Abstract
Methylenetetrahydrofolate reductase (MTHFR) is essential for DNA biosynthesis and the epigentic process of DNA methylation. It has been reported that abnormal DNA methylation contributes to the pathogenesis of congenital anomalies. There were many published case control studies assessing the associations of MTHFR C677T polymorphism with risks of nosyndromic cleft lip with and without palate (nsCL/P), but with inconsistent results. To derive a more precise estimation of the relationship, a meta-analysis was performed. Eligible articles were identified by search of databases including PubMed, Science Direct, Google Scholar and Springer Link up to December, 2015. Finally, a total of 22 studies with 3724 nsCL/P cases and 5275 controls were included in the present meta-analysis. Odds ratios (ORs) with corresponding 95% confidence intervals (95% CIs) were pooled to assess the association. Subgroup analysis based on ethnicity was also performed. All statistical analyses were done by MIX program. Meta-analysis results suggested that MTHFR C677T polymorphism contributed to the increased nsCL/P risk in overall population using four genetic models except homozygote model (for T vs. C: OR = 1.24, 95% CI = 1.1–1.4; for TT + CT vs. CC: OR = 1.29, 95% CI = 1.04–1.59; for CT vs. CC: OR = 1.26, 95% CI = 0.98–1.63; for TT vs. CC: OR = 1.02, 95% CI = 0.74–1.4; for TT vs. CT + CC: OR = 1.36, 95% CI = 1.05–1.74). In conclusion, results of present meta-analysis suggested that MTHFR C677T polymorphism is significantly associated with nonsyndromic orofacial cleft.
Keywords: nsCL/P, MTHFR, C677T, Folate, Meta-analysis, Polymorphism
Introduction
Nonsyndromic cleft lip with or without cleft palate (nsCL/P) is a common congenital defect with the prevalence rate of 1 in 300–2000 birth depending upon ethnicity, and socioeconomic status [1–3]. Twin and family studies suggested that genetic factors play an important role in the etiology of nsCL/P [4]. The risk of recurrence in first-degree relatives of affected persons is about 40-folds greater than in the general population, which also suggests a strong genetic component [5–7]. Its frequency is highest in Asian and Native American populations of Asian genetic origin, intermediate in Caucasian population and the lowest in African and African-American populations [8].
Perinatal intake of folic acid and multivitamins is suggested to provide protection from neural tube defects (NTD) and nsCL/P birth defects [9–17]. Several studies reported that perinatal supplementation of folic acid reduces the risk of neural tube defects (NTD) [18, 19] led to speculation that folic acid supplementation might also reduces the risk of craniofacial closure defects such as nsCL/P [9–11, 20].
A number of observational studies [10, 11, 19, 21–23] although not all have reported that folate deficiency during conception period results in nsCL/P in offspring. Some studies have found a decline in cleft rates since food fortification with folic acid began [24], although others have not found a change [25]. These observations have led investigators to look for associations between facial clefts and folate enzyme genes, including methylenetetrahydrofolate reductase (MTHFR) [26–35].
MTHFR enzyme catalyzes the reduction of 5, 10-methylenetetrahydrofolate to 5-methyltetrahydrofolate (5-THF), which donates methyl group for the conversion of homocysteine to methionine. Several polymorphisms were reported in MTHFR gene, out of which C677T variant is most clinically important polymorphism. The C677T variant has been associated with decreased activity of MTHFR, and increased homcysteine plasma level [36]. A hypofunctional MTHFR leads to lower S-adenosyl-l-methionine levels and consequently to hypomethylation. MTHFR C677T polymorphism is reported as risk factor for several diseases and disorders like- neural tube defects [37], Down syndrome [36, 38], congenital heart defects [39], and cardiovascular diseases etc. Globally, the prevalence of 677T allele ranged from 24.1 to 64.3% among Europeans, 2–48% among North Americans, 7% among South Americans, 0–35.5% among Africans and 2–63.1% in Asians [40–44].
Since the first report in 1998 that the MTHFR C677T variant genotype was found more commonly among nsCL/P cases than controls [45], several case–control and epidemiological studies have been published to determine the exact role of MTHFR gene in the etiology of oral clefts. However, the results remain conflicting rather than conclusive. So, to shed some light on this association present meta-analysis was carried out of all available studies relating the C677T polymorphism of the MTHFR gene to the risk of nsCL/P.
Methods
Identification of Studies
Literature search for eligible studies was conducted on PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Google Scholar (http://scholar.google.com), Science Direct (http://www.sciencedirect.com), and Springer Link (http://link.springer.com) databases up to December 31, 2015, using keywords “methylenetetrahydrofolate reductase”, “MTHFR”, “C677T” and “cleft lip and palate”.
