Abstract
RNA has been found to play an ever-increasing role in a variety of biological processes. The function of most non-coding RNA molecules depends on their structure. Comparing and classifying macromolecular 3D structures is of crucial importance for structure-based function inference and it is used in the characterization of functional motifs and in structure prediction by comparative modeling. However, compared to the numerous methods for protein structure superposition, there are few tools dedicated to the superimposing of RNA 3D structures. Here, we present SupeRNAlign (v1.3.1), a new method for flexible superposition of RNA 3D structures, and SupeRNAlign-Coffee—a workflow that combines SupeRNAlign with T-Coffee for inferring structure-based sequence alignments. The methods have been benchmarked with eight other methods for RNA structural superposition and alignment. The benchmark included 151 structures from 32 RNA families (with a total of 1734 pairwise superpositions). The accuracy of superpositions was assessed by comparing structure-based sequence alignments to the reference alignments from the Rfam database. SupeRNAlign and SupeRNAlign-Coffee achieved significantly higher scores than most of the benchmarked methods: SupeRNAlign generated the most accurate sequence alignments among the structure superposition methods, and SupeRNAlign-Coffee performed best among the sequence alignment methods.
INTRODUCTION
Comparison is the most fundamental research technique in biology. Knowledge of similarity between biomolecules enables clustering them, grouping them into families, inferring their evolutionary history, detecting functional motifs and thus predicting the mechanism of their action (1). Comparison of protein structures has led to the development of commonly used databases and hierarchical structural classifications, such as SCOP (2) and CATH (3). To assess the similarities between two three-dimensional structures, they typically need to be superimposed in space—that is, one of them positioned over the other, so that the best fit is obtained. Superposition of homologous (evolutionarily related) structures reveals correspondence between evolutionarily conserved residues and motifs that may be indicative of common biological functions. Frequent tasks that benefit from a global superposition of macromolecular structures include homology modeling, structural classification, and function prediction. In the case of protein structures, a large number of algorithms have been proposed to address this task (4,5) with different levels of success (6). In general, the most difficult problem to solve in this context is the accurate superposition of structures that exhibit conformational changes and cannot be aligned without introducing flexibility (7,8).
The continuous growth of experimentally solved RNA tertiary structures has led to the emergence of computational methods for measuring the similarity between them. However, compared with protein structure superposition tools, there are still relatively few programs that allow for superimposing entire RNA molecules. This is because RNA and protein molecules are folded differently, and superposition methods developed for protein structures do not take into account key features of RNA structures. In particular, secondary structure in RNA depends on long-range interactions and base-pairs involve residues that are often very remote in sequence. Besides, RNA molecules have more modular 3D structure than proteins, and they present local 3D motifs (often composed of multiple fragments of the RNA chain), which behave as semi-independent rigid bodies that move with respect to each other during conformational changes. Frequently, the aim of superimposing RNA structures is the detection of common local structural features. In such cases, algorithms specifically intended for solving this problem are not useful for global superpositions, because even in closely related structures, minor shifts at the base of a domain may significantly alter the available conformations leading to a divergent result.
Superposition tools employ various methods and algorithms. They include two major types. Programs of one type reduce 3D structures to 1D sequence alignments, representing nucleotides by some local structural features. The resulting representation allows the utilization of existing sequence alignment algorithms. Methods of the second type operate on 3D coordinates. They find a local structural superposition and extend it to obtain a larger alignment. Importantly, programs for RNA structure superposition are typically based on rigid body structural alignment; therefore, they do not take into account hinges, internal rearrangements, or the potential flexibilities of secondary structure elements.
Among the methods of the first type (alignment approaches), PRIMOS (9) represents nucleotides as backbone torsion angles and may be considered the first RNA structural alignment tool. In like manner, DIAL encodes dihedral angles, nucleotide type and nucleotide base-pairing (10). SARA employs a unit-vector approach, where a vector-based simplified representation of selected atoms within nucleotides are used to find equivalent structural elements by means of dynamic programming. The similarity of the vectors is assessed with a unit-vector root mean square (URMS) approach (11). The required secondary structure information is provided by an external program, 3DNA (12). SARA-Coffee is the enhancement of SARA, combining its algorithm with R-Coffee (13) to produce multiple-sequence alignments using both sequence and structural data (14). LaJolla uses an n-gram model to generate hash tables for analyzing similar derived sequence (15). A similar discretized structural alphabet underlies an algorithm used in iPARTS (16). It is worth mentioning that programs of this category do not optimize the global superposition in terms of distances between corresponding atoms; i.e., to minimize root-mean-square deviation (RMSD).
Among the methods of the second type (structural superposition), ARTS aligns two structures based on structurally similar tuples of phosphate atoms of consecutive base pairs (called quadrats) (17). It also requires the secondary structure to be inferred by 3DNA from coordinates of the structures to be superimposed. R3D Align finds the maximum clique of the local alignment graph, which leads to finding the optimal alignment of the structures. The local alignment graph is based on distinguishing 4-nt neighborhood clusters that are similar in both structures (18). Another tool has been developed by the authors of R3D Align that employs R3D Align to detect local conformational differences between homologous RNA molecules (19). The algorithm used in SETTER divides the structures into generalized secondary structure units (GSSU) and calculates the optimal transformations between all of them (20). A novel approach is to apply elastic shape analysis (ESA), which treats the RNA backbones as three-dimensional curves (21). This method was used in RASS, which compares two structures by calculating the distance between their representations in an infinite-dimensional topological space of curves (22). Rclick is based on the CLICK algorithm (23), adapted to processing RNA structures. It matches cliques of points (representative atoms of the residues), finding structurally similar residues, and performs a 3D least-squares fit using these equivalences (24).
In this study, we address the problem of the accuracy of the tools for global superposition of RNA structures. We developed a benchmark for assessing the performance of the structural superpositions and the resulting sequence alignments by way of various measures. We evaluated the performance of ten available programs, based on experimentally determined structures of homologous RNA molecules. In addition to structural measures (RMSD and derivatives), we used sequence-based scores (e.g., SPS) as measures of the accuracy of structural alignments. These measures assess the similarity of the superposition-derived sequence alignments returned by the benchmarked programs and reference alignments generated from manually-curated covariance models from the Rfam database. Moreover, we look into the structural characteristics that account for the differences in accuracy and identify the features that may be important for the interpretation of a superposition obtained with the benchmarked tools.
