Abstract
Structured RNAs such as ribozymes must fold into specific 3D structures to carry out their biological functions. While it is well-known that architectural features such as flexible junctions between helices help guide RNA tertiary folding, the mechanisms through which junctions influence folding remain poorly understood. We combine computational modeling with single molecule Förster resonance energy transfer (smFRET) and catalytic activity measurements to investigate the influence of junction design on the folding and function of the hairpin ribozyme. Coarse-grained simulations of a wide range of junction topologies indicate that differences in sterics and connectivity, independent of stacking, significantly affect tertiary folding and appear to largely explain previously observed variations in hairpin ribozyme stability. We further use our simulations to identify stabilizing modifications of non-optimal junction topologies, and experimentally validate that a three-way junction variant of the hairpin ribozyme can be stabilized by specific insertion of a short single-stranded linker. Combined, our multi-disciplinary study further reinforces that junction sterics and connectivity are important determinants of RNA folding, and demonstrates the potential of coarse-grained simulations as a tool for rationally tuning and optimizing RNA folding and function.
INTRODUCTION
Helical junctions are ubiquitous architectural features of nucleic acids. They can be classified into different groups based on the number of helical arms and by the presence of additional nucleotides at the branch point. A variety of junctions, such as bulges, three-way and four-way junctions (3WJ and 4WJ), connect constituent base-paired helices (1–4). Many studies have demonstrated that such junctions encode specific conformational preferences that can exert a powerful influence on DNA and RNA tertiary folding and stability (1,5–7). Although this topic has been studied for decades, junction-induced inter-helical bending in RNA is still not sufficiently understood for effective utilization in rational RNA structure design. Here, we investigate the contribution of junction topology (8) in the folding and function of the model hairpin ribozyme using coarse-grained simulations and single molecule fluorescence microscopy. These tools allow complementary prediction and direct measurement, respectively, of the impact of junction-induced inter-helical bends on folding, and lead us to test their relevance to catalytic activity of the hairpin ribozyme.
The hairpin ribozyme is a small self-cleaving RNA enzyme found in the negative strand of tobacco ringspot virus satellite RNA, where it is responsible for generating unit-length circular satellite RNA (3,9–13). This naturally occurring ribozyme is a classical model of a tertiary structured RNA, consisting of two primary structural elements: a central conserved four-way junction (4WJ), and two internal loops (A and B) that form long-range tertiary interactions to establish the active site (Figure 1) (14). Multiple studies have dissected the role of this central junction in hairpin ribozyme folding, and have shown that the natural 4WJ is the ideal topology for promoting fast, stable ‘docking’ of the A and B loops (6,15,16). By contrast, other junction topologies, such as a 3WJ or 2WJs with bulges of various sizes, render the ribozyme more amenable to gene therapeutic applications against external substrates and increase cleavage rate (due to more rapid product release) (17). However, these alternative topologies exhibit significantly reduced docking stabilities (ΔΔG = 4–8 kJ/mol) (6,15,18). The folding stability of the 4WJ hairpin ribozyme variant has been primarily attributed to favorable interhelical stacking interactions formed by the 4WJ in the docked conformation, with differences in folding entropy also playing a potential role (19,20). Still, the driving forces through which different junctions affect folding have remained poorly understood.
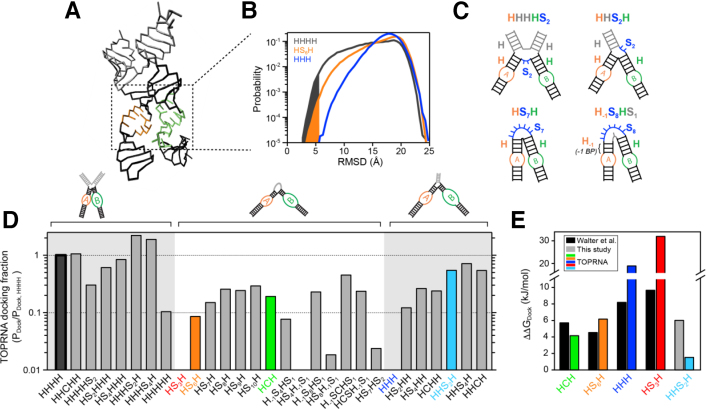
Figure 1.
Junction topological constraints are key determinants of docking propensity. (A) The 4WJ (HHHH) hairpin ribozyme in its docked conformation (catalytic core boxed), shown in its TOPRNA coarse-grained representation. The A and B loops used for RMSD calculations are colored orange and green, respectively. (B) RMSD distribution of the A and B loops from their native docked conformation for three ribozyme variants, with the docking probability (Pdock) highlighted as the area under the curve. (C) Depiction of the HS junction nomenclature using four example hairpin variants. (D) Docking probabilities relative to HHHH of different junctions computed from TOPRNA simulations. Junctions of particular interest are colored. (E) Comparison between ΔΔGdock computed from TOPRNA simulations and by prior bulk FRET experiments (6). ΔΔGdock values are computed relative to the HHHH ribozyme variant. Bars are colored as in panel D. The experimental ΔΔGdock value for the HHS2H junction was computed as ΔΔGdock(HHS2H, HHH) + ΔΔGdock(HHH, HHHH), where ΔΔGdock(HHS2H, HHH) is the free energy difference between the HHS2H and HHH junctions measured by smFRET, and ΔΔGdock(HHH, HHHH) is the previously measured difference (6).
Recent work by us and others have emphasized the importance of topological constraints posed by the steric and connectivity properties of junctions in RNA tertiary folding (20–26). Studies of a variety of model systems, including 2WJs and the 4WJ in tRNA, have shown that these basic topological constraints strongly limit the set of three-dimensional conformations accessible to an RNA molecule. By using optimal junction topologies, natural RNAs harness topological constraints to prevent formation of non-native tertiary contacts, and tune the entropic cost of tertiary folding. Taken together, these prior studies suggest that topological constraints may be an important consideration for de novo structure design efforts. Unfortunately, topological constraints are highly system dependent, and their effects on folding—and particularly their potential entropic consequences—are challenging to predict a priori. For this purpose, we previously developed the TOPRNA coarse-grained molecular dynamics model, which allows measuring how a pre-defined secondary structure constrains RNA tertiary folding. Enthalpic interactions such as stacking (27), tertiary hydrogen bonding, and electrostatics (28,29) are ignored, allowing TOPRNA to efficiently sample the range of topologically accessible tertiary conformations. Conformations that are geometrically strained and hence entropically disfavored are sampled with lower probability. Thus, comparisons between simulations of different RNA constructs allow identification of junctions that entropically favor or disfavor a folded conformation. While the deliberate simplifications of the TOPRNA model render it insensitive to nucleotide substitutions that, for example, disrupt a tertiary interaction, previous work has shown that TOPRNA can rationalize the observed effects of junction modifications on tertiary folding stability (22–24).
