Abstract
ProCos (Protein Composition Server, script version), one of the machine learning techniques, was used to classify nitrilases as aliphatic and aromatic nitrilases. Some important feature vectors were used to train the algorithm, which included pseudo-amino acid composition (PAAC) and five-factor solution score (5FSS). This clearly differentiated into two groups of nitrilases, i.e., aliphatic and aromatic, achieving maximum sensitivity of 100.00%, specificity of 90.00%, accuracy of 95.00% and Mathew Correlation Coefficient (MCC) of about 0.90 for the pseudo-amino acid composition. On the other hand, five-factor solution score achieved a sensitivity of 96.00%, specificity of 84.00%, accuracy of 90.00% and Mathew Correlation Coefficient (MCC) of about 0.81. The total count of aliphatic amino acids, Ala (A), Gly (G), Leu (L), Ile (I), Val (V), Met (M) and Pro (P), was found to be higher, i.e., 42.7 in case of aliphatic nitrilases, whereas it was 40.1 in aromatic nitrilases. On the other hand, aromatic amino acids, Tyr (Y), Trp (W), His (H) and Phe (F) number, were found to be higher, i.e., 12.7 in aromatic nitrilases as compared to aliphatic nitrilases which was 10.7. This approach will help in predicting a nitrilase as aromatic or aliphatic nitrilase based on its amino acid sequence. Access to the scripts can be done logging onto GitHub using keyword ‘Nitrilase’ or ‘https://github.com/rover2380/Nitrilase.git’.
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1102-9) contains supplementary material, which is available to authorized users.
Keywords: Aliphatic nitrilase, Aromatic nitrilase, Amino acid composition, Protein composition server (ProCos)
Introduction
Nitrilases are the enzymes which catalyze the hydrolysis of various nitriles into corresponding acid and ammonia. These enzymes have been well identified and characterized in plants, bacteria and fungi, and are engaged as an industrially important biocatalyst for the production of bulk and fine chemicals. For example, mandelonitrile could be hydrolyzed to optically pure (R)-(-)- mandelic acid, which is widely used for the production of semisynthetic cephalosporins, penicillins, antitumor agents, and anti-obesity agents (Wang et al. 2014). Researchers have revealed that nitrilases play a vital role in various biological processes and plant–microbe interaction, but despite their valuable importance they are relatively less explored for their metabolic functions.
Nitrilases differ variably in substrate specificities and find wide application in the transformation of a range of nitriles to acids (Sharma et al. 2006, 2012; Bhatia et al. 2014). Previous studies have revealed that nitrilases are specific for aromatic nitriles while nitrile hydratase has affinity towards aliphatic nitriles, but in light of rapidly growing information regarding nitrile metabolizing enzymes, various aspects have to be reconsidered (Mylerova and Martinkova 2003). Because of the established fact that amino acids are responsible for protein structure and function (Yeom et al. 2008; Liu et al. 2013), they are found to play a significant role in classifying nitrilases as aliphatic or aromatic.
With the exponential growth in the quantity of biological data in past years, there has been an impressive progress in computational biology. In silico analysis and various machine learning techniques are being applied for knowledge generation from the data. The machine learning approach is one such area of programming computers to optimize the performance criterion using example data or past results. The genome-based discoveries being continually increased, the possibility of finding novel sources of nitrilases has also increased tremendously (Gong et al. 2013; Kaplan et al. 2011). The annotation with functional assignments for their respective classes through various wet lab techniques is time consuming and labor intensive, which makes machine learning to be effectively used to complement them by saving time, money and labor (Pant et al. 2011). ProCoS script version is one such machine learning algorithm that has recently become prominent for in silico analysis, as they have a high dimensionality and accuracy in prediction of results not only for protein–protein complexes but also for enzyme classification (Rishishwar et al. 2010). Amino acid composition is a predictive feature vector for classification of various classes of proteins on the basis of their substrate specificity and position specificity (Kumar et al. 2011; Sharma et al. 2009).
The present article aims to serve for an insightful categorization and classification of nitrilases using script version of the ProCoS. The peptide composition features have been used for making pseudo-amino acid composition (PAAC) and five-factor solution score (5FSS) models in the present study.
