Abstract
Pre-miRNA-499 gene is associated with autoimmune disease. Mir-449 rs3746444 polymorphism is inconsistent for rheumatoid arthritis (RA). This study aimed to investigate association of mir-499 rs3746444 polymorphism with RA activity and severity in Egyptian population. The study population was conducted as case control study in 100 RA patients diagnosed according to the American College of Rheumatology classification criteria for RA, and the control group included 100 healthy subjects who were age-and sex-matched to the RA group. Different genotypes were assessed using polymerase chain reaction–restriction fragment length polymorphism. 95% Confidence interval and odds ratio were defined to assess the strength of association. Regarding patients, thirty-three patients carried TT genotype, fifty-three patients carried TC genotype and fourteen patients carried CC genotype. So the frequency of the minor C allele in RA patients was significantly higher than the control subjects (P = 0.037). TC, CC genotypes and C allele frequencies were significantly associated with disease severity as they had high rheumatoid factor (55.78 µIU/ml) and anti-cyclic citrullinated peptide (Anti-CCP) antibody (297.32 µIU/ml). Moreover, the heterozygote TC had more severe and more active form of the disease compared with homozygote CC or TT as they had high Anti-CCP antibody, and disease activity score 28 (score 5). Our work suggests that C allele of Pre-miRNA rs3746444 polymorphism contributes to heritability of susceptibility to RA compared to T allele. This polymorphism was associated with the activity and severity of the disease.
Keywords: MicroRNA-499, Polymerase chain reaction–restriction fragment length polymorphism, rs3746444 polymorphism, Rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is an autoimmune disorder characterized by chronic inflammation of the joint [1]. Inflammatory cytokines, including IL-1b and tumor necrosis factor (TNF)-a, play a critical role in the pathogenesis of RA. Treatment with blockage of these cytokines has clinical efficacy in RA patients [2]. The etiology of RA is unknown, but genetic factors are thought to be important in the pathogenesis and progress of RA [3]. In many studies [4] the genetic differences, which may result in functional and biological variations, in many genes/loci have been demonstrated to be involved with the development of RA.
RNA interference is mediated by short noncoding RNAs called miRNA, which arise from far longer, genomically encoded nuclear transcript and negatively regulate gene expression at the post-transcriptional level [5]. Bioinformatic data states that a single miRNA could bind to hundreds targets of mRNA these targets could be associated with the regulation of almost every biological process [6]. Compelling evidence indicates that miRNAs play a key role in such fundamental process as early development, cell proliferation, differentiation, apoptosis, cell fate determination, stress resistance, signal transduction and organ development [7]. In some studies [8] it has been found that miRNA modulate T cell selection and receptor sensitivity and also Treg cell development [9], which suggests that miRNA may be associated with the development of autoimmune disease. Some targets of mir-499 were IL-17RB, IL-23a, IL-2Rβ, IL-6, IL-2, and IL-18R. IL-6 can trigger the synthesis of C-reactive protein (CRP) and fibrinogen through the liver [10] and IL-17RB, IL-23a, IL-2Rβ, IL-6, IL-2, and IL-18R play critical roles in the pathogenesis of RA [11, 12]. Therefore, mir-499 can affect the production of CRP and inflammation in RA. Another target of miR-499 was Peptidyl arginine deiminase type IV (PADI4) gene, which encodes PADI4 enzyme. PADI4 enzyme produces autoantigene citrullinated peptides recognized by anti-cyclic citrullinated peptide (anti-CCP) [13] through posttranslational citrullination by deimination of arginine residues to citrullines in the presence of calcium ions [14]. Therefore miR-499 can regulate the production of anti-CCP antibody by regulating PADI4 gene expression. Therefore, mir-499 may play an important role in RA.
Small variation in miRNAs may have an influence on thousands of target mRNAs and result in various functional consequences. The SNPs can affect the functions of miRNA which can underlie differences in our susceptibility to disease [15]. Since SNPs located in mature miRNA regions may directly affect both the binding to target mRNAs and the maturation process of the pre-miRNA, whereas SNPs located in other regions of pre-miRNA may affect the maturation of pre-miRNA [16]. In this study we hypothesized that pre-miRNA rs3746444 SNP may be involved in the occurrence and development of RA. To prove this hypothesis, we genotyped selected SNP (rs3746444 T/C). The aim of this study was to investigate the influence of pre-miRNA rs3746444 polymorphism on the susceptibility to RA in Egyptian population.
