Abstract
DNA mismatch repair (MMR) is a highly-conserved DNA repair mechanism, whose primary role is to remove DNA replication errors preventing them from manifesting as mutations, thereby increasing the overall genome stability. Defects in MMR are associated with increased cancer risk in humans and other organisms. Here, we characterize the interaction between MMR and a proofreading-deficient allele of the human replicative DNA polymerase delta, PolδD316A;E318A, which has a higher capacity for strand displacement DNA synthesis than wild type Polδ. Human cell lines overexpressing PolδD316A;E318A display a mild mutator phenotype, while nuclear extracts of these cells exhibit reduced MMR activity in vitro, and these defects are complemented by overexpression or addition of exogenous human Exonuclease 1 (EXO1). By contrast, another proofreading-deficient mutant, PolδD515V, which has a weaker strand displacement activity, does not decrease the MMR activity as significantly as PolδD316A;E318A. In addition, PolδD515V does not increase the mutation frequency in MMR-proficient cells. Based on our findings, we propose that the proofreading activity restricts the strand displacement activity of Polδ in MMR. This contributes to maintain the nicks required for EXO1 entry, and in this manner ensures the dominance of the EXO1-dependent MMR pathway.
INTRODUCTION
High fidelity DNA replication is essential to maintain a low genomic mutation rate and to maintain the viability and fitness of all cells and organisms. Genomic mutation rate is determined by three factors/processes and their relative efficiency/accuracy: (i) base-selection/insertion by replicative DNA polymerases during semi-conservative DNA replication; (ii) the intrinsic proofreading 3′-exonuclease of the replicative DNA polymerases and (iii) post-replicative DNA mismatch repair (MMR) (1,2).
In eukaryotic cells MMR occurs in three steps. The first step is mismatch recognition by MutSα (MSH2/MSH6) or MutSβ (MSH2/MSH3), where MutSα preferentially recognizes single base mismatches and small insertions/deletions (indels) and MutSβ preferentially recognizes large indels. In the second step, additional MMR proteins and MMR accessory factors, including MutLα (MLH1/PMS2), are recruited to the site of the mismatch, and newly-synthesized DNA including the mismatch is excised by exonuclease 1 (EXO1). Subsequently, the ssDNA gap is filled-in by DNA polymerase delta (Polδ) and ligated by DNA ligase I. Proliferative cell nuclear antigen (PCNA) and its clamp loader replication factor C (RFC) are required during mismatch recognition, excision and resynthesis (3–5). Cells with defects in MMR generally display a mutator phenotype and increased susceptibility to cancer (6–8).
Human MMR has been reconstituted in vitro using purified proteins and single mismatch-containing heteroduplex DNA. In the in vitro MMR assay, a nick/gap 5′ or 3′ to the mismatch is the strand discrimination signal, and it is required for correct and efficient MMR (9,10). In eukaryotic cells, EXO1 is the only excision nuclease known to carry out DNA excision during MMR (11). EXO1 belongs to the Rad2 gene family, and it is a 5′-3′ exonuclease (12). In the current MMR model, when the strand discrimination signal is a nick 3′ to the mismatch, the latent endonuclease associated with MutLα introduces a 5′ nick, where EXO1 initiates DNA excision and removes the mismatch (13,14). Deletion of EXO1 in mouse and yeast cells results in a weaker mutator phenotype than deletion of MSH2, and leaves significant residual MMR activity (15–17). This suggests that cells express an alternate EXO1-independent MMR mechanism/pathway (19).
Polδ carries out lagging strand DNA replication in eukaryotic cells and may play a role in replication of the leading strand in yeast, implicating its prominent role in genomic replication (18–20). In mammalian cells, Polδ is comprised of four subunits, with the largest subunit, p125, encoding polymerase and 3′- exonuclease activity (21). In mice, defects in the polymerase or exonuclease activities of Polδ increase the mutation rate and the incidence of cancer (22–25). In humans, recent studies show that germline mutations in the proofreading domain of Polδ predispose to cancer (26–28). Polδ also plays a role in DNA base excision repair (BER), nucleotide excision repair (NER), and double-strand break (DSB) repair (29–34) as well as translesion synthesis (TLS) and maturation of Okazaki fragments (35–37). The 3′- exonuclease activity of Polδ is also implicated in BER, TLS, compensation for exonuclease-deficient Polymerase α (Polα) and regulation of strand displacement (30,35,38–40).
The strand displacement activity of yeast Polδ is well characterized (17) and may complement the MMR defect in EXO1-deficient cells. It is also thought that the strand displacement activity of Polδ is negatively regulated by the Polδ 3′- exonuclease activity (37,40–42). Consistent with this, in human cells, an alternative form of Polδ, Polδ3, that lacks the p12 subunit possesses stronger 3′- exonuclease activity and weaker strand displacement activity than wild type Polδ (43). Furthermore, Polδ is required for the D-loop extension in homologous recombination, which is highly-dependent on strand displacement (44). Yeast cells depleted for EXO1 that have a defect in the 3′- exonuclease of Polδ display lower viability and higher mutation rate than wild type, which is consistent with the proposed role of the Polδ 3′-exonuclease in MMR (23,45). Therefore, we investigated the role of 3′-5′ exonuclease (proofreading) activity of human Polδ in MMR to expand our understanding of Polδ in MMR in addition to the basic function in resynthesis of the excision gap. Our results suggest that the proofreading activity of Polδ plays a role in shunting MMR to an EXO1-dependent excision pathway as opposed to directly participating in gap formation via its 3′-5′ exonuclease activity.
MATERIALS AND METHODS
Construction of plasmids
The plasmid expressing wild type Polδ was constructed by sub-cloning the coding region of POLD1, encoding p125 human Polδ from plasmid pVL1393-P125. A mutation was introduced to change the last codon of the p125 open reading frame in POLD1 from ‘stop’ to a glycine codon, generating the construct pcDNA3.1A (–)-POLD1-WT-his6. This plasmid generates wild type p125 with a terminal 6x histidine tag, and it was subsequently mutagenized to introduce D316A and E318A substitution mutations. The resulting plasmid is referred to as pcDNA3.1A (-)-POLD1-D316A;E318A-his6. The QuikChange site-directed mutagenesis kit (Stratagene) was used according to manufacturer's instructions. The DNA sequences of the entire coding regions of POLD1 and mutated POLD1 were verified by sequencing (Macrogen, Korea). Primers used for mutagenesis were as follows: pcDNA3.1A (-)-POLD1 stop codon substitution: Forward, 5′ ACCTGAGGCCTGGGGACATATGGGATCCGA, Reverse, 5′ TCGGATCCCATATGTCCCCAGGACTCAGGT. Primers for D316A and E318A: Forward, 5′ TGCTCAGCTTCGCTATCGCGTGCGCCGGCCGCAAA, Reverse, 5′ TTTGCGGCCGGCGCACGCGATAGCGAAGCTGAGCA.
Transfection, preparation of nuclear extracts, quantitative real-time PCR and western blotting
Approximately 8 × 105 HeLa cells (Clontech) were seeded in T75 flasks (Sigma). After 24 h, cells were transfected with 7 μg plasmid DNA using Polyjet transfection reagent (SignaGen Laboratories) according to the manufacturer's instructions. The nuclear extracts were prepared 24 h after transfection, as previously described (46). To obtain the nuclear extracts from HeLa cells treated with siEXO1 and overexpressing Polδ or Polδ mutant proteins, approximately 1.25 × 106 cells were seeded in T75 flasks. After 24 h, cells were transfected with 40 nM (final concentration) siRNA (Life technology) using DharmaFECT 1 transfection reagent. After 43 h, cells were confluent and split 1/3 in T75 flasks. Five hours later, cells were transfected with plasmids overexpressing Polδ or Polδ mutant proteins. After 24 h, cells were harvested for nuclear extract and RNA extraction for qPCR analysis. RNA extraction was performed using NucleoSpin RNA kit from MACHEREY-NAGEL and total cDNA was obtained using Superscript III reverse transcriptase (Life Technologies) and Oligo(dT)12–18 primers. Approximately 3 ng cDNA was used for quantitative real-time PCR using the ABI StepOnePlus system. Protein concentrations were determined by Bradford assay. 12% Polyacrylamide gels (Expedeon), polyvinylidene difluoride membranes (Biorad), were used for Western blotting. The following antibodies were used: hMSH2 (1:500, CalBiochem.), hMLH1 (1:500, Santa Cruz), PCNA (1:500, Santa Cruz), hPolδ subnunit-A (1:500, Santa Cruz), RPA32-pS33 (Bethyl, A300–246A, 1/1000), RPA2/RPA32 (Abcam, ab2175, 1/500), and Actin (1:500, NeoMarkers). The siRNA for EXO1 knockdown: 5′ UAGUGUUUCAGGAUCAACAUCAUCU 3′, control LUC siRNA Control: 5′ CGUACGCGGAAUACUUCGAUU 3′. The following primers were used for quantitative real-time PCR: EXO1-Forward, 5′ CAC ATCTCCGCGAGACAGAG; EXO1-Reverse, 5′ GGTGCCAAATTAACTACCTCTCA; βactin-Forward, 5′ CATGTACGTTGCTATCCAGGC; βactin-Reverse: CTCCTTAATGTCACGCACGAT.