Materials and methods
Dataset
The amino acid sequences of the nitrilases were downloaded from the ExPASy (http://www.expasy.org/sprot/) proteomic server and NCBI website. Nitrilases on the basis of their substrate specificity are distributed into two sets, i.e., positive (aliphatic nitrilase) and negative (aromatic nitrilase) dataset. Fifty amino acid sequences were considered in the study for both the datasets (Tables 1 and 2). Test and training sets were designed from a fivefold cross-validation scheme to create a model for the classification of a new sequence of nitrilase. The script used is accessible both as an applet and as a server, which is designed in Java and the server works on Perl-PHP backbone deposited in GitHub (https://github.com/rover2380/Nitrilase.git). The minimum input requirement for the analysis is the protein sequences in fasta format and output can be achieved in the form of tables.
Table 1.
Aliphatic nitrilases with their accession and amino acid number
| Aliphatic nitrilases | |||
|---|---|---|---|
| S. no | Name of the microorganism | Accession number | Length (amino acid) |
| 1 | Rhodococcus rhodochrous K22 | gi|417382 | 383 |
| 2 | Rhodococcus rhodochrous J1 | gi|417384 | 366 |
| 3 | Nocardia sp. C-14-1 | gi|60280369 | 381 |
| 4 | Synechococcus sp. ATCC 27144 | WP_011243013 | 334 |
| 5 | Polaromonas naphthalenivorans | gi|500125486 | 353 |
| 6 | Rhizobium leguminosarum bv. viciae 3841 | gi|116255137 | 340 |
| 7 | Variovorax paradoxus EPS | gi|315596504 | 344 |
| 8 | Burkholderia sp. BT03 | gi|495013900 | 356 |
| 9 | Danaus plexippus F2 | gi|357616093 | 389 |
| 10 | Comamonas testosterone | gi|1082009 | 354 |
| 11 | Sorangium cellulosum So0157-2 | gi|521469000 | 342 |
| 12 | Rhizoctonia solani 123E | gi|660965364 | 364 |
| 13 | Polycyclovorans algicola | gi|659838894 | 362 |
| 14 | Rhizobium leguminosarum | gi|659064095 | 348 |
| 15 | Methylobacterium sp. L2-4 | gi|657247605 | 358 |
| 16 | Bosea sp. 117 | gi|657241356 | 350 |
| 17 | Bradyrhizobium sp. th.b2 | gi|656043203 | 360 |
| 18 | Azospirillum halopraeferens | gi|655966390 | 354 |
| 19 | Bradyrhizobium elkanii | gi|654889008 | 354 |
| 20 | Rhizobium sp. JGI 0001019-L19 | gi|655350271 | 348 |
| 21 | Burkholderia mimosarum | gi|654755069 | 350 |
| 22 | Amycolatopsis taiwanensis | gi|654475327 | 346 |
| 23 | Variovorax sp.P21 | gi|654178860 | 350 |
| 24 | Agrobacterium rhizogenes ATCC 15834 | gi|653181208 | 350 |
| 25 | Saccharomonospora viridis DSM 43017 | ACU96985 | 331 |
| 26 | Mesorhizobium loti | gi|652688040 | 348 |
| 27 | Acidovorax oryzae | gi|651303417 | 344 |
| 28 | Achromobacter xylosoxidans | gi|651250268 | 345 |
| 29 | Variovorax paradoxus | gi|648592180 | 350 |
| 39 | Methylobacterium sp. 88A | gi|648483839 | 363 |
| 31 | Burkholderia kururiensis | gi|648430021 | 359 |
| 32 | Pseudomonas syringae B728a | WP_011266126 | 336 |
| 33 | Methylopila sp. 73B | gi|519032254 | 350 |
| 34 | Sphingopyxis alaskensis | WP_011541682 | 338 |
| 35 | Bradyrhizobium sp. ORS278 | WP_011927383 | 337 |
| 36 | Xanthobacter sp. 126 | gi|635631313 | 352 |
| 37 | Colletotrichum fioriniae PJ7 | gi|615443311 | 362 |
| 38 | Oligotropha carboxidovorans OM5 | gi|209874119 | 354 |
| 39 | Methylibium petroleiphilum PM1 | gi|124258961 | 357 |
| 40 | Marinomonas ushuaiensis DSM 15871 | gi|575464044 | 344 |
| 41 | Betaproteobacteria bacterium MOLA814 | gi|557914537 | 367 |
| 42 | Cupriavidus sp. WS | gi|519051014 | 356 |
| 43 | Methylopila sp. M107 | gi|519021908 | 352 |
| 44 | Methyloversatilis universalis | gi|519007573 | 345 |
| 45 | Teredinibacter turnerae | gi|518436209 | 349 |
| 46 | Shimwellia blattae ATCC 29907 | WP_002439083 | 342 |
| 47 | Burkholderia gladioli | gi|503455327 | 373 |
| 48 | Starkeya novella | gi|502933508 | 357 |
| 49 | Serratia sp. M24T3 | gi|497320793 | 342 |
| 50 | Janthinobacterium sp. Marseille | gi|501028829 | 355 |
Table 2.
