Abstract
Inhibition of the Akt kinase activates HPV16 late gene expression by reducing HPV16 early polyadenylation and by activating HPV16 late L1 mRNA splicing. We identified ‘hot spots’ for RNA binding proteins at the early polyA signal and at splice sites on HPV16 late mRNAs. We observed that hnRNP L was associated with sequences at all HPV16 late splice sites and at the early polyA signal. Akt kinase inhibition resulted in hnRNP L dephosphorylation and reduced association of hnRNP L with HPV16 mRNAs. This was accompanied by an increased binding of U2AF65 and Sam68 to HPV16 mRNAs. Furthermore, siRNA knock-down of hnRNP L or Akt induced HPV16 gene expression. Treatment of HPV16 immortalized keratinocytes with Akt kinase inhibitor reduced hnRNP L binding to HPV16 mRNAs and induced HPV16 L1 mRNA production. Finally, deletion of the hnRNP L binding sites in HPV16 subgenomic expression plasmids resulted in activation of HPV16 late gene expression. In conclusion, the Akt kinase inhibits HPV16 late gene expression at the level of RNA processing by controlling the RNA-binding protein hnRNP L. We speculate that Akt kinase activity upholds an intracellular milieu that favours HPV16 early gene expression and suppresses HPV16 late gene expression.
INTRODUCTION
Human papillomaviruses (HPV) are small DNA viruses with a strict tropism for human epithelial cells (1). A subset of the HPVs has tropism for mucosa and are mainly sexually transmitted (2). The vast majority of these HPV infections are asymptomatic and persist for 18–24 months before they are cleared by the immune system of the host (3). In rare cases, sexually transmitted HPVs of high-risk type may establish chronic persistent infections that last for years or decades. These infections may cause cervical lesions that can progress to cervical cancer (4).
More than 99% of all cervical cancers contain HPV DNA (5). Epidemiological studies have established that the most common high-risk HPV type is HPV16, and that HPV16 is present in ∼50% of the >500 000 cases of cervical cancer diagnosed annually worldwide (6). Many questions regarding the role of HPV16 in cancer are still unanswered (7). For example, it is still not entirely clear how HPV16 can persist for decades in the presence of a functional immune system. We speculate that a strict control of HPV16 gene expression contributes to its ability to hide from the immune system. It is therefore of interest to investigate how HPV16 gene expression is regulated (8,9).
The HPV16 late genes L1 and L2 encode highly immunogenic proteins. Consequently, expression of L1 and L2 is delayed until the HPV16 infected cells reach the upper layers of the stratified mucosal epithelium (10). Most likely, this is necessary for the virus to remain undetected by the immune system long enough to be sexually transmitted, but it may also contribute to long-term persistence and development of cancer. We speculate that inhibition of HPV16 L1 and L2 expression is a prerequisite for development of HPV16-induced cancer (8).
The switch from the early to the late HPV16 gene expression program includes a promoter switch, a polyA site switch and a shift in HPV16 alternative mRNA splicing (8,10). The HPV16 replication and transcription factor E2, may contribute to this switch by down regulating the HPV16 early promoter (11–13). In addition to cellular RNA binding proteins, HPV16 E2 contributes directly to the control of HPV16 RNA processing by interfering with HPV16 RNA polyadenylation (14). HPV16 E2 has also been shown to affect HPV16 RNA processing indirectly by activating expression of splicing factors (15), possibly a result of E2 binding to cellular DNA (16). Cellular factors such as CTCF may also control both transcription and processing of HPV mRNAs (17). Much of HPV16 gene regulation occurs at the level of RNA processing (8,15,18) and HPVs produce a large number of alternatively spliced and polyadenylated mRNAs (9). A number of cis acting RNA elements controlling HPV16 and HPV18 splices sites and polyA signals have been identified and characterized (8), and a review of all identified HPV RNA binding proteins has recently been published (19).
The PI3K/Akt/mTOR signalling pathway is intimately linked to HPV infection and is activated in response to HPV infection, a feature also characteristic of HPV-infected cervical cancer cells (20). At the molecular level, the HPV E7 protein directly activates Akt (21,22) and in cervical cancer cells this pathway is further stimulated by amplifications and mutations of the PI3K gene (23,24). Also HPV E5 and E6 stimulate the PI3K/Akt/mTOR signalling pathway (25–27). Activation of the PI3K/Akt/mTOR signalling pathway promotes cell proliferation and inhibits apoptosis and differentiation (28), all features of the early stages of the HPV life cycle as well as of HPV-induced cancer, but is counteractive for entry into late stages of the viral life cycle. We speculate that inhibition of the Akt kinase is necessary to complete the HPV life cycle, to re-enter cellular differentiation and to turn on HPV late gene expression. In HPV-infected cells, inhibition of the PI3K/Akt pathway is probably a consequence of the shut-down of the HPV early promoter by E2, which reduces expression of the pro-mitotic E6 and E7 proteins. If high expression or activity of the PI3K/Akt pathway is fixed by mutations in the HPV16 infected cells, HPV16 may be unable to exit the early stage of the viral life cycle. The inability of the HPV16 infected cell to differentiate and enter the late phase of the viral life cycle may enhance the risk of progression to cancer. As a result of its key role in the control of HPV gene expression and development of cervical cancer, the PI3K/Akt/mTORC pathway may be a therapeutic target for cervical cancer (29).
There are numerous examples in the literature of the Akt kinase controlling splicing reactions, often through SR proteins (30–34) and/or SRPK1 (35–37) or Clk/sty (38). Interestingly, the Akt kinase pathway has also been linked to splicing regulation of bovine papillomavirus type 1 in cells lacking splicing factor SRSF1 (ASF/SF2) (39). The connection between the Akt kinase, SR proteins and HPV16, remains to be investigated. Proteins Sam68, hnRNP A1 and hnRNP A2 have been shown previously to regulate splicing also of the HPV16 early E6/E7 mRNAs, a function controlled via the Erk1/2 pathway (40). One may speculate that the major splicing regulatory factors are controlled by multiple kinases, perhaps at different stages of the HPV16 replication cycle.
Here, we show that hnRNP L binds HPV16 mRNAs and inhibits HPV16 late gene expression. hnRNP L is under the control of the cellular Akt kinase and inhibition of Akt reduces hnRNP L binding to HPV16 mRNAs and activates HPV16 late gene expression. We have identified hot spots for many RNA binding proteins including hnRNP L at key splice sites on the HPV16 mRNAs. Deletion of the hnRNP L binding sites activated HPV16 late gene expression. In conclusion, we have identified hnRNP L as a novel regulator of HPV16 RNA processing.
MATERIALS AND METHODS
Cell lines
The C33A2 cell line will be described in detail elsewhere. Briefly, C33A cells were transfected with subgenomic HPV16 plasmid pBELsLuc (Figure 1A) that contains a gene segment encoding poliovirus 2A internal ribosome entry site together with the Metridia longa secreted luciferase (sLuc) gene in the L1 coding region of HPV16. Induction of HPV16 late gene expression induces production of sLuc in the cell culture medium (41). The 3310 cell line has been described previously and was generated by transfection of normal human primary keratinocytes with full-length HPV16 genome plasmid pHPV16AN (42,43). The 3310 cells were cultured in EpiLife Medium supplemented with Human Keratinocyte Growth Supplement (Gibco Thermo Fisher Science). Propagation of neonatal human epidermal keratinocytes has been described previously (43). For differentiation of the 3310 cells, CaCl2 was added into the culture medium at a concentration of 2.5 mM, followed by incubation for the indicated time periods. 293T, HeLa and C33A2 cells were cultured in Dulbecco's modified Eagle medium (GE Healthcare Life Science HyClone Laboratories) with 10% heat-inactivated foetal bovine calf serum (GE Healthcare Life Sciences HyClone Laboratories) and penicillin/streptomycin (Gibco Thermo Fisher Science).
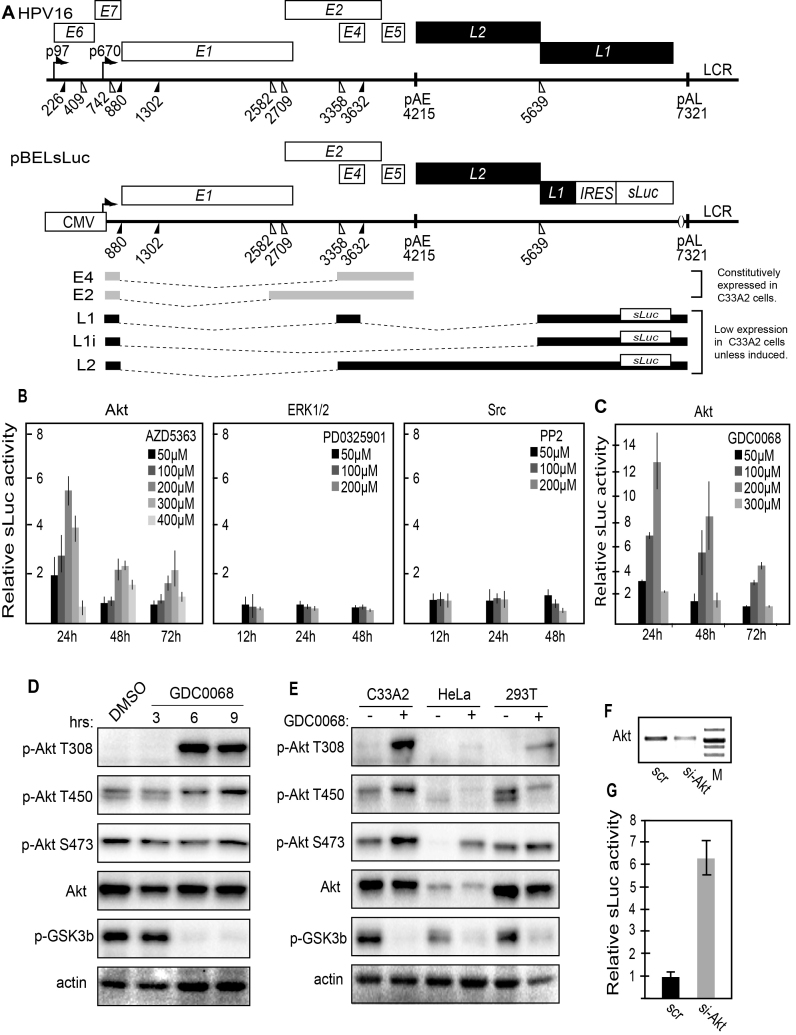
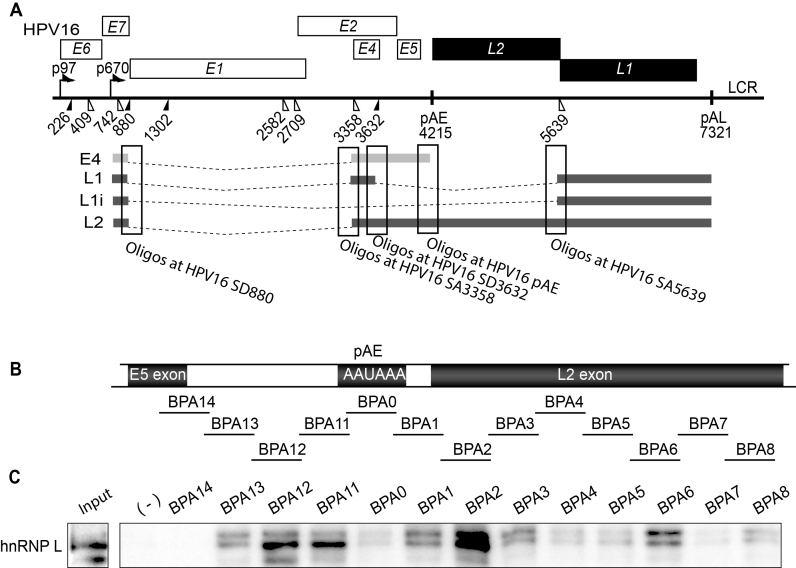
Figure 1.
(A) Schematic representation of the HPV16 genome. Rectangles represent open reading frames, promoters p97 and p670 are indicated as arrows, filled and open triangles represent 5′- and 3′-splices sites respectively, HPV16 early and late polyA signals pAE and pAL are indicated. Below the HPV16 genome, a schematic representation of the pBELsLuc reporter plasmid stably integrated in the genome of the C33A2 cells. Transcription of the HPV16 sequences in the pBELsLuc plasmid is driven by the human cytomegalovirus promoter (CMV). The sLuc gene inserted into the L1 region is indicated and is preceded by the poliovirus 2A internal ribosome entry site (IRES). HPV16 E2 and E4 mRNAs mRNAs produced by C33A2 cells are indicated in light grey and HPV16 late mRNAs encoding sLuc that can be induced in this reporter cell line are indicated in black. (B and C) Fold induction of secreted luciferase enzyme activity (sLuc) in the cell culture medium of reporter cell line C33A2 treated with various concentrations of the indicated inhibitors over DMSO treated cells. sLuc activity was monitored at the indicated time points. sLuc activity is displayed as fold over DMSO-treated C33A2 cells at the various time points. (B) Results show the effect on C33A2 cells of inhibitors to the cellular kinases Akt (AZD5363), ERK1/2 (PDO325901) and Src (PP2). (C) Induction of sLuc activity by the Akt kinase inhibitor GDC0068. (D). Western blot of the Akt kinase, various phosphorylated versions of the Akt kinase and phosphorylated GSK3b in C33A2 cells in the absence or presence of Akt kinase inhibitor GDC0068 (100 uM) at different time points. (E) Western blot of the same kinases as in (D) in various cell lines treated with Akt kinase inhibitor GDC0068. Note that Akt kinase inhibitor GDC0068 locks the Akt kinase in an inactive, but phosphorylated state appearing as a band detected with antibodies to phosphorylated forms of Akt. (F) RT-PCR on total RNA extracted from C33A2 cells of the Akt1 mRNA in C33A2 cells transfected with siRNAs to the Akt1 kinase (si-Akt), compared to cells transfected with scrambled siRNAs (scr). (G) Relative sLuc activity induced by transfection of C33A2 cells with siRNAs to the Akt1 kinase (si-Akt), compared to cells transfected with scrambled siRNAs (scr).
Drug treatment of cells
Cell culture medium was replaced with medium containing indicated concentrations of inhibitors for indicated time periods. Src inhibitors: PP2 (S7008, Selleck chemicals) and Saracatinib (S1006, Selleck chemicals). Erk 1/2 inhibitors: PD0325901 (S1036, Selleck chemicals) and U0126 (S1102, Selleck chemicals). Pan-Akt inhibitors: AZD5363 (S8019, Selleck chemicals), GDC-0068 (S2808, Selleck chemicals), Afusertib (S7521, Selleck chemicals). Pan-PI3K inhibitors: GDC-0941, BKM120, Wortamannin, and LY294002. PDK-1 inhibitors: GSK2334470 (S7078, Selleck chemicals) and BX-795 (S7521, Selleck chemicals). mTORC1/2 inhibitors: INK128 (S2811, Selleck chemicals) and AZD8055 (S1555, Selleck chemicals). mTORC1specific inhibitors: Everolimus (07741, SIGMA Aldridge) and Temsirolimus (PZ0020, SIGMA Aldridge). SRPK inhibitor: SRPIN340 (S7270, Selleck chemicals). All inhibitors were dissolved in DMSO, and DMSO in the absence of inhibitor was used as a control in all experiments.
In vitro phosphorylation
GST and GST-hnRNP L were washed three times with the kinase buffers (50mM Tris-HCl pH7.5, 0.1% b-mercaptoethanol, 0.1 mM EDTA, 0.1mM EGTA, 10 mM MgCl2). In vitro phosphorylation of the fusion proteins was carried out at 30°C for 20 min in 25 μl kinase buffer containing 0.9 μCi [γ-32P]-ATP and indicated amount of recombinant Akt1 (ab79792). The reaction was stopped by adding SDS sample buffer. Then samples were subjected to SDS-PAGE and analyzed by autoradiography.