In the current work, we apply coarse-grained simulations and single molecule Förster resonance energy transfer (smFRET) experiments to systematically explore the role of junction topology in hairpin ribozyme folding. Our results support that geometric accessibility of the folded conformation plays a vital role in RNA folding and demonstrate the utility of TOPRNA as a useful tool in RNA structure design.
MATERIALS AND METHODS
Junction nomenclature
We use the HS notation (30) to concisely abbreviate different junction topologies of the hairpin ribozyme (Figure 1C). The junction components are indicated as H (helix), S (strand, or unpaired nucleotides), and C (a ‘cut’ or break in junction connectivity), listed beginning with helix A (containing loop A), and moving in the clockwise direction around the junction, ending with the connection between helix B (containing loop B) and helix A. Subscripts are used to denote the nucleotide length of S components (e.g. S2 corresponds to a two-nucleotide long single-strand). In addition to exploring modifications of the junction, we explored the effects of altering the number of base pairs separating loops A and B from the junction. In the canonical hairpin ribozyme, helix A and helix B are four and five base pairs long, respectively (in other words, four and five base pairs separate loop A and loop B from the junction). We use subscripts to indicate the number of base pairs that have been added (e.g. H+1) or deleted (e.g. H–1) in helix A and/or B with respect to the canonical reference (Figure 1C).
TOPRNA simulations
Simulations were run using the TOPRNA coarse-grained model. TOPRNA models of the hairpin ribozyme were built using the toprnaCreate.pl Perl utility (available from http://brooks.chem.lsa.umich.edu). The ‘fromc’ flag in the Perl script enforced the correct initial positions (PDB ID 1M5K) (3) of the loop regions when generating the molecule. Dihedral constraints (‘cons’ flags in the Perl script) held the A and B loop regions in their native conformation so that no rearrangements of the loop regions are required for docking. The sequence of each simulated molecule is listed in Supplementary Table S1.
Each construct was simulated for 200 μs across 80 WExplore (31) replicas at a temperature of 300 K. All simulations were run using the CHARMM software (32). To determine the docking probability, Pdock, we used the RMSD of the sugar sites of loop regions A and B from the active conformation (from the crystal structure, PDB ID 1M5K) (3). We restricted calculation of the RMSD to the loop regions since these loops form the active site in the docked state and were identical across all simulations (maintaining consistency of the order parameter among constructs). We defined docked conformations as all states where RMSD <5.5 Å. The relevant information obtained from Pdock is whether one topology increases/decreases docking propensity, and whether this increase/decrease is more or less dramatic than in other junction topologies. Repeating Pdock calculations using larger RMSD cutoffs of 9 and 15 Å yielded comparable conclusions, supporting that our predictions did not depend on a parameter specific to the simulation (not shown).
RNA purification and labeling
A three-piece hairpin ribozyme construct (with strands RzA, RzB and non-cleavable substrate, NCS) was used to assemble the three-way junction hairpin ribozyme. RzA: Cy5-AAA UAG AGA AGC GAA CCA GAG AAA CAC ACG CCA AA-C7-NH2; RzB: 5′-Biotin AUA UAU UUG GCG UGG UAC AUU ACC UGG UAC GAG UUG AC-3′; or RzBm: 5′-Biotin AUA UAU UUG GCG UGG UAC AUU ACC UGG UUUAC GAG UUG AC-3′; NCS: 5′- GUC AAC UCG UUC GC (2′OMeA) GUC CUA UUU-3′. The 2′-O-methyl-adenosine (2′OMeA) modification at the cleavage site prevents cleavage during structural probing by smFRET; an unmodified A residue replaced this nucleotide for cleavage assays. RNAs were purchased from the Keck Foundation Resource Laboratory at the Yale University School of Medicine. The 2′-hydroxyl protection groups were deprotected following the manufacturer's protocol (http://medicine.yale.edu/keck/oligo/services/protocols/RNA.aspx). The RNA was purified by denaturing gel electrophoresis (20% (w/v) polyacrylamide and 8 M urea). The RNAs were eluted using the crush-and-soak method overnight at 4°C in 0.5 M NaOAc (pH 5.3) and 0.1 mM EDTA, followed by chloroform extraction, ethanol precipitation, and C18 reverse-phase HPLC (buffer of 20% acetonitrile, 80% 100 mM TEAA pH 7.0). The 3′-end C7 amino linker (C7-NH2) was labeled with Cy3 succinimidyl-ester (GE Healthcare) in labeling buffer (0.1 M sodium carbonate, pH 8.5) overnight at room temperature. This step was followed by ethanol precipitation and further C18 reverse-phase HPLC to remove free dye (33). RNA concentrations were measured using a NanoDrop and calculated after background subtraction.
Single molecule FRET
Single molecule experiments were performed according to previously described protocols (34,35). The ribozyme strands (RzA and RzB) were heated to 90°C in standard buffer (50 mM Tris–HCl, pH 7.5, 12 mM MgCl2) for 45 seconds before cooling to room temperature over 15 min. The annealed strands were diluted to 25–50 pM and immobilized onto a fused silica slide through a biotin-streptavidin interaction. Ribozyme assembly was completed by adding 400 nM NCS strand in standard buffer, with the 2′OMeA modification at the cleavage site preventing cleavage. Experiments were performed in standard buffer with an oxygen scavenger system composed of 43 nM protocatechuate dioxygenase, 4.3 mM protocatechuate and 4.3 mM Trolox to reduce the photobleaching of the fluorophores. We excited the donor fluorophore (Cy3) using a home-built total internal reflection microscope with a green laser (532 nm, ∼12 mW at the slide). We separated emission from the donor and acceptor using a dichroic mirror (Chroma 610DCXR), filtered them individually (Chroma HQ580–60M and HQ655LP), and detected them as side-by-side images on an intensified CCD camera (I-Pentamax, Princeton Instruments).