Aromatic nitrilases with their accession and amino acid number
| Aromatic nitrilases | |||
|---|---|---|---|
| S. no | Name of the microorganism | Accession number | Length (amino acid) |
| 1 | Pantoea sp. AS-PWVM4 | gi|544758631 | 328 |
| 2 | Elizabethkingia | gi|544938496 | 318 |
| 3 | Fodinicurvata sediminis | gi|550981872 | 310 |
| 4 | Thalassospira lucentensis | gi|550982983 | 311 |
| 5 | Rhizobium leguminosarum bv. trifolii WSM1325 | gi|240856665 | 330 |
| 6 | Cellulophaga algicola DSM 14237 | gi|319421185 | 316 |
| 7 | Maricaulis maris MCS10 | gi|114340126 | 310 |
| 8 | Pseudomonas sp. GM41 | gi|576708726 | 324 |
| 9 | Burkholderia sp. BT03 | gi|576730682 | 328 |
| 10 | Morganella morganii subsp. morganii KT | gi|455420318 | 338 |
| 11 | Rubellimicrobium mesophilum DSM 19309 | gi|598658225 | 319 |
| 12 | Tomitella biformata | gi|640112707 | 324 |
| 13 | Pedobacter jeongneungensis | gi|640722764 | 318 |
| 14 | Flexithrix dorotheae | gi|648518461 | 314 |
| 15 | Sediminispirochaeta bajacaliforniensis | gi|648603114 | 316 |
| 16 | Niabella soli DSM 19437 | gi|570745400 | 321 |
| 17 | Butyrivibrio sp. MC2021 | gi|651408280 | 310 |
| 18 | Dyadobacter alkalitolerans | gi|651643084 | 314 |
| 19 | Arenibacter latericius | gi|652415782 | 316 |
| 20 | Maribacter antarcticus | gi|652759557 | 316 |
| 21 | Chryseobacterium sp. UNC8MFCol | gi|653122843 | 319 |
| 22 | Meiothermus chliarophilus | gi|654421979 | 314 |
| 23 | Sphingobacterium thalpophilum | gi|654603925 | 318 |
| 24 | Desulfatibacillum aliphaticivorans | gi|654863925 | 307 |
| 25 | Parabacteroides gordonii | gi|655317710 | 317 |
| 26 | Pseudonocardia spinosispora | gi|655591302 | 310 |
| 27 | Stappia stellulata | gi|656017004 | 316 |
| 28 | Rhodococcus aetherivorans | gi|657826219 | 322 |
| 29 | Marssonina brunnea sp. MB_m1 | gi|597582433 | 321 |
| 30 | Pseudomonas pseudoalcaligenes CECT:5344 | gi|652791517 | 324 |
| 31 | Burkholderia multivorans CGD1 | WP_006401663 | 307 |
| 32 | Thalassiosira pseudonana | EED91795 | 320 |
| 33 | Saccharomyces cerevisiae RM11-1a | EDV09642 | 322 |
| 34 | Ajellomyces dermatitidis ER-3 | EEquation 85041 | 297 |
| 35 | Scheffersomyces stipitis ATCC 58785 | XP_001385512 | 307 |
| 36 | Methanosarcina mazei BAA-159 | WP_011033178 | 307 |
| 37 | Arabidopsis thaliana | AEE77890 | 346 |
| 38 | Bacillus sp. OxB-1 | AB028892 | 339 |
| 39 | Synechocystis sp. PCC6803 | gi|1001835 | 346 |
| 40 | Aeribacillus pallidus | gi|111054396 | 323 |
| 41 | Runella slithyformis | WP_013931053 | 310 |
| 42 | Pseudomonas entomophila L48 | WP_011534641 | 307 |
| 43 | Shewanella sediminis HAW-EB3 | ABV35137 | 317 |
| 44 | Microscilla marina ATCC 23134 | WP_002693358 | 304 |
| 45 | Janthinobacterium sp. Marseille | WP_012080333 | 316 |