Secreted luciferase assay and CAT ELISA
The Metridia longa secreted luciferase activity in the cultured medium of the C33A2 cells was monitored with the help of the Ready To Glow secreted luciferase reporter assay according to the instructions of the manufacturer (Clontech Laboratories). Briefly, 50 μl of cell culture media were mixed with 5 μl of secreted luciferase substrate in reaction buffer, and the luminescence was monitored in a Tristar LB941 luminometer (Berthold Technologies). Cell extracts were prepared from transfected cells according to the protocol from the CAT enzyme-linked immunosorbent assay (ELISA) kit (Roche). All cell extracts were appropriately diluted to obtain an absorbance reading in the linear range of the CAT ELISA assay. CAT values relative to the positive control plasmid pCMVCAT16 (rCAT) were calculated as described previously (41).
RNA extraction and RT-PCR
Nuclear RNA was extracted at 24 h posttransfection. Total RNA was extracted using TRI Reagent (SIGMA Aldrich Life Science) and Direct-zol RNA MiniPrep (ZYMO Research) according to the manufacturer's protocol. Cytoplasmic RNA was extracted as described previously (41). 1μg of total RNA were reverse transcribed in a 20 μl reaction at 37°C by using M-MLV Reverse Transcriptase (Invitrogen) and random primers (Invitrogen) according to the protocol of the manufacturer. One microliter of cDNA was subjected to PCR amplification. E4 mRNAs spliced from SD880 to SA3358 were amplified with primers 773s and E4as, E2 mRNAs with primers 773s and E2as, L2 mRNAs with primers Set3-F and Set3-R and L1 and L1i mRNAs with primers 773s and L1as. All late mRNAs (L1, L1i and L2 mRNAs) were collectively PCR-amplified with primers L1–2-F and L1–2-R. See Figure 2A for location of RT-PCR primers and Supplementary Table S1 for their sequences. For quantitation of L1 mRNA by RT-qPCR, primers E4sVar and L1as were used (Supplementary Table S1).
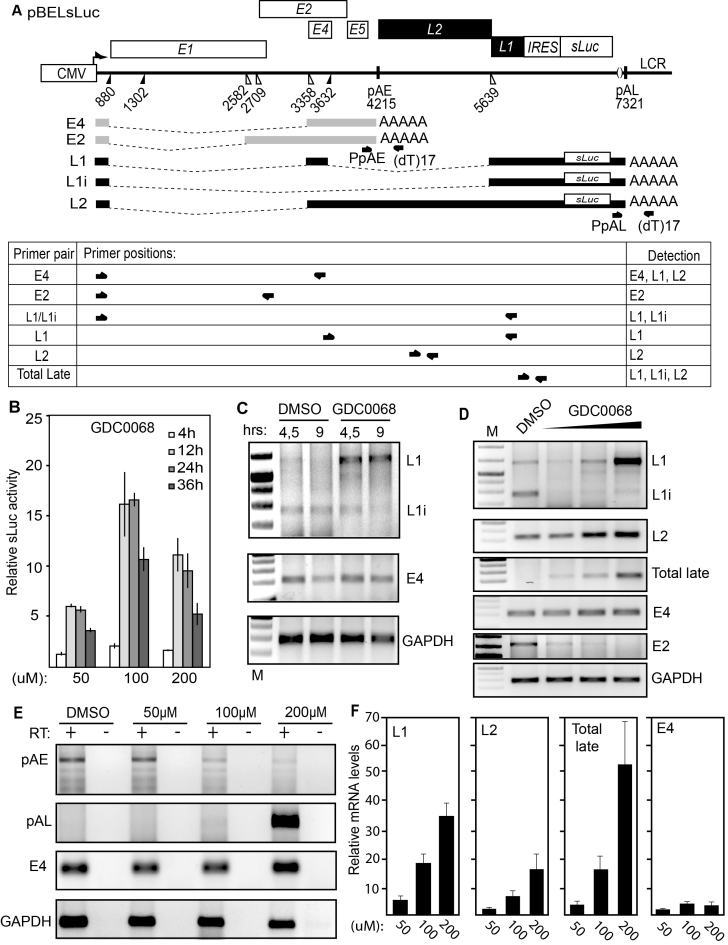
Figure 2.
(A) Schematic representation of the pBELsLuc reporter plasmid stably integrated in the genome of the C33A2 reporter cell line. Transcription of the HPV16 sequences in the pBELsLuc plasmid is driven by the human cytomegalovirus promoter (CMV). The sLuc gene inserted into the L1 region is indicated and is preceded by the poliovirus 2A internal ribosome entry site (IRES). HPV16 E2 and E4 mRNAs mRNAs produced by C33A2 cells are indicated in light grey (E2 and E4 mRNAs) and HPV16 late mRNAs encoding sLuc that can be induced in this reporter cell line are indicated in black. The location of RT-PCR primers is indicated below. (B) Secreted luciferase enzyme activity (sLuc) in the cell culture medium of reporter cell line C33A2 treated with various concentrations of the Akt kinase inhibitor GDC0068. sLuc activity was monitored at the indicated time points. sLuc activity is displayed as fold over DMSO-treated C33A2 cells at the various time points. (C) RT-PCR on total RNA extracted from the C33A2 reporter cell line treated with DMSO alone or Akti kinase inhibitor GDC0068 for 4.5hrs or 9hrs. The RT-PCR primers detected HPV16 L1/L1i-, E4- or GAPDH-mRNAs. (D) RT-PCR on total RNA extracted from the C33A2 reporter cell line treated with DMSO alone or various concentrations of Akt kinase inhibitor GDC0068 (50-, 100- or 200uM) for 9hrs. The RT-PCR primers detected HPV16 L1/L1i-, L2-, total late (L2 + L1 +L1i), E4-, E2- or GAPDH-mRNAs. (E) A 3′-RACE assay on total RNA extracted from C33A2 cells. Primers were F-Set2 and (dT)17-P3 for pAE and sLuc-S-inner and (dT)17-P3 for pAL (for primers see Supplementary Table S1). Spliced HPV16 E4 mRNAs and cellular GAPDH mRNAs are also shown. (F) RT-qPCR on the cDNA samples used for RT-PCR in Figure 2D.
Real-time quantitative PCR (qPCR) of cDNAs
qPCR was performed on 1 μl of cDNA prepared as described above in a MiniOpticon (Bio-Rad) using the Sso Advanced SYBR Green Supermix (Bio-Rad) according the manufacturer's instructions. Primers were those described above, and all cDNA quantitations were normalized to GAPDH mRNA levels. GAPDH cDNA was amplified with primers GAPDH-F and GAPDH-R (Supplementary Table S1).
Protein extraction, Western blotting and immunoprecipitation
Western blotting was performed as described previously (14). Briefly, cells were lysed in RIPA buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 1 mM DTT and protease inhibitors) and subjected to SDS-PAGE. Proteins were transferred onto nitrocellulose membranes, blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween 20, and stained with antibodies to the indicated proteins. For a list of the antibodies see Supplementary Table S3. Secondary antibodies conjugated with horseradish peroxidase were used, and proteins were detected using the Clarity Western ECL Substrate (Bio Rad) or the Super Signal West Femto chemiluminescence substrate (Pierce).
Plasmids
The sequences encoding the Rous sarcoma virus (RSV) long terminal repeat (LTR), HPV16 sequences and the simian virus 40 (SV40) polyadenylation signal that are present in plasmids pE4EL1M, pE4EL1, pE4D and pE4DL1D (shown schematically in Figure 10B) were synthesized by Eurofins Genomics and cloned into the pEX-A2 vector. To generate pBSpD1MCAT, pBSpD2MCAT and pBSpD3MCAT, site-directed ligase-independent mutagenesis (SLIM) was performed on pBSp5MCAT as described previously (41). Mutagenesis primer sequences are available on request.
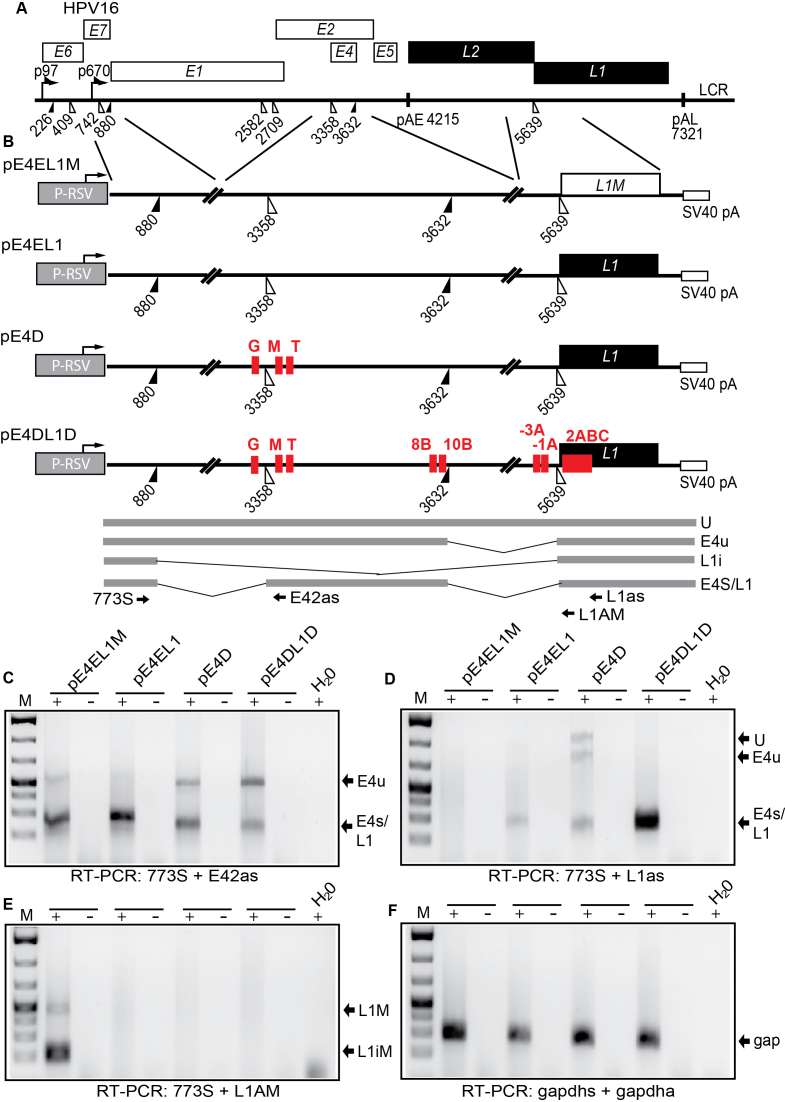
Figure 10.
(A) Schematic representation of the HPV16 genome. Rectangles represent open reading frames, promoters p97 and p670 are indicated as arrows, filled and open triangles represent 5′- and 3′-splices sites respectively, HPV16 early and late polyA signals pAE and pAL are indicated. (B) Schematic representations of reporter plasmids pE4EL1M, pE4EL1, pE4D and pE4DL1D. Lines indicate segments from the HPV16 genome that have been inserted into the ‘HPV16 mini-constructs’. The plasmids encode the Rous sarcoma virus (RSV) long terminal repeat (LTR) promoter followed by HPV16 sequences that encompass HPV16 splice sites SD880, SA3358, SD3632 and SA5639 and the simian virus 40 (SV40) polyadenylation signal (SV40 pA). The hnRNP L binding sites mapped to oligos G, M, T, 8B, 10B, -3C, -1A, and 2 are indicated. Binding site ‘2’ consist of the overlapping binding sites 2 2A, 2B and 2C). The bindings sites refer to the mapping of hnRNP L binding sites in Figures 3 and 4 and supplementary figures S2 and S3. C33A cells were transfected with the various plasmids and cytoplasmic RNA was extracted and subjected to cDNA synthesis and RT-PCR with the oligonucleotides shown in (B) and indicated below the gels. (C) RT-PCR with primers 773S and E42s detecting mRNAs spliced between SD880 and SA3358. (D) RT-PCR with primers 773S and L1as detecting all spliced HPV16 mRNAs. (E) RT-PCR with primers 773S and L1AM specifically detecting mRNAs spliced to SA5639 in plasmid pE4EL1M that contains a mutant L1 sequence in which previously described splicing silencer elements have been inactivated (49). (F) RT-PCR with primers gapdhs and gapdha detecting spliced gapdh mRNAs. gap; gapdh mRNAs.
Plasmid transfections and intracellular UV cross-linking and immunoprecipitation of RNA–protein complexes
293T cells grown in 10-cm dishes were transfected for 24 h with Turbofect (Thermo Fisher Scientific) according to the manufactures’ instructions, washed with ice-cold PBS and UV-irradiated at 150 mJ/cm2 in a Bio-link cross-linker (Biometra). Cells were lysed in 1 ml of RIPA buffer (described above) and were incubated on ice for 30 min with occasional vortexing to lyse cells. For immunoprecipitations, 1 μg of the indicated antibody or normal mouse IgG (#12-371, Millipore) were incubated for 2 h at 4°C in 0.5 ml of lysate. Twenty μl (0.6 mg) of Dynabeads Protein G (#10004D, Invitrogen) were blocked with 1% BSA in RIPA buffer for 0.5 h, washed three times, and then added to the antibody–protein mixture followed by incubation for 1 h at 4°C. The beads were washed three times with RIPA buffer and RNA was eluted by phenol/chloroform extraction. The RNA was ethanol-precipitated and dissolved in 20 μl of water. Ten microliters of immunoprecipitated RNA was reverse transcribed using M-MLV reverse transcriptase (Invitrogen) and random primers (Invitrogen) according to the protocol of the manufacturer. Two microliter of cDNA were subjected to PCR amplification using primers 773s and E4as (Supplementary Table S1) as described above.
siRNA library and siRNA transfections
A custom-made siRNA DHARlibrary consisting siGENOME SMARTpools of four siRNAs to each mRNA was ordered from GE Healthcare Dharmacon. Each well in the 96-well plates contained 0.1 nmol of siRNA pool, including one well with a scrambled control siRNA. The siRNA library was transfected in triplicates at three independent occasions into C33A2 cells grown in 96-well plates using DharmaFECT1 (GE Healthcare Dharmacon) according to the instructions of the manufacturer. Cell culture medium was harvested at 72 h posttransfection and sLuc activity was monitored as described above. Additional siRNAs to hnRNP L, hnRNP A1, hnRNP A2B1 and Akt1, as well as scrambled negative control siRNAs, were purchased from GE Healthcare Dharmacon as siRNA SMARTpools to confirm screening results and to perform RNA analysis by transfection of C33A2 cells in 6-well plates. Transfections were conducted with DharmaFECT1 (GE Healthcare Dharmacon) and cell culture medium or cells were harvested 72 h posttransfection.
ssDNA/RNA–protein pull-down assay
Nuclear extracts were prepared from subconfluent HeLa cells or C33A2 cells according to the procedure described by Dignam et al. (44). Biotin-labeled ssDNA oligos and 2′-O-Me-RNA oligos were purchased from Eurofins Genomics and biotin-labeled ssRNA oligos were purchased from SIGMA Aldrich (Supplementary Table S2). To pull down proteins, HeLa nuclear extract was mixed with streptavidin-coated magnetic beads (Dynabeads M-280 Streptavidin, Invitrogen) bound to biotin-labeled ssDNA or RNA oligonucleotides in 500 μl of binding buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 0.5% Triton X-100) (41). After incubation with rotation for 1 h at room temperature, beads were washed five times in binding buffer. Proteins were eluted by boiling of the beads in SDS-PAGE loading buffer and subjected to SDS-PAGE followed by Western blot analysis with the indicated antibodies (Supplementary Table S3).
