Abstract
Targeted differentiation of human induced pluripotent stem cells (hiPSCs) using only chemicals would have value-added clinical potential in the regeneration of complex cell types including cardiomyocytes. Despite the availability of several chemical inhibitors targeting proteins involved in signaling pathways, no bioactive synthetic DNA-binding inhibitors, targeting key cell fate-controlling genes such as SOX2, are yet available. Here, we demonstrate a novel DNA-based chemical approach to guide the differentiation of hiPSCs using pyrrole–imidazole polyamides (PIPs), which are sequence-selective DNA-binding synthetic molecules. Harnessing knowledge about key transcriptional changes during the induction of cardiomyocyte, we developed a DNA-binding inhibitor termed PIP-S2, targeting the 5′-CTTTGTT-3′ and demonstrated that inhibition of SOX2–DNA interaction by PIP-S2 triggers the mesoderm induction in hiPSCs. Genome-wide gene expression analyses revealed that PIP-S2 induced mesoderm by targeted alterations in SOX2-associated gene regulatory networks. Also, employment of PIP-S2 along with a Wnt/β-catenin inhibitor successfully generated spontaneously contracting cardiomyocytes, validating our concept that DNA-binding inhibitors could drive the directed differentiation of hiPSCs. Because PIPs can be fine-tuned to target specific DNA sequences, our DNA-based approach could be expanded to target and regulate key transcription factors specifically associated with desired cell types.
INTRODUCTION
Human induced pluripotent stem cells (human iPSCs; hiPSCs) are a promising resource for regenerative medicine, drug discovery, and disease modeling, because they can differentiate into all three germ layers and avoid the ethical issues associated with the use of human embryonic stem cells (ESCs). Directed differentiation of hiPSCs has been achieved by modulating signaling pathways with various growth factors and cytokines to mimic natural organ development (1,2). Differentiation of hiPSCs using small molecule inhibitors has been favored because they are transgene-free, cost-effective and are readily applicable (3–6). In principle, these synthetic molecules modulate cell fate-regulating signaling pathways by binding to specific proteins and inhibiting specific receptor–ligand interactions or enzymatic activity. However, these compounds might target additional signaling components, and the requirement of multiple modulators for directed differentiation also complicates the issues.
Since the transcriptional network eventually dictates cell fate specification, the direct regulation of specific gene expression could be an effective strategy to control the differentiation of hiPSCs. High-throughput sequencing studies have been revealing several cell fate-modulating transcription factors (TFs) and their key regulatory motifs (7–9). Until now, strategies to modulate key TFs still largely rely on exogenous genetic materials and hence are not favored for clinical use. Consequently, there is a need to develop convenient-to-use DNA-binding inhibitors with defined compositions for cell fate-regulating TFs. For this purpose, we explored whether we could harness the chemical biology of nucleic acids to construct a DNA-binding inhibitor for a TF named SOX2, known as a negative regulator of mesoderm induction of hiPSCs (10,11). Hairpin pyrrole–imidazole polyamides (PIPs) are a class of synthetic molecules composed of N-methylpyrrole units (Py) and N-methylimidazole units (Im) (12). PIPs can bind to the DNA minor groove in a sequence-selective manner (anti-parallel Py/Py pairs recognize A•T or T•A base pairs and Im/Py pairs recognize G•C base pairs) (13) and their selectivity and affinity are similar to those of naturally occurring TFs (14). PIPs have been reported to repress transcription by inhibiting DNA–TF interactions in sequence-specific manners (15–19). PIPs have moderate cell permeability and nuclear localizing properties without the need for delivery reagents (19,20). Moreover, PIPs can be generated readily using automated solid phase peptide synthesizers.
Until now, PIPs have not been shown to differentiate hiPSCs, but here we demonstrate that a PIP targeting the consensus-binding motif of the SOX2 TF could significantly guide the differentiation of hiPSCs into mesoderm. Cardiomyocytes were chosen as the target cell type because the differentiation progression and regulatory mechanisms leading to the generation of cardiomyocytes are well studied (21,22). PIP-induced mesoderm generated spontaneously contracting cardiomyocytes upon treatment with a Wnt/β-catenin signaling inhibitor. This study serves as the first proof-of-concept study validating the application of a synthetic DNA-binding inhibitor to induce directed differentiation of hiPSCs.
MATERIALS AND METHODS
PIP synthesis
All PIPs used in this study were synthesized as described previously (18,23). Briefly, Fmoc solid-phase synthesis of designed PIP structures was performed on a PSSM-8 (Shimadzu), then cleavage was carried out with 1 ml of 3,3′-diamino-N-methyldipropylamine (Dp) for 3 h at 45°C. The resulting crude PIPs were purified by flash column chromatography. The purified PIPs were then characterized by HPLC and MALDI-TOF/MS. Further detailed information about PIP synthesis is provided in supplementary information.
T m assay
PIPs (12.5 μM) were mixed with DNA duplex (2.5 μM) in 100 μl mixture containing sodium cacodylate (10 mM, pH 7.0) and sodium chloride (10 mM) containing 2.5% (v/v) of dimethylformamide (DMF). The sequence of DNA duplex used is 5′-GGCACTTTGTTATGCGG-3′, where the underlined region indicates binding site of SOX2. Annealing was performed by decreasing the temperature of the mixture from 90°C to 25°C (−1°C/min). DNA melting was assessed by measuring the absorbance of mixture at λ = 260 nm from 25°C to 90°C (1°C/min, 1 measurement/°C) on V-650 UV-VIS spectrometer (Jasco).
Electrophoretic mobility shift assay (EMSA)
Recombinant protein of DNA-binding domain of SOX2 (high mobility group; HMG) was prepared as previously described (24,25). PIPs were incubated with the DNA duplex (5′–GGCACTTTGTTATGCGG–3′, 30 pmol) in 9 μl mixture containing sodium cacodylate (13.3 mM, pH 7.0) and sodium chloride (13.3 mM) for at least 1 h at room temperature. Human recombinant SOX2 (90 pmol) was then added to attain 12 μl of total volume and the mixture was incubated for 1 h at room temperature. 2.5 μl of loading dye (#B7025S; New England Biolabs) was added and 10 μl was loaded to 20% polyacrylamide gel (acrylamide:bis-acrylamide = 29:1, 1× TBE). The gel was run at 120 V for first 30 min, followed by 180 V for 150 min. DNA was stained with ethidium bromide and visualized. The band intensities of SOX2–DNA complex were determined using ImageJ software (https://imagej.nih.gov/ij/).