Data Extraction
The following information were collected from studies: first author family name, year of publication, name of journal, country name, ethnicity, number of cases and controls, numbers of different MTHFR genotypes in nsCL/P cases and controls.
Inclusion Criteria
The study inclusion criteria were that (1) the study should be case–control study, (2) study should be published as full papers, and (3) complete information of different MTHFR genotype number should be reported in the study. The study exclusion criterion were (1) only cases were studied, (2) review papers, editorial, letter to editor and (3) containing overlapping data and (4) no enough information to estimate OR with 95% CI.
Statistical Analysis
Odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were used as the measure of effect to evaluate the strength of association between the MTHFR C677T polymorphism and nsCL/P risk. The meta-analysis examined the association between C677T polymorphism and nsCL/P using all five genetic models like- allele contrast/additive (T vs. C), homozygote (TT vs. CC), recessive (TT vs. CT + CC), dominant (TT + CT vs. CC) and co-dominant (CT vs. CC) models. The pooled OR was performed by using both fixed effects (FE) (Mantel–Haenszel) and random effects (DerSimonian and Laird) models [46, 47]. The heterogeneity between studies was tested by the Q-statistic [48, 49] and was quantified with the I2 metric, which is independent of the number of studies in the meta-analysis [50, 51]. I2 takes values between 0 and 100% with higher values denoting greater degree of heterogeneity [52]. When there is higher heterogeneity between studies, then the pooled OR is preferably estimated using the random effects model. Further subgroup analysis according to ethnicity and sensitivity analysis were performed to explore potential heterogeneity. To assess the study quality, control population of individual study was tested for Hardy–Weinberg equilibrium (HWE) using an online program available at https://ihg.gsf.de/cgi-bin/hw/hwa1.pl.
Publication Bias
An estimate of publication bias was carried out by the Begg’s funnel plot, in which the standard error of log (OR) and precision of each study was plotted against its log (OR) [53]. The funnel plot asymmetry was further assessed by the method of Egger’s linear regression test [54]. All analyses were performed by MIX version 1.7 [55]. A p value less than 0.05 was considered statistically significant.
Results
Present meta-analysis was carried out according to the Moose guidelines. The article search concerning the association of MTHFR C677T polymorphism with nsCL/P resulted in 76 studies in total. After examination of abstracts, 24 studies were excluded, which were review, editorials and comments etc. Full text of remaining 52 articles were examined and again 30 article were excluded. Out of 30 articles, sixteen articles were irrelevant for present meta-analysis and two were published meta-analyses on the same topic, 2 articles were not case control study and in 7 studies maternal MTHFR C677T polymorphism was reported. Flow diagram of study selection was given in Fig. 1.
Fig. 1.
Flow chart of the selection of studies
Selected Studies
Twenty-two case-control eligible studies on MTHFR C677T polymorphism and nsCL/P were identified through literature search and selection based on the inclusion and exclusion criteria and included in the present meta-analysis [17, 26, 27, 29, 30, 33, 34, 45, 56–69]. All these twenty-two studies were performed in different countries—Argentina [45], Brazil [27, 56, 60], China [64], France [17], Germany [58], India [62, 68], Ireland [61], Italy [29, 59], Mexico [69], Netherlands [34], Thailand [33], Turkey [66, 67], Ukraine [65], and USA [26, 30, 57, 63]. Details of included studies are given in Table 1.
Table 1.
