Abstract
Type II restriction endonucleases (REases) form a large and highly diverse group of enzymes. Even REases specific for a common recognition site often vary in their oligomeric structure, domain organization and DNA cleavage mechanisms. Here we report biochemical and structural characterization of the monomeric restriction endonuclease UbaLAI, specific for the pseudosymmetric DNA sequence 5′-CC/WGG-3′ (where W = A/T, and ‘/’ marks the cleavage position). We present a 1.6 Å co-crystal structure of UbaLAI N-terminal domain (UbaLAI-N) and show that it resembles the B3-family domain of EcoRII specific for the 5′-CCWGG-3′ sequence. We also find that UbaLAI C-terminal domain (UbaLAI-C) is closely related to the monomeric REase MvaI, another enzyme specific for the 5′-CCWGG-3′ sequence. Kinetic studies of UbaLAI revealed that it requires two recognition sites for optimal activity, and, like other type IIE enzymes, uses one copy of a recognition site to stimulate cleavage of a second copy. We propose that during the reaction UbaLAI-N acts as a handle that tethers the monomeric UbaLAI-C domain to the DNA, thereby helping UbaLAI-C to perform two sequential DNA nicking reactions on the second recognition site during a single DNA-binding event. A similar reaction mechanism may be characteristic to other monomeric two-domain REases.
INTRODUCTION
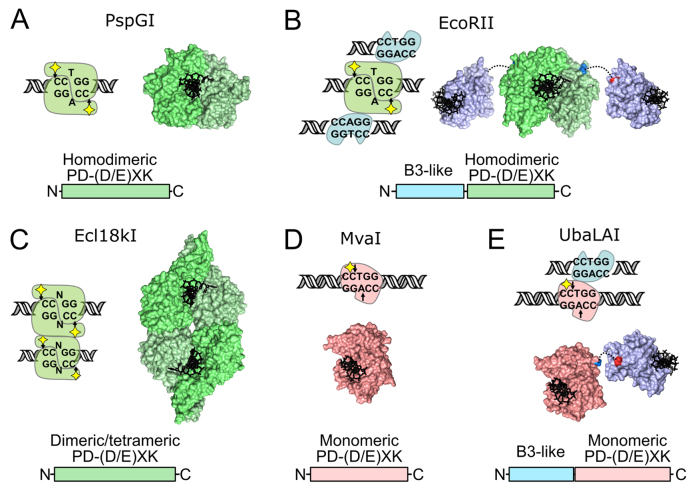
During evolution, protein domains are used as building blocks that can be combined in different arrangements, resulting in enzymes with altered activity, specificity or entirely new functions (1). A remarkable illustration of this process is provided by Type II restriction endonucleases (REases), one of the largest and most diverse groups of enzymes. At present, >4000 REases specific for more than 400 recognition sites were identified in different bacteria and archaea species (2). Despite a similar function, i.e. cleavage of DNA within or close to short (4–8 bp) specific recognition sites, REases, even those specific for the same DNA sequence (isoschizomers), often differ dramatically in their domain organization, oligomeric structure and DNA cleavage mechanisms. Notable examples are enzymes specific for the 5′-GGCGCC-3′ sequence: of 7 biochemically characterized REases, 5 different reaction mechanisms could be discerned (3). Another example, backed up by extensive structural data, is enzymes specific for a set of closely related pseudopalindromic recognition sequences 5′-CCNGG-3′ (Ecl18kI), 5′-CCWGG-3′ (PspGI, EcoRII, MvaI) and 5′-CCSGG-3′ (BcnI), where N stands for any nucleotide, W = T, A, and S = G, C (Figure 1). As revealed by the co-crystal structure, PspGI is a one-domain homodimeric enzyme that forms a symmetric complex with a single copy of the 5′-/CCWGG-3′ site (‘/’ marks the cleavage position), placing two catalytic centers in the proximity of two scissile phosphodiester bonds (Figure 1A, (4)). Such mode of DNA binding and cleavage is historically considered as normal for Type II REases. Another homodimeric enzyme, EcoRII, is a two-domain protein capable of simultaneous binding of three copies of the 5′-/CCWGG-3′ recognition site, one by the PspGI-like dimer of the C-terminal domains, and two more by the N-terminal effector domains that are structurally related to the B3 domains of plant transcription factors (Figure 1B (5,6)). Only one DNA site is cleaved (the one bound by the C-terminal domains), the other two sites helping to activate the enzyme to cleave the first copy (7). EcoRII and other REases containing two functionally distinct types of DNA binding clefts, one capable of DNA cleavage and the other (or others) of activator sequence(s) binding are classified as Type IIE enzymes (8). In contrast, the one-domain Type IIF enzyme Ecl18kI forms a homotetrameric complex with two copies of the 5′-/CCNGG-3′ sequence, and simultaneously cuts both copies of the recognition site in each of the two PspGI-like primary dimers (Figure 1C (9,10)). Finally, BcnI (5′-CC/SGG-3′), and the closely related REase MvaI (5′-CC/WGG-3′) are monomeric one-domain enzymes containing a single catalytic center (Figure 1(D) (11,12)). To accomplish cleavage of double-stranded DNA these enzymes must first bind the recognition sequence in one orientation, nick the first DNA strand, then re-bind the nicked recognition site in the opposite orientation and nick the second DNA strand (13). Surprisingly, biochemical studies of BcnI and preliminary characterization of MvaI suggest that both these enzymes, despite having a single catalytic center, are highly efficient REases, capable of cleaving both strands of the recognition site during a single binding event (14). It was proposed that the switch in enzyme orientation on the nicked site must involve enzyme hopping and sliding on the DNA without dissociation into the bulk solution, as now is confirmed for BcnI using single molecule techniques (15).
Figure 1.
Diversity of REases specific for the 5′-CCNGG-3′ and 5′-CCWGG-3′ recognition sites. In all panels catalytic PD-(D/E)XK domains in the central base pair flipping enzymes PspGI, EcoRII and Ecl18kI (36) are colored in different shades of green, monomeric MvaI-like PD-(D/E)XK domains are red, B3-like domains are blue. Yellow diamonds in cartoons denote the catalytic center. (A) PspGI (recognition sequence 5′-/CCWGG-3′) is an orthodox homodimeric REase that binds a single DNA copy (PDB ID 3bm3 (4)). (B) EcoRII (5′-/CCWGG-3′) is a homodimeric type IIE enzyme, capable of simultaneous binding of three recognition sites. One is cleaved by the PspGI-like dimer of the catalytic C-domains, while two others, one per each EcoRII-N effector domain, stimulate cleavage of the first site (PDB IDs 3hqf and 3hqg (5,7)). (C) Ecl18kI (5′-/CCNGG-3′) is a type IIF enzyme, which forms a tetramer on the DNA and simultaneously cuts both recognition sites (PDB ID 2fqz (9,10)). (D) MvaI (5′-CC/WGG-3′), like the related enzyme BcnI (5′-CC/SGG-3′), is a monomeric enzyme that uses a single catalytic center to cleave sequentially the first and then the second DNA strands (PDB ID 2oaa (11,12,14)). (E) UbaLAI (5′-CC/WGG-3′) is a monomeric REase consisting of an MvaI-like catalytic domain (red) and an EcoRII-N-like effector domain (blue, PDB ID 5o63). Structure of the UbaLAI-C domain is a model built using Modeller (37) and an MvaI-UbaLAI-C alignment generated with HHpred (38).
In this study, we present biochemical and structural characterization of REase UbaLAI (5′-CC/WGG-3′), isolated from an environmental DNA sample. We find that UbaLAI is a two-domain enzyme, which consists of structural domains found in two other REases specific for the 5′-CCWGG-3′ DNA sequence: the C-terminal domain of UbaLAI (UbaLAI-C) is similar to MvaI, while the N-terminal domain (UbaLAI-N) is related to the effector domain of EcoRII (Figure 1E). We perform structural analysis of the N-terminal accessory domain and show that UbaLAI, being an unusual fusion of building blocks found in other REases, is a monomeric Type IIE enzyme that employs a DNA cleavage mechanism that is distinct from the PspGI, EcoRII, Ecl18kI and MvaI/BcnI mechanisms described above. We propose that the UbaLAI-N domain acts as a handle that tethers the monomeric UbaLAI-C domain to the DNA, thereby helping UbaLAI-C to perform two sequential DNA nicking reactions on the second recognition site during a single DNA-binding event. We believe that the reaction mechanism deduced here for UbaLAI may be valid for other monomeric two-domain REases, for example DpnI and Sau3AI.
MATERIALS AND METHODS
Protein expression and purification
Expression and purification of full-length UbaLAI and its N-terminal domain were performed as described in the Supplementary Data section.
Oligonucleotides
All synthetic oligonucleotides used for cloning, site-specific mutagenesis, protein crystallization and DNA binding studies were purchased from Metabion (Germany). Oligoduplexes used in this study are listed in Supplementary Table S1.