RESULTS
Inhibition of the cellular Akt-kinase induces HPV16 late gene expression
The C33A2 reporter cell line developed by us is derived from C33A cells and contains a stably integrated subgenomic HPV16 plasmid termed pBELsLuc (Figure 1A). This plasmid has the secreted luciferase (sLuc) reporter gene inserted in the late region of HPV16 (Figure 1A). C33A2 cells constitutively express HPV16 early genes whereas HPV16 late genes are suppressed. The C33A2 cells were treated with various concentrations of inhibitors to the cellular kinases Akt, ERK1/2 and src. The results revealed that only the Akt kinase inhibitor AZD5363 induced HPV16 late gene expression, monitored here as a time- and inhibitor dose-dependent increase in secreted luciferase enzymatic activity in the cell culture medium of the C33A2 cells (Figure 1B and Supplementary Figure S1A). To confirm these results, the C33A2 cells were treated with two additional inhibitors of the Akt kinase: GDC0068 and afuresertib. While afuresertib did not induce sLuc production (data ot shown), kinase inhibitor GDC0068 strongly induced HPV16 late gene expression (Figure 1C). Next we confirmed that the Akt inhibitor GDC0068 inhibited the Akt kinase in C33A2 cells. As can be seen in Figure 1D and 1E, the Akt-kinase is locked in a hyperphosphorylated but inactive complex so characteristic for Akt inhibitor GDC0068 (45) as it is added to the C33A2 cells (Figure 1D and E). Interestingly, Akt kinase inhibitor GDC0068 appeared to act primarily on Akt kinase tyrosine T308 and to a lower extent on T450 and S473 (Figure 1D and E). Similar results were obtained with HeLa and 293T cells, although HeLa cells in particular contain less Akt than the other two cell lines (Figure 1E). Further evidence that the Akt kinase was indeed inhibited by GDC0068 in C33A2 cells was obtained by Western blotting of phosphorylated GSK3b, a direct target of the Akt kinase (Figure 1D and E). Inhibition of Akt by GDC0068 caused a significant drop in the levels of phosphorylated GSK3b (p-GSK3b) (Figure 1D and E), strongly indicating that the kinase activity of Akt had been inhibited. Similar results were obtained with all three cell lines (Figure 1E). The effect of Akt inhibitor GDC0068 was time dependent and fully developed already after 6hrs, but not 3hrs, of treatment as evidenced by the appearance of phosphorylated but inactive Akt kinase (Figure 1D), and the reduction of phosphorylated GSK3b (Figure 1D). Taken together, we concluded that inhibition of the Akt kinase induced HPV16 late gene expression in the C33A2 reporter cell line, indicating that HPV16 late gene expression is regulated by the Akt kinase.
Since the Akt kinase is part of the PI3K-Akt kinase signal transduction pathway, we also analysed the effects on HPV16 gene expression of inhibitors to PI3K itself: GDC00941, BKM120, Wortmanin and LY294002 (Supplementary Figure S1B). Two of these inhibitors also induced sLuc production (GDC00941, BKM120), but to a lower extent than did Akt inhibitor GDC0068 (Supplementary Figure S1B). The GSK 2334470 and BX-795 inhibitors of PDK1, another kinase upstream of Akt, showed a relatively minor effect on sLuc production (Supplementary Figure S1C).
Next, we analysed inhibitors to kinases located downstream of Akt in the signal transduction pathways: mTORC1 and 2. Both dual inhibitors to mTORC1 and 2 (INK128, AZD8055) activated sLuc production, whereas inhibitors specific for mTORC1 (Everolimus, Temsirolimus) had no effect on HPV16, suggesting that mTORC2 accounted for the major HPV16 inhibitory activity. However, the induction was relatively minor compared to the Akt inhibitor GDC0068 (Supplementary Figure S1E). We also investigated if inhibitors to the SR-protein-specific kinase SRPK (SRPIN340) affected HPV16 late gene expression. However, we did not observe any effects on sLuc production with this inhibitor (Supplementary Figure S1D). Combined, our results indicated that the Akt kinase played an important role in the control of HPV16 late gene expression and suggested that the Akt kinase suppresses HPV16 late gene expression. To confirm that the Akt kinase inhibits HPV16 late gene expression we knocked down Akt in C33A2 cells by siRNA transfection (Figure 1F). As can be seen from the results, knock down of Akt induced HPV16 late gene expression (Figure 1G). We concluded that the Akt kinase contributes to suppression of HPV16 late gene expression.
Inhibition of the Akt kinase reduces HPV16 early polyadenylation, causes read-through into the late region and activates late L1 mRNA splicing
Next we analysed the effect of the GDC0068 Akt kinase inhibitor on HPV16 mRNA levels and mRNA processing by RT-PCR and RT-qPCR using HPV16 specific primers (Figure 2A). Concomitantly with an induction of sLuc activity in the cell culture medium (Figure 2B), the levels of HPV16 L1/L1i mRNAs increased in a time- (Figure 2C) and GDC0068 dose-dependent manner (Fig 2D). The L2 mRNA levels increased as well (Figure 2D), and a robust increase in HPV16 late gene expression was seen with RT-PCR primers that detect all late (L1, L1i and L2) mRNAs (Total Late) (Figure 2D). In contrast, the levels of E4 mRNAs were only marginally affected and the E2 mRNAs decreased (Figure 2C and D). Interestingly, treatment of the cells with the Akt inhibitor also caused a clear shift in the L1 mRNA splicing pattern from L1i mRNAs to L1 mRNAs (the L1 mRNA contains the internal E4 exon, while L1i does not) (Figure 2A, C and D). Taken together, these results suggested that HPV16 splice sites SA2709, SA3358, SD3632 and SA5639 were under control of the Akt kinase (see Figure 2A). The induction of all late HPV16 mRNAs and the L1 and L2 mRNAs individually, as well as the relatively minor effect on the E4 mRNAs, were confirmed by RT-qPCR (Figure 2F). The induction of L1 mRNAs, and the shift from L1i to L1 mRNAs, demonstrated that utilization of HPV16 splice sites SA3358, SD3632 and SA5639 increased. In addition, the induction of HPV16 L2 mRNA levels indicated that read-through occurred at pAE, suggesting that the effect of the Akt kinase inhibitor included inhibition of polyadenylation at pAE (Figure 2A, D and F). To investigate the effects of the Akt kinase inhibitor on polyadenylation further, a 3′-RACE experiment was performed to monitor polyadenylation at HPV16 early and late polyA signals pAE and pAL. The results revealed that polyadenylation at pAE was reduced and polyadenylation at pAL was increased in an Akt-inhibitor concentration-dependent manner (Figure 2E). We concluded that inhibition of the Akt kinase reduced polyadenylation activity at HPV16 pAE, which resulted in read-through into the HPV16 late region, and de-repressed the late HPV16 splice sites SD3632 and SA5639.
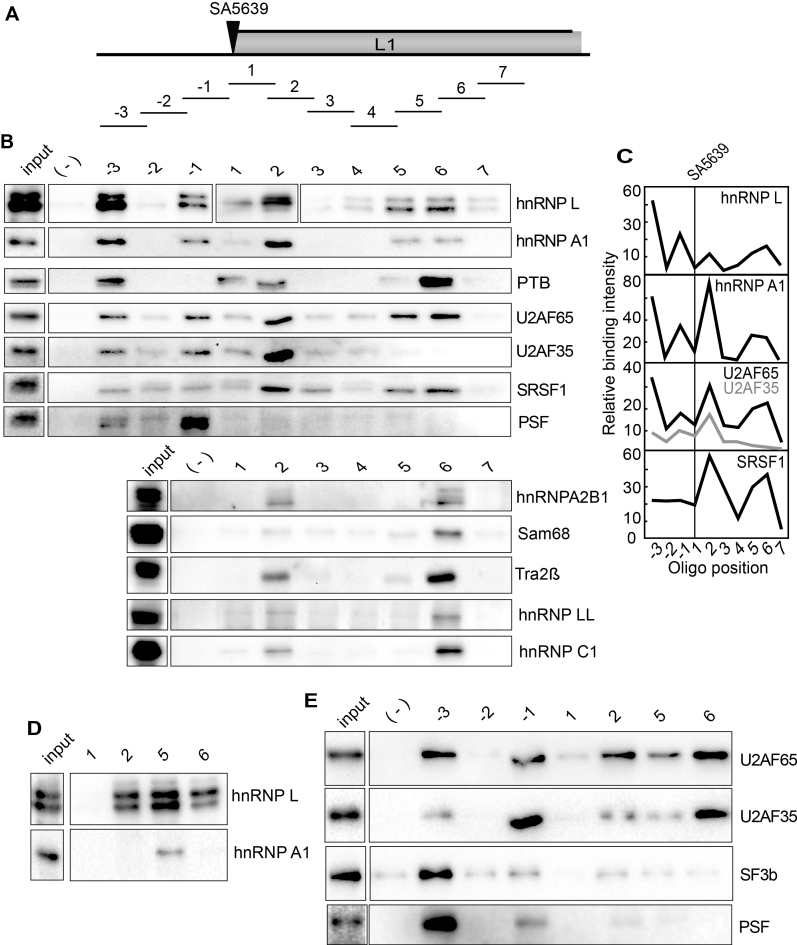
Identification of ‘hot spots’ for RNA-binding proteins at HPV16 late splice sites SA3358 and SD3632 – high prevalence of hnRNP L interactions
Since our results demonstrated that splicing of L1 mRNAs and polyadenylation at pAE were affected by the Akt inhibitor, we wished to identify RNA binding proteins that served as targets for Akt and participated in the regulation of HPV16 late gene expression under control of the Akt kinase. To identify RNA binding proteins that interact with HPV16, we used a pull-down assay with biotinylated oligonucleotides bound to streptavidin coated magnetic beads. We have previously shown that ssDNA oligos can be used instead of RNA oligos in this type of experiments at approximately 10% of the cost for RNA oligos (41). We confirmed these findings by comparing ssDNA and ssRNA oligos ability to pull down the RNA binding protein hnRNP L from nuclear extracts, in a sequence specific manner (Supplementary Figure S2A and B). ssDNA and ssRNA oligos with AC-repeats pulled down hnRNP L, whereas ssDNA oligos of GT and RNA oligos of GU did not (Supplementary Figure S2A and B). A previously described hnRNP L binding site pulled down hnRNP L (46), while the antisense version of this oligo did not (Supplementary Figure S2A and B), further demonstrating specificity of the pull-down assay. The pull down of hnRNP L is also dose dependent (Supplementary Figure S2C). With these results as a foundation, we used ssDNA oligos to identify RNA binding proteins that interact with HPV16 sequences at HPV16 key splice sites and polyA signals. Such factors could potentially be regulated by the Akt kinase. First, we used multiple oligos encompassing exon E4 of HPV16 in an effort to identify RNA binding proteins that regulate HPV16 splice sites SA3358 and SD3632 that are both used for production of L1 mRNAs (Figure 3A). We found that oligos spanning the E4 exon were associated with many different RNA binding proteins (Figure 3A). Interestingly, there appears to be ‘hot spots’ for RNA binding proteins immediately adjacent to splice sites SA3358 and SD3632, e.g. binding of many proteins to oligos -2 to 2 at SA3358, and oligos 8–10 at SD3632 (Figure 3A and B). To the credit of our method, we found hnRNP D binding at SD3632 and SRSF1 inside exon 4 (Figure 3A and B), as previously reported by us (41,43,47). Tra2b pull down coincided with SRSF1 (Figure 3A and B), probably as both are SR proteins. Furthermore, U2AF65 and U2AF35 are primarily binding to the polypyrimidine tract upstream of 3′-splice site SA3358 (oligos-2, -1 and 1), as expected for U2AF65/35 (Figure 3A and B). Several proteins appeared to bind next to the HPV16 splice sites SA3358 and SD3632 such as hnRNP L, hnRNP A1, Sam68, PTB, PTB-associated splicing factor (PSF) and hnRNP D (Figure 3A and B). One protein of particular interest was hnRNP L since its binding appeared to cluster with binding of positive splicing factor U2AF65/35, but in a manner that seemed to suggest a mutually exclusive binding pattern (Figure 3A and B). However, the hnRNP L-like protein hnRNP LL was only weakly detected in the pull down samples (Figure 3A). We therefore investigated hnRNP L further. Pull downs were first performed pairwise with sense and antisense oligos, confirming sequence-specific pull down of hnRNP L, while hnRNP A1 did not interact with these oligos at all, as expected (Supplementary Figure S2D). A detailed mapping of sites of interaction for hnRNP L, hnRNP A1 and U2AF65 using shorter oligos showed that their interactions did not directly overlap, suggesting that they were not competing for the same binding sites (Figure 3C). hnRNP L interacted primarily with sequences downstream of SA3358 (Figure 3A and B). In contrast, binding of hnRNP A1 showed a preference for sequences immediately upstream of SA3358 (Figure 3A and B), which suggested competition with U2AF65, but the fine mapping of binding revealed non-overlapping binding sites for hnRNP A1 and U2AF65 (Figure 3C). Although variation in pull down efficiency was observed with oligo 1 in Figure 3, the following results support the presence of an hnRNP L binding site in oligo 1 (albeit a weak site): mapping with shorter oligos identified a binding site, albeit weak, for hnRNP L within oligo M (Figure 3C), and RNA- or 2′-O-Me RNA-versions of oligo 1 pulled down hnRNP L (Supplementary Figure S2).
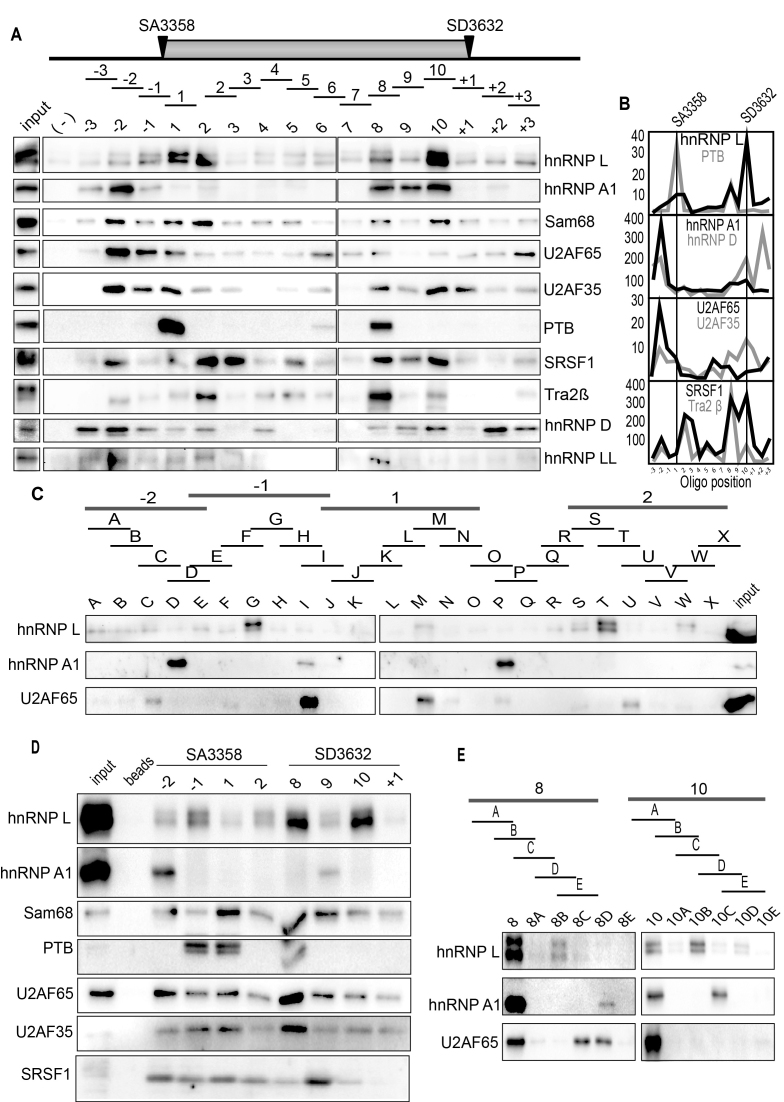
Figure 3.