Characteristics of twenty-two studies included in the present meta-analysis
| Study | Ethnicity | Country | Case/control | Case genotype | Control genotypes | P value of HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | |||||
| Shaw et al. (1998) | Caucasian | USA | 310/383 | 143 | 127 | 40 | 156 | 178 | 49 | 0.87 |
| Tolarova et al. (1998) | Caucasian | Argentina | 111/106 | 43 | 49 | 19 | 46 | 52 | 8 | 0.19 |
| Gasper et al. (1999) | Caucasian | Brazil | 77/103 | 30 | 39 | 8 | 49 | 49 | 5 | 0.09 |
| Wyszynski et al. (2000) | Caucasian | USA | 259/327 | 114 | 109 | 36 | 129 | 154 | 44 | 0.85 |
| Martinelli et al. (2001) | Caucasian | Italy | 116/106 | 64 | 22 | 30 | 46 | 43 | 17 | 0.20 |
| Blanton et al. (2002) | Caucasian | USA | 75/50 | – | – | – | – | – | – | – |
| Grunert et al. (2002) | Caucasian | Germany | 66/184 | 34 | 26 | 6 | 90 | 69 | 25 | 0.05 |
| Shoteresuk et al. (2003) | Asian | Thailand | 109/202 | 84 | 25 | 0 | 154 | 46 | 2 | 0.47 |
| van Roij et al. (2003) | Caucasian | Netherlands | 105/128 | 54 | 45 | 6 | 70 | 54 | 4 | 0.09 |
| Gasper et al. (2004) | Caucasian | Brazil | 644/269 | 327 | 269 | 48 | 213 | 17 | 39 | 0.00 |
| Pezzetti et al. (2004) | Caucasian | Italy | 110/289 | 28 | 58 | 24 | 95 | 151 | 43 | 0.17 |
| Brandalize et al. (2007) | Caucasian | Brazil | 114/100 | 49 | 46 | 19 | 45 | 41 | 14 | 0.35 |
| Chevrier et al. (2007) | Caucasian | France | 168/148 | 54 | 81 | 33 | 66 | 60 | 22 | 0.17 |
| Mills et al. (2008) | Caucasian | Ireland | 492/1599 | 217 | 221 | 54 | 715 | 721 | 163 | 0.34 |
| Ali et al. (2009) | Asian | India | 323/214 | – | – | – | – | – | – | – |
| Guo et al. (2009) | Asian | China | 96/103 | 19 | 53 | 24 | 22 | 57 | 24 | 0.27 |
| Sozen et al. (2009) | Caucasian | USA | 179/138 | 81 | 80 | 18 | 66 | 65 | 7 | 0.07 |
| Chorna et al. (2011) | Caucasian | Ukraine | 33/50 | 12 | 17 | 4 | 22 | 26 | 2 | 0.09 |
| Semic-Jusufagic et al. (2012) | Asian | Turkey | 56/76 | 25 | 28 | 3 | 44 | 24 | 8 | 0.10 |
| Kumari et al. (2013) | Asian | India | 467/469 | 327 | 125 | 15 | 364 | 100 | 5 | 0.52 |
| Aslar et al. (2013) | Asian | Turkey | 80/125 | 13 | 57 | 10 | 59 | 62 | 4 | 0.01 |
| Estandia-Ortega et al. (2014) | Caucasian | Mexico | 132/370 | 39 | 55 | 38 | 143 | 172 | 55 | 0.77 |
Characteristics of Included Studies
The studies were published between 1998 and 2014. Two studies [56, 67] departed from HWE. Two studies did not reported MTHFR genotypes in cases and controls [30, 62], reported only allele numbers in cases and controls. The smallest case sample size was 3365 and highest sample size was 64,456.
In included studies, total cases were 3274 with CC (1757), CT (1532) and TT (435), and controls were 5275 with CC (2594), CT (2141), and TT (540). In controls genotype percentage of CC, CT and TT were 49.2, 40.59 and 10.24% respectively. In cases genotype percentage of CC, CT and TT were 47.18, 41.14 and 11.68% respectively. Six studies did not show any association [26, 29, 30, 33, 57, 58] and odds ratio was above one in other sixteen studies.
Meta-Analysis
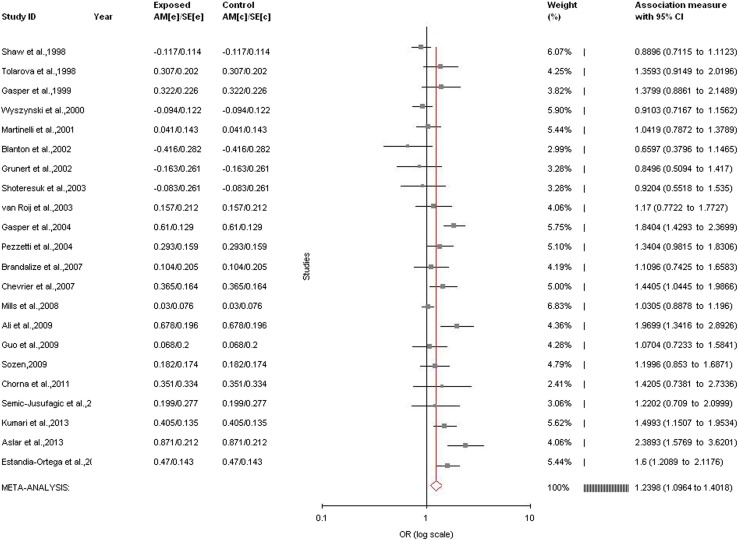
Allele contrast meta-analysis (T vs. C) of twenty-two studies indicated a strong significant association between MTHFR C677T polymorphism and nsCL/P susceptibility with both fixed (OR = 1.20, 95% CI = 1.12–1.28, p < 0.0001) and random (OR = 1.24, 95% CI = 1.1–1.4, p = 0.0006) effect models (Table 2, Fig. 2).