Culture and maintenance of hiPSCs
All cell cultures were performed at 37°C with 5% CO2. 201B7 iPS cell line was provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. After being cultured in SNL feeder/20% Knockout Serum Replacement (KSR) condition, hiPSCs were transferred and routinely maintained on Matrigel (hESC-qualified; Corning) with mTeSR1 (Stemcell Technologies) medium supplemented with 0.5× penicillin/streptomycin (Nacalai Tesque). Cells were passaged using the dissociation reagent containing 0.5 mM ethlendiaminetetraacetic acid disodium salt (EDTA•2Na) in Dulbecco's phosphate-buffered saline without calcium/magnesium (D-PBS(−); Nacalai Tesque). 2.5 μM of Rock inhibitor Y-27632 (Wako) was supplemented to the medium for one day after passage.
Mesoderm induction
On day −2, hiPSCs of passage 50–90 were detached with 0.5 mM EDTA•2Na/D-PBS(−) to attain small cell clumps. Cells were suspended in mTeSR1 that is supplemented with 0.5× penicillin/streptomycin and 2.5 μM Y-27632 and then were seeded onto Matrigel-coated culture dishes/plates (typically 0.2–1.0 × 104 cells/cm2). On day 0 the medium was replaced with differentiation medium (DM) constituted with Advanced RPMI 1640 medium (Thermo Fisher Scientific) and 0.2% fetal bovine serum (FBS; Sigma) and 1× l-glutamine (Wako) with/without 2 μM PIP. 0.1% dimethyl sulfoxide (DMSO) was used as a vehicle. On day 1, the medium was exchanged with the same composition. Typically mesoderm-like cells emerged on day 4 or 5. To evaluate the induction efficiency by PIP-S2, the combination of PIP-S2 and Matrigel overlay was used where the chilled DM supplemented with Matrigel (one aliquot in 25 ml medium according to the product information) and PIP-S2 was used with medium exchange on day 1.
For the CHIR99021-mediated mesoderm induction, cells were seeded with the same protocol and incubated with daily medium exchange until the cells become 30% confluent. Then the medium was replaced with DM supplemented with 2 μM of CHIR99021 (Wako) and the cultured cells were incubated for 2 days (5).
Cardiomyocytes generation
After performing mesoderm induction as described above, the medium was replaced with DM supplemented with 5 μM IWP-2 (Wako). Afterward, on day 7 the medium was replaced with DM without any additive and exchanged once in 2–4 days. For immunocytochemistry analysis of the individual cardiomyocytes, the spontaneously contracting cluster was picked up and dissociated by TrypLE Select (Thermo Fisher Scientific). The cells were re-seeded onto a cover glass (Matsunami Glass) coated with Matrigel (hESC-qualified; Corning).
RT-qPCR
Total RNA was extracted by RNeasy Mini Kit (Qiagen) according to the manufacturer's manual. 100–500 ng of total RNA was reverse transcribed into cDNA by ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo) and qPCR was performed using THUNDERBIRD SYBR qPCR Mix (Toyobo) in LightCycler 480 (Roche Diagnostics GmbH). Cp value was determined by the second derivative maximum method and relative RNA amount was calculated by the ΔΔCp method. P-value was calculated by one-sample t-test using data from independent experiments. The sequences of the primers are listed in Supplementary Table S1.
Immunocytochemistry
For immunocytochemical analysis, except for primary antibody incubation, all the following incubations were performed at room temperature. Cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.5% Triton X-100 for 10 min and blocked with 5% bovine albumin serum (BSA; Nacalai Tesque) for 30 min. Afterward, the cells were incubated with primary antibody in 1% BSA for overnight at 4°C and incubated with secondary antibody in 1% BSA for 50 min. Nuclei were stained with 1 μg/ml Hoechst 33342. Antibodies used are listed in Supplementary Table S2.
Flow cytometry
Cells were detached by treating TrypLE Select (Thermo Fisher Scientific) for 7–10 min at 37°C, followed by chemical fixation with 4% paraformaldehyde for 10 min at 37°C and permeabilization with 90% methanol for 30 min at 4°C. Then, the cells were incubated with primary antibody for 60 min at room temperature followed by incubation with secondary antibody for 30 min at room temperature. FACS Aria II (BD Bioscience) was used to detect fluorescence. Antibodies used are listed in Supplementary Table S2.
Microarray
After extracting total RNA as above, the quality of RNA was assessed with Bioanalyzer (Agilent Technologies) using Agilent RNA 6000 Pico Kit (Agilent Technologies). Biotinylated fragmented cDNA was prepared by GeneChip WT PLUS Reagent Kit (Affymetrix) and hybridized to Human Gene 2.1 ST Array Strip (Affymetrix) using GeneChip Hybridization, Wash, and Stain Kit (Affymetrix). Fluidics and scanning were performed in GeneAtlas System (Affymetrix) using GeneChip Hybridization, Wash, and Stain Kit (Affymetrix) and the obtained raw data set was normalized and summarized by Expression Console (Affymetrix). Ingenuity Pathway Analysis (IPA; Qiagen) was used for upstream analysis.
Z-score for gene A in sample i (Z(A, i)) was calculated by the following formula:
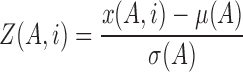 |
where x(A, i) is the normalized signal intensity of gene A in sample i, μ(A) is the mean of x(A), σ(A) is the standard deviation of x(A).
RESULTS
Design and synthesis of a DNA-binding inhibitor of SOX2
The target selection of our proof-of-concept study to construct the DNA-binding inhibitor arose conceptually from the well-known mechanism of cardiomyocyte differentiation. Differentiation of hiPSCs into cardiomyocytes comprises of two major stages: mesoderm induction and cardiomyocyte specification (21,22). The first stage of mesoderm induction is typically achieved by activating the Wnt/β-catenin and TGF-β signaling pathways (4,26,27). The repression of SOX2 downstream of these pathways is a critical event in this stage (22). SOX2 is a key TF that is highly expressed in pluripotent stem cells, and is one of the essential components for generation of both mouse and human iPSCs (28,29). In hiPSCs, SOX2 is responsible for maintaining pluripotency and neuroectodermal lineage specification and prevents differentiation into mesodermal and endodermal lineages (30). Consistent with this idea, knockdown of SOX2 is known to induce meso/endoderm induction from hiPSCs (10,22). In addition, it has been also demonstrated that the inhibiting transcriptional activity of SOX2 in mESCs enhances the appearance of early neuronal and cardiac progenitors (31). Because short interfering (si)RNA-mediated gene knockdown involves problematic handling and stability issues, there is a need to develop DNA-binding chemical inhibitors.