DNA binding experiments
DNA binding was assessed using the electrophoretic mobility shift assay (EMSA). 33P-labeled DNA (final concentration typically 1–10 nM) was incubated with the protein (final concentrations, depending on the protein, were in the range from 0.2 nM to 10 μM) for 15 min in 20 μl of the binding buffer containing 40 mM Tris-acetate (pH 8.3 at 25°C), 0.1 mg/ml BSA, 5 mM calcium acetate and 10% (v/v) glycerol. Free DNA and protein-DNA complexes were separated by electrophoresis through 8% polyacrylamide gels (29:1 acrylamide/bisacrylamide) in 40 mM Tris-acetate, pH 8.3 for 1.5–3.0 hours at 5 V/cm. In some experiments both binding and electrophoresis buffers were devoid of calcium, but instead were supplemented with 1 mM EDTA; a subset of EMSA experiments was also performed with a mixture of short (16 bp) radiolabeled oligoduplex DNA and a longer (205 bp) unlabeled PCR fragment containing a single UbaLAI recognition site, or a 164 bp PCR fragment devoid of UbaLAI recognition sequences. Radiolabeled DNA and protein-DNA complexes were detected and quantified using the ‘Cyclone’ phosphorimager and OptiQuant software (Packard Instruments).
Plasmids and DNA fragments
Supercoiled plasmids pEcoRII-1, pEcoRII-2 and pEcoRII-3, containing a single, two and three 5′-CCWGG-3′ sites were purified as published (7). The 205 bp PCR fragment containing a single UbaLAI/EcoRII recognition site used in some EMSA experiments was as published (16). The 164 bp PCR fragment lacking UbaLAI recognition sites was obtained by PCR amplification of a pUC19 region using primers 5′-CCGCTTACAGACAAGCTGTGACCGTCTCCGGCCGCTGCATG-3′ and 5′- CCGAAAAGTGCCACCTGACGTCTAAGTCCGGCCGCTTATCATGAC-3′.
Reactions with supercoiled DNA
Cleavage reactions were performed at 25°C with 2 nM plasmid DNA and various enzyme concentrations (typically, 0.2 nM for steady-state reactions and 2–20 nM for single-turnover reactions) in Buffer Y (33 mM Tris-acetate, pH 8.0 at 25°C, 66 mM potassium acetate, 10 mM magnesium acetate and 0.1 mg/ml BSA). Reactions were initiated by addition of diluted protein to the mixture of other reaction components. 25 μl aliquots were removed at timed intervals, quenched by adding 8 μl of non-denaturing loading dye solution (50% v/v glycerol, 75 mM EDTA, bromphenol blue) and electrophoresed through agarose. Densitometric analysis of ethidium-bromide-stained gels was performed using ImageJ software.
Data analysis
The preparations of plasmids used in this study contained up to 25% of randomly nicked form. Assuming that randomly nicked DNA is equivalent to the intact substrate, we have corrected the experimentally determined amounts of supercoiled (SC) and nicked (OC) DNA as described in ref. (14). Data points in the graphs are presented as mean values from several independent experiments ±1 standard deviation. Steady-state reaction rates were determined from linear fits to the initial DNA cleavage data. Single turnover reactions were evaluated by fitting a single exponential function to the SC DNA cleavage data. All fitting procedures used KyPlot 2.0 beta 14 software. Determined rate constants are presented as the optimal value ± 1 standard error.
Crystallization and structure determination
UbaLAI-N domain in the buffer (20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM DTT) was mixed with the 9 bp oligoduplex 9SP (Supplementary Table S1) in ratio 1:1.2 (final protein concentration 4.5–5.5 mg/ml). Crystals were grown using the sitting drop vapor diffusion method at 19°C by mixing 0.6 μl of the complex solution with 0.4 μl of the crystallization buffer (0.15 M Na-citrate pH 5.6, 16–22% PEG3350). Crystals belonging to the space group P212121 appeared after 1 day. Native crystals were very sensitive to soaking in heavy atom solutions. UbaLAI-N domain contains only one (first) methionine, therefore Se-Met phasing could not be used directly. To overcome this problem we have introduced three methionines by site-directed mutagenesis (L24M, L53M, L95M: the ‘3Met’ mutant). UbaLAI-N-3Met crystallized under the same conditions as wt protein, and the crystals were much more robust and survived soaking. Hg derivative of UbaLI-N-3Met crystals was prepared by soaking crystals in 1 mM ethylmercury chloride for 2 days. The crystals were cryo-protected by adding the crystallization solution supplemented with 15% ethylene glycol, and flash frozen in liquid nitrogen. Data collection and structure refinement are described in the Supplementary Data section.
RESULTS
Restriction endonuclease UbaLAI
Restriction endonuclease UbaLAI, specific for the DNA sequence 5′-CC/WGG-3′ (W stands for A or T, ‘/’ marks the cleavage position) was isolated from an environmental DNA sample (see Supplementary Material for details). Sequence analysis of the protein revealed that its C-terminal part (domain UbaLAI-C, 165–405 residues) is similar to its isoschizomer R.MvaI (Supplementary Figure S1B), and the N-terminal part (domain UbaLAI-N, 1–164 residues) shares weak sequence similarities with the effector domain of EcoRII, another enzyme specific for the 5′-CCWGG-3′ target site (Figure 1(E) and Supplementary Figure S1C). Since UbaLAI-C-like single-domain enzymes MvaI and BcnI are efficient monomeric REases, our aim was to understand the role of the additional N-terminal domain in UbaLAI, and to determine the mechanism of double-stranded DNA cleavage by this enzyme.
DNA binding by wt UbaLAI
MW of purified UbaLAI determined by gel-filtration is 33 kDa (Supplementary Figure S3A). It is consistent with the theoretic mass of UbaLAI monomer, 46.2 kDa. Gel-filtration experiments performed with an equimolar mixture of wt UbaLAI and 16 bp cognate DNA 16SP revealed two peaks in gel-filtration elution profile, one matching apo-UbaLAI, and the second (with an apparent MW of 74 kDa) matching the theoretic MW of UbaLAI monomer bound to two 16SP duplexes (66.2 kDa, Supplementary Figure S3B).
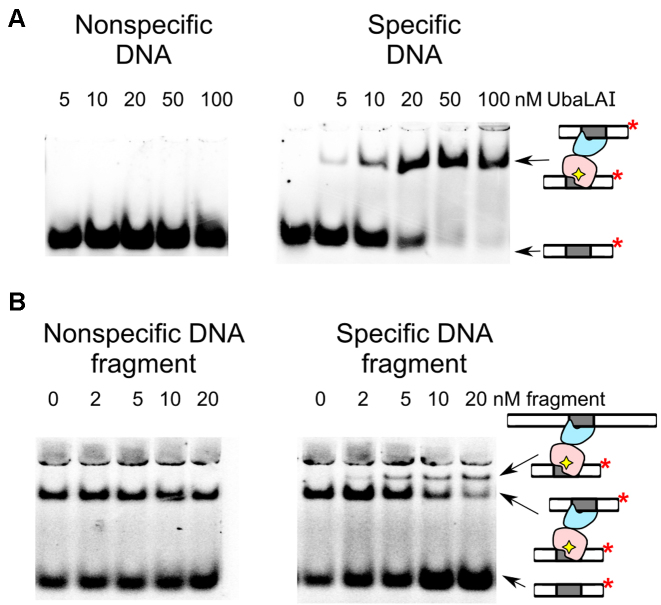
The DNA binding of UbaLAI was also investigated using electrophoretic mobility shift assay (EMSA). Under standard EMSA conditions, UbaLAI forms a single complex with cognate DNA and no complexes with non-specific DNA (Figure 2A). The specific UbaLAI–DNA complex was further examined by following an approach used previously to characterize DNA binding by the tetrameric endonuclease SfiI (17). If a protein can bind two segments of DNA at the same time, binding mixtures containing two DNA molecules of different length can yield three types of complexes: with two short, two long and one additional complex containing one molecule of each. These complexes can be separated by EMSA.
Figure 2.