Table 2.
Summary estimates for the odds ratio (OR) of MTHFR C677T in various allele/genotype contrasts, the significance level (p value) of heterogeneity test (Q test), and the I2 metric and publication bias p value (Egger and Beggs Tests)
| Genetic models | Fixed effect OR (95% CI), p | Random effect OR (95% CI), p | Heterogeneity p value (Q test) | I2 (%) | Publication bias (p of Egger’s test) | Publication bias (p value of Beggs test) |
|---|---|---|---|---|---|---|
| All studies | ||||||
| Allele contrast (T vs. C) | 1.2 (1.12–1.28), <0.0001 | 1.24 (1.1–1.4), 0.0006 | <0.0001 | 66.26 | 0.40 | 1 |
| Co-dominant (Ct vs. CC) | 1.24 (1.13–1.37), <0.0001 | 1.26(0.98–1.63), 0.007 | <0.0001 | 83.34 | 0.32 | 0.16 |
| Homozygote (TT vs. CC) | 0.8 (0.69–0.91), 0.001 | 1.02 (0.74–1.4), 0.09 | <0.0001 | 75.14 | 0.03 | 0.04 |
| Dominant (TT + CT vs. CC) | 1.26 (1.15–1.38), <0.0001 | 1.29 (1.04–1.59), 0.02 | <0.0001 | 78.17 | 0.50 | 0.34 |
| Recessive (TT vs. CT + CC) | 1.24 (1.07–1.43), 0.002 | 1.36 (1.05–1.74), 0.02 | 0.0004 | 59.32 | 0.18 | 0.14 |
| Asian studies | ||||||
| Allele contrast (T vs. C) | 1.5 (1.27–1.74), <0.0001 | 1.49 (1.1–1.9), 0.006 | 0.02 | 63.39 | 0.63 | 1 |
| Co-dominant (Ct vs. CC) | 1.5(1.21–1.88), 0.0002 | 1.6 (1.03–2.5), 0.03 | 0.02 | 66.85 | 0.6 | 0.8 |
| Homozygote (TT vs. CC) | 1.32 (0.81–2.1), 0.3 | 1.2 (0.52–2.7), 0.65 | 0.07 | 53.49 | 0.48 | 1 |
| Dominant (TT + CT vs. CC) | 1.55 (1.25–1.92), <0.0001 | 1.6 (1.01–2.5), 0.04 | 0.008 | 70.68 | 0.72 | 0.46 |
| Recessive (TT vs. CT + CC) | 1.5 (0.98–2.3), 0.06 | 1.5 (−0.67 to 3.38),0.31 | 0.05 | 56.81 | 0.89 | 1 |
| Caucasian studies | ||||||
| Allele contrast (T vs. C) | 1.15 (1.06–1.23), 0.0002 | 1.17 (1.03–1.33), 0.01 | 0.0005 | 62.31 | 0.55 | 0.093 |
| Co-dominant (Ct vs. CC) | 1.19 (1.06–1.32), 0.002 | 1.17 (0.85–1.59), 0.32 | <0.0001 | 85.5 | 0.50 | 0.28 |
| Homozygote (TT vs. CC) | 0.75 (0.65–0.87), 0.0002 | 0.98 (0.68–1.42), 0.92 | <0.0001 | 78.19 | 0.02 | 0.09 |
| Dominant (TT + CT vs. CC) | 1.20 (1.1–1.33), 0.0003 | 1.2 (0.95–1.5), 0.1 | <0.0001 | 79.74 | 0.74 | 0.55 |
| Recessive (TT vs. CT + CC) | 1.21 (1.04–1.4), 0.01 | 1.3 (1.0–1.72), 0.04 | 0.0008 | 61.94 | 0.13 | 0.13 |
Fig. 2.