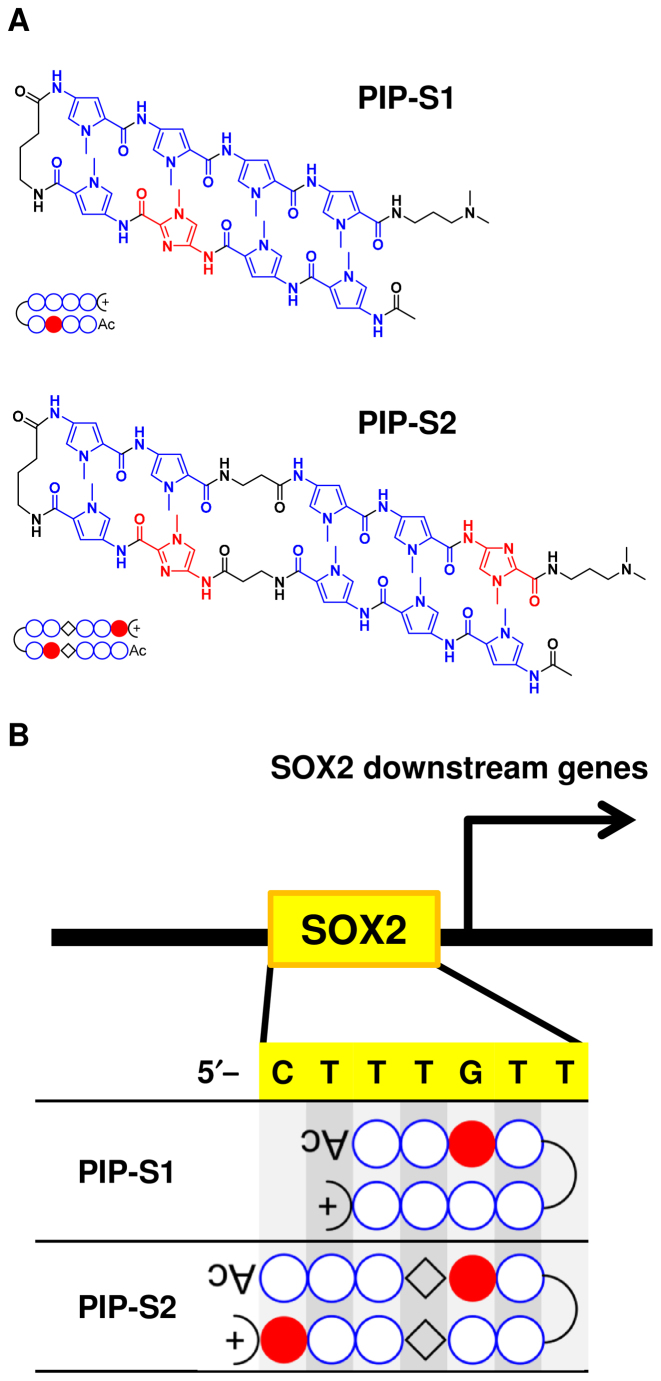
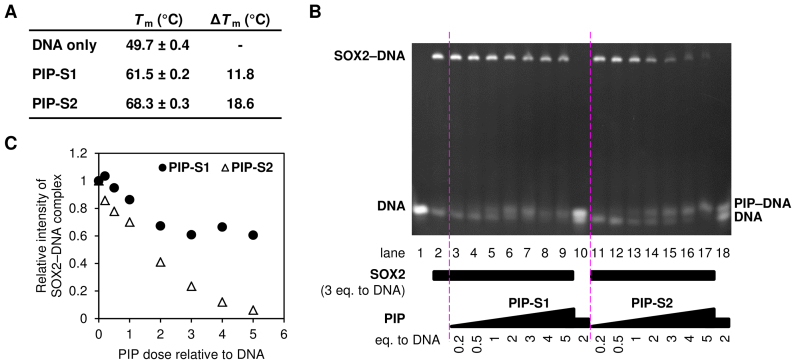
Based on these facts, we designed two hairpin DNA-binding PIPs (PIP-S1 and PIP-S2) to inhibit SOX2 function by occupying its target DNA sequence (5′-CTTTGTT-3′) for mesoderm induction (Figure 1) (32). PIP-S1 has a typical 4–4 ring architecture targeting the 5′-CTTTGTT-3′ sequence. PIP-S2 has an extended architecture with increased recognition specificity to target 5′-CTTTGTT-3′. The anticipated binding mode of the two PIPs is shown in Figure 1B. To confirm the binding, we used a template DNA with the following sequence 5′-GGCACTTTGTTATGCGG-3′. A melting temperature (Tm) assay (33) confirmed the binding of both the PIPs to target DNA as Tm shifts of 11.8°C and 18.6°C were observed for PIP-S1 and PIP-S2, respectively (Figure 2A). An electrophoretic mobility shift assay (EMSA) carried out to study SOX2–DNA interaction showed that increasing concentrations of both the PIPs decreased the amount of the SOX2–DNA complex (Figure 2B and C). Following this concept, PIP-S2 inhibited SOX2 binding more effectively than did PIP-S1. At the same concentration as SOX2 (3 eq. against DNA), PIP-S2 reduced the band intensity of the SOX2–DNA complex by >70% (Figure 2B and C, lane 15), confirming its superior binding affinity to SOX2. PIP-S1 showed the plateau in its effect at higher concentrations (Figure 2B and C, lanes 7–9). One possible reason for this plateau could be the formation of aggregates at the higher concentration of PIP-S1 (34).
Figure 1.
The design of pyrrole–imidazole polyamides (PIPs) targeting the binding DNA sequence of SOX2. (A) Chemical structure of hairpin PIPs, namely PIP-S1 and PIP-S2. (B) Targeting mode of designed PIPs to the consensus sequence of a SOX2 binding site (5′–CTTTGTT–3′).
Figure 2.
PIP-S2 is a potent inhibitor of SOX2–DNA interaction. (A) Tm assay of PIPs. Means ± standard deviations from three measurements are indicated. (B) EMSA of PIPs. DNA (2.5 μM) with increasing concentrations of each PIP was incubated with 7.5 μM of SOX2 (three times the amount of DNA) for 1 h. DNA and the SOX2–DNA complex were separated by polyacrylamide gel electrophoresis and stained with ethidium bromide. (C) Inhibition of SOX2–DNA interaction by PIPs. The band intensities of SOX2–DNA complex in the Figure 2B were determined using ImageJ software (https://imagej.nih.gov/ij/). The intensity in lane 2 was set to 1.0.
