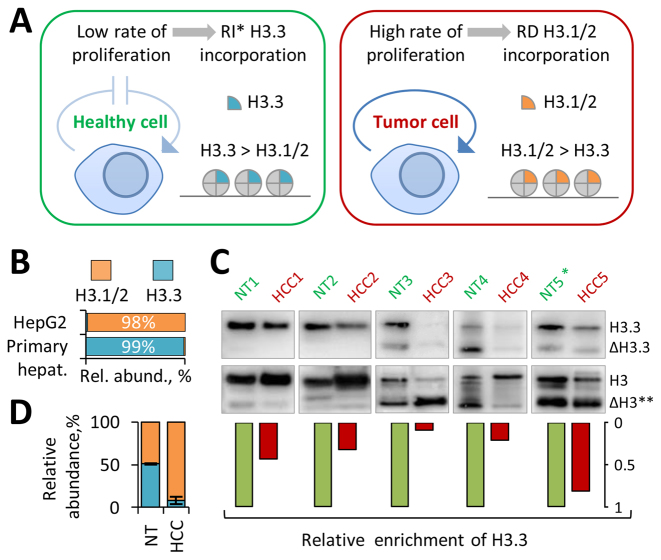
Given our finding that age-dependent replacement of H3.1/2 with H3.3 in mouse cells is not restricted to non-dividing post-mitotic tissues, such as heart and brain, but also occurs in slow-dividing mitotic tissues, such as liver and kidney, we next hypothesized that the relative abundance of H3 variants might reflect the proliferating status of the latter ones. Specifically, we hypothesized that, while the chromatin of slow-dividing tissues would be preferentially marked by H3.3 when these tissues are in homeostasis, i.e. have a low cell turnover rate, the level of H3.3 will decrease markedly if they become abnormally proliferating, such in the case of cancer (Figure 2A). Given the fact that each time the cell divides it doubles the amount of histones by expressing canonical H3 isoforms, we hypothesized that active cell proliferation would lead to overall dilution of the H3.3 variant with H3.1/2 isoforms being incorporated into chromatin through the replication-dependent (RD) nucleosome assembly pathway. To test this we first compared the relative abundance of H3 variants in primary human hepatocytes derived from healthy human liver and hepatocellular carcinoma cell line HepG2/C3A using quantitative MS analysis. In agreement with our hypothesis, H3.3 represented over 99% of the total H3 pool in primary human hepatocytes (Figure 2B), which was consistent with our results obtained from mouse liver, while the relative abundance of H3.3 in the HepG2/C3A cell line was only 1.5%. To evaluate whether the relative abundance of H3 variants can be potentially used as a biomarker of abnormal cell proliferation activity in vivo, we next compared the relative abundance of H3.3 between human hepatocellular carcinoma (HCC) tissues and adjacent non-tumor (NT) liver tissues obtained from five individual patients. Western blot analysis with H3.3-specific antibodies demonstrated pronounced enrichment of H3.3 in NT samples as compared to HCC samples in four out of five cases (Figure 2C). These results were further validated by targeted MS analysis that showed that the relative abundance of H3.3 was about five times higher in NT samples as compared to HCC samples (Figure 2D). Notably, the pair of tumor and adjacent non-tumor tissues that demonstrated similar levels of H3.3 was obtained from the patient diagnosed with liver cirrhosis, a condition that was previously associated with elevated hepatocytes proliferation activity (52). Collectively, these results suggest that the relative abundance of H3 variants can be used as a biomarker for abnormal cell proliferation activity in vivo and highlight its potential utility as a tumor biomarker.
Figure 2.
H3.3 is enriched in non-tumor human hepatocytes but is depleted in hepatocarcinoma cells (A) Schematic model depicting the putative difference in H3 variant dynamics between healthy tissues with low cell proliferation rates (such as liver) and their highly proliferating tumor counterparts. * RI – replication-independent. (B) Relative abundance of histone H3 variants in primary human hepatocytes and C3A/HepG2 cell line as revealed by middle-down MS analysis. (C) Immunoblot analysis of H3.3 level in whole-cell extracts from human hepatocellular carcinoma (HCC) tissues and adjacent non-tumor (NT) liver tissues derived from five individual patients. Representative western blot pictures and semi-quantitative data are presented. Total H3 was used as loading control and for normalization. (*) Cirrhosis liver; (**) proteolytically processed H3 proteoforms (42,46). (D) Relative abundance of H3 variants in pooled HCC and pooled NT samples as revealed by quantitative targeted bottom-up MS analysis.
Accumulation of H3.3 with age is accompanied by profound changes in the global levels of H3 methyl modifications
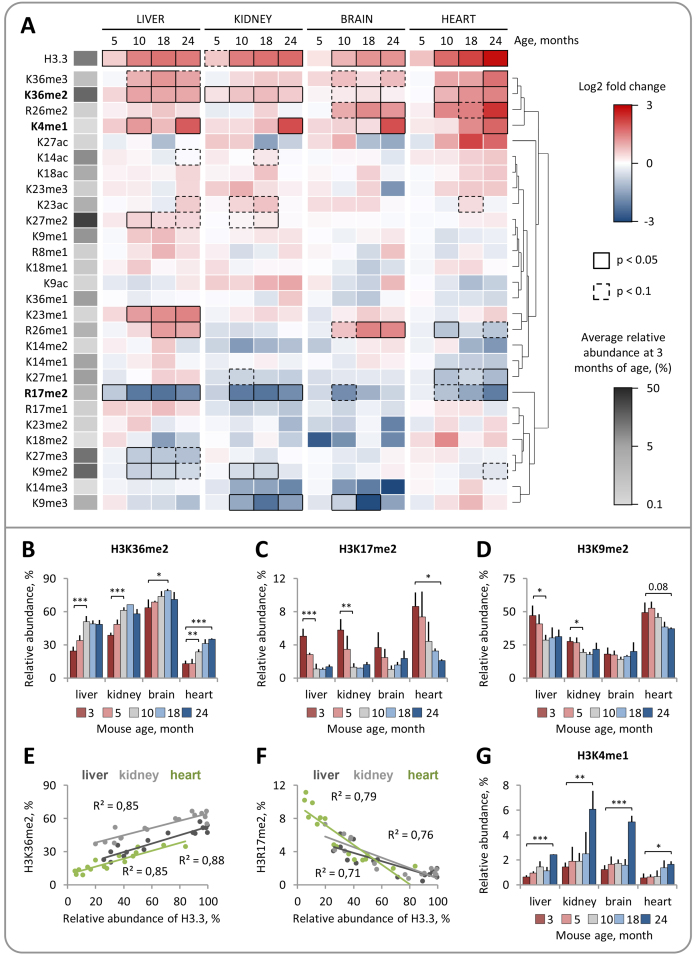
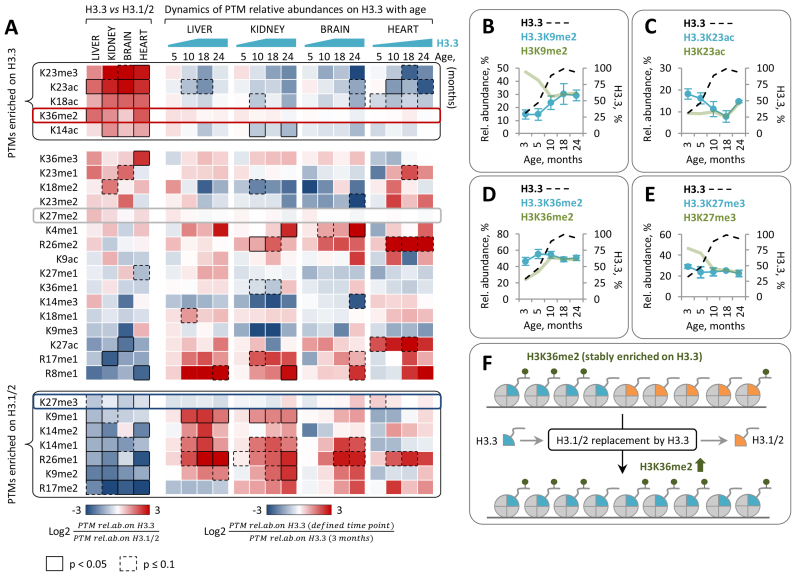
There is increasing evidence that H3.3 is functionally distinct from its canonical counterparts H3.1 and H3.2 (36). The amino acid sequence variations among the H3 variants were recently found to directly affect the deposition and recognition of PTMs on H3 N-terminal tails. In Arabidopsis thaliana, the histone methyltransferase ATXR5 selectively monomethylates K27 on histone H3.1 but not on the H3.3 variant (38). In mammals, candidate tumor suppressor ZMYND11 selectively recognizes K36me3 on H3.3 and co-localizes with H3.3K36me3 within gene bodies (35,37). These findings raise the possibility that histone H3 variant replacement may affect the levels of H3 PTMs and their functional readout. To explore this idea, we investigated whether progressive replacement of H3.1/2 isoforms with H3.3 in mouse cells is accompanied by changes in the global levels of H3 PTMs. We quantified the relative abundances of 28 distinct H3 PTMs at 5 time points across the mouse lifespan using middle-down MS (Supplementary Table S1, Supplementary Figure S3). Quantitative MS analysis revealed a number of H3 PTMs, including methylation at distinct lysine and arginine residues, whose relative abundances were significantly altered with age (Figure 3A). Remarkably, the relative abundances of several PTMs exhibited a similar pattern of change in all analyzed tissues and these changes were concurrent with an accumulation of H3.3. The relative abundance of H3K36me2 in liver and kidney gradually increased by ∼110% and ∼60%, respectively, from 3 to 10 months of age and remained relatively stable thereafter (Figure 3B). In the heart, the relative abundance of H3K36me2 demonstrated an increasing trend over the entire time course consistent with a continuous accumulation of H3.3. A similar increase with age was observed for the relative abundance of H3K36me3 (Supplementary Figure S4A). By contrast, the relative abundance of H3R17me2, a PTM which was previously associated with transcriptional activation (53,54), gradually decreased during the first 10 months of animal life in liver, kidney and brain and during the 24 months in the heart (Figure 3C). A similar decreasing trend was observed for the relative abundance of H3K9me2, which dropped by 30% and 40% by the age of 10 months in kidney and liver tissues, respectively, and by 24% by the age of 24 months in heart (Figure 3D); the relative abundance of H3K9me2 in brain remained at the stable level throughout the mouse lifespan. Notably, the relative abundances of two age-regulated PTMs, H3R17me2 and H3K36me2, were strongly correlated with the relative abundance of H3.3, suggesting that changes in the levels of these PTMs might be directly associated with accumulation of H3.3 (Figure 3E and F).