(A) Upper panel: Schematic drawing of HPV16 exon 4 and the 35-nucleotide, biotinylated ssDNA oligos (overlapping by 5-nucleotides) used in pull down assays. Location of HPV16 3′-splice site SA3358 and 5′-splice site SD3632 are indicated. Lower panel: Pull downs of cellular factors from nuclear extracts using the indicated ssDNA oligos covering the E4 exon of HPV16 followed by Western blot analysis using antibodies to proteins indicated to the right. (-); mock pull downs using streptavidin beads in the absence of oligo. (B) Quantitation of some of the Western blots of the pull downs in (A). (C) Upper panel: Schematic drawing of shorter oligos (A-X) designed to better map binding sites for hnRNP L, hnRNP A1 and U2AF65. Lower panel: Western blots for hnRNP L, hnRNP A1 and U2AF65 of on proteins pulled down by the shorter biotinylated ssDNA oligos. (D) Western blots of indicated proteins on pull downs using biotinylated ssRNA oligos of a subset of the ssDNA oligos shown in (A). (E) Upper panel: Schematic drawing of shorter oligos (A–E) of the two original 35-nucleotide oligos 8 and 10 located near HPV16 late 5′-splice site SD3632. Lower panel: Western blots for hnRNP L, hnRNP A1 and U2AF65 of on proteins pulled down by the shorter biotinylated ssDNA oligos.
hnRNP L also interacted strongly with sequences upstream of HPV16 late 5′-splice site SD3632 (Figure 3A and E). Also in this case were binding sites for hnRNP A1, hnRNP L and U2AF65 distinct (Figure 3E). Lastly, we confirmed that these, and additional splicing factors such as Sam68, PTB and SF3b, also interacted with RNA versions of the same oligos (Figure 3D). Indeed, binding was more efficient with RNA oligos, but the specificity was the same: see pull downs with oligos -2, -1, 1 and 2 at SA3358 and oligos 8, 9, 10 and +1 at SD3632 (Figure 3D). Reduced pull down efficiency, but retained specificity, was observed when 2′-O-Me-RNA oligos were used (Supplementary Figure S2E). We concluded that hnRNP L in particular showed strong binding at, and downstream of HPV16 3′-splice site SA3358, and upstream of HPV16 5′- splice site SD3632, which suggested a regulatory role for hnRNP L in HPV16 late L1 mRNA processing.
Identification of ‘hot spots’ for RNA-binding proteins at HPV16 late L1 3′-splice site SA5639 – high prevalence of hnRNP L interactions
Since HPV16 late L1 mRNAs also use 3′-splice site SA5639 (Figure 4A), and since we have previously shown that it is suppressed by downstream elements that interact with hnRNP A1 and other yet unidentified factors (48–50), we also performed pull down experiments with sequences spanning 3′-splice site SA5639 (Figure 4A and B). Interestingly, also at SA5639 did we identify ‘hot spots’ for RNA binding proteins both upstream and downstream of SA5639 (oligos -3, -1, 2, 5 and 6) (Figure 4B and C). These oligos interacted with multiple RNA binding proteins in addition to hnRNP L, hnRNP A1, U2AF65 and U2AF35, such as SRSF1, PSF, hnRNP A2B1, Sam68, Tra2b, hnRNP LL and hnRNP C1 (Figure 4B). We found that U2AF65/35 binds to the polypyrimidine tract of 3′-splice site SA5639, as expected (Figure 4B and C). In sharp contrast to pull downs of U2AF65/35 at the efficiently used 3′-splice sites SA3358 in the E4 exon, U2AF65/35 at the suppressed 3′-splice site SA5639 was pulled down with the same oligos as hnRNP L and hnRNP A1 (Figure 4B and C), and additional splicing inhibitory factors such as PTB and PSF (Figure 4B). This correlation was true also for shorter oligos used for fine mapping of pull downs at SA5639 (Supplementary Figure S3A and B). Since SA5639 encodes a relatively long and uninterrupted polypyrimidine tract (in contrast to SA3358), U2AF65 interacted with relatively many of the shorter oligos used for mapping (Supplementary Figure S3A and B). Taken together, we concluded that pull downs with oligos at HPV16 3′-splice site SA5639 yielded different results than pull downs with oligos from HPV16 3′-splice site SA3358. Our results suggested that hnRNP L, hnRNP A1, PSF and PTB could interfere negatively with the recognition of SA5639 by interacting or interfering with U2AF65/35. Key pull down results were confirmed with RNA oligos (Figure 4D and E). Reduced pull down efficiency, but retained specificity, was observed when 2′-O-RNA oligos were used (Supplementary Figure S3C), but for all oligos, the pull-down results were corroborated with RNA oligos.
Figure 4.
(A) Schematic drawing of the HPV16 region around late 3′-splice site SA5639 and the 35-nucleotide biotinylated ssDNA oligos (overlapping by 5-nucleotides) used in pull down assays. Location of HPV16 3′-splice site SA5639 is indicated. (B) Upper and lower panels show pull downs of cellular factors from nuclear extracts using the indicated ssDNA oligos covering the region of HPV16 late 3′-splice site SA5639 followed by Western blot analysis using antibodies indicated to the right. (–) mock pull downs using streptavidin beads in the absence of oligo. (C) Quantitation of some of the Western blots of the pull downs in (B). (D and E) Western blots of indicated proteins on pull downs using biotinylated ssRNA oligos of a subset of the ssDNA oligos shown in (B).
We have previously shown that RNA sequences downstream of SA5639, located within the L1 coding region, suppress HPV16 late 3′-splice site SA5639 (48–50). To investigate if the pull-down of hnRNP L and hnRNP A1 were specific for HPV16 L1 sequences with splicing inhibitory activity, we also performed pull downs with wild type and mutant versions of oligos 1, 2, 3, 4 and 5 from the L1 coding region (Supplementary Figure S3D). The mutant L1 sequences represent sequences that were shown previously to be inactive as splicing silencer elements downstream of splice site SA5639 (48–50). As can be seen in Supplementary Figure S3D, hnRNP A1 binds specifically to the L1 wt sequence (Supplementary Figure S3D) (as we reported previously (48–50)) and so did hnRNP L (Supplementary Figure S3D), demonstrating that binding of both hnRNP A1 and hnRNP L correlates with splicing inhibition. We concluded that interactions of hnRNP L with sequences at HPV16 late splice site SA5639 correlated with splicing inhibition, warranting further analysis of hnRNP L.
hnRNP L interacts with sequences upstream and downstream of HPV16 pAE
Since treatment of C33A2 cells with Akt inhibitor GDC0068 caused a significant reduction in polyadenylation at HPV16 early polyA signal pAE (Figure 2E), we investigated if hnRNP L interacts with sequences at the HPV16 early polyA signal pAE. We performed pull-down experiments using oligos spanning the early UTR, the early polyA signal pAE and downstream sequences (Figure 5A and B). We have shown previously that the HPV16 early UTR and the downstream L2 coding region contain sequences that affect polyadenylation at pAE (51–54). Interestingly, hnRNP L is efficiently pulled down by oligos on either side of the AAUAAA polyA signal itself (Figure 5B and C), suggesting that also pAE could be controlled by hnRNP L.
Figure 5.
(A) Schematic representation of the HPV16 genome. Rectangles represent open reading frames, promoters p97 and p670 are indicated as arrows, filled and open triangles represent 5′- and 3′-splices sites respectively, HPV16 early and late polyA signals pAE and pAL are indicated. The regions on the HPV16 mRNAs from which the ssDNA and ssRNA oligos are derived are boxed. (B) Schematic drawing of the HPV16 region around early polyA signal pAE and the 35-nucleotide, biotinylated ssDNA oligos (overlapping by 5-nucleotides) used in pull down assays. Location of HPV16 early polyA signal pAE is indicated as well as E5 and L2 coding regions. (C) Pull downs of cellular factors from nuclear extracts using the indicated ssDNA oligos spanning HPV16 pAE followed by Western blot for hnRNP L. (-); mock pull downs using streptavidine beads in the absence of oligo.
Reduced pull down of hnRNP L and hnRNP A1 and increased pull down of U2AF65 with extracts from Akt kinase inhibitor treated cells
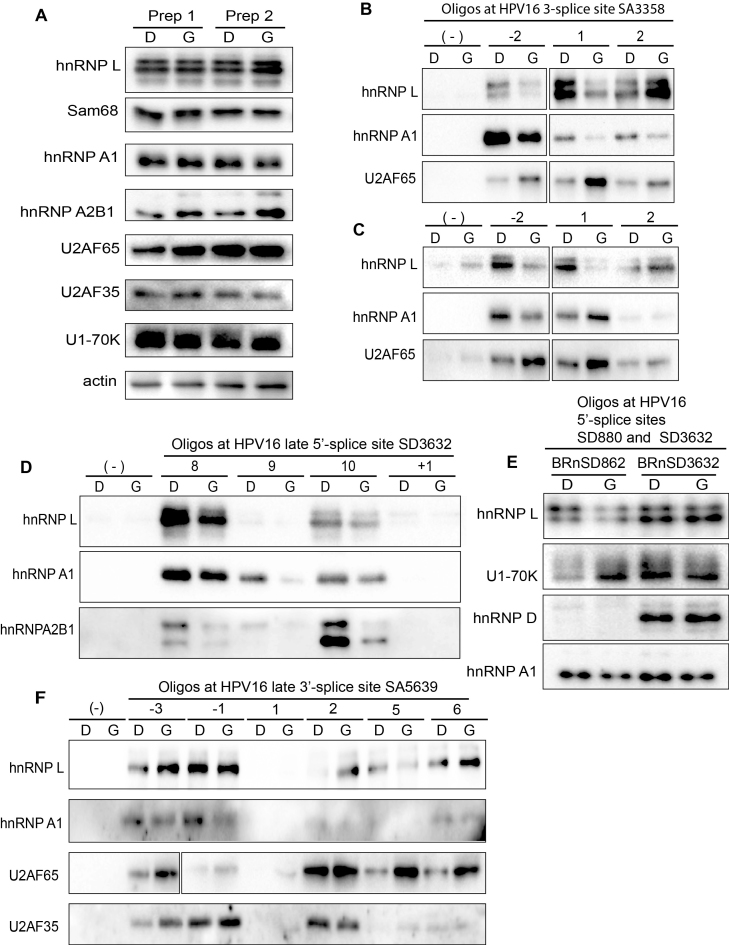
Having identified hot spots for RNA binding proteins at HPV16 splice sites used for production of L1 and L2 mRNAs, as well as at the HPV16 early polyA signal, we wished to investigate if pull down of these factors by their target sequences was affected by inhibition of the Akt kinase. We were particularly interested in hnRNP L since this protein apparently binds at strategic positions at a number of HPV16 RNA processing sites. In addition, we investigated core splicing factors such as U2AF65/35 and U1snRNP (U1–70K) and splicing regulatory factors hnRNP A1, hnRNP A2B1, hnRNP D and SRSF1 that we have shown previously are involved in control of HPV16 gene expression (48–50,55). Nuclear extracts were prepared from DMSO or Akt kinase inhibitor treated C33A2 cells and were shown to contain similar levels of these RNA binding proteins (Figure 6A). Pull downs with selected oligos at HPV16 3′-splice site SA3358 revealed that interactions of hnRNP L and hnRNP A1 with oligos upstream (oligo -2) or directly at the splice site (oligo 1) were reduced in extracts prepared from Akt kinase inhibitor-treated cells (Figure 6B). (For an overview of the location of the oligos see Figure 5A.) In contrast, interactions of U2A65 with these same oligos increased (Figure 6B). Similar results were obtained using RNA oligos (Figure 6C). However, interactions of hnRNP L with oligos downstream of SA3358 were either unaffected by Akt inhibitor GDC0068, or increased, but that did not substantially affect binding of U2AF65 (Figure 6B and C). Since splicing to SA3358 is connected to polyadenylation at pAE (56), we also investigated if interactions of hnRNP L with oligos at pAE was affected by Akt kinase inhibition. Pull down of hnRNP L with oligos at pAE was reduced in extracts from cells treated with Akt kinase inhibitor, but to a relatively low extent (Supplementary Figure S4A and B). We concluded that treatment of cells with Akt kinase inhibitor GDC0068 resulted in reduced binding of hnRNP L and hnRNP A1 to specific sites upstream of, or at SA3358, and that this resulted in increased binding of positive splicing factor U2AF65 to SA3358.
Figure 6.
(A) Western blots of various RNA binding proteins in two independent preparations of nuclear extracts (Prep 1 and Prep 2) prepared from C33A2 cells treated with DMSO (D) or 100 uM Akt kinase inhibitor GDC0068 (G) for 3 h. Western blots of indicated proteins pulled down with biotinylated ssDNA oligos (B) or RNA oligos (C) representing sequences at the HPV16 3′-splice site SA3358 in the E4-coding exon. (D) Western blots of indicated proteins pulled down with biotinylated ssDNA oligos representing sequences at the HPV16 late 5′-splice site SD3632 in the E4-coding exon. (E) Western blots of indicated proteins pulled down with biotinylated ssRNA oligos spanning HPV16 5′-splice sites SD880 (BRnSD862) and SD3632 (BRnSD3632). (F) Western blots of hnRNP L, hnRNP A1, U2AF65 or U2AF35 pulled down from nuclear extracts using ssDNA oligos at HPV16 late splice site SA5639. For all pull downs in the figure, nuclear extracts prepared from C33A2 cells treated with DMSO (D) or Akt kinase inhibitor GDC0068 (G) were used.
Reduced pull down of hnRNP L and hnRNP A1 with sequences upstream of HPV16 late splice site SD3632 with extracts prepared from Akt kinase inhibitor treated cells
Next we investigated interactions between hnRNP L and hnRNP A1 and RNA oligos at HPV16 late 5′-splice site SD3632 in the absence or presence of Akt kinase inhibitor. The results revealed that interactions between hnRNP L, hnRNP A1 and hnRNP A2B1 and oligos immediately upstream of SD3632 (oligos 8, 9, 10) also decreased in extracts from Akt inhibitor GDC0068-treated cells (Figure 6D), suggesting a role for these proteins in the control of HPV16 late 5′-splice site SD3632. At 5′-splice sites, core splicing factor U1snRNP, rather than U2AF65, defines the splice site. We therefore attempted to pull down U1snRNP, monitored here with Western blotting using antibody to the U1–70K protein. We used a number of ssDNA and RNA oligos spanning the two major HPV16 5′-splice sites SD880 and SD3632 (Supplementary Figure S4C). For HPV16 5′-splice site SD880, one RNA oligo (BRnSD862) and three partially overlapping ssDNA oligos (BSD856, BSD865, BSD876) were used (Supplementary Figure S4C). For HPV16 5′-splice site SD3632, two RNA oligos (BRnSD3632 and BRnSD3632opt) and two ssDNA oligos (BSD3632 and BSD3632opt) were used (Supplementary Figure S4C). U1snRNP binding to these oligos was monitored here by Western blotting to U1–70K, a component of U1snRNP. While none of the ssDNA oligos pulled down U1–70K (Supplementary Figure S4D), all RNA oligos did (Supplementary Figure S4D). We concluded that ssDNA oligos cannot replace RNA oligos in pull downs of U1snRNP. Optimization of the U1snRNA binding site in SD3632 as in RNA oligo BRnSD3632op increased pull down of U1–70K, as expected (Supplementary Figure S4D), which demonstrated specificity in these pull-down experiments. Next we investigated pull down of U1–70K by the HPV16 5′-splice sites SD880 and SD3632 in cell extracts from DMSO or Akt inhibitor GDC0068-treated C33A2 cells. Pull down of U1–70K by BRnSD862 that spans HPV16 5′-splice site SD880 increased in cell extract obtained from Akt inhibitor GDC0068-treated cells (Figure 6E), followed by a decrease in hnRNP L binding (Figure 6E). These results suggested that hnRNP L may control splicing from HPV16 5′-splice site SD880. However, binding of U1–70K to BRnSD3632 (representing HPV16 late 5′-splice site SD3632) appeared to be relatively unaffected by inhibition of the Akt kinase (Figure 6E). However, sequences upstream of SD3632 interacted also with hnRNP L and hnRNP A1 (Figure 6E), and binding of these factors was reduced after inhibition of Akt (Figure 6E). It is reasonable to speculate that these upstream sequences control binding of U1snRNP to SD3632 (41,56). We concluded that inhibition of the Akt kinase reduced binding of hnRNP L and hnRNP A1 to sequences upstream of SD3632 and immediately at SD880, and that this resulted in increased binding of U1snRNP to at least SD880.