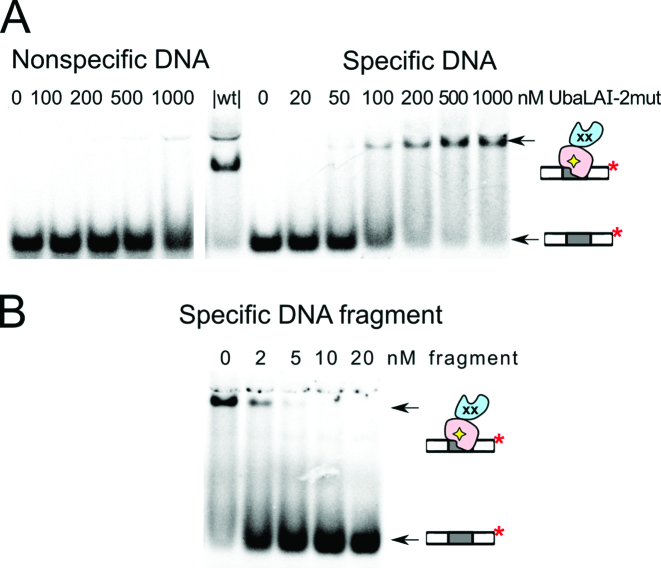
UbaLAI binding analysed by electrophoretic mobility shift (EMSA) assay. (A) EMSA experiments were performed in a pH 8.3 Tris-acetate buffer supplemented with 5 mM Ca2+ (see Materials and Methods for details). The reactions contained 33P-labeled duplex (10 nM), either the non-specific 16NSP (left-hand gel) or the specific 16SP (right-hand gels), and wt UbaLAI. The protein concentrations (in terms of nM monomer) were as indicated by each lane. (B) Stoichiometry of the specific UbaLAI-DNA complex. The reactions contained 10 nM cognate 33P-labeled oligoduplex 16SP, 20 nM wt UbaLAI enzyme, and the unlabeled DNA fragment that either contains (right-hand gel) or does not contain (left-hand gel) an UbaLAI recognition site. The concentrations of the DNA fragments are noted above the relevant lanes. The cartoons on the right-hand side of the gels denote radiolabeled DNA and various types of radiolabeled complexes detected in the experiments.
To test this possibility, specific UbaLAI-DNA complex was first formed by incubating radiolabeled 16 bp oligoduplex with UbaLAI. This complex was then titrated with increasing amounts of 205 bp unlabeled DNA fragment, that also contains an UbaLAI recognition site. When 205 bp DNA was added to the UbaLAI–oligoduplex complex, a fraction of the radiolabeled DNA became incorporated into a novel complex of lower electrophoretic mobility (Figure 2B). No such changes were observed when titration was performed with a non-specific DNA fragment (Figure 2B).
Since the electrophoretic mobility of a specific radiolabeled DNA bound to UbaLAI can be altered by adding a specific unlabeled DNA, the UbaLAI protein must be able to bind both the labeled and unlabeled DNA at the same time. Thus, in agreement with gel-filtration results, the UbaLAI-DNA complex resolved by EMSA must contain two copies of specific DNA bound to an UbaLAI monomer. We assume that one specific DNA is bound by the UbaLAI-N domain, and the second by the UbaLAI-C domain (Figure 1E).
DNA cleavage by wt UbaLAI
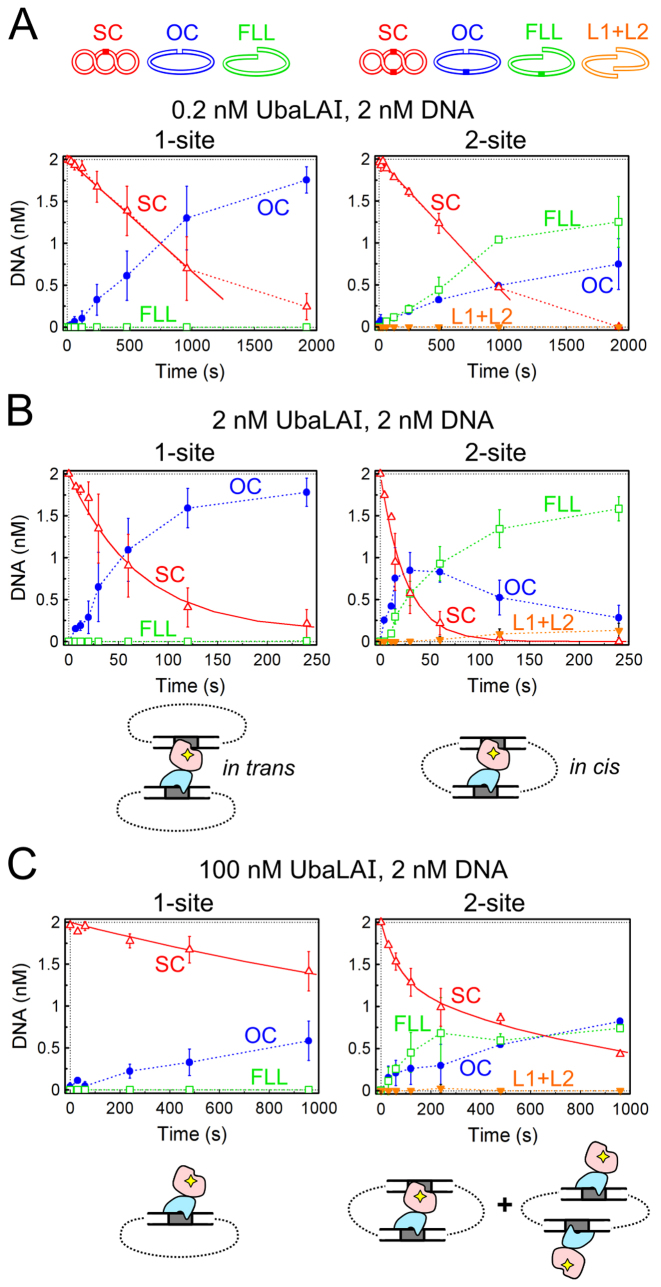
REases capable of simultaneous interaction with two recognition sites, including Type IIE and Type IIF enzymes, are less active when bound to one site, and more active when bound to both recognition sites. This is best manifested by comparing enzyme activities on supercoiled plasmid substrates with either one or two copies of the specific recognition site (18–20). To find out if interaction of UbaLAI with two recognition sites enhances its catalytic activity, we used plasmids pEcoRII-1 and pEcoRII-2 (7), containing one and two EcoRII/UbaLAI recognition sites with identical surrounding sequences. Cleavage reactions were performed under three sets of reaction conditions: (i) substrate excess conditions (0.2 nM enzyme and 2 nM plasmid DNA, Figure 3A); (ii) reactions with equimolar UbaLAI and plasmid concentrations (2 nM each, Figure 3B) and (iii) enzyme excess conditions (2 nM DNA, 100 nM enzyme, Figure 3C).
Figure 3.
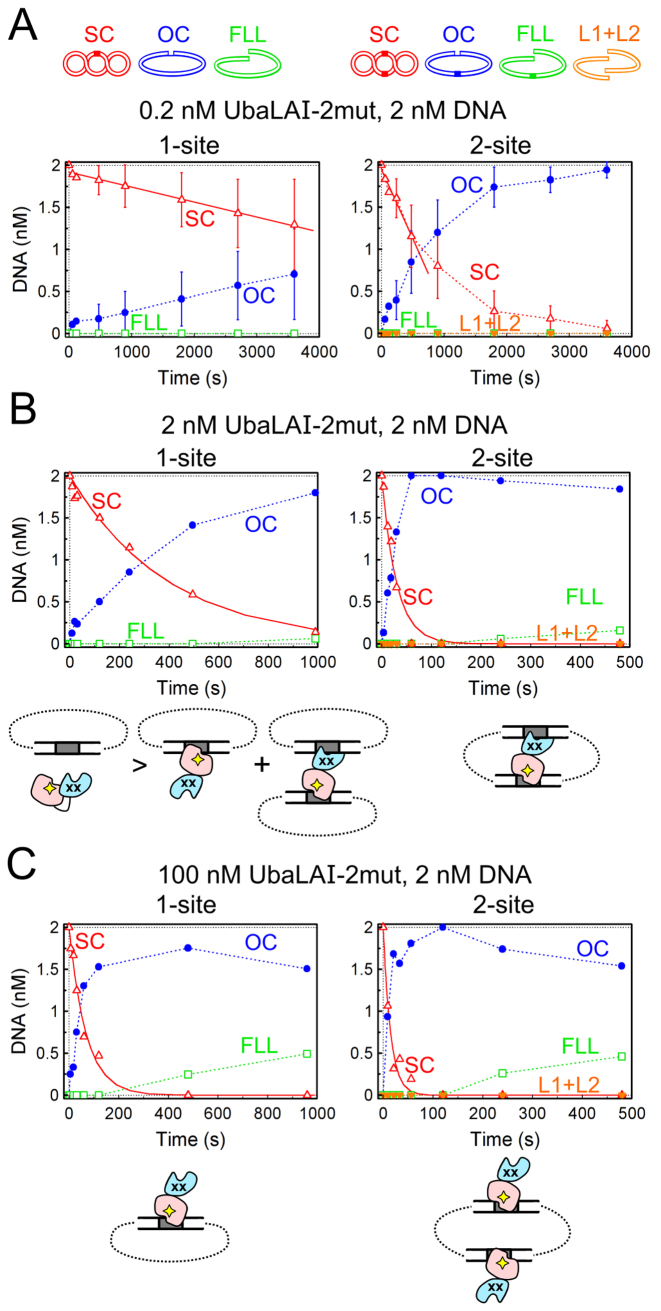
UbaLAI reactions on plasmids with one and two recognition sites. The cartoons above panel (A) illustrate DNA forms that can exist during UbaLAI reactions on plasmids with one or two UbaLAI sites: supercoiled DNA (SC, marked by open red triangles in the graphs); open circular (nicked) DNA (OC, filled blue circles); linear DNA cut at one UbaLAI site (FLL, open green squares); and, only with the 2-site plasmid, linear DNA cut at both UbaLAI sites (L1+L2, inverted orange triangles); OC and FLL forms of the 2-site plasmid may also contain an additional nick. All data points are presented as mean values from 2–7 independent experiments ±1 σ. All reactions were performed with 2 nM of either 1-site plasmid pEcoRII-1 (left-hand graphs) or 2-site plasmid pEcoRII-2 (right-hand graphs) as described in Materials and Methods. (A) Steady-state reactions, performed with 2 nM DNA and 0.2 nM enzyme. Continuous lines are linear fits to the initial SC DNA cleavage data that yielded DNA cleavage rates (optimal value ± SEM) of 0.0013 ± 0.0003 nM s−1 and 0.0016±0.0004 nM s−1 for the 1-site and 2-site plasmids, respectively. (B) Reactions performed with equimolar enzyme and DNA (2 nM each) concentrations. Continuous lines are single exponential fits to the SC DNA cleavage data that yielded the observed first order rate constants of 0.014 ± 0.002 s−1 and 0.040 ± 0.003 s−1 for the 1-site and the 2-site plasmids, respectively. Cartoons below the graphs schematically depict the synaptic complexes formed by UbaLAI with 1- and 2-site plasmids. Red shapes mark the catalytic UbaLAI-C domain, the blue shapes mark the UbaLAI-N domain. (C) Reactions performed under enzyme excess conditions (100 nM UbaLAI, 2 nM DNA). Continuous lines are single (left-hand graph) and double (right-hand gel) exponential fits to the SC DNA cleavage data that yielded the observed first order rate constant of 0.00038 ± 0.00002 s−1 for the left-hand graph, and 0.014 ± 0.003 s−1 (amplitude 35 ± 6%) and 0.0011 ± 0.0002 s−1 (amplitude 65±6%) rate constants for the right-hand graph. Cartoons below the graphs schematically depict the non-productive UbaLAI–DNA complexes formed under these conditions (all specific sites are occupied by the non-catalytic UbaLAI-N domains, blue shapes).
