Abstract
Complications arising from antibiotic-resistant bacteria are becoming one of the key issues in modern medicine. Members of drug-resistant Enterobacteriaceae spp. include opportunistic pathogens (e.g., Salmonella spp.) that are among the leading causes of morbidity and mortality worldwide. Overgrowth of these bacteria is considered a hallmark of intestinal dysbiosis. Microcins (small antimicrobial peptides) produced by some gut commensals can potentially cure these conditions by inhibiting these pathogens and have been proposed as a viable alternative to antibiotic treatment. In this proof-of-concept work, we leverage this idea to develop a genetically engineered prototype probiotic to inhibit Salmonella spp. upon exposure to tetrathionate, a molecule produced in the inflamed gut during the course of Salmonella infection. We developed a plasmid-based system capable of conferring the ability to detect and utilize tetrathionate, while at the same time producing microcin H47. We transferred this plasmid-based system to Escherichia coli and demonstrated the ability of the engineered strain to inhibit growth of Salmonella in anaerobic conditions while in the presence of tetrathionate, with no detectable inhibition in the absence of tetrathionate. In direct competition assays between the engineered E. coli and Salmonella, the engineered E. coli had a considerable increase in fitness advantage in the presence of 1 mM tetrathionate as compared to the absence of tetrathionate. In this work, we have demonstrated the ability to engineer a strain of E. coli capable of using an environmental signal indicative of intestinal inflammation as an inducing molecule, resulting in production of a microcin capable of inhibiting the organism responsible for the inflammation.
Keywords: microbiome, engineering, probiotics, Salmonella, tetrathionate, microcins, Escherichia coli Nissle
Graphical Abstract
Medical complications related to the emergence of antibiotic-resistant bacteria are a major issue in modern healthcare due to the resulting increase in morbidity, mortality, length of hospitalization, and related healthcare costs.1 The CDC estimates that, every year, more than 2 million people acquire multidrug-resistant infections, resulting in over 23 000 related deaths.2 Enterobacteriaceae spp. include opportunistic pathogens (Carbapenem-resistant Klebsiella spp., Fluoroquino-lone-resistant Salmonella spp., adherent–invasive Escherichia coli) that are among the leading causes of morbidity and mortality worldwide.3,4 According to the CDC, drug-resistant Salmonella spp. are responsible for more than 100 000 infections, while their nonresistant counterparts already account for 1.2 million infections and 450 deaths in the US every year.2 It was recently demonstrated that microcin production by specific gut bacteria can ameliorate these conditions by inhibiting certain pathogens.5 However, in these organisms, microcin production is dependent upon a narrow range of environmental conditions6,7 and regulatory mechanisms which are, in many cases, not fully understood.5
Engineered microbes as diagnostic tools or therapeutics have recently been proposed as “smart” alternatives to traditional drugs. For the former, recent remarkable examples have included engineering microbes to sense cancer or inflammation deriving from the production of reactive oxygen species by the host.8,9 For the latter, major examples have included the construction of nonpathogenic E. coli that kill Pseudomonas aeruginosa by means of microcins or pyocins, elicited through this pathogen’s quorum-sensing signaling system.10–12
A recent report from Sassone-Corsi et al. demonstrated that the production of microcins from E. coli strain Nissle 1917 (EcN) can therapeutically displace pathogenic Enterobacteriaceae species in specific environmental conditions (e.g., iron limitation).5 Our group independently verified that the class IIb sideromycin, microcin H47 (MccH47),13 a high molecular mass, post-translationally modified peptide, originally isolated from E. coli strain H47, can inhibit Salmonella growth in vitro. Therefore, rather than relying on the wild-type regulatory mechanisms, we sought to develop a probiotic with a specifically designed induction system for MccH47.
During gut inflammation, reactive oxygen species produced by the host react with luminal thiosulfate, resulting in production of tetrathionate.14 Salmonella species utilize the gene products of the ttr operon (ttrRSBCA), which provide this pathogen with the ability to utilize tetrathionate as a terminal electron acceptor, conferring a growth advantage over the competing microbiota during inflammation conditions.14 The ttrBCA genes of Salmonella, encoding the three subunits of tetrathionate reductase, have previously been transferred to E. coli, resulting in functional tetrathionate reductase activity.14 The ttrBCA promoter is positively regulated by TtrR in the presence of tetrathionate and by Fnr under anoxic conditions.14,15 Very recently, the ttrRS genes of Salmonella, encoding the tetrathionate sensor and response regulator, were chromosomally integrated into E. coli strain NGF-1 and served as a functional tetrathionate detection probiotic in a mouse model of inflammation for over 6 months.9 In light of these advances, we developed an engineered strain of EcN harboring a plasmid-based system carrying mchAXIBCDEF and ttrRSBCA, capable of producing MccH47 in response to environmental tetrathionate, resulting in the ability to inhibit and out-compete Salmonella during in vitro experiments.
