Summary
Influenza D virus (IDV) is a newly described influenza type of the Orthomyxoviridae virus family that was first isolated from diseased swine in 2011 and has subsequently been detected in cattle around the world in 2014. In addition, serological evidence for IDV infection in humans has been recently established. Despite all the progress, the full range of susceptible hosts for this novel virus has yet to be determined, but includes swine, bovine, small ruminants and human. This study was designed to determine if equine is a possible host to this newly emerging influenza virus. Three hundred and sixty-four equine serum samples were collected in 2015 from 141 farms within the Midwestern United States. Serum samples were examined using hemagglutination inhibition (HI) assay against two established IDV lineages (D/OK and D/660) and one IDV-related human ICV lineage (C/JHB). Results of this study showed 44 (44 of 364, 12%) samples positive for antibodies against D/OK, 39 (39 of 364, 11%) samples positive for antibodies against D/660, and 41 (41 of 364, 11%) samples positive for antibodies against C/JHB. A subset of these samples was further confirmed via microtitre neutralization (MN) assay. Our data demonstrated that horses are susceptible to two lineages of IDV, and that these viruses were present in equine populations throughout multiple Midwestern states of the United States. These findings continue to support the need for further surveillance of IDV viruses in agricultural species to work towards a better understanding of the full host range and natural reservoirs of influenza D virus.
Keywords: equine, horse, influenza virus, serology, type C, type D
1 | INTRODUCTION
The Orthomyxoviridae virus family has three influenza genera, A, B and C, which are classified according to antigenic differences in their nucleoprotein and matrix proteins (Palese & Shaw, 2007). The fourth genus of influenza, named influenza D, has been recently described (https://www.cdc.gov/flu/about/viruses/types.htm). Influenza D (IDV) represents a novel type of virus more closely related to influenza C (ICV) than influenza A (IAV) or influenza B (IBV). IDV uses bovine as its primary reservoir, and has been isolated from cattle herds from multiple countries including the following: China, France, Italy and the United States (Chiapponi et al., 2016; Ducatez, Pelletier, & Meyer, 2015; Hause et al., 2013, 2014; Jiang et al., 2014). Susceptibility to infection by this novel virus has also been demonstrated in swine, sheep, goats, guinea pigs and ferrets (Hause et al., 2013, 2014; Quast et al., 2015; Sreenivasan et al., 2015). In addition, serological evidence for IDV infection in humans has been recently established (White, Ma, McDaniel, Gray, & Lednicky, 2016).
The worldwide prevalence and broad species tropism of this new influenza virus represents a growing potential threat to humans and other agricultural species. Many species are vulnerable to influenza infection, including humans; therefore, it is important to identify other potential hosts of this novel type. The primary goals of this study were to investigate the seroprevalence of IDV in American equine populations by conducting multiple serological surveys, as well as studying the antibody prevalence in this animal host of human ICV, which is closely related to IDV.
2 | MATERIALS AND METHODS
2.1 | Cell culture, reference serum and virus production
Madin–Darby canine kidney (MDCK) cells (ATCC) were cultured using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum (PAA Laboratories Inc., Dartmouth, MA, USA) and 1% streptomycin and penicillin (Life Technologies, Carlsbad, CA, USA). Isolated from bovine and swine presenting respiratory disease symptoms, influenza D/bovine/Oklahoma/660/2013 (D/660) and D/swine/Oklahoma/1334/2011 (D/OK) were grown on MDCK cells at 0.01 MOI and incubated at 37°C with ~5% CO2 for at least 5 days. D/OK and D/660 are representative strains of two antigenic lineages of IDV, D/OK being of swine origin and D/660 being of cattle origin. These two viruses have been found to be antigenically different lineages of IDV, likely due to their host divergence (Hause et al., 2014). Influenza C/human/Johannesburg/1/1966, originally isolated from humans, was produced in the same manner.
