Abstract
Collie Eye Anomaly (CEA) encompasses a spectrum of different ophthalmic phenotypes from clinically inconsequential choroidal hypoplasia to blindness from coloboma of the optic nerve head (ONH). A previous study found a 7.8kb deletion in intron 4 of the NHEJ1 gene to be associated with CEA. A genetic test based on this association is recommended for many breeds, including the Nova Scotia Duck Tolling Retriever (NSDTR). Collection of ONH coloboma affected NSDTR showed lack of concordance of the NHEJ1 intronic deletion with ONH coloboma. Using genome-wide single nucleotide polymorphism (SNP) genotyping in 7 ONH coloboma affected NSDTR cases and 47 unaffected NSDTR controls with no ophthalmic signs, one SNP, located on chromosome 7, demonstrated genome-wide significance. However, high genomic inflation may have confounded the results. Therefore, the genome-wide association study was repeated using EMMAX to control for population structure in the cohort of 7 cases and 47 controls. However, no regions of the genome were significantly associated with ONH coloboma. These results failed to document significant association to the CEA locus. Due to the complex genetic etiology of ONH coloboma, the NHEJ1 intronic deletion test results should be carefully considered when make breeding decisions. If the goal is to select for visually competent dogs, our data suggest that eye examinations of puppies would be more effective as a guide in selection of breeding pairs than relying solely on currently available genetic tests.
Keywords: Collie Eye Anomaly, optic nerve head coloboma, dog, genome-wide association, inherited, coloboma
Introduction
Vertebrate eye development is a complex process involving multiple embryonic tissue types.[1] A multitude of ocular disorders can result from improper induction, migration, proliferation, or differentiation of ocular tissues during embryonic development. A coloboma (G. kolobōma, lit., the part taken away in mutilation, fr. koloboō, to dock, mutilate) is an absence of normal ocular tissue and can involve the eyelid, uvea, lens, sclera, retina, or optic nerve.[2] Inferior uveal, retinal, or optic nerve head (ONH) colobomas are typically due to failure of the choroidal fissure to close, while colobomatous malformations at other locations are typically due to abnormal differentiation of the outer layer of the optic cup.[3] In human patients, the prevalence of coloboma is 2–14 per 100,000 live births and can occur in association with other ocular anomalies, such as microphthalmia.[4, 5] Mutations in multiple genes critical to eye development including PAX6, SOX2, RAX, BMP7, SHH, and ALDH1A3 have been identified in human patients with coloboma.[6]
In dogs, the most well studied syndrome associated with coloboma is collie eye anomaly (CEA), a congenital, heritable ocular disorder that occurs commonly in the Rough and Smooth Collie and Shetland Sheepdog.[7–9] A survey study investigating CEA in Rough Collies in Norway found 40.8% of dogs examined to be affected with CEA, of which 93.7% had choroidal hypoplasia and 18.9% had coloboma.[9] Similarly in Swedish Rough Collies, the prevalence of choroidal hypoplasia was found to be 68.1% and the prevalence of coloboma to be 12.4%.[10] This syndrome primarily affects the posterior segment of the eye and varies in both the severity of its clinical manifestations and the ophthalmic lesions present.[11] The two primary ocular lesions are temporal choroidal hypoplasia and coloboma of the ONH; although tortuous retinal blood vessels, retinal detachment, and intraocular hemorrhage have also been variably described.[7] Dogs with CEA can range from mildly to moderately affected individuals with no vision impairment; however, dogs presenting with ONH coloboma are often most severely affected and at risk of retinal detachment and blindness.[11]
In 2003, Lowe et al. used linkage mapping to localize a 3.9 cM locus associated with choroidal hypoplasia on chromosome 37.[11] Parker and colleagues then used fine-mapping techniques in the region to identify a 7.8kb deletion in intron 4 of the NHEJ1 gene, termed the cea locus, that segregated with the specific phenotype of choroidal hypoplasia across multiple dog breeds in an autosomal recessive manner.[12] While their mapping classified cases from controls by the presence or absence of choroidal hypoplasia, respectively, the authors included ONH coloboma as part of “the cea extended phenotype.” To our knowledge, there are no published reports of the co-occurrence of the different ocular phenotypes associated with CEA and the NHEJ1 intronic deletion. In addition to the traditional herding breeds, other breeds with “a CEA-like phenotype” (it is unclear from the publication if this means choroidal hypoplasia or optic nerve coloboma) were found to concordantly segregate the NHEJ1 intronic deletion, including the Lancashire Heeler, Longhaired Whippet, and Nova Scotia Duck Tolling Retriever (NSDTR).[12] The reporting of the NHEJ1 intronic deletion causing CEA in multiple breeds resulted in the development of a PCR-based genetic test being made available to breeders and dog owners.[12, 13] Currently, the Orthopedic Foundation for Animals (http://www.ofa.org) lists the CEA mutation test as available for the Australian Shepherd, Bearded Collie, Border Collie, Boykin Spaniel, Collie, Lancashire Heeler, Miniature American Shepherd, Shetland Sheepdog, Silken Windhound, and NSDTR.