RESULTS AND DISCUSSION
Construction and Analysis of a Plasmid-Based System for the Inducible Production of Microcin H47
Plasmid pJPMcH47 was developed for the L-rhamnose dependent production of MccH47 and constructed by Gibson Assembly (see Methods). pJPMcH47 contains all mch genes of E. coli H47 (mchAXIBCDEF), with the mchXIB genes immediately downstream of the rhaPBAD promoter (Figure 1A). The aim of this design is to specifically regulate production of the MccH47-precursor (MchB), on the basis of L-rhamnose concentration. For the purpose of in vitro assays, we developed E. coli strain NEB10β pJPMcH47 and EcN pJPMcH47 and then assessed the ability of each strain to inhibit Salmonella enterica subsp. Enterica serovar Typhimurium (hereafter referred to as S. Typhimurium) and E. coli strain DH5α.
Figure 1.
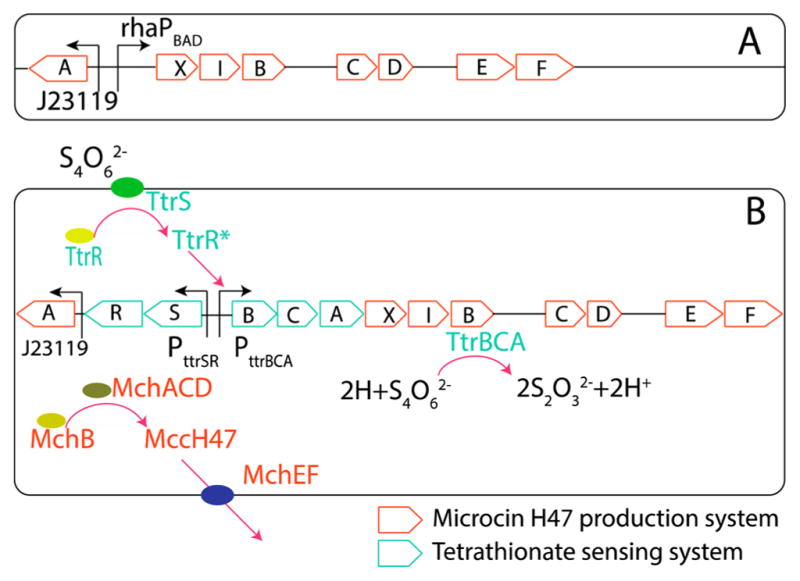
(A) Plasmid pJPMcH47 for the L-rhamnose-induced production of MccH47. (B) Plasmid pttrMcH47 for the tetrathionate-induced production of MccH47. Orange indicates genes and reactions needed for MccH47 production; cyan indicates genes and reactions related to tetrathionate sensing and utilization.
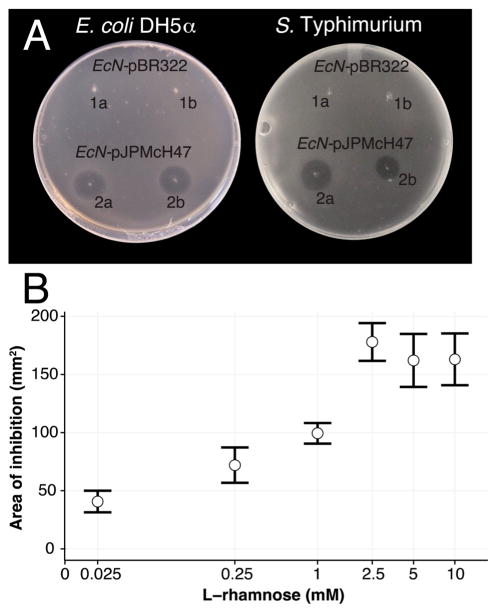
Inhibition assays were based on previous work with microcin J25 by Delgado et al.,16 where inhibition was evaluated visually by measuring a zone of inhibition in susceptible lawns grown on agar plates previously stabbed with a strain carrying pJPMcH47 and inactivated with chloroform and UV. Both EcN pJPMcH47 (Figure 2A) and E. coli NEB10β pJPMcH47 (Figure S1) were capable of inhibiting both S. Typhimurium and E. coli DH5α. As expected, on the basis of the nature of the L-rhamnose dependent induction system utilized,17 inhibition increases with increasing L-rhamnose concentration, while EcN WT showed no inhibition (Figure 2B). Taken together, these results demonstrate that the MccH47 produced by our engineered strains is the causative agent in the inhibition of S. Typhimurium. This is in line with reported literature, showing that MccH47 produced from EcN WT is unable to inhibit S. Typhimurium even during iron starvation,5,6 while MccH47 produced from our donor E. coli strain H47 is able to inhibit S. Typhimurium.6,13 One reason could be the lack of post-translational modifications in MccH47 derived from EcN WT,18 potentially due to the lack of mchA, a necessary gene for mature MccH47 antibacterial activity. Another possibility could be the observed divergence in amino acid sequence for genes mchE and mchF between EcN and E. coli H47.6
Figure 2.