MATERIALS AND METHODS
A new method of superposition
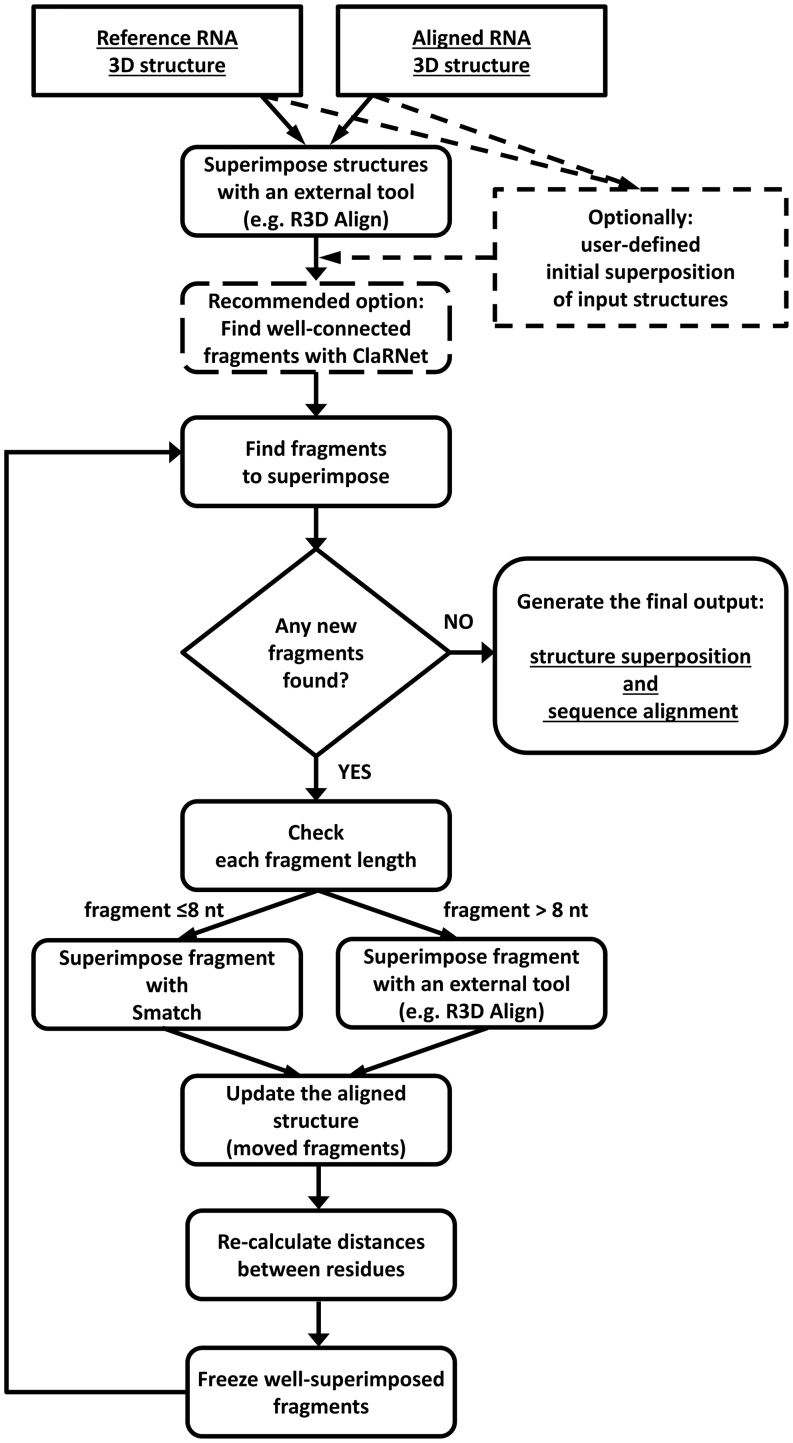
We developed a new method, called SupeRNAlign, for flexible pairwise superposition of RNA 3D structures. SupeRNAlign iteratively superimposes the RNA structures and splits them into fragments to maximize the local fit. In this work, we define a fragment of RNA 3D structure as a set of at least four ribonucleotide residues, physically connected with each other (covalently or non-covalently), which is used as an independent unit of structural superposition. A detailed illustration of SupeRNAlign workflow is presented in Figure 1.
Figure 1.
SupeRNAlign workflow. Optional steps are indicated with dashed lines.
As an input, SupeRNAlign takes two RNA structures in the PDB format (further called the ‘reference structure’ and ‘aligned structure’, respectively). The first superposition is performed on the entire structure. SupeRNAlign may split the aligned structure into fragments, using a special procedure implemented in a software tool ClaRNet developed in our laboratory (see the following section). Alternatively, it can omit ClaRNet and use only the general procedure described below, which is also used in all subsequent iterations. According to our preliminary tests, SupeRNAlign used with ClaRNet performed slightly better (Supplementary Table S5) and ultimately the benchmark presented in this work provides results obtained for the variant, in which the first division of the aligned structure into fragments is performed with ClaRNet, followed by the general procedure (based on detection of close fits) in further iterations. For difficult cases of superposition, we recommend testing the use of SupeRNAlign with and without ClaRNet and inspecting the outcome with respect to the biological relevance of the structural correspondence as well as sequence alignment. However, expert inspection of output is beyond the scope of this work, which focuses on automated superposition and alignment.
Rigid body superpositions are carried out by different tools, depending on the size of fragments analyzed. The original structures, as well as their fragments longer than eight residues, are superimposed by an external tool: thus far we developed interfaces for ARTS, SETTER, LaJolla, R3D Align and SARA. Based on tests described in this work we have ultimately selected R3D Align, because it proved to score best among the tools for rigid-body superposition tested in our benchmark. However, we included results for all these aligners coupled with SupeRNAlign in the Supplementary Materials (Supplementary Figure S8). Superposition of fragments shorter than nine residues (continuous or non-continuous) is done by a specialized tool Smatch developed in our laboratory. A detailed description of this tool is presented in our earlier work (25), Supplementary Materials (Section 4). Briefly, Smatch iteratively inspects all possible rigid body transformations to structures to find the one that results in a minimal RMSD value calculated for P, C1' and C4' atoms. Since this task is computationally very expensive, the search is optimized using two improvements: guiding the search using alignments of 3-atom pivots and usage of k-dimensional trees data structure to obtain efficient geometric queries.