Figure 3.
Middle-down mass spectrometry reveals distinct, dynamic changes in the relative abundances of individual H3 PTMs during mouse lifespan. (A) Dynamic changes in H3 PTM abundances in somatic mouse tissues with age (3–24 months). The heat map shows hierarchical clustering of the log2 transformed fold changes in H3 PTM abundances at indicated time points relative to those at 3 months of age. Significantly up- and down-regulated PTMs (Limma test with Benjamini-Hochberg correction for multiple comparisons) are indicated by black boxes (continuous line for P < 0.05; dashed line for P < 0.1) as shown in the color key. (B–D) Relative abundances of H3 PTMs that demonstrated a similar pattern of change with age in different tissues. The level of H3K36me2 increases during the adulthood (months 3–10) (B) and correlates with the relative abundance of H3.3 (E). The level of H3R17me2 decreases during the adulthood (months 3–10) (C) and correlates with the relative abundance of H3.3 (F). The level of H3K9me2 decreases during the adulthood (months 3–10) in liver and kidney tissues (D). H3K4me1 is upregulated in tissues derived from 24-month-old mice (G). Error bars indicate SD (n = 2–4 biological replicates).
Unlike H3 PTMs that were up- or down-regulated concurrently with an accumulation of H3.3, H3K4me1 displayed a pattern of change, which was strongly associated with the late aging period. The level of H3K4me1 remained relatively stable during the first 18 months of mouse life but increased by 3–4-fold in all analyzed tissues derived from 24-month-old animals (Figure 3G). The drastic increase in the level of H3K4me1 in aged mice can potentially be explained by the well-documented age-dependent decline in DNA methylation in rodents, which is, in turn, associated with the appearance of H3K4me1 (55,56).
Some H3 PTMs, such as H3K9me3, H3R26me1 and H3K27me2/me3, were only regulated in one or two tissues or demonstrated a distinct pattern of change between tissues, suggesting the cell type-specific character of these chromatin rearrangements (Figure 3A; Supplementary Figure S4). Finally, in contrast to methylation, the level of H3 acetylation remained relatively stable across the entire mouse lifespan, indicating that this type of modification is less susceptible to age-dependent changes in chromatin organization (Figure 3A; Supplementary Figure S4). The relative abundances of H3K14ac, H3K18ac and H3K23ac demonstrated a modest (<1.6-fold), yet insignificant, increase, while the relative abundances of H3K9ac and H3K27ac oscillated between 1% to 3% with no consistent pattern of change (Supplementary Figure S4, Supplementary Table S1). Collectively, these results provide the first comprehensive quantitative characterization of dynamic changes in relative abundances of individual H3 PTMs during the mouse lifespan and demonstrate that histone methylation, which is traditionally viewed as a relatively stable epigenetic mark (57), undergoes profound changes in somatic tissues with age concurrently with progressive replacement of H3.1/2 by the H3.3.
A distinct dynamics of PTM levels on H3.3 throughout lifespan suggests a causative relationship between H3 variant replacement with age and accumulation of H3K36me2
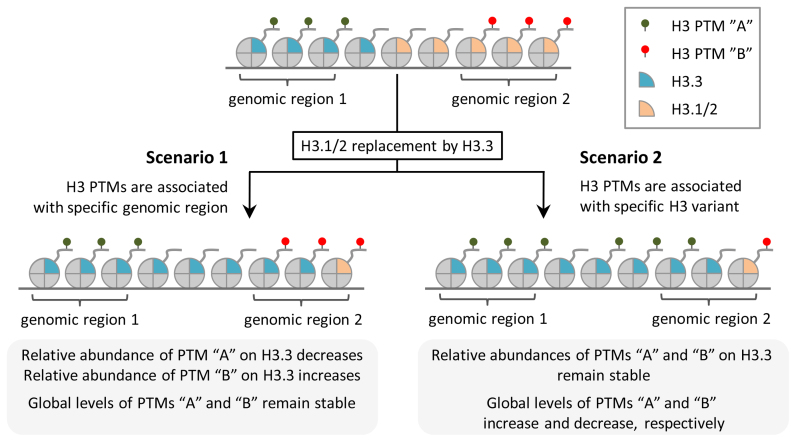
Given our finding that H3.3 accumulation is concurrent with profound changes in the relative abundances of several H3 methyl PTMs, we next hypothesized that H3 variant replacement might be directly involved in modulating the levels of these PTMs with age. If replacement of canonical H3.1/2 isoforms by the H3.3 variant alters the PTMs pattern present on chromatin, then we presumed that the following criteria must be fulfilled: (i) the relative abundance of PTMs should be different between H3.3 and H3.1/2; (ii) the relative abundance of PTMs on H3.3 should be independent of H3.3 level in the chromatin and H3.3 genomic distribution. The latter is important to distinguish between two possible scenarios (Figure 4). First, given the fact that H3.3 variant and H3 PTMs are not uniformly distributed across the genome, H3.3 can exhibit distinct levels of certain PTMs (different from those for H3.1/2) due to its preferential localization within genomic regions that are specifically enriched (depleted) in these PTMs independent of which H3 variant is deposited at these regions. This scenario relies on the fact that H3.3 tend to associate with active chromatin regions exhibiting fast H3 turnover, such as promoters and enhancers (58). If this scenario is correct, then the relative abundances of the PTMs on H3.3 would be dependent on its genomic distribution and would change concurrently with its genome-wide spread with age (Figure 4, ‘scenario 1’). The global levels of PTMs on total H3 would, in contrast, remain stable and would be unaffected by the H3 variant replacement with age. The second scenario implies that a certain PTM(s) can be specifically enriched/depleted on H3.3 due to the functional distinctness between H3 variants and regardless of their genomic distributions. For example, a certain enzyme can preferentially modify only one of the H3 variants, as was recently shown for H3.1-specific methyltransferase ATXR5 (38). If this scenario is correct, then the relative abundance of the PTM(s) on H3.3 would be stable and independent of its distribution across the genome and its level in the chromatin (Figure 4, ‘scenario 2’). In this case, H3 variant replacement would directly affect the global level of the PTM(s).
Figure 4.