Single molecule data were processed and analyzed as previously described (34–36). Each trajectory was analyzed using MATLAB codes to identify single molecules using the following selection criteria: Molecules with stable fluorescence emission, anti-correlated donor–acceptor emission intensities and single step photobleaching were counted. Apparent FRET was calculated from the intensities of the donor divided by donor plus and acceptor fluorophore. Each selected FRET trajectory was binned into a FRET histogram. Histograms of over 100 molecules were combined and fitted with a sum of Gaussian functions using Microcal Origin (36). The number of peaks in the histogram characterizes the number of conformational states. The values of ΔGdock were calculated by converting the FRET histograms into values of ΔGdock using the following equations:
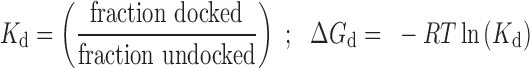 |
Kinetic information was calculated from single molecule measurements by dwell time analysis of individual states. Consistent with the aggregate histogram analysis, each molecule's trajectory showed a two-state system. Discrete FRET states were determined through hidden Markov modeling and dwell time histograms were constructed for docking and undocking transitions (HMM). Previous single molecule studies have shown that the hairpin ribozyme exhibits heterogeneous docking and undocking kinetics (34,35,37). Therefore, following these studies, docking and undocking transitions were fit with a double-exponential function using Microcal Origin:
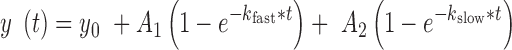 |
kfast and kslow represent apparent fast and slow rate constants, respectively, for each transition, and A1 and A2 represent the total number of fast and slow transitions. To facilitate comparisons between different ribozyme architectures, rather than simply use single-exponential fits, we report a single kundock or kdock rate constant more rigorously calculated by taking the weighted average of kfast and kslow:
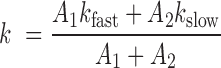 |
Cleavage assays
Cleavage reactions were performed under single-turnover conditions in standard buffer (50 mM Tris–HCl, pH 7.5, 12 mM MgCl2) at room temperature. Ribozyme strands RzA and RzB (for design HHH) or RzBm (for design HHS2H) were heated at 90°C for 45 seconds in the absence of MgCl2, followed by slow cooling over 15 min at room temperature in the presence of MgCl2. 5′-32P-labeled cleavable substrate was prepared by phosphorylation with T4 polynucleotide kinase and [γ-32P]-ATP. The 5′-32P-labeled substrate (<1 nM final concentration; with an unmodified A at the cleavage site) was mixed with the ribozyme at 200 nM final concentration. Aliquots of the reaction were removed at different time points and quenched with an equal volume of loading buffer (60 mM EDTA and 95% formamide). The 5′ cleavage product (P) was separated from uncleaved substrate (S) by denaturing PAGE (8 M urea, 20% (w/v)), and was quantified and normalized to the sum of the substrate and product bands using a Typhoon 9410 Variable Mode Imager (GE Healthcare Life Sciences) with IMAGEQUANT software (Molecular Dynamics). Following the fitting approach used to derive docking kinetics, the fraction cleaved over time was globally fit with a double-exponential of the form:
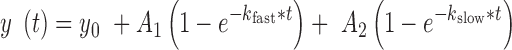 |
where (A1 + A2) is the final extent of cleavage and kfast and kslow are the apparent fast and slow rate constants, respectively (34,35,37). The fast- and slow-cleaving ribozyme populations were previously found to correlate with the distinct (un)docking propensities observed in the single molecule data, where in particular slower undocking leads to faster cleavage and vice versa (34,35).
RESULTS
Topological constraints help explain hairpin ribozyme folding propensity
To understand the potential contributions of junction topological constraints to hairpin ribozyme tertiary folding, we performed TOPRNA (21) simulations of a series of fixed ribozyme architectures previously characterized by bulk FRET assays (6). Comprehensive computational sampling of folded (docked) and various undocked conformations was achieved using the enhanced sampling algorithm WExplore (31). We measured the root mean square deviation (RMSD) of sugar sites in loop regions A and B from the corresponding sites in the crystal structure (3), defining proximity to the docked conformation (Figure 1b). Docked states were defined to be those with RMSD <5.5 Å, giving rise to a probability for docking, Pdock, among all conformations sampled.
Significantly, our simulations revealed large variations in Pdock across the different junction topologies, implying that varied topological resistance of juxtaposing loops A and B can significantly affect hairpin ribozyme folding propensity (Figure 1B and D). The natural HHHH architecture possessed the highest Pdock (absolute Pdock = 3.9 × 10−3), consistent with experimental studies indicating that it is the most favorable for docking (6,15,16,19). To facilitate comparison, we therefore consider Pdock of other architectures relative to that of the HHHH. The HCH and HS6H variants sample the docked conformation with roughly one-tenth the probability of the HHHH architecture (orange and green bars, Figure 1D). By comparison, the HS3H and HHH architectures have relative Pdock values of 3 × 10−6 and 5 × 10−4, respectively (out of range in Figure 1D), indicating that topological constraints impose a very strong entropic penalty on docking.
Qualitatively, the HHHH > HCH > HS6H > HHH > HS3H trend we observe in Pdock matches the experimental trend (6,19). To provide a more quantitative comparison between our simulations and experiment, we converted Pdock to a free energy, ΔΔGdock, which reflects the entropic cost posed by topological constraints on adopting the docked conformation relative to the HHHH topology:
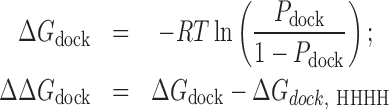 |
where R is the ideal gas constant and T is 300 K. As shown in Figure 1E, the TOPRNA derived ΔΔGdock values of 4–6 kJ/mol for the HCH and HS6H variants are in excellent agreement with experiment (estimated error from replicate simulations is ΔΔGdock ≈ 2 kJ/mol). This agreement suggests that much of the difference in docking stability between the HHHH, HCH and HS6H variants can be explained by different topological accessibilities. We note that this quantitative agreement is consistent with studies of other RNAs, which have shown that ΔG values measured by TOPRNA represent real contributions to the free energy landscape of RNA tertiary folding (21,23,24).