Forest plot for the association between MTHFR C677T polymorphism and nsCL/P for allele contrast model (T vs. C) with random effect model in total studies
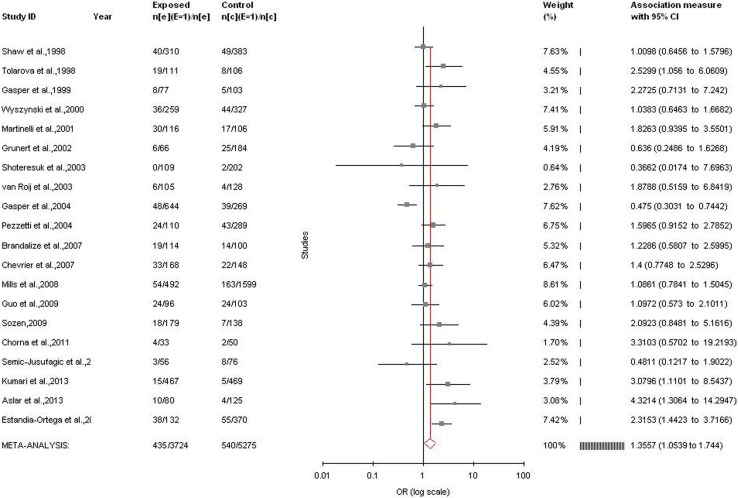
Significant association was detected between the MTHFR C677T polymorphism and the susceptibility to nsCL/P using other genetic models except homozygote model adopting random effect model (for CT vs. CC (co-dominant/heterozygote): OR = 1.26, 95%CI = 0.99–1.63, p = 0.007; for TT + CT vs. CC (dominant): OR = 1.29, 95% CI = 1.04–1.59, p = 0.02; for TT vs. CC (homozygote): OR = 1.02, 95% CI = 0.74–1.4, p = 0.09; for TT vs. CT + CC (recessive): OR = 1.36, 95% CI = 1.05–1.74, p = 0.02) (Table 2; Fig. 3). Significant association was also found in fixed effect models using all genetic models except homozygote (for CT vs. CC: OR = 1.24, 95%CI = 1.13–1.37, p < 0.0001; for TT + CT vs. CC: OR = 1.26, 95% CI = 1.15–1.38, p < 0.0001; for TT vs. CC: OR = 0.80, 95% CI = 0.69–0.91, p = 0.001; for TT vs. CT + CC: OR = 1.24, 95% CI = 1.07–1.43, p = 0.002; for) (Table 2).
Fig. 3.
Forest plot for the association between MTHFR C677T polymorphism and nsCL/P for recessive model (TT vs. CT + CC) with random effect model in total studies
A significant between studies heterogeneity was existed in allele contrast (Pheterogeneity < 0.0001, Q = 62.24, I2 = 66.26%, t2 = 0.05, z = 3.4), genotype homozygote (Pheterogeneity < 0.0001, Q = 76.44, I2 = 75.14%, t2 = 0.37, z = 0.13), dominant (Pheterogeneity < 0.0001, Q = 87.02, I2 = 78.17%, t2 = 0.17, z = 2.34) and recessive (Pheterogeneity = 0.002, Q = 46.71, I2 = 59.32%, t2 = 0.16, z = 2.37) comparisons.
Sensitivity Analysis
Sensitivity analysis was performed by exclusion of the studies with small sample size and studies deviated from Hardy–Weinberg equilibrium. Genotypes distribution in control population of two studies 56,67 was not in HW equilibrium and sample size in one study 65 was less than 50. Exclusion of these three studies, heterogeneity between studies was decreased (Pheterogeneity = 0.003, Q = 38.35, I2 = 53.07%, t2 = 0.03, z = 2.7) and OR was also decreased (T vs. C: OR = 1.17, 95% CI = 1.04–1.22, p = 0.0003).
Subgroup Analysis
Sub-group analysis based on ethnicity was also performed. In Asian population (number of studies = 6; 808/975 cases/controls), allele contrast meta-analysis showed significant association adopting both fixed (ORT vs. C = 1.5; 95% CI = 1.27–1.74; p = < 0.0001; I2 = 63.39%; Pheterogeneity = 0.02; PPb = 0.63) and random (ORT vs. C = 1.49; 95% CI = 1.1–1.94; p = 0.006) effect models. Combined mutant genotypes also showed significant association with fixed (ORTT + CT vs. CC = 1.55; 95% CI = 1.25–1.9; p < 0.0001; I2 = 70.68%; Pheterogeneity = 0.008; PPb = 0.72) and random (ORTT + CT vs. CC = 1.6; 95% CI = 1.01–2.5; p = 0.04) effect models (Table 2).
Results of Caucasian studies (number of studies = 16; 2916/4300 cases/controls) meta-analysis also indicated significant association with both fixed (ORT vs. C = 1.15; 95% CI = 1.06–1.23; p = 0.0002) and random (ORT vs. C = 1.17; 95% CI = 1.03–1.33; p = 0.01) effect models. However, higher between studies significant heterogeneity was also observed (I2 = 62.31%; Pheterogeneity = 0.0005). The combined mutant genotype showed statistically significant association with fixed effect model (ORTT + CT vs. CC = 1.20; 95% CI = 1.1–1.33; p = 0.0003; I2 = 79.74%; Pheterogeneity < 0.0001) and insignificant association with random effect (ORTT + CT vs. CC = 1.2; 95% CI = 0.95–1.5; p = 0.1) (Table 2).