Individuals within the NSDTR breed have been documented to have a “CEA-like phenotype” that is concordant with the NHEJ1 intronic deletion.[12] The genetic test is currently required by the Nova Scotia Duck Tolling Retriever Club of the United Kingdom (http://www.toller-club.co.uk) and recommended by the Nova Scotia Duck Tolling Retriever Club in the United States (http://nsdtrc-usa.org/) and Netherlands (http://www.tollertales.nl/). However, the identification of a dog with an optic nerve coloboma that was not homozygous for the NHEJ1 mutation was brought to our laboratory’s attention. This initiated further investigation into the concordance of the NHEJ1 intronic deletion with ONH coloboma in the NSDTR, as well as the genetic basis of the disease in the breed. As additional sample collection showed that the NHEJ1 intronic deletion did not segregate with ONH coloboma in the NSDTR, genome-wide association analysis was performed to investigate a separate locus responsible for this phenotype. After correction for population stratification, no regions of the genome were significantly associated with ONH coloboma, further supporting that the previously identified NHEJ1 intronic deletion is not solely responsible for all ONH colobomas in this breed.
Materials & Methods
Phenotyping
Client selected board-certified veterinary ophthalmologists examined NSDTRs with suspected ophthalmic signs. Veterinary clinical records, Canine Eye Registration Foundation (CERF) reports and/or Orthopedic Foundation for Animals Eye Certification Registry (CAER--http://www.ofa.org/eye_overview.html) or European College of Veterinary Ophthalmologist (ECVO) certificate of eye examination reports were submitted for affected dogs in the United States and Europe, respectively, along with EDTA whole blood samples. NSDTR controls and relatives of cases had a record of a clear CERF/CAER exam, as documented in the Orthopedic Foundation for Animals database (offa.org).
Canine Sample DNA Extraction
The DNA was extracted from EDTA whole blood samples using Gentra Puregene DNA purification extraction kit (Qiagen, Valenica, CA). Collection of canine samples was approved by the University of California, Davis Animal Care and Use Committee (protocol # 18561).
Collie Eye Anomaly Genotyping
Thirty-five NSDTR were genotyped for the 7.8kb deletion implicated in CEA, as described by Parker et al.[12] This included seven cases affected with ONH coloboma and their relatives when available (sires, dams, siblings, etc.). Each PCR reaction consisted of 13.9µl water, 2µl 10X buffer with MgCl2, 1µl dNTP, 1µl of each primer, 0.1µl AmpliTaq Gold, and 1µl of genomic DNA. Cycling conditions were as follows: 94°C for 12 minutes; 35 cycles 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 2 minutes; 72°C for 20 minutes; 4° hold. Results were visualized on a 2% agarose gel with ethidium bromide.
Genome-wide Association Study
Genome-wide single nucleotide polymorphism (SNP) genotyping was performed using the Illumina Canine HD 174,000 SNP array (Illumina, San Diego, CA) for 7 ONH coloboma cases and 47 controls with no reported ophthalmic disease (clear CERF report). SNPs were pruned from analysis if their minor allele frequency was <5% and the call rate <90%. Chi-square association analysis, odds ratios, Bonferroni adjustments, multidimensional scaling (MDS), and quantile-quantile (QQ) plot and genomic inflation calculations were performed in Plink.[14] The GWAS was repeated using the above criteria in Golden Helix (Golden Helix, Inc. Bozeman, MT) with EMMAX (single locus mixed model) to control for the underlying population structure of samples used.