By contrast, the ΔΔGdock values measured by TOPRNA for the HHH and HS3H junctions are significantly greater than the experimentally observed values (8 versus 19 kJ/mol, and 10 versus 32 kJ/mol, respectively; Figure 1E; Supplementary Table S2). The high ΔΔGdock predicted by TOPRNA reflects the extremely low probability of sampling docked conformations in our simulations, and indicates that these ribozymes can only dock under significant strain. This is intuitive when one considers that docking requires a parallel positioning of the A and B helices, and hence the linker connecting the helices must be able to span across two helices; neither the single helix of the HHH junction nor the 3-nt bulge of the HS3H junction is long enough. The experimental observation that the HHH and HS3H junctions dock with ΔΔGdock ≈ 10 kJ/mol therefore likely reflects base pair melting or some other conformational change of the secondary structure that mediates docking. In other words, our simulations report on a fixed secondary structure, whereas real RNAs can adopt a higher energy secondary structure in order to form otherwise inaccessible tertiary interactions. Supporting that such a tradeoff occurs, studies have observed fraying of the junction base pairs upon docking of the hairpin ribozyme (38,39). We also note that HS3H ribozymes are inactive, suggesting that the ΔΔGdock = 10 kJ/mol measured by FRET may underreport the effect of this junction on docking (40,41). Regardless, we emphasize that our simulations do a good job of explaining the docking variation among different junctions, and hence argues for an essential role of topological constraints in hairpin ribozyme folding.
Systematic screen of junction architectures discovers stabilizing modifications
To better understand how topological constraints affect hairpin ribozyme docking, and to identify potential novel stabilizing modifications, we repeated our simulations for an additional 42 junction variants. We systematically explored the folding propensity of the 2WJ, 3WJ and 4WJ junction architectures, introducing changes such as extending the junction circumference by introducing single-stranded joiners, cutting (disconnecting) joiners, and extending helices (Figure 1D).
For the 4WJ architecture, we observed an interesting dependence of Pdock on junction connectivity. Cutting the junction (HHCHH) has no effect on Pdock, which indicates that steric rather connectivity constraints are responsible for the favorable docking properties of the HHHH topology. Notably, the Pdock of the HHCHH variant is also 5-fold higher than the HCH junction, which has identical connectivity constraints. Thus, steric constraints posed by the two ‘non-functional’ junction helices exclude significant conformational space, biasing the junction towards docking. Modifying the connectivity of the junction via insertion of single-stranded linkers either penalizes docking (HHHHSn or HSnHHH) or promotes (HHHSnH) docking, depending on the location of the insertion. Entropy drives these linkers to adopt semi-extended conformations, which in turn biases the junction towards or away from docking-component conformations; the location-dependence of this effect highlights the anisotropy of the topological constraints on the junction. We also tested the effect of inserting an additional helix into the junction (creating a 5WJ; abbreviated HHHHH). While the extra helix extends the circumference of the junction, the sterics and rigidity of this helix pose additional constraints that substantially decrease docking propensity (Figure 1D). Lastly, we explored the importance of the 4WJ in juxtaposing the A and B loops in the correct rotational and translational register for docking by deleting base pairs in the respective helices (Supplementary Figure S1). Individual deletions have only minor effects on Pdock, while shortening both helices increases Pdock 2-fold. Thus, while a HHHH topology biases the junction towards docking-competent conformations, it is not particularly optimized for orienting the A and B loops in the correct register. Experimental studies have also observed that the hairpin ribozyme is tolerant of shortening the A and B helices (42).
For the 2WJ architecture, increasing the length of the bulge linker leads to modest increases in Pdock (Figure 1D), matching trends observed in cleavage assays (40,41) (Supplementary Table S2). This increase derives partly from a relaxation of connectivity constraints, which reduce entropic strain experienced by the shorter HS6H junction. Bulges longer than eight nucleotides also have higher Pdock than the HCH junction (which lacks connectivity constraints), and hence as observed for HHHSnH topologies, linker entropy can bias the junction towards docked conformations. We also tested the hypothesis that docking of the HS3H variant is mediated by base pair melting at the junction. Strikingly, melting one base pair in the A helix (H-1S4HS1) results in a 104-fold increase in Pdock, yielding a docking propensity comparable to the HS6H junction (Figure 1D). Again highlighting the anisotropy of the topological constraints on the junction, melting a base pair in the B helix does not yield comparable increases in Pdock. This observation is consistent with the experimentally observed fraying of the A helix, but not B helix (38,39). For the less constrained HCH and HS7H junctions, melting a base pair in the A helix results in modest increases in Pdock, whereas melting in the B helix disfavors docking. Similar to the 4WJ, changing the register of the A and B loops via deletion of helix base pairs modestly improves docking (H-1S7H and H-1S7H-1; Supplementary Figure S1). However, adding base pairs to either the A or B helices decreases Pdock 50-fold. This deletion/addition asymmetry arises from the chirality of the helices. Additional base pairs rotate the A and B loops away from one another, causing the A and B helices to sterically block docking. By contrast, deleting base pairs effectively rotates the loops towards each other, alleviating steric constraints.
Lastly, we explored modifications of the 3WJ architecture with the hope of identifying stabilizing variants. Strikingly, our simulations predict that inserting a 2-nt single-strand adjacent to the B helix (HHS2H) increases Pdock by 1000-fold, resulting in a docking propensity greater than any 2WJ and within 50% of the natural HHHH junction (blue, Figure 1d). Inserting a single-stranded linker adjacent to the A helix (HS2HH) also significantly increases Pdock, but to a lesser degree than HHS2H. A similar asymmetry is observed when the junction connectivity is cut (HCHH versus HHCH). This asymmetry is consistent with the trend observed in 4WJs, and indicates that steric constraints exerted by the ‘non-functional’ helix play a central role in biasing the junction towards docking-competent conformations. Increasing the length of the inserted linkers from 2-nt to 4-nt modestly boosts Pdock relative to the cut junctions, indicating that longer linkers can promote docking via their ‘entropic spring’ behavior (Figure 1D, Supplementary Table S2).
Taken together, our simulations indicate that complex steric and connectivity constraints posed by the hairpin ribozyme junction strongly modulate folding propensity. Importantly, our results help explain prior experimental observations, while also providing predictive insight into potential new junction topologies. In particular, sterics appear to be responsible for the favourable docking properties of the natural HHHH junction, while connectivity constraints limit docking for other topologies. With the HHS2H design, we show that these connectivity constraints can be alleviated while still harnessing the volume excluding properties of additional junction helices, yielding significant increases in docking propensity.
smFRET validates that the HHS2H modification increases docking
We next sought to validate the prediction from our simulations that docking should be stabilized in the HHS2H variant. Notably, previous attempts to rationally improve docking of the 3WJ hairpin ribozyme architecture were unsuccessful (6), making HHS2H junctions a particularly interesting target for experimental follow-up. Another study concluded that catalytic turnover of the hairpin ribozyme is unaffected by a HHS2H modification, contrary to our prediction (18) (Supplementary Table S2). However, this study relied on a multiple-turnover kinetics assay that convolutes several molecular steps, including substrate binding and release, making it impossible to infer docking efficiency or unimolecular catalytic rates. Therefore, we pursued our own validation experiments on the HHS2H junction.