| 46 | Burkholderia cepacia J2315 | WP_006483427 | 307 |
| 47 | Bordetella bronchiseptica | WP_003808910 | 310 |
| 48 | Geodermatophilus obscurus ATCC 25078 | WP_012946300 | 260 |
| 49 | Nocardiopsis dassonvillei ATCC 23218 | WP_013156158 | 280 |
| 50 | Streptomyces albus J1074 | WP_003950974 | 315 |
Features
Amino acid composition (AAC)
The amino acid frequency was calculated for both the datasets of proteins (aliphatic and aromatic nitrilases). Calculation of amino acid frequencies gives the value of the occurrence of that amino acid in the particular protein sequence. The fraction of the twenty amino acids was calculated using the following equation:
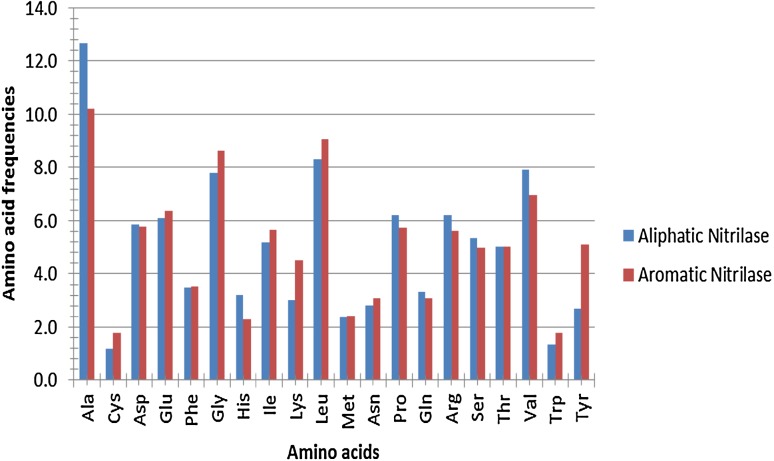
This gives a significance of a particular amino acid. The script takes an input of 20 vectors corresponding to twenty amino acids. Figure 1 shows that the amino acid frequencies of aromatic and aliphatic nitrilases are different, so they can be easily distinguished.
Fig. 1.
Comparison of amino acid frequencies of aliphatic and aromatic nitrilases using ProCoS
Dipeptide composition (DPC)
Dipeptide composition was calculated for all the 20 × 20 (400) combinations of amino acid. It gives significance to the combination of amino acids. The fraction of each dipeptide was calculated using the following equation:
Tripeptide composition (TPC)
Tripeptide composition was also calculated like amino acid and dipeptide composition, thus generating all 20 × 20 × 20 (8000) feature vectors for training and testing datasets.
Pseudo-amino acid composition (PAAC)
The use of simple amino acid composition feature misses the important information in order of amino acid present in the peptide. Keeping this in view, the following information is incorporated with the help of PAAC as mentioned by Chou (2001). The feature vectors built according to this concept contains the frequency of 20 amino acids followed by their respective order information. Web server for calculation of PAAC had been proposed which calculates the respective feature (Shen and Chou 2008).
Split amino acid composition (SAAC)
Peptides were split into three parts to compute split amino acid composition of each part of protein separately. In this way, a vector of dimension 60 (3 × 20) was created instead of 20 in case of amino acid composition. In SAAC, each protein was divided into three parts like: (1) 20 amino acids of the N terminus, (2) 20 amino acids of the C–terminus, and (3) remaining protein length after removing 20 amino acids from N– and C– terminus.
Hybrid model 1
First hybrid model was made by combining the feature vectors of amino acid composition and dipeptide composition (AAC + DPC) giving us 420 vectors (20 + 400) for training and testing dataset.