Results
Phenotyping
CERF/CAER or ECVO certificates or medical records were available for seven ONH coloboma affected NSDTRs. Three cases were affected with an ONH coloboma OD, one was affected OS, and three were affected OU. Four of these cases were classified as affected with CEA by the examining veterinary ophthalmologist - either by checking the CEA box along with the CEA-associated coloboma box on CERF/CAER/ECVO forms or specifically mentioning it in the medical records. Additional phenotype information for each case is summarized in Table 1.
Table 1.
Phenotype information for NSDTR cases affected with ONH coloboma;
| Case | ONH Coloboma |
Age at Diagnosis (years) |
NHEJ1 Deletion Status |
OD | OS | OU | Additional Phenotype Information |
|---|---|---|---|---|---|---|---|
| Case 1 | OU | 1 | +/+ | Distichiasis* | |||
| Case 2 | OS | 1 | +/cea | CEA, partial retinal detachment, ONH hypoplasia | |||
| Case 3 | OD | 1 | +/+ | Retro-lental pigment dots | CEA*, punctual aplasia*, distichiasis* | ||
| Case 4 | OD | 1 | cea/cea | Persistent hyaloid remnant | Iris to iris persistent pupillary membrane | Distichiasis | |
| Case 5 | OD | 2 | cea/cea | Multifocal retinal dysplasia | |||
| Case 6 | OU | 8 | cea/cea | Microphthalmos, lateral strabismus | CEA | ||
| Case 7 | OU | 5 | +/cea | CEA |
: observed in at least one eye, specific eye not mentioned.
NHEJ1 Intronic Deletion Genotyping
Seven NSDTRs affected with ONH coloboma were genotyped by PCR for the 7.8kb NHEJ1 intron 4 deletion previously associated with CEA. Three cases were homozygous for the 7.8kb deletion, 2 were heterozygous, and 2 were homozygous for the wild type allele (Fig. 1, Table 1). In order to investigate segregation of the CEA locus within the breed, an additional 28 unaffected, but related, NSDTRs were also genotyped for the 7.8kb deletion: 8 were heterozygous for the NHEJ1 intronic deletion, while the remaining 20 were homozygous for the wild type allele.
Figure 1.
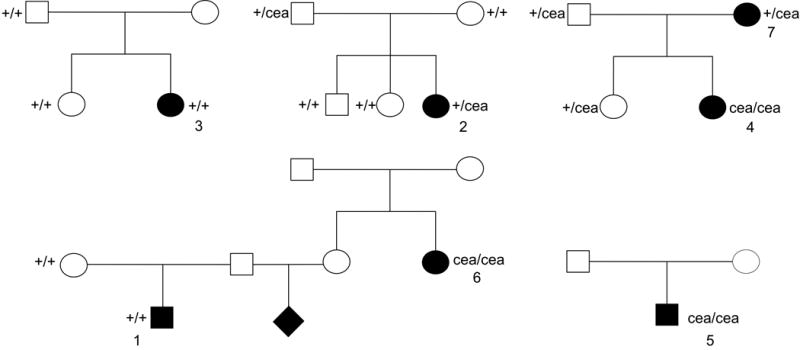
Abbreviated pedigrees of collected NSDTRs affected with ONH coloboma highlight the lack of segregation of the CEA mutation with the ONH coloboma phenotype. A circle denotes female cases and controls, while males are denoted by a square. Black filled shapes represent the cases, while unfilled shapes represent unaffected, CERF clear controls. Numbers correspond to case descriptions presented in Table 1.