(A) Inhibition assays comparing the effect of EcN WT and EcN pJPMcH47 against lawns of pathogenic S. Typhimurium (left) and E. coli DH5α (right) on minimal media, supplemented with 22 mM L-rhamnose and 0.2 mM 2,2′-dipyridyl (see Methods). (B) Measurement of inhibition area in the S. Typhimurium lawn as a function of L-rhamnose concentration on LB agar plates supplemented with variable L-rhamnose (0.025–10 mM) and 0.2 mM 2,2′-dipyridyl. 2,2′-Dipyridyl addition was necessary for maximum S. Typhimurium inhibition by MccH47 (see Methods and Figure S2).
Construction and Analysis of a Plasmid-Based Tetrathionate-Detection System
Plasmid pttrMcH47 (Figure 1B) was developed to confer utilization of tetrathionate reductase activity and tetrathionate dependent production of MccH47 and was constructed by Gibson Assembly (see Methods). Plasmid pttrMcH47 contains all genes of the ttr operon from S. Typhimurium (ttrRSBCA) and all genes necessary for mature MccH47 production, immunity, and secretion (mchAXIBCDEF). mchXIB is encoded immediately downstream of ttrA resulting in cotranscription along with ttrBCA from the ttrBCA promoter. To assess successful tetrathionate-induced MccH47 production, we compared the inhibitory effect of EcN pttrMcH47 against E. coli DH5α in variable conditions (Figure 3). We observed that MccH47 dependent inhibition increases in iron limited conditions (Figure 3A,B). This is consistent with previous studies that have shown that species susceptible to inhibition by MccH47 express catecholate siderophore receptors iroN/fui, cir, and fepA,6 which serve as the mode of entry into susceptible cells. Importantly, this should not constitute an issue with respect to the functioning of our construct in vivo, as it is known that the iron limitation is a typical feature of a host’s inflamed gut.18 When testing for tetrathionate addition in iron limitation, we observe the expected emergence of inhibition only in the presence of tetrathionate (Figure 4A). Supplementation to the media with 0.1% D-glucose reduced inhibition across all experimental conditions (Figure 3A,C), which is noteworthy in light of published reports which utilize glucose supplementation alongside PttrRS-based induction and MccH47 production.9,19
Figure 3.
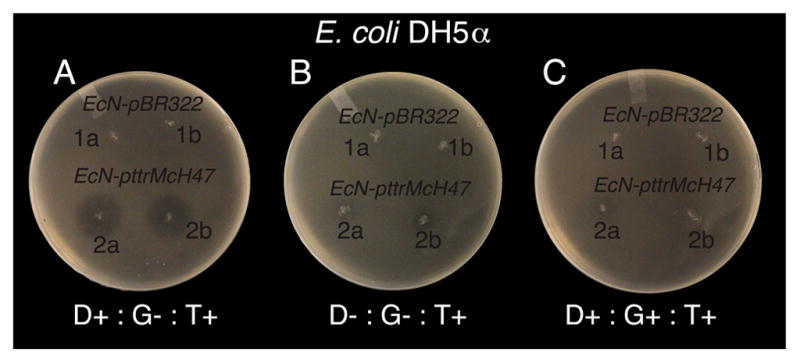
Characterization of MccH47-based inhibition of E. coli DH5α by EcN pttrMcH47, with EcN pBR322 control, as a function of 0.2 mM 2,2′-dipyridyl (D), 0.1% D-Glucose (G), and 1 mM of potassium tetrathionate (T). + stands for compound present; – stands for compound absent.
Figure 4.