Active HPV16 5′-splice site SD880 is associated with SRSF1 whereas suppressed splice site SD3632 is associated with hnRNP D
Pull-down of U1–70K was unexpectedly more efficient with BRnSD3632, which represents the suppressed HPV16 splice site SD3632, compared with BRnSD862, which represents the more active HPV16 splice site SD880 (Figure 6E). SD880 is located in the E1 coding region and is used for both early and late HPV16 mRNAs. Analysis of a number of splicing factors pulled down by oligos BRnSD862 and BRnSD3632 identified one protein, SRSF1, that preferentially interacted with active HPV16 5′-splice site SD880 (Supplementary Figure S4E). In contrast, splicing inhibitory factor hnRNP D interacted specifically with the suppressed HPV16 5′-splice site SD3632 (Figure 6E), as previously reported by us (41). Taken together, our results suggested that the intrinsic activity of SD880 and SD3632 correlated with binding of splicing enhancing protein SRSF1 to the former, and binding of splicing inhibitory factor hnRNP D to the latter, and that the activity of these HPV16 5′-splice sites could potentially be regulated by hnRNP L and hnRNP A1 binding either at, or nearby these splice sites.
Inhibition of the Akt kinase results in increased binding of U2AF65 at HPV16 SA5639
Next, we investigated if the Akt kinase inhibitor GDC0068 affected protein binding at HPV16 late 3′-splice site SA5639, including previously identified downstream splicing silencers in the L1-coding region (8,9,48,49). The major effect of the Akt inhibitor treatment of C33A2 cells was an increased binding of positive splicing factor U2AF65 to oligos representing sequences both upstream and downstream of SA5639 (Figure 6F). Although this correlated with less binding of hnRNP L to some oligos (Figure 6F), the major effect of the Akt inhibitor was reduced binding of hnRNP A1 upstream and downstream of SA5639 (Figure 6F). Reduced binding of hnRNP A2B1 was also observed (Supplementary Figure S4F). We concluded that inhibition of the Akt kinase resulted in increased binding of U2AF65 at HPV16 late 3′-splice site SA5639, and that this correlated with reduced binding of hnRNP A1 and hnRNP A2B1 to adjacent RNA sequences.
Reduced binding of hnRNP L, hnRNP A1, hnRNP A2B1 and hnRNP C1, and increased binding of U2AF65, Sam68 and PSF, to HPV16 mRNAs in cells treated with Akt kinase inhibitor GDC0086
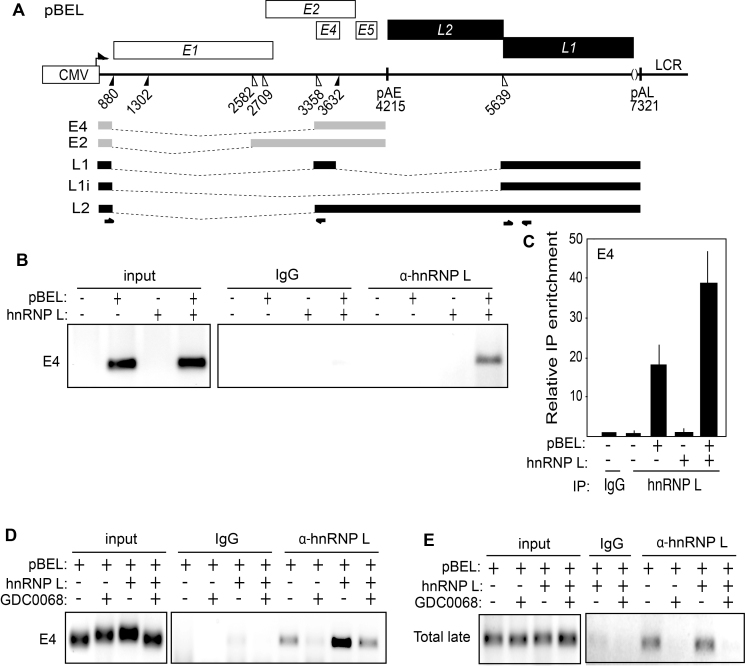
To investigate if the reduced interactions of hnRNP L, hnRNP A1 and hnRNP A2B1 with HPV16 oligos observed with extracts from cells treated with Akt kinase inhibitor GDC0068 also occurred in living cells, we performed CLIP experiments in which living cells treated with DMSO alone or Akt kinase inhibitor GDC0068 were subjected to UV cross linking of proteins to RNA followed by immunoprecipitation of RNA–protein complexes. We transfected 293T cells with pBEL plasmid (Figure 7A), in the absence or presence of hnRNP L expressing plasmid, followed by UV-irradiation and immunoprecipitation of RNA–protein complexes with anti-hnRNP L antibody. RT-PCR- and RT-qPCR-experiments revealed that hnRNP L is associated with HPV16 mRNAs in living cells and that this interaction is enhanced when hnRNP L is overexpressed by transient transfection (Figure 7B and C). Furthermore, treating the transfected cells with Akt kinase inhibitor GDC0068 resulted in reduced interactions between hnRNP L and the HPV16 mRNAs spliced between SD880 and SA3358 (E4) (Figure 7D and Supplementary Figure S5A and B). Similar results were obtained with HPV16 late mRNAs using RT-PCR primers that detected all HPV16 late mRNAs (L2, L1 and L1i mRNAs) (Total late) (Figure 7E). We also observed that the interaction of hnRNP L with HPV16 mRNAs in pBEL transfected 293T cells was negatively affected by GDC0068 (Figure 7D and E). When the association of hnRNP L with HPV16 mRNAs decreased after Akt kinase inhibitor GDC0068 treatment, so did binding of other RNA binding factors with inhibitory effects on HPV16 late gene expression and splicing such as hnRNP A1, hnRNP A2B1 and hnRNP C1/C2 (Supplementary Figure S5C and D). In contrast, interactions with other RNA-binding proteins such as U2AF65, PSF and Sam68 increased (Supplementary Figure S5C and D). We concluded that interactions with several hnRNP-proteins, including hnRNP L decreased in response to inhibition of the Akt kinase, while interactions with other proteins increased, suggesting that loss of binding of the various hnRNPs including hnRNP L paves the way for binding to HPV16 mRNAs of proteins with positive effect on HPV16 late gene expression.
Figure 7.
(A) Schematic representation of the HPV16 subgenomic expression plasmid pBEL. Transcription of the HPV16 sequences in the pBEL plasmid is driven by the human cytomegalovirus promoter (CMV). Rectangles represent open reading frames. Filled and open triangles represent 5′- and 3′-splices sites respectively, and HPV16 early and late polyA signals are indicated as pAE and pAL respectively. The HPV16 E4 and E2 mRNAs produced by pBEL upon transfection of mammalian cells is indicated in light grey. HPV16 late mRNAs L1, L1i and L2 that can be induced from the pBEL reporter plasmid are indicated in black. Positions of HPV16 RT-PCR primers are indicated. (B) Left panel: RT-PCR of HPV16 E4 mRNAs spliced from SD880 to SA3358 in 293T cells transfected with or without HPV16 subgenomic plasmid pBEL in the absence or presence of hnRNP L expressing plasmid pCMV-hnRNP L. The transfected cells were subjected to UV irradiation before harvest as detailed in Materials and methods for CLIP assay. Right panel: The extracts from the transfected cells were subjected to immunoprecipitation with IgG or anti-hnRNP L antibody followed by RNA extraction and RT-PCR with primers that detect E4 mRNAs spliced from SD880 to SA3358. (C) RT-qPCR on the RNA analysed in (B). (D and E) RT-PCR of HPV16 E4 mRNAs spliced from SD880 to SA3358 in 293T cells transfected with HPV16 subgenomic plasmid pBEL in the absence or presence of hnRNP L expressing plasmid pCMV-hnRNP L and in the absence or presence of 100uM Akt kinase inhibitor GDC0068 (GDC0068). The transfected cells were subjected to UV irradiation and extracts from the transfected cells were prepared and subjected to immunoprecipitation with IgG or anti-hnRNP L antibody followed by RNA extraction and RT-PCR with primers that detect HPV16 E4 mRNAs (D) or all HPV16 late mRNAs (Total late: HPV16 L2, L1 and L1i mRNAs).
Inhibition of Akt causes dephosphorylation of hnRNP L and reduces interactions of hnRNP L with hnRNP C1 and Sam68, but not with U2AF65
Since treatment of cells with the Akt inhibitor affected binding to HPV16 mRNAs of hnRNP L, hnRNP A1 and hnRNP C1 negatively, and binding to U2AF65 and Sam68 positively, one may speculate that the phosphorylation of at least one of these proteins must be affected by Akt inhibitor treatment. As can be seen in Figure 8A, immunoprecipitation with anti-phospho-epitope antibody followed by hnRNP L western blotting, or vice versa, revealed that hnRNP L is phosphorylated in C33A2 cells and that reduced levels of hnRNP L-phosphorylation were observed in cells treated with Akt inhibitor (Figure 8A). Low levels of hnRNP L parylation and acetylation were also detected, but these modifications were not affected by Akt inhibitor treatment of the cells (Figure 8A). Since our results indicated that hnRNP was phosphorylated by Akt, we performed an in vitro phosphorylation assay with recombinant Akt kinase and GST-hnRNP L that was expressed in bacteria and purified (Supplementary Figure S6A–C). As can be seen from the results, Akt kinase phosphorylated GST-hnRNP L in vitro (Figure 8B). This phosphorylation was blocked in the presence of Akt inhibitor GDC0068 (Figure 8B). We concluded that hnRNP L can be phosphorylated by the Akt kinase.
Figure 8.
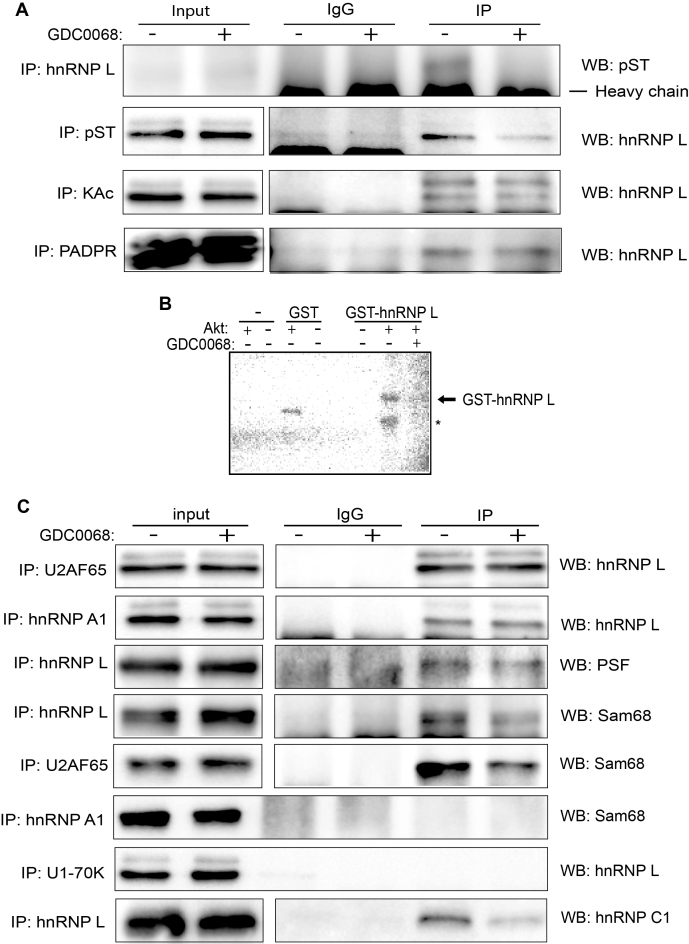
(A) Cell extracts from untreated (DMSO) (–) or 100uM Akt kinase inhibitor GDC0068-treated (+) C33A2 cells were subjected to immunoprecipitation with IgG or the indicated antibodies to hnRNP L, pST (serine/threonine phosphorylation), KAc (lysine acetylation) or PADPR (polyADP ribose), followed by Western blotting with antibodies to pST or hnRNP L. (B) Autoradiograph of in vitro phosphorylation reactions with recombinant Akt kinase in the presence of 32P-ATP and purified GST or GST-hnRNP L. Reactions were performed in the absence or presence of Akt inhibitor GDC0068 (1 uM). (C) Cell extracts from DMSO-treated (–) or Akt kinase inhibitor GDC0068-treated (+) C33A2 cells were subjected to immunoprecipitation with IgG or the indicated antibodies to U2AF65, hnRNP A1, hnRNP L or U1–70K, followed by Western blotting with antibodies to hnRNP L, PSF, Sam68 or hnRNP C1.
In an effort to explain how hnRNP L could control HPV16 3′-splice sites, we investigated if hnRNP L interacted with U2AF65 in C33A2 cells. Indeed, the results revealed that hnRNP L binds well to U2AF65 (Figure 8C), thereby connecting the splicing regulatory protein hnRNP L to the splicing factor U2AF65. In addition, hnRNP L interacted with hnRNP A1, an RNA-binding protein also associated with splicing inhibition, but these interactions were not affected by inhibition of the Akt kinase (Figure 8C). hnRNP L also interacted with splicing factors PSF and Sam68 (Figure 8C), proteins that displayed enhanced binding to HPV16 mRNAs in the presence of Akt inhibitor (Supplementary Figure S7A and S7B). Interactions between hnRNP L and Sam68 or PSF were reduced in the presence of Akt inhibitor (Figure 8C), suggesting that these interactions could contribute to the Akt kinase inhibitor induction of HPV16 late gene expression. The interaction between hnRNP L and Sam68 decreased over time in Akt kinase inhibitor treated cells (Supplementary Figure S7A), and was specific for hnRNP L since Sam68 did not interact with hnRNP A1 (Figure 8B). However, Sam68 also coimmunoprecipitated with U2AF65, an interaction that was reduced in response to Akt inhibitor treatment (Figure 8C). Since the interactions between Sam68 and the other factors could potentially be affected by Sam68 phosphorylation, we monitored Sam68 phosphorylation in the absence or presence of Akt kinase inhibitor, but phosphorylation of Sam 68 could not be detected (Supplementary Figure S7B). However, when we analysed various posttranslational modifications of Sam68, we found that Sam68 was ubiquitinated in response to Akt inhibition (Supplementary Figure S7B), suggesting that ubiquitination of Sam68 could interfere with its binding to hnRNP L and/or U2AF65. The reduced association of Sam68 with hnRNP L and U2AF65 could be either caused by hnRNP L dephosphorylation, or increased Sam68 ubiquitination, or both. Taken together, our results suggested that posttranslational modifications of hnRNP L and Sam68 that occurred in response to Akt kinase inhibition resulted in dissociation of these two factors from a splicing inhibitory complex. As a consequence, less hnRNP L, and more U2AF65 and Sam68 interacted with HPV16 mRNAs.