Based on the rigid-body superposition(s) obtained in a given iteration, which takes into account the whole aligned structure or all its substructures, a sequence alignment (relation between residues from the two structures) is inferred by pdb3aln—a tool described in the Scoring section, together with a sequence of distances (in Å) between the aligned residues. Then, the distances are smoothed by a moving average (window of five residues). Segments that contain at least four residues superimposed below a given threshold (median distance for the superposition or 5.5 Å, whichever is lower), are ‘frozen’ and—since they are already well-aligned—excluded from further superpositions. The remaining part of the aligned structure is split into ‘free’ fragments, separated in sequence by the ‘frozen’ regions. To preserve the secondary structure, ‘free’ fragments (i.e., those that remain to be superimposed) that share Watson–Crick-paired residues are combined with each other. The sequence alignment inferred from the superposition is used to determine the correspondence between fragments in the aligned structure and the reference structure.
Each of the fragments that remain ‘free’ after an iteration of structure superposition, splitting and freezing, are selected for processing in a new iteration, namely each fragment is superimposed on its counterpart from the reference structure, and potentially subjected to another round of splitting and freezing. Successfully superimposed fragments (or sub-fragments) of the aligned structure are frozen and merged with fragments frozen in the earlier iteration(s). When all the fragments have been processed, the global alignment and the list of pairwise distances for all residues are updated. After each iteration, the exit condition is tested: if all residues have been frozen or the arrangement of fragments has not changed since the last iteration, the processing ends and structural superpositions and sequence alignments are returned. A user can obtain the final solution as well as all intermediate superpositions and corresponding alignments—though this is not a default option.
To further improve the quality of sequence alignments yielded by SupeRNAlign, we enabled its coupling with T-Coffee (26), which uses structural alignment data produced by our program to generate a T-Coffee library file—similarly to SARA-Coffee (13). The advantage of this approach is that the two input structures are flexibly fitted to each other, which improves the likelihood of identifying the correct correspondence between homologous residues. Using the T-Coffee requires generating a biologically reasonable sequence alignment, without relying solely on the geometry of superposition, hence the combination of SupeRNAlign with T-Coffee is expected to generate better sequence alignments than SupeRNAlign alone. We benchmarked this method, called SupeRNAlign-Coffee or shortly SA-Coffee, using T-Coffee with the R-Coffee mode.
Improved fragmentation of the input structure with ClaRNet
Optionally, just in the first iteration, SupeRNAlign may use a dedicated procedure for splitting the aligned structure into fragments, which utilizes ClaRNet. The purpose of this tool is to divide the 3D RNA structure into substructures that are highly interconnected and relatively well packed. Such structures are likely to be rather rigid even if the entire structure undergoes conformational rearrangements, such as movements of independently folded domains with respect to each other. ClaRNet takes as an input a PDB file and processes it using ModeRNA (27) to standardize the names and numbering of residues; in particular modified residues are substituted with their unmodified counterparts. Subsequently, it uses ClaRNA (28) to identify and classify pairwise contacts between residues comprising the structure. The interactions recognized by ClaRNA include canonical and non-canonical base pairing, stacking, base-phosphate and base-ribose interactions. The detected contacts are then evaluated and those with a ClaRNA score above a threshold of 0.6 are considered further. ClaRNA also takes into account all covalent bonds between residues adjacent in sequence. The RNA molecule at this point can be represented as a graph, with residues as nodes and interactions as edges (Supplementary Figure S1). Since the purpose of the program is to break selected covalent bonds between regions that are well connected, due to canonical base pairs and other interactions, the procedure assigns variable weights to different types of edges: 0.5 for covalent bonds, 2 for canonical base pairs and 1 for all other interactions. The key step in the procedure is the identification of the most densely connected clusters of residues using the Markov Cluster Algorithm (29) with the inflation parameter set to 1.3, which has been established by a trial and error method. The final result is the division of the input structure into substructures; in the context of this work, these substructures are passed to SupeRNAlign for further processing, and can be subdivided further, based on the algorithm described earlier. A full list of ClaRNet parameters is provided in Supplementary Material (Supplementary Table S6).
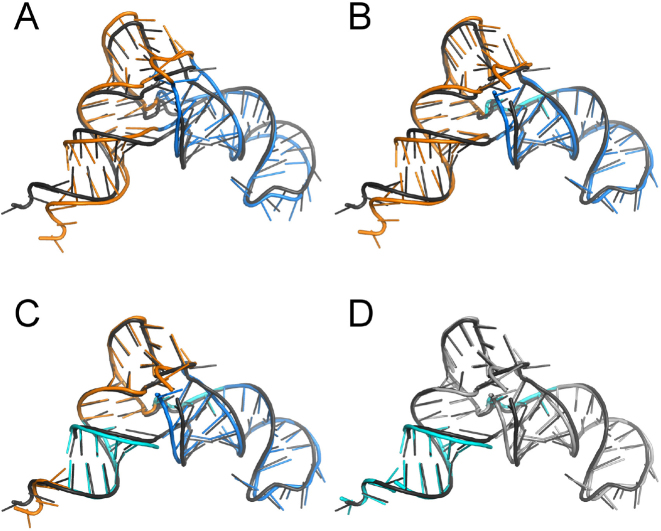
An example of superposition of two tRNA structures by SupeRNAlign is presented in Figure 2. A more complex example—a pair of 16S rRNA molecules (PDB codes: 1FJG and 2AW7) – is presented in Supplementary Figure S5 (graph for 2AW7) and Supplementary Figure S6 (SupeRNAlign workflow). Initially, a global superposition of the reference structure and aligned structure is performed. At this stage, the aligned structure may optionally be divided by ClaRNet into substructures (indicated by different colors in Figure 2, panel A). Each of these fragments is superimposed on its corresponding segment in the reference structure, with the correspondence defined by the initial alignment resulting from the first superposition. The series of distances between corresponding residues (according to the current alignment) in the reference and aligned structures is smoothed to avoid excessive fragmentation of the structure. Each of the distances is then compared to the threshold value and the given pair of residues is classified as well-superimposed or poorly-superimposed. Well-superimposed regions (indicated in cyan in Figure 2) are ‘frozen’ and excluded from further processing. The remaining ‘free’ fragments are then subject to separate superpositions and the result is incorporated into a global superposition. This procedure is repeated iteratively until either all fragments are considered ‘well-superimposed’ or no improvement is observed.