Publication Bias
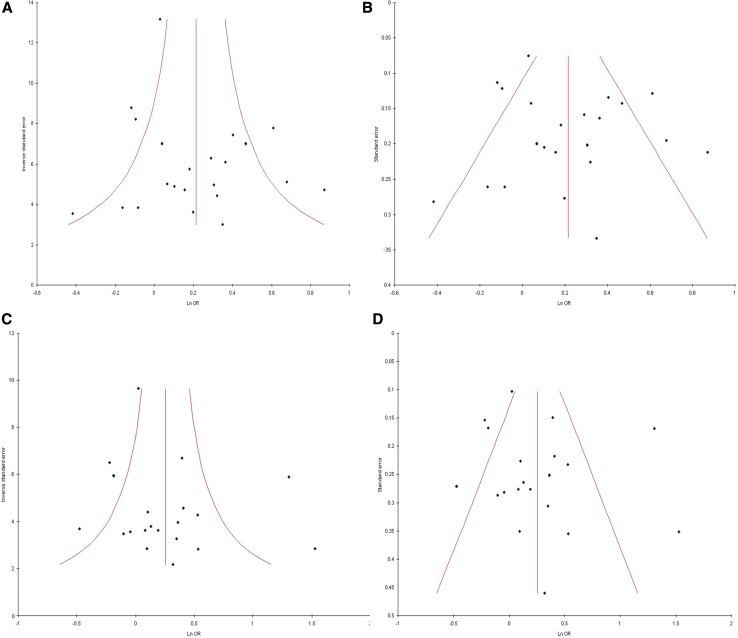
Except homozygote model publication bias could not be observed in all genetic models by Begg’s and Egger’s test. Funnel plots did not reveal any evidence of asymmetry, and except homozygote model p value were more than 0.05 (Begg’s p = 1.0, Egger’s p = 0.40 for T vs. C; Begg’s p = 0.04, Egger’s p = 0.03 for TT vs. CC; and Begg’s p = 0.16, Egger’s p = 0.32 for CT vs. CCA; Begg’s p = 0.34, Egger’s p = 0.50 for TT + AC vs. CC; Begg’s p = 0.14, Egger’s p = 0.18 for TT vs. CT + CC) (Table 2; Fig. 4).
Fig. 4.
Funnel plots A. precision versus OR (T vs. C), B. standard error versus OR (T vs. C), C. precision versus OR (TT + CT vs. CC), D. standard error versus OR (TT + CT vs. CC)
Discussion
A number of population-based epidemiological studies have suggested that inadequate intake of folic acid [12, 14, 16, 57, 70, 71], or exposure to folic acid antagonists [72] during pregnancy might predispose to orofacial clefts. Conversely, multivitamin supplementation during pregnancy may reduce the risk of orofacial clefts. Folic acid and its derivatives are essential for DNA synthesis and methylation which are required for normal cell division and gene expression during fetal development. Genetic polymorphism in folate/homocysteine pathway genes like MTHFR and MTRR increases concentration of homocysteine (hyperhomocysteinemia), which has been identified as risk factors for certain congenital defects (e.g. NTD, congenital heart defects) and late-age disorders (e.g. cardiovascular, cancers) [73].
There are strong experimental evidences showing that folate deficiency and/or dysfunctional MTHFR gene is risk factor for nsCL/P like- (1) in murine model, knockout of MTHFR gene induced apoptosis in embryonic palatal mesenchymal cells (MEPM) and prevent growth [74], (2) supplementation with folic acid was sufficient to reverse the teratogenic effect of the silenced MTHFR gene (3) exposure to folic acid antagonists [72] during pregnancy might predispose to orofacial clefts, (4) number of population-based epidemiological studies have suggested that inadequate intake of folic acid [12, 14, 16, 57, 70, 71], during pregnancy might predispose to orofacial clefts and (5) Munger et al. [75] showed that folic acid could reduce the risk of facial clefts in mouse models [75].
Meta-analysis is a useful strategy for elucidating genetic factors in different diseases/disorders. Several meta-analysis were published which evaluated risk of MTHFR polymorphism for different disease and disorders- like- congenital heart defects [39], Down syndrome [76]; recurrent pregnancy loss [77]; stroke [78]; psychiatric disorders [79–81]; Alzheimers disease [82]; and cancer [83–86].