Heteroduplex substrates and in vitro mismatch repair assay
The heteroduplexes were prepared as described previously (47). Briefly, CSH50 bacteria were infected with M13mp2 phage, and single-strand phage DNA (+) was precipitated from cleared culture supernatant. The replicative form (RF) phage DNA was harvested from NR9099 bacteria infected with M13mp2Δ2 phage and purified by CsCl gradient centrifugation. RF DNA was linearized with restriction enzymes AvaII or Bsu36I. Heteroduplexes were prepared by annealing 2-fold molar excess of ssDNA to denatured RF DNA linearized with Bsu36I or AvaII. The resulting nicked heteroduplex was purified by gel electrophoresis and isolated from a gel slice using Qiaquick gel extraction kit (Qiagen). MMR assay was performed as previously described (47). Briefly, the standard in vitro MMR assay (50 μL) was performed in buffer containing 30 mM Hepes (pH 7.8), 7 mM MgCl2,4 mM ATP, 200 μM each CTP, GTP,UTP, 100 μM each dATP, dGTP, dTTP, dCTP, 40 mM creatine phosphate, 100 μg/ml phosphokinase, 15 mM sodium phosphate buffer (pH 7.5), 5 ng heteroduplex DNA and 100 μg nuclear extract protein. The reactions were incubated at 37°C for 1 h, terminated by adding 50 μl stop mix (2 mg proteinase K, 2% SDS, 50 mM EDTA pH 8.0) and incubated for 30 min at 37°C. Subsequently, repair products were precipitated by adding 60 μl precipitation mix (0.71 mg Escherichia coli tRNA/ml, 1.7 M ammonium acetate) and purified by phenol/chloroform extraction. The repair products were transformed into competent NR9162 cells (MMR-deficient), which were plated with CSH50 cells and X-gal on minimal medium plates containing IPTG. Correctly repaired phage generate blue plaques while unrepaired phage generate mixed plaques. MMR efficiency is calculated as follows: 100 x [1 – (ratio of mixed plaques in assay X)/(ratio of mixed plaques in control)], where the control is untreated. In most assays, >500 plaques were counted per assay.
Mismatch-provoked excision was quantified using the same DNA substrate and assay buffer as the MMR assay, except that 25 ng of substrate was used per reaction, aphidicolin was added, dNTPs were omitted and reaction product was identified by its sensitivity to cleavage by restriction enzymes. Repair product was purified as described above and incubated with EcoRI and BamHI in the presence of 30 μg/ml RNase A for 2 h. Reaction products were visualized on agarose gels stained with ethidium bromide. The intensity of each band was measured using ImageJ, and the relative excision capacity was calculated for each lane from the intensity of the upper band/total intensity × 100%. To determine the capacity of nuclear extract to degrade the mismatch containing strand in vitro, the MMR reactions was performed as before with/without dNTPs and aphidicolin as indicated. Loading buffer containing SDS was added to stop the reaction after 1 h, samples were boiled at 95°C, and loaded on SDS-PAGE gel and analyzed by western blot. The intensity of each band was measured by ImageJ and normalized to the value of total RPA.
Flow cytometry
Cells were trypsinized, washed twice with PBS and 5 × 105 cells were transferred to FACS tubes and fixed in 2 ml ice-cold 70% ethanol for at least 30 min on ice or stored at –20°C. Cells were spun down, washed with PBS and resuspended in 400 μl PBS. 50 μL RNase A (1 mg/ml) and 20 μl propidium iodide (1 mg/ml, Sigma) were added and the cells were incubated in the dark for at least 30 min at room temperature. Cells were analyzed using the BD FACSCalibur system and the results were analyzed using ModFit LT software.
Clonogenic assay
Cells overexpressing wild type Polδ, mutant Polδ or knockdown of MLH1, EXO1 were trypsinized and seeded into 6-well plates 24 or 48 h after transfection, respectively. Cells were transfected with siRNA and after 24 h transfected with mutant Polδ plasmids to obtain siEXO1 knockdown + mutant Polδ cells, which were seeded into six-well plates after another 24 h. Cells were grown in six-well plates for one day before being exposed to 10 μM O6-Benzylguanine to inactivate MGMT. After 1 h incubation with O6-Benzylguanine, cells were treated with various concentrations of N-methyl-N′-nitro-N-nitrosoguanidine (MNNG). After 8 days, cells were stained with 0.5% crystal violet in 20% ethanol. Only colonies containing >100 cells were counted.
HPRT mutation assay
Three days after transfection, HCT116 (MMR-deficient) cells were trypsinized and reseeded into 10 cm dishes in growth medium containing 5 μg/ml 6-thioguaine (6-TG) (Sigma: A4882) at density of 4.5 × 105 cells/dish in triplicate. In parallel, 300 cells were seeded into a six-well plate in medium lacking 6-TG. After 10–12 days, cells were stained and counted. Cells in six-well plates were used to measure the plating efficiency. The mutant frequency was calculated as the ratio of the cloning efficiency with 6-TG to the cloning efficiency without 6-TG. For HCT116+Chr3 (MMR-proficient), cells were transfected every three days for 15 days, and then seeded into at least six 15 cm dishes with 1.5 × 106 cells/dish containing 5 μg/ml 6-TG. In parallel, 400 cells were seeded into two wells of a six-well plate in medium lacking 6-TG. The mutation frequency was calculated as for HCT116 cells.
dNTP incorporation assay
MMR assay was performed as described, except H3-labeled dATP and dTTP substituted for dATP and dTTP. At multiple time points, aliquots of the reaction were spotted on DEAE Filtermat (Perkin Elmer 1450-522) and dried thoroughly. Next, the DEAE Filtermat was washed 3× in 5% NaH2PO4 2× in MilliQ water, and dried overnight. The next day, MeltiLex solid scintillator (Perkin Elmer 1450=441) was added to the Filtermat at 80°C and it was incubated at room temperature. The amount of H3 isotope per spot was estimated by scintillation counting in a MicroBeta2 Plate Counter (Perkin Elmer 2450-0010) for 5 min.