Figure 2.
Graphical illustration of the SupeRNAlign workflow, using as an example a pair of two tRNA(Asn) molecules (PDB code: 3KFU, reference structure shown in dark grey; and PDB code: 4WJ4, aligned structure shown in other colors). (A) First round: result of superposition of two RNA structures treated as rigid bodies; the aligned structure is then analyzed by ClaRNet and two substructures identified are colored blue and orange. (B) Second round: result of independent superposition of two fragments of the aligned structure identified by ClaRNet onto the corresponding fragments of the reference structure; a fragment identified as ‘well superimposed’ and frozen for further iterations is indicated in cyan, while the remaining fragments will continue being subjected to superposition. (C) Third round: result of superposition of fragments that remained ‘free’ after the previous iteration, an additional region in the CCA stem is found to be ‘well superimposed’ and is colored in cyan, regions that remain above the threshold of ‘good superposition’ remains shown in blue and orange colors. (D) The final superposition, in which the single-stranded CCA terminus (in the bottom left corner) is superimposed well and colored in cyan, while the superposition of other ‘free’ fragments (now shown in gray) does not improve according to SupeRNAlign; this superposition is used to generate the final sequence alignment.
Based on testing, ClaRNet yields statistically significant improvement in alignments (Wilcoxon test results can be found in Supplementary Table S5). As stated above, it contributes to the alignment quality by detecting compact elements that may constitute potential structurally and evolutionarily conserved domains. However, the deterioration of mutual position of structural segments can be sometimes observed for molecules, which exhibit different secondary structures or in which the chain is interrupted. Therefore, we advise to run the program in both modes, with and without ClaRNet, analyze the superpositions visually and evaluate the biological relevance of results obtained.
Benchmarked programs
From the available programs for RNA structure comparison, we selected those that yield a superposition (either PDB file(s) or rotation/translation data) and/or a sequence alignment given two PDB files with single-chain RNA structures. Therefore, DIAL could not be tested because it did not meet these requirements (the Web server was available online, but in our hands it has not returned any results). Likewise, PRIMOS, which was mentioned in the Introduction, is available neither as a downloadable program nor as a Web server. Although RASS was available at the time of writing this manuscript, in our hands it was unable to return results for any of the pairs from the benchmarking set. We have informed the authors of RASS and DIAL about the inability to obtain results of our superpositions. According to the authors’ responses, these problems could not be solved in reasonable time, and hence both programs were excluded from our comparison. Table 1 presents the tools included in the benchmark with a list of algorithms employed by each of them. Eight of them are available as standalone applications, running on Linux and usually also on MS Windows (especially those written in multi-platform languages, like Python or Java). Two tools (iPARTS and Rclick) are available only as Web servers; in these cases, we wrote scripts for parsing results directly from the respective websites of the servers. R3D Align, SARA-Coffee, and SupeRNAlign are available both as standalone tools and as Web servers.
Table 1. Programs analyzed in this work.
| Name | Ref. | URL (http://) | Application type | Language | SSU rRNA5 | LSU rRNA6 | Output data | Algorithm |
|---|---|---|---|---|---|---|---|---|
| ARTS | (17) | bioinfo3d.cs.tau.ac.il/ARTS/index.html | Standalone & server | Compiled | + | + | Two pdb files2 | Structurally similar tuples of P atoms |
| SARA | (11) | structure.uib.es/sara/ | Standalone & server | Python | – | – | Single pdb file3 | Unit-vector structural representation |
| LaJolla | (15) | lajolla.sourceforge.net | Standalone | Java | – | – | List of pdb files4 | n-gram query-target matching |
| R3D Align | (18) | rna.bgsu.edu/r3dalign/ | Standalone & server | MATLAB1 | + | + | Single pdb file3 | ‘Maximum-clique’ |
| SETTER | (20) | setter.projekty.ms.mff.cuni.cz | Standalone & server | Compiled | + | + | Rotation/ translation data | Generalized secondary structure units |
| iPARTS | (16) | genome.cs.nthu.edu.tw/iPARTS/ | Server | N/A | – | – | Single .pdb file3 | Discretized structural alphabet |
| SARA-Coffee | (14) | tcoffee.crg.cat/apps/tcoffee/do:saracoffee | Standalone & server | Compiled | – | – | Sequence alignment | SARA + R-Coffee |
| Rclick | (24) | mspc.bii.a-star.edu.sg/minhn/rclick.html | Server | N/A | + | – | Two .pdb files2 | Clique matching, based on CLICK |
| Supe RNAlign | this work | genesilico.pl/supernalign/ | Standalone & server | Python | + | + | Single .pdb file3 | Iterative superpositions of structural fragments |
| Supe RNAlign-Coffee | this work | genesilico.pl/supernalign/ | Standalone | Python | + | + | Sequence alignment | SupeRNAlign + R-Coffee |
1Can be executed under GNU Octave.
2Each output file contains one structure.
3The output file contains both superimposed structures.
4Each output file contains one structure; multiple superposition models are produced.
5Program did (‘+’) or did not align (‘–’) SSU rRNA structures in the specified time (12 h).
6Program did (‘+’) or did not align (‘–’) LSU rRNA structures in the specified time (12 h).
Benchmarking set
We created groups of homologous and non-redundant RNA structures, using two datasets: Rfam (30) and RNA 3D Hub (31). Two RNA structures were considered homologous if and only if their sequences belong to the same Rfam family, and non-redundant if they are not members of one class at the given resolution cut-off in RNA 3D Hub.