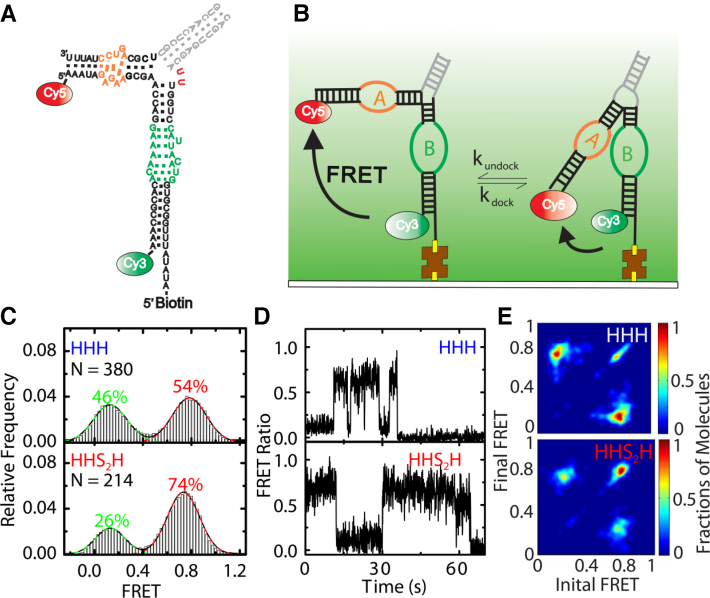
We performed smFRET experiments using prism-based total internal reflection fluorescence (TIRF) microscopy to dissect the thermodynamics and kinetics of docking for both the HHS2H with HHH ribozymes. With our fluorophore labeling scheme, we monitored the undocked and docked conformations as low and high FRET states, respectively (Figure 2A and B) (34,35,43). FRET probability distributions comprising several hundred molecules under standard ribozyme conditions (50 mM Tris–HCl, pH 7.5, 12 mM MgCl2 at room temperature) reveal two populations, as expected, with a significant stabilization of docked state from 54% to 74% from HHH to HHS2H (Figure 2C). When converting these fractions to the free energy of docking, we find that ΔGdock = –0.38 ± 0.17 kJ/mol for the HHH junction, in good agreement with prior measurements (6,19). Importantly, for the HHS2H junction, ΔGdock = –2.55 ± 0.21 kJ/mol, indicating that the additional linker stabilizes docking by –2.17 kJ/mol. This value is less than the –17.3 kJ/mol stabilization predicted by our simulations, but as discussed above, TOPRNA substantially overestimates the ΔGdock of the HHH junction, and therefore quantitative agreement was not expected. As an alternative comparison, our simulations predict that the ΔΔGdock = 1.5 kJ/mol between the HHS2H and HHHH junctions; combining our measurements with prior experimental data, we derive ΔΔGdock = 6.0 kJ/mol between the HHS2H and HHHH junctions (Figure 1E). Thus, TOPRNA modestly over-predicts the stability of the HHS2H ribozyme. One potential explanation is that base-stacking interactions not considered by TOPRNA stabilize the undocked conformation of HHS2H junction. Nevertheless, we emphasize that the -2 kJ/mol stabilization measured here is significant, and is comparable to the stability differences between ‘good’ and ‘poor’ docking 2WJ and 4WJ variants.
Figure 2.
Adding a 2-nucleotide loop to a 3WJ stabilizes loop A:loop B docking as monitored by smFRET. (A) Schematic representation of the secondary structure, sequence and FRET labeling of the HHS2H hairpin ribozyme; the two red U residues are not present in the HHH design. (B) The 3WJ hairpin ribozyme was immobilized on a quartz slide via a biotin-streptavidin linkage and imaged by TIRF microscopy. (C) FRET histogram analysis revealing two major FRET states fitted with Gaussian distributions. (D) Representative trajectories of dynamic single HHH and HHS2H hairpin ribozyme molecules. (E) Heat map of the transition occupancy density plots (TODP) for HHH and HHS2H constructs (35). TODP plots are scaled by the fraction of all the molecules within a population that exhibit a transition between high- and low-FRET. The population on the diagonal represents the fraction of static molecules that do not interconvert within the time window of their FRET trajectory.
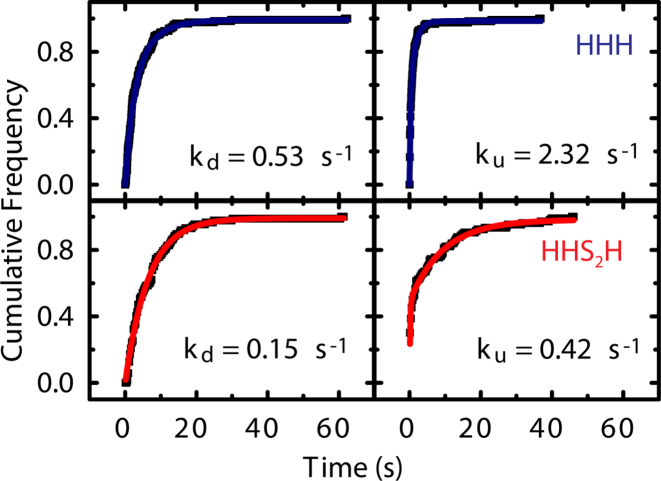
We also examined the docking kinetics of the two junctions. Representative FRET time trajectories of single ribozyme molecules reveal slower switching dynamics between the two FRET states for the relaxed HHS2H ribozyme (Figure 2D). In both variants, dynamic and static high-FRET states are observed, with the fraction of static high-FRET molecules increasing from 29% in the HHH ribozyme to 44% in the HHS2H ribozyme (Figure 2E). A cumulative dwell time analysis of the dynamic trajectories found weighted average docking (kdock) and undocking rate constants (kundock) of 0.53 s−1 and 2.32 s−1, respectively, for HHH; and 0.15 s−1 and 0.42 s−1, respectively, for HHS2H (Figure 3) (36). This difference in kinetics indicates that the two ribozymes have different docking transition states. The slower undocking rate of the HHS2H variant is consistent with our prediction that the ribozyme should form stronger tertiary interactions due to lower junction strain or more docking-conducive overall architecture. However, it is perhaps surprising that the HHS2H junction ribozyme also exhibits slower docking kinetics, since our simulations suggest that the HHH junction must undergo some form of local melting or conformational change in order to dock. One possibility is that the HHH junction secondary structure is never fully formed, and hence there is no ‘melting’ barrier to docking. Alternatively, the HHS2H junction may adopt a particularly stable ‘undocked’ conformation with a high-energy barrier that slows docking. While interesting, we note that our data are unable to resolve the cause of these differences in kinetics.