To better promote viral growth, DMEM maintenance media was supplemented using 1 μg/ml tolylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin (Sigma, St. Louis, MO, USA) and 1% penicillin and streptomycin (Life Technologies, Carlsbad, CA, USA). Virus titre (TCID50 per ml) was measured using both the Reed and Muench’s method (Reed & Muench, 1938) and the hemagglutination assay as described in the W.H.O. standard manual (www.who.int/influenza/en/).
Reference serum samples were generated against the three influenza virus strains via rabbit immunization in lab at SDSU (IACUC # 16-027A) and at the Covance Research Products. Covance produced the D/OK reference serum, while the D/660 and C/JHB serum were produced at SDSU. All groups used a nearly identical method of antibody generation involving hyperimmunization of immune-competent rabbits with UV-inactivated virus of the desired lineage formulated in adjuvant. The rabbits were immunized via both the intramuscular and subcutaneous routes. Serum was purified from blood drawn from the rabbits periodically throughout the generation procedure.
2.2 | Serology
Hemagglutination inhibition (HI) and MDCK-based microneutralization (MN) assays were employed as described in the WHO standard manual and previous literature to detect antibody titres (Ran et al., 2015) (www.who.int/influenza/en/), with minor revisions to the MN assay for use with ICV and IDV. Our revision for the MN assay was that virus was cultured on MDCK cells in 96-wells plates in the presence of serially diluted serum for 120 hr (5 days) to ensure that the slow-growing ICV and IDV viruses could reach a detectable titre level if they hadn’t been neutralized by the antibodies present in the serum. For the assessment of titre, Turkey red blood cells (Lampire Biological Laboratories, Pipersville, PA, USA) were used in both the HI and MN assays. For both the MN and HI assays, a titre of 40 was used as the threshold to describe a positive sample in accordance with the WHO protocols (www.who.int/influenza/en/) and an additional confirmation study (Wood et al., 2012). HI and MN assays were tested in duplicate, and HI or MN titres were described as the reciprocal of the final serum dilution that blocks viral ability to agglutinate red blood cells or inhibit viral replication, respectively. All samples were assayed in three separate experiments, and the antibody titres were determined as the mean of these triplicate data.
2.3 | Serum sample collection
Three hundred and sixty-four equine samples were collected during the summer of 2015 from farms and ranches in six states: Iowa (IA), Minnesota (MN), North Dakota (ND), Nebraska (NE), South Dakota (SD), and Wyoming (WY). These states occupy a generally north-central location of the continental United States of America and share numerous borders with each other. These samples were collected through the Animal Disease Research and Diagnostic Laboratory at South Dakota State University (SDSU). A further 100 equine samples were collected during the summer of 2016 again through the ADRDL at SDSU. The 100 samples were collected from horses from farms or ranches in Iowa (IA), Minnesota (MN), Nebraska (NE) and South Dakota (SD). Anti-sera against two IDV lineages (D/OK and D/660) and one human ICV (C/JHB) was generated from rabbits via immunization as described above, and collected for use as antibody controls and for the purpose of testing cross-reactivity between ICV and IDV.
3 | RESULTS AND DISCUSSION
We first investigated the potential cross-reactivity between IDV and ICV by the HI assay. Rabbit reference anti-sera to two IDV lineages, Influenza D/swine/OK/1334/2011 (D/OK) and Influenza D/bovine/660/2013 (D/660), and one human ICV, Influenza C/Victoria/2/2012 (C/Vic), as well as negative-control sera, were tested. Two IDV lineage-representative viruses, D/OK and D/660 as above, and one human ICV, C/Vic as above, were used in the HI cross-reactivity assay. As summarized in Table 1, the anti-serum for the D/660 strain was equally reactive to the D/660 virus and the D/OK virus with an HI titre of 1,280 against both, while having no detectable reactivity with C/Vic. Furthermore, the anti-serum generated against the D/OK strain was more specific to the D/OK virus, with a HI titre of 2,560, but was also cross-reactive with D/660 with a titre of 640. The anti-serum generated against C/Vic was specific to the C/Vic virus with a HI titre of 2,560 with no detectable activity with either IDV lineage. These results confirmed the specificity of our HI assay in the detection of anti-IDV antibodies, demonstrating that it could be used for screening of equine serum samples for antibodies to IDV and ICV. As described below, the possibility of a similar cross-reactive relationship between D/OK and D/660 within equine populations is observed.