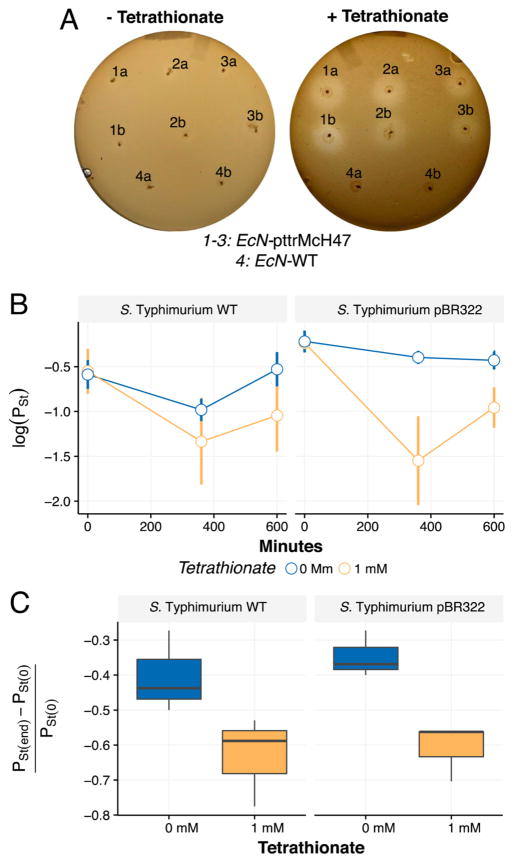
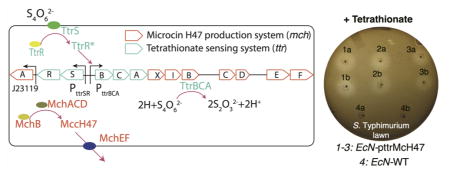
Tetrathionate-induced inhibition by EcN pttrMcH47 MccH47 production. (A) Inhibition assays comparing the ability of three separate clones of EcN pttrMcH47 and EcN WT to inhibit growth of S. Typhimurium in the absence (left) and presence (right) of 1 mM potassium tetrathionate (a,b indicate duplicate stabs).·(B) Results from in vitro ecological competition experiments, showing the proportion of S. Typhimurium WT (left) or S. Typhimurium pBR322 (right) over time. (C) S. Typhimurium fitness estimation obtained from competition experiments (see ref 21). Supplementation of 1 mM potassium tetrathionate leads to production of MccH47 and a statistically significant reduction (p < 0.05) in S. Typhimurium fitness.
Analysis of Tetrathionate-Induced Inhibition
We proceeded to test the tetrathionate-induced inhibition in vitro against S. Typhimurium. First, solid media inhibition assays were carried out anaerobically in LB agar, again supplemented with 0.2 mM 2,2′-dipyridyl, with and without 1.5 mM potassium tetrathionate. In media containing potassium tetrathionate, EcN pttrMcH47 was capable of inhibiting S. Typhimurium while EcN WT did not (inhibition zone not detected) (Figure 4A) confirming the ability of our construct to inhibit S. Typhimurium at a physiologically relevant disease state.14
While static plate assays demonstrate the functional capability of the constructs, these experiments do not account for the effect of competition for growth nutrients. Therefore, we performed competition experiments between EcN pttrMcH47 and either S. Typhimurium WT or S. Typhimurium pBR322. Assays were conducted anaerobically, in LB broth supplemented with 0.2 mM 2,2′-dipyridyl, in the presence or absence of 1 mM potassium tetrathionate, and supplemented with 100 μg/mL carbenicillin when both strains contained resistance. In experiments to analyze growth rate, we determined 1 mM of potassium tetrathionate to be a plausible concentration resulting in no significant fitness cost due to tetrathionate addition (Figure S3). Competition experiments were initiated with a S. Typhimurium (pBR322)–EcN pttrMcH47 ratio of approximately 1:1 (Figure 4B). Bacteria were grown for a total of 10 h, and 10-fold dilutions were plated onto MacConkey agar for colony enumeration. For both strains, we followed Lenski et al.20 and our previous work21,22 to determine relative fitness as
| (1) |
where Pi(t) is the proportion of strain i at time t. We performed linear regression analysis and fitted the model WSE ~ 1 + Tet + Tag where Tet and Tag are two “dummy” variables, with Tet indicating absence/presence of tetrathionate and Tag indicating use of S. Typhimurium WT or S. Typhimurium pBR322. Results of linear regression show a significant decrease in S. Typhimurium fitness at 1 mM potassium tetrathionate irrespective of S. Typhimurium WT or S. Typhimurium pBR322 (p < 0.02). We also observe that the inhibitory effect is reduced when competition is performed against S. Typhimurium WT in a media lacking antibiotic addition (p < 0.05). However, because we do not detect significant differences in growth curve dynamics in isolation for any of the strains used in this experiment, neither in the presence nor in the absence of tetrathionate (Figure S3), we speculate that this reduced competitiveness against S. Typhimurium WT may be due to plasmid loss in the absence of antibiotic. When testing for an increase in S. Typhimurium proportion observed at 600 min compared to 360 min for the EcN pttrMch47 vs S. Typhimurium WT, we do not observe statistical significance (p > 0.05) between the two data points. We acknowledge but cannot explain the recovery by S. Typhimurium pBR322 from 360 to 600 min. Our results confirm the ability of EcN pttrMcH47 to suppress S. Typhimurium growth beyond what is obtained in direct competition experiments in an environment without the supplementation of tetrathionate (Figure 4C). The increased competitive advantage of EcN pttrMcH47 over S. Typhimurium in an environment supplemented with tetrathionate is particularly important considering recent work which has indicated that tetrathionate in the lumen of the inflamed gut provides a growth advantage for S. Typhimurium over the rest of the competing microbiota.4