PIP-S2 induces mesoderm from hiPSCs
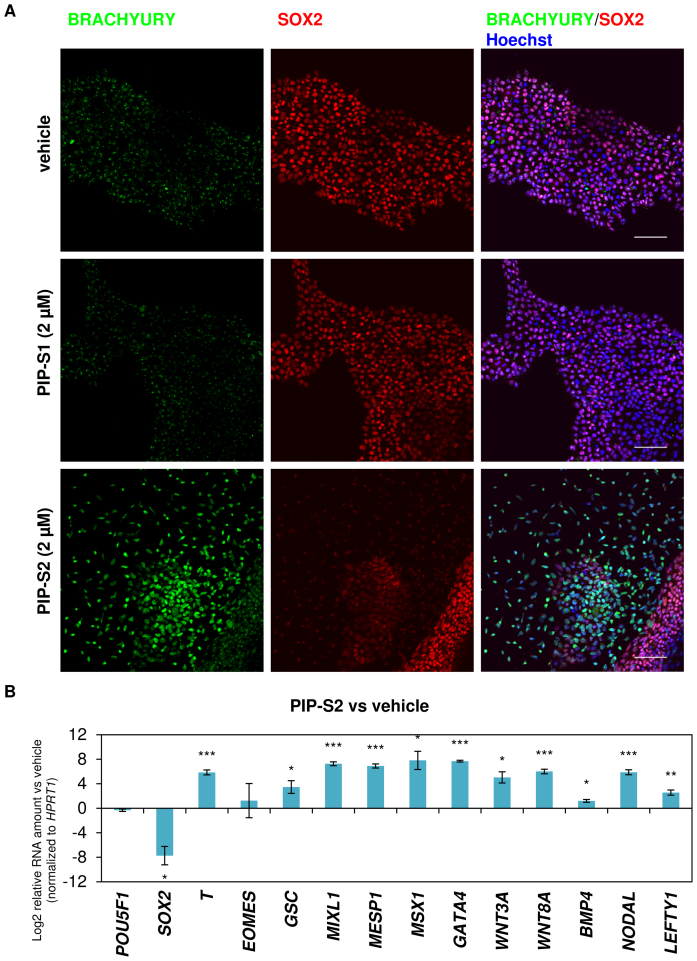
To evaluate the bioactivity of the designed PIPs, after initial optimization studies, 2 μM of PIP-S1 and PIP-S2 were added to the RPMI 1640-based differentiation medium used for culturing hiPSCs (Supplementary Figure S1). Immunocytochemical studies for the mesoderm marker BRACHYURY (T) revealed that PIP-S1-treated hiPSCs had a background level of BRACHYURY expression similar to that observed with vehicle-treated hiPSCs. On the other hand, PIP-S2 caused remarkable upregulation of BRACHYURY protein expression in hiPSCs (Figure 3A, Panel BRACHYURY). Because SOX2 expression is known to be repressed immediately following mesoderm induction (22), the effects of these PIPs on the expression of SOX2 were evaluated. As we expected, BRACHYURY-positive cells induced by PIP-S2 suppressed the level of the SOX2 protein (Figure 3A, Panel SOX2). Consistently, expression of SOX2 was maintained in hiPSCs treated with vehicle and PIP-S1 (Figure 3A, Panel SOX2). Quantitative reverse transcription polymerase chain reaction (RT–qPCR) analysis further validated that PIP-S2 treatment triggered significant induction (P < 0.001) of BRACHYURY (T) and significant repression (P < 0.05) of SOX2 gene expression (Figure 3B, Bars T and SOX2). It is important to note here that the different bioactivity between PIP-S1 and PIP-S2 is consistent with the binding data attained with the in vitro Tm assays and EMSA discussed above. Therefore, it is rational to propose that mesoderm induction is caused by the relatively higher binding affinity of PIP-S2 than that of PIP-S1 toward the target sequence. To further evaluate the effect of PIP-S2 on transcription programs associated with mesoderm specification, endogenous expression of other mesoderm marker genes was evaluated. Again, PIP-S2 significantly (P < 0.05) activated mesoderm/mesendoderm-associated marker genes (GSC, MIXL1, MSX1) and cardiac mesoderm-related marker genes (MESP1 and GATA4) (Figure 3B). PIP-S2 also triggered significant (P < 0.05) upregulation of genes related to Wnt/β-catenin (WNT3A and WNT8A) and TGFβ (BMP4, NODAL and LEFTY1) signaling pathways, which are essential for the induction of mesoderm (Figure 3B) (26). Thus, we successfully developed PIP-S2 as a DNA-binding inhibitor of SOX2 capable of significantly inducing marker genes associated with cardiac mesoderm specification in hiPSCs.
Figure 3.
PIP-S2 induces the mesodermal lineage in hiPSCs. (A) Immunohistochemistry of the mesoderm marker BRACHYURY and pluripotency/ectoderm marker SOX2 in PIP-treated 201B7-hiPSCs on day 5. Scale bars; 100 μm. (B) RT-qPCR analysis of 201B7-hiPSCs treated with PIP-S2 for 5 days. Relative RNA amount (vehicle on day 5 vs PIP-S2 on day 5) is shown in logarithmic scale. Means ± the standard errors of the mean (SEM) from three independent experiments are indicated. *P < 0.05, **P < 0.01, ***P < 0.001.
Genome-wide gene expression analysis validates sequence-specific inhibition of SOX2 and its function as the mechanism behind the PIP-S2-mediated induction of mesoderm
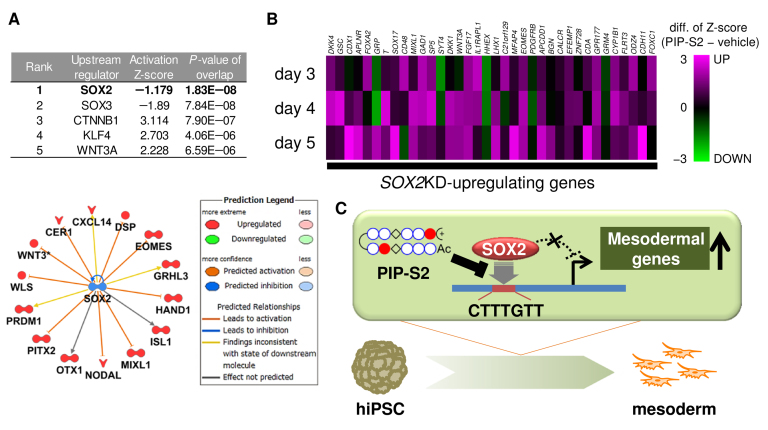
To investigate the possible mechanism behind the induction of cardiac mesoderm by our DNA-binding inhibitor, we performed genome-wide gene expression studies of the PIP-S2-treated hiPSCs and compared their expression profile with that observed in vehicle-treated hiPSCs. Upstream analysis of the differentially expressed genes at day 3 (PIP-S2 vs vehicle) predicted SOX2 as the most significant upstream regulator (P = 1.38 × 10−8; Figure 4A) and suggested the inhibition of SOX2 and not its activation (Figure 4A). However, the Z-score of -1.18 was not significant enough (>2 or <−2 is significant). To compare the bioactivity of PIP-S2 and its siRNA, we extracted the expression profile of a series of genes reported to be activated/repressed by SOX2 knockdown (22). For most genes activated by SOX2 knockdown, we observed the upregulation by PIP-S2 (27, 27, 32 out of 36 genes on day 4, 5, 6, respectively; Figure 4B, Supplementary Table S3). On the other hand, the genes downregulated by SOX2 knockdown showed lesser correlation than those by PIP-S2 (17, 15, 11 out of 26 genes on day 4, 5, 6, respectively; Supplementary Figure S2, Supplementary Table S4). The reason could be attributed to the intrinsic smaller expression changes for SOX2 knockdown-repressing genes (Supplementary Table S4). Analysis of overlapping genes in the top 100 regulated transcripts also highlighted the significant correlation for the upregulated genes between PIP-S2 and SOX2 knockdown, but not for the downregulated genes (Supplementary Figure S3). Overall, the generated heat map inferred that the expression pattern observed with PIP-S2 treatment was roughly consistent with the effect of siRNA-mediated SOX2 knockdown, especially in the upregulated genes. Thus, these genome-wide gene analyses implied the inhibition of SOX2 by PIP-S2 as a plausible mechanism behind the induction of mesoderm (Figure 4C), thereby validating the efficacy of our strategy.