RESULTS
Polδ 3′-5′ proofreading activity affects MMR activity
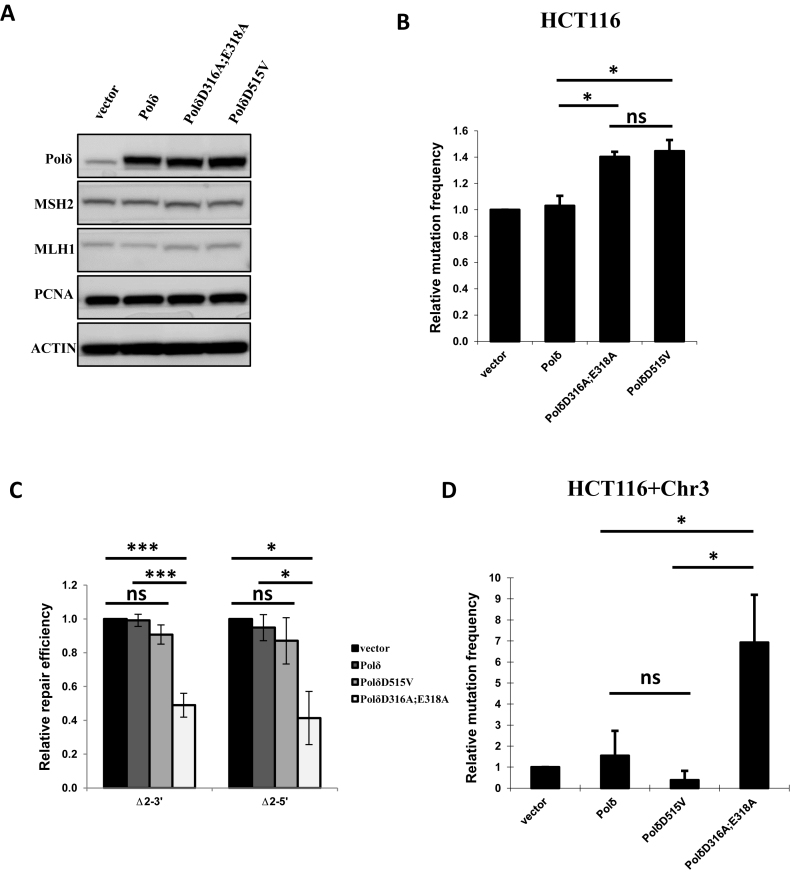
Eukaryotic DNA Polδ consists of four subunits. Subunit A (POLD1, p125) is the catalytic subunit, which has intrinsic DNA polymerase and 3′-5′ exonuclease activities encoded by three highly-conserved exonuclease motifs (19). Previous studies of yeast DNA Polδ indicate that alanine substitution mutations of D321 and E323 (D321A;E323A) in the catalytic subunit inactivates Polδ exonuclease but leaves the DNA polymerase activity intact (48,49). Here, we constructed a plasmid to overexpress the equivalent mutant of human Polδ subunit-A, PolδD316A;E318A, and characterized the mutant protein with particular focus on the role of Polδ exonuclease in MMR (Supplementary Figure S1). The mutant protein was overexpressed in cells carrying the expression construct, as confirmed by the western blot in Figure 1A. As expected, overexpression of the mutant Polδ in cells expressing endogenous wild type Polδ resulted in a dominant-negative phenotype, and the mutation frequency increased when PolδD316A;E318A was expressed in MMR-deficient HCT116 cells (Figure 1B). Western blot analysis indicated that overexpression of wild type or mutant Polδ does not affect the expression of MSH2, MLH1 or PCNA (Figure 1A).
Figure 1.
In vitro MMR. (A) HeLa nuclear extracts from cells carrying empty vector (vector), wild type Polδ, PolδD316A;E318A or PolδD515V were analyzed by western blot. Western blot data are presented for MSH2, MLH1 and PCNA. (B) Mutation frequency in HCT116 (MLH1-/-) cells overexpressing PolδD316A;E318A or PolδD515V. (C)In vitro MMR assay using 2 nt indel heteroduplex DNA substrates Δ2–3′ or Δ2–5′ (see Materials and Methods). (D) As in B except using chromosome 3 complemented HCT116 (HCT116+Chr3) cells. The data are the mean ± SD of three independent experiments. *P< = 0.05; **P< = 0.01; ***P< = 0.005.
MMR assays were performed in vitro using nuclear extracts from cells overexpressing PolδD316A;E318A (see methods for details). The heteroduplex DNA substrate carries a 2 nt indel mismatch (Δ2) and a nick 5′ or 3′ to the mismatch, referred to as Δ2–5′ and Δ2–3′ heteroduplex DNA substrates, respectively. These substrates were chosen based on a previous study showing that nuclear extracts from EXO1-deficient murine ES cells have significantly higher MMR activity on small indel mismatches than on single-base mismatches (15). We observe that nuclear extracts expressing mutant PolδD316A;E318A had 2-fold lower MMR activity than extracts from cells overexpressing either PolδD515V, wild type Polδ or transfected with empty vector. Similar results were obtained with Δ2–5′ and Δ2–3′ heteroduplex DNA substrates (Figure 1C), and no significant differences in MMR activity were detected between assays using extracts from cells overexpressing wild type Polδ or extracts from HeLa cells carrying empty vector (Figure 1C).
We next wondered whether the lower MMR activity detected in samples containing the PolδD316A;E318A mutant was due to cell cycle alterations. We analyzed cell cycle distribution by flow cytometry (50) and found no differences between HeLa cells carrying empty vector and HeLa cells overexpressing PolδD316A;E318A or wildtype Polδ (Supplementary Figure S2).
MMR activity was also investigated in HeLa cells overexpressing PolδD515V, a p125 variant for which the corresponding yeast mutant PolδD520V is reported to be exonuclease-deficient (51) (Supplementary Figure S1). In vitro MMR assay using extracts from HeLa cells expressing human PolδD515V mutant protein showed that MMR activity was not significantly lower than control assays with wild type Polδ (Figure 1C). This result suggests that loss of Polδ exonuclease per se does not interfere with MMR efficiency, but that a specific property of human PolδD316A;E318A plays a role in lowering MMR efficiency during in vitro MMR. In this regard, it is worth noting that yeast PolδD321A;E323A displays stronger strand displacement activity than yeast PolδD520V, and it is reasonable to propose that the same is true for the corresponding human Polδ variants PolδD316A;E318A and PolδD515V (42).
The relative mutation frequencies at endogenous HPRT was investigated in MMR-proficient HCT116+Chr3 cells expressing PolδD316A;E318A or PolδD515V. The results showed that HPRT mutation frequency was significantly higher in cells expressing PolδD316A;E318A than in cells expressing wild type Polδ or PolδD515V or in control cells carrying empty vector (Figure 1D). In contrast, relative HPRT mutation frequency was significantly higher than the control in MMR-deficient HCT116 cells expressing either PolδD316A;E318A or PolδD515V (Figure 1B). These results are consistent with the hypothesis that PolδD316A;E318A interacts with components of the MMR pathway and may inhibit normal MMR in cells in which it is overexpressed.
Mismatch-provoked excision is not affected by PolδD316A; E318A
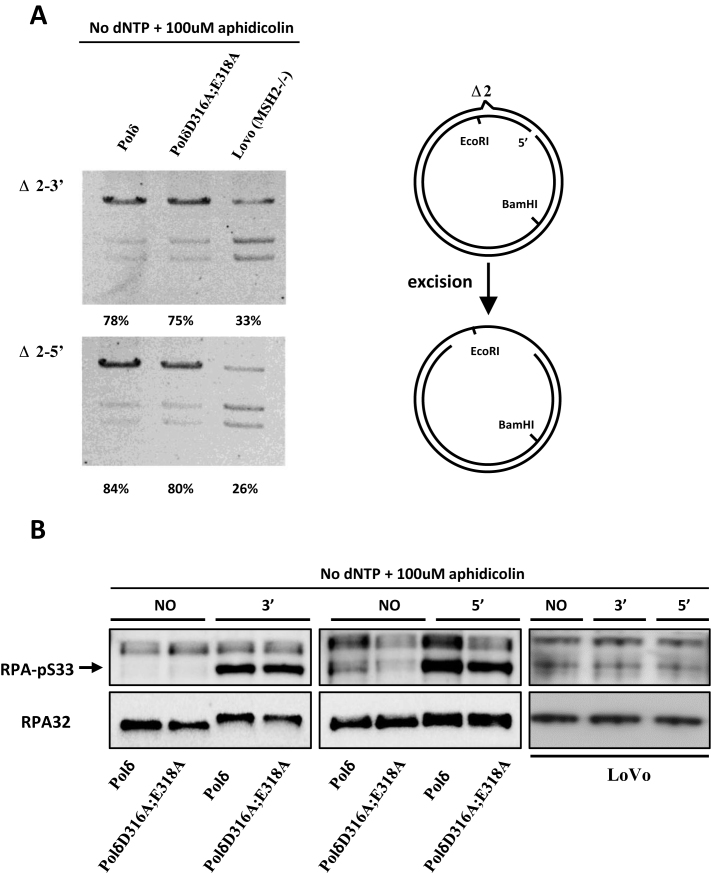
We excluded from our analysis the first repair step, mismatch recognition as it has been shown to be carried out by the MutS heterodimers independently of the replicative polymerase. We then analyzed in vitro the second step, mismatch provoked excision using nuclear extracts obtained from cells overexpressing either wild type Polδ or PolδD316A;E318A mutant. Mismatch-provoked DNA excision was quantified in nuclear extracts of cells overexpressing wild type Polδ or PolδD316A;E318A. The results of this in vitro assay suggest that the excision step of in vitro MMR is not negatively affected by the overexpression of PolδD316A;E318A (Figure 2A).
Figure 2.