TABLE 1.
Cross-reactivity of influenza C and D viruses by HI Assay
| D/OK | D/660 | C/Vic | |
|---|---|---|---|
| D/660 anti-serum | 1,280 | 1,280 | <10 |
| D/OK anti-serum | 2,560 | 640 | <10 |
| C/Vic anti-serum | <10 | <10 | 2,560 |
Bovine, swine and small ruminant species are known to host IDV (Hause et al., 2013, 2014; Quast et al., 2015). To determine if the equine population is susceptible to IDV, we tested 364 horse serum samples gathered in 2015 from six states (IA, MN, ND, NE, SD, WY), as well as 100 more samples gathered in 2016 from four states (IA, MN, NE, SD). All of the above samples were also analyzed for the presence of ICV antibodies. All samples were tested for virus-specific antibodies via the standard hemagglutinin inhibition assay (HI) in triplicate. Any serum sample with an average antibody titre greater than or equal to 40 was considered positive for antibodies against that given virus species.
Table 2 details a subset of the HI data from serum samples collected in 2015. Serum samples were grouped by farm of origin, and any farm that was the origin of more than one serum sample and had at least one serum sample test positive for antibodies against at least one of the viral lineages is included in the table. Of the 141 farms that serum was collected from, 47 of them are included in this table. HI data are provided for all three virus lineages tested, and is detailed by one column containing the number of positive serum samples originating from that farm followed by the total number of samples originating from that farm. A second column details the average titre value of the positive samples originating from that farm. The state of the United States in which the farm resides is also included with the serological data. Of the 230 individual serum samples included in the table, 36 tested positive for antibodies against D/OK, 28 tested positive for antibodies against D/660 and 37 tested positive for antibodies against C/JHB.
TABLE 2.
Equine serological surveillance of Influenza virus types C and D in 2015
| Farm ID | Location | D/OK | D/660 | C/JHB | |||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Positive\Total | Average Titre | Positive\Total | Average Titre | Positive\Total | Average Titre | ||
| 1 | SD | 6\42 | 51.7 | 3\42 | 46.7 | 14\42 | 60 |
|
| |||||||
| 2 | SD | 0\13 | – | 0\13 | – | 2\13 | 40 |
|
| |||||||
| 3 | SD | 1\11 | 80 | 1\11 | 40 | 0\11 | – |
|
| |||||||
| 4 | SD | 0\9 | – | 1\9 | 60 | 2\9 | 60 |
|
| |||||||
| 5 | SD | 2\9 | 40 | 0\9 | – | 0\9 | – |
|
| |||||||
| 6 | SD | 1\7 | 40 | 0\7 | – | 0\7 | – |
|
| |||||||
| 7 | SD | 1\7 | 45 | 0\7 | – | 1\7 | 40 |
|
| |||||||
| 8 | SD | 1\6 | 60 | 0\6 | – | 0\6 | – |