Figure 4.
PIP-S2 induces mesoderm via the inhibition of SOX2. (A) Upstream regulator analysis of >2 fold up-/downregulated genes in PIP-S2-treated 201B7-hiPSCs compared with vehicle-treated 201B7 on day 3. Rank 1–5 upstream regulators based on P values are shown. (B) Heat map of genes upregulated by the SOX2 knockdown in hESCs. Microarray studies were performed for the PIP-S2- or vehicle-treated cells on days 3, 4 and 5. Values represent the differences in Z-scores between the expression level in PIP-S2-treated hiPSCs and that in vehicle-treated cells. (C) Schematic illustration of the plausible mechanism for mesoderm induction by PIP-S2.
To confirm that the effect of PIP-S2 is sequence-specific, we designed and synthesized another PIP (PIP-C) of the same length but with a different chemical structure as a control (Supplementary Figure S4). A melting temperature (Tm) assay for PIP-C showed a Tm shift of 13.1°C, suggesting the lower affinity to the template DNA than that towards PIP-S2 (Supplementary Figure S5). EMSA showed that the SOX2-inhibiting ability of the PIP-C was worse than that of PIP-S2 (Supplementary Figure S6). Consistent with the pattern observed with in vitro binding studies, PIP-C also could not repress SOX2 gene expression nor activate mesoderm-associated T expression (Supplementary Figure S7). Hence, sequence-specific recognition of 5′–CTTTGTT–3′ by PIP-S2 appears to be essential for the bioactivity.
PIP-S2-induced cardiac mesoderm further differentiates into functional cardiomyocytes upon supplementation with a Wnt/β-catenin signaling inhibitor
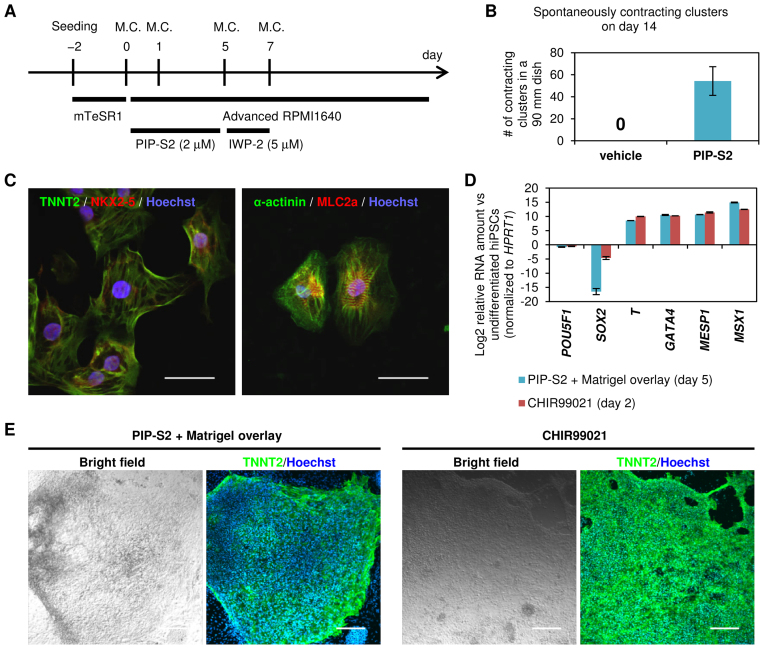
Because targeted differentiation using chemicals alone is believed to have clinical benefits, we screened for the optimal small molecule that might complement the bioactivity of PIP-S2 and differentiate the PIP-S2-induced mesoderm into functional cardiomyocytes. Cardiac specification, the second stage followed by mesoderm induction, is known to be driven upon the inhibition of Wnt/β-catenin signaling (4). Hence, after incubating 201B7 hiPSCs with PIP-S2 for 5 days, the cells were exposed to Wnt/β-catenin signaling inhibitor IWP-2 for 2 more days (days 5–7). After this short-term exposure to IWP-2, the cells were further incubated in the differentiation medium with exchange every 2–4 days (Figure 5A). Around day 12, several cells began to spontaneously contract (Supplementary Movie S1 and S2), indicating the generation of functional cardiomyocytes. Counting the generated cell clusters corroborated that sequential treatment with PIP-S2 and IWP-2 successfully generated several pulsating clusters, whereas the treatment of hiPSCs without PIP-S2 did not generate any pulsating clusters (Figure 5B). Immunocytochemistry also confirmed the expression of the main marker proteins TNNT2, NKX2–5, α-actinin and MLC2A to validate the typical characteristics of functional cardiomyocytes (Figure 5C). Furthermore, the expression patterns of TNNT2 and α-actinin indicated the presence of sarcomeres, the fundamental structures of myocytes. These independent lines of evidence imply that the cardiomyocytes generated by PIP-S2/IWP-2 have typical structural and functional characteristics of cardiomyocytes.
Figure 5.