Mismatch-provoked excision in HeLa cells expressing PolδD316A;E318A. (A) Mismatch provoked excision assay was performed as described in Materials and Methods using the nuclear extracts indicated. The extent of DNA excision was estimated by measuring susceptibility/resistance to cleavage by EcoRI, whose recognition sequence lies in between the DNA excision initiation site and the mismatch (schematic diagram right). Reaction products were digested with EcoRI and BamHI, separated by agarose gel electrophoresis and visualized by staining with ethidium bromide. The intensity of each band was quantified using ImageJ, and the relative excision capacity was calculated as the intensity of the slowest migrating (largest) reaction product per lane /total intensity per lane × 100. Nuclear extracts from MSH2-deficient LoVo cells were used as the negative control. (B) Western blot of pS33-RPA32 was performed to monitor amount of ssDNA generated during in vitro MMR. Briefly, MMR assay was performed as described in Materials and Methods with no substrate (NO), Δ2–3′ (3′) or Δ2–5′ (5′) substrate in the reaction. The MMR was terminated by adding SDS containing loading buffer and boiling at 95°C for 10 min. Western blot was performed as described in Materials and Methods. Aphidicolin was included and dNTPs were omitted as indicated for inhibition of polymerase synthesis of Polδ. The total RPA32 was used as control.
The single strand DNA (ssDNA) binding protein replication protein A (RPA) is required to complete MMR (9,52–54). Phosphorylation of RPA by ssDNA-activated ATR is essential in DNA damage or replication stress induced signaling pathways (55,56). Furthermore, it has been shown that RPA is phosphorylated gradually during MMR in vitro and the phosphorylation is essential for the resynthesis step (57). Here, phosphorylation of RPA at pS33 in combination with addition of aphidicolin and absence of dNTPs was used as a measure for the extent of ssDNA tracts generated during MMR (Figure 2B). In MMR reactions lacking dNTPs and including amphidicolin, the level of phosphorylated RPA32 was comparable between nuclear extracts containing either wild type Polδ or PolδD316A;E318A (Figure 2B). The LoVo extract was used as a control, showing no increase of the phosphorylated RPA level after incubation with MMR substrates (Figure 2B). These results suggest that mismatch-provoked DNA excision is independent of the integrity of the Polδ proofreading activity.
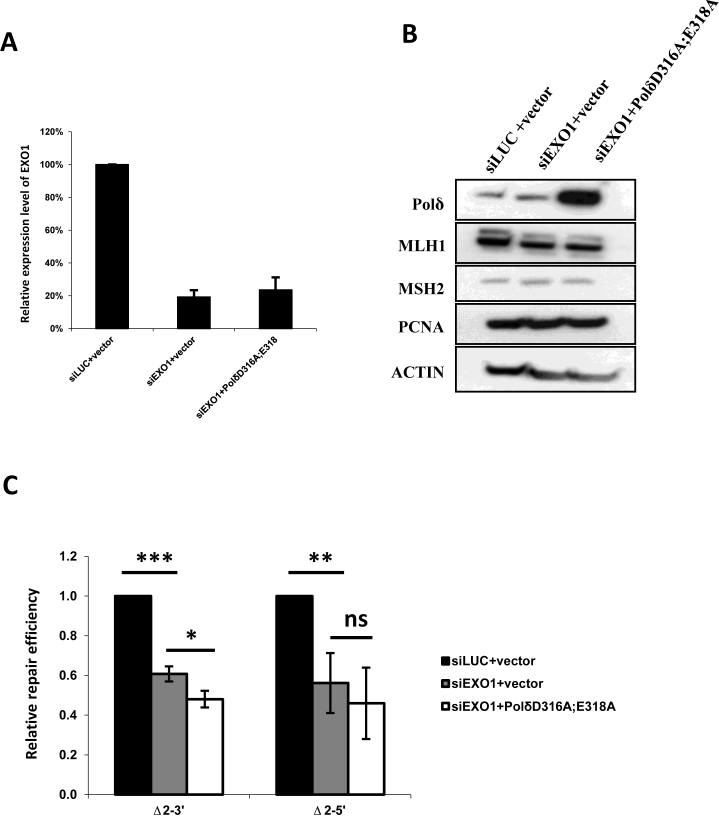
Depletion of EXO1 is not synergistic with PolδD316A; E318A in inhibiting in vitro MMR activity
It has been shown that depletion of yeast EXO1 (5′-3′ exo) in 3′-5′ exonuclease-deficient yeast Polδ has a strong synergetic effect on the mutation rate in S. cerevisiae (23,45). Furthermore, human EXO1 is the only known exonuclease known to function in eukaryotic MMR to date. Therefore, we tested whether depletion of human EXO1 is synergistic with human PolδD316A;E318A in the in vitro MMR assay. No synergetic effect on MMR was detected (Figure 3C). It was also confirmed that expression of MMR proteins and cell cycle progression were not altered under any of the conditions tested (Figure 3B and Supplementary Figure S1B). The above results demonstrate that PolδD316A;E318A has an identical effect on in vitro MMR activity when the Δ2–3′ and the Δ2–5′ DNA substrates are used in the MMR assay. Together, the results support the notion that the 3′-5′ exonuclease of Polδ does not directly participate in MMR by degrading error-containing DNA from 3′-5′ in human cells, but that the roles of EXO1 and the 3′-5′ exonuclease of Polδ in MMR may be partially redundant.
Figure 3.
Effect of depletion of EXO1 on MMR in cells overexpressing PolδD316A;E318A. (A) Efficiency of EXO1 knockdown was confirmed by qPCR, and (B) overexpression of PolδD316A;E318A mutant was confirmed by western blot. Besides, no alternations in expression levels of MLH1, MSH2 and PCNA was observed among nuclear extracts from cells with Luciferase siRNA and empty vector (siLUC+vector), cells with EXO1 siRNA and empty vector (siEXO1+vector) and cells with EXO1 siRNA and expressing PolδD316A;E318A (siEXO1+ PolδD316A;E318A). (C) in vitro MMR assay was performed as described in Materials and Methods. No strong synergetic effect on MMR activity when EXO1 depletion and expressing PolδD316A;E318A were combined in the nuclear extracts. The data represent the mean ± SD of three independent experiments. *P ≤ 0.05; **P ≤ 0.01.
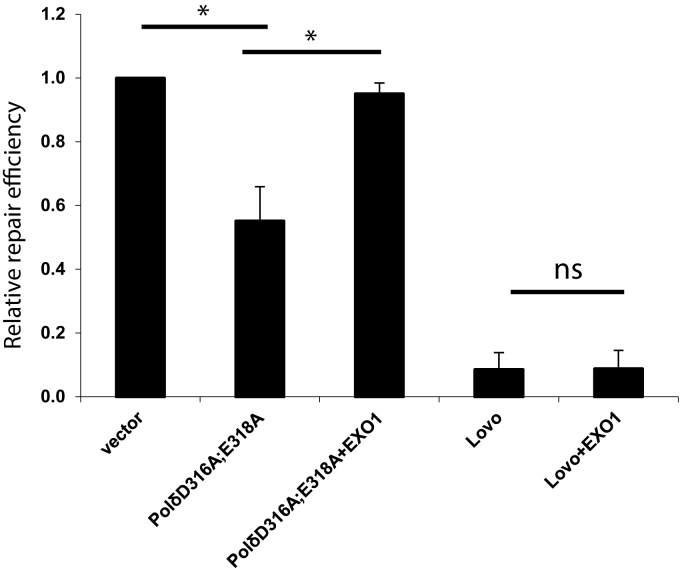
MMR defect in cells expressing PolδD316A; E318A is complemented by exogenous EXO1
As mentioned above, exonuclease-deficient yeast PolδD321A;E323A displays stronger strand displacement activity than wild type yeast Polδ or exonuclease-deficient yeast PolδD520V (37,42). Noting that previous studies suggest one MMR pathway that depends on strand displacement activity of Polδ (17), we propose that, by analogy to yeast PolδD321A;E323A, human PolδD316A;E318A has more potent strand displacement activity than wild type human Polδ and that this inhibits MMR by deregulation of EXO1-dependent excision pathway in human cells. This hypothesis was tested by performing MMR assays as described above in the presence or absence of exogenous purified human EXO1. The results show that exogenous EXO1 complements the MMR defect associated with overexpression of PolδD316A;E318A (Figure 4). A control experiment showed no effect on MMR when EXO1 was added to MMR-deficient extracts of LoVo (MSH2−/−) cells (Figure 4). Our results also show that addition of EXO1 to extracts expressing PolδD515V did not rescue the modest MMR defect (data not shown). Based on these results, we propose that DNA strand displacement by PolδD316A;E318A reduces access of EXO1 to nicks 5′ to the mismatch, thereby inhibiting mismatch-provoked DNA excision by EXO1. In this context, exogenous EXO1 increases the ability of EXO1 to compete for access to nicks in the non-template DNA strand, and restores the normal balance between DNA excision and DNA strand displacement during MMR.