Materials and Methods
Study Population
A case control study was conducted on 200 subjects. They were divided into 100 RA patients and 100 healthy subjects who were age and sex matched to the RA group. Patients were selected from the Outpatient Clinic of the Suez Canal university Hospital. Diagnosis of RA was done according to the ACR criteria of classification of RA [17]. All RA patients were assessed by disease activity score 28 (DAS28).
Classification criteria for RA (score-based algorithm: score of categories A–D.(
A score of ≥6/10 is needed for classification of a patient as having definite RA and patients with a score of <6/10 are not classifiable as having RA.
Joint Involvement (Swollen or Tender Joint)
If patients have 1 large joint (shoulders, elbows, hips, knees, and ankles) is affected, will take 0 score, with 2–10 large joints are affected will take 1 score, with 1–3 small joints (metacarpophalangeal joints, proximal interphalangeal joints, second through fifth metatarsophalangeal joints, thumb interphalangeal joints, and wrists) are affected with or without involvement of large joints will take 2 score and with 4–10 small joints (with or without involvement of large joints) will take 3 score.
Serology
If patients have negative RF and negative Anti-CCP antibody they will take 0 score. Patients have Low-positive RF or low-positive Anti-CCP antibody will take 2 score. High-positive RF or high-positive Anti-CCP antibody give patients 3 score.
Acute-Phase Reactants
Patients have Normal CRP and normal ESR will take 0 score while patient have abnormal CRP or abnormal ESR so they will take 1 score.
Duration of Symptoms
Patients have symptoms for <6 weeks so they will take 0 score while patients have symptoms for ≥6 weeks, they will take 1 score.
The study groups excluded patients with hepatitis C, diabetes mellitus, seronegative spondyloarthropathy (e.g. ankylosing spondylitis, reactive arthritis, psoriatic arthritis and inflammatory bowel disease), thyroid disease, Behcet’s disease, sarcoidosis, systemic lupus erythematous (SLE) and dyslipidemia.
The present study was conducted according to the principles of the Declaration of Helsinki, and all the participants provided written informed consent, following a protocol approved by the Suez Canal University Research Ethics Committee. Anti-CCP antibody, rheumatoid factor (RF), CRP and erythrocyte sedimentation rate (ESR) were measured for patient subjects.
Laboratory Measurements
Peripheral blood (6 ml) was drawn of which a portion was collected on EDTA and used for DNA extraction. Serum was separated from the remaining part of the blood samples and used for assessment of the following parameters:
Anti-CCP antibody was measured by enzyme linked immune sorbent assay (ORG 601 Anti-CCP high sensitive).
RF and CRP were measured using automated chemistry method (Cobas 6000) while ESR was measured using Westergren Sedimentation Rate.
Genomic DNA Extraction and Genotyping
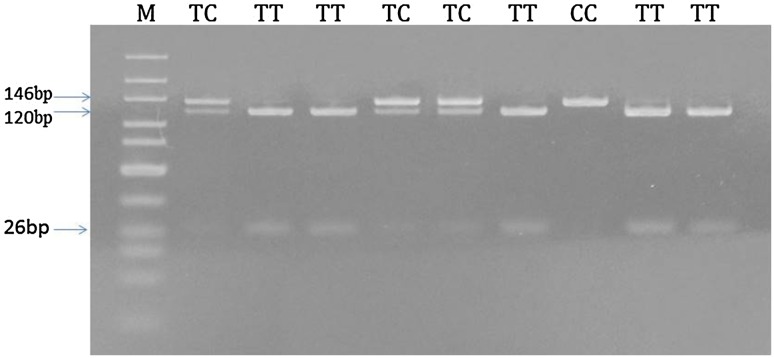
Genomic DNA was extracted from 300 µl whole blood collected in EDTA anticoagulated tubes using the Wizard genomic DNA purification kit (Promega, Madison, USA) according to the manufacturer instructions. The pre-miRNA-499 rs3746444 polymorphism was performed by polymerase chain reaction-based restriction fragment length polymorphism (PCR-RFLP). Primers that were used in the assay of 146 base pairs (bp) are: forward primer 5′-CAAAGTCTTCACTTCCCTGCCA-3′ and a reverse primer 5′-GATGTTTAACTCCTCTCCACGTGATC-3′ [18]. The PCR reaction was conducted in a total volume of 25 μl containing 1 μl genomic DNA (~100 ng/μl), l μl of each primer (10 pmol/μl), 12.5 μl Go Taq® Green Master Mix (2×) (Promega, Madison, USA) and 9.5 μl DNase-free water. DNA was denatured at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 45 s and extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. Thermal cycling was conducted in Eppendorf Mastercycler® machine (Eppendorf AG, Hamburg, Germany). The PCR products of pre-miRNA-499 gene fragment were digested with 5 units of BcII restriction enzymes (Promega, Madison, USA) at 37 °C overnight, which only cut the T allele producing fragments of 120 and 26 bp while C allele appeared as 146 bp. The cleavage products were electrophoresed on 12% polyacrylamide gel (Fig. 1).