Two possible scenarios for the relationship between H3 variant replacement and H3 PTMs dynamics. Scenario 1: H3 PTMs are associated with specific chromatin regions regardless of which H3 variant is deposited at these regions (left panel). Replacement of canonical H3.1/2 isoforms by the H3.3 variant does not affect global levels of H3 PTMs. Relative abundances of PTMs on H3.3 change consistently with an accumulation of H3.3. Scenario 2: H3 PTMs are associated with H3.3 variant (right panel). Replacement of canonical H3.1/2 isoforms by the H3.3 variant is associated with changes in global levels of H3 PTMs. Relative abundances of PTMs on H3.3 remain stable after H3.3 accumulation.
To test whether H3.3 meets the criteria described above we determined the relative abundances of PTMs on H3 variants at 5 time points across the mouse lifespan using middle-down MS (Supplementary Table S2). In a first step, we compared the relative abundances of PTMs between H3 variants in tissues derived from young mice where H3.3 represented 10–40% of the total H3 pool (Figure 5A, left panel). In agreement with previous reports, we found that H3.3 exhibits 2–8 times higher level of acetylation at K14, K18 and K23 compared to H3.1/2 (Supplementary Figure S5) (51,59). H3.3 was also enriched in K23me3 and K36me2. In contrast, the two well-known heterochromatin PTMs, K9me2 and K27me3, as well as transcription-associated arginine methylations, R17me2 and R26me1, were enriched on canonical H3.1/2 isoforms (Figure 5A, left panel; Supplementary Figure S5).
Figure 5.
Relative abundances of PTMs on H3.3 exhibit distinct pattern of change with age. (A) Left panel: the heat map shows the difference in the relative abundances of PTMs between H3.3 and H3.1/2 isoforms in tissues derived from 3-month-old mice. Differentially abundant PTMs (paired t-test with Benjamini-Hochberg correction for multiple comparisons) are indicated by black boxes (continuous line for P < 0.05; dashed line for P < 0.1) as shown in the color key. Right panel: the heat map shows the log2 fold change in the relative abundances of PTMs on H3.3 at the indicated time points as compared to those at 3 months of age. Data shown in (B–E) are for liver tissue. (B) Relative abundance of H3.3K9me2 increases with age consistently with an accumulation of H3.3. (C) Relative abundance of H3.3K23ac decreases with age consistently with an accumulation of H3.3. (D–E) Relative abundances of H3.3K36me2 (D) and H3.3K27me3 (E) remain stable independently of mouse age and H3.3 relative abundance. For H3.3 PTM dynamics in kidney, brain and heart tissues see Supplementary Figure S6. (F) A putative model illustrating the causal relation between H3 variant replacement with age and increase in the global level of H3K36me2.
To gain insight into potential causes of the differences in PTM levels between H3 variants, we examined whether and how the relative abundances of PTMs on H3.3 change with increasing H3.3 level and mouse age. The relative abundances of the majority of PTMs on H3.3 showed significant changes with age-dependent accumulation of H3.3 (Figure 5A, right panel), suggesting that these PTMs are likely associated with specific chromatin regions rather than with H3.3 or H3.1/2 variants themselves (Figure 4, ‘scenario 1’). The relative abundances of PTMs that were enriched on H3.3 compared to H3.1/2 at 3 months of age, such as K23ac and K23me3, decreased on H3.3 consistently with the increase in the level of this histone variant with age (Figure 5A and C; Supplementary Figure S6). By contrast, the relative abundances of PTMs that were depleted on H3.3 at 3 months of age, such as K9me2 and R26me1, showed an increase on H3.3 with age (Figure 5A and B; Supplementary Figure S6). Collectively, these findings suggest that the differences in the relative abundances of K9me1/me2, K14me1/me2/ac, K18ac, K23me3/ac and K26me1 between H3.1/2 and H3.3 variants at 3 months of age were likely arising from the difference in their genomic distributions. Interestingly, the relative abundances of 3 out of 28 measured PTMs on H3.3, including K36me2, K27me2 and K27me3, remained stable throughout the entire mouse lifespan and independent of the H3.3 level in chromatin (Figure 5A, D–E). This, together with the fact that K36me2 was highly (up to 4-fold) enriched on H3.3 as compared to H3.1/2 (Supplementary Figure S5), indicate that the higher level of K36me2 on H3.3 is likely not dictated by the H3.3 genomic distribution but rather can be attributed to the specific property of the H3.3 variant itself (Figure 4, ‘scenario 2’). We, therefore, propose that the increase in the global level of H3K36me2 with age is likely driven by the progressive replacement of H3.1/2 with the H3.3 variant (Figure 5F). Unlike K36me2, K27me2 and K27me3 exhibited relatively similar relative abundances on both H3 variants (≤1.8 - fold difference). K27me2 demonstrated a modest, yet insignificant, enrichment on H3.3, while K27me3 was enriched on H3.1/2. The most profound difference in the levels of these PTMs between H3 variants was observed in liver chromatin, where the relative abundances of K27me2 and K27me3 on H3.3 were 1.7-times higher and 1.8-times lower, respectively, as compared to H3.1/2. Consistently with this, replacement of H3.1/2 by the H3.3 in liver was associated with a partial switch from tri- to dimethylation state at K27 (Supplementary Figures S3 and S4). However, the levels of H3K27me2 and H3K27me3 in other tissues remained relatively stable throughout the lifespan. Collectively, these findings suggest a causative relationship between the H3 variant replacement with age and the dynamic changes in the global level of H3 methylation, including an increase in the level of H3K36me2 observed in liver, kidney and heart chromatin and a decrease in the level of K27me3 observed in liver chromatin.
H3 variant replacement with age is associated with profound changes in combinatorial H3 PTMs
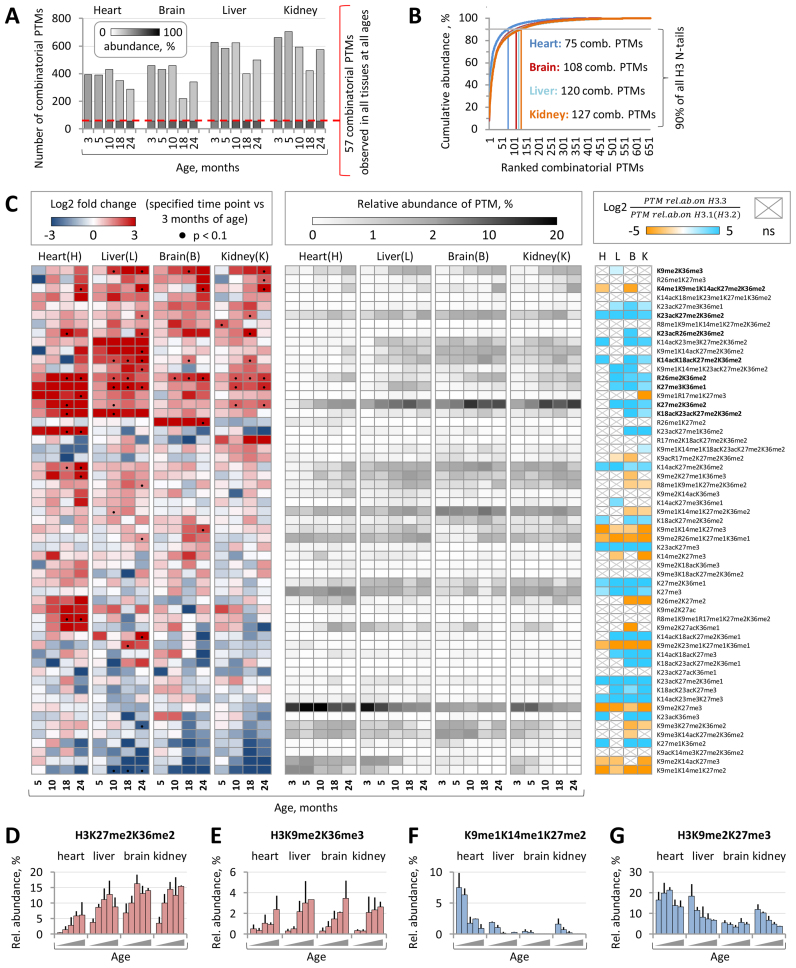
The N-terminal tails of histone proteins are often decorated by multiple PTMs that form distinct combinatorial modification patterns (60). Importantly, different combinations of coexisting PTMs can promote distinct biological outcomes indicating that at least some of the regulatory effects of histone PTMs are mediated by their cooperative action. However, our knowledge about the combinatorial histone modification landscape and its temporal changes during mammalian development and aging still remains very limited. To fill this gap, we took advantage of our middle-down MS platforms and performed an extensive quantitative analysis of the dynamic changes in combinatorial H3 PTMs throughout mouse lifespan. We identified and quantified from 200 to over 600 distinct combinatorial PTMs present on the H3 N-terminal tails in four mouse tissues at five different ages (Figure 6A). Among those, 57 H3 PTMs were reproducibly detected in all analyzed biological samples. Many of these PTMs were highly abundant (relative abundance > 1%) and together accounted for 53–76% of all quantified H3 peptides depending on tissue type and mouse age (Figure 6A). The top five most abundant combinatorial H3 PTMs at 3 months of age included H3K9me2K27me3, H3K9me1K14me1K27me2K36me2, H3K27me3 (given the fact that this single modification reflects specific H3 modification state where all residues except for K27me3 are unmodified (un), i.e. K4unK9unK14unK23unK27me3K36un, we considered it along with combinatorial PTMs), H3K27me2K36me2 and H3K9me2K14acK27me3 (Supplementary Figure S7A). Among those, H3K9me2K27me3 represented the most abundant combinatorial PTM in all analyzed tissues (relative abundance ranging from 5% to 18%) except for the brain. In the brain, the most abundant combinatorial PTM was H3K9me1K14me1K27me2K36me2 with the relative abundance of 8% (Supplementary Figure S7C). Notably, approximately 100 most abundant combinatorial PTMs in each tissue accounted for 90% of all quantified H3 peptides, while the remaining several hundred lower abundant PTMs accounted for only 10% of the H3 peptides (Figure 6B). On one hand, these data illustrate the great complexity of combinatorial PTM patterns present on the H3 N-terminal tails; on the other hand, it indicates that the vast majority of chromatin is decorated by a very limited number of abundant combinatorial H3 modifications.