Figure 3.
Kinetic analysis of the time spent in the undocked and the docked states for HHH (top panel) and HHS2H (bottom panel) to derive the weighted-average rate constants kd(ock) and ku(ndock), respectively (see Materials and Methods).
The HHS2H ribozyme has enhanced catalytic activity
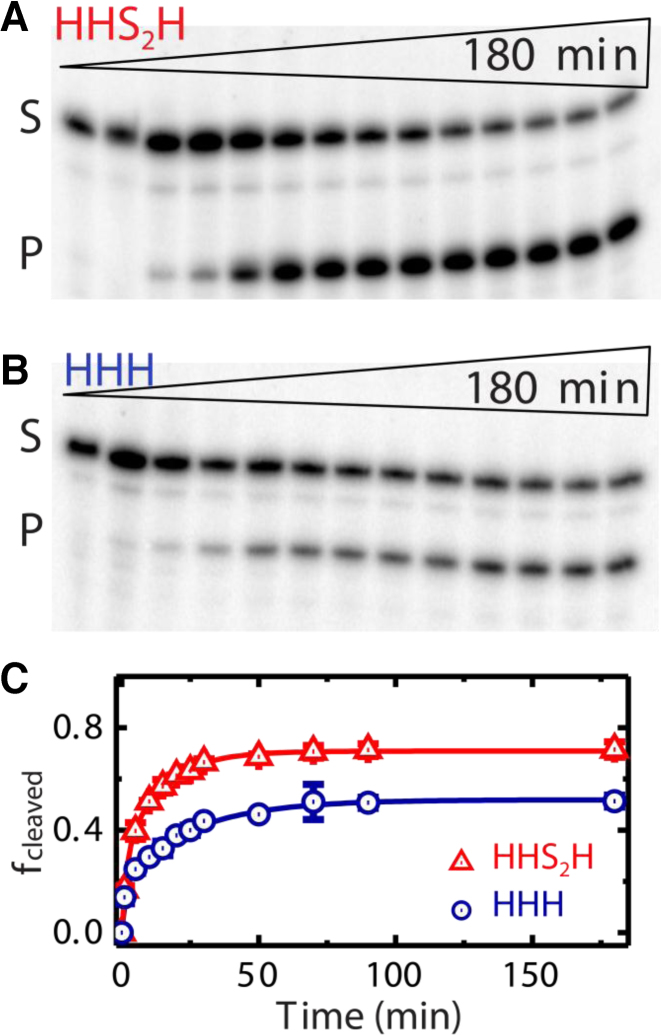
We finally tested whether the stabilized docking of the HHS2H ribozyme translates into enhanced catalytic function by performing cleavage assays and quantifying the fractions of substrate (S) and cleaved product (P) over time (Figure 4). For the HHH variant, the fraction cleaved increased with significant biphasic kinetics, where the rate constants of the fast cleaving fraction (A1 = 0.16 ± 0.02) and the slowly cleaving fraction (A2 = 0.33 ± 0.01) are kfast = 0.71 min−1 and kslow = 0.04 min−1; respectively (34,35,44). For the HHS2H variant, the data reveal a significant increase (>20%) in the final product fraction due to a higher fraction of molecules cleaving in the fast phase (A1 = 0.29 ± 0.03, with an unchanged rate constant of kf = 0.71 min−1), combined with an 8% increase in a slow fraction (A2 = 0.41 ± 0.02) cleaving ∼2-fold faster (ks = 0.08 min−1). Thus, the increased docking stability of the HHS2H junction as predicted by TOPRNA and observed by smFRET translates into increased functional activity of the ribozyme.
Figure 4.
Increased docking of the predicted design leads to enhanced ribozyme cleavage. (A) Gel electrophoretic analysis of the fraction of intact substrate (S) and cleaved product (P) as a function of time (0−180 min) for the HHS2H design. (B) Gel electrophoretic analysis of the fraction of intact substrate (S) and cleaved product (P) as a function of time (0−180 min) for the unmodified HHH design. (C) Fraction of cleaved product, fcleaved, as a function of time. The HHS2H ribozyme (kfast = 0.71 min−1, Afast = 29%; kslow = 0.078 min−1, Aslow = 41%) generates significantly more product than the HHH ribozyme (kfast = 0.71 min−1, Afast = 16%; kslow = 0.056 min−1, Aslow = 33%) under otherwise identical conditions. Error bars were calculated from at least two independent assays.
DISCUSSION
Previous studies have highlighted the importance of topological constraints encoded by secondary structure in the folding and dynamics of several RNA systems (20–26). In our current work, we sought to test the relevance of topological constraints in the folding of the hairpin ribozyme and, further, to harness these constraints to rationally improve tertiary folding. We rely on simulations of the TOPRNA coarse-grained model, which samples the ensemble of molecular conformations accessible to a fixed secondary structure (21). Tertiary interactions, inter-helical stacking, and electrostatics are ignored, and hence the probability at which a given conformation is sampled reflects the (primarily entropic) cost posed by topological constraints. Significantly, simulations of hairpin ribozymes with different junction architectures revealed a strong variation in the topological accessibility of the docked conformation, matching previously measured trends in docking stability (6,16,19). For the HCH and HS6H junction variants, TOPRNA quantitatively predicts the experimentally measured difference in docking free energy relative to the HHHH junction (6). Our simulations also corroborate the observation that the ribozyme is tolerant of base pair deletions in the A and B helices (42), and provides a physical explanation for why helix A but not helix B frays in the 2WJ hairpin ribozyme (38,39). Collectively, this agreement strongly argues that topological constraints imposed by the central junction are a key contributor to the docking stability of the hairpin ribozyme.