GWAS
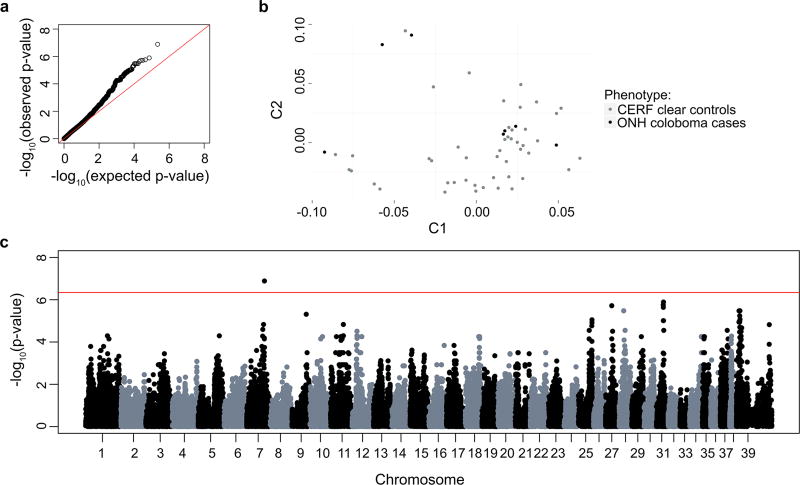
To determine a region of the genome associated with ONH coloboma in the Nova Scotia Duck Tolling Retriever, genome-wide association analysis was performed using 7 confirmed ONH coloboma cases and 47 CERF clear controls. Before pruning, there were 173,662 markers genotyped. 60,424 SNPs failed a minor allele frequency test (MAF <0.05) and 3536 SNPs failed a genotyping rate of 90%, leaving 109,702 SNPs for chi square analysis. Genomic inflation was elevated at 1.32697 due to population stratification, as shown in the QQ plot in Fig. 2a, despite well-matched cases and controls based on MDS (Fig. 2b). Only 1 SNP was genome-wide significant with a pBonferroni<0.05 (chr7:66,174,129: praw=1.295×107, pBonferroni=0.01421) (Fig. 2c).
Figure 2.
a) QQ plot shows −log10 of the expected versus observed p-values plotted for each SNP. b) MDS plot of samples used in GWAS with 7 ONH coloboma cases and 47 controls with no ophthalmic signs (clear CERF report). c) Manhattan plot of −log10 of the raw p-values (y-axis) for each of the genotyped SNPs by chromosome (x-axis) for 7 ONH coloboma cases and 47 controls. The red line indicates genome-wide significance based on a Bonferroni correction. A single genome-wide significant SNP is present on chromosome 7.
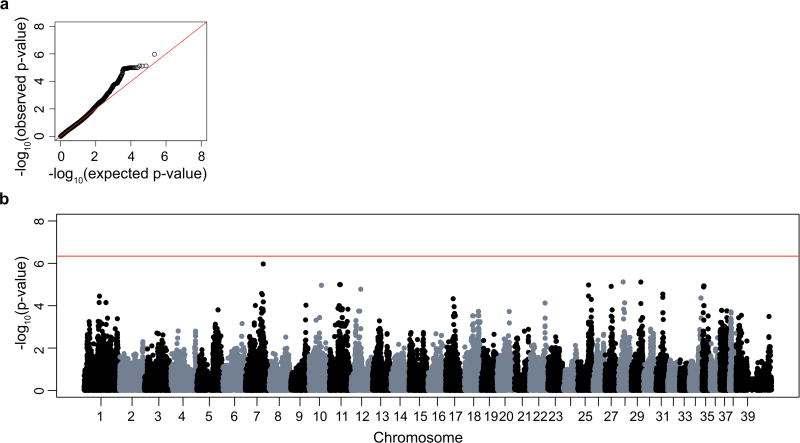
To control for the underlying population structure of the genotyped samples, the GWAS was repeated in Golden Helix using EMMAX. After SNP quality control, 109,394 SNPs remained for analysis. While population stratification was improved (see QQ plot, Fig. 3a), the single SNP on chromosome 7 was no longer significantly associated and no other SNPs demonstrated genome-wide significance (Fig 3b). It is interesting to note that neither GWAS yielded an association to chromosome 37, where the CEA locus was originally mapped.
Figure 3.
a) QQ plot shows −log10 of the expected versus observed p-values plotted for each SNP after correction for population structure using EMMAX. b) Manhattan plot of −log10 of the raw p-values (y-axis) for each of the genotyped SNPs by chromosome (x-axis) for 7 ONH coloboma cases and 47 controls after correction for population structure of genotyped samples. The red line indicates genome-wide significance based on a Bonferroni correction. There were no SNPs with genome-wide significant association.