First, we downloaded a full set of RNA 3D structure classes defined in RNA 3D Hub version 1.89 at a resolution cut-off of 4.0 Å. We used a dataset with reduced redundancy to retain only biologically relevant variation among the RNAs analyzed and to ignore variation due to different experimental conditions. Then, for each PDB code representative of a class, we downloaded the corresponding PDB file from the PDB (32). From a total of 876 classes, we selected only those which could be used for the purpose of this benchmark; i.e., were not a sole structurally characterized member of an RNA family. All RNA sequences (after replacing modified residues with their unmodified counterparts with ModeRNA) within each PDB file were extracted using ModeRNA libraries (27). Identical sequences within one PDB file were removed, and for each RNA sequence, a Rfam family was matched using cmscan from the Infernal 1.0.2 package (33) on Rfam covariance models. In the case of multiple matches, the family with a longer matching region or broader definition was assigned. Multiple sequences within one Rfam family were aligned with Infernal's cmalign and the resulting alignments were considered as true references. In the case of 16S rRNA, we manually searched for all non-redundant structures at http://www.rcsb.org and aligned their sequences using SSU-ALIGN (http://eddylab.org/software/ssu-align/). In the case of the large ribosomal subunit RNA (LSU_rRNA), we used the alignment from the Comparative RNA Web database (34), to which we aligned sequences corresponding to compared rRNA structures with MAFFT (35) (option ‘–add’).
Programming tools
The scripts used in this benchmark were written in Python (data collection) and R (statistical analysis and visualization). A 64-bit virtual machine with Ubuntu 14.04 (GNU/Linux 4.4.0-62) onboard and Python 2.7.6 and R 3.2.3 installed was employed to run the benchmark. For the processing of PDB files, Biopython 1.65 (36) and ModeRNA 1.7.1 Python libraries were used.
Scoring
To assess the accuracy of superpositions performed by the benchmarked programs, we used three independent measures of similarity.
Alignment-based root-mean-square deviation (RMSD) measures the mean structural deviation based on the reference alignment, and therefore is independent of the sequence alignment inferred from the superposition. It is a root mean square deviation of distances between residues aligned in the reference alignments. We chose the phosphorus (P) atoms as a reference point for each nucleotide residue. Though RMSD is often used, simple changes in a short stem-loop (due to flexibility) can lead to large RMSD values, because this measure is dominated by the amplitude of the deviation from the reference. On the other hand, errors in the backbone can appear negligible because the amplitude of the misorientation is small (37). Therefore, other measures of RNA similarity should also be considered.
Sum-of-pairs score (SPS) is dependent on the sequence alignments generated from 3D superpositions; i.e., it measures the quality of sequence alignments obtained from 3D superpositions, as compared to the reference ones (38). SPS was used in previous benchmarks (39,40) and yields a normalized similarity between two alignments (from 0 for completely distinct alignments to 1 for identical alignments), iterating over columns and scoring each column that is identical in both alignments. SPS counts the correctly aligned residue pairs and is commonly used in assessing the performance of multiple sequence alignment algorithms. Here, as the references, we used sequence alignments based on covariance models for RNA families deposited in the Rfam database (30). Given a pair of the test and reference sequence alignment, we calculated sum-of-pairs score (SPS) (41).
Another score, 3SP (Secondary Structure Sum of Pairs) (42), is similar to SPS in that it calculates the number of pairs matching the reference alignment, but also secondary structure of the aligned RNAs is taken into account: the alignment is penalized for each Watson-Crick pair in the second structure not matching a pair in the first structure (i.e., paired residues not aligned together). In this case, X3DNA (43) was used to determine secondary structures.
Sequence alignments were inferred from superimposed structures using the pdb3aln software tool developed in our laboratory (available as a part of the SupeRNAlign software package), based on a modified Smith-Waterman algorithm. In pdb3aln, the penalty is calculated as the reciprocal of the distance between each pair of residues from both RNA structures. R3D Align returns not only a global structural superposition, but also a sequence alignment generated from local superpositions. These sequence alignments were included in the comparison and the results obtained were indicated as R3D Align* (with an asterisk).
Analysis
Within each RNA family, pairwise superpositions were made for all structures. For each superposition, we inferred a pairwise sequence alignment and calculated its deviation from the reference alignment. To reduce the overrepresentation of superpositions of structures from large families and to avoid the risk that a program would achieve a high overall score, making accurate superpositions just for one large family, median scores for the families were calculated for each program analyzed. For these median scores, exploratory analysis of distributions (calculation of variances and quartiles) was performed. Wilcoxon test was used to compare overall scores for all programs in our benchmark and to establish whether the differences between their performance were statistically significant.
RESULTS
To assess the performance of the ten programs listed in Table 1, we tested them on a non-redundant set of pairs of 3D structures of homologous RNA molecules. With 151 structures from 32 families, 1734 pairwise superpositions were examined (Supplementary Table S1). Due to the technical limitations, some programs failed to align some of the structures (for instance, only SETTER, ARTS, R3D Align, SupeRNAlign and SupeRNAlign-Coffee were able to process LSU rRNAs).
Although structural superposition is commonly used in comparing evolutionarily related RNA molecules, there is no gold standard of assessment of the superposition algorithms. For this reason, in addition to the commonly used root-mean-square deviation (RMSD; calculated for P atoms), we also used the Sum-of-Pairs Score (SPS) to measure the ability to reconstruct the reference sequence alignments of the homologous molecules (39,40). A high SPS and a low alignment-based RMSD correspond to accurate superpositions. In the test set, they negatively correlate with each other across all calculations (Spearman's rank correlation coefficient, ρ, of -0.83, P-value < 0.01).
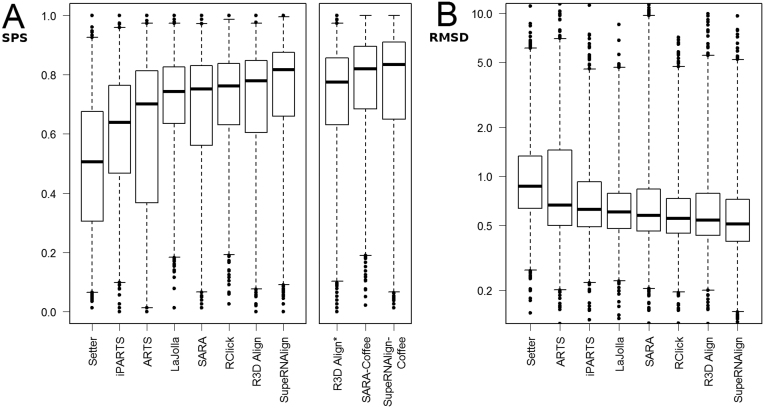
In terms of SPS measure in both structure and sequence-based methods, our programs scored best with SPS 0.82 and 0.84, respectively for SupeRNAlign and SupeRNAlign-Coffee. Similarly, as measured by 3SP, SupeRNAlign and SupeRNAlign-Coffee scored 0.81 and 0.84, respectively (see Supplementary Table S1 and Figure 3). For structure-based methods, reference alignment-based RMSD was used to evaluate the quality of superposition. According to the entire benchmarked structures set, SupeRNAlign and R3D Align scored best with median RMSD 4.23 and 4.51.