Figure 4.
EXO1 complements MMR defect in nuclear extracts expressing PolδD316A;E318A. The in vitro MMR assay was performed as described in Materials and Methods and the nuclear extracts were the same as used in Figure 1. Purified human EXO1 (2.5 nM) was added to the reaction as indicated. The Δ2–3′ heteroduplex was used in the MMR assays. The data represent the mean ± SD of three independent experiments. *P ≤ 0.05; **P ≤ 0.01.
PolδD316A; E318A can induce aberrant dNTP incorporation during MMR in vitro
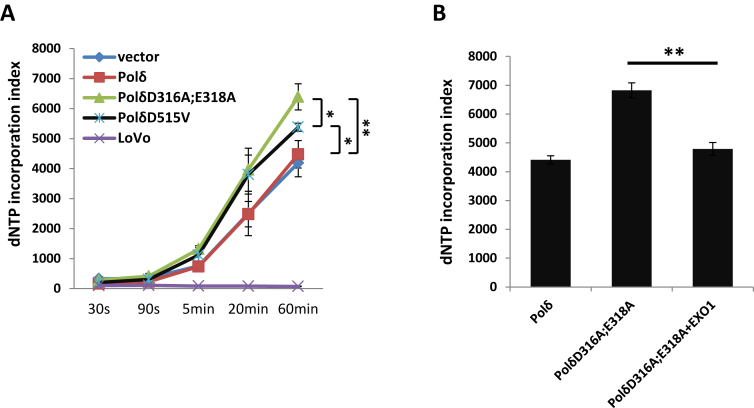
We suggested above that PolδD316A;E318A may disturb the EXO1-dependent excision pathway by the enhanced strand displacement activity of the polymerase. Therefore, it is also possible that PolδD316A;E318A could alter the nature of dNTP incorporation during MMR. The following experiment tests whether PolδD316A;E318A influences the fidelity of DNA replication at the dNTP incorporation step during the DNA resynthesis step of MMR. To investigate this, in vitro MMR assays were performed in the presence of 3H-labeled dATP and dTTP and incorporation of 3H was quantified at several time points during the assay (see Materials and Methods). Surprisingly, although the extracts with PolδD316A;E318A has decreased MMR activity (Figure 1C), dNTP incorporation was higher than in extracts from cells overexpressing wild type Polδ. As expected, negligible dNTP incorporation was detected in extracts prepared from MMR-deficient LoVo cells (Figure 5A). Furthermore, in MMR assays performed with extracts from cells expressing PolδD515V, which does not show significantly decreased MMR activity (Figure 1B), dNTP incorporation was also higher than in control assays, but not as high as in extracts from cells overexpressing PolδD316A;E318A at the 60 min time point (Figure 5A). Interestingly, when exogenous EXO1 was added to the assay, 3H incorporation by PolδD316A;E318A at 60 min decreased (Figure 5B). This result suggests that increased strand displacement correlates with increased dNTP incorporation during MMR and both are counteracted/decreased by exogenous EXO1.
Figure 5.
Effect of PolδD316A;E318A on incorporation of dNTPs during in vitro MMR. In panel (A) dNTP incorporation was analyzed as described in Methods and Materials. The Δ2–5′ heteroduplex was used as substrate. The asterisk indicates the P value of the pairwise comparison of extracts with PolδD316A;E318A, with PolδD515V, and with wild type Polδ overexpression. In panel (B), dNTP incorporation was measured 60 min after start of the MMR reaction. The data are the mean ± SD of three independent experiments. *P ≤ 0.05; **P ≤ 0.01.
Cells overexpressing PolδD316A;E318A are sensitive to SN1 DNA methylating agent MNNG
MNNG is an SN1 DNA methylating agent that adds an O6-methyl group to deoxyguanine in DNA. The methyl group of O6MeG is subject to direct repair (i.e. removal of the O6-methyl group from O6-methylguanine) by methylguanine methyl transferase (MGMT) (58). If O6MeG is not repaired it can cause O6meG:T/C mismatches that are recognized by MMR. However, because MMR is strand-specific, the MMR machinery is trapped in a futile cycle when it tries to repair O6MeG residues on the template strand, leading to persistent gaps/nicks that ultimately are converted to potentially lethal double stranded breaks (DSBs) (59,60). An alternative model suggests that DNA damage response factors are recruited to the O6meG:T/C mismatches and directly signal the downstream checkpoint factors (61). Both models involve MMR activity (62,63). Another consequence of the futile cycle induced by unrepaired O6 –methylguanine residues is that MMR-deficient cells are more resistant to killing by MNNG than MMR-proficient cells.
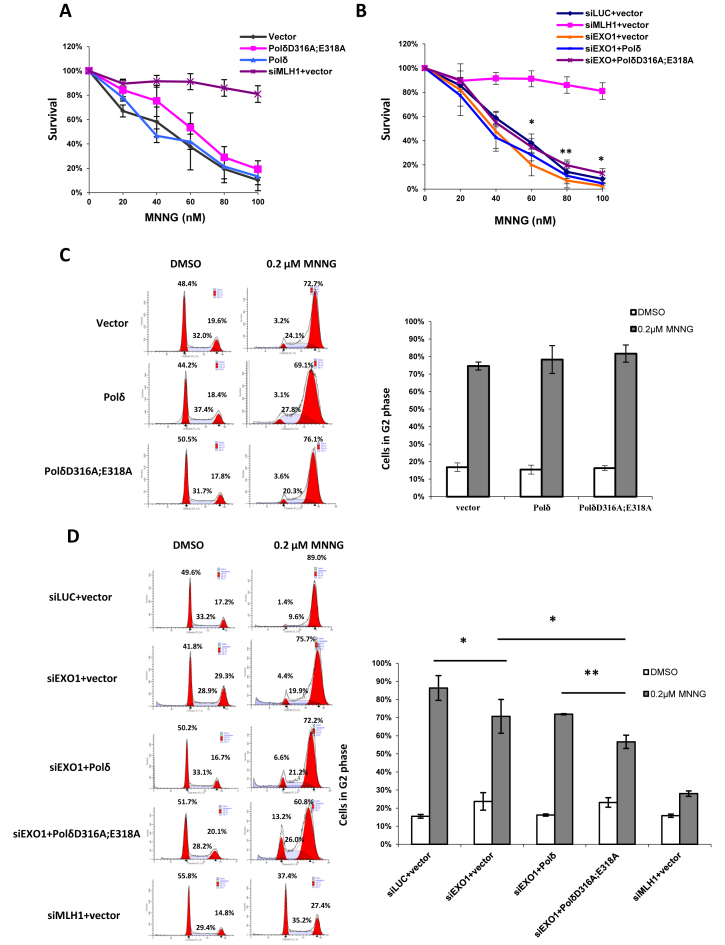
Here, cells overexpressing PolδD316A;E318A or wild type Polδ were exposed to MNNG, and the response was compared using a clonogenic survival assay. The results show comparable susceptibility to killing by MNNG in cells expressing PolδD316A;E318A or wild type Polδ (Figure 6A), indicating that PolδD316A;E318A does not affect the formation of the nicks on the DNA strand by MMR machinery after MNNG treatment. The same result was obtained after depletion of EXO1 (siLUC) (Figure 6B). This is in contrast to previous reports, which showed that EXO1-depleted or knockout EXO1 MEF cells were more resistant to MNNG than control cells (60,64); but, the result obtained here is similar to results in S. pombe (65). The discrepancy may reflect use of two different assays: clonogenic survival assay (longer in duration) and cell viability in culture (shorter in duration) or another difference in the cells used in the two studies. Interestingly, overexpression of PolδD316A;E318A in EXO1-depleted cells slightly increased resistance to MNNG, with statistically significant increase observed at 60, 80 and 100 nM MNNG (Figure 6B).
Figure 6.