Four meta-analyses were published so far investigated MTHFR polymorphisms as risk factor for cleft lip and palate [87–90]. Out of four meta-analyses, two were investigated MTHFR A1298C polymorphism as risk factor [88, 89] whereas other two meta-analyses ncluded C677T polymorphism [87, 90]. Verkleij-Hagoort et al. [87] published a meta-analysis of eight studies and reported no association (OR = 1.0; 95% CI = 0.9–1.2). In a recent meta-analysis, Zhao et al. [90] carried out meta-analysis of nine Asian studies and reported significant association between C677T polymorphism and CL/P risk (OR = 1.41; 95% CI = 1.23–1.61). Previous meta-analyses [87, 90] included only eight and nine case–control studies, which were too little to confirm the association between MTHFR C677T polymorphism and nsCL/P risk. Hence to provide the more comprehensive assessment, present meta-analysis was carried out to update the existing meta-analyses by including studies published in the interim not included in the previous meta-analyses. Present meta-analysis has included data from twenty-two case control studies with 3592 nsCL/P cases and 4905 controls.
The present meta-analysis showed a higher heterogeneity between studies, which may be due to differences in the ethnicity of studied populations and study design. There are few limitations in the present meta-analysis like crude ORs used in without adjustment; other factors such as folate intake etc. were not considered. nsCL/P may be due to several other gene polymorphisms involved in the homocysteine and folate metabolic pathway which were not considered in this meta-analysis. In addition, present meta-analysis has strength also like (1) absence of publication bias and (2) inclusion of larger number of studies.
In conclusion, present meta-analysis indicated that MTHFR gene C677T polymorphism is associated with the higher risk of nsCL/P. Simultaneously, subgroup analyses based on ethnicity further confirmed this association. In future, well designed studies with larger samples are necessary to validate association between this polymorphism and role of micronutrient factors like folate and B12 in the susceptibility of nsCL/P. However, results of the present meta-analysis should be interpreted cautiously, owing to the higher heterogeneity among studies.
Acknowledgements
The author is highly grateful to Leon Bax (Chief Scientific Officer at BiostatXL, UMC Utrecht) for his valuable suggestions which help her in statistical analysis.
Abbreviations
- nsCL/P
Nonsyndromic cleft lip with or without cleft palate
- MTHFR
Methylenetetrahydrofolate reductase
Compliance with Ethical Standards
Conflict of interest
The author declares that she has no conflict of interest.
References
- 1.Croen LA, Shaw GM, Wasserman CR, Tolarova MM. Racial and ethnic variations in the prevalence of orofacial clefts in California 1983–1992. Am J Med Genet. 1998;79(1):42–47. doi: 10.1002/(SICI)1096-8628(19980827)79:1<42::AID-AJMG11>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 2.Vanderas AP. Incidence of cleft lip, cleft palate, and cleft lip and palate among races: a review. Cleft Palate J. 1987;24:216–225. [PubMed] [Google Scholar]
- 3.Clark JD, Mossey PA, Sharp L, Little J. Socioeconomic status and orofacial clefts in Scotland, 1989 to 1998. Cleft Palate Craniofac J. 2003;40:481–485. doi: 10.1597/1545-1569(2003)040<0481:SSAOCI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 4.Wyszynski DF, Beaty TH. Review of the role of potential teratogens in the origin of human nonsyndromic oral clefts. Teratology. 1996;53:309–317. doi: 10.1002/(SICI)1096-9926(199605)53:5<309::AID-TERA5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 5.Lie RT, Wilcox AJ, Skjaerven R. A population-based study of the risk of recurrence of birth defects. N Engl J Med. 1994;331:1–4. doi: 10.1056/NEJM199407073310101. [DOI] [PubMed] [Google Scholar]
- 6.Lie RT, Wilcox AJ, Skjaerven R. Survival and reproduction among males with birth defects and risk of recurrence in their children. JAMA. 2001;285:755–760. doi: 10.1001/jama.285.6.755. [DOI] [PubMed] [Google Scholar]
- 7.Skjaerven R, Wilcox AJ, Lie RT. A population-based study of survival and childbearing among female subjects with birth defects and the risk of recurrence in their children. N Engl J Med. 1999;340:1057–1062. doi: 10.1056/NEJM199904083401401. [DOI] [PubMed] [Google Scholar]