Figure 3.
A comparison of the accuracy of benchmarked methods. These boxplots show the distribution of scores (A, sum-of-pairs; B, RMSD (in Å, shown in logarithmic scale) obtained by the RNA superposition methods. Boxes mark quartiles (Q1, median, Q3); whiskers stretch from 1st to 99th percentile; outliers are shown as dots.
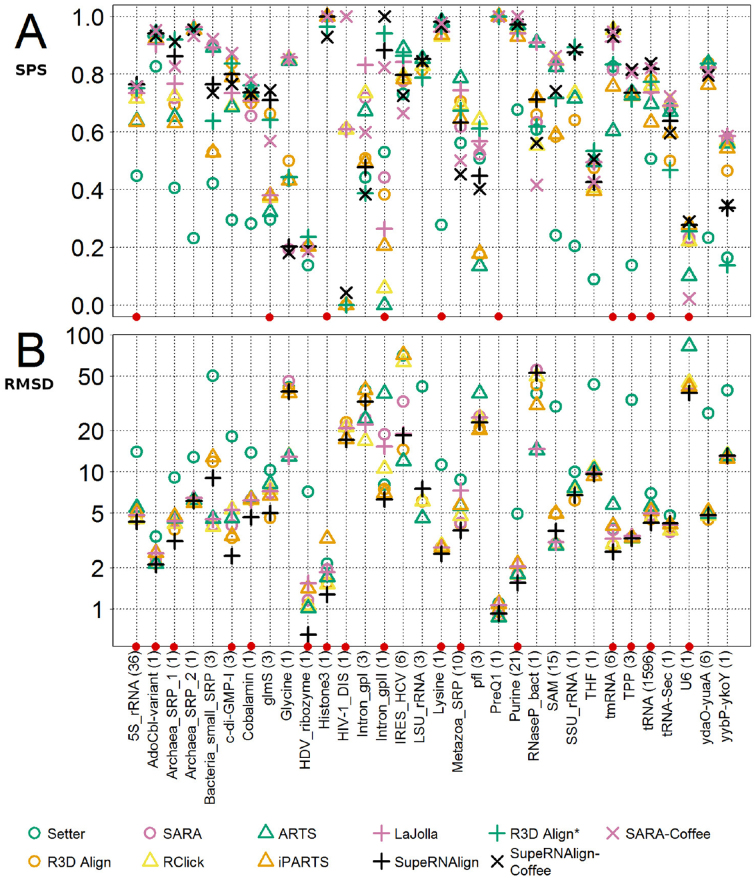
To avoid introducing bias resulting from the differences between the sample sizes of RNA family groups (in particular for the very large group of tRNA structures), for each program, apart from one median score, we calculated medians of scores obtained for pairs belonging to one RNA family. Median scores for the benchmarked programs within each RNA familiy are shown in Figure 4 and Supplementary Table S1 (according to SPS score) and Supplementary Table S2 (according to 3SP score). Among the structure-based methods, the alignments returned by SupeRNAlign and R3D Align yielded the best score according to the SPS metrics with median SPS 0.76 and 0.70, respectively. Among sequence-based methods, SARA-Coffee and SupeRNAlign-Coffee scored best with SPS 0.79 and 0.74 respectively. According to the median RMSD calculated for benchmarked RNA families, SupeRNAlign performed better than other structure-based methods with RMSD 4.76.
Figure 4.
A comparison of the accuracy of benchmarked methods within RNA families. The plots show scores (A, sum-of-pairs; B, RMSD (in Å, logarithmic scale) obtained by the benchmarked programs for each RNA family. Each symbol represents the median value of score for the particular family—different programs are marked with colors and symbols. SupeRNAlign and SupeRNAlign-Coffee are denoted in black. The families where either SupeRNAlign or SupeRNAlign-Coffee performed best are marked with red dots. The families are sorted alphabetically, and this sorting order is consistent with the order in the tables to facilitate comparison of results.
Some differences in the programs’ performance may also be noted for particular RNA families. While most of the algorithms scored well (with an SPS above 0.9) for superpositions within AdoCbl variant RNA (RF01689), histone 3′ UTR stem–loop (RF00032), lysine riboswitches (RF00168), and purine riboswitches (RF00167), they failed to produce accurate superpositions (aligning accurately at least 50% of the residues) for the hepatitis delta virus ribozyme (RF00094) U6 spliceosomal RNA (RF00026) and glycine riboswitch (RF00504). One reason for those discrepancies might be the difference in structure sizes. For instance, the difference in structure sizes for the lysine riboswitch (RF00168) is 7%, whilst in case of the glycine riboswitch (RF00504) structures obtained from the PDB database, the size differences are up to 94% (one of the structures—3OWW—contains only the glycine-sensing domain). Another possible reason is the identity of the compared sequences. In the families where the mean sequence identity is low, there is a visible drop in the performance of benchmarked programs; e.g. in the representatives of cyclic di-GMP-I riboswitch family (RF01051) the mean sequence identity is 65%, and in hepatitis delta virus ribozyme (RF00094) the mean identity is 29%. The median scores for each family are shown in Figure 4.
As can be deduced from the average scores for different RNA families, the superposition accuracies of the methods correlate with each other over the sets of structures (Supplementary Figure S4). Spearman's correlation coefficients for the pairwise comparisons among the programs are highest between R3D Align and SupeRNAlign (ρ = 0.91) and Rclick and LaJolla (ρ = 0.92). One possible reason is that these programs use rather similar algorithms. For R3D Align and SupeRNAlign the relationship is obvious, since SupeRNAlign is based on superpositions generated with R3D Align. However, Rclick uses different underlying principles than the LaJolla algorithm.