Under steady-state reaction conditions (Figure 3A) the only product of UbaLAI reaction on 1-site plasmid was nicked (open circular, OC) DNA. This implies that during a single enzyme-DNA encounter, the UbaLAI molecule nicks just one DNA strand, and then dissociates from the nicked intermediate. In the presence of substrate excess, the released UbaLAI then associates and nicks another intact plasmid, resulting in accumulation of nicked DNA. In contrast, the major reaction product accumulating from the reaction start in the 2-site plasmid cleavage reaction is DNA with a single double strand break (full-length linear, FLL, Figure 3A). This suggests that on a 2-site substrate UbaLAI is able to cleave both DNA strands of one recognition site during a single binding event.
Comparison of 1-site and 2-site plasmid cleavage at equimolar enzyme to substrate ratio (Figure 3B) indicates that the 1-site plasmid is cleaved slowly (rate constant 0.014 s−1), the major product being nicked DNA, while the 2-site plasmid is cleaved 3-fold faster (rate constant 0.040 s−1), with consecutive formation of nicked DNA and a product with a single double-strand break. Such cleavage pattern is expected for a Type IIE enzyme, where one specific site stimulates catalytic activity of the enzyme at the second site, leaving the first site intact. But, unlike the previously characterized dimeric Type IIE enzymes, e.g. NaeI and EcoRII, UbaLAI is a monomer. Presumably, UbaLAI binds the first site via the non-catalytic UbaLAI-N (effector) domain, which then stimulates cleavage of the second site by the catalytic UbaLAI-C domain. On a 2-site plasmid, such optimal complex (cartoons in Figure 3B) may be formed by bridging two recognition sites in cis, a favorable type of interaction; in the case of the 1-site plasmid, a synaptic complex would require an unfavorable type of interaction, i.e. bridging of two supercoiled plasmids in trans, resulting in less efficient cleavage.
An increase in enzyme concentration (50-fold enzyme excess over the plasmids, Figure 3C) inhibited both 1-site and 2-site plasmid cleavage reactions. Apparently, this is due to binding of individual UbaLAI monomers to every available UbaLAI recognition site (cartoons in Figure 3C), thereby precluding formation of synaptic complexes either in cis (2-site DNA) or in trans (1-site DNA).
Characterization of the UbaLAI-N domain
In order to understand the roles of individual UbaLAI domains, we have attempted to clone, purify and characterize separate UbaLAI-N and UbaLAI-C domains. UbaLAI-N domain (1–164 residues of the wt protein) with a C-terminal HisTag was successfully expressed in Escherichia coli and purified (see Materials and Methods for details). As expected, gel-filtration experiments were consistent with a monomeric oligomeric state of UbaLAI-N and a 1:1 DNA binding stoichiometry (Supplementary Figure S4). EMSA experiments performed both in the presence and in the absence of Ca2+ ions (Supplementary Figure S5A, B) demonstrated that UbaLAI-N forms a tight complex with specific DNA (KD approximately equal to 0.04 nM), but no complex was observed with nonspecific DNA (Supplementary Figure S5A and B).
Structure of the UbaLAI-N domain
To elucidate DNA recognition mechanism of UbaLAI, we have solved a crystal structure of UbaLAI DNA recognition domain (UbaLAI-N) in complex with 9 bp oligoduplex containing 5′-CCWGG-3′ recognition sequence at 1.6 Å resolution (Figure 4A). The UbaLAI-N used for structural studies contained 3 leucine-methionine substitutions (mutations L24M, L53M and L95M) in the hydrophobic core of the protein that had no significant effect on DNA binding by the domain (Supplementary Figure S5C). Originally designed for phasing purposes using SeMet-substituted protein, these mutations had a positive effect on crystal stability, enabling soaking experiments and phasing using the SIRAS protocol (see Materials and Methods for details).
Figure 4.
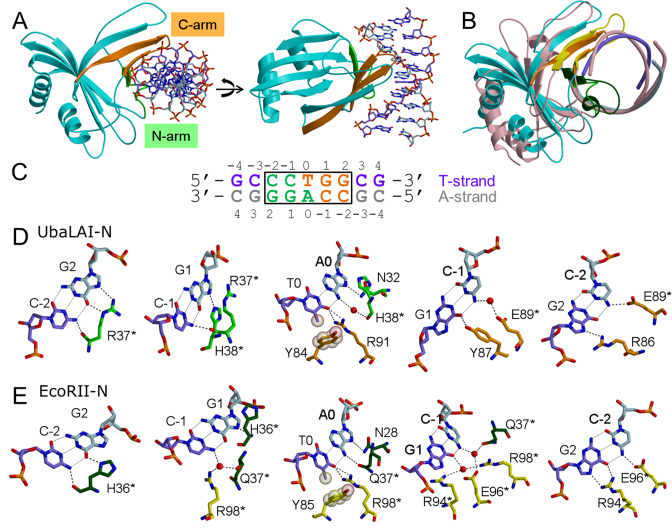
UbaLAI-N–DNA complex structure. (A) Two different views of UbaLAI-N–DNA complex structure. The secondary structure elements of the N-arm are shown in green; those belonging to the C-arm are shown in orange, the T-strand of DNA is shown in violet. (B) Superposition of UbaLAI-N and EcoRII-N DNA complexes. UbaLAI-N structure is colored as in (A), EcoRII-N–DNA complex is colored pink. N-arm and C-arm of EcoRII-N are colored dark green and yellow, respectively. (C) Oligonucleotide used for crystallization of UbaLAI-N. Recognition sequence is boxed, DNA bases recognized by residues from N-arm and C-arm are shown in green and orange letters, respectively. (D) Recognition of the 5′-CCTGG-3′ sequence by UbaLAI-N. Panels for individual base pairs are arranged following the T-strand in 5′-3′ direction. Residues belonging to the N- and C-arms are colored in green and orange, respectively. (E) Recognition of the 5′-CCTGG-3′ sequence by EcoRII-N. Residues belonging to the N- and C-arms are colored in dark green and yellow, respectively.
In the crystal UbaLAI-N domain binds DNA as a monomer. It is composed of eight β strands making a pseudo barrel decorated by two α helices (Figure 4A). The overall fold of the domain belongs to SCOP double-split β -barrel fold, DNA binding pseudobarrel domain superfamily (21). The structure of the UbaLAI-N domain is similar to the DNA recognition domains of restriction endonucleases EcoRII (PDB ID 1na6, DALI Z-score 10.6, Figure 4B), BfiI (3zi5, 10.0), 2c1l and NgoAVII (4crt, 5.3), and to B3 domains of plant transcription factors, including auxin response factors 1 (4ldx, 9.4) and 5 (4ldu, 9.3), DNA binding domain of RAV1 (1wid, 6.3), and hypothetical A. thaliana protein AT1G16640 (1yel, 6.0).