Fig. 1.
Detection of pre-miRNA-499 (rs3746444) genotypes by polyacrylamide gel electrophoresis. M: 10 bp marker. The CC had one band at 146, TT genotype yielded two bands at 120 and 26 bp, while the TC produced three bands at 146, 120, and 26 bp
Statistical Analysis
Statistical comparison of clinical and demographic data between patient and control was done by student t test. The genotype and allele frequencies of polymorphism in control and RA patients were analyzed using Chi square test. Hardy–Weinberg’s equilibrium was evaluated using Chi square test. One way ANOVA test was used to analyze the associations of genotypes with diagnostic tests in RA patients followed by Tukey’s test for multiple comparisons. Statistical analysis was performed using SPSS ver. 17 program (Chicago, Illinois, United States). A P value of <0.05 was considered to be statistically significant. All data were presented as mean ± standard deviation (SD).
Results
The clinical characteristics and demographic data of RA patients and the control group are summarized in Table 1. Both groups are age and sex matched. RA patient have a significantly higher family history than the control group.
Table 1.
Clinical characteristics and demographic data of the study population
| Variables | Control (100) | Cases (100) |
|---|---|---|
| Age (mean ± SD) | 39.98 ± 12.13 | 41.7 ± 8.5 |
| Sex (female/male) | 85/15 | 86/14 |
| Family history of RA | 3 | 48* |
| Duration of the disease (recent\old) | 29/71 | |
* P < 0.05 significantly different from normal control
A Rheumatoid arthritis patient had high RF values(mean ± SD = 42.71 ± 16.21) in which three patients of them show negative RF and 97 show positive RF. Eighty of positive RF subgroup show high RF+ and 17 patient show low RF+ . Also RA patients show high Anti-CCP value (mean ± SD = 237.28 ± 92.28). No patient shows a negative Anti-CCP antibody and all of them show high Anti-CCP+ values. Also they had high ESR (mean ± SD = 29.31 ± 10.14), CRP (mean ± SD = 27.71 ± 10.26) and DAS28 (mean ± SD = 4.496 ± 1.163).
Allele Frequencies and Genotype Distribution of Pre-miRNA-499 rs3746444 Polymorphisms in Control and RA Patients
The genotype frequencies of both polymorphisms were in accordance with Hardy–Weinberg equilibrium.
As shown in Table 2; the minor C allele of pre-miRNA-499 rs3746444 polymorphism was more frequent in the RA group than in healthy control group, whereas the major T allele was more frequent in healthy control group than in RA group. These differences were significant (P < 0.037). Additionally TC genotype is significantly associated with the incidence of RA in the study population compared to the TT genotype (P < 0.032).
Table 2.