To investigate how hnRNP L could possibly control the 5′-splice sites of HPV16, we attempted co-immunoprecipitations of hnRNP L and U1snRNP (U1–70K), but interactions between the two were not detected (Figure 8C). We have previously reported that hnRNP C1 is involved in the regulation of the HPV16 late 5′-splice site SD3632 and that it binds to the HPV16 early UTR (51,52), suggesting that hnRNP C1 could be involved in SD3632 and pAE regulation in response to Akt kinase inhibition. Indeed, interactions between hnRNP L and hnRNP C1 were detected (Figure 8C), and these interactions were reduced in the presence of Akt kinase inhibitor (Figure 8C). These results suggested that hnRNP L could affect HPV16 early polyadenylation and splicing through interactions with hnRNP C1. Taken together, our results demonstrated that hnRNP L, hnRNP A1, hnRNP C1, Sam68 and U2AF65 are associated with each other in C33A2 cells. Inhibition of the Akt kinase alters the composition of this complex, which results in decreased binding of hnRNP L, hnRNP A1, hnRNP A2B1 and hnRNP C1 to HPV16 mRNAs while binding of U2AF65/35 and Sam68 to HPV16 mRNAs increases. This results in inhibition of HPV16 early polyA signal and activation of HPV16 splice sites, in particular the late splice sites SD3632 and SA5639, thereby activating HPV16 late gene expression.
siRNA mediated knock down of hnRNP L induces HPV16 late L1 and L2 gene expression
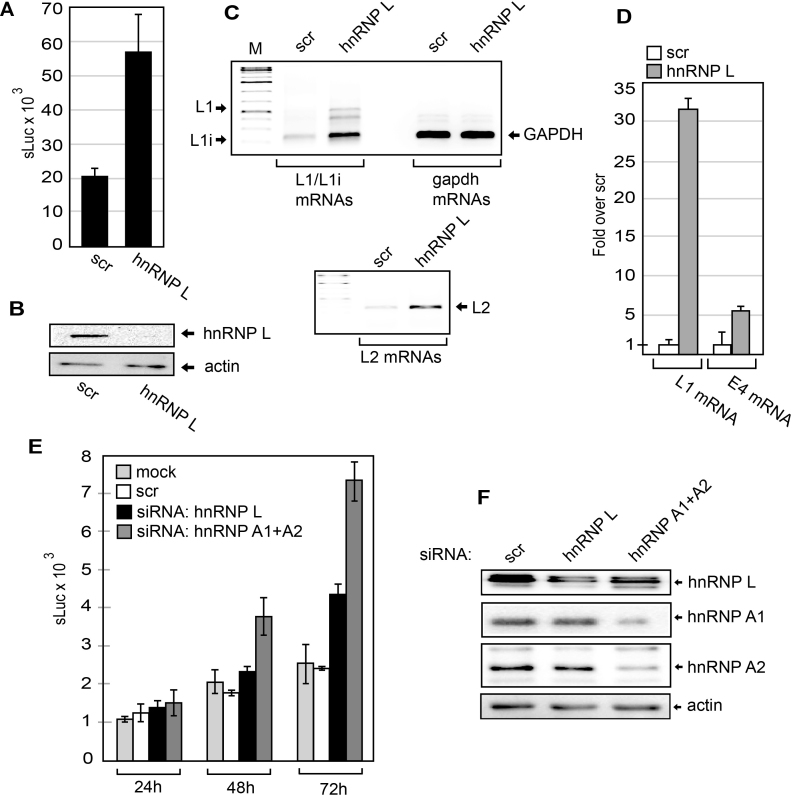
If hnRNP L indeed is a suppressor of HPV16 late gene expression, siRNA knock down of hnRNP L should activate HPV16 late gene expression. In addition to hnRNP L, we had identified various RNA binding proteins binding to ‘hot spots’ at HPV16 splices sites and polyA signal. To investigate their role in HPV16 gene regulation, we had siRNAs synthesized to the majority of all hnRNP proteins and SR proteins (see Materials and Methods) and transfected them into reporter cell line C33A2 in a 96-well format. The most significant de-repression of HPV16 late gene expression observed with siRNAs to a single protein was with siRNAs towards hnRNP L (Supplementary Figure S8). Re-ordering of the siRNA and transfection in larger scale verified the induction of sLuc (Figure 9A) by the siRNA mediated knock down of hnRNP L (Figure 9B). Analysis of HPV16 mRNAs in C33A2 cells after hnRNP L knock down revealed a significant upregulation of HPV16 L1 and L2 mRNAs (Figure 9C – upper and lower panels). RT-qPCR showed an approximately 30-fold increase in L1 mRNA levels, whereas E4 mRNA levels increased by a factor of five (Figure 9D). We were surprised that the majority of the siRNAs to single hnRNPs or SR-proteins had very weak or no effect on HPV16 gene expression (Supplementary Figure S8). In particular, hnRNP A1 that we have shown previously binds to splicing silencer elements in HPV16 L1 (49). However, both members of the SR protein- and hnRNP protein-families may have overlapping functions, as has been shown for hnRNP A1 and A2B1, and hnRNP H and hnRNP F. We therefore knocked down hnRNP A1 and hnRNP A2B1 individually and combined. The results revealed that knock down of both hnRNP A1 and hnRNP A2B1 showed a greater induction of HPV16 late gene expression than individual knock downs (Figure 9E and F), suggesting that they have overlapping function. We concluded that hnRNP L is one of the single most important cellular inhibitor of HPV16 late gene expression at the level of RNA processing.
Figure 9.
(A) sLuc activity in the cell culture medium of C33A2 cells transfected with scrambled siRNAs or siRNAs to hnRNP L from a different batch than the siRNA library. (B) Western blot of hnRNP L in C33A2 cells transfect with scrambled siRNA or siRNA to hnRNP L. (C) RT-PCR of HPV16 L1/L1i mRNAs, GAPDH mRNAs (upper panel) and HPV16 L2 mRNAs (lower panel) in C33A2 cells transfect with scrambled siRNAs or siRNAs to hnRNP L. (D) RT-qPCR of HPV16 L1 or E4 mRNAs in C33A2 cells transfect with scrambled siRNAs or siRNAs to hnRNP L. (E) sLuc activity in the cell culture medium of C33A2 cells at different time points after mock transfection or transfection with scrambled siRNA or siRNA to hnRNP A1+A2 or hnRNP L. (F) Western blot of hnRNP L, hnRNP A1, hnRNP A2 or actin in C33A2 cells transfected with scrambled siRNA (scr) or siRNAs to hnRNP L or hnRNP A1 + hnRNP A2.
Deletion or mutation of hnRNPL binding sites activates HPV16 late mRNA splicing
In order to determine the functional significance of the hnRNP L binding sites we utilised an HPV16 mini-exon plasmid (pE4EL1) (Figure 10A and B) in which we could determine the effect of hnRNP L binding sites on HPV16 mRNA splicing. Plasmid pE4EL1 encodes the following HPV16 splice sites SD880, SA3358, SD3632 and SA5639 (Figure 10B). The wild type plasmid is efficiently spliced between SD880 and SA3358 (Figure 10C), as expected, but not between the two late splice sites SD3632 and SA5639 as they are suppressed by adjacent splicing silencer elements (Figure 10D). Control plasmid pE4EL1M, in which previously identified splicing silencers in the L1 coding region had been inactivated by multiple point mutations as previously described, produced similar levels of E4s mRNAs as pE4EL1 (Figure 10C), but in addition high levels of spliced L1i mRNAs, as expected (Figure 10E). Deletion of the hnRNP L binding sites at SA3358 in pE4EL1, as in plasmid pE4D (Figure 10B), resulted in reduced splicing to SA3358 and the appearance of mRNAs unspliced between SD880 and SA3358 (Figure 10C). These mRNAs were also detected by primers 773s and L1as (Figure 10D). These results indicating that the deleted sequences G, M and T were required for efficient splicing to SA3358.
Next we deleted all identified hnRNP L binding sites in the pE4EL1 plasmid, resulting in pE4DL1D (Figure 10B). The pE4DL1D produced less of the E4s/L1 mRNA and more of the mRNAs utilizing HPV16 spliced site SA5639 such as the E4u and the L1i mRNAs (Figure 10C and D). Presumably due to the increased competition from SA5639 as the inhibitory hnRNP L binding sites at SA5639 had been deleted. Similar levels of spliced gapdh mRNAs were present in all samples (Figure 10F). We concluded that sequences to which hnRNP L binds at SA3358 (G, M, T) have a positive effect on splicing to SA3358, whereas hnRNP L binding sites at SD3632 appeared to play an inhibitory role (Figure 8B and 10B). In contrast, hnRNP L binding sites at SA5639 (-3, -1A, 2ABC) appeared to be largely inhibitory.
Since HPV16 late 5′-splice site SD3632 is also suppressed by hnRNP D and these binding sites are very close to the hnRNP L binding sites, we wished to determine if the hnRNP L binding site contributes to inhibition of SD3632. To this end we used a previously described CAT reporter plasmid named pBspliceMCAT that only contains the HPV16 late splice sites SD3632 and SA5639 (Supplementary Figure S9A). CAT production from pBspliceMCAT in transfected cells reflects the usage of SD3632 (Supplementary Figure S9B). We introduced deletions at the hnRNP L binding site identified with oligo 10 and fine mapped by oligo 10B (Supplementary Figure S9C). The results revealed that CAT production increased as the hnRNP L binding site was deleted, indicating that utilisation of HPV16 SD3632 increased (Supplementary Figure S9C). Since the hnRNP L binding site is so close to two previously identified inhibitory hnRNP D binding sites, we also introduced point mutations in the hnRNP L binding site to interfere as little as possible with the two hnRNP D binding sites (Supplementary Figure S9D). Also these nucleotide substitutions in the hnRNP L binding site resulted in enhanced splicing from SD3632 to SA5639 (Supplementary Figure S9D), supporting the idea that hnRNP L contributes to suppression of SD3632. Combined, our results indicated that mutational inactivation of hnRNP L binding sites at HPV16 late splice sites SD3632 and SA5639 enhanced HPV16 late gene expression.
Inhibition of Akt induces HPV16 late gene expression in HPV16-immortalized keratinocytes in the absence or presence of cell differentiation
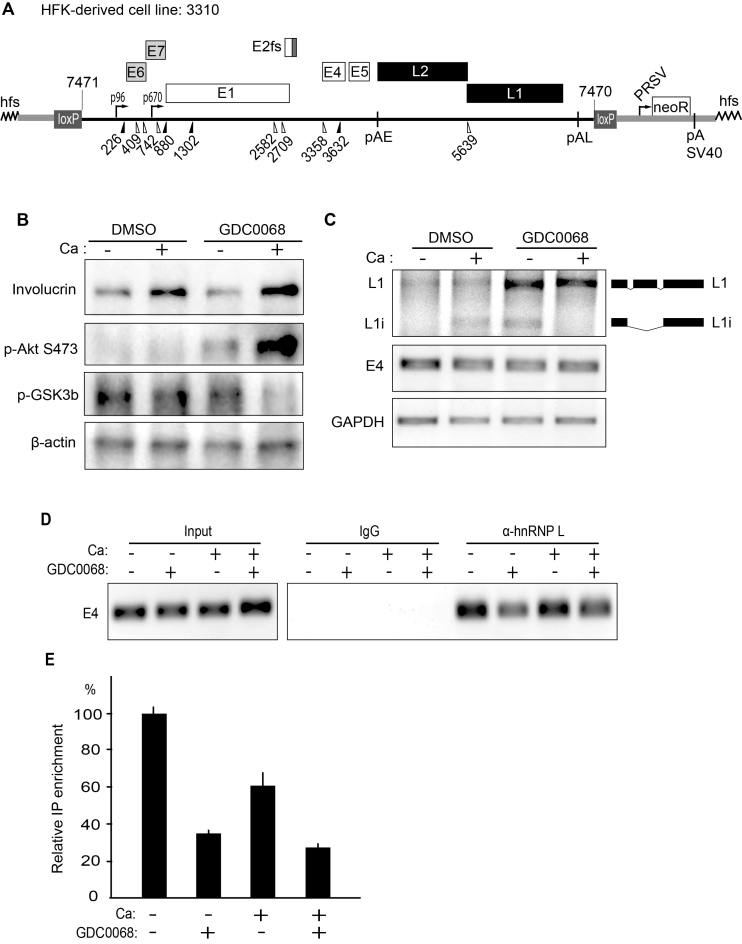
We wished to investigate if inhibition of the Akt kinase also activated HPV16 late gene expression in a differentiating environment, in cells that can be induced to differentiate. To this end we used 3310 cells, a human keratinocyte cell line that we had immortalized by transfection with a complete HPV16 genome (42). The HPV16 genome is integrated in such a manner that all HPV16 genes can be expressed (Figure 11A). Incubation of these cells with CaCl2 induced cell differentiation as evidenced by production of differentiation marker involucrin (Figure 10B). Addition of the Akt kinase inhibitor enhanced involucrin production (Figure 11B), and at the same time locking the Akt-kinase in a hyperphosphorylated inactive state (p-Akt S473), which is characteristic for Akt inhibitor GDC0068, and caused inhibition of GSK3 phosphorylation (Figure 11B) (45). More importantly, while the calcium-induced differentiation activated a relatively low production of HPV16 L1 mRNAs in 3310 cells (Figure 11C), inhibition of the Akt kinase, strongly induced HPV16 L1 mRNA levels, while E4 mRNA levels were relatively unaffected, as were GAPDH mRNA levels (Figure 11C). We concluded that inhibition of the Akt kinase enhanced cell differentiation by calcium and efficiently induced HPV16 late gene expression in HPV16-immortalized human keratinocytes.
Figure 11.
(A) Schematic representation of HPV16 plasmid pHPV16ANE2fs that is integrated in the genome of the transfected and immortalized human keratinocyte cell line 3310 (42). Open reading frames are represented as rectangles, the early and late promoters p97 and p670 are indicated as arrows, and splice sites as triangles. The long control region (LCR) and the early and late polyA signals pAE and pAL are indicated. Two loxP sites and two SphI restriction sites flanking the HPV16 genome are indicated, as is the RSV-neoR-SV40pA cassette. The E2 frame-shift mutation is indicated. (B) Western blot analysis of differentiation marker involucrin, phosphorylated Akt kinase (p-Akt S473) and the Akt phosphorylation substrate p-GSK3b and b-actin in the absence (DMSO) or presence of Akt inhibitor GDC0068, and in the absence (–) or presence (+) of Ca-induced differentiation. (C) RT-PCR of HPV16 L1, L1i, E4 and GAPDH mRNAs in the absence (DMSO) or presence of Akt inhibitor GDC0068, and in the absence (–) or presence (+) of Ca-induced differentiation. (D) Upper left panel: RT-PCR of HPV16 E4 mRNAs spliced from SD880 to SA3358 in 3310 cells grown in the absence (DMSO) or presence of Akt inhibitor GDC0068, and in the absence (–) or presence (+) of Ca-induced differentiation. Upper right panel: extracts from UV irradiated 3310 cells grown in the absence (DMSO) or presence of Akt inhibitor GDC0068, and in the absence (–) or presence (+) of Ca-induced differentiation were subjected to immunoprecipitation with IgG or anti-hnRNP L antibody followed by RNA extraction and RT-PCR with primers that detect E4 mRNAs spliced from SD880 to SA3358. (E) RT-qPCR on the HPV16 E4 mRNAs spliced from SD880 to SA3358 shown in (D).
Next we investigated if this induction of HPV16 late gene expression was accompanied by a reduced association of HPV16 mRNAs with hnRNP L. We therefore performed a CLIP experiment in which we immunoprecipitated hnRNP L-RNA complexes from 3310 cells and performed RT-PCR on the HPV16 mRNAs. The results revealed that hnRNP L was associated with HPV16 mRNAs in DMSO treated cells in the absence or presence of calcium-induced cell differentiation (Figure 11D). Induction of cell differentiation with calcium did not significantly affect the interaction between hnRNP L and HPV16 mRNAs, whereas incubation with Akt kinase inhibitor GDC0068 significantly reduced the interaction between hnRNP L and HPV16 mRNAs (Figure 11D). Taken together, our results demonstrate that inhibition of the Akt kinase in the absence or presence of human keratinocyte differentiation, reduces the association of hnRNP L with HPV16 mRNAs and activates HPV16 late gene expression. These results suggested that both differentiation and inhibition of Akt are required for activation of HPV16 late gene expression.