DNA recognition by UbaLAI-N
UbaLAI-N interacts with its pseudo-symmetric recognition sequence 5′-CCWGG-3′ asymmetrically (Figure 4), forming different contacts with the ‘T’ strand (the DNA strand containing a T at the central position of the recognition sequence) and the ‘A’ strand (the DNA strand with central A, Figure 4C). In the UbaLAI-N–DNA complex structure ∼1750 Å2 of protein surface is buried at the protein–DNA interface. Contacts to DNA in the major groove are made by two structural elements, called N-arm and C-arm in analogy with the restriction endonuclease EcoRII (5).
N-arm comprises residues 31–43 and folds into a loop and a β strand (Figure 4A). C-arm (residues 75–94) forms a β hairpin structure, which is a part of the pseudo-barrel. Residues from N- and C-arms make direct and water-mediated contacts to one-half of the CCTGG sequence: N-arm recognizes the first two C:G base pairs and the central A base, while C-arm recognizes the central T base and the last two G:C base pairs (Figure 4C and D). Overall, UbaLAI-N makes 11 direct contacts and 2 water-mediated hydrogen bonds to DNA bases.
In particular, R37 from the N-arm makes contacts to both bases of the C-2:G2 base pair involving main chain and side chain atoms and accepts hydrogen bond from N4 atom of cytosine C-1 from the adjacent base pair. H38 NE2 atom donates hydrogen bond to N7 atom of G1, and its main chain carbonyl oxygen atom makes water-mediated contact to N6 atom of adenine A0. A0 O6 atom accepts hydrogen bond from ND2 atom of N32 (Figure 4D).
Side chain of Y84 from the C-arm makes van der Waals interaction to the methyl group of T0. The NH1 and NH2 atoms of R91 make hydrogen bonds to the thymine O4 atom. O6 atom of guanine G1 (G1:C-1 bp) accepts hydrogen bond from hydroxyl group of Y87. Side chain of E89 makes a water-mediated hydrogen bond to N4 atom of cytosine C-1 and a direct hydrogen bond to N4 atom of C-2 (G2:C-2 bp). N7 of G2 accepts hydrogen bond from NH1 atom of R86 (Figure 4D).
UbaLAI variants with an impaired UbaLAI-N domain
We have made expression vectors for the UbaLAI-C domain with either N- or C-terminal affinity tags. Unfortunately, the yield of the soluble protein in all cases was negligible, and we therefore could not purify and characterize an isolated UbaLAI-C domain.
As an alternative approach we have generated full-length UbaLAI variants with an impaired UbaLAI-N domain, and compared its biochemical properties to the wt enzyme. In the first iteration, we have replaced two UbaLAI-N residues, E89 and R91, implicated in base-specific contacts (Figure 4D), with alanines, both in the context of the full-length UbaLAI (henceforth referred to as the double mutant UbaLAI-2mut), and the separate UbaLAI-N domain. EMSA experiments revealed that the E89A+R91A mutant of the UbaLAI-N domain retained residual DNA binding ability (KD for specific DNA is approx. 200 nM, or 5000-fold lower than observed with wt UbaLAI-N, Supplementary Figure S5A and D). To generate an UbaLAI variant with a completely disabled N-domain, we have introduced two additional mutations, R37A and H38A, thereby generating a quadruple (R37A + H38A + E89A + R91A) mutant of the full-length UbaLAI (henceforth referred to as UbaLAI-4mut), and a corresponding variant of the UbaLAI-N domain. EMSA experiments with the quadruple UbaLAI-N mutant confirmed complete loss of its specific DNA binding ability (Supplementary Figure S5E).
DNA binding by the full-length UbaLAI-2mut and UbaLAI-4mut proteins was investigated by a standard gel-shift assay. Both mutants formed complexes with specific DNA (Figure 5A and Supplementary Figure S7), but the electrophoretic mobility of these complexes was significantly lower than mobility of the complex formed by wt UbaLAI (Figure 2A). Using EMSA with DNA fragments of different length we have confirmed that UbaLAI-2mut-DNA binding stoichiometry in the specific complex is 1:1 (no additional radiolabeled complex was formed upon titration of the UbaLAI-2mut-DNA complex with a long unlabeled DNA fragment, Figure 5B). We presume that the single DNA in this complex is bound by the unperturbed UbaLAI-C domain. The higher mobility of the UbaLAI-2×DNA complex in comparison to the UbaLAI-2mut-1×DNA complex is most likely due to the fact that the extra negative charge of the second DNA molecule outweighs the increase in complex size; similar differences in electrophoretic mobilities were also observed with other REases, e.g. Bse634I (18).
Figure 5.
DNA binding by the E89A+R91A UbaLAI mutant UbaLAI-2mut. (A) EMSA experiments were performed as in Figure 2A with 33P-labeled specific 16SP (right-hand gel) or non-specific 16NSP (left-hand gel) DNA, and the UbaLAI-2mut protein. The protein concentrations (in terms of nM monomer) were as indicated by each lane. Lane ‘|wt|’ contained 10 nM 16SP DNA and 50 nM wt UbaLAI. (B) Stoichiometry of the specific UbaLAI-2mut-DNA complex. The reactions contained 10 nM cognate 33P-labeled oligoduplex 16SP, 100 nM wt UbaLAI enzyme, and the unlabeled DNA fragment that contains an UbaLAI recognition site. The concentrations of the DNA fragments are noted above the relevant lanes. The cartoons in panels (A) and (B) depict the single radiolabeled complex detected in the experiments.
The DNA cleavage reactions of both UbaLAI-2mut (Figure 6) and UbaLAI-4mut (Supplementary Figure S8) were also affected by the mutations in the UbaLAI-N domain. Under substrate excess conditions (Figure 6A) the sole reaction product formed by UbaLAI-2mut in 1-site and 2-site plasmid cleavage reactions was nicked DNA. Thus, unlike the wt enzyme, UbaLAI-2mut is unable to complete cleavage of both DNA strands of the 2-site plasmid during a single binding event, but instead dissociates from the nicked intermediate.
Figure 6.
UbaLAI-2mut reactions on plasmids with one and two recognition sites. As in Figure 3, cartoons above the graphs depict DNA forms that can exist during UbaLAI reactions on plasmids with one and two UbaLAI sites. (A) Steady-state reactions, performed with 2 nM DNA and 0.2 nM UbaLAI-2mut. Continuous lines are linear fits to the initial SC DNA cleavage data that yielded DNA cleavage rates (optimal value ± SEM) of 0.00018 ± 0.00008 nM s−1 and 0.0016 ± 0.0006 nM s−1 for the 1-site and 2-site plasmids, respectively. (B) Reactions performed with equimolar enzyme and DNA (2 nM each) concentrations. Continuous lines are single exponential fits to the SC DNA cleavage data that yielded the observed first order rate constants of 0.0025 ± 0.0002 s−1 and 0.034 ± 0.004 s−1 for the 1-site and the 2-site plasmids, respectively. (C) Reactions performed under enzyme excess conditions (100 nM UbaLAI-2mut, 2 nM DNA). Single exponential fits yielded the observed first order rate constants of 0.015 ± 0.002 s−1 and 0.07 ± 0.01 s−1 for the 1-site and the 2-site plasmids, respectively. Cartoons below the graphs in panels (B) and (C) schematically depict the protein–DNA complexes formed by UbaLAI-2mut with 1- and 2-site plasmids.
The differences were less pronounced in the reactions performed with equimolar UbaLAI-2mut and DNA concentrations (Figure 6B): UbaLAI-2mut displayed a preference for the 2-site substrate, and generated the linear reaction product via nicked intermediate. But in stark contrast to the wt enzyme, we did not observe any inhibition of DNA cleavage under enzyme excess conditions (compare Figure 3B and C and Figure 6B and C). The catalytic activity of UbaLAI-4mut, the enzyme variant with a completely disabled UbaLAI-N domain, increased steadily with increasing enzyme concentrations (up to 200-fold enzyme excess over the substrate, the highest enzyme excess used in our experiments) on both 1-site and 2-site plasmid substrates (Supplementary Figure S8).
DISCUSSION
DNA recognition by UbaLAI-N and EcoRII N-domains
We show here that Type IIE enzymes UbaLAI and EcoRII use similar N-terminal effector domains specific for the 5′-CCWGG-3′ sequence. Despite the fact that UbaLAI-N and EcoRII-N domains share only 18% sequence identity (Supplementary Figure S1C), their overall structures are very similar (Figure 4B): 134 of 173 residues of the EcoRII N-terminal domain (3hqf) could be aligned with UbaLAI-N with r.m.s.d. of 3.1 Å (DALI server (22)). The loops of the N-arms adopt slightly different conformations in both domains; the C-arms overlay better, but the loop between the strands in the C-arm of UbaLAI-N is shorter (Supplementary Figure S6). Both domains make identical contacts with the central T0:A0 base pairs employing overlaying residues (Figure 4D and E, Supplementary Figure S6). Contacts to the cytosines C-1 and C-2 made by the C-arms are also identical, and involve residues E89 and E96 of UbaLAI and EcoRII, respectively.