Allele frequencies and genotype distribution of pre-miRNA-499 (rs3746444) polymorphism in the control group and rheumatoid arthritis patients
| Control n = 100 |
Patients n = 100 |
P | OR (95% CI) | |
|---|---|---|---|---|
| T allele | 139 | 119 | 0.037*a | 0.645 (0.427–0.974) |
| C allele | 61 | 81 | ||
| Genotype | ||||
| TT | 49 | 33 | ||
| TC | 41 | 53 | 0.032*b | 0.521 (0.286–0.950) |
| CC | 10 | 14 | 0.117c | 0.481 (0.191–1.211) |
Comparisons were performed with the Chi square test
CI confidence interval, OR odds ratio
* Significant difference at P < 0.05
* Significant difference at P < 0.05
aC versus T
bTC versus TT
cCC versus TT
We also investigated the relation of different genotypes of pre-miRNA-499 rs3746444 polymorphism with diagnostic tests of the study population.
Table 3 shows that, CC genotype of mir-499 (rs3746444) polymorphism had higher RF than TC and TT genotype carriers (P = 0.001, 0.015) and higher Anti-CCP value than TT genotype (P = 0.001). While TC genotype showed higher Anti-CCP value than CC and TT genotype (P = 0.001). Also TC had a higher value of CRP than CC genotype (P = 0.01). Regarding DAS-28 there was a significant difference between different genotypes in which TC genotype showed higher value than CC and TT genotypes (P = 0.001, 0.015). There was no significant difference in ESR level between different genotypes.
Table 3.
The relationship between the values of diagnostic tests of the study population with different genotypes of pre-miRNA-499 (rs3746444) polymorphism
| Carrier of CC n = 14 |
Carrier of TC N = 53 |
Carrier of TT n = 33 |
|
|---|---|---|---|
| RFa | 55.78 ± 13.32 | 42.58 ± 17.94* | 37.36 ± 10.68* |
| Anti-CCP antibodyb | 211.06 ± 62.24 | 279.32 ± 91.64* | 139.92 ± 50.61*,** |
| ESRc | 29.35 ± 10.44 | 29.31 ± 10.14 | 30.51 ± 9.81 |
| CRPd | 20.85 ± 5.2 | 29.64 ± 11.7* | 25.78 ± 7.78 |
| DAS28e | 3.42 ± 1.08 | 4.86 ± 1.13* | 4.209 ± 0.87** |
Data are presented as mean ± SD. Comparisons were performed with one way ANOVA test followed by the Tukey’s test for multiple comparison
* Significant difference from CC genotype carriers at P < 0.05
** Significant difference from TC genotype carriers at P < 0.05
aRheumatoid factor
bAnti-cyclic citrullinated
cPeptide indicates erythrocyte sedimentation rate
dC-reactive protein
eDisease activity score
Discussion
RA is a chronic inflammatory disease that may involve extra-articular organs in addition to involving joints. Genetic and environmental factors are implicated in the pathogenesis of RA. RA is considered a clinically heterogeneous condition with a wide spectrum of clinical manifestation, great variability in severity and disease progression, and different responses to a range of therapies [19]. MiRNAs are small non-coding RNAs of 18–23 nucleotides, and their function in cellular physiology, development, and disease is to negatively regulate the expression of protein-coding genes [20]. It has been proposed that binding of specific miRNA to the 3′UTR of its target mRNA inhibits gene expression.
Various studies have showed that RA was significantly associated with the HLA-DRB1 alleles encoding the Shared epitope (SE) and signal transducer and activator of transcription 4 (STAT4). Some studies has been confirmed that Asian RA sufferers have SE conserved across the HLADRB1 and STAT4 [21, 22]. One target of mir-449 is regulatory factor X 4(RFX4) which can affect the expression of HLADRB1, so mir-449 can control HLADRB1 by regulating REX4. The PADI4 gene was another target of miR-499 which encodes PADI4 enzyme. PADI4 mRNA was found in synovial tissue of RA patients and hematological cells, and was remarkably overexpressed in the blood of RA patients [23]. Moreover PADI4 enzyme produces autoantigenes citrullinated peptides recognized by anti-CCP [13] by citrullination through deimination of arginine to citrulline in the presence of calcium ions [14]. Therefore miR-499 can control the production of anti-CCP antibody by regulating PADI4 gene expression. Therefore, mir-499 may play an important role in the pathogenesis and the severity of RA.
Pre-micro RNA-499 rs3746444 T/C polymorphism is found to be associated with other diseases as hepatocellular carcinoma [24], myocardial infarction [25] and pediatric acute lymphoblastic leukemia [26].