Figure 6.
Combinatorial H3 PTM landscape undergoes dynamic changes with age. (A) The total numbers of distinct combinatorial H3 PTMs identified and quantified by the middle-down MS in four mouse tissues at the five indicated time points. 57 combinatorial PTMs were reproducibly detected in all biological samples. Relative abundances of H3 proteins marked by one of the 57 common or by one of remaining PTMs in each sample are indicated by grayscale shading as shown in the color key. (B) The cumulative relative abundance of combinatorial H3 PTMs ranked according to their abundance. (C) Left panel: the heat map shows the log2 fold change in relative abundances of 57 common combinatorial PTMs at the indicated time points as compared to those at 3 months of age. Statistically significant differences (P < 0.1, Limma test with Benjamini-Hochberg correction for multiple comparisons) are indicated by black dots. Central panel: the heat map shows the relative abundances of 57 common H3 combinatorial PTMs at the indicated time points. Right panel: the heat map shows the difference in average relative abundance of 57 common H3 combinatorial PTMs between H3.3 and H3.1/2 isoforms in tissues derived from 3- and 5-month-old mice. H3.3- and H3.1/2-enriched PTMs (P < 0.05, paired t-test with Benjamini–Hochberg correction for multiple comparisons) are shown in cyan and orange, respectively; non-significant differences are indicated by gray crosses. Relative abundances of H3K27me2K36me2 (D) and H3K9me2K36me3 (E) increase with age in all analyzed tissues, while the relative abundance of H3K9me1K14me1K27me2. (F) decreases with age. The relative abundance of H3K9me2K27me3 (G) decreases in liver and kidney chromatin.
We next investigated whether and how the relative abundances of combinatorial H3 PTMs change with age. We focus on 57 PTMs that were reproducibly quantified by MS in all analyzed biological samples. Among those, nine PTMs demonstrated significant increase with age in at least two different tissues (Figure 6C). The most profound increase in the PTM level between 3- and 24-month-old mice was observed for H3K27me2K36me2 which relative abundance raised from ∼4% to 9% and from 4% to 15% in liver and kidney tissues, respectively, from 7% to 14% in brain and from 0.4% to 6% in heart (Figure 6D). A similar pattern of change was observed for H3R26me2K36me2 and H3K27me3K36me1, which relative abundances demonstrated a several fold increase with age in all analyzed tissues. Interestingly, while the relative abundance of H3K27me3K36me1 in liver increased from 0.2% at 3 months of age to 2.4% at 10 months (Figure 6C), the relative abundance of H3K27me3 demonstrated the opposite pattern of change and dropped from 46% to 23% (Supplementary Figure S4). A similar discordance was observed for H3K9me2K36me3 whose relative abundance showed 5–10 times increase with age in all analyzed tissues (Figure 6E) and H3K9me2 whose relative abundance dropped during the same period by 20–34% (Figure 3D). These results indicate that the relative abundances of individual H3 PTMs and combinatorial H3 PTMs involving these individual PTMs can exhibit distinct dynamics with age. Other combinatorial PTMs that exhibited a significant increase in abundance with age included H3K4me1K9me1K14acK27me2K36me2, H3K23acR26me2K36me2, H3K23acK27me2K36me2, H3K18acK23acK27me2K36me2 and H3K14acK18acK27me2K36me2. Of note, five out of nine combinatorial PTMs that were upregulated with age contained both K27me2 and K36me2, indicating a drastic increase in the level of K27me2K36me2-modified H3 N-terminal tails in the chromatin of middle-aged and old mice (this observation is discussed below in more detail). In contrast to PTMs listed above, the relative abundances of H3K27me1K36me2, H3K9acK14me3K27me2K36me2, H3K9me2K14acK27me3 and H3K9me1K14me1K27me2 demonstrated consistent decrease with age in at least three different tissues (Figure 6C and F). Similarly, the relative abundance of H3K9me2K27me3 dropped from 18% and 12% at 3 months of age to 7% and 4% at 24 months in liver and kidney tissues, respectively, while, in heart and brain, it oscillated in a range of 13–21% and 3–6%, respectively, with no clear pattern (Figure 6D). Collectively, these changes resulted in a prominent reorganization of the combinatorial H3 PTM landscape in the chromatin of aged mice. In contrast to young mice, where the most abundant combinatorial PTM was H3K9me2K27me3, the chromatin of 24 months old animals was preferentially decorated by H3K27me2K36me2 that represented the most abundant combinatorial PTM in all tissues except for heart (Supplementary Figure S7B). In the heart, the most abundant combinatorial PTM was H3K9me2K27me3 (Supplementary Figure S7D). The other abundant PTMs in the chromatin of aged mice included H3K9me2K27me3K36me1, H3K9me1K14me1K27me2K36me2 and H3K27me3.
We next aimed to evaluate the possible causal relationship between H3 variant exchange and dynamic changes in combinatorial H3 PTM landscape with age. Following the same logic as above (Figure 4), we first compared the relative abundances of combinatorial PTMs between H3 variants in tissues isolated from 3 months old mice. We identified 85 and 58 combinatorial PTMs that were significantly enriched on H3.3 and H3.1/2, respectively, in at least one mouse tissue (Supplementary Figure S8). Among those, 10 and 8 PTMs demonstrated consistent enrichment on H3.3 and H3.1/2, respectively, in all analyzed tissues. Next, we evaluated whether and how the relative abundances of these combinatorial PTMs on H3.3 changes with mouse age concurrent with an accumulation of H3.3. Most of the combinatorial PTMs that were specifically enriched (depleted) on H3.3 in chromatin of young mice—when H3.3 occupied only very limited part of the chromatin—lost the H3.3-enriched (depleted) profile when H3.3 spread to nearly entire chromatin with age (Supplementary Figures S9–S12). These data indicate that similar to the individual H3 PTMs, the majority of combinatorial H3 PTMs are likely associated with specific genomic regions (chromatin domains) rather than with H3.3 or H3.1/2 variants themselves (Figure 4, scenario 1). Interestingly, several PTMs, such as H3K27me2K36me2, R26me2K36me2 and H3K27me3K36me1, remained stably enriched on H3.3 throughout the entire mouse lifespan and independent of H3.3 abundance in the chromatin (Supplementary Figures S9–S12), indicating that they might be directly associated with H3.3 regardless of where it deposited in the genome (Figure 4, scenario 2). These results suggest a causative link between the dynamic increase in the global levels of these combinatorial PTMs with age and accumulation of H3.3. In total, six out of nine PTMs that were significantly upregulated with age also demonstrated specific enrichment on H3.3 in at least two different tissues (Figure 6C, right panel). The quantitative information on combinatorial H3 PTM abundances for each analyzed mouse tissue is publicly available in the CrossTalkDB repository at http://crosstalkdb.bmb.sdu.dk. Collectively, our findings indicate that the relative abundances of combinatorial H3 PTMs undergo dynamic changes during mouse lifespan and suggest that some of these changes might be driven by the H3 variant replacement with age.