Hybrid model 2
Second hybrid model was made by combining split amino acid feature to the hybrid 1 (AAC + DPC + SAAC) feature resulting in 480 (20 + 400 + 60) feature vectors for SVM.
Machine learning using script version of ProCos (Protein composition server)
The present study uses the script version which has been implemented and is a supervised machine learning algorithm. The idea behind using the script is the classification which attaches the feature vector with each sample (this case its peptide) to represent those points in a high dimensional feature space and then assigning the points into a particular category (positive or negative class) on the basis of an optimal separating hyperplane. The script training most preciously gives a global solution to optimize the hyperplane, thus avoiding the problem of overfitting of the data to one another class.
Cross-validation and evaluation parameter
A fivefold cross-validation for validating pseudo-amino acid composition (PAAC) and five-factor solution score (5FSS) model predictors was used. The performance of all the models was evaluated by the following standard parameter method:
-
Sensitivity or coverage of positive examples: It is the percent of aromatic nitrilase proteins correctly predicted.
-
Specificity or coverage of negative examples: It is the percent of aliphatic nitrilase proteins correctly predicted aliphatic nitrilase.
-
Accuracy: It is the percentage of correctly predicted proteins (aromatic and aliphatic proteins).
Mathew’s correlation coefficient (MCC): It is considered to be the most robust parameter of any class prediction method. MCC equal to 1 is regarded as perfect prediction while 0 for completely random prediction.
where TP and TN are truly or correctly predicted aliphatic and aromatic nitrilases. FP and FN are wrongly predicted aliphatic and aromatic nitrilases.
Results
The script written is a powerful applet and a classification tool that has become increasingly popular in various machine learning applications. Machine learning approach is considered to be one of the vital subfields of artificial intelligence which is more concerned with the development of techniques and methods that enable the computer to learn. The present study classifies nitrilases on the basis of their amino acid composition which is responsible for their substrate specificity, stability and selectivity. The model developed by machine learning technique is used to differentiate between the two groups of nitrilases. The total count of aliphatic amino acids, i.e., alanine (A), glycine (G), leucine (L), isoleucine (I), valine (V), methionine (M) and proline (P), was found to be higher, i.e., 42.7 in case of aliphatic nitrilase as compared to aromatic nitrilases which is 40.1 (Fig. 1). On the other hand, aromatic amino acids, tyrosine (Y), tryptophan (W), histidine (H) and phenylalanine (F) number, were found to be higher, i.e., 12.7 as when compared to aliphatic nitrilases which were 10.7.
For aliphatic and aromatic class of nitrilases, machine was trained using ProCoS, each with a different type of kernel (linear, polynomial, radial basis and sigmoid). The output with the best training results was considered with high sensitivity, specificity, accuracy and Mathew’s correlation coefficient which has been summarized in Table 3 (detailed information provided as supplementary data S1-S7).
Table 3.
Performance of the models based on vectors for amino acid composition (AAC), dipeptide composition (DPC), split amino acid composition (SAAC), pseudo-amino acid composition (PAAC), tripeptide composition (TPC), hybrid 1 (AAC + DPC) and hybrid 2 (AAC + DPC + SAAC), respectively, Matthews correlation coefficient (MCC), rate of false prediction (RFP)
| Model | Sensitivity | Specificity | Accuracy | MCC | RFP |
|---|---|---|---|---|---|
| AAC | 90.00 | 93.88 | 91.92 | 0.84 | 6.25 |
| DPC | 94.00 | 91.84 | 92.93 | 0.86 | 7.84 |
| SAAC | 92.00 | 81.63 | 86.87 | 0.74 | 16.36 |
| PAAC | 100.00 | 90.00 | 95.00 | 0.90 | 9.09 |
| TPC | 94.00 | 92.00 | 93.00 | 0.86 | 7.84 |
| hyb1 | 96.00 | 87.76 | 91.92 | 0.84 | 11.11 |
| hyb2 | 92.00 | 93.88 | 92.93 | 0.86 | 6.12 |
Sensitivity, specificity and accuracy are in percentage (in bold and italics are the maximum accuracy and MCC)
Amino acid composition (AAC)
A sensitivity of 90.00%, specificity of 93.88%, accuracy of 91.92% and MCC of about 0.84 for AAC was achieved which clearly indicates the difference between the two classes of nitrilase, i.e., aliphatic and aromatic nitrilases but with the rate of false prediction (RFP) of 6.25.