Outlook
The use of microcins for the treatment of enterobacterial colitis can be regarded as a novel and potent alternative to antibiotics.23 Here, we have developed a genetically engineered E. coli that produces a S. Typhimurium-inhibiting microcin in response to tetrathionate, a molecule resulting from the inflammation of the intestine, a hallmark of a Salmonella infection.4 Using in vitro assays, we have shown that EcN pttrMcH47 not only prevents S. Typhimurium growth in static agar inhibition assays but also significantly reduces S. Typhimurium fitness in pairwise ecological competition experiments.
While current focus on engineered probiotics (with respect to gut pathogen infections) has been mostly directed toward disease sensing and reporting,8,9 our work provides a first proof-of-concept toward using a genetically engineered living therapeutic to induce a specific microbiome correction during the course, or at the onset of, a particular disease state.
The realization that microcins play a crucial role in gut microbiome ecology5 opens a new door for the treatment and prevention of a range of gut pathogens. As new microcins or related antimicrobial peptides with narrow target specificity are further discovered and characterized,24 our work provides a demonstration of a plausible delivery method for timely gut pathogen eradication with minimal off-target effects, as compared to broad spectrum antibiotics.
To obtain a proof-of-concept system that would ensure a fully functional inflammation sensing–pathogen killing phenotype, without the need of any extra strain-specific chromosomal features, we included all the necessary genes for tetrathionate sensing and MccH47 production into a single DNA construct. We then transferred this system to EcN, an FDA-approved probiotic that has been shown to be amenable toward engineered probiotic applications.8,11 For illustrative purposes and in line with recent studies on the construction and preliminary testing of engineered bacteria,8,10,11,25 we opted for a plasmid-based approach. However, for translation to clinics, as well as for trials using in vivo models, future work will be performed to achieve stable chromosomal integration. This may involve using a background genome with promising characteristics regarding host colonization and retention of recombinant genes, such as E. coli NGF-1.9
A possible issue that is common to biocontrol through supplementation of native and engineered bacteria comes from the emergence of mutations that may alter the regulation or function of the system. However, in light of recent work that demonstrated wild-type stability and mutational rate of the ttr operon in an engineered E. coli,9 we expect our probiotic to properly work over the time frame of a Salmonella infection. Moreover, future efforts to properly control for this possible issue could lead to the development of a probiotic that autolyzes within a certain time frame after supplementation.26 As such, utilizing this method, it is possible to envision a single probiotic bacterium capable of colonizing a host, sensing a disease state or particular pathogen, and eradicating the disease-causing organism without the need for physician intervention or use of broad-spectrum antibiotics.
METHODS
Microbial Strains, Media, and Growth Conditions
Strains used in this study include Escherichia coli strain NEB10β (New England Biolabs. Ipswich, MA), E. coli strain DH5α (New England Biolabs. Ipswich, MA), E. coli strain Nissle 1917 (gift from),21 and/or Salmonella enterica subsp. enterica; serovar Typhimurium ATCC 29630 (purchased from ATCC, Manassas, VA). Plasmid constructs developed in this work were first transformed by electroporation into E. coli NEB10β cells and then to E. coli Nissle (EcN). Oligonucleotides used in this study are listed in Table 1. All media and additional reagents listed in this study were purchased from Sigma-Aldrich, St. Louis, MO, unless otherwise indicated.