DISCUSSION
Here, we show that the Akt kinase inhibits HPV16 late gene expression at the level of RNA processing by controlling phosphorylation status and RNA-binding-ability of hnRNP L. Inhibition of the Akt kinase resulted in dephosphorylation of hnRNP L in C33A2 cells, and induction of HPV16 late gene expression as a result of altered HPV16 polyadenylation and splicing. The ability of Akt to control gene expression at the level of RNA processing through hnRNP L is not without precedence as it has been shown previously to occur also in non-small cell lung cancer (NSCLC) cells (57). Vu et al described reduced binding of dephosphorylated hnRNP L to caspase 9 mRNAs (57). In accordance with their results in NSCLC cells, we observed reduced binding of hnRNP L to HPV16 mRNAs upon dephosphorylation. Results published previously have shown that phosphorylation of hnRNP L by CaMKIV inhibits U2A65, suggesting that control of hnRNP L phosphorylation is a common mechanism for splicing regulation (58). Even if hnRNP L is phosphorylated by different kinases, the effects of hnRNP L phosphorylation appears to converge on U2AF65. The Akt inhibitors that induce HPV16 late gene expression are ATP competitive inhibitors of Akt. In contrast, PI3K- and PDK-inhibitors act on kinases upstream of Akt. They did not induce HPV16 late gene expression to the same extent as the Akt inhibitors GDC-0068 and AZD5363, suggesting that both PI3K and PDK1 activates Akt in the C33A cells. We also showed that Akt can phosphorylate hnRNP L in vitro, supporting the idea that direct phosphorylation of hnRNP L by Akt results in increased binding of hnRNP L to HPV16 mRNAs and suppression of HPV16 late gene expression. Not all Akt-kinase inhibitors functioned equally well, an observation that may be explained by differences in stability or uptake of the various molecules in cells or cell culture. We are not sure why the relatively high concentration of Akt inhibitor GDC0068 is needed to induce HPV16 late gene expression in C33A2 cells, but in cells other factors than affinity of the inhibitor for the target enzyme affect the efficiency of the inhibitor. For example, the bioavailability and activity of kinase inhibitors in cells depend on uptake, half-life and efflux of the inhibitor. Furthermore, the intracellular concentrations of various phosphatases may affect the inhibitory activity of a kinase inhibitor. Regarding GDC0068 used here, which is a highly selective ATP competitive pan-Akt inhibitor, it has been shown to inhibit other kinases belonging to AGC kinase family in vitro, but at least for PKA and PKC (two kinases that have been shown to phosphorylate RNA-binding proteins), GDC0068 had >100-fold higher selectivity for Akt than for PKA and PKC. It is therefore conceivable that GDC0068 inhibits other kinases as well, and that other RNA binding proteins than hnRNP L are phosphorylated by Akt, but we believe that our results combined support our conclusions that Akt phosphorylates hnRNP L and that both Akt and hnRNP L play important roles in the control of HPV16 late gene expression at the level of RNA processing. In addition, cervical cancers have been shown to carry mutations in genes coding for factors in the PI3K/Akt pathway and the C33A cell line is no exception with mutations in PI3KCA. One may speculate that such mutations affect the efficacy of some of the kinase inhibitors.
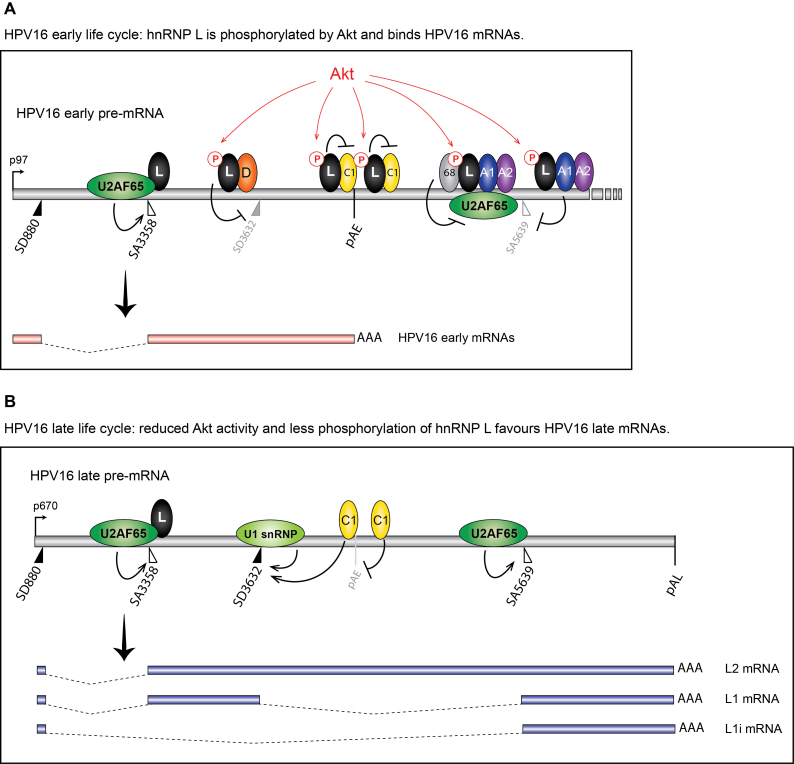
Previously published CLIP-seq results have shown that hnRNP L binds at both excluded and included exons on cellular mRNAs, suggesting a multifunctional role of hnRNP L in splicing regulation (59). On repressed exons, hnRNP L was binding upstream of both 5′- and 3′-splice sites (59). On included exons, hnRNP L interacted with sequences downstream of the 5′-and 3′-splice sites. It was suggested that hnRNP L suppresses 3′-splice sites by binding to the polypyrimidine tract upstream of the invariable AG-dinucleotide of the 3′-splice site. On the other hand, hnRNP L binding to intronic sequences downstream of 5′- splice sites correlated with splicing activation (59). At the active HPV16 3′-splice site SA3358, binding of hnRNP L is primarily downstream of SA3358 (Figure 3). Binding of hnRNP L upstream of SA3358 occurred with lower efficiency, which suggested a positive or neutral role of hnRNP L on HPV16 SA3358 (Figure 12).
Figure 12.
Model for the control of HPV16 late gene expression by Akt-kinase regulated RNA binding hnRNP-proteins. (A) At efficiently used HPV16 3′-splice site SA3358, hnRNP L (L) binds primarily downstream of the splice site in the exonic sequences and does not interfere with U2AF65/35 binding upstream of SA3358. This binding pattern suggest that hnRNP L does not inhibit SA3358, and is consistent with a positive role for hnRNP L on SA3358. At the same time, hnRNP L binds to splicing inhibitory sequences upstream of SD3632, adjacent to hnRNP D (D) binding sites that have been shown previously to suppress SD3632, which suggest a splicing inhibitory role for hnRNP L at SD3632. In addition, hnRNP L is pulled down by oligos that also bind to hnRNP C (C1) in the HPV16 early UTR, and hnRNP L interacts with hnRNP C. We suggest a model in which Akt-phosphorylated hnRNP L binds at HPV16 SA3358, SD3632 and pAE and that this prevents utilisation of HPV16 late 5′-splice site SD3632 and favour utilisation of HPV16 3′-splice site SA3358 and the HPV16 early polyadenylation signal pAE. At the other suppressed HPV16 late splice site SA5639, hnRNP L binds at both upstream and downstream sequences. The interactions of Sam68 (68), hnRNP A1 (A1), hnRNP A2 (A2) and hnRNP L (L) with sequences at HPV16 SA5639 is consistent with an inhibitory function of these proteins on splicing, most likely by inhibiting binding of U2AF65 at SA5639. This scenario represents the early state of the HPV16 life cycle. (B) As the HPV16 infected cells differentiate and Akt kinase activity is reduced, hnRNP L is dephosphorylated and binding to HPV16 mRNAs is reduced. We suggest a model for activation of HPV16 late gene expression in which interactions between hnRNP L and splicing silencer sequences at SD3632, the early UTR and with the hnRNP C are reduced or lost. Inhibition of Akt kinase dephosphorylates hnRNP L, thereby freeing hnRNP C at the early UTR, allowing it to interfere with hnRNP D at SD3632, which results in recognition of HPV16 SD3632 by U1snRNP activation of HPV16 L1 mRNA production. We have previously shown that overexpression of hnRNP C activates SD3632 in an HPV16 early UTR-dependent manner (41). Similarly, lost binding of hnRNP L at HPV16 late 3′-splice site SA5639 allows this splice site to interact with splicing factor U2AF65, thereby activating SA5639. Since each hnRNP L molecule can interact simultaneously with at least two RNA binding sites, and two RNA-bound hnRNP L proteins can interact with each other (68), one may speculate that multiple binding sites for hnRNP L enhances the inhibitory effect on HPV16 late splice site SA5639. The overall result of Akt inhibition and hnRNP L dephosphorylation, is that binding of hnRNP L to HPV16 mRNAs decreases and U2AF65 binding increases. This scenario represents the late state of the HPV16 life cycle. A1, hnRNP A1; A2, hnRNP A2; C1, hnRNP C; D, hnRNP D; L, hnRNP L; 68, Sam68; pAE, HPV16 early polyadenylation signal; pAL, HPV16 late polyadenylation signal.
Binding of hnRNP L to the suppressed HPV16 late 5′-splice site SD3632 occurs upstream of SD3632, with little or no binding downstream of SD3632, which suggests a repressive function of hnRNP L on HPV16 SD3632. hnRNP L binds preferentially to weak 5′-splice sites to execute hnRNP L-dependent splicing regulation (60). The efficient pull down of hnRNP L with oligos surrounding the suppressed HPV16 late 5′-splice site SD3632 supports this idea. Interestingly, hnRNP D was also efficiently pulled down by these sequences, confirming our previously published results that hnRNP D inhibits SD3632 (41). hnRNP D interacted specifically with SD3632, and not with SD880, while hnRNP L was more efficiently pulled down by the suppressed 5′-splice site SD3632 than the more active HPV16 5′-splice site SD880. Mutational inactivation of the hnRNP L binding sites activated late gene expression, which supports this idea. Taken together, our data indicate that hnRNP L contributes to suppression of HPV16 late mRNA splicing by interacting with SD3632 together with hnRNP D (Figure 12). However, other regulatory sequences are present in HPV16 exon E4 (43,46,47,56) but hnRNP L stands out in that it binds to multiple strategic sites at HPV16 splice sites SA3358, SD3632 and SA5639. hnRNP L binds both upstream and downstream of the suppressed HPV16 late 3′-splice site SA5639. The binding pattern at SA5639 is consistent with an inhibitory function of hnRNP L on L1 mRNA splicing with multiple binding sites upstream of SA5639 (59). Deletion of the hnRNP L binding sites at SA5639 activated HPV16 late gene expression, which supported our conclusion. Suppression of HPV16 late 5′-splice site SD3632 appears to be performed by hnRNP L and hnRNP D while suppression of late 3′-splice site SA5639 appears to be a result of hnRNP L, hnRNP A2/B1 and hnRNP A1 binding to splicing silencer elements (48–50). Combined knock down of hnRNP A1 and A2B1 induced HPV16 late gene expression. Interestingly, hnRNP L was also found in these complexes with hnRNP A1 and A2B1. Several articles describe an inhibitory role of hnRNP L in splicing (57,61,62). Interactions between hnRNP A1 and hnRNP L have been shown to repress exon 4 on the CD45 mRNAs, by suppressing a 5′-splice site (63). We concluded that hnRNP A1, hnRNP A2B1, hnRNP D and hnRNP L suppress HPV16 late mRNA splicing and that hnRNP L appeared to be the single most important factor (Figure 12).
hnRNP L binding has been reported to overlap with predicted microRNA binding sites in the 3′-UTR of cellular mRNAs (59). However, no microRNA binding sites have been identified in the HPV16 UTRs, nor have any been predicted, suggesting that hnRNP L binding at HPV16 pAE may have another function. hnRNP L binding at HPV16 pAE is not restricted to the UTR but is also detected immediately downstream of pAE, supports the idea of a function distinct from competition with microRNAs. Previously published results on the ASAHI mRNAs suggested that an internal polyA signal was used more frequently after hnRNP L knock-down (64). However, on these mRNAs the downstream polyA signal was the default choice, whereas in HPV16, the upstream polyA signal termed pAE is the most efficiently used polyA site, and the downstream late polyA signal pAL is suppressed (8,9). Utilization of HPV16 pAE is controlled by upstream UTR sequences as well as downstream elements (8,52–54), a property that is conserved in HPV31 (65,66). We have previously shown that hnRNP C binds the HPV16 early UTR and controls splicing and polyadenylation ((51) and data not shown). Here we show that hnRNP C and hnRNP L are interacting with each other in C33A2 cells, and that binding of hnRNP L to hnRNP C is reduced in the presence of Akt inhibitor, while interactions between hnRNP L and a number of other RNA binding proteins such as hnRNP A1 and U2AF65 is unaffected (Figure 8C). Our results therefore suggest that hnRNP L is involved in HPV16 polyadenylation regulation. A complex consisting of hnRNP L and hnRNP C binds to exon 10 on cellular MUSK mRNAs and prevents exon inclusion (67), suggesting that the hnRNP C-hnRNP L interaction could also include splicing regulation. Multiple hnRNP L proteins binding to an RNA molecule can also interact with each other to facilitate binding (68). Taken together, our results support a model in which binding of hnRNP L on each side of pAE promotes HPV16 early polyadenylation and suppresses HPV16 late splice site SD3632 (Figure 12). Phosphorylated hnRNP L may bind to hnRNP C1 and mask its polyadenylation inhibitory effects as well as its ability to activate upstream HPV16 late splice site SD3632. Upon hnRNP L dephosphorylation, hnRNP L and hnRNP C1 dissociate (Figure 8C) and hnRNP C1 is free to inhibit HPV16 early polyadenylation and to activate HPV16 late 5′-splice site SD3632 as described by us previously (51). Our results suggest that hnRNP L indirectly inhibits HPV16 late gene expression by promoting HPV16 early polyadenylation.
In addition to binding of hnRNP A1 and hnRNP L to exonic splicing silencer sequences in L1 downstream of SA5639, these proteins were also found in complexes upstream of SA5639, overlapping the polypyrimidine tract of this 3′-splice site (Figure 4). This is in stark contrast to the efficiently used 3′-splice site SA3358, at which U2AF65 binds efficiently upstream of SA3358, as expected, whereas binding of hnRNP L was undetectable (Figure 3). Furthermore, at SA3358, U2AF65 and U2AF35 are pulled-down together, supporting the idea that they form a splicing-active complex at SA3358, in the absence of hnRNP L. In contrast, oligos located upstream of SA5639, some of which covered the polypyrimidine tract, pulled down U2AF65/U2AF35 together with hnRNP L and hnRNP A1. These results suggested that hnRNP L and/or hnRNP A1 were either competing with splicing factor U2A65 for its binding sites upstream of SA5639, or binding to the U2AF65 protein to inhibit its function. Competition between hnRNP L and U2AF65 has been suggested to account for the inhibitory effect of hnRNP L on splicing (69). Position-dependent effects of hnRNP L have been described (70), supporting the idea that hnRNP L may have different effects on the efficiently used SA3358 and the suppressed SA5639. From our results it appeared that oligos located upstream of SA5639 and that efficiently pulled down hnRNP L also efficiently pulled down U2AF65. These results suggested that hnRNP L and U2AF65 interacted with each other. Indeed, co-immunoprecipitations revealed that hnRNP L interacted with U2AF65 (Figure 8). However, these protein-protein interactions were unaffected by the inhibition of Akt kinase. A more likely mechanism for the inhibition of HPV16 late 3′-splice site SA5639 is that hnRNP L and U2AF65 form a splicing inactive complex at SA5639 and that this complex was affected by reduced binding of hnRNP L to HPV16 mRNA in the Akt inhibitor treated cells (Figure 12). This is not without precedence since signal regulated RNA occupancy of U2AF has been described (71).
Regarding the role of hnRNP L in the HPV16 replication cycle we note that expression of hnRNP L shows an inverse correlation to cervical cell differentiation and thereby also to HPV16 late gene expression (72), which is consistent with an inhibitory role for hnRNP L in HPV16 late gene expression during the HPV16 life cycle. Cervical cancer cells also express high levels of hnRNP L (72) and hnRNP L appears to be required for the oncogenic process also in oral squamous cell carcinoma (OSCC) cells (73). Since HPV16 late gene expression is suppressed in cancer cells, these observations support the idea that hnRNP L functions as a suppressor of HPV16 late gene expression. Our results also suggest that a state of inhibition of HPV16 late gene expression is partially maintained by the Akt kinase. Indeed, our experiments showed that inhibition of the Akt kinase was an efficient way of inducing HPV16 late gene expression in HPV16 immortalized (but not transformed) human keratinocytes (Figure 11).