|
| |||||||
| 9 | SD | 0\5 | – | 0\5 | – | 1\5 | 40 |
|
| |||||||
| 10 | SD | 1\5 | 40 | 0\5 | – | 0\5 | – |
|
| |||||||
| 11 | SD | 2\5 | 80 | 2\5 | 50 | 0\5 | – |
|
| |||||||
| 12 | SD | 1\5 | 45 | 0\5 | – | 0\5 | – |
|
| |||||||
| 13 | SD | 0\5 | – | 0\5 | – | 1\5 | 40 |
|
| |||||||
| 14 | SD | 1\5 | 80 | 1\5 | 80 | 0\5 | – |
|
| |||||||
| 15 | MN | 2\4 | 70 | 2\4 | 80 | 0\4 | – |
|
| |||||||
| 16 | SD | 0\4 | – | 0\4 | – | 2\4 | – |
|
| |||||||
| 17 | SD | 0\4 | – | 1\4 | 40 | 0\4 | – |
|
| |||||||
| 18 | NE | 0\4 | – | 0\4 | – | 1\4 | 40 |
|
| |||||||
| 19 | MN | 4\4 | 70 | 4\4 | 60 | 0\4 | – |
|
| |||||||
| 20 | SD | 0\4 | – | 0\4 | – | 1\4 | 40 |
|
| |||||||
| 21 | NE | 1\4 | 50 | 0\4 | – | 0\4 | – |
|
| |||||||
| 22 | SD | 0\4 | – | 1\4 | 45 | 0\1 | – |
|
| |||||||
| 23 | SD | 1\4 | 60 | 1\4 | 80 | 0\4 | – |
|
| |||||||
| 24 | SD | 1\4 | 60 | 1\4 | 40 | 0\4 | – |
|
| |||||||
| 25 | SD | 1\4 | 40 | 1\4 | 45 | 0\4 | – |
|
| |||||||
| 26 | SD | 0\4 | – | 0\4 | – | 1\4 | 40 |
|
| |||||||
| 27 | NE | 0\3 | – | 0\3 | – | 0\3 | – |
|
| |||||||
| 28 | SD | 1\3 | 40 | 0\3 | – | 0\3 | – |
|
| |||||||
| 29 | SD | 0\3 | – | 0\3 | – | 1\3 | 40 |
|
| |||||||
| 30 | SD | 1\3 | 60 | 2\3 | 70 | 0\3 | – |
|
| |||||||
| 31 | SD | 0\3 | – | 1\3 | 80 | 0\3 | – |
|
| |||||||
| 32 | SD | 0\3 | – | 1\3 | 40 | 0\3 | – |
|
| |||||||
| 33 | SD | 1\2 | 40 | 0\2 | – | 0\2 | – |
|
| |||||||
| 34 | WY | 0\2 | – | 0\2 | – | 1\2 | 40 |
|
| |||||||
| 35 | IA | 0\2 | – | 0\2 | – | 1\2 | 80 |
|
| |||||||
| 36 | SD | 1\2 | 60 | 0\2 | – | 1\2 | 40 |
|
| |||||||
| 37 | SD | 0\2 | – | 0\2 | – | 2\2 | 60 |
|
| |||||||
| 38 | SD | 1\2 | 40 | 0\2 | – | 0\2 | – |
|
| |||||||
| 39 | SD | 0\2 | – | 0\2 | – | 1\2 | 80 |
|
| |||||||
| 40 | SD | 1\2 | 40 | 0\2 | – | 0\2 | – |
|
| |||||||
| 41 | SD | 1\2 | 60 | 1\2 | 80 | 0\2 | – |
|
| |||||||
| 42 | SD | 1\2 | 80 | 1\2 | 80 | 1\2 | 40 |
|
| |||||||
| 43 | SD | 1\2 | 60 | 1\2 | 40 | 0\2 | – |
|
| |||||||
| 44 | SD | 0\2 | – | 0\2 | – | 1\2 | 40 |
|
| |||||||
| 45 | SD | 0\2 | – | 0\2 | – | 1\2 | 80 |
|
| |||||||
| 46 | SD | 1\2 | 80 | 1\2 | 80 | 0\2 | – |
|
| |||||||
| 47 | SD | 0\2 | – | 1\2 | 40 | 1\2 | 40 |
|
| |||||||
| Total | 36\230 | 28\230 | 37\230 | ||||
Only farms with at least one serum sample positive to at least one of the listed three viruses were included. This table does not include the farms that only had one sample. Each farm ID is accompanied by its location by state within the USA. For each farm, the number of samples positive to the virus over the number of total samples and the average HI titre of the positive samples are shown. A “–” indicates the samples were negative to the virus.