Effect of PolδD316A;E318A on susceptibility to killing by MNNG. (A and B) HeLa cells were transfected and seeded as described in Materials and Methods. After one day, cells were treated with 10 μM O6-Benzylguanine and MNNG at the concentration indicated. Colonies were counted after 8 days after crystal violet staining. The percentage of survival for each drug concentration was calculated as number of colonies after MNNG treatment / number of colonies without MNNG treatment. The asterisk above the error bar indicates the P value between group siEXO1+Polδ and group siEXO1+PolδD316A;E318A. (C and D) Cells were seeded into T75 flasks one day after transfection. After 24 hours, cells were treated with 10 μM O6-Benzylguanine and 0.2 μM MNNG and incubated for another two days before subject to FACS analysis. The data represent the mean ± SD of three independent experiments. *P ≤ 0.05.
It has also been reported that MNNG induced MMR-dependent G2 arrest is delayed, and is observed in the second G2 after exposure to MNNG (66–68). A similar effect was observed here after first cell cycle, in that cell survival rate was similar after exposure of cells expressing PolδD316A;E318A or wild type Polδ (Figure 6C). Furthermore, murine EXO1-depleted cells were slightly resistant to MNNG-induced G2 arrest (60). Interestingly, overexpression of PolδD316A;E318A in EXO1-depleted cells decreased MNNG induced G2 arrest (Figure 6D). These results are in agreement with the results of clonogenic survival assay described above and support the hypothesis that PolδD316A;E318A and depletion of EXO1 synergistically increase resistance to MNNG-induced G2 arrest, although PolδD316A;E318A alone does not increase resistance to MNNG.
DISCUSSION
Here, we report that overexpression of 3′-5′ exonuclease-deficient human PolδD316A;E318A, which is equivalent to yeast PolδD312A;E323A (69), leads to decreased MMR activity in vitro and an elevated mutation frequency in chromosome 3-complemented HCT116 MMR-proficient cells. These results demonstrate an intriguing interaction between the 3′-5′ exonuclease of Polδ and the MMR pathway in human cells.
In eukaryotic cells, DNA polymerases ϵ (Polϵ) and δ are the primary replicative polymerases on the leading and lagging DNA strands, respectively. In mice, defects in the 3′-exonuclease of Polϵ or Polδ increase mutation frequency as much as tenfold (70). In this study, overexpression of PolδD316A;E318A or PolδD515V in HCT116 MMR-deficient cells also increases mutation rate, which demonstrates a dominant-negative effect on endogenous wild type Polδ. Dominant-negative effects have also been observed when defective alleles of human Polϵ or Polα were overexpressed in cells carrying the corresponding wild type replicative DNA polymerase (71,72).
Although Polδ or Polϵ are both involved in genome replication, the mutation spectra of proofreading-deficient Polδ and Polϵ are distinct. Firstly, mutation rate in Polδ exonuclease-deficient yeast strains is higher than Polϵ exonuclease-deficient strains (73,74). Secondly, proofreading-deficient Polϵ and Polδ mice exhibit mostly distinct, but overlapping tissue specific tumor phenotypes. For instance, nodal lymphomas and histiocytic sarcomas are prevalent in Polϵ mutant mice, while thymic lymphomas and skin papillomas/sarcomas are frequent in Polδ mutant mice. Furthermore, the mutation rate in Polδ mutant mice is higher than in Polϵ mutant mice, with more frameshift mutations and microsatellite instability (MSI) resembling that of MMR-deficiency (75,76). These results suggest that the proofreading activity of Polδ possesses unique and more prominent function in genome stability maintenance than Polϵ. Although this difference between Polϵ and Polδ can be explained by the involvement of Polδ in Okazaki fragment maturation, we decided to investigate if the proofreading activity of Polδ is also involved in MMR.
Previously, it was reported that in yeast, proofreading-deficiency of Polδ but not Polϵ increases mutation rate of d(CA)n repeat sequences 5–10-fold, though not as severely as the increase in mutation rate caused by deletion of MSH2 (48). In addition, specific germline mutations in the exonuclease domains of both Polϵ and Polδ have been recently suggested to be involved in the development of colorectal adenomas and colorectal cancer (CRC), which is the predominant cancer type in MMR-deficient patients (26–28,77). It was also shown that CRCs caused by proofreading-deficient Polϵ are MSI-negative, while the Polδ proofreading-deficient sporadic CRCs were reported MSI-positive (26,77,78). These results suggest that cells carrying a 3′-exonuclease-deficient allele of Polδ and cells carrying defects in MMR share characteristics at the cellular level (24). It has been suggested that proofreading activity of Polδ is involved in regulating MMR activity in mice (24). These data strongly suggest a role for the proofreading activity of Polδ in MMR.
The 3′- exonuclease activity of Polδ can act in trans on DNA mismatches introduced by Polα, indicating that 3′- exonuclease of Polδ can correct the errors in the DNA strand independently from its polymerase activity (38). Together with the existence of an EXO1-independent MMR pathway, we initially proposed that apart from resynthesizing the new DNA strand in MMR, Polδ may also take part in MMR by degrading the mismatch in the 3′-5′ direction, when it carries a functional 3′-exonuclease activity. However, this hypothesis is not consistent with the observation that excess PolδD316A;E318A inhibits in vitro MMR to a similar extent on 5′-nicked and 3′-nicked heteroduplex DNA substrates (Figure 1). Interestingly, we show that EXO1 depletion only partly decreases MMR activity in cells expressing mutant Polδ suggesting that EXO1 and Polδ participate in the same pathway.
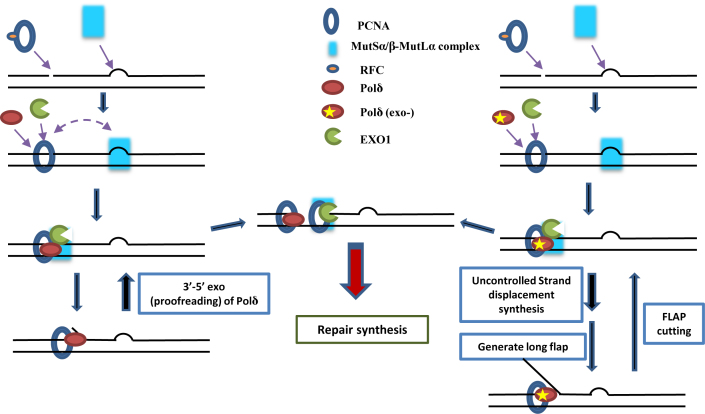
In addition, mismatch induced excision capacity was not altered in vitro in assays with using nuclear extracts from cells overexpressing PolδD316A;E318A. Based on the idea that strand displacement activity of Polδ is negatively regulated by its 3′ -exonuclease (37), which implies that the strand displacement activity of PolδD316A;E318A is likely to be upregulated, we hypothesized that PolδD316A;E318A competes with and/or inhibits EXO1-mediated DNA excision during MMR in vitro. This could reflect steric hindrance at the nick due to long PolδD316A;E318A-generated 5′-flap structures (Figure 7). Consistent with this idea, addition of exogenous purified EXO1 to the PolδD316A;E318A nuclear extracts complemented the in vitro MMR defect. This suggests that an increase in the level of EXO1 restored a balance between EXO1-dependent excision of mismatches and Polδ-dependent DNA strand displacement upstream of the DNA mismatch, either through direct EXO1-dependent inhibition of PolδD316A;E318A-catalyzed DNA strand displacement or by EXO1-dependent removal of 5′-flaps generated by PolδD316A;E318A (79).
Figure 7.
Model of the role of Polδ during MMR. Briefly, PolδD316A;E318A competes with and/or inhibits EXO1-mediated DNA excision during MMR in vitro.
Previous studies provide evidence that there are subtle differences in MMR on the leading and lagging strands of the DNA replication fork. Kunkel and colleagues showed that the MMR efficiency is higher for errors generated by DNA Polα than for errors generated by DNA Polϵ or Polδ (80–82). This conclusion is supported by evidence that EXO1 preferentially excises mismatches generated by Polα (83) and that MMR preferentially repairs 8-oxo-G•dA on the lagging strand (84). One explanation of these findings is that the discontinuous nature of lagging strand replication increases accessibility of MMR enzymes and EXO1 to the nascent DNA. Therefore, MMR on the leading strand may be strictly-dependent on the latent endonuclease activity of MutLα and/or MMR that initiates at MutLα-generated nicks may be inherently less efficient than MMR initiating at the 5′ end of an Okazaki fragment, and this could be at least in part because of interference from 5′-flaps generated by Polδ –dependent strand displacement activity.