To statistically evaluate the prediction accuracy of a method compared to another, scores obtained with each method were compared against each other using the paired Wilcoxon test. To compensate for errors resulting from multiple comparisons, FDR correction was used (44). For those comparisons with p-values larger than 0.01, the benchmark fails to demonstrate that there is a significant difference in superposition accuracies between the two methods with a low Type I error. Results of the test show that SupeRNAlign-Coffee scores significantly better than all programs except SARA-Coffee in terms of SPS (Supplementary Table S3). For RMSD, SupeRNAlign scores significantly better than other programs (Supplementary Table S4).
We measured the computation times for one representative structure from each of the analyzed families and compared the performance of benchmarked programs (Supplementary Figure S7). Since SupeRNAlign requires multiple superpositions, it is obvious that its running time is longer compared to other programs.
DISCUSSION
We have developed a structural superposition tool for RNA molecules, SupeRNAlign, which combines global superposition with the detection of fragments that can be superimposed locally. As a measure of accuracy of the resulting superpositions, the SPS measure was used, calculated for the sequence alignments generated from the superimposed structures, with respective alignments from Rfam as references. The principle underlying this approach was that a good superposition should yield a sequence alignment that agrees well with the curated alignment that takes into account conservation in the whole RNA family. Additionally, RMSD was used to give insight into the accuracy of superpositions generated by different programs.
In general, most of the benchmarked programs scored equally well in certain categories (no statistically significant differences in the overall score); however, there were clear differences in their performance within several RNA families. From the detailed results (Figure 4 and Supplementary Table S1), one can conclude that, for example, SupeRNAlign-Coffee scored much better than other tools for group II intron domain 5 (RF00029), with a median SPS score of 1.0, while SupeRNAlign outperformed other programs in superimposing transfer-messenger RNAs (RF00023) with a median SPS score of 0.95. Moreover, some programs could not cope with some of the structures, either failing to do the superposition (because of the large chain length) or yielding unsatisfactory results. Some factors—like running speed—were not taken into account in this benchmark, but for some users they may play an important role, for instance when superimposing very large structures. To superimpose two or more RNA structures with the best results, it would be beneficial to compare the results of several programs to avoid local errors, and—if necessary—to re-align the structures with respect to conserved regions. Such amendments can be currently done only by human experts, which makes such an exercise beyond the scope of our analysis. Nonetheless, our tools can be of course used as part of a manually-amended superposition.
Benchmarks of SupeRNAlign against other methods showed, on average, that the most accurate results are achieved when the algorithm can detect small conformational changes between the structures. Typically, hinge points in multibranch loops are regions where small conformational changes can yield drastic differences in RMSD, even though the overall topology is essentially unchanged. Permitting a greater flexibility in conformations within the superposition algorithm allows some level of better melding of the reference structure with the target structure. As these benchmarks have shown, the strength of our approach is evident, especially when aligning similar structures with minor structural discrepancies, which cannot be optimally aligned as rigid bodies; like the comparison of domain 5 structures from two group II introns: 1R2P_A and 2F88_A, where SupeRNAlign scored higher than other methods available.
SupeRNAlign can be very useful in applications that demand consideration of comparative structure analysis and sequence alignment, and which have to deal with conformational rearrangements in RNA structures. The potential uses of SupeRNAlign include evolutionary analyses (e.g.,detection of conserved substructures between remotely related RNA structures that combine well-alignable and poorly-alignable fragments), preparation of structures to be used as templates in multiple-template comparative modeling, and clustering of structure predictions obtained with different modeling methods or for different homologous sequences. In particular, as demonstrated in this work, flexible superpositions obtained with SupeRNAlign often have lower RMSDs than the best superpositions that can be obtained by rigid body fitting. As a result, a higher fraction of homologous residues is superimposed well and can be used for identifying a structurally conserved core, which can be confidently modeled by a template-based (comparative) modeling method. On the other hand, structural elements that cannot be superimposed even at the level of fragments indicate evolutionary variation and suggest that an expert modeler should either select just one structural template for modeling of a given region or consider folding the given region in a template-free mode. In the long-run, the practical utility of SupeRNAlign can be tested, and possibly new uses can be invented in the framework of community-wide experiments such as RNA Puzzles (45,46).
AVAILABILITY
SupeRNAlign is written in Python and is available for download and installation as a standalone tool (from https://bitbucket.org/cosi1/supernalign) or can be accessed through the Web server at http://genesilico.pl/supernalign/. This website is free and open to all users and there is no login requirement.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the authors of the benchmarked programs for their help: David Hoksza (SETTER), Emidio Capriotti (SARA), Anton Petrov and Ryan Rahrig (R3D Align), Kristian Rother (LaJolla), and Chin Lung Lu (iPARTS). We also thank Astha, Catarina Almeida, Grzegorz Chojnowski, Stanisław Dunin-Horkawicz and Marcin Magnus for critical reading and comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Polish National Science Centre [2012/04/A/NZ2/00455 to J.M.B., 2014/15/B/ST6/05082, 2013/09/B/NZ2/00121 to W.D.]; Foundation for Polish Science (FNP) [TEAM/2009-4/2 to J.M.B. and ‘Ideas for Poland’ fellowships to J.M.B.]; European Research Council (ERC) [261351 to J.M.B]. Computing power was provided by IIMCB, funded by EU structural funds [POIG.02.03.00–00-003/09 to J.M.B.]. Funding for open access charge: Polish National Science Centre [2012/04/A/NZ2/00455 to J.M.B.].
Conflict of interest statement. Janusz M. Bujnicki is an Executive Editor of Nucleic Acids Research.