Farms only represented by one serum sample are not detailed on this table, but the data are as follows: of the 74 single-sample farms, 16 serum samples were positive for antibodies against at least one virus. Of these, eight were positive for antibodies against D/OK, 11 were positive for antibodies against D/660 and four were positive for antibodies against C/JHB.
It should be noted that of 57 total positive samples to IDVs for 2015 serum samples, 23 (23 of 364; 6.3%) of them were positive for both IDV lineages (D/OK and D/660), while 21 (21 of 364; 5.8%) and 13 (13 of 364; 3.6%) tested positive for only D/OK or D/660 lineage, respectively. Based on the cross-reactivity data of two IDV lineages generated against reference rabbit anti-serum (Table 1), we speculated that the 23 horses with detectable antibodies to both lineages were infected largely by D/660- or D/660-like viruses because D/660 anti-sera recognized both lineages equally well in the HI assay. Furthermore, our observation that 21 horses possessed antibodies only to D/OK seemed to indicate that these animals were infected by D/OK- or D/OK-like viruses because rabbit anti-serum specific for D/OK has a 4-fold higher HI antibody titre to the D/OK than D/660 (Table 1). Interestingly, we also found 13 horses only seropositive to the D/660 lineage. We suspect that these animals may have been exposed to a D/660-like virus, which may have evolved some mutations abolishing epitopes common to the two lineages tested (D/660 and D/OK). A recent study in Japan has provided some preliminary evidence supporting for the existence of a potential third lineage of IDV (Murakami et al., 2016). It is also possible that the samples positive for both lineages of IDV could be exhibiting super-infection or co- infection of both D/660 and D/OK. This is difficult to determine due to both the cross-reactive nature of these viral lineages as well as our lack of temporal data related to the samples collected. These hypotheses will be investigated in future studies. In summary, the results of our serology study demonstrated that multiple lineages of IDV already infected horses in the Midwestern region, and that there is possibly some element of super-infection, co-infection and/or cross-reactivity between the two IDV lineages studied, or there is the potential for the presence of an unidentified IDV lineage related to both D/OK and D/660 circulating among horses.
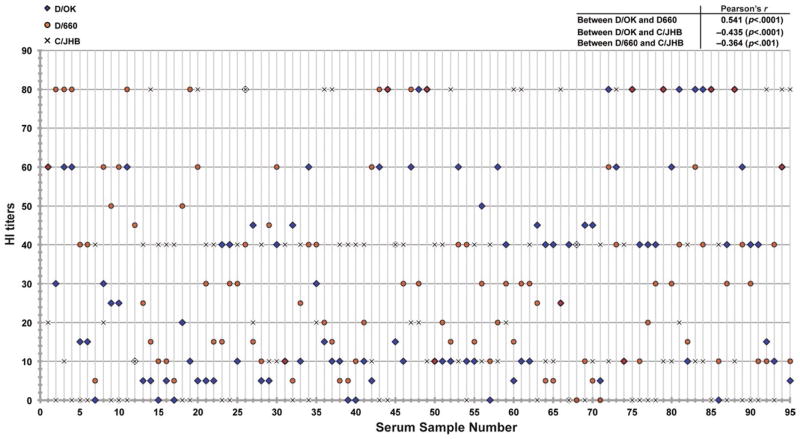
In Figure 1, we graphed a 2-D scatter plot containing all 2015 samples from the HI trials that tested positive for at least one of the two IDVs (D/OK and D/660) and one ICV. The correlation coefficients show that there is strong positive correlation between the antibody titres of the two IDV lineage viruses (D/660-D/OK), with an R value of 0.541 (p < .0001). Interestingly, we also found negative correlations between the antibody titres found for our ICV strain and the two IDV strains, with an R of −0.364 (p < .0001) between C/JHB and D/660, and an R of −0.435 (p < .001) between C/JHB and D/OK.
FIGURE 1.