We propose a model (Figure 7) where Polδ is loaded by PCNA at a pre-existing nick and Polδ inserts 1–2 nt generating a short 5′-flap. In this scenario, the strand displacement activity is limited by the regulatory effect of Polδ 3′-5′ exonuclease. After EXO1 entry and excision downstream of the nick, the polymerase activity of Polδ is further restricted by binding of unphosphorylated RPA (57). When Polδ lacks 3′-exonuclease, strand displacement activity generates longer 5′-flaps, which are substrates of FEN1 or DNA2, but not EXO1. The consequences are that EXO1 fails to play the role of MMR excision nuclease, and the efficiency of MMR decreases significantly. This model is supported by the demonstration that dNTP incorporation increases during in vitro MMR even when MMR efficiency decreases (Figure 5).
An alternative explanation could be based on our data presented in Figure 1D, which show that overexpression of PolD D316A;E318A, but not PolD D515V, increases mutation frequency in MMR-proficient cells. One interpretation is that PolD D316A;E318A is error-prone and that PolD D515V is error-free. This interpretation is supported by the results in Figure 5 showing that both PolD mutants have increased strand displacement as well as increased nucleotide incorporations.
The yeast homologs of human PolδD321A;E323A and PolδD520V are defective in 3-exonuclease activity (51). Interestingly, the impact of human PolδD515V on in vitro MMR appears to be considerably weaker than the impact of PolδD316A;E318A. In particular, MMR activity is not decreased and the dNTP incorporation during MMR in presence of PolδD515V is less than for PolδD316A;E318A, and PolδD515V does not increase mutation frequency in MMR-proficient cells. These results suggest that the nature of the defect in Polδ 3-exonuclease differs in these two Polδ variants. Previous studies also show that yeast PolδD520V has a weaker mutator phenotype than PolδD321A;E323A with or without a secondary mutation in RAD27 (51). More importantly, in vitro assays showed that the strand displacement activity of PolδD520V is weaker than the strand displacement activity of PolδD321A;E323A (42). These data implicate the existence of a threshold of the proofreading ability or the strand displacement ability of Polδ, which determines whether certain mutation affecting Polδ proofreading activity is able to influence the MMR pathway.
In summary, the present study supports the hypothesis that the mismatches in a DNA heteroduplex are removed/replaced during in vitro MMR either by EXO1-dependent DNA excision (followed DNA resynthesis in a second step) or in a single step by Polδ strand displacement (Figure 7). Our results suggest that a balance between excision by EXO1 and Polδ strand displacement is maintained during normal MMR that favors DNA excision by EXO1, and that this balance requires a functional Polδ 3′-exonuclease.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dmitry A. Gordenin for valuable suggestions and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nordea-fonden; China Scholarship Council (CSC); United States National Institutes of Health (NIH) National Cancer Institute (NCI) [R01 CA164944]. Funding for open access charge: Nordea-fonden.
Conflict of interest statement. None declared.
REFERENCES
- 1. Modrich P., Lahue R.. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996; 65:101–133. [DOI] [PubMed] [Google Scholar]
- 2. Li G.M. Mechanisms and functions of DNA mismatch repair. Cell research. 2008; 18:85–98. [DOI] [PubMed] [Google Scholar]
- 3. Jascur T., Boland C.R.. Structure and function of the components of the human DNA mismatch repair system. Int. J. Cancer. 2006; 119:2030–2035. [DOI] [PubMed] [Google Scholar]
- 4. Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996; 10:1433–1442. [DOI] [PubMed] [Google Scholar]
- 5. Schofield M.J., Hsieh P.. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003; 57:579–608. [DOI] [PubMed] [Google Scholar]
- 6. Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R.. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1994; 75:1027–1038. [DOI] [PubMed] [Google Scholar]
- 7. Bronner C.E., Baker S.M., Morrison P.T., Warren G., Smith L.G., Lescoe M.K., Kane M., Earabino C., Lipford J., Lindblom A. et al. . Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994; 368:258–261. [DOI] [PubMed] [Google Scholar]
- 8. Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003; 21:1174–1179. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Yuan F., Presnell S.R., Tian K., Gao Y., Tomkinson A.E., Gu L., Li G.M.. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005; 122:693–705. [DOI] [PubMed] [Google Scholar]
- 10. Constantin N., Dzantiev L., Kadyrov F.A., Modrich P.. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005; 280:39752–39761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tishkoff D.X., Amin N.S., Viars C.S., Arden K.C., Kolodner R.D.. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998; 58:5027–5031. [PubMed] [Google Scholar]
- 12. Szankasi P., Smith G.R.. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 1992; 267:3014–3023. [PubMed] [Google Scholar]
- 13. Dzantiev L., Constantin N., Genschel J., Iyer R.R., Burgers P.M., Modrich P.. A defined human system that supports bidirectional mismatch-provoked excision. Mol. Cell. 2004; 15:31–41. [DOI] [PubMed] [Google Scholar]
- 14. Kadyrov F.A., Dzantiev L., Constantin N., Modrich P.. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006; 126:297–308. [DOI] [PubMed] [Google Scholar]
- 15. Wei K., Clark A.B., Wong E., Kane M.F., Mazur D.J., Parris T., Kolas N.K., Russell R., Hou H. Jr, Kneitz B. et al. . Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003; 17:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amin N.S., Nguyen M.N., Oh S., Kolodner R.D.. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 2001; 21:5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kadyrov F.A., Genschel J., Fang Y., Penland E., Edelmann W., Modrich P.. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bell S.P., Dutta A.. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002; 71:333–374. [DOI] [PubMed] [Google Scholar]
- 19. Burgers P.M. Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 2009; 284:4041–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson R.E., Klassen R., Prakash L., Prakash S.. A major role of DNA polymerase delta in replication of both the leading and lagging DNA strands. Mol. Cell. 2015; 59:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie B., Mazloum N., Liu L., Rahmeh A., Li H., Lee M.Y.. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry. 2002; 41:13133–13142. [DOI] [PubMed] [Google Scholar]
- 22. Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M., Kunkel T.A.. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005; 280:29980–29987. [DOI] [PubMed] [Google Scholar]
- 23. Morrison A., Sugino A.. The 3′→5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet.: MGG. 1994; 242:289–296. [DOI] [PubMed] [Google Scholar]
- 24. Goldsby R.E., Hays L.E., Chen X., Olmsted E.A., Slayton W.B., Spangrude G.J., Preston B.D.. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:15560–15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Venkatesan R.N., Treuting P.M., Fuller E.D., Goldsby R.E., Norwood T.H., Gooley T.A., Ladiges W.C., Preston B.D., Loeb L.A.. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol. Cell. Biol. 2007; 27:7669–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Church D.N., Briggs S.E., Palles C., Domingo E., Kearsey S.J., Grimes J.M., Gorman M., Martin L., Howarth K.M., Hodgson S.V. et al. . DNA polymerase epsilon and delta exonuclease domain mutations in endometrial cancer. Hum. Mol. Genet. 2013; 22:2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palles C., Cazier J.B., Howarth K.M., Domingo E., Jones A.M., Broderick P., Kemp Z., Spain S.L., Guarino E., Salguero I. et al. . Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013; 45:136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rayner E., van Gool I.C., Palles C., Kearsey S.E., Bosse T., Tomlinson I., Church D.N.. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer. 2016; 16:71–81. [DOI] [PubMed] [Google Scholar]
- 29. Araujo S.J., Tirode F., Coin F., Pospiech H., Syvaoja J.E., Stucki M., Hubscher U., Egly J.M., Wood R.D.. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 2000; 14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 30. Parsons J.L., Preston B.D., O’Connor T.R., Dianov G.L.. DNA polymerase delta-dependent repair of DNA single strand breaks containing 3′-end proximal lesions. Nucleic Acids Res. 2007; 35:1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G. et al. . Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010; 37:714–727. [DOI] [PubMed] [Google Scholar]
- 32. Brocas C., Charbonnier J.B., Dherin C., Gangloff S., Maloisel L.. Stable interactions between DNA polymerase delta catalytic and structural subunits are essential for efficient DNA repair. DNA Repair. 2010; 9:1098–1111. [DOI] [PubMed] [Google Scholar]
- 33. Li X., Stith C.M., Burgers P.M., Heyer W.D.. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol. Cell. 2009; 36:704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Longley M.J., Pierce A.J., Modrich P.. DNA polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem. 1997; 272:10917–10921. [DOI] [PubMed] [Google Scholar]