Table 1.
Strains, Plasmids, and Oligonucleotides
| relevant characteristics/sequence (5′–3′) | source or reference | |
|---|---|---|
| strains | ||
| Escherichia coli Nissle 1917 | 27 | |
| Salmonella Typhimurium | Salmonella enterica subsp. enterica serovar Typhimurium ATCC 29630 | ATCC |
| Escherichia coli NEB10β | Δ(ara-leu) 7697 araD139 fhuA ΔlacX74 galK16 galE15 e14-ϕ80dlacZΔM15 recA1 relA1 endA1 nupG rpsL (StrR) rph spoT1 Δ(mrr-hsdRMS-mcrBC) | New England Biolabs |
| Escherichia coli DH5α | F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | New England Biolabs |
| plasmids | ||
| pBR322 | pMB1, ApR, TcR | New England Biolabs |
| pLJV3 | pUC57, CFP, mchA, rhaPBAD | this study; synthesized by General Biosystems |
| pJPMcH47 | rhaPBAD, mchAXIBCDEF | this study |
| pttrMcH47 | ttrRSBCA, mchAXIBCDEF | this study |
| pEX2000 | pBR322, mchAXIBCDEF | 29 |
| oligonucleotides | ||
| pBR322FWD | GGATTATTTCTATTTAAAATGAGGCCCTTTCGTCTTCAAGAATTCT | |
| pBR322REV | GAGATAGCGGTAGCTAACTAGACGTCAGGTGGCACTTTTCG | |
| pLJV3FWD | TTACCAGACCTCACCCAGACTTCATTACGACCAGTCTAAAAAGCGCCTG | |
| pLJV3REV | GAAAAGTGCCACCTGACGTCTAGTTAGCTACCGCTATCTCCAACGTGC | |
| pEX2000FWD | TTTAGACTGGTCGTAATGAAGTCTGGGTGAGGTCTGGTAAGA | |
| pEX2000REV | CTTGAAGACGAAAGGGCCTCATTTTAAATAGAAATAATCCTGTCAACAGTTCTCAACG | |
| pJPMcH47FWD | AAAAATCGAGCGTATATAACGTCTGGGTGAGGTCTGGTAAGA | |
| pJPMcH47REV | TCTGTTGGTTTGATCTGGCGGGATGTGACGATCGTTGACAGC | |
| SentFWD | TTACCAGACCTCACCCAGACGTTATATACGCTCGATTTTTGCCGGC | |
| SentREV | TGTCAACGATCGTCACATCCCGCCAGATCAAACCAACAGAA | |
pJPMcH47 and pttrMcH47 were constructed using standard methods for Gibson Assembly28 and the Gibson Assembly Master Mix (New England Biolabs. Ipswich, MA). To construct pJPMcH47, a linear version of pBR322 was produced by polymerase chain reaction using primers pBR322FWD and pBR322REV, and select regions of pLJV3 (encoding rhaPBAD, CFP, mccF, and mchA) and pEX2000 (encoding mchXIBCDEF) were amplified using primer sets pLJV3FWD/pLJV3REV and pEX2000FWD/pEX2000REV, respectively. pttrMcH47 was constructed by amplification of pJPMcH47 using primers pJPMcH47FWD/pJPMcH47REV and amplification of the ttrRSBCA operon of S. Typhimurium using primers SentFWD/SentREV. ttrRSBCA was included in the plasmid as opposed to just ttrRS because in Riglar et al.9 the constructed sensor strain, when in direct competition with Salmonella, only detected tetrathionate if the Salmonella was incapable of tetrathionate utilization (ΔttrR).