Discussion
Collie eye anomaly is a complex phenotype, potentially including a number of different ophthalmic structures including the retina, sclera, choroid, and lens.[7] Despite the heterogeneous presentation, choroidal hypoplasia is considered the most consistent sign of CEA, even though severe cases of CEA often have concurrent ONH abnormalities.[17] It has been suggested by others that these 2 phenotypes are inherited separately.[10, 18] Further complicating accurate phenotyping and diagnosis, choroidal hypoplasia may be masked in a dog older than 3 months of age due to retinal pigmentation.[9] In this work we were not able to evaluate the dogs for choroidal hypoplasia as puppies so the separation of these phenotypes could not be evaluated in this cohort. This is in part due to the nature of the pre-breeding ocular evaluation forms used in North America and abroad. The ability of veterinary ophthalmologists to check a box termed CEA rather than the specific clinical phenotypes contributes to continued misevaluation of the true nature of the disease.
A “cea-like phenotype” has been previously documented in the NSDTR, whose basis is supported by anecdotal breed history involving collie ancestors.[12] However, presence of ONH coloboma cases in the breed that are not homozygous for the autosomal recessive mutation associated with CEA is contradictory. Of the 7 NSDTR ONH coloboma cases genotyped for the NHEJ1 intronic deletion, only 3 had concordant genotypes (Fig. 1). The remaining 4 were either heterozygous for the deletion or homozygous for the wild type allele, suggesting that ONH coloboma in the NSDTR can occur independently of the documented NHEJ1 intronic deletion. Additionally, of the four ONH coloboma cases specifically stated to be associated with CEA by the examining veterinarian, only one was homozygous for the NHEJ1 intronic deletion. Since NSDTR breeders have been using this test for some time to avoid producing animals homozygous for the NHEJ1 intronic deletion, it is impossible to know what the association and penetrance are of this mutation with respect to either choroidal hypoplasia or ONH coloboma. Choroidal hypoplasia by itself has not been proven to result in blindness or clinically detectable visual deficits, therefore there is little justification to select against it per se unless it is associated with ONH coloboma.[18] It is the authors’ opinion that further work is required to determine the correlation and penetrance of the NHEJ1 intronic deletion with any visual deficits prior to using this test to make breeding decisions.
Concordance of the NHEJ1 intronic deletion with the CEA phenotype has similarly been investigated in Danish Rough Collies and Shetland Sheepdogs.[19] Fredholm et al found that in Danish Rough Collies the CEA mutation is not prognostic for choroidal hypoplasia. However, they did find concordance of the mutation with choroidal hypoplasia in the Shetland Sheepdog. Given that ONH colobomas and choroidal hypoplasia are often presented as more severe and less severe forms of CEA, respectively, lack of concordance of homozygosity for the NHEJ1 intronic deletion with lesions that could impact vision is concerning. Even before association of the NHEJ1 intronic deletion with CEA, breeders were unsuccessful in the use of strict selective breeding practices to prevent choroidal hypoplasia and ONH coloboma. Specifically, in Swedish Rough Collies, incidences of choroidal hypoplasia and ONH coloboma were documented before and after practices were in place (i.e. ophthalmic examination before 12 weeks of age to avoid breeding of “go-normal” dogs) to limit breeding to dogs with only mild to moderate choroidal hypoplasia and without coloboma.[10] Strict selection based on phenotype did not alter the prevalence of coloboma in the Rough Collie population, however, the prevalence of choroidal hypoplasia increased. The differential changes in ONH coloboma and choroidal hypoplasia despite selection supports that CEA is not a simple, monogenic autosomal recessive disease.
As a result of the lack of association of the NHEJ1 intronic deletion with ONH coloboma, and previous literature suggesting two separate modes of inheritance, genome-wide association analysis was performed on a cohort of ONH coloboma affected NSDTRs.[10, 18] In an initial GWAS using 7 cases and 47 controls, there was 1 SNP with a genome-wide significant association. However, there was considerable population stratification, as demonstrated by the elevated genomic inflation factor and stratified QQ plot (Fig. 2), which potentially obscured a true association within false positive associations.[20] To help correct for this, a second association analysis was performed using EMMAX to account for the underlying population structure. The population stratification was improved (Fig. 3a); however, there were no SNPs with a genome-wide significant association with ONH coloboma (Fig. 3b). The lack of association of ONH coloboma to any region of the genome, including chromosome 37 where CEA has been previously associated, supports that the NHEJ1 intronic deletion is not responsible for ONH coloboma in the NSDTR.