Dipeptide composition (DPC)
This model performed better than AAC with sensitivity of 94.00%, specificity of 91.84%, accuracy of 92.93% and MCC of 0.86. RFP was found to be more than AAC, i.e., 7.84, respectively.
Split amino acid composition (SAAC)
This model gave sensitivity of 92.00%, specificity of 81.63%, accuracy of 86.87% and MCC of 0.74, but the RFP was high with the value of 16.36.
Tripeptide composition (TPC)
he model based on TPC feature achieved sensitivity of 94.00%, specificity of 92.00%, accuracy of 93.00% and MCC of 0.86 with the RFP of 7.84.
Pseudo-amino acid composition (PAAC)
Model based on PAAC feature vector achieved the highest sensitivity of 100.00%, specificity of 90.00%, accuracy of 95.00% and MCC of 0.90 and the RFP of 9.09, respectively (Tables 3 and 4). Among all the models, this model has the maximum accuracy and MCC so we considered this feature model as the best out of all models built yet in this study for nitrilase classification.
Table 4.
Performance of ProCos model using pseudo-amino acid calculation (PAAC) and five-factor solution score (5FFSS) features
| Threshold | PAAC | 5FFSS | ||||||
|---|---|---|---|---|---|---|---|---|
| Sn | Sp | Acc | Mcc | Sn | Sp | Acc | Mcc | |
| − 0.1 | 100.00 | 90.00 | 95.00 | 0.90 | 96.00 | 84.00 | 90.00 | 0.81 |
| 0.0 | 96.00 | 90.00 | 93.00 | 0.86 | 92.00 | 86.00 | 89.00 | 0.78 |
| 0.1 | 94.00 | 92.00 | 93.00 | 0.86 | 90.00 | 88.00 | 89.00 | 0.78 |
Sn sensitivity, Sp specificity, Acc accuracy, Mcc Matthews correlation coefficient
Discussion
As the next generation DNA sequencing (NGS) techniques have become cheaper and more efficient in yielding sequence data in a short time, the number of sequences in the public domain has increased significantly but still important annotations are missing (Chakravorty and Hegde 2017). Experimental validation of every uncharacterized, putative and hypothetical sequence may not be possible with the same pace (Rottig et al. 2010) and assigning functions to all the predicted genes/proteins would be time and cost ineffective (Kim et al. 2013). The characterized set of sequences deposited in the gene/protein databases for nitrilases is fewer in number; therefore, automated computational methods are needed to assign a putative function to uncharacterized sequences reliably (Mills et al. 2015). To the best of our knowledge, no study has been carried out for reliable classification of nitrilases as aliphatic or aromatic.
Previous analysis has confirmed that functional annotation between a test sequence and annotated sequence is above 60%, below which the probability of predicting the function of the test to the query sequence is rather low (Tian et al. 2003; Arakaki et al. 2009; Rottig et al. 2010). It has been inferred in the past that low sequence similarities (below 30%) have resulted in more of paralogs with the query sequence instead of orthologs (Chen and Jeong 2000). Nitrilases with sequence identity as low as 27% with that of characterized nitrilase retained true nitrilase activity if the catalytic triad was found to be conserved (Kaushik et al. 2012). Overall data in the present study share average value of more than 30% identity and conserved catalytic triad. This has led us to infer that sequences retain true nitrilase activity with identity as low as 27% and catalytic triad is conserved throughout. This information will be helpful for the analysis and to predict the models to gain insights into the mechanism of enzyme–substrate specificity as reported in the past (Stachelhaus et al. 1999; Challis et al. 2000; Sharma et al. 2017). Substrate range for nitrilases is rather broad including aliphatic, aromatic and arylnitriles which depends on the groups attached to the side chain (Gong et al. 2012). Characteristics of residues surrounding the active site and the presence of specific amino acids increase the probability for predicting the substrate affinity of nitrilases.