The scatter plot comparing viral HI antibody titres among the three tested viruses. The figure is plotted by arranging all positive samples collected in 2015 on the x-axis and HI antibody titres on the y-axis, which illustrates antibody titres of each sample against all three viruses. Pearson correlation analysis was used to determine correlations between sample titres among the three viruses. The derived correlation efficiencies among each virus’s measured HI antibody titres (C/JHB-D/660, D/660-D/OK, and C/JHB-D/OK) are listed in the figure. A high positive R value denotes a strong positive correlation, while a negative R value describes a negative correlation. [Colour figure can be viewed at wileyonlinelibrary.com]
In addition to 2015 serum samples, we also collected 100 horse serum samples in 2016 to determine the presence of IDV- or ICV- specific antibodies. Compared with 2015 serum samples deriving from aged horses, 2016 serum samples were largely from young ponies in SD and the region. Our result showed three (three of 100, 3%) samples positive for antibodies against D/OK, two (two of 100, 2%) samples positive for antibodies against D/660 and eight (eight of 100, 8%) samples positive for antibodies against C/JHB (data not shown). The discrepancies observed in the overall seroprevalence between the samples in 2015 and in 2016 may be attributed to a much smaller samples size consisting of samples taken from a much narrower population of horses. The 2016 samples represent both fewer states and a much smaller total number of farms.
To further verify the results observed in the HI assays, the MN assay was used for a select subset of samples. Samples were chosen such that we could confirm both positive and negative results: MN sample groups were made of samples positive for multiple viruses, positive for just one virus and positive for no viruses. As shown in Table 3 (2015 samples) and Table 4 (2016 samples), the MN assay confirms the results found via HI assay. The titre threshold for confirming a positive sample was considered anything greater than or equal to 40 (Quast et al., 2015; Ran et al., 2015). Based on the MN results, the HI seroprevalence results appear to be accurate for all three virus lineages, confirming the presence of IDV and ICV within American equine populations. It should be noted that despite the overall agreement between HI and MN assays, several outliers were observed. For example, three samples (IDs# 4, 6, 21) in Table 3 and one sample (IDs# 19, 23), tested positive in the HI assay, did not contain measurable neutralizing antibodies in the MN assay. The discordance may be caused by suboptimal quality of these samples after the long-term storage process. We also found that some samples (IDs 4, 5, 17, 18, 21, and 22 in Table 3 and ID# 71 in Table 4) tested negative by HI, turned out to be positive in the MN assay. This inconsistency is likely due to more sensitive nature the MN assay offers in the antibody detection. Another possibility is that the MN assay could detect other functional antibodies within the horse serum, such as those blocking virus–cell fusion, which could not be detected by HI assay.
TABLE 3.
Microtitre neutralization confirmation of the HI titres observed for serum samples in 2015
| Sample ID | D/OK | D/660 | C/JHB | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| HI | MN | HI | MN | HI | MN | |
| 1 | 80 | 80 | 80 | 160 | – | – |
|
| ||||||
| 2 | – | – | – | – | – | – |
|
| ||||||
| 3 | 80 | 80 | 80 | 160 | – | – |
|
| ||||||
| 4 | 40 | – | 40 | 40 | – | 80 |
|
| ||||||
| 5 | – | – | – | 40 | – | – |
|
| ||||||
| 6 | 40 | – | – | – | – | – |
|
| ||||||
| 7 | – | – | – | – | – | – |
|
| ||||||
| 8 | 80 | 160 | 80 | 80 | – | – |
|
| ||||||
| 9 | 80 | 40 | – | – | – | – |
|
| ||||||
| 10 | – | – | – | – | – | – |
|
| ||||||
| 11 | – | – | – | – | – | – |
|
| ||||||
| 12 | – | – | – | – | – | – |
|
| ||||||
| 13 | – | – | – | – | – | – |
|
| ||||||
| 14 | – | – | – | – | – | – |
|
| ||||||
| 15 | – | – | – | – | – | – |
|
| ||||||
| 16 | – | – | – | – | – | – |
|
| ||||||
| 17 | – | – | – | 40 | – | – |
|
| ||||||
| 18 | – | – | – | 40 | – | – |
|
| ||||||
| 19 | – | – | – | – | – | – |
|
| ||||||
| 20 | 60 | 40 | 80 | 40 | – | – |
|
| ||||||
| 21 | 40 | – | – | 40 | 80 | – |
|
| ||||||
| 22 | – | 80 | 60 | 40 | – | – |
A “–” indicates the samples were negative to the virus.