While it has been suggested that mapping of simple autosomal recessive disease alleles in the dog requires 20 cases and 20 controls, there have been numerous examples of dog GWAS studies successfully producing actionable data utilizing many fewer samples.[21–30] If ONH coloboma was a simple, autosomal recessive trait, like CEA is purported to be, it should be identifiable using genome-wide association techniques given the length of linkage disequilibrium in the breed, the number of cases, and the number of controls used in this study.[23, 24]
Given the lack of concordance of the NHEJ1 intronic deletion with ONH coloboma, as well as the lack of evidence that ONH coloboma is inherited in a simple autosomal recessive manner, the CEA mutation test should not be used to direct breeding decisions for ONH coloboma in the NSDTR until a clear understanding of the mode of inheritance and the frequency and severity of visual deficits associated with the NHEJ1 intronic deletion are available.
Acknowledgments
NIH T35 OD010956 (Summer Biomedical Research Fellowship)
NIH 5 T32 OD010931 (Predoctoral Institutional Grant, YEAR Program)
CCAH grant 2012-03-F
Maxine Adler Endower Chair Funds
Contributor Information
Emily A. Brown, Department of Population Health and Reproduction, School of Veterinary Medicine, University of California—Davis.
Sara M. Thomasy, Department of Surgical and Radiological Sciences, School of Veterinary Medicine, University of California—Davis.
Christopher J. Murphy, Department of Surgical and Radiological Sciences, School of Veterinary Medicine, Department of Ophthalmology & Vision Science, School of Medicine, University of California—Davis.
Danika L. Bannasch, Department of Population Health and Reproduction, School of Veterinary Medicine, University of California—Davis, Davis, CA 95616, dlbannasch@ucdavis.edu.
References
- 1.Chow RL, Lang RA. Early eye development in vertebrates. Annual review of cell and developmental biology. 2001;17(1):255–96. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Dubielzig R, Ketring K, McLellan G, et al. Congenital, developmental, or hereditary abnormalities in animals. In: Edwards R, Rodenhuis J, Welh L, editors. Veterinary Ocular Pathology. Elsevire Saunders; London: 2010. pp. 32–3. [Google Scholar]
- 3.Gelatt KN, Gilger BC, Kern TJ. Veterinary ophthalmology. John Wiley & Sons; 2012. [Google Scholar]
- 4.Skalicky SE, White AJ, Grigg JR, et al. Microphthalmia, anophthalmia, and coloboma and associated ocular and systemic features: understanding the spectrum. JAMA ophthalmology. 2013;131(12):1517–24. doi: 10.1001/jamaophthalmol.2013.5305. [DOI] [PubMed] [Google Scholar]
- 5.Williamson KA, FitzPatrick DR. The genetic architecture of microphthalmia, anophthalmia and coloboma. European journal of medical genetics. 2014;57(8):369–80. doi: 10.1016/j.ejmg.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Reis LM, Semina EV. Conserved genetic pathways associated with microphthalmia, anophthalmia, and coloboma. Birth Defects Research Part C: Embryo Today: Reviews. 2015;105(2):96–113. doi: 10.1002/bdrc.21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett K, Stades F. Collie eye anomaly in the Shetland sheepdog in the Netherlands. Journal of Small Animal Practice. 1979;20(6):321–9. doi: 10.1111/j.1748-5827.1979.tb06731.x. [DOI] [PubMed] [Google Scholar]
- 8.Roberts S. The collie eye anomaly. Journal of the American Veterinary Medical Association. 1969;155(6):859. [PubMed] [Google Scholar]
- 9.Bjerkås E. Collie eye anomaly in the rough collie in Norway. Journal of Small Animal Practice. 1991;32(2):89–92. [Google Scholar]
- 10.Wallin-Håkanson B, Wallin-Hakanson N, Hedhammar Å. Influence of selective breeding on the prevalence of chorioretinal dysplasia and coloboma in the rough collie in Sweden. Journal of Small Animal Practice. 2000;41(2):56–9. doi: 10.1111/j.1748-5827.2000.tb03163.x. [DOI] [PubMed] [Google Scholar]
- 11.Lowe JK, Kukekova AV, Kirkness EF, et al. Linkage mapping of the primary disease locus for collie eye anomaly. Genomics. 2003;82(1):86–95. doi: 10.1016/s0888-7543(03)00078-8. [DOI] [PubMed] [Google Scholar]