In the present analysis, the script is used to classify the amino acid composition and their dominance in aliphatic and aromatic nitrilases which is responsible for differences in substrate affinity. Cysteine acts as a nucleophile for substrate attack and is activated due to the deprotonation of sulfhydryl group of cysteine by glutamic acid (Zang et al. 2014). Glutamic acid acts as a general base, whereas lysine as general acid (Martinkova and Kren 2010). The aliphatic amino acid alanine (A) also plays a significant role in overall activity of nitrilases (Sharma et al. 2009; Kaushik et al. 2012). Glycine (G), leucine (L), isoleucine (I), valine (V), methionine (M) and proline (P) are other important amino acids which support the aliphaticity of nitrilases. On the other hand, aromatic substrate affinity for some nitrilases is due to tyrosine (Y), tryptophan (W), histidine (H) and phenylalanine (F) which are found to be higher in aromatic nitrilases. These amino acids create aromatic-rich environment near the catalytic centre of nitrilases which prefer aromatic substrates (Liu et al. 2013; Zang et al. 2014). The present data clearly define the role of amino acids for the substrate specificity determination which will further play a significant role in mutational studies of nitrilases to achieve better stability, specificity and reactivity.
Conclusion
The article focuses on the use of the script based method for classification of aliphatic and aromatic group of nitrilases. The results clearly exhibited that the algorithm can be used as a tool to classify nitrilases as aliphatic and aromatic class. The overall accuracy achieved by writing the following script is 95.00%. These machine learning techniques can be used to predict different features of the gene/protein and selection of these algorithms for the prediction of gene/protein function.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the Department of Biotechnology, New Delhi for the continuous support to the Bioinformatics Centre, Himachal Pradesh University, Summer Hill, Shimla, India.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-018-1102-9) contains supplementary material, which is available to authorized users.
Contributor Information
Ruchi Verma, Email: ruchi1st2002@gmail.com.
Tek Chand Bhalla, Phone: +91-177-2832154, Email: bhallatc@rediffmail.com.
References
- Arakaki AK, Huang Y, Skolnick J. EFICAz2: enzyme function inference by a combined approach enhanced by machine learning. BMC Bioinform. 2009;10:107. doi: 10.1186/1471-2105-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia SK, Mehta PK, Bhatia RK, Bhalla TC. Optimization of arylacetonitrilase production from Alcaligenes sp. MTCC 10675 and its application in mandelic acid synthesis. Appl Microbiol Biot. 2014;98:83–94. doi: 10.1007/s00253-013-5288-9. [DOI] [PubMed] [Google Scholar]
- Chakravorty S, Hegde M. Gene and variant annotation for mendelian disorders in the era of advanced sequencing technologies. Annu Rev Genom Hum Genet. 2017;18:229–256. doi: 10.1146/annurev-genom-083115-022545. [DOI] [PubMed] [Google Scholar]
- Challis GL, Ravel J. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol Lett. 2000;187:111–114. doi: 10.1111/j.1574-6968.2000.tb09145.x. [DOI] [PubMed] [Google Scholar]
- Chen R, Jeong SS. Functional prediction: identification of protein orthologs and paralogs. Prot Sci. 2000;9:2344–2353. doi: 10.1110/ps.9.12.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CK. Prediction of protein cellular attributes using pseudo amino acid composition. Proteins Struct Funct Genet. 2001;43:246–255. doi: 10.1002/prot.1035. [DOI] [PubMed] [Google Scholar]
- Gong JS, Lu ZM, Li H, Shi JS, Zhou ZM, Xu ZH. Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Fact. 2012;11:142. doi: 10.1186/1475-2859-11-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JS, Lu ZM, Li H, Zhou ZM, Shi JS, Xu ZH. Metagenomic technology and genome mining: emerging areas for exploring novel nitrilases. Appl Microbiol Biot. 2013;97:6603–6611. doi: 10.1007/s00253-013-4932-8. [DOI] [PubMed] [Google Scholar]