Global levels of H3 methylation at K9, K27 and K36 are strongly correlated with each other
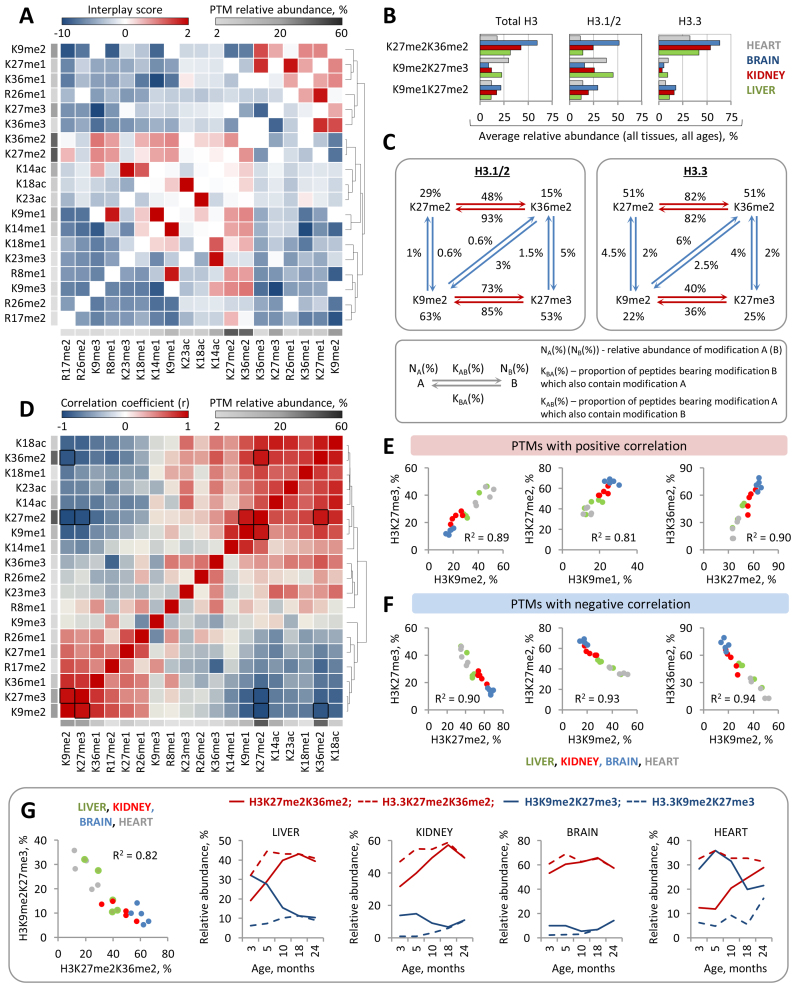
Histone PTMs are known to affect each other by attracting or repelling different histone modifying enzymes, such as modification ‘writers’ and ‘erasers’. For example, methylation of H3K36 was found to antagonize PRC2-mediated methylation of H3K27 (61). Along with other examples, histone PTM crosstalk is emerging as an important regulatory mechanism for modulating chromatin states and transcriptional activity (62). Yet, very few studies have investigated the functional relation between distinct histone PTMs on the large scale. Hence, we next aimed to evaluate the possible crosstalk between various H3 N-terminal PTMs using the quantitative data on combinatorial H3 PTMs defined by middle-down MS and our previously described mathematical model (45). Briefly, we compared the observed co-frequencies of binary histone PTMs to their expected co-frequencies calculated based on the assumption that the two modifications in each pair are independent of each other. The expected co-frequency of binary modification AB was therefore calculated by multiplying the relative abundances of modification A by modification B. To evaluate the crosstalk between two histone PTMs we used interplay score, calculated as log2(observed frequency of binary PTM/expected frequency of binary PTM), where highly negative interplay values indicate that the two PTMs are mutually exclusive, while positive interplay values suggest codependency of the two PTMs (for more details see Materials and Methods). Using this model we evaluated the possible crosstalk between 19 distinct H3 PTMs representing the major acetylation and methylation events (relative abundance ≥ 2%) on the H3 N-terminal tail (Figure 7A). Our analysis confirmed a number of previously described PTM crosstalks, such as the mutually exclusive relation between H3K9me3, H3K27me3 and H3K36me3, and revealed several previously uncharacterized relationships between H3 PTMs, such as the positive interplays between H3K9me2 and H3K36me3 and between H3K9me3 and H3K14ac (Figure 7A). The latter two examples are particularly interesting as they involve PTMs that are generally believed to have the opposite effect on gene expression, where H3K9me2 and H3K9me3 are associated with repressed chromatin state and H3K14ac and H3K36me3 are associated active loci. Although the functional significance of co-occurrence of H3K9me3 and H3K14ac on the same H3 tails is unclear, a recent ChIP-seq study in mouse embryonic stem cells (mESCs) showed that H3K14ac colocalizes with H3K9me3 on a subset of inactive promoters, suggesting that the combination of these PTMs might be involved in the dynamic transcriptional regulation (63). Although unexpected, the co-enrichment of K9me2 and K36me3 on the same H3 tails was consistent with a previous study showing that H3K36me2 exhibits profound enrichment at different heterochromatin domains marked by H3K9me2 (64).
Figure 7.
Analysis of histone modification crosstalk reveals a strong correlation between global levels of H3 methyl marks. (A) The heat map shows hierarchical clustering of interplay scores between H3 N-terminal PTMs. PTMs that tend to colocalize together (positive interplay values) or demonstrate mutually exclusive profile (negative interplay values) are indicated by red and blue colors, respectively; the average PTMs abundances are indicated by grayscale shading as shown in the color key. (B) Co-existence frequencies of the top three most abundant binary PTMs on total H3 and their co-existence frequencies on H3.3 and H3.1/2. (C) The scheme depicts a co-occurrence network between individual H3 PTMs involved in the top two most abundant binary PTMs specified in (B). (D) The heat map shows hierarchical clustering of Pearson's correlation coefficients between relative abundances of H3 N-terminal PTMs among all analyzed histone samples. Positive and negative correlations are indicated by red and blue colors, respectively, as shown in the color key; Pairs with strong correlation (|R| > 0.9) are outlined by black boxes. The average PTM abundances are shown as in (A). (E) Scatter plots highlight pairs of H3 PTMs whose relative abundances exhibit strong positive or negative (F) correlation. (G) Relative abundances of the two binary PTMs, H3K9me2K27me3 and H3K27me2K36me2, exhibit strong negative correlation, indicating the mutually exclusive relation between them. In line with the distinct stable levels of H3K9me2K27me3 and H3K27me2K36me2 on H3.3 and H3.1/2, H3 variant replacement with age is associated with profound changes in the global levels of these binary PTMs.
Notably, the majority of PTM pairs demonstrated the similar interplay values in all analyzed tissues and in all age groups indicating that PTM crosstalk on H3 is conserved between different somatic cell types as well as during postnatal mammalian development (Supplementary Figure S13). We also observed a close correlation between the interplay values for PTM pairs present on H3.1/2 and H3.3 suggesting that the functional relationship between H3 PTMs is largely independent of which H3 variants they are deposited on (Supplementary Figure S14).
We next aimed to investigate in more detail the relation between H3 PTMs that form the most abundant combinations occupying the majority of H3 tails. The top three pairs of PTMs that showed the highest co-frequencies were K27me2K36me2, K9me2K27me3 and K9me1K27me2 (Figure 7B). Expectedly, these PTM pairs involved highly abundant individual PTMs that exhibited positive interplay between each other. Consistently with the difference in the relative abundances of individual PTMs between H3 variants, the co-frequency for K27me2K36me2 was higher on H3.3 as compared to that on H3.1/2, while K9me2K27me3 was, in contrast, more abundant on H3.1/2. Interestingly, despite the fact that the PTMs in these pairs exhibited positive interplay with each other on both H3 variants, the absolute overlap between them, i.e the proportions of H3 tails modified with PTM ‘A’ that also bear PTM ‘B’, demonstrated substantive difference between H3.3 and H3.1/2 (Figure 7C, Supplementary Figure S15). Specifically, while about 80–90% of all K27me2-modified H3.3 tails were also bearing K36me2, and vice versa, about 80% of K36me2-modified H3.3 tails were bearing K27me2, the prominent overlap between these PTMs were partially disrupted on H3.1/2. In the case of H3.1/2, about 90% of all K36me2-modified histone tails were bearing K27me2, but only about 50% K27me2-modified tails were bearing K36me2. These results indicate that while K27me2 and K36me2 nearly always appear together on H3.3, a substantial proportion of H3.1/2 bear K27me2 without K36me2. Notably, we observed very little overlap between binary PTMs K9me2K27me3 and K27me2K36me2, i.e. both K9me2 and K27me3 very rarely coexisted with either K27me2 or K36me2 and vice versa (Figure 7C).