- 12.Parker HG, Kukekova AV, Akey DT, et al. Breed relationships facilitate fine-mapping studies: A 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Research. 2007;17(11):000. doi: 10.1101/gr.6772807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizukami K, Chang HS, Ota M, et al. Collie eye anomaly in Hokkaido dogs: case study. Veterinary ophthalmology. 2012;15(2):128–32. doi: 10.1111/j.1463-5224.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeRoy G, Rickards B, Flint SJ. The Double Bromodomain Proteins Brd2 and Brd3 Couple Histone Acetylation to Transcription. Molecular Cell. 2008;30(1):51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka J, Swigut T, Milne TA, et al. WDR5 Associates with Histone H3 Methylated at K4 and Is Essential for H3 K4 Methylation and Vertebrate Development. Cell. 2005;121(6):859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 17.Mason T, Cox K. Collie eye anomaly. Australian veterinary journal. 1971;47(2):38–40. doi: 10.1111/j.1751-0813.1971.tb02101.x. [DOI] [PubMed] [Google Scholar]
- 18.Curtis R. Retinal diseases in the dog and cat: an overview and update. Journal of Small Animal Practice. 1988;29(7):397–415. [Google Scholar]
- 19.Fredholm M, Larsen RC, Jönsson M, et al. Discrepancy in compliance between the clinical and genetic diagnosis of choroidal hypoplasia in Danish Rough Collies and Shetland Sheepdogs. Animal Genetics. 2016;47(2):250–2. doi: 10.1111/age.12405. [DOI] [PubMed] [Google Scholar]
- 20.Balding DJ. A tutorial on statistical methods for population association studies. Nature reviews Genetics. 2006;7(10):781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nature reviews Genetics. 2008;9(9):713–25. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- 22.Vernau KM, Runstadler JA, Brown EA, et al. Genome-Wide Association Analysis Identifies a Mutation in the Thiamine Transporter 2(SLC19A3) Gene Associated with Alaskan Husky Encephalopathy. PloS one. 2013;8(3):e57195. doi: 10.1371/journal.pone.0057195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf ZT, Leslie EJ, Arzi B, et al. A LINE-1 insertion in DLX6 is responsible for cleft palate and mandibular abnormalities in a canine model of Pierre Robin sequence. PLoS Genet. 2014;10(4):e1004257. doi: 10.1371/journal.pgen.1004257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf ZT, Brand HA, Shaffer JR, et al. Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genet. 2015;11(3):e1005059. doi: 10.1371/journal.pgen.1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safra N, Bassuk AG, Ferguson PJ, et al. Genome-wide association mapping in dogs enables identification of the homeobox gene, NKX2–8, as a genetic component of neural tube defects in humans. PLoS Genet. 2013;9(7):e1003646. doi: 10.1371/journal.pgen.1003646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forman OP, De Risio L, Mellersh CS. Missense mutation in CAPN1 is associated with spinocerebellar ataxia in the Parson Russell Terrier dog breed. PloS one. 2013;8(5):e64627. doi: 10.1371/journal.pone.0064627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahonen SJ, Arumilli M, Lohi H. A CNGB1 frameshift mutation in Papillon and Phalene dogs with progressive retinal atrophy. PloS one. 2013;8(8):e72122. doi: 10.1371/journal.pone.0072122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng R, Farias F, Johnson G, et al. A truncated retrotransposon disrupts the GRM1 coding sequence in Coton de Tulear dogs with Bandera's neonatal ataxia. Journal of veterinary internal medicine. 2011;25(2):267–72. doi: 10.1111/j.1939-1676.2010.0666.x. [DOI] [PubMed] [Google Scholar]
- 29.Ahonen SJ, Kaukonen M, Nussdorfer FD, et al. A novel missense mutation in ADAMTS10 in Norwegian elkhound primary glaucoma. PloS one. 2014;9(11):e111941. doi: 10.1371/journal.pone.0111941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owczarek-Lipska M, Jagannathan V, Drögemüller C, et al. A frameshift mutation in the cubilin gene (CUBN) in Border Collies with Imerslund-Gräsbeck syndrome (selective cobalamin malabsorption) PloS one. 2013;8(4):e61144. doi: 10.1371/journal.pone.0061144. [DOI] [PMC free article] [PubMed] [Google Scholar]