In conclusion, we show that inhibition of the Akt kinase induces HPV16 late gene expression. Inhibition of Akt results in inhibition of HPV16 early polyadenylation and activation of HPV16 late L1 mRNA splicing. These effects are mediated at least partly by the RNA-binding protein hnRNP L. Binding of hnRNP L to HPV16 mRNAs favors HPV16 early mRNA processing and prevents HPV16 late mRNA splicing. Inhibition of Akt and loss of hnRNP L phosphorylation results in less hnRNP L-binding to HPV16 mRNAs, which results in activation of HPV16 late mRNA processing. We speculate that inhibition of the Akt kinase is a prerequisite for induction of HPV16 late gene expression (Figure 12).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr K.W. Lynch for providing us with the GST-hnRNP L plasmid.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council-Medicine [VR2015-02388]; Swedish Cancer Society [CAN2015/519]; Gunnar Nilsson Cancer Research Trust Fund. Funding for open access charge; Swedish Research Council-Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chow L.T., Broker T.R., Steinberg B.M.. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010; 118:422–449. [DOI] [PubMed] [Google Scholar]
- 2. Schiffman M., Doorbar J., Wentzensen N., de Sanjose S., Fakhry C., Monk B.J., Stanley M.A., Franceschi S.. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers. 2016; 2:16086. [DOI] [PubMed] [Google Scholar]
- 3. Bulkmans N.W., Berkhof J., Bulk S., Bleeker M.C., van Kemenade F.J., Rozendaal L., Snijders P.J., Meijer C.J. POBASCAM Study Group . High-risk HPV type-specific clearance rates in cervical screening. Br. J. Cancer. 2007; 96:1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mighty K.K., Laimins L.A.. The role of human papillomaviruses in oncogenesis. Rec. Results Cancer Res. 2014; 193:135–148. [DOI] [PubMed] [Google Scholar]
- 5. Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N.. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999; 189:12–19. [DOI] [PubMed] [Google Scholar]
- 6. Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L. et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009; 10:321–322. [DOI] [PubMed] [Google Scholar]
- 7. Langsfeld E., Laimins L.A.. Human papillomaviruses: research priorities for the next decade. Trends Cancer. 2016; 2:234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johansson C., Schwartz S.. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat. Rev. Microbiol. 2013; 11:239–251. [DOI] [PubMed] [Google Scholar]
- 9. Schwartz S. Papillomavirus transcripts and posttranscriptional regulation. Virology. 2013; 445:187–196. [DOI] [PubMed] [Google Scholar]
- 10. Kajitani N., Satsuka A., Kawate A., Sakai H.. Productive lifecycle of human papillomaviruses that depends upon squamous epithelial differentiation. Front. Microbiol. 2012; 3:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McBride A.A. The papillomavirus E2 proteins. Virology. 2013; 445:57–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thierry F. Transcriptional regulation of the papillomavirus oncogenes by cellular and viral transcription factors in cervical carcinoma. Virology. 2009; 384:375–379. [DOI] [PubMed] [Google Scholar]
- 13. Kadaja M., Silla T., Ustav E., Ustav M.. Papillomavirus DNA replication—from initiation to genomic instability. Virology. 2009; 384:360–368. [DOI] [PubMed] [Google Scholar]
- 14. Johansson C., Somberg M., Li X., Backström Winquist E., Fay J., Ryan F., Pim D., Banks L., Schwartz S.. HPV-16 E2 contributes to induction of HPV-16 late gene expression by inhibiting early polyadenylation. EMBO J. 2012; 13:3212–3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graham S.V. Human papillomavirus E2 protein: linking replication, transcription, and RNA processing. J. Virol. 2016; 90:8384–8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vosa L., Sudakov A., Remm M., Ustav M., Kurg R.. Identification and analysis of papillomavirus E2 protein binding sites in the human genome. J. Virol. 2012; 86:348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pentland I., Parish J.L.. Targeting CTCF to control virus gene expression: a common theme amongst diverse DNA viruses. Viruses. 2015; 7:3574–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia R., Zheng Z.M.. Regulation of bovine papillomavirus type 1 gene expression by RNA processing. Front. Biosci. 2009; 14:1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kajitani N., Schwartz S.. RNA binding proteins that control human papillomavirus gene expression. Biomolecules. 2015; 5:758–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henken F.E., Banerjee N.S., Snijders P.J., Meijer C.J., De-Castro Arce J., Rosl F., Broker T.R., Chow L.T., Steenbergen R.D.. PIK3CA-mediated PI3-kinase signalling is essential for HPV-induced transformation in vitro. Mol. Cancer. 2011; 10:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pim D., Massimi P., Dilworth S.M., Banks L.. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene. 2005; 24:7830–7838. [DOI] [PubMed] [Google Scholar]
- 22. Menges C.W., Baglia L.A., Lapoint R., McCance D.J.. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 2006; 66:5555–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma Y.Y., Wei S.J., Lin Y.C., Lung J.C., Chang T.C., Whang-Peng J., Liu J.M., Yang D.M., Yang W.K., Shen C.Y.. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000; 19:2739–2744. [DOI] [PubMed] [Google Scholar]
- 24. Bertelsen B.I., Steine S.J., Sandvei R., Molven A., Laerum O.D.. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: frequent PIK3CA amplification and AKT phosphorylation. Int. J. Cancer. 2006; 118:1877–1883. [DOI] [PubMed] [Google Scholar]
- 25. Vande Pol S.B., Klingelhutz A.J.. Papillomavirus E6 oncoproteins. Virology. 2013; 445:115–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roman A., Munger K.. The papillomavirus E7 proteins. Virology. 2013; 445:138–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DiMaio D., Petti L.M.. The E5 proteins. Virology. 2013; 445:99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Calautti E., Li J., Saoncella S., Brissette J.L., Goetinck P.F.. Phosphoinositide 3-kinase signaling to Akt promotes keratinocyte differentiation versus death. J. Biol. Chem. 2005; 280:32856–32865. [DOI] [PubMed] [Google Scholar]
- 29. Wu J., Chen C., Zhao K.N.. Phosphatidylinositol 3-kinase signaling as a therapeutic target for cervical cancer. Curr. Cancer Drug Targets. 2013; 13:143–156. [DOI] [PubMed] [Google Scholar]
- 30. White E.S., Sagana R.L., Booth A.J., Yan M., Cornett A.M., Bloomheart C.A., Tsui J.L., Wilke C.A., Moore B.B., Ritzenthaler J.D. et al. Control of fibroblast fibronectin expression and alternative splicing via the PI3K/Akt/mTOR pathway. Exp. Cell Res. 2010; 316:2644–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang K., Patel N.A., Watson J.E., Apostolatos H., Kleiman E., Hanson O., Hagiwara M., Cooper D.R.. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 2009; 150:2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blaustein M., Pelisch F., Tanos T., Munoz M.J., Wengier D., Quadrana L., Sanford J.R., Muschietti J.P., Kornblihtt A.R., Caceres J.F. et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 2005; 12:1037–1044. [DOI] [PubMed] [Google Scholar]
- 33. Patel N.A., Kaneko S., Apostolatos H.S., Bae S.S., Watson J.E., Davidowitz K., Chappell D.S., Birnbaum M.J., Cheng J.Q., Cooper D.R.. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J. Biol. Chem. 2005; 280:14302–14309. [DOI] [PubMed] [Google Scholar]
- 34. Shultz J.C., Goehe R.W., Wijesinghe D.S., Murudkar C., Hawkins A.J., Shay J.W., Minna J.D., Chalfant C.E.. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res. 2010; 70:9185–9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Z., Qiu J., Liu W., Zhou Y., Plocinik R.M., Li H., Hu Q., Ghosh G., Adams J.A., Rosenfeld M.G. et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell. 2012; 47:422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang P., Zhou Z., Hu A., Ponte de Albuquerque C., Zhou Y., Hong L., Sierecki E., Ajiro M., Kruhlak M., Harris C. et al. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol. Cell. 2014; 54:378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanidas I., Polytarchou C., Hatziapostolou M., Ezell S.A., Kottakis F., Hu L., Guo A., Xie J., Comb M.J., Iliopoulos D. et al. Phosphoproteomics screen reveals akt isoform-specific signals linking RNA processing to lung cancer. Mol. Cell. 2014; 53:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Prasad J., Colwill K., Pawson T., Manley J.L.. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 1999; 19:6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu X., Mayeda A., Tao M., Zheng Z.M.. Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 2003; 77:2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosenberger S., De-Castro Arce J., Langbein L., Steenbergen R.D.M., Rösl F.. Alternative splicing of human papillomavirus type-16 E6/E6* early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:7006–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li X., Johansson C., Glahder J., Mossberg A.K., Schwartz S.. Suppression of HPV-16 late L1 5′-splice site SD3632 by binding of hnRNP D proteins and hnRNP A2/B1 to upstream AUAGUA RNA motifs. Nucleic Acids Res. 2013; 22:10488–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansson C., Jamal Fattah T., Yu H., Nygren J., Mossberg A.K., Schwartz S.. Acetylation of intragenic histones on HPV16 correlates with enhanced HPV16 gene expression Virology. 2015; 482:244–259. [DOI] [PubMed] [Google Scholar]
- 43. Li X., Johansson C., Cardoso-Palacios C., Mossberg A., Dhanjal S., Bergvall M., Schwartz S.. Eight nucleotide substitutions inhibit splicing to HPV-16 3′-splice site SA3358 and reduce the efficiency by which HPV-16 increases the life span of primary human keratinocytes. PLoS One. 2013; 8:e72776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dignam J.D., Lebovitz R.M., Roeder R.G.. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983; 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin J., Sampath D., Nannini M.A., Lee B.B., Degtyarev M., Oeh J., Savage H., Guan Z., Hong R., Kassees R. et al. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin. Cancer Res. 2013; 19:1760–1772. [DOI] [PubMed] [Google Scholar]
- 46. Jia R., Liu X., Tao M., Kruhlak M., Guo M., Meyers C., Baker C.C., Zheng Z.M.. Control of the papillomavirus early-to-late switch by differentially expressed SRp20. J. Virol. 2009; 83:167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Somberg M., Schwartz S.. Multiple ASF/SF2 sites in the HPV-16 E4-coding region promote splicing to the most commonly used 3′-splice site on the HPV-16 genome. J. Virol. 2010; 84:8219–8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao X., Fay J., Lambkin H., Schwartz S.. Identification of a 17-nucleotide splicing enhancer in HPV-16 L1 that counteracts the effect of multiple hnRNP A1-binding splicing silencers. Virology. 2007; 369:351–363. [DOI] [PubMed] [Google Scholar]
- 49. Zhao X., Rush M., Schwartz S.. Identification of an hnRNP A1 dependent splicing silencer in the HPV-16 L1 coding region that prevents premature expression of the late L1 gene. J. Virol. 2004; 78:10888–10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao X., Schwartz S.. Inhibition of HPV-16 L1 expression from L1 cDNAs correlates with the presence of hnRNP A1 binding sites in the L1 coding region. Virus Genes. 2008; 36:45–53. [DOI] [PubMed] [Google Scholar]
- 51. Dhanjal S., Kajitani N., Glahder J., Mossberg A.K., Johansson C., Schwartz S.. Heterogeneous nuclear ribonucleoprotein C proteins interact with the human papillomavirus type 16 (HPV16) early 3′-untranslated region and alleviate suppression of HPV16 Late L1 mRNA splicing. J. Biol. Chem. 2015; 290:13354–13371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao X., Öberg D., Rush M., Fay J., Lambkin H., Schwartz S.. A 57 nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2 and PTB. J. Virol. 2005; 79:4270–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oberg D., Collier B., Zhao X., Schwartz S.. Mutational inactivation of two distinct negative RNA elements in the human papillomavirus type 16 L2 coding region induces production of high levels of L2 in human cells. J. Virol. 2003; 77:11674–11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oberg D., Fay J., Lambkin H., Schwartz S.. A downstream polyadenylation element in human papillomavirus type 16 encodes multiple GGG-motifs and interacts with hnRNP H. J. Virol. 2005; 79:9254–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosenberger S., De-Castro Arce J., Langbein L., Steenbergen R.D., Rosl F.. Alternative splicing of human papillomavirus type-16 E6/E6* early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:7006–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rush M., Zhao X., Schwartz S.. A splicing enhancer in the E4 coding region of human papillomavirus type 16 is required for early mRNA splicing and polyadenylation as well as inhibition of premature late gene expression. J. Virol. 2005; 79:12002–12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vu N.T., Park M.A., Shultz J.C., Goehe R.W., Hoeferlin L.A., Shultz M.D., Smith S.A., Lynch K.W., Chalfant C.E.. hnRNP U enhances caspase-9 splicing and is modulated by AKT-dependent phosphorylation of hnRNP L. J. Biol. Chem. 2013; 288:8575–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu G., Razanau A., Hai Y., Yu J., Sohail M., Lobo V.G., Chu J., Kung S.K., Xie J.. A conserved serine of heterogeneous nuclear ribonucleoprotein L (hnRNP L) mediates depolarization-regulated alternative splicing of potassium channels. J. Biol. Chem. 2012; 287:22709–22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rossbach O., Hung L.H., Khrameeva E., Schreiner S., Konig J., Curk T., Zupan B., Ule J., Gelfand M.S., Bindereif A.. Crosslinking-immunoprecipitation (iCLIP) analysis reveals global regulatory roles of hnRNP L. RNA Biol. 2014; 11:146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shankarling G., Cole B.S., Mallory M.J., Lynch K.W.. Transcriptome-wide RNA interaction profiling reveals physical and functional targets of hnRNP L in human T cells. Mol. Cell. Biol. 2014; 34:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cole B.S., Tapescu I., Allon S.J., Mallory M.J., Qiu J., Lake R.J., Fan H.Y., Fu X.D., Lynch K.W.. Global analysis of physical and functional RNA targets of hnRNP L reveals distinct sequence and epigenetic features of repressed and enhanced exons. RNA. 2015; 21:2053–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Loh T.J., Cho S., Moon H., Jang H.N., Williams D.R., Jung D.W., Kim I.C., Ghigna C., Biamonti G., Zheng X. et al. hnRNP L inhibits CD44 V10 exon splicing through interacting with its upstream intron. Biochim. Biophys. Acta. 2015; 1849:743–750. [DOI] [PubMed] [Google Scholar]
- 63. Chiou N.T., Shankarling G., Lynch K.W.. hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol. Cell. 2013; 49:972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hung L.H., Heiner M., Hui J., Schreiner S., Benes V., Bindereif A.. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008; 14:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Terhune S.S., Hubert W.G., Thomas J.T., Laimins L.A.. Early polyadenylation signals of human papillomavirus type 31 negatively regulate capsid gene expression. J. Virol. 2001; 75:8147–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Terhune S.S., Milcarek C., Laimins L.A.. Regulation of human papillomavirus 31 polyadenylation during the differentiation-dependent life cycle. J. Virol. 1999; 73:7185–7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nasrin F., Rahman M.A., Masuda A., Ohe K., Takeda J., Ohno K.. HnRNP C, YB-1 and hnRNP L coordinately enhance skipping of human MUSK exon 10 to generate a Wnt-insensitive MuSK isoform. Sci. Rep. 2014; 4:6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang W., Zeng F., Liu Y., Zhao Y., Lv H., Niu L., Teng M., Li X.. Crystal structures and RNA-binding properties of the RNA recognition motifs of heterogeneous nuclear ribonucleoprotein L: insights into its roles in alternative splicing regulation. J. Biol. Chem. 2013; 288:22636–22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Heiner M., Hui J., Schreiner S., Hung L.H., Bindereif A.. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010; 7:56–64. [DOI] [PubMed] [Google Scholar]
- 70. Motta-Mena L.B., Heyd F., Lynch K.W.. Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol. Cell. 2010; 37:223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tisserant A., Konig H.. Signal-regulated Pre-mRNA occupancy by the general splicing factor U2AF. PLoS One. 2008; 3:e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 73. Jia R., Li X., Yu C., Fan M., Guo J.. The splicing factor hnRNP C regulates expression of co-stimulatory molecules CD80 and CD40 in dendritic cells. Immunol. Lett. 2013; 153:27–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.