- 35. Fazlieva R., Spittle C.S., Morrissey D., Hayashi H., Yan H., Matsumoto Y.. Proofreading exonuclease activity of human DNA polymerase delta and its effects on lesion-bypass DNA synthesis. Nucleic Acids Res. 2009; 37:2854–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCulloch S.D., Kokoska R.J., Garg P., Burgers P.M., Kunkel T.A.. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta. Nucleic Acids Res. 2009; 37:2830–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stith C.M., Sterling J., Resnick M.A., Gordenin D.A., Burgers P.M.. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 2008; 283:34129–34140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pavlov Y.I., Frahm C., Nick McElhinny S.A., Niimi A., Suzuki M., Kunkel T.A.. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol.: CB. 2006; 16:202–207. [DOI] [PubMed] [Google Scholar]
- 39. Prindle M.J., Loeb L.A.. DNA polymerase delta in DNA replication and genome maintenance. Environ. Mol. Mutagen. 2012; 53:666–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garg P., Stith C.M., Sabouri N., Johansson E., Burgers P.M.. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004; 18:2764–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin Y.H., Ayyagari R., Resnick M.A., Gordenin D.A., Burgers P.M.. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003; 278:1626–1633. [DOI] [PubMed] [Google Scholar]
- 42. Garg P., Burgers P.M.. How the cell deals with DNA nicks. Cell Cycle. 2005; 4:221–224. [PubMed] [Google Scholar]
- 43. Lin S.H., Wang X., Zhang S., Zhang Z., Lee E.Y., Lee M.Y.. Dynamics of enzymatic interactions during short flap human Okazaki fragment processing by two forms of human DNA polymerase delta. DNA Repair. 2013; 12:922–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sebesta M., Burkovics P., Juhasz S., Zhang S., Szabo J.E., Lee M.Y., Haracska L., Krejci L.. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA Repair. 2013; 12:691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tran H.T., Gordenin D.A., Resnick M.A.. The 3′→5′ exonucleases of DNA polymerases delta and epsilon and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999; 19:2000–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schanz S., Castor D., Fischer F., Jiricny J.. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5593–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thomas D.C., Umar A., Kunkel T.A.. Measurement of heteroduplex repair in human cell extracts. Methods. 1995; 7:187–197. [Google Scholar]
- 48. Jin Y.H., Garg P., Stith C.M., Al-Refai H., Sterling J.F., Murray L.J., Kunkel T.A., Resnick M.A., Burgers P.M., Gordenin D.A.. The multiple biological roles of the 3′→5′ exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol. Cell. Biol. 2005; 25:461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tran H.T., Degtyareva N.P., Gordenin D.A., Resnick M.A.. Genetic factors affecting the impact of DNA polymerase delta proofreading activity on mutation avoidance in yeast. Genetics. 1999; 152:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edelbrock M.A., Kaliyaperumal S., Williams K.J.. DNA mismatch repair efficiency and fidelity are elevated during DNA synthesis in human cells. Mutat. Res. 2009; 662:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jin Y.H., Obert R., Burgers P.M., Kunkel T.A., Resnick M.A., Gordenin D.A.. The 3′→5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5122–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Prakash S., Prakash L.. Nucleotide excision repair in yeast. Mutat. Res. 2000; 451:13–24. [DOI] [PubMed] [Google Scholar]
- 53. Ramilo C., Gu L., Guo S., Zhang X., Patrick S.M., Turchi J.J., Li G.M.. Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Molecular and cellular biology. 2002; 22:2037–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Genschel J., Modrich P.. Functions of MutLalpha, replication protein A (RPA), and HMGB1 in 5′-directed mismatch repair. J. Biol. Chem. 2009; 284:21536–21544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McGowan C.H., Russell P.. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 2004; 16:629–633. [DOI] [PubMed] [Google Scholar]
- 56. Lindsey-Boltz L.A., Reardon J.T., Wold M.S., Sancar A.. In vitro analysis of the role of replication protein A (RPA) and RPA phosphorylation in ATR-mediated checkpoint signaling. J. Biol. Chem. 2012; 287:36123–36131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guo S., Zhang Y., Yuan F., Gao Y., Gu L., Wong I., Li G.M.. Regulation of replication protein A functions in DNA mismatch repair by phosphorylation. J. Biol. Chem. 2006; 281:21607–21616. [DOI] [PubMed] [Google Scholar]
- 58. Pieper R.O. Understanding and manipulating O6-methylguanine-DNA methyltransferase expression. Pharmacol. Ther. 1997; 74:285–297. [DOI] [PubMed] [Google Scholar]
- 59. Mojas N., Lopes M., Jiricny J.. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007; 21:3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stojic L., Mojas N., Cejka P., Di Pietro M., Ferrari S., Marra G., Jiricny J.. Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev. 2004; 18:1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang Y., Qin J.. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kat A., Thilly W.G., Fang W.H., Longley M.J., Li G.M., Modrich P.. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aquilina G., Biondo R., Dogliotti E., Bignami M.. Genetic consequences of tolerance to methylation DNA damage in mammalian cells. Carcinogenesis. 1993; 14:2097–2103. [DOI] [PubMed] [Google Scholar]
- 64. Schaetzlein S., Chahwan R., Avdievich E., Roa S., Wei K., Eoff R.L., Sellers R.S., Clark A.B., Kunkel T.A., Scharff M.D. et al. . Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2470–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kuntz K., O’Connell M.J.. Initiation of DNA damage responses through XPG-related nucleases. EMBO J. 2013; 32:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kaina B., Ziouta A., Ochs K., Coquerelle T.. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex-, Mex+ and methylation-tolerant mismatch repair compromised cells: facts and models. Mutat. Res. 1997; 381:227–241. [DOI] [PubMed] [Google Scholar]
- 67. Cejka P., Stojic L., Mojas N., Russell A.M., Heinimann K., Cannavo E., di Pietro M., Marra G., Jiricny J.. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003; 22:2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mastrocola A.S., Heinen C.D.. Nuclear reorganization of DNA mismatch repair proteins in response to DNA damage. DNA Repair. 2010; 9:120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gary R., Park M.S., Nolan J.P., Cornelius H.L., Kozyreva O.G., Tran H.T., Lobachev K.S., Resnick M.A., Gordenin D.A.. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 1999; 19:5373–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lange S.S., Takata K., Wood R.D.. DNA polymerases and cancer. Nat. Rev. Cancer. 2011; 11:96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tanaka S., Cao K., Niimi A., Limsirichaikul S., Miao H.Q., Nakamura N., Murate T., Hasegawa Y., Takahashi T., Suzuki M.. Functions of base selection step in human DNA polymerase alpha. DNA Repair. 2010; 9:534–541. [DOI] [PubMed] [Google Scholar]
- 72. Agbor A.A., Goksenin A.Y., LeCompte K.G., Hans S.H., Pursell Z.F.. Human Pol epsilon-dependent replication errors and the influence of mismatch repair on their correction. DNA Repair. 2013; 12:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Morrison A., Bell J.B., Kunkel T.A., Sugino A.. Eukaryotic DNA polymerase amino acid sequence required for 3′—-5′ exonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 1991; 88:9473–9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Simon M., Giot L., Faye G.. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991; 10:2165–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Albertson T.M., Ogawa M., Bugni J.M., Hays L.E., Chen Y., Wang Y., Treuting P.M., Heddle J.A., Goldsby R.E., Preston B.D.. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:17101–17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Prolla T.A., Christie D.M., Liskay R.M.. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol. Cell. Biol. 1994; 14:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Briggs S., Tomlinson I.. Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J. Pathol. 2013; 230:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Keijzers G., Bohr V.A., Rasmussen L.J.. Human exonuclease 1 (EXO1) activity characterization and its function on flap structures. Biosci. Rep. 2015; 35:e00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pavlov Y.I., Mian I.M., Kunkel T.A.. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr. Biol.: CB. 2003; 13:744–748. [DOI] [PubMed] [Google Scholar]
- 81. Nick McElhinny S.A., Kissling G.E., Kunkel T.A.. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta}. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21070–21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lujan S.A., Williams J.S., Pursell Z.F., Abdulovic-Cui A.A., Clark A.B., Nick McElhinny S.A., Kunkel T.A.. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012; 8:e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Liberti S.E., Larrea A.A., Kunkel T.A.. Exonuclease 1 preferentially repairs mismatches generated by DNA polymerase alpha. DNA Repair. 2013; 12:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pavlov Y.I., Newlon C.S., Kunkel T.A.. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell. 2002; 10:207–213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.