Solid Media Inhibition Assays
Inhibition assays in solid media were designed and carried out on the basis of previous work by Delgado et al.16 Briefly, E. coli strains were grown overnight on LB agar plates, supplemented with carbenicillin (100 μg/mL) when relevant. Individual colonies were then used to inoculate 3 mL of LB broth, supplemented with carbenicillin (100 μg/mL) when relevant. 1 μL of liquid culture was then used to create an agar stab in solid media and incubated at 37 °C, either aerobically or anaerobically, for 24 h. Postincubation, cells were inactivated with chloroform and UV. Molten 3% agar was then added to an overnight culture of susceptible cells to a final concentration of 0.75%, and then, 3 mL of the mixture was overlaid on top of the inactivated agar stab plates and allowed to solidify. Plates were then incubated aerobically overnight at 37 °C, and inhibition halos were observed. When inhibition halo size was a relevant characteristic, ImageJ software (https://imagej.nih.gov/ij/) was utilized. For solid medium inhibition assays of S. Typhimurium and E. coli DH5α by EcN pJPMcH47, agar stabs were made in M9 minimal salts supplemented with 0.1 mM CaCl2, 2 mM MgSO4, 0.2 mM 2,2′-dipyridyl, and 0.4% L-rhamnose. For variable L-rhamnose concentration inhibition experiments, culture stabs of EcN pJPMcH47 were made in LB agar supplemented with 0.2 mM 2,2′-dipyridyl and L-rhamnose, ranging from 0.25 μM to 10 mM. All aspects of inhibition assays utilizing plasmid pJPMcH47 were performed aerobically.
For solid medium inhibition assays of S. Typhimurium by EcN pttrMcH47, culture stabs in LB agar supplemented with 0.2 mM 2,2′-dipyridyl and 1.0 or 1.5 mM (see main text) potassium tetrathionate were incubated anaerobically at 37 °C for 24 h in Oxoid anaerobic jars with anaerobic atmosphere generation bags. Notably, potassium tetrathionate was not added to the LB agar medium until the temperature had reached 50 °C and was prepared immediately before introduction to the media and sterilized by filtration using a 0.22 μm filter membrane. Upon removal from the jars, cells were immediately inactivated, overlaid with S. Typhimurium culture in 0.75% agar, and incubated aerobically overnight.
Because microcin H47 mode of action relies on functional catecholate siderophore receptors iroN, cir, and fepA,6 2,2′-dipyridyl addition was essential to render S. Typhimurium maximally susceptible to inhibition by our probiotic. Control experiments using the laboratory strain E. coli DH5a allowed us to rule out necessity of iron limitation for the MccH47 production by our constructs (Figure 3A,B).
Liquid Media Competition Assays
Competition assays were carried out in triplicate for each experimental condition in a Forma Scientific Model 1025 anaerobic chamber. LB was allowed to equilibrate in anaerobic conditions overnight, prior to initiation of any competition assays. 3 mL of LB was distributed to individual test tubes, with a final concentration of 0.2 mM 2,2′-dipyridyl and both 1 mM potassium tetrathionate and 100 μg/mL carbenicillin, as indicated. Potassium tetrathionate was prepared immediately before inoculation and sterilized by filtration using a 0.22 μm filter membrane. Individual colonies of EcN pttrMcH47 and S. Typhimurium and S. Typhimurium pBR322 were taken from LB agar plates and incubated aerobically overnight at 37 °C in 3 mL of LB, supplemented with 100 μg/mL carbenicillin for plasmid retention when relevant. Liquid cultures were then transferred to the anaerobic chamber, and ~105 cells of each culture were transferred into media for experimental conditions for analysis of relative fitness in competition, on the basis of the presence of tetrathionate. Competition assays were set up to compete EcN pttrMcH47 with S. Typhimurium and EcN pttrMcH47 with S. Typhimurium pBR322, in which case 100 μg/mL carbenicillin was supplemented to the media. Cultures were serially diluted and plated onto MacConkey Agar for colony enumeration.
Supplementary Material
Acknowledgments
V.B. acknowledges support from the National Institute of Allergy and Infectious Disease (Grant R15-AI112985-01A1) and the National Science Foundation (Grant 1458347). V.B., M.W.S., C.J.B., and B.A.M. acknowledge support from the UMass President Science and Technology Office. B.A.M. acknowledges support from NIH DK056754. J.D.P. acknowledges support through a UMassD Distinguished Doctoral Fellowship. The authors acknowledge Aunnesha Bhowmick, Lindsey Ly, and Shakti Bhattarai for help with experiments and constructive discussion.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfec-dis.7b00114.