Cardiomyocyte induction from hiPSCs using PIP-S2. (A) Scheme of the differentiation process. Mesodermal lineage induced by PIP-S2 was exposed to 5 μM of IWP-2 (a Wnt/β-catenin inhibitor) for 2 days followed by culture in the medium without any additive. M.C.; medium change. (B) Spontaneously contracting clusters were counted on day 14. The mean ± SEM of three 90 mm culture dishes is indicated. (C) Immunohistochemistry of cardiomyocyte markers in differentiated 201B7-hiPSCs with PIP-S2. On day 16, spontaneously contracting clusters were picked up, dissociated, and plated on cover glasses. Staining was performed on the next day. Scale bars; 50 μm. (D) RT–qPCR analysis of 201B7-hiPSCs treated with PIP-S2 + Matrigel overlay (day 5) and CHIR99021 (day 2). Relative RNA amount (vs undifferentiated 201B7-hiPSCs) is shown in logarithmic scale. Means ± the standard deviation of the mean (SDM) from multiple culture wells (three each for PIP-S2 treatment and two each for CHIR99021 treatment) in one experiments are indicated. (E) Immunocytochemistry of cardiomyocyte-specific marker TNNT2 in the differentiated cardiomyocytes by the treatment of PIP-S2 + Matrigel overlay (day 20) and CHIR99021 (day 15). Scale bars; 200 μm.
Matrigel overlay approach is known to promote epithelial–mesenchymal transition (EMT) in the mesoderm induction stage (35). Accordingly, our optimization studies also showed that Matrigel overlay could enhance PIP-S2-mediated mesoderm induction (data not shown). The efficiency of mesoderm induction was then evaluated with this optimized protocol along with the standard protocol using a GSK3β inhibitor CHIR99021 (4,5). Flow cytometry analysis showed that PIP-S2 produced 44.7% of SOX2 (−)/BRACHYURY (+) population after 5 days treatment which was ∼47% lower than the population attained with CHIR99021 for 2 days (Supplementary Figure S8). However, RT-qPCR studies revealed the comparable expression level between CHIR99021 and PIP-S2-treated cells and T, GATA4, MESP1 and MSX1 were up regulated and POU5F1 was down regulated (Figure 5D, bars T, GATA4, MESP1, MSX1 and POU5F1). Interestingly, SOX2 down-regulation was more pronounced in PIP-S2-treated cells than that observed in CHIR99021-treated cells (Figure 5D, Bars SOX2). To evaluate the efficacy of PIP-S2 to induce cardiomyocytes, we carried out the standard protocol with IWP2-treatment as mentioned before. Immunocytochemistry analysis confirmed the clusters of TNNT2 (+) cardiomyocytes with PIP-S2 treatment (Figure 5E, Panel PIP-S2). However, the efficiency was still lower than CHIR99021-mediated standard protocol (Figure 5E, Panel CHIR99021). The relatively lower efficiency of cardiomyocyte induction with PIP-S2 treatment than that with CHIR99021 could be attributed to the lack of homogeneity of differentiating cells. Nevertheless, it is important to note here that this observation is also consistent with the previous reports showing that SOX2 does not specifically inhibit cardiac mesoderm but inhibit the formation of mesendoderm, including endoderm and other mesoderm lineages (10,22). Therefore, it is not straightforward to compare the efficiency of CHIR99021 and PIP-S2. Taken together, the data suggest the potential of our programmable DNA-binding inhibitor to inhibit specific transcription factors of interest. Hence, a combinatorial approach to target alternate transcription factors could enhance the efficiency of cardiomyocyte generation.
DISCUSSION
Cardiomyocytes are the critical components that make up the atria and ventricle of the heart. It is difficult to repair damaged cardiomyocytes as they lose their capacity for proliferation and regeneration shortly after birth, making them unavailable later in life (21). Many cardiomyocytes could in theory be generated from human blastocyst-derived pluripotent ESCs. Cellular reprogramming strategies like the use of hiPSCs are more promising because they overcome the problems and ethical dilemmas associated with human ESCs.
Transgenic approaches such as the introduction of exogenous TFs associated with cardiac cells and gene knockdown by siRNAs have shown success in modulating target gene expression directly and in inducing the directed differentiation of hiPSCs into cardiomyocytes (4,10). However, such transgenic approaches intrinsically have the risk of ‘foreign’ genome integration. Alternative strategies such as the direct transduction of proteins, instead of protein-coding DNA vectors, could overcome this risk. Accordingly, a recent study demonstrated a bacterial injection-based protein delivery of Gata4, Mef2c and Tbx5 as a transgene-free method for the introduction of cardiomyocyte-specific TFs (36). Nevertheless, time-consuming material preparation processes and the intrinsic chemically undefined composition of such proteins are still roadblocks to achieving clinical translation. In this regard, synthetic molecule-based gene modulation is to be desired because synthetic molecules could overcome the technical obstacles associated with the approaches mentioned above. Recently, our group reported a PIP-based epigenetic activator capable of activating nervous system genes in hiPSCs (37). Although the expression of neural genes was demonstrated, the repression of pluripotency markers and subsequent differentiation of hiPSCs into a terminal cell type were not achieved. In the present study, we have characterized PIP-S2 as a first DNA-binding inhibitor of SOX2, which is an essential pluripotency TF (Figure 2B and C). Interestingly, PIP-S2 treatment successfully activated the expression of the main mesoderm-specific markers including BRACHYURY, GSC, MIXL1, MSX1, MESP1 and GATA4 to suggest a shift in the transcriptional program from a pluripotent to a cardiac mesodermal state (Figure 3). When incubated with a Wnt/β-catenin signaling inhibitor, the derived mesoderm could be differentiated successfully into cardiomyocytes, which is one of the terminal cell types in the mesodermal lineage. To our knowledge, this work reports the first DNA-binding synthetic molecule capable of guiding the differentiation of hiPSCs into a particular cell lineage.
Although PIPs are potent DNA-binding molecules thanks to their unique programmable properties, they have not been applied to achieve targeted differentiation of pluripotent stem cells. In a previous study, the authors tried inducing differentiation of mouse embryonal carcinoma (EC) cells and mouse ESCs by the conventional 4–4 ring module hairpin PIPs targeting the binding sequence of OCT4, but did not demonstrate remarkable bioactivities (38). In the present study, we tested a hairpin PIP with a 4–4 ring module and an extended 6–6 ring module, with a β-alanine substitute in one of the rings to reduce the rigidity of the PIP (39,40). As a result, we demonstrated that the 6–6 module PIP (PIP-S2), but not the 4–4 module (PIP-S1), successfully guided the differentiation of hiPSCs into mesoderm. The dissociation constant (KD) of a typical 4–4 module PIP is 10−7–10−8 M (41), whereas the 6–6 module PIP has a KD of 10−8–10−9 M according to previous studies (42). Importantly, the binding affinity of the 6–6 module PIP to the SOX2-target DNA sequence is similar to the binding affinity of SOX2 whose KD is 5.3 × 10−9 M (43). Therefore, it is reasonable to assume that the extension of PIP enhanced its affinity to the target sequence. The extension might also contribute to enhanced specificity. Because PIPs with extended modules apparently recognize longer sequences, they are expected to have higher specificity to targeted sequences in the genome. Therefore, the extension of a PIP is a significant step for attaining any intended bioactivity.