Residues making contacts to other bases do not overlay upon superposition of the structures. Nevertheless, they use the same principle of recognition. For example, the first base pair C-2:G2 is recognized by a single amino acid (R37 and H36 of UbaLAI and EcoRII, respectively), which makes contacts with C-2 using main chain carbonyl oxygen and recognizes G2 through hydrogen bonds with the side chain atoms. The same residues are also involved in the recognition of the second base pair, but the contacts are different. Guanines of the last G2:C-2 base pair are recognized by non-overlaying arginines (R86 (UbaLAI) and R94 (EcoRII)), cytosines in both complexes are recognized by matching glutamates E89 and E96 of UbaLAI and EcoRII, respectively (Figure 4D and E, Supplementary Figure S6). Contacts to guanine of G1:C-1 base pair are also different in both domains.
Taken together, the overall similarity of UbaLAI-N and EcoRII-N structures, including conservation of many base-specific contacts to the common 5′-CCWGG-3′ recognition site, suggest they both descent from a common ancestor. On the other hand, extensive changes acquired by the domains in UbaLAI and EcoRII demonstrate extreme adaptability of this DNA recognition domain to different structural contexts and reaction mechanisms of these enzymes.
The interplay between UbaLAI-N and UbaLAI-C domains
DNA binding and cleavage data presented in our study suggest that UbaLAI domains UbaLAI-N and UbaLAI-C do not act independently. Indeed, the calculated KD for the UbaLAI-N–DNA interaction, 0.04 nM (Supplementary Figure S5A), is much lower than concentration of full-length UbaLAI required to bind an appreciable fraction of cognate DNA in EMSA experiments (Figure 2A; KD for the wt UbaLAI–DNA interaction was not calculated here due to the UbaLAI-2×DNA stoichiometry of the specific complex). This implies that the C-terminal UbaLAI domain partially interferes with DNA binding by the N-terminal domain.
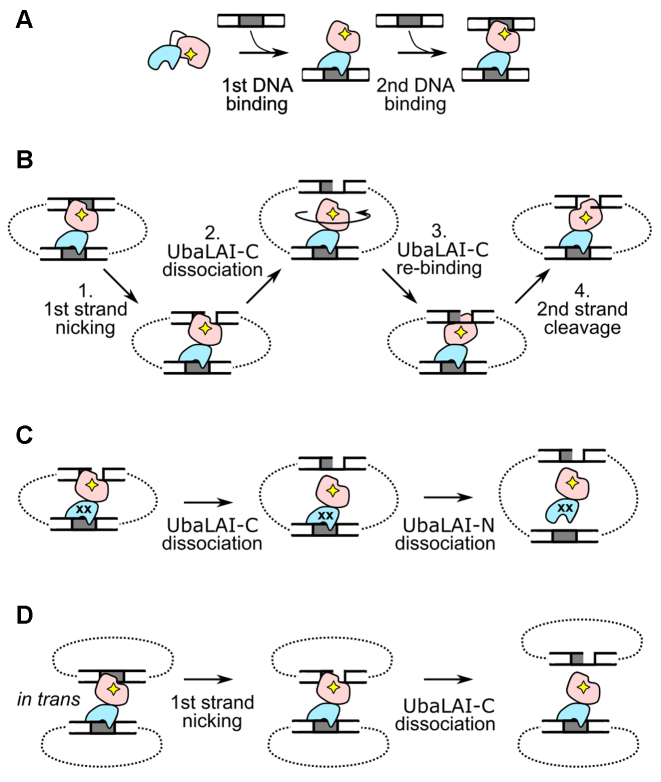
A plausible model of UbaLAI-DNA interaction is summarized in Figure 7A. We propose that wt UbaLAI binds the first DNA site primarily via its high-affinity N-terminal non-catalytic domain. Such an interaction may in turn trigger DNA binding by the catalytic UbaLAI-C domain (Figure 7A), leading to formation (even at nanomolar enzyme/DNA concentrations) of a stable catalytically competent synaptic in cis complex on the 2-site plasmid, or a less stable in trans complex on the 1-site plasmid (Figure 3B). Based on the apo-structure, a similar ‘autoinhibition’ model was previously proposed for EcoRII. In the apo-form, the non-catalytic EcoRII-N domains block the DNA binding/catalytic center formed by the catalytic EcoRII-C domains; upon DNA binding, the EcoRII-N domains unblock the DNA binding cleft formed by the EcoRII-C domains, thereby enabling DNA cleavage (6).
Figure 7.
UbaLAI reaction mechanism. (A) UbaLAI–DNA association. The first DNA site is bound via the non-catalytic UbaLAI-N domain. This triggers binding and cleavage of the second DNA site by the catalytic UbaLAI-C domain. (B) The proposed UbaLAI reaction mechanism (‘sequential nicking via tethered activation’) on 2-site DNA, involving the ‘swiveling’ catalytic UbaLAI-C domain and the UbaLAI-N domain that tethers the catalytic domain to the multi-site DNA. (C) Proposed mechanism for the UbaLAI-2mut reaction on the 2-site DNA. The complex dissociates before UbaLAI-C can re-bind in the opposite orientation and complete the reaction. (D) Wt UbaLAI reaction on 1-site DNA. The in trans synaptic complex dissociates after nicking the first DNA strand.
DNA binding cooperativity
Formation of a single UbaLAI-2×DNA complex in EMSA experiments is also indicative of highly cooperative DNA binding by wt UbaLAI, when attachment of the first DNA site stimulates binding of the second site. Otherwise, one would expect formation of two different UbaLAI–DNA complexes, one with a single DNA (a low mobility complex, as observed with UbaLAI-2mut, Figure 5A), and one with two DNAs (the higher mobility complex, Figure 2A). Indeed, two different protein-DNA complexes are resolved in the case of REase BfiI, an enzyme that shows little cooperativity in DNA binding (19). Under enzyme excess conditions (e.g. 100 nM wt UbaLAI, 2 nM DNA), the preferential UbaLAI binding to DNA via the N-terminal domain results in significant inhibition of both 1-site and 2-site DNA cleavage reactions (Figure 3C), as in this case the productive synaptic complexes are replaced by complexes where all recognition sites are occupied by protein molecules bound via the non-catalytic UbaLAI-N domain (cartoons in Figure 3C).
Cooperative binding of two recognition sites by wt UbaLAI may also explain relatively small differences between the nicking rates observed on the 1- and 2-site plasmids under substrate excess conditions (almost equal nicking rates, but different products, Figure 3A), and with equimolar enzyme and substrate concentrations (approx. 3-fold difference, Figure 3B). Presumably, under these conditions wt UbaLAI bridges a significant fraction of the 1-site plasmids in trans (cartoons in Figure 3B), despite the fact that this type of protein-DNA complex is less stable. Efficient nicking of the 1-site substrate discriminates UbaLAI from other enzymes that tether two recognition sites, e.g. SfiI and Bse634I, which both cut the 1-site plasmids >10-fold slower than the optimal 2-site substrates (18,20).
The situation is quite different with the UbaLAI-2mut enzyme. Presumably, the residual DNA binding ability of mutant UbaLAI-N is still sufficient to ‘authorize’ DNA binding by the UbaLAI-C domain, resulting in formation of a catalytically competent in cis synaptic complex on the 2-site substrate. Indeed, under equimolar enzyme and DNA conditions UbaLAI-2mut cuts the 2-site plasmid with a comparable rate to the wt enzyme (Figure 6B). However, in the case of 1-site plasmid, impaired DNA binding by the mutant N-domain and loss of DNA binding cooperativity precludes formation of the less stable in trans synaptic complexes, resulting in very slow 1-site plasmid cleavage at low enzyme concentrations (the 1-site plasmid is cleaved ∼10-fold slower than the 2-site plasmid, Figure 6B; most of the 1-site plasmid under these conditions may also remain enzyme-free, as depicted in Figure 6B). But in stark contrast to wt UbaLAI, an increase in UbaLAI-2mut concentration does not inhibit, but instead accelerates DNA cleavage, especially in the case of the 1-site plasmid (Figure 6C). We attribute this lack of inhibition to preferential binding of UbaLAI-2mut to DNA via the catalytic UbaLAI-C domain, thereby forming productive complexes (Figure 6C). Such direct interaction of UbaLAI-2mut with the DNA via the C-domain bypasses the ‘authorization’ by UbaLAI-N, and therefore requires higher enzyme concentrations. Exclusive use of this ‘unauthorized’ DNA binding pathway by the UbaLAI-4mut enzyme explains why its activity is detectable only at high protein concentrations (Supplementary Figure S8).