A comprehensive screen of alternative architectures revealed the primary mechanisms through which topological constraints influence the hairpin ribozyme, and enabled us to identify novel stabilizing modifications. Interestingly, the most important factor in the superior docking propensity of the natural HHHH architecture appears to be steric constraints imposed by the two additional ‘non-functional’ helices of the central junction. These helices exclude a large fraction of the ‘undocked’ conformational space, and hence bias the junction towards docked conformations. Connectivity constraints penalize other architectures, most notably the 3WJ HHH variant, as the circumference of the junction is too short to accommodate a parallel arrangement of the A and B helices. By strategically placing a linker adjacent to the B helix with the HHS2H modification, these connectivity constraints can be alleviated while simultaneously taking advantage of the favorable excluded volume properties of a third helix, dramatically improving docking propensity. Our smFRET data validate this TOPRNA-predicted stabilization of docking, and we further show that enhanced docking translates into increased catalytic activity (Figure 4). Our data thus demonstrate that less favorable junction architectures, such as that of the 3WJ, can be redesigned and predictably improved by minimizing the folding penalty posed by topological constraints.
The predictive power of TOPRNA simulations in understanding differences in hairpin ribozyme docking is consistent with prior studies on diverse RNAs, including 2WJs, tRNA, the Azoarcus ribozyme (21–24). While this predictive power may be surprising given the dramatic simplifications inherent to the TOPRNA model, we believe it can be understood in the context of the additive RNA free energy landscape. Topological constraints posed by the secondary structure specify a basal entropic component of the free energy landscape, which is then layered by the typically dominant contributions of electrostatics, base-stacking and tertiary hydrogen-bonding interactions (21). However, when comparing two different architectures of the hairpin ribozyme, the electrostatics and tertiary interactions components of the free energy will be comparable. Thus, on balance, the difference between two architectures will reduce to the difference in topological accessibility of the docked conformation. This argument is convenient, but is obviously limited. Notably, TOPRNA significantly over-predicts the docking penalties of the HS3H and HHH junctions, which likely fold through pathways involving secondary structure changes that violate the assumptions of our model. Folding of any RNA is dominated by enthalpic terms such as tertiary interactions and electrostatics (45–47), and understanding folding and stability requires a consideration of these components. In short, TOPRNA is a complementary tool, not a substitute, for understanding the complexities of RNA folding.
In designing a stably folding RNA, it is widely recognized that optimizing tertiary interactions and electrostatics is essential (48). The role of junctions, and by implicit extension topological constraints, has also long been appreciated, and is commonly taken into account during de novo design by selecting junction architectures observed in natural RNAs (49,50). However, physics-based tools for rationally designing new junctions have been lacking. Our study suggests that TOPRNA may be a broadly useful tool for such purposes. The simplifying assumptions of the TOPRNA model yield a dramatic improvement in conformational sampling over traditional simulation methods, rendering TOPRNA ideal for rapid RNA mutational screening. While we used WExplore to achieve comprehensive sampling, simulations using straightforward temperature replica exchange methods gave actionable answers in 2–3 days (∼500 cpu hours per junction; unpublished data). TOPRNA is freely available and is packaged with the TOPRNAcreate utility, which makes it easy to rapidly build models of alternative junction topologies. Due to its impact on folding and functional activity, junction conformational flexibility should become a vital consideration when interpreting the effect of junction topology on folding propensity, with important implications for applications such as designing RNAs with novel properties or as building blocks for RNA nanostructures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Chun for her help in preparing the RNA constructs, A. Dickson for assistance in implementing the WExplore methodology, and J. Widom for help editing the manuscript.
Footnotes
Present address: Anthony M. Mustoe, Department of Chemistry, University of North Carolina, Chapel Hill, NC 27599-3290, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM062357 to N.G.W.]; National Science Foundation [CHE1506273 to C.L.B.III]; National Science Foundation graduate research fellowship (to A.M.M.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ouellet J., Melcher S., Iqbal A., Ding Y., Lilley D.M.J.. Structure of the three-way helical junction of the hepatitis C virus IRES element. RNA. 2010; 16:1597–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cui Y., Kong D., Ghimire C., Xu C., Mao H.. Mutually exclusive formation of G-Quadruplex and i-Motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016; 55:2291–2299. [DOI] [PubMed] [Google Scholar]
- 3. Rupert P.B., Massey A.P., Sigurdsson S.T., Ferré-D’Amaré A.R.. Transition state stabilization by a catalytic RNA. Science. 2002; 298:1421–1424. [DOI] [PubMed] [Google Scholar]
- 4. Lescoute A., Westhof E.. Topology of three-way junctions in folded RNAs. RNA. 2006; 12:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mustoe A.M., Bailor M.H., Teixeira R.M., Brooks C.L. III, Al-Hashimi H.M.. New insights into the fundamental role of topological constraints as a determinant of two-way junction conformation. Nucleic Acids Res. 2012; 40:892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walter N.G., Burke J.M., Millar D.P.. Stability of hairpin ribozyme tertiary structure is governed by the interdomain junction. Nat. Struct. Biol. 1999; 6:544–549. [DOI] [PubMed] [Google Scholar]
- 7. Zacharias M., Hagerman P.J.. Bulge-induced bends in RNA: quantification by transient electric birefringence. J. Mol. Biol. 1995; 247:486–500. [DOI] [PubMed] [Google Scholar]
- 8. Bailor M.H., Mustoe A.M., Brooks C.L. III, Al-Hashimi H.M.. Topological constraints: using RNA secondary structure to model 3D conformation, folding pathways, and dynamic adaptation. Curr. Opin. Struct. Biol. 2011; 21:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berzal-Herranz A., Joseph S., Chowrira B.M., Butcher S.E., Burke J.M.. Essential nucleotide sequences and secondary structure elements of the hairpin ribozyme. EMBO J. 1993; 12:2567–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nesbitt S., Hegg L.A., Fedor M.J.. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997; 4:619–630. [DOI] [PubMed] [Google Scholar]
- 11. Hampel A., Cowan J.A.. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997; 4:513–517. [DOI] [PubMed] [Google Scholar]
- 12. Murray J.B., Seyhan A.A., Walter N.G., Burke J.M., Scott W.G.. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998; 5:587–595. [DOI] [PubMed] [Google Scholar]