In the present study, we analyzed the correlation between genetic polymorphisms in pre-miRNA-499 rs3746444 polymorphism and susceptibility to RA in a sample of the Suez Canal area in the Egyptian population.
In this study, RA male and female patients showed association with the minor allele of mir-499 (rs3746444) polymorphism. These results are in agreement with Hashemi et al. study [27].
We also studied the relation between mir-449 rs3746444 and the level of anti-CCP antibody, RF, CRP and ESR, which are usually used to diagnose and evaluate the RA. There was a significant difference between different genotypes carriers of SNPs rs3746444 polymorphism in RF, Anti-CCP, CRP and DAS28. The genotype CC in rs3746444 greatly increased the level of RF compared with the genotype TC and TT. Also CC genotype has higher level of Anti-CCP than TT genotype. This contrasts Yang et al. study [28] however our study is with agreement with this study concerning that TC genotype carriers greatly increased the level of anti-CCP antibody compared with other genotypes. So TC, CC genotypes and C allele frequencies in Pre-miRNA-499 rs3746444 polymorphism were significantly associated with disease severity. These increased level of Anti-CCP antibody in TC genotype carriers suggest that SNPs rs3746444 polymorphism may be a candidate biomarker for predicting joint damage in RA patients and influence the severity of the disease. Also TC genotype carriers showed higher level of CRP than CC as in Yang et al. study [7]. DAS 28 was significantly higher in TC than CC and TT genotypes. Moreover, the heterozygote TC had more severe and more active form of the disease compared with homozygote CC or TT as they had high Anti-CCP antibody and DAS 28. Over all, the heterozygote in rs3746444 in RA exhibited higher levels of CRP, Anti-CCP antibody and DAS 28 than the homozygote; therefore, the heterozygote needed more effective treatment than the homozygote [7]. There is no significant difference in ESR level between different genotype.
In conclusion, our results confirm a genetic association of SNPs rs3746444 polymorphism with RA in Egyptian population. This study demonstrated that genotype TC in rs3746444 greatly increased the levels of CRP, DAS28 and Anti-CCP. All results suggest that SNP rs3746444 may play an important role in RA activity and severity. Researchers are just beginning to understand the relationship between miRNA expression patterns and functions in different diseases. Therefore, further characterization of the SNPs of miRNAs would improve our understanding of miRNA biogenesis and the potential contribution of these SNPs to disease etiology and progression.
Acknowledgements
We gratefully acknowledge Afaf Zakaria and Maha Abdel Fattah (assistant lecturers at rheumatology department faculty of medicine Suez Canal University) for their help in RA patient’s diagnosis.
Abbreviations
- Mir
micro RNA
- SNPs
Single nucleotide polymorphisms
- PCR–RFLP
Polymerase chain reaction restriction fragment length polymorphism
- RF
Rheumatoid factor
- Anti-CCP
Anti-cyclic citrullinated peptide
Author contributions
All authors contributed equally.
Compliance with Ethical Standards
Conflict of interest
None of the authors have any conflicts of interest.