Mechanism of double-stranded DNA cleavage
Structural homologs of UbaLAI-C, REases BcnI and MvaI, are monomeric single-domain REases (Figure 1D) that form an asymmetric complex with their recognition sites (11,12). To cut double-stranded DNA they must first associate with the recognition sequence in one orientation, nick the first DNA strand, re-bind the nicked intermediate in the opposite orientation, and nick the second strand. Intriguingly, the switch in enzyme orientation occurs without dissociation into bulk solution, but instead via sliding and hopping of the protein on the DNA. This enables cleavage of both DNA strands during a single enzyme-DNA association event (14).
DNA cleavage kinetics of wt UbaLAI under substrate excess conditions indicates that wt UbaLAI, despite being a monomeric REase with a single catalytic center, is also able to introduce a double-strand break during a single binding event (linear DNA product accumulates from the start of the steady-state 2-site plasmid cleavage reaction, Figure 3A). But for this UbaLAI requires (i) the presence of two specific sites in cis, as no linear product is formed in the steady-state reaction on a 1-site plasmid (Figure 3A); and (ii) an intact N-terminal DNA binding domain, as UbaLAI-2mut enzyme variant cuts only one DNA strand prior to dissociation, both on the 1-site and the 2-site plasmid substrates (Figure 6A). The UbaLAI reaction mechanism consistent with the above requirements is depicted in Figure 7B. First, UbaLAI forms a complex with two recognition sites in cis, and the UbaLAI-C domain nicks the first recognition site. Next, UbaLAI-C dissociates from the nicked intermediate, but remains tethered to the nicked substrate via the UbaLAI-N domain, bound to the second recognition site. This enables the UbaLAI-C domain to re-bind the nicked site in the opposite orientation and cut the remaining strand. The reaction is then completed by dissociation of UbaLAI-N from the intact recognition site, the slowest reaction step. The lower limit for the latter step can be inferred from the UbaLAI steady-state reactions (Figure 3A): since the reaction rate observed with 0.2 nM enzyme is 0.0016 nM s−1, the lower limit for UbaLAI turnover rate is 0.008 s−1 (0.0016 nM s−1/0.2 nM; the real turnover rate of UbaLAI might be higher, since part of the enzyme under our reaction conditions could remain in the DNA-free state). This rate constant is indeed considerably lower than the observed first-order DNA cleavage rate constants observed in the single turnover reactions (0.04 s−1, Figure 3B).
This ‘sequential nicking via tethered activation’ mechanism would also be efficient on multi-site DNA substrates: while UbaLAI-N keeps the enzyme tethered to one site, UbaLAI-C may nick and eventually cleave some of the remaining recognition sites. Indeed, the 3-site DNA (plasmid pEcoRII-3, as described in (7)) cleavage kinetics was similar to experiments performed on 2-site DNA, with the only difference that now two types of products (DNA with 1 and 2 double strand breaks) accumulated in the reactions (Supplementary Figure S9).
The mechanism in Figure 7B predicts that if the UbaLAI-N–DNA interaction time was shorter than the time required for UbaLAI-C to change its orientation on the DNA, this would result in complete dissociation of UbaLAI from the nicked reaction product. We believe that the latter happens with the UbaLAI-2mut protein. Residual DNA binding ability of UbaLAI-N domain of this protein (more than 1000-fold decrease in affinity in comparison to wt UbaLAI-N, Supplementary Figure S5D) is sufficient to form an in cis synaptic complex on 2-site DNA, but this complex completely dissociates before UbaLAI-C can complete cleavage of the second DNA strand (Figure 7C).
The mechanism in Figure 7B is also in good agreement with accumulation of nicked products in the 1-site DNA cleavage reactions. In this case wt UbaLAI may form a synaptic complex in trans, but after cleavage of the first DNA strand and dissociation of the UbaLAI-C domain, UbaLAI is no longer physically linked to the nicked DNA molecule (Figure 7D).
A ‘sequential nicking via tethered activation’ mechanism similar to UbaLAI may also be employed by other monomeric two-domain Type II REases, consisting of one catalytic/DNA binding and one DNA binding domain, for example DpnI and Sau3AI. Structural studies of DpnI revealed that it consists of the N-terminal PD-(D/E)XK domain that binds one Dam methylated 5′-Gm6ATC-3′ site, an cuts one DNA strand after ‘A’, and the C-terminal non-catalytic winged helix (wH) domain that binds the second 5′-Gm6ATC-3′ site (23). A possible role of the DpnI wH in anchoring the catalytic domain to the substrate via the second Dam site was discussed by Siwek et al. (24).
Sau3AI (recognition sequence 5′-/GATC-3′) is a biochemically characterized Type IIE enzyme, which consists of two PD-(D/E)XK domains, both structurally related to MutH and BcnI/MvaI (25). According to sequence analysis, only the N-terminal Sau3AI domain contains an intact catalytic center, while the catalytically inactive C-terminal domain acts as an effector domain (26). Though the originally proposed Sau3AI reaction mechanism involved transient dimerization (25), recent structural studies of apo-Sau3AI performed by Xu et al. are consistent with the monomeric oligomeric state of Sau3AI (PDB IDs 2reu and 4pxg (27)). We therefore propose that Sau3AI may also employ an UbaLAI-like reaction mechanism, as depicted in Figure 7B.
A similar mechanism to UbaLAI was also described for the Type IIS enzyme BfiI, which contains a B3-like DNA recognition domain specific for the 5′-ACTGGG-3′ DNA sequence attached to a nonspecific nuclease that cuts DNA 5/4 nucleotides downstream from the recognition site (28,29). Despite being a homodimer, BfiI contains a single catalytic center at the interface of the nuclease domains, and uses it for sequential nicking of the two DNA strands. Throughout these reactions, which involve swiveling of the catalytic center between DNA strands of opposite polarity, the catalytic domains of BfiI remain tethered to the DNA via the DNA recognition domain (30).
Building blocks of UbaLAI
UbaLAI consists of two structural domains that are recurring in various DNA binding proteins and enzymes. The UbaLAI-N domain is structurally related to the B3 domains of plant transcription factors; similar domains are also widespread in various REases, including Type IIE enzymes (e.g. EcoRII (5)), Type IIS enzymes (e.g. BfiI (31)) and unusual ATP-driven enzymes (e.g. NgoAVII (32)). The second building block, the monomeric MutH/MvaI-like PD-(D/E)XK catalytic domain, is found in different structural contexts and performing different functions. For example, MutH is a nicking endonuclease involved in DNA repair that binds the hemimethylated GATC sequence in one orientation (33), while BcnI and MvaI are single-domain restriction endonucleases that bind their recognition sites in both orientations, and cleave both DNA strands at the target site in a reaction involving enzyme re-orientation via sliding and hopping (14). Apparently, the MvaI-like catalytic domain of UbaLAI has no such DNA sliding/hopping ability. It therefore acts as a fusion to the second DNA binding domain and employs a reaction mechanism that is unique among structurally and biochemically characterized REases specific for the 5′-CCWGG-3′ recognition site. In addition, the two domain structure and preference for multi-site substrates in the case of UbaLAI provides extra functionality—the ‘safety-catch’ that prevents host DNA cleavage if a single unmodified site becomes available. Such requirement for multiple recognition sites is widespread among many families of evolutionary unrelated Type II REases, implying the importance of this ‘safety catch’ mechanism in the function and spread of restriction-modification systems (17,34,35).
Taken together, the specific case of UbaLAI, and the more general case of restriction enzymes possessing various combinations of structural domains, demonstrate how recombination of different protein building blocks during evolution may yield a myriad of possible reaction mechanisms an functions.
ACCESSION NUMBERS
Coordinates and structure factors of UbaLAI-N complex with DNA are deposited under PDB ID 5o63. The DNA fragment encoding for the UbaLAI restriction-modification system was submitted to GenBank under accession number MF085529.
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge Kotryna Temčinaitė for the help with construction of the pBAD24_UbaLAI vector and optimization of UbaLAI expression conditions, and Dr Dmitrij Golovenko for the help with diffraction data collection. Authors also thank Dr Donald Comb (New England Biolabs) for the pACYC_M. PspGI plasmid, and Dr Gleb Bourenkov at EMBL/DESY P13 beamline at Petra III storage ring, Hamburg, Germany, for the invaluable help with the beamline operation.