- 13. Walter N., Burke J.. The hairpin ribozyme: structure, assembly and catalysis. Curr. Opin. Chem. Biol. 1998; 2:303. [DOI] [PubMed] [Google Scholar]
- 14. Walter N.G., Perumal S.. The small ribozymes: common and diverse features observed through the FRET lens. Springer Ser. Biophys. 2009; 13:103–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan E., Wilson T.J., Nahas M.K., Clegg R.M., Lilley D.M.J., Ha T.. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9308–9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Z.Y., Wilson T.J., Maxwell K., Lilley D.M.. The folding of the hairpin ribozyme: dependence on the loops and the junction. RNA. 2000; 6:1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller S., Appel B., Krellenberg T., Petkovic S.. The many faces of the hairpin ribozyme: structural and functional variants of a small catalytic RNA. IUBMB Life. 2012; 64:36–47. [DOI] [PubMed] [Google Scholar]
- 18. Komatsu Y., Shirai M., Yamashita S., Ohtsuka E.. Construction of hairpin ribozymes with a three-way junction. Bioorg. Med. Chem. 1997; 5:1063–1069. [DOI] [PubMed] [Google Scholar]
- 19. Klostermeier D., Millar D.P.. Helical junctions as determinants for RNA folding: origin of tertiary structure stability of the hairpin ribozyme †. Biochemistry. 2000; 39:12970–12978. [DOI] [PubMed] [Google Scholar]
- 20. Laing C., Schlick T.. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009; 390:547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mustoe A.M., Al-Hashimi H.M., Brooks C.L. III. Coarse grained models reveal essential contributions of topological constraints to the conformational free energy of RNA bulges. J. Phys. Chem. B. 2014; 118:2615–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mustoe A.M., Brooks C.L. III, Al-Hashimi H.M.. Topological constraints are major determinants of tRNA tertiary structure and dynamics and provide basis for tertiary folding cooperativity. Nucleic Acids Res. 2014; 42:11792–11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mustoe A.M., Liu X., Lin P.J., Al-Hashimi H.M., Fierke C.A., Brooks C.L. III. Noncanonical secondary structure stabilizes mitochondrial tRNA(Ser(UCN)) by reducing the entropic cost of tertiary folding. J. Am. Chem. Soc. 2015; 137:3592–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mustoe A.M., Al-Hashimi H.M., Brooks C.L. III. Secondary structure encodes a cooperative tertiary folding funnel in the Azoarcus ribozyme. Nucleic Acids Res. 2016; 44:402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bailor M.H., Sun X., Al-Hashimi H.M.. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010; 327:202–206. [DOI] [PubMed] [Google Scholar]
- 26. Chu V.B., Lipfert J., Bai Y., Pande V.S., Doniach S., Herschlag D.. Do conformational biases of simple helical junctions influence RNA folding stability and specificity. RNA. 2009; 15:2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walter A.E., Turner D.H., Kim J., Lyttle M.H., Müller P., Mathews D.H., Zuker M.. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Draper D.E., Grilley D., Soto A.M.. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005; 34:221–243. [DOI] [PubMed] [Google Scholar]
- 29. Misra V.K., Draper D.E.. The linkage between magnesium binding and RNA folding. J. Mol. Biol. 2002; 317:507–521. [DOI] [PubMed] [Google Scholar]
- 30. Lilley D.M., Clegg R.M., Diekmann S., Seeman N.C., Kitzing V.E., Hagerman P.J.. A nomenclature of junctions and branchpoints in nucleic acids. Nucleic Acids Res. 1995; 23:3363–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dickson A., Brooks C.L. III. WExplore: hierarchical exploration of high-dimensional spaces using the weighted ensemble algorithm. J. Phys. Chem. B. 2014; 118:3532–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brooks B.R., Brooks C.L. III, Mackerell A.D., Nilsson L., Petrella R.J., Roux B., Won Y., Archontis G., Bartels C., Boresch S. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009; 30:1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walter N.G. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods. 2001; 25:19–30. [DOI] [PubMed] [Google Scholar]
- 34. Zhuang X., Kim H., Pereira M.J.B., Babcock H.P., Walter N.G., Chu S.. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002; 296:1473–1476. [DOI] [PubMed] [Google Scholar]
- 35. Rueda D., Bokinsky G., Rhodes M.M., Rust M.J., Zhuang X., Walter N.G.. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10066–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blanco M., Walter N.G.. Analysis of complex single-molecule FRET time trajectories. Methods Enzymol. 2010; 472:153–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Esteban J.A., Walter N.G., Kotzorek G., Heckman J.E., Burke J.M.. Structural basis for heterogeneous kinetics: reengineering the hairpin ribozyme. Proc. Natl. Acad. Sci. U.S.A.. 1998; 95:6091–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Butcher S.E., Burke J.M.. Structure-mapping of the hairpin ribozyme. Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme-substrate complex. J. Mol. Biol. 1994; 244:52–63. [DOI] [PubMed] [Google Scholar]
- 39. Walter N.G., Yang N., Burke J.M.. Probing non-selective cation binding in the hairpin ribozyme with Tb(III). J. Mol. Biol. 2000; 298:539–555. [DOI] [PubMed] [Google Scholar]
- 40. Komatsu Y., Koizumi M., Nakamura H., Ohtsuka E.. Loop-size variation to probe a bent structure of a hairpin ribozyme. J. Am. Chem. Soc. 1994; 116:3692–3696. [Google Scholar]
- 41. Feldstein P.A., Bruening G.. Catalytically active geometry in the reversible circularization of ‘mini-monomer’ RNAs derived from the complementary strand of tobacco ringspot virus satellite RNA. Nucleic Acids Res. 1993; 21:1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pinard R., Lambert D., Pothiawala G., Major F., Burke J.M.. Modifications and deletions of helices within the hairpin ribozyme-substrate complex: an active ribozyme lacking helix 1. RNA. 2004; 10:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu S., Bokinsky G., Walter N.G., Zhuang X.. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic ‘fingerprinting’. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12634–12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paudel B.P., Rueda D.. Molecular crowding accelerates ribozyme docking and catalysis. J. Am. Chem. Soc. 2014; 136:16700–16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brion P., Westhof E.. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997; 26:113–137. [DOI] [PubMed] [Google Scholar]
- 46. Lipfert J., Doniach S., Das R., Herschlag D.. Understanding nucleic acid-ion interactions. Annu. Rev. Biochem. 2014; 83:813–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Butcher S.E., Pyle A.M.. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011; 44:1302–1311. [DOI] [PubMed] [Google Scholar]
- 48. Grabow W.W., Jaeger L.. RNA self-assembly and RNA nanotechnology. Acc. Chem. Res. 2014; 47:1871–1880. [DOI] [PubMed] [Google Scholar]
- 49. Boerneke M.A., Dibrov S.M., Hermann T.. Crystal-structure-guided design of self-assembling RNA nanotriangles. Angew. Chem. Int. Ed. 2016; 55:4097–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chworos A., Severcan I., Koyfman A.Y., Weinkam P., Oroudjev E., Hansma H.G., Jaeger L.. Building programmable jigsaw puzzles with RNA. Science. 2004; 306:2068–2072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.