References
- 1.Smolen JS, Aletaha D, Koeller M, Weisman MH, Emery P. New therapies for treatment of rheumatoid arthritis. Lancet. 2007;370:1861–1874. doi: 10.1016/S0140-6736(07)60784-3. [DOI] [PubMed] [Google Scholar]
- 2.Lipsky PE, van der Heijde DM, St Clair EW, Furst DE, Breedveld FC, Kalden JR, et al. Infliximab and methotrexate in the treatment of rheumatoid arthritis. N Engl J Med. 2000;343:1594–1602. doi: 10.1056/NEJM200011303432202. [DOI] [PubMed] [Google Scholar]
- 3.Perricone C, Ceccarelli F, Valesini G. An overview on the genetic of rheumatoid arthritis: a never-ending story. Autoimmun Rev. 2011;10(10):599–608. doi: 10.1016/j.autrev.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 4.Thabet MM, Wesoly J, Slagboom PE, Toes RE, Huizinga TW. FCRL3 promoter 169 CC homozygosity is associated with susceptibility to rheumatoid arthritis in Dutch Caucasians. Ann Rheum Dis. 2007;66:803–806. doi: 10.1136/ard.2006.064949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 6.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 7.Yang B, Chen J, Li Y, Zhang J, Li D, Huang Z, et al. Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Hum Immunol. 2012;73:101–106. doi: 10.1016/j.humimm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Investig. 2003;111:1805–1812. doi: 10.1172/JCI200318921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying B, Shi Y, Pan X, Song X, Huang Z, Niu Q, et al. Association of polymorphisms in the human IL-10 and IL-18 genes with rheumatoid arthritis. Mol Biol Rep. 2011;38:379–385. doi: 10.1007/s11033-010-0119-x. [DOI] [PubMed] [Google Scholar]
- 12.Yuan FL, Hu W, Lu WG, Li X, Li JP, Xu RS, et al. Targeting interleukin-21 in rheumatoid arthritis. Mol Biol Rep. 2011;38:1717–1721. doi: 10.1007/s11033-010-0285-x. [DOI] [PubMed] [Google Scholar]
- 13.Schellekens GA, Visser H, de Jong BA, van den Hoogen FH, Hazes JM, Breedveld FC, et al. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum. 2000;43:155–163. doi: 10.1002/1529-0131(200001)43:1<155::AID-ANR20>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Chavanas S, Méchin MC, Takahara H, Kawada A, Nachat R, Serre G, et al. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene. 2004;330:19–27. doi: 10.1016/j.gene.2003.12.038. [DOI] [PubMed] [Google Scholar]
- 15.Loktionov A. Common gene polymorphisms, cancer progression and prognosis. Cancer Lett. 2004;208(1):1–33. doi: 10.1016/j.canlet.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 17.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, 3rd, et al. Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010;69:1580–1588. doi: 10.1136/ard.2010.138461. [DOI] [PubMed] [Google Scholar]
- 18.Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Investig. 2008;118(7):2600–2608. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orozco G, Rueda B, Martin J. Genetic basis of rheumatoid arthritis. Biomed Pharmacother. 2006;60:656–662. doi: 10.1016/j.biopha.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 21.Van der Helm-van Mil AHM, Huizinga TWJ. Advances in the genetics of rheumatoid arthritis point to subclassification into distinct disease subsets. Arthritis Res Ther. 2008;10:205. doi: 10.1186/ar2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji JD, Lee WJ, Kong KA, Woo JH, Choi SJ, Lee YH, et al. Association of STAT4 polymorphism with rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep. 2010;37(1):141–147. doi: 10.1007/s11033-009-9553-z. [DOI] [PubMed] [Google Scholar]
- 23.Harney SM, Meisel C, Sims AM, Woon PY, Wordsworth BP, Brown MA. Genetic and genomic studies of PADI4 in rheumatoid arthritis. Rheumatology. 2005;44:869–872. doi: 10.1093/rheumatology/keh614. [DOI] [PubMed] [Google Scholar]
- 24.Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, Yang SF, et al. MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS ONE. 2014;9(2):e89930. doi: 10.1371/journal.pone.0089930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Hong H, Chen L, Shi X, Chen Y, Weng Q. Association of microRNA polymorphisms with the risk of myocardial infarction in a Chinese population. Tohoku J Exp Med. 2014;233(2):89–94. doi: 10.1620/tjem.233.89. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez-Camino A, Lopez-Lopez E, Martin-Guerrero I, Piñan MA, Garcia-Miguel P, Sanchez-Toledo J, et al. Noncoding RNA-related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr Res. 2014;75(6):767–773. doi: 10.1038/pr.2014.43. [DOI] [PubMed] [Google Scholar]
- 27.Hashemi M, Eskandari-Nasab E, Zakeri Z, Atabaki M, Bahari G, Jahantigh M, et al. Association of pre-miRNA-146a rs2910164 and pre-miRNA-449 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol Med Rep. 2013;7(1):287–291. doi: 10.3892/mmr.2012.1176. [DOI] [PubMed] [Google Scholar]
- 28.Yang B, Zhang JL, Shi YY, Li DD, Chen J, Huang ZC, et al. Association study of single nucleotide polymorphisms in pre-miRNA and rheumatoid arthritis in a Han Chinese population. Mol Biol Rep. 2011;38:4913–4919. doi: 10.1007/s11033-010-0633-x. [DOI] [PubMed] [Google Scholar]