Although the interplay score provides a useful metric for predicting possible functional cross-talk between histone PTMs, it has a major limitation in terms of positive predictive value. The fact the two PTMs co-localize on the same histone tail much more or much less often than if they were distributed in chromatin independent of each other does not necessarily imply that they exhibit positive or negative functional crosstalk with each other. In order to further evaluate the functional relationship between distinct H3 PTMs, we performed a correlation analysis between the global levels of H3 PTMs using the quantitative MS data obtained from the analysis of different mouse tissues at different ages (Figure 7D). We presumed that if two PTMs exhibit positive (or negative) crosstalk, i.e. one PTM facilitate the deposition (or removal) of the other one, then the relative abundances of these PTMs should be positively (or negatively) correlated. We identified six pairs of PTMs which relative abundances were closely correlated (R > 0.9) with each other (Figure 7E and F), suggesting that one PTM in each pair might be functionally affecting the other one. Of note, the pairs of PTMs that demonstrated positive correlation, including H3K9me2-H3K27me3, H3K9me1-H3K27me2 and H3K27me2-H3K36me2, were identical to the top three most abundant binary H3 PTMs described above. Unlike PTMs that demonstrated codependency, some of the abundant PTMs seemed to compete with each other for occupying the H3 N-terminal tail. Specifically, a strong negative correlation was found between the relative abundances of H3K27me2 and H3K27me3, H3K9me2 and H3K27me2 and between H3K9me2 and H3K36me2 (Figure 7F). This was in agreement with the fact that these PTMs are almost never colocalized together on the same H3 molecules (Figure 7C). Interestingly, a similar negative correlation was observed between the relative abundances of binary PTMs H3K9me2K27me3 and H3K27me2K36me2 (Figure 7G). In line with the fact these two binary PTMs almost never overlap with each other (Figure 7C) and together occupy approximately 50% of all H3 N-terminal tails (which corresponds to 50–100% of the genome), these results suggest that they might serve as specific PTM signatures separating the genome into two types of distinct chromatin states or non-overlapping chromatin domains. Notably, both H3K27me2 and H3K36me2 demonstrated higher co-occurrence frequencies with acetyl PTMs compared to H3K9me2 and H3K27me3. Furthermore, the relative abundances of H3K27me2 and H3K36me2 were positively correlated with the level of H3 acetylation, while the relative abundances of K9me2 and K27me3 demonstrated a negative correlation with acetyl PTMs (Figure 7D). Hence, we speculate that H3K27me2K36me2 might serve as a specific PTM signature of the open permissive chromatin state, while H3K9me2K27me3 might represent a PTM signature of the condensed restrictive chromatin state. The latter is in agreement with previous observations showing that H3K9me2 and H3K27me3 colocalize on the repressed chromatin (65). Notably, in line with a stable enrichment of K9me2K27me3 and K27me2K36me2 on H3.1/2 and H3.3, respectively, H3 variant replacement with age was associated with a decrease in the level of K9me2K27me3-marked H3 tails and with an increase in the level of K27me2K36me2-marked H3 tails (Figure 7G). This finding, together with the results described above, suggests that H3 variant replacement with age might play a causative role in the dynamic changes in the H3 modification landscape during mouse lifespan.
DISCUSSION
Here, we provide the first comprehensive characterization of the dynamic changes in histone H3 variant composition and H3 modification landscape throughout the lifespan in mice. The most prominent changes are summarized in Table 2. Although it is well appreciated that histone variant H3.3 replaces canonical H3.1/2 isoforms with age in mouse brain tissue (31,49), the lifelong dynamics of H3 variant replacement in other tissue types has remained undefined. We show that, similar to its enrichment profile in the mouse brain, H3.3 exhibit age-dependent genome-wide accumulation in other somatic tissues, including liver, kidney and heart (Table 2). Using quantitative middle-down MS we demonstrate that H3.3 accumulation rates vary between tissues. In liver, kidney and brain, H3.3 progressively replaces H3.1/2 isoforms during the adulthood and reaches saturation levels, i.e. ≈99% of the total H3 pool, by the age of 18 months, while in heart the relative abundance of H3.3 increases throughout life and raises only up to 76% of total H3 by the age of 24 months. Given our finding that heart chromatin exhibits specific H3 PTM profile which was previously associated with slow H3 turnover rate (50), we speculate that the global H3 variant replacement kinetics might be at least partially modulated by the cell-type specific H3 modification landscape. Together with previous studies (31,32,49), our results indicate that progressive age-dependent accumulation of H3.3 variant and consequential loss of canonical H3.1/2 isoforms constitute a conserved feature of chromatin organization in slow and non-dividing cells. Although the physiological relevance of H3 variant replacement with age remains largely undefined, a recent study revealed the essential role of H3.3 in regulating pro-longevity transcriptional programs in C. elegans (66). Furthermore, lifelong H3.3 turnover was found to be involved in the regulation of neuronal activity-dependent gene expression in mouse brain (31). Given our finding that H3.3 also accumulates in mouse liver, kidney and heart, it will be of high relevance to investigate whether H3.3 plays a role in the lifelong regulation of gene expression in these tissues.
Table 2. A summary of the changes in H3 variant composition and H3 modification abundances with age. * ‘-‘ indicates that there is no clear pattern of change or no significant difference.
| Changes in the relative abundances of H3 variants with age | |||||
| H3 variant | Change with age | Tissues | Note | ||
| H3.3 | Increase | Liver, kidney, brain, heart | Enriched in non-tumor human liver tissue | ||
| H3.1/2 | Decrease | Liver, kidney, brain, heart | Enriched in human hepatocellular carcinoma tissue | ||
| Changes in the relative abundances of H3 PTMs with age | |||||
| Modification | Change with age (total H3) | H3 variant-specific enrichment | Change with age (on H3.3) | Tissue(s) | |
| Individual PTMs | K36me2 | Increase | H3.3 | Stable abundance | Liver, kidney, brain, heart |
| K36me3 | Increase | - * | - | Liver, brain, heart | |
| K27me2 | Increase | - | Stable abundance | Liver, Kidney | |
| R26me2 | Increase | - | - | Brain, heart | |
| K4me1 | Increase | - | - | Liver, kidney, brain, heart | |
| R26me1 | Increase | H3.1/2 | Increase | Liver, brain | |
| Decrease | H3.1/2 | Increase | Heart | ||
| R17me2 | Decrease | H3.1/2 | - | Liver, kidney, brain, heart | |
| K27me1 | Decrease | - | - | Heart | |
| K27me3 | Decrease | H3.1/2 | Stable abundance | Liver | |
| K9me2 | Decrease | H3.1/2 | Increase | Liver, kidney | |
| K9me3 | Decrease | - | - | Kidney, brain | |
| Binary PTMs | K27me2K36me2 | Increase | H3.3 | Stable abundance | Liver, kidney, heart |
| K9me2K27me3 | Decrease | H3.1/2 | Stable abundance | Liver, heart | |
| Combinatorial PTMs | K9me2K36me3 | Increase | H3.3 (liver) | - | Liver, kidney, brain |
| K4me1K9me1K14ac-K27me2K36me2 | Increase | H3.1/2 (heart, brain) | - | Liver, kidney, brain, heart | |
| K23acK27me2K36me2 | Increase | H3.3 | - | Liver, Kidney | |
| K23acR26me2K36me2 | Increase | H3.3 (brain) | Increase (brain) | Liver, kidney, heart | |
| K14acK18acK27me2K36me2 | Increase | H3.3 | - | Liver, kidney, brain | |
| R26me2K36me2 | Increase | H3.3 | Stable/increase | Liver, kidney, brain, heart | |
| K27me3K36me1 | Increase | H3.3 | Stable/increase | Liver, kidney | |
| K27me2K36me2 | Increase | H3.3 | Stable/increase | Liver, kidney, heart | |
| K18acK23acK27me2K36me2 | Increase | H3.3 (kidney, brain) | - | Liver, heart | |
| K9me1K14me1K27me2 | Decrease | H3.1/2 | - | Liver | |
In contrast to its genome-wide accumulation profile in slow and non-dividing cells, H3.3 represents only a small fraction of the total H3 pool in proliferating cell culture models (59). Here we show that H3.3 is also highly depleted in human hepatocarcinoma tissue compared to adjacent non-tumor liver tissue. The difference in the relative abundances of H3.3 between actively proliferating tumor and non/slow-proliferating non-tumor liver cells can be explained by the fact that the level of H3.3 in chromatin gets diluted during each round of cell division due to the replication-coupled expression of canonical H3 and its deposition into newly synthesized DNA strands. We, therefore, propose that the relative abundance of H3.3 can be used as a biomarker of abnormal cell proliferation activity in vivo. The remarkable difference in H3.3 abundance between slow-proliferating non-tumor and rapidly proliferating tumor cells is particularly interesting in light of the recent findings demonstrating the existence of histone variant-specific PTM readers, such as the candidate tumor suppressor protein ZMYND11 (35) and the transcription factor ZMYND8 (67). It would be, therefore, important to investigate whether and how the decline in H3.3 level in cancer cells affects their transcriptional programs and chromatin properties.