TABLE 4.
Microtitre neutralization confirmation of the HI titres observed for serum samples in 2016
| Sample ID | D/OK | D/660 | C/JHB | |||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| HI | MN | HI | MN | HI | MN | |
| 6 | 60 | 160 | 80 | 160 | – | – |
|
| ||||||
| 11 | – | – | – | – | – | – |
|
| ||||||
| 12 | – | – | – | – | – | – |
|
| ||||||
| 13 | 50 | 80 | 40 | 80 | – | – |
|
| ||||||
| 19 | – | – | – | – | 40 | – |
|
| ||||||
| 21 | – | – | – | – | 40 | 80 |
|
| ||||||
| 23 | – | – | – | – | 40 | – |
|
| ||||||
| 25 | – | – | – | – | 60 | 40 |
|
| ||||||
| 30 | – | – | – | – | 40 | 160 |
|
| ||||||
| 32 | – | – | – | – | 60 | 80 |
|
| ||||||
| 71 | – | 80 | 40 | 40 | – | – |
|
| ||||||
| 91 | – | – | – | – | 40 | 40 |
|
| ||||||
| 92 | – | – | – | – | – | – |
|
| ||||||
| 93 | – | – | – | – | – | – |
A “–” indicates the samples were negative to the virus.
In summary, we presented serological evidence that equines are susceptible to IDV and ICV infections. A previous study had showed that ICV antibodies were present in horses (Ditchfield & Macpherson, 1965). Based on these findings, horses should be added into the host range of this novel influenza virus that already includes bovines, swine, sheep and goats (Chiapponi et al., 2016; Ducatez et al., 2015; Ferguson et al., 2016; Hause et al., 2013, 2014; Jiang et al., 2014; Quast et al., 2015). Periodically, spillover of IDV from cattle (primary reservoir) to horses where two species live in close proximity probably within the same farm may cause IDV infection of horses as observed in this study. Future surveillance of IDV in horse populations is needed, as well as more serological investigations into other species potentially susceptible to influenza viruses to determine the total host range of IDV.
Impacts.
We found serological evidence that equine is susceptible to infection by newly emerged influenza D virus. Based on this observation, horses should be added into the host range of this novel zoonotic influenza virus that already includes bovines, swine, sheep, and goats and human.
Multiple lineages of influenza D viruses co-circulated in horse populations of the Midwest United Stated.
Our findings continue to support the need for further surveillance of IDV viruses in agricultural species to work towards a better understanding of the full host range and natural reservoirs of influenza D virus.
Acknowledgments
Funding information
South Dakota Agricultural Experimental Station, Grant/Award Number: 3AH477; South Dakota BioSNTR Foundation
The authors would like to thank fellow lab colleagues in Dr. Feng Li’s lab for technical and personal help and the Animal Disease Research and Diagnostic Lab at SDSU for the procurement and preparation of samples. This work was partially supported by an EPSCOR undergraduate research grant through the South Dakota BioSNTR Foundation and SDSU AES 3AH477.
Footnotes
CONFLICT OF INTEREST
None.