- Kaplan O, Bezouska K, Malandra A, Vesela AB, Petrıckova A, Felsberg J, Rinagelova A, Kren V, Martinkova L. Genome mining for the discovery of new nitrilases in filamentous fungi. Biotechnol Lett. 2011;33:309–312. doi: 10.1007/s10529-010-0421-7. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Mohan U, Banerjee UC. Exploring residues crucial for nitrilase function by site directed mutagenesis to gain better insight into sequence-function relationships. Int J Biochem Biotechnol. 2012;3:384–391. [PMC free article] [PubMed] [Google Scholar]
- Kim M, Lee KH, Yoon SW, Kim BS, Chun J, Yi H. Analytical tools and databases for metagenomics in the next-generation sequencing era. Genom Inform. 2013;11:102–113. doi: 10.5808/GI.2013.11.3.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Bhalla TC. In silico analysis of amino acid sequences in relation to specificity and physiochemical properties of some aliphatic amidases and kynurenine formamidases. J Bioinform Seq Anal. 2011;3:116–123. [Google Scholar]
- Liu H, Gao Y, Zhang M, Qiu X, Cooper AJ, Niu L, Teng M. Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2. Acta Crystallogr D Biol Crystallogr. 2013;69:1470–1481. doi: 10.1107/S0907444913009347. [DOI] [PubMed] [Google Scholar]
- Martinkova L, Kren V. Biotransformations with nitrilases. Curr Opin Chem Biol. 2010;14:130–137. doi: 10.1016/j.cbpa.2009.11.018. [DOI] [PubMed] [Google Scholar]
- Mills CL, Beuning PJ, Ondrechen MJ. Biochemical functional predictions for protein structures of unknown or uncertain function. Comput Struct Biotechnol J. 2015;13:182–191. doi: 10.1016/j.csbj.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylerova V, Martinkova L. Synthetic applications of nitrile converting enzymes. Curr Org Chem. 2003;7:1–17. [Google Scholar]
- Pant B, Pant K, Pardasani KR. Multiclass SVM model for prediction and classification of ribonucleases. Int J Integr Biol. 2011;12:44–49. [Google Scholar]
- Rishishwar L, Mishra N, Pant B, Pant K, Pardasani KR. ProCoS—PROtein COmposition Server. Bioinformation. 2010;5:227. doi: 10.6026/97320630005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottig M, Rausch C, Kohlbacher O. Combining structure and sequence information allows automated prediction of substrate specificities within enzyme families. PLoS Comput Biol. 2010 doi: 10.1371/journal.pcbi.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NN, Sharma M, Kumar H, Bhalla TC. Nocardia globerula NHB-2: bench scale production of nicotinic acid. Process Biochem. 2006;41:2078–2081. doi: 10.1016/j.procbio.2006.04.007. [DOI] [Google Scholar]
- Sharma N, Kushwaha R, Sodhi JS, Bhalla TC. In silico analysis of amino acid sequences in relation to specificity and physiochemical properties of some microbial nitrilases. J Proteom Bioinform. 2009;2:185–192. doi: 10.4172/jpb.1000076. [DOI] [Google Scholar]
- Sharma NN, Sharma M, Bhalla TC. Nocardia globerula NHB-2 nitrilase catalysed biotransformation of 4-cyanopyridine to isonicotinic acid. AMB Express. 2012;2:25. doi: 10.1186/2191-0855-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Thakur N, Raj T, Savitri, Bhalla TC. Mining of microbial genomes for the novel sources of nitrilases. Biomed Res Int. 2017;14:2017. doi: 10.1155/2017/7039245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HB, Chou KC. PseAAC: a flexible web-server for generating various kinds of protein pseudo amino acid composition. Anal Biochem. 2008;373:386–388. doi: 10.1016/j.ab.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol. 2003;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jing R, Hua Y, Fu Y, Dai X, Huang L, Menglong L. Classification of multi-family enzymes by multi-label machine learning and sequence-based descriptors. Anal Methods. 2014;17:6832–6840. doi: 10.1039/C4AY01240B. [DOI] [Google Scholar]
- Yeom SJ, Kim HJ, Lee JK, Kim DE, Oh DK. An amino acid at position 142 in nitrilase from Rhodococcus rhodochrous ATCC 33278 determines the substrate specificity for aliphatic and aromatic nitriles. Biochem J. 2008;415:401–407. doi: 10.1042/BJ20080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yin B, Wang C, Jiang S, Wang H, Wei YD. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803. J Struct Biol. 2014;188:93–101. doi: 10.1016/j.jsb.2014.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.