REFERENCES
- 1. Atkinson H.J., Morris J.H., Ferrin T.E., Babbitt P.C.. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009; 4:e4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murzin A.G., Brenner S.E., Hubbard T., Chothia C.. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995; 247:536–540. [DOI] [PubMed] [Google Scholar]
- 3. Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M.. CATH–a hierarchic classification of protein domain structures. Structure. 1997; 5:1093–1108. [DOI] [PubMed] [Google Scholar]
- 4. Holm L., Sander C.. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995; 20:478–480. [DOI] [PubMed] [Google Scholar]
- 5. Zhang Y., Skolnick J.. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005; 33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wohlers I., Malod-Dognin N., Andonov R., Klau G.W.. CSA: comprehensive comparison of pairwise protein structure alignments. Nucleic Acids Res. 2012; 40:W303–W309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Friederich M.W., Vacano E., Hagerman P.J.. Global flexibility of tertiary structure in RNA: yeast tRNAPhe as a model system. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagerman P.J. Flexibility of RNA. Annu. Rev. Biophys. Biomol. Struct. 1997; 26:139–156. [DOI] [PubMed] [Google Scholar]
- 9. Duarte C.M., Wadley L.M., Pyle A.M.. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003; 31:4755–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferre F., Ponty Y., Lorenz W.A., Clote P.. DIAL: a web server for the pairwise alignment of two RNA three-dimensional structures using nucleotide, dihedral angle and base-pairing similarities. Nucleic Acids Res. 2007; 35:W659–W668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Capriotti E., Marti-Renom M.A.. RNA structure alignment by a unit-vector approach. Bioinformatics. 2008; 24:i112–i118. [DOI] [PubMed] [Google Scholar]
- 12. Lu X.J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilm A., Higgins D.G., Notredame C.. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008; 36:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Di Tommaso P., Bussotti G., Kemena C., Capriotti E., Chatzou M., Prieto P., Notredame C.. SARA-Coffee web server, a tool for the computation of RNA sequence and structure multiple alignments. Nucleic Acids Res. 2014; 42:W356–W360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bauer R.A., Rother K., Moor P., Reinert K., Steinke T., Bujnicki J.M., Preissner R.. Fast structural alignment of biomolecules using a hash table, N-grams and string descriptors. Algorithms. 2009; 2:692–709. [Google Scholar]
- 16. Wang C.W., Chen K.T., Lu C.L.. iPARTS: an improved tool of pairwise alignment of RNA tertiary structures. Nucleic Acids Res. 2010; 38:W340–W347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dror O., Nussinov R., Wolfson H.. ARTS: alignment of RNA tertiary structures. Bioinformatics. 2005; 21(Suppl. 2):ii47–ii53. [DOI] [PubMed] [Google Scholar]
- 18. Rahrig R.R., Leontis N.B., Zirbel C.L.. R3D Align: global pairwise alignment of RNA 3D structures using local superpositions. Bioinformatics. 2010; 26:2689–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rahrig R.R., Zirbel C.L.. Detecting conformational differences between RNA 3D structures. Jp. J. Biostat. 2015; 12:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoksza D., Svozil D.. Efficient RNA pairwise structure comparison by SETTER method. Bioinformatics. 2012; 28:1858–1864. [DOI] [PubMed] [Google Scholar]
- 21. Laborde J., Robinson D., Srivastava A., Klassen E., Zhang J.. RNA global alignment in the joint sequence-structure space using elastic shape analysis. Nucleic Acids Res. 2013; 41:e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He G., Steppi A., Laborde J., Srivastava A., Zhao P., Zhang J.. RASS: a web server for RNA alignment in the joint sequence-structure space. Nucleic Acids Res. 2014; 42:W377–W381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen M.N., Tan K.P., Madhusudhan M.S.. CLICK–topology-independent comparison of biomolecular 3D structures. Nucleic Acids Res. 2011; 39:W24–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen M.N., Verma C.. Rclick: a web server for comparison of RNA 3D structures. Bioinformatics. 2015; 31:966–968. [DOI] [PubMed] [Google Scholar]
- 25. Chojnowski G., Walen T., Piatkowski P., Potrzebowski W., Bujnicki J.M.. Brickworx builds recurrent RNA and DNA structural motifs into medium- and low-resolution electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 2015; 71:697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Notredame C., Higgins D.G., Heringa J.. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000; 302:205–217. [DOI] [PubMed] [Google Scholar]
- 27. Rother M., Rother K., Puton T., Bujnicki J.M.. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 2011; 39:4007–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walen T., Chojnowski G., Gierski P., Bujnicki J.M.. ClaRNA: a classifier of contacts in RNA 3D structures based on a comparative analysis of various classification schemes. Nucleic Acids Res. 2014; 42:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Dongen S., Abreu-Goodger C.. Using MCL to extract clusters from networks. Methods Mol. Biol. 2012; 804:281–295. [DOI] [PubMed] [Google Scholar]
- 30. Burge S.W., Daub J., Eberhardt R., Tate J., Barquist L., Nawrocki E.P., Eddy S.R., Gardner P.P., Bateman A.. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013; 41:D226–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leontis N., Zirbel C.. Leontis N, Westhof E. RNA 3D Structure Analysis and Prediction. 2012; 27, Berlin Heidelberg: Springer; 281–298. [Google Scholar]
- 32. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nawrocki E.P., Kolbe D.L., Eddy S.R.. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009; 25:1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M. et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002; 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katoh K., Standley D.M.. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013; 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009; 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kufareva I., Abagyan R.. Methods of protein structure comparison. Methods Mol. Biol. 2012; 857:231–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carrillo H., Lipman D.. The multiple sequence alignment problem in biology. SIAM J. Appl. Math. 1988; 48:1073–1082. [Google Scholar]
- 39. Gardner P.P., Wilm A., Washietl S.. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005; 33:2433–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torarinsson E., Lindgreen S.. WAR: Webserver for aligning structural RNAs. Nucleic Acids Res. 2008; 36:W79–W84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thompson J.D., Plewniak F., Poch O.. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999; 27:2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kemena C., Bussotti G., Capriotti E., Marti-Renom M.A., Notredame C.. Using tertiary structure for the computation of highly accurate multiple RNA alignments with the SARA-Coffee package. Bioinformatics. 2013; 29:1112–1119. [DOI] [PubMed] [Google Scholar]
- 43. Colasanti A.V., Lu X.J., Olson W.K.. Analyzing and building nucleic acid structures with 3DNA. J. Vis. Exp. 2013; e4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjamini Y., Hochberg Y.. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 1995; 57:289–300. [Google Scholar]
- 45. Cruz J.A., Blanchet M.F., Boniecki M., Bujnicki J.M., Chen S.J., Cao S., Das R., Ding F., Dokholyan N.V., Flores S.C. et al. RNA-Puzzles: A CASP-like evaluation of RNA three-dimensional structure prediction. RNA. 2012; 14:610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miao Z., Adamiak R.W., Blanchet M.F., Boniecki M., Bujnicki J.M., Chen S.J., Cheng C., Chojnowski G., Chou F.C., Cordero P. et al. RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA. 2015; 21:1066–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.