Footnotes
Present address: Jelena Čalyševa, EMBL Heidelberg, Meyerhofstraße 1, 69117 Heidelberg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Council of Lithuania [MIP-81/2010 to G.S.]. Funding for open access charge: Research Council of Lithuania.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bashton M., Chothia C.. The generation of new protein functions by the combination of domains. Structure. 2007; 15:85–99. [DOI] [PubMed] [Google Scholar]
- 2. Roberts R.J., Vincze T., Posfai J., Macelis D.. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015; 43:D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gowers D.M., Bellamy S.R.W., Halford S.E.. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004; 32:3469–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szczepanowski R.H., Carpenter M.A., Czapinska H., Zaremba M., Tamulaitis G., Siksnys V., Bhagwat A.S., Bochtler M.. Central base pair flipping and discrimination by PspGI. Nucleic Acids Res. 2008; 36:6109–6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Golovenko D., Manakova E., Tamulaitiene G., Grazulis S., Siksnys V.. Structural mechanisms for the 5′-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII. Nucleic Acids Res. 2009; 37:6613–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou X.E., Wang Y., Reuter M., Mücke M., Krüger D.H., Meehan E.J., Chen L.. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 2004; 335:307–319. [DOI] [PubMed] [Google Scholar]
- 7. Tamulaitis G., Sasnauskas G., Mucke M., Siksnys V.. Simultaneous binding of three recognition sites is necessary for a concerted plasmid DNA cleavage by EcoRII restriction endonuclease. J. Mol. Biol. 2006; 358:406–419. [DOI] [PubMed] [Google Scholar]
- 8. Roberts R.J., Belfort M., Bestor T., Bhagwat A.S., Bickle T.A., Bitinaite J., Blumenthal R.M., Degtyarev S.K., Dryden D.T.F., Dybvig K. et al. . A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003; 31:1805–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bochtler M., Szczepanowski R.H., Tamulaitis G., Grazulis S., Czapinska H., Manakova E., Siksnys V.. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006; 25:2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaremba M., Owsicka A., Tamulaitis G., Sasnauskas G., Shlyakhtenko L.S., Lushnikov A.Y., Lyubchenko Y.L., Laurens N., Van Den Broek B., Wuite G.J.L. et al. . DNA synapsis through transient tetramerization triggers cleavage by Ecl18kI restriction enzyme. Nucleic Acids Res. 2010; 38: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sokolowska M., Kaus-Drobek M., Czapinska H., Tamulaitis G., Szczepanowski R.H., Urbanke C., Siksnys V., Bochtler M.. Monomeric restriction endonuclease BcnI in the Apo form and in an asymmetric complex with target DNA. J. Mol. Biol. 2007; 369:722–734. [DOI] [PubMed] [Google Scholar]
- 12. Kaus-Drobek M., Czapinska H., Sokolowska M., Tamulaitis G., Szczepanowski R.H., Urbanke C., Siksnys V., Bochtler M.. Restriction endonuclease MvaI is a monomer that recognizes its target sequence asymmetrically. Nucleic Acids Res. 2007; 35:2035–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kostiuk G., Sasnauskas G., Tamulaitiene G., Siksnys V.. Degenerate sequence recognition by the monomeric restriction enzyme: Single mutation converts BcnI into a strand-specific nicking endonuclease. Nucleic Acids Res. 2011; 39:3744–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sasnauskas G., Kostiuk G., Tamulaitis G., Siksnys V.. Target site cleavage by the monomeric restriction enzyme BcnI requires translocation to a random DNA sequence and a switch in enzyme orientation. Nucleic Acids Res. 2011; 39:8844–8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kostiuk G., Dikić J., Schwarz F.W., Sasnauskas G., Seidel R., Siksnys V.. The dynamics of the monomeric restriction endonuclease BcnI during its interaction with DNA. Nucleic Acids Res. 2017; 45:5968–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shlyakhtenko L.S., Gilmore J., Portillo A., Tamulaitis G., Siksnys V., Lyubchenko Y.L.. Direct visualization of the EcoRII−DNA triple synaptic complex by atomic force microscopy. Biochemistry. 2007; 46:11128–11136. [DOI] [PubMed] [Google Scholar]
- 17. Embleton M.L., Williams S.A., Watson M.A., Halford S.E.. Specificity from the synapsis of DNA elements by the Sfi I endonuclease. J. Mol. Biol. 1999; 289:785–797. [DOI] [PubMed] [Google Scholar]
- 18. Zaremba M., Sasnauskas G., Urbanke C., Siksnys V.. Conversion of the tetrameric restriction endonuclease Bse634I into a dimer: oligomeric structure-stability-function correlations. J. Mol. Biol. 2005; 348:459–478. [DOI] [PubMed] [Google Scholar]
- 19. Lagunavicius A., Sasnauskas G., Halford S.E., Siksnys V.. The metal-independent type IIs restriction enzyme BfiI is a dimer that binds two DNA sites but has only one catalytic centre. J. Mol. Biol. 2003; 326:1051–1064. [DOI] [PubMed] [Google Scholar]
- 20. Bilcock D.T., Daniels L.E., Bath A.J., Halford S.E.. Reactions of type II restriction endonucleases with 8-base pair recognition sites. J. Biol. Chem. 1999; 274:36379–36386. [DOI] [PubMed] [Google Scholar]
- 21. Fox N.K., Brenner S.E., Chandonia J.-M.. SCOPe: Structural Classification of Proteins–extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 2014; 42:D304–D309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mierzejewska K., Siwek W., Czapinska H., Kaus-Drobek M., Radlinska M., Skowronek K., Bujnicki J.M., Dadlez M., Bochtler M.. Structural basis of the methylation specificity of R.DpnI. Nucleic Acids Res. 2014; 42:8745–8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siwek W., Czapinska H., Bochtler M., Bujnicki J.M., Skowronek K.. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI. Nucleic Acids Res. 2012; 40:7563–7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedhoff P., Lurz R., Lüder G., Pingoud A.. Sau3AI, a monomeric type II restriction endonuclease that dimerizes on the DNA and thereby induces DNA loops. J. Biol. Chem. 2001; 276:23581–23588. [DOI] [PubMed] [Google Scholar]
- 26. Bujnicki J.M. A model of structure and action of Sau3AI restriction endonuclease that comprises two MutH-like endonuclease domains within a single polypeptide. Acta Microbiol. Pol. 2001; 50:219–231. [PubMed] [Google Scholar]
- 27. Xu C.-Y., Yu F., Xu S.-J., Ding Y., Sun L.-H., Tang L., Hu X.-J., Zhang Z.-H., He J.-H.. Crystal structure and function of C-terminal Sau3AI domain. Biochim. Biophys. Acta. 2009; 1794:118–123. [DOI] [PubMed] [Google Scholar]
- 28. Zaremba M., Urbanke C., Halford S.E., Siksnys V.. Generation of the BfiI restriction endonuclease from the fusion of a DNA recognition domain to a non-specific nuclease from the phospholipase D superfamily. J. Mol. Biol. 2004; 336:81–92. [DOI] [PubMed] [Google Scholar]
- 29. Sasnauskas G., Halford S.E., Siksnys V.. How the BfiI restriction enzyme uses one active site to cut two DNA strands. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:6410–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sasnauskas G., Zakrys L., Zaremba M., Cosstick R., Gaynor J.W., Halford S.E., Siksnys V.. A novel mechanism for the scission of double-stranded DNA: BfiI cuts both 3′-5′ and 5′-3′ strands by rotating a single active site. Nucleic Acids Res. 2010; 38:2399–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Golovenko D., Manakova E., Zakrys L., Zaremba M., Sasnauskas G., Gražulis S., Siksnys V.. Structural insight into the specificity of the B3 DNA-binding domains provided by the co-crystal structure of the C-terminal fragment of BfiI restriction enzyme. Nucleic Acids Res. 2014; 42:4113–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tamulaitiene G., Silanskas A., Grazulis S., Zaremba M., Siksnys V.. Crystal structure of the R-protein of the multisubunit ATP-dependent restriction endonuclease NgoAVII. Nucleic Acids Res. 2014; 42:14022–14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee J.Y., Chang J., Joseph N., Ghirlando R., Rao D.N., Yang W.. MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage. Mol. Cell. 2005; 20:155–166. [DOI] [PubMed] [Google Scholar]
- 34. Zaremba M., Sasnauskas G., Siksnys V.. The link between restriction endonuclease fidelity and oligomeric state: A study with Bse634I. FEBS Lett. 2012; 586:3324–3329. [DOI] [PubMed] [Google Scholar]
- 35. Gasiunas G., Sasnauskas G., Tamulaitis G., Urbanke C., Razaniene D., Siksnys V.. Tetrameric restriction enzymes: Expansion to the GIY-YIG nuclease family. Nucleic Acids Res. 2008; 36:938–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamulaitis G., Zaremba M., Szczepanowski R.H., Bochtler M., Siksnys V.. Nucleotide flipping by restriction enzymes analyzed by 2-aminopurine steady-state fluorescence. Nucleic Acids Res. 2007; 35:4792–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Webb B., Sali A.. Comparative protein structure modeling Using MODELLER. Current Protocols in Protein Science. 2016; 86, Hoboken: John Wiley & Sons, Inc; 2.9.1–2.9.37. [DOI] [PubMed] [Google Scholar]
- 38. Alva V., Nam S.-Z., Söding J., Lupas A.N.. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016; 44:W410–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.