Despite the fact that H3.3 differs from the canonical H3.2 and H3.1 by only four and five amino acids, respectively, it was hypothesized to directly affect chromatin organization (68). Consistently with this hypothesis, H3.3 was shown to play an important role in maintaining a decondensed chromatin state and high levels of the open chromatin modifications—H3K36me2 and H4K16ac—in fertilized mouse zygotes (69). Our study provides further evidence that H3.3 can be involved in modulating the histone modification landscape. We show that the relative abundances of a number of H3 PTMs, including both individual and combinatorial modifications, undergo dynamic changes with age, concurrently with an accumulation of H3.3 (Table 2). Remarkably, a subset of age-regulated PTMs, including K36me2, exhibit stable enrichment on H3.3, suggesting that the increase in their levels during the lifespan might be directly associated with accumulation of H3.3. Given the recent identification of H3.1 specific K27 methyltransferase in Arabidopsis thaliana (38), we speculate that the stable lifelong enrichment of K36me2 on H3.3 may be due to preferential methylation of H3.3 by an H3 variant-specific yet undefined H3K36 methyltransferase.
Next, we provide a large-scale evaluation of the relationship between 19 distinct H3 N-terminal PTMs. By comparing co-frequencies for pairwise combinations of these PTMs defined by MS with their co-frequencies calculated based on the assumption that the two PTMs in each pair are independent of each other, we identify a number of modifications demonstrating either mutually exclusive relation or co-enrichment. Interestingly, the relative abundances of several PTMs co-enriched of the same H3 tails, including H3K9me2-H3K27me3 and H3K27me2-H3K36me2 pairs, show strong positive correlation with each other, suggesting that one PTMs in each pair might facilitate the formation of the other one. This notion is in agreement with a recent study, demonstrating that the loss of PRC2, and subsequent loss of H3K27me3, reduces the amount of H3K9me2 present on the inactive X chromosome (Xi) (70). The relative abundances of several mutually exclusive PTMs, such as H3K9me2 and H3K36me2, in contrast, demonstrate strong negative correlation. Similar negative correlation is observed between the two most abundant binary PTMs H3K27me2K36me2 and H3K9me2K27me3. We speculate these two binary PTMs might represent PTM signatures of the two functionally distinct chromatin states where H3K27me2K36me2 marks permissive chromatin and H3K9me2K27me3 marks inactive chromatin. Interestingly, K27me2K36me2 is stably enriched on H3.3, while K9me2K27me3 is enriched on H3.1/2 and H3 variant replacement with age is associated with a dynamic increase in the K27me2K36me2 level and a decrease in the K9me2K27me3 level in the chromatin (Table 2). In line with the above, these results further support the notion that H3 variant replacement can be directly involved in modulating the histone H3 modification landscape with age. In summary, our study provides the first comprehensive characterization of the dynamic changes in histone H3 PTM levels during mouse lifespan and links these changes to the progressive age-dependent accumulation of the histone variant H3.3.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Bente Finsen, Institute of Molecular Medicine, University of Southern Denmark, Odense, Denmark, for providing C57Bl6 mice. We thank associate professors Krzysztof Wrzesinski and Stephen J. Fey for providing the C3A/HepG2 cell line. We thank Professor Anja Groth at University of Copenhagen and Center for Epigenetics for critical reading of this manuscript and helpful comments.
Footnotes
Present address: Andrey Tvardovskiy, Institute of Functional Epigenetics, Helmholtz Center Munich, 85764 Neuherberg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
VILLUM Center for Bioanalytical Sciences (VILLUM Foundation); Center for Epigenetics from the Danish National Research Foundation (DNRF) [#82]. Funding for open access charge: SDU [95-310-33125-01400].
Conflict of interest statement. None declared.
REFERENCES
- 1. Timp W., Feinberg A.P.. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer. 2013; 13:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lund A.H., van Lohuizen M.. Epigenetics and cancer. Genes Dev. 2004; 18:2315–2335. [DOI] [PubMed] [Google Scholar]
- 3. Portela A., Esteller M.. Epigenetic modifications and human disease. Nat. Biotechnol. 2010; 28:1057–1068. [DOI] [PubMed] [Google Scholar]
- 4. Cheung P., Lau P.. Epigenetic regulation by histone methylation and histone variants. Mol. Endocrinol. 2005; 19:563–573. [DOI] [PubMed] [Google Scholar]
- 5. Zhang Y., Reinberg D.. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001; 15:2343–2360. [DOI] [PubMed] [Google Scholar]
- 6. Huang H., Sabari B.R., Garcia B.A., Allis C.D., Zhao Y.M.. SnapShot: histone modifications. Cell. 2014; 159:458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang H., Lin S., Garcia B.A., Zhao Y.. Quantitative proteomic analysis of histone modifications. Chem. Rev. 2015; 115:2376–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tarakhovsky A. Tools and landscapes of epigenetics. Nat. Immunol. 2010; 11:565–568. [DOI] [PubMed] [Google Scholar]
- 9. Musselman C.A., Lalonde M.E., Cote J., Kutateladze T.G.. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012; 19:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bannister A.J., Kouzarides T.. Regulation of chromatin by histone modifications. Cell Res. 2011; 21:381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung P., Allis C.D., Sassone-Corsi P.. Signaling to chromatin through histone modifications. Cell. 2000; 103:263–271. [DOI] [PubMed] [Google Scholar]
- 12. Gorisch S.M., Wachsmuth M., Toth K.F., Lichter P., Rippe K.. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005; 118:5825–5834. [DOI] [PubMed] [Google Scholar]
- 13. Lee D.Y., Hayes J.J., Pruss D., Wolffe A.P.. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993; 72:73–84. [DOI] [PubMed] [Google Scholar]
- 14. Alabert C., Barth T.K., Reveron-Gomez N., Sidoli S., Schmidt A., Jensen O.N., Imhof A., Groth A.. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015; 29:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barth T.K., Imhof A.. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 2010; 35:618–626. [DOI] [PubMed] [Google Scholar]
- 16. Huang C., Xu M., Zhu B.. Epigenetic inheritance mediated by histone lysine methylation: maintaining transcriptional states without the precise restoration of marks. Philos. Trans. Roy. Soc. B. 2013; 368:20110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maze I., Noh K.M., Soshnev A.A., Allis C.D.. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 2014; 15:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Polo S.E., Roche D., Almouzni G.. New histone incorporation marks sites of UV repair in human cells. Cell. 2006; 127:481–493. [DOI] [PubMed] [Google Scholar]
- 19. Lyons S.M., Cunningham C.H., Welch J.D., Groh B., Guo A.Y., Wei B., Whitfield M.L., Xiong Y., Marzluff W.F.. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res. 2016; 44:9190–9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Skene P.J., Henikoff S.. Histone variants in pluripotency and disease. Development. 2013; 140:2513–2524. [DOI] [PubMed] [Google Scholar]
- 21. Ausio J. Histone variants–the structure behind the function. Brief. Funct. Genomic Proteomic. 2006; 5:228–243. [DOI] [PubMed] [Google Scholar]
- 22. Jang C.W., Shibata Y., Starmer J., Yee D., Magnuson T.. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015; 29:1377–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Udugama M., FT M.C., Chan F.L., Tang M.C., Pickett H.A., JD R.M., Mayne L., Collas P., Mann J.R., Wong L.H.. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015; 43:10227–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elsasser S.J., Noh K.M., Diaz N., Allis C.D., Banaszynski L.A.. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015; 522:U240–U323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nashun B., Hill P.W., Smallwood S.A., Dharmalingam G., Amouroux R., Clark S.J., Sharma V., Ndjetehe E., Pelczar P., Festenstein R.J. et al. . Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell. 2015; 60:611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ricketts M.D., Frederick B., Hoff H., Tang Y., Schultz D.C., Singh Rai T., Grazia Vizioli M., Adams P.D., Marmorstein R.. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 2015; 6:7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lewis P.W., Elsaesser S.J., Noh K.M., Stadler S.C., Allis C.D.. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elsaesser S.J., Allis C.D.. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harb. Symp. Quant. Biol. 2010; 75:27–34. [DOI] [PubMed] [Google Scholar]