Unlike the well-studied differentiation routes to achieve cardiomyocytes, several challenges remain to attain other cell types from hiPSCs, which include the poor efficiency, need for transfection of exogenous TFs, and complex culture techniques (44,45). Over the past decade, loss-/gain-of-function studies and high-throughput sequencing studies such as ChIP-seq have identified the critical function of cell type-specific TFs and their key regulatory motifs in controlling lineage specification (7–9,46–48). Because PIPs can be designed to target preferred DNA sequences, the strategy demonstrated in this study could be expanded for rationally designing PIPs to match corresponding regulatory sequence motifs and differentiate hiPSCs into any cell type. Additionally, PIPs are amenable to the conjugation with other chemical components including epigenetic modulators such as histone deacetylase (HDAC) inhibitors and histone acetyltransferase (HAT) activators, resulting in ‘epigenetic switches’ which activate selective genes through induction of histone acetylation (49–53). Combined use of these ‘epigenetic switches’ along with simple PIPs like PIP-S2 as DNA-binding inhibitors will achieve control over more complex cell fates in our future studies. Overall, this work serves as a pioneering study for the use of gene-targeting synthetic molecules in regenerative medicine.
ACCESSION NUMBERS
Microarray data have been deposited with the Gene Expression Omnibus under accession number GSE97621.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS [16H06356 to H.S., 15J02111 to T.J., 16K12896 to G.N.P.]; Suzuken Memorial Foundation [16-077 to G.N.P.]; Kyoto University SPIRITS (to G.N.P.); NRF [2016R1A2B2008081 to K.K.K.]. Funding for open access charge: KAKENHI JSPS [16H06356].
Conflict of interest statement. None declared.
REFERENCES
- 1. Laflamme M.A., Chen K.Y., Naumova A. V, Muskheli V., Fugate J. a, Dupras S.K., Reinecke H., Xu C., Hassanipour M., Police S. et al. . Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007; 25:1015–1024. [DOI] [PubMed] [Google Scholar]
- 2. Carpenter M.K., Inokuma M.S., Denham J., Mujtaba T., Chiu C.-P., Rao M.S.. Enrichment of neurons and neural precursors from human embryonic stem cells. Exp. Neurol. 2001; 172:383–397. [DOI] [PubMed] [Google Scholar]
- 3. Minami I., Yamada K., Otsuji T.G., Yamamoto T., Shen Y., Otsuka S., Kadota S., Morone N., Barve M., Asai Y. et al. . A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012; 2:1448–1460. [DOI] [PubMed] [Google Scholar]
- 4. Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P.. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. et al. . Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014; 11:855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L.. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009; 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parikh A., Wu J., Blanton R.M., Tzanakakis E.S.. Signaling pathways and gene regulatory networks in cardiomyocyte differentiation. Tissue Eng. Part B. Rev. 2015; 21:377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He A., Kong S.W., Ma Q., Pu W.T.. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. et al. . Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008; 133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 10. Wang Z., Oron E., Nelson B., Razis S., Ivanova N.. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012; 10:440–454. [DOI] [PubMed] [Google Scholar]
- 11. Rao J., Greber B.. Concise review: signaling control of early fate decisions around the human pluripotent stem cell state. Stem Cells. 2016; 35:277–283. [DOI] [PubMed] [Google Scholar]
- 12. Dervan P.B., Edelson B.S.. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003; 13:284–299. [DOI] [PubMed] [Google Scholar]
- 13. Trauger J.W., Baird E.E., Dervan P.B.. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996; 382:559–561. [DOI] [PubMed] [Google Scholar]
- 14. Carlson C.D., Warren C.L., Hauschild K.E., Ozers M.S., Qadir N., Bhimsaria D., Lee Y., Cerrina F., Ansari A.Z.. Specificity landscapes of DNA binding molecules elucidate biological function. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4544–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gearhart M.D., Dickinson L., Ehley J., Melander C., Dervan P.B., Wright P.E., Gottesfeld J.M.. Inhibition of DNA binding by human estrogen-related receptor 2 and estrogen receptor alpha with minor groove binding polyamides. Biochemistry. 2005; 44:4196–4203. [DOI] [PubMed] [Google Scholar]
- 16. Nickols N.G., Szablowski J.O., Hargrove A.E., Li B.C., Raskatov J.A., Dervan P.B.. Activity of a Py-Im polyamide targeted to the estrogen response element. Mol. Cancer Ther. 2013; 12:675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Olenyuk B.Z., Zhang G.-J., Klco J.M., Nickols N.G., Kaelin W.G., Dervan P.B.. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:16768–16773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Syed J., Pandian G.N., Sato S., Taniguchi J., Chandran A., Hashiya K., Bando T., Sugiyama H.. Targeted suppression of EVI1 oncogene expression by sequence-specific pyrrole-imidazole polyamide. Chem. Biol. 2014; 21:1370–1380. [DOI] [PubMed] [Google Scholar]
- 19. Lai Y., Fukuda N., Ueno T., Matsuda H., Saito S., Matsumoto K., Ayame H., Bando T., Sugiyama H., Mugishima H. et al. . Synthetic pyrrole-imidazole polyamide inhibits expression of the human transforming growth factor-beta1 gene. J. Pharmacol. Exp. Ther. 2005; 315:571–575. [DOI] [PubMed] [Google Scholar]
- 20. Best T.P., Edelson B.S., Nickols N.G., Dervan P.B.. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:12063–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burridge P.W., Keller G., Gold J.D., Wu J.C.. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012; 10:16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rao J., Pfeiffer M.J., Frank S., Adachi K., Piccini I., Quaranta R., Araúzo-Bravo M., Schwarz J., Schade D., Leidel S. et al. . Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016; 18:1–13. [DOI] [PubMed] [Google Scholar]
- 23. Sawatani Y., Kashiwazaki G., Chandran A., Asamitsu S., Guo C., Sato S., Hashiya K., Bando T., Sugiyama H.. Sequence-specific DNA binding by long hairpin pyrrole-imidazole polyamides containing an 8-amino-3, 6-dioxaoctanoic acid unit. Bioorganic Med. Chem. 2016; 24:3603–3611. [DOI] [PubMed] [Google Scholar]
- 24. Yamamoto S., De D., Hidaka K., Kim K.K., Endo M., Sugiyama H.. Single molecule visualization and characterization of Sox2-Pax6 complex formation on a regulatory DNA element using a DNA origami frame. Nano Lett. 2014; 14:2286–2292. [DOI] [PubMed] [Google Scholar]