Figure S1, E. coli Nissle 1917 pBR322 and E. coli NEB10β pJPMcH47 inhibition of E. coli DH5α and S. Typhimurium; Figure S2, E. coli Nissle 1917 pBR322 and E. coli Nissle 1917 pJPMcH47 inhibition of E. coli DH5α and S. Typhimurium in the presence and absence of 0.2 mM 2,2′-dipyridyl; Figure S3, growth curves in LB media under anaerobic conditions with 0.2 mM 2,2′-dipyridyl (PDF)
References
- 1.Medina E, Pieper DH. Tackling Threats and Future Problems of Multidrug-Resistant Bacteria. In: Stadler M, Dersch P, editors. How to Overcome the Antibiotic Crisis. Springer International Publishing; New York: 2016. pp. 3–33. http://dx.doi.org/10.1007/82_2016_492. [DOI] [PubMed] [Google Scholar]
- 2.CDC. Antibiotic Resistance Threats in the United States, 2013. CDC; Atlanta, GA: 2013. [Google Scholar]
- 3.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, Bäumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology. 2003;149:2557–2570. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 7.Behrens HM, Six A, Walker D, Kleanthous C. The therapeutic potential of bacteriocins as protein antibiotics. Emerg Top Life Sci. 2017;1:65–74. doi: 10.1042/ETLS20160016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daeffler KNM, Galley JD, Sheth RU, Ortiz-Velez LC, Bibb CO, Shroyer NF, Britton RA, Tabor JJ. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol Syst Biol. 2017;13:923. doi: 10.15252/msb.20167416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, Silver PA. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol. 2017;35:653–658. doi: 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SSJ, Poh CL, Chang MW. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, Chang MW. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta S, Bram EE, Weiss R. Genetically Programmable Pathogen Sense and Destroy. ACS Synth Biol. 2013;2:715–723. doi: 10.1021/sb4000417. [DOI] [PubMed] [Google Scholar]
- 13.Laviña M, Gaggero C, Moreno F. Microcin H47, a chromosome-encoded microcin antibiotic of Escherichia coli. J Bacteriol. 1990;172:6585–6588. doi: 10.1128/jb.172.11.6585-6588.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 16.Delgado MA, Vincent PA, Farías RN, Salomón RA. YojI of Escherichia coli functions as a microcin J25 efflux pump. J Bacteriol. 2005;187:3465–3470. doi: 10.1128/JB.187.10.3465-3470.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haldimann A, Daniels LL, Wanner BL. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vassiliadis G, Destoumieux-Garzón D, Lombard C, Rebuffat S, Peduzzi J. Isolation and Characterization of Two Members of the Siderophore-Microcin Family, Microcins M and H47. Antimicrob Agents Chemother. 2010;54:288–297. doi: 10.1128/AAC.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azpiroz MF, Rodríguez E, Laviña M. The Structure, Function, and Origin of the Microcin H47 ATP-Binding Cassette Exporter Indicate Its Relatedness to That of Colicin V. Antimicrob Agents Chemother. 2001;45:969–972. doi: 10.1128/AAC.45.3.969-972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-Term Experimental Evolution in Escherichia coli. I Adaptation and Divergence During 2,000 Generations. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 21.Bucci V, Nadell, Xavier JB. The Evolution of Bacteriocin Production in Bacterial Biofilms. Am Nat. 2011;178:E162–E173. doi: 10.1086/662668. [DOI] [PubMed] [Google Scholar]
- 22.Nadell CD, Bucci V, Drescher K, Levin SA, Bassler BL, Xavier JB. Cutting through the complexity of cell collectives. Proc R Soc London, Ser B. 2013;280:20122770. doi: 10.1098/rspb.2012.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotter PD, Ross RP, Hill C. Bacteriocins — a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 24.Zschüttig A, Zimmermann K, Blom J, Goesmann A, Pöhlmann C, Gunzer F. Identification and Characterization of Microcin S, a New Antibacterial Peptide Produced by Probiotic Escherichia coli G3/10. PLoS One. 2012;7:e33351. doi: 10.1371/journal.pone.0033351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holowko MB, Wang H, Jayaraman P, Poh CL. Biosensing Vibrio cholerae with Genetically Engineered Escherichia coli. ACS Synth Biol. 2016;5:1275–1283. doi: 10.1021/acssynbio.6b00079. [DOI] [PubMed] [Google Scholar]
- 26.Camacho EM, Mesa-Pereira B, Medina C, Flores A, Santero E. Engineering Salmonella as intracellular factory for effective killing of tumour cells. Sci Rep. 2016;6:srep30591. doi: 10.1038/srep30591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart E, Deng L, Yeliseyev V, Delaney ML, Liu Q, Olle B, Stein RR, Honda K, Bry L, Gerber GK. MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol. 2016;17:1–17. doi: 10.1186/s13059-016-0980-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 29.Gaggero C, Moreno F, Laviña M. Genetic analysis of microcin H47 antibiotic system. J Bacteriol. 1993;175:5420–5427. doi: 10.1128/jb.175.17.5420-5427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.