References
- Chiapponi C, Faccini S, De Mattia A, Baioni L, Barbieri I, Rosignoli C, … Foni E. Detection of influenza D virus among Swine and Cattle, Italy. Emerging Infectious Diseases. 2016;22:352–354. doi: 10.3201/eid2202.151439. https://doi.org/10.3201/eid2202.151439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield J, Macpherson LW, Zbitnew A. Upper respiratory disease in thouroughbred horses: Studies of its viral etiology in the toronto area, 1960 to 1963. Canadian Journal of Comparative Medicine and Veterinary Science. 1965;29:18–22. [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Pelletier C, Meyer G. Influenza D virus in cattle, France, 2011–2014. Emerging Infectious Diseases. 2015;21:368–371. doi: 10.3201/eid2102.141449. https://doi.org/10.3201/eid2102.141449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L, Olivier AK, Genova S, Epperson WB, Smith DR, Schneider L, … Wan XF. Pathogenesis of influenza D virus in cattle. Journal of Virology. 2016;90:5636–5642. doi: 10.1128/JVI.03122-15. https://doi.org/10.1128/JVI.03122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Collin EA, Liu R, Huang B, Sheng Z, Lu W, … Li F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. mBio. 2014;5:e00031–00014. doi: 10.1128/mBio.00031-14. https://doi.org/10.1128/mbio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause BM, Ducatez M, Collin EA, Ran Z, Liu R, Sheng Z, … Li F. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathogens. 2013;9:e1003176. doi: 10.1371/journal.ppat.1003176. https://doi.org/10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang WM, Wang SC, Peng C, Yu JM, Zhuang QY, Hou GY, … Chen JM. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes. 2014;49:493–496. doi: 10.1007/s11262-014-1107-3. https://doi.org/10.1007/s11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
- Murakami S, Endoh M, Kobayashi T, Takenaka-Uema A, Chambers JK, Uchida K, … Horimoto T. Influenza D virus infection in herd of cattle, Japan. Emerging Infectious Diseases. 2016;22:1517–1519. doi: 10.3201/eid2208.160362. https://doi.org/10.3201/eid2208.160362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw M. In: Orthomyxoviridae: The viruses and their replication. 5. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Vol. 2. Philadelphia, PA: Fields virology; 2007. pp. 1647–1689. [Google Scholar]
- Quast M, Sreenivasan C, Sexton G, Nedland H, Singrey A, Fawcett L, … Li F. Serological evidence for the presence of influenza D virus in small ruminants. Veterinary Microbiology. 2015;180:281–285. doi: 10.1016/j.vetmic.2015.09.005. https://doi.org/10.1016/j.vetmic.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Z, Shen H, Lang Y, Kolb EA, Turan N, Zhu L, … Ma W. Domestic pigs are susceptible to infection with influenza B viruses. Journal of Virology. 2015;89:4818–4826. doi: 10.1128/JVI.00059-15. https://doi.org/10.1128/JVI.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. American Journal of Epidemiology. 1938;27:493–497. https://doi.org/10.1093/oxfordjournals.aje.a118408. [Google Scholar]
- Sreenivasan C, Thomas M, Sheng Z, Hause BM, Collin EA, Knudsen DE, … Li F. Replication and transmission of the novel bovine influenza D virus in a guinea pig model. Journal of Virology. 2015;89:11990–12001. doi: 10.1128/JVI.01630-15. https://doi.org/10.1128/JVI.01630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SK, Ma W, McDaniel CJ, Gray GC, Lednicky JA. Serologic evidence of exposure to influenza D virus among persons with occupational contact with cattle. Journal of Clinical Virology. 2016;81:31–33. doi: 10.1016/j.jcv.2016.05.017. https://doi.org/10.1016/j.jcv.2016.05.017. [DOI] [PubMed] [Google Scholar]
- Wood JM, Major D, Heath A, Newman RW, Höschler K, Stephenson I, … Zambon MC. Reproducibility of serology assays for pandemic influenza H1N1: Collaborative study to evaluate a candidate WHO International Standard. Vaccine. 2012;30:210–217. doi: 10.1016/j.vaccine.2011.11.019. https://doi.org/10.1016/j.vaccine.2011.11.019. [DOI] [PubMed] [Google Scholar]