- 25. Saha A., Kizaki S., De D., Endo M., Kim K.K., Sugiyama H.. Examining cooperative binding of Sox2 on DC5 regulatory element upon complex formation with Pax6 through excess electron transfer assay. Nucleic Acids Res. 2016; 44:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paige S.L., Osugi T., Afanasiev O.K., Pabon L., Reinecke H., Murry C.E.. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010; 5:e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang M., Schulte J.S., Heinick A., Piccini I., Rao J., Quaranta R., Zeuschner D., Malan D., Kim K.-P., Röpke A. et al. . Universal cardiac induction of human pluripotent stem cells in two and three-dimensional formats: implications for in vitro maturation. Stem Cells. 2015; 33:1456–1469. [DOI] [PubMed] [Google Scholar]
- 28. Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 29. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S.. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872. [DOI] [PubMed] [Google Scholar]
- 30. Zhang S., Cui W.. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells. 2014; 6:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. De D., Jeong M.-H., Leem Y.-E., Svergun D.I., Wemmer D.E., Kang J.-S., Kim K.K., Kim S.-H.. Inhibition of master transcription factors in pluripotent cells induces early stage differentiation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chew J., Loh Y., Zhang W., Chen X., Tam W., Yeap L., Li P., Ang Y., Lim B., Robson P. et al. . Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005; 25:6031–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Meier J.L., Montgomery D.C., Dervan P.B.. Enhancing the cellular uptake of Py-Im polyamides through next-generation aryl turns. Nucleic Acids Res. 2012; 40:2345–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hargrove A.E., Raskatov J.A., Meier J.L., Montgomery D.C., Dervan P.B.. Characterization and solubilization of pyrrole-imidazole polyamide aggregates. J. Med. Chem. 2012; 55:5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J., Klos M., Wilson G.F., Herman A.M., Lian X., Raval K.K., Barron M.R., Hou L., Soerens A.G., Yu J. et al. . Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res. 2012; 111:1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bai F., Ho Lim C., Jia J., Santostefano K., Simmons C., Kasahara H., Wu W., Terada N., Jin S.. Directed differentiation of embryonic stem cells Into cardiomyocytes by bacterial injection of defined transcription factors. Sci. Rep. 2015; 5:15014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei Y., Pandian G.N., Zou T., Taniguchi J., Sato S., Kashiwazaki G., Vaijayanthi T., Hidaka T., Bando T., Sugiyama H.. A multi-target small molecule for targeted transcriptional activation of therapeutically significant nervous system genes. ChemistryOpen. 2016; 5:517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hsu C.F. Completion of a programmable DNA-binding small molecule library. 2009; Calif. Inst. Technol; Diss. (Ph.D.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Turner J.M., Swalley S.E., Baird E.E., Dervan P.B.. Aliphatic/aromatic amino acid pairings for polyamide recognition in the minor groove of DNA. J. Am. Chem. Soc. 1998; 120:6219–6226. [Google Scholar]
- 40. Watanabe T., Shinohara K.. Double β-alanine substitutions incorporated in 12-ring pyrrole-imidazole polyamides for lengthened DNA minor groove recognition. Adv. Tech. Biol. Med. 2016; 4:175. [Google Scholar]
- 41. Anandhakumar C., Li Y., Kizaki S., Pandian G.N., Hashiya K., Bando T., Sugiyama H.. Next-generation sequencing studies guide the design of pyrrole-imidazole polyamides with improved binding specificity by the addition of beta-alanine. ChemBioChem. 2014; 15:2647–2651. [DOI] [PubMed] [Google Scholar]
- 42. Matsuda H., Fukuda N., Ueno T., Tahira Y., Ayame H., Zhang W., Bando T., Sugiyama H., Saito S., Matsumoto K. et al. . Development of gene silencing pyrrole-imidazole polyamide targeting the TGF-beta1 promoter for treatment of progressive renal diseases. J. Am. Soc. Nephrol. 2006; 17:422–432. [DOI] [PubMed] [Google Scholar]
- 43. Takayama Y., Clore G.M.. Impact of protein/protein interactions on global intermolecular translocation rates of the transcription factors Sox2 and Oct1 between DNA cognate sites analyzed by z-exchange NMR spectroscopy. J. Biol. Chem. 2012; 287:26962–26970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abujarour R., Valamehr B.. Generation of skeletal muscle cells from pluripotent stem cells: advances and challenges. Front. Cell Dev. Biol. 2015; 3:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noguchi T.K., Ninomiya N., Sekine M., Komazaki S., Wang P.-C., Asashima M., Kurisaki A.. Generation of stomach tissue from mouse embryonic stem cells. Nat. Cell Biol. 2015; 17:984–993. [DOI] [PubMed] [Google Scholar]
- 46. Tomaru Y., Hasegawa R., Suzuki T., Sato T., Kubosaki A., Suzuki M., Kawaji H., Forrest A.R.R., Hayashizaki Y., Shin J.W. et al. . A transient disruption of fibroblastic transcriptional regulatory network facilitates trans-differentiation. Nucleic Acids Res. 2014; 42:8905–8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bouchard M., Souabni A., Mandler M., Neubüser A., Busslinger M.. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002; 16:2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raposo A.A.S.F., Vasconcelos F.F., Drechsel D., Marie C., Johnston C., Dolle D., Bithell A., Gillotin S., van den Berg D.L.C., Ettwiller L. et al. . Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 2015; 10:1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pandian G.N., Nakano Y., Sato S., Morinaga H., Bando T., Nagase H., Sugiyama H.. A synthetic small molecule for rapid induction of multiple pluripotency genes in mouse embryonic fibroblasts. Sci. Rep. 2012; 2:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Han L., Pandian G.N., Junetha S., Sato S., Anandhakumar C., Taniguchi J., Saha A., Bando T., Nagase H., Sugiyama H.. A synthetic small molecule for targeted transcriptional activation of germ cell genes in a human somatic cell. Angew. Chem. Int. Ed. 2013; 52:13410–13413. [DOI] [PubMed] [Google Scholar]
- 51. Pandian G.N., Taniguchi J., Junetha S., Sato S., Han L., Saha A., AnandhaKumar C., Bando T., Nagase H., Vaijayanthi T. et al. . Distinct DNA-based epigenetic switches trigger transcriptional activation of silent genes in human dermal fibroblasts. Sci. Rep. 2014; 4:3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han L., Pandian G.N., Chandran A., Sato S., Taniguchi J., Kashiwazaki G., Sawatani Y., Hashiya K., Bando T., Xu Y. et al. . A synthetic DNA-binding domain guides distinct chromatin-modifying small molecules to activate an identical gene network. Angew. Chemie Int. Ed. 2015; 54:8700–8703. [DOI] [PubMed] [Google Scholar]
- 53. Pandian G.N., Sugiyama H.. Nature-inspired design of smart biomaterials using the chemical biology of nucleic acids. Bull. Chem. Soc. Jpn. 2016; 89:843–868. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.