- 29. Szenker E., Lacoste N., Almouzni G.. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 2012; 1:730–740. [DOI] [PubMed] [Google Scholar]
- 30. Lin C.J., Koh F.M., Wong P., Conti M., Ramalho-Santos M.. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell. 2014; 30:268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maze I., Wenderski W., Noh K.M., Bagot R.C., Tzavaras N., Purushothaman I., Elsasser S.J., Guo Y., Ionete C., Hurd Y.L. et al. . Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015; 87:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Urban M.K., Zweidler A.. Changes in nucleosomal core histone variants during chicken development and maturation. Dev. Biol. 1983; 95:421–428. [DOI] [PubMed] [Google Scholar]
- 33. Rogakou E.P., Sekeri-Pataryas K.E.. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 1999; 34:741–754. [DOI] [PubMed] [Google Scholar]
- 34. Saade E., Pirozhkova I., Aimbetov R., Lipinski M., Ogryzko V.. Molecular turnover, the H3.3 dilemma and organismal aging (hypothesis). Aging Cell. 2015; 14:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J. et al. . ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014; 508:263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Szenker E., Ray-Gallet D., Almouzni G.. The double face of the histone variant H3.3. Cell Res. 2011; 21:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo R., Zheng L., Park J.W., Lv R., Chen H., Jiao F., Xu W., Mu S., Wen H., Qiu J. et al. . BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell. 2014; 56:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jacob Y., Bergamin E., Donoghue M.T.A., Mongeon V., LeBlanc C., Voigt P., Underwood C.J., Brunzelle J.S., Michaels S.D., Reinberg D. et al. . Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science. 2014; 343:1249–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Molden R.C., Garcia B.A.. Middle-down and top-down mass spectrometric analysis of co-occurring histone modifications. Curr. Protoc. Protein Sci. 2014; 77:1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beck H.C., Nielsen E.C., Matthiesen R., Jensen L.H., Sehested M., Finn P., Grauslund M., Hansen A.M., Jensen O.N.. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics: MCP. 2006; 5:1314–1325. [DOI] [PubMed] [Google Scholar]
- 41. Sidoli S., Schwammle V., Ruminowicz C., Hansen T.A., Wu X., Helin K., Jensen O.N.. Middle-down hybrid chromatography/tandem mass spectrometry workflow for characterization of combinatorial post-translational modifications in histones. Proteomics. 2014; 14:2200–2211. [DOI] [PubMed] [Google Scholar]
- 42. Tvardovskiy A., Wrzesinski K., Sidoli S., Fey S.J., Rogowska-Wrzesinska A., Jensen O.N.. Top-down and middle-down protein analysis reveals that intact and clipped human histones differ in post-translational modification patterns. Mol. Cell. Proteomics. 2015; 14:3142–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Young N.L., DiMaggio P.A., Plazas-Mayorca M.D., Baliban R.C., Floudas C.A., Garcia B.A.. High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics. 2009; 8:2266–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pesavento J.J., Mizzen C.A., Kelleher N.L.. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem. 2006; 78:4271–4280. [DOI] [PubMed] [Google Scholar]
- 45. Schwammle V., Aspalter C.M., Sidoli S., Jensen O.N.. Large scale analysis of co-existing post-translational modifications in histone tails reveals global fine structure of cross-talk. Mol. Cell Proteomics. 2014; 13:1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howe C.G., Gamble M.V.. Enzymatic cleavage of histone H3: a new consideration when measuring histone modifications in human samples. Clin. Epigenetics. 2015; 7, doi:10.1186/s13148-014-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sidoli S., Garcia B.A.. Characterization of individual histone posttranslational modifications and their combinatorial patterns by mass spectrometry-based proteomics strategies. Methods Mol. Biol. 2017; 1528:121–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mohammad F., Weissmann S., Leblanc B., Pandey D.P., Hojfeldt J.W., Comet I., Zheng C.Q., Johansen J.V., Rapin N., Porse B.T. et al. . EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 2017; 23:483-492. [DOI] [PubMed] [Google Scholar]
- 49. Pina B., Suau P.. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 1987; 123:51–58. [DOI] [PubMed] [Google Scholar]
- 50. Zee B.M., Levin R.S., DiMaggio P.A., Garcia B.A.. Global turnover of histone post-translational modifications and variants in human cells. Epigenet. Chromatin. 2010; 3, doi:10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McKittrick E., Gaften P.R., Ahmad K., Henikoff S.. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Delhaye M., Louis H., Degraef C., LeMoine O., Deviere J., Gulbis B., Jacobovitz D., Adler M., Galand P.. Relationship between hepatocyte proliferative activity and liver functional reserve in human cirrhosis. Hepatology. 1996; 23:1003–1011. [DOI] [PubMed] [Google Scholar]
- 53. Wu J., Xu W.. Histone H3R17me2a mark recruits human RNA polymerase-associated factor 1 complex to activate transcription. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5675–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bauer U.M., Daujat S., Nielsen S.J., Nightingale K., Kouzarides T.. Methylation at arginine 17 of histone H3 is linked to gene activation. Embo Rep. 2002; 3:39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fernandez A.F., Bayon G.F., Urdinguio R.G., Torano E.G., Garcia M.G., Carella A., Petrus-Reurer S., Ferrero C., Martinez-Camblor P., Cubillo I. et al. . H3K4me1 marks DNA regions hypomethylated during aging in human stem and differentiated cells. Genome Res. 2015; 25:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson V.L., Smith R.A., Ma S., Cutler R.G.. Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 1987; 262:9948–9951. [PubMed] [Google Scholar]
- 57. Barth T.K., Imhof A.. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem. Sci. 2010; 35:618–626. [DOI] [PubMed] [Google Scholar]
- 58. Kraushaar D.C., Jin W., Maunakea A., Abraham B., Ha M., Zhao K.. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013; 14:R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hake S.B., Garcia B.A., Duncan E.M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D.P., Allis C.D., Hunt D.F.. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006; 281:559–568. [DOI] [PubMed] [Google Scholar]
- 60. Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 61. Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B.. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011; 286:7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee J.S., Smith E., Shilatifard A.. The language of histone crosstalk. Cell. 2010; 142:682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L.. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012; 13:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chantalat S., Depaux A., Hery P., Barral S., Thuret J.Y., Dimitrov S., Gerard M.. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011; 21:1426–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rougeulle C., Chaumeil J., Sarma K., Allis C.D., Reinberg D., Avner P., Heard E.. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell. Biol. 2004; 24:5475–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Piazzesi A., Papic D., Bertan F., Salomoni P., Nicotera P., Bano D.. Replication-independent histone variant H3.3 controls animal lifespan through the regulation of pro-longevity transcriptional programs. Cell Rep. 2016; 17:987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Adhikary S., Sanyal S., Basu M., Sengupta I., Sen S., Srivastava D.K., Roy S., Das C.. Selective recognition of H3.1K36 dimethylation/H4K16 acetylation facilitates the regulation of all-trans-retinoic acid (ATRA)-responsive genes by putative chromatin reader ZMYND8. J. Biol. Chem. 2016; 291:2664–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hake S.B., Allis C.D.. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6428–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lin C.J., Conti M., Ramalho-Santos M.. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013; 140:3624–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Escamilla-Del-Arenal M., da Rocha S.T., Spruijt C.G., Masui O., Renaud O., Smits A.H., Margueron R., Vermeulen M., Heard E.. Cdyl, a new partner of the inactive X chromosome and potential reader of H3K27me3 and H3K9me2. Mol. Cell. Biol. 2013; 33:5005–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.