- 8.Bhaskar LVKS, Murthy J, Venkatesh Babu G. Polymorphisms in genes involved in folate metabolism and orofacial clefts. Arch Oral Biol. 2011;56:723–737. doi: 10.1016/j.archoralbio.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Tolarova M. Orofacial clefts in Czechoslovakia. Incidence, genetics, and prevention of cleft lip and palate over a 19-year period. Scand J Plast Reconst Surg Hand Surg. 1987;21:19–25. doi: 10.3109/02844318709083574. [DOI] [PubMed] [Google Scholar]
- 10.Tolarova M, Harris J. Reduced recurrence of orofacial clefts after periconceptional supplementation with high-dose folic acid and multivitamins. Teratology. 1995;51:71–78. doi: 10.1002/tera.1420510205. [DOI] [PubMed] [Google Scholar]
- 11.Shaw GM, O’Malley CD, Wasserman CR, Tolarova MM, Lammer EJ. Maternal periconceptional use of multivitamins and reduced risk for conotruncal heart defects and limb deficiencies among offspring. Am J Med Genet. 1995;59:536–545. doi: 10.1002/ajmg.1320590428. [DOI] [PubMed] [Google Scholar]
- 12.Loffredo LC, Souza JM, Feitas JA, Mossey PA. Oral clefts and vitamin supplementation. Cleft Palate Craniofac J. 2001;38:76–83. doi: 10.1597/1545-1569(2001)038<0076:OCAVS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell LE, Murray JC, O’Brien S, Christensen K. Retinoic acid receptor alpha gene variants, multivitamin use, and liver intake as risk factors for oral clefts: a population based case-control study in Denmark. Am J Epidemiol. 2003;158(1):1991–1994. doi: 10.1093/aje/kwg102. [DOI] [PubMed] [Google Scholar]
- 14.van Rooij IA, Ocké MC, Straatman H, Zielhuis GA, Merkus HM, Steegers-Theunissen RP. Periconceptual folate intake by supplement and food reduces the risk of nonsyndromic cleft lip with or without cleft palate. Prev Med. 2004;39:689–694. doi: 10.1016/j.ypmed.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 15.Krapels IP, Vermeij-Keers C, Muller M, de Klein A, Steegers- Theunissen RP. Nutrition and genes in the development of orofacial clefting. Nutr Rev. 2006;64:280–288. doi: 10.1111/j.1753-4887.2006.tb00211.x. [DOI] [PubMed] [Google Scholar]
- 16.Badovinac RL, Werler WM, Williams PL, Kelsey KT, Hayes C. Folic acid-containing supplement consumption during pregnancy and risk for oral clefts: a meta-analysis. Birth Def Res (Part A) 2007;79:8–15. doi: 10.1002/bdra.20315. [DOI] [PubMed] [Google Scholar]
- 17.Chevrier C, Perret C, Bahuan M, Zhu H, Nelva A, Herman C, et al. Fetal and maternal MTHFR C677T genotype, maternal folate intake and the risk of nonsyndromic oral clefts. Am J Med Genet A. 2007;143:248–257. doi: 10.1002/ajmg.a.31462. [DOI] [PubMed] [Google Scholar]
- 18.MRC Vitamin Study Research Group Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. Lancet. 1991;338:131–137. doi: 10.1016/0140-6736(91)90133-A. [DOI] [PubMed] [Google Scholar]
- 19.Czeizel AE, Dudas I. Prevention of the first occurrence of neural tube defects by periconceptional vitamin supplementation. N Engl J Med. 1992;327:1832–1835. doi: 10.1056/NEJM199212243272602. [DOI] [PubMed] [Google Scholar]
- 20.Rosenquist TH, Finell RH. Genes, folate and homocysteine in embryonic development. Proc Nutr Soc. 2001;60:1–9. [PubMed] [Google Scholar]
- 21.Czeizel AE. Periconceptional folic acid containing multivitamin supplementation. Eur J Obstet Gynecol Reprod Biol. 1998;78:151–161. doi: 10.1016/S0301-2115(98)00061-X. [DOI] [PubMed] [Google Scholar]
- 22.Itikala PR, Watkins ML, Mulinare J, Moore CA, Liu Y. Maternal multivitamin use and orofacial clefts in offspring. Teratology. 2001;63:79–86. doi: 10.1002/1096-9926(200102)63:2<79::AID-TERA1013>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Boyles AL, Wilcox A, Taylor JA, Meyer K, Fredriksen A, Ueland PM, et al. Folate and one-carbon metabolism gene polymorphisms and their associations with oral facial clefts. Am J Med Genet A. 2008;146A:440–449. doi: 10.1002/ajmg.a.32162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yazdy MM, Honein MA, Xing J. Reduction in orofacial clefts following folic acid fortification of the U.S. grain supply. Birth Defects Res A Clin Mol Teratol. 2007;79:16–23. doi: 10.1002/bdra.20319. [DOI] [PubMed] [Google Scholar]
- 25.Ray JG, Meier C, Vermeulen MJ, Wyatt PR, Cole DE. Association between folic acid food fortification and congenital orofacial clefts. J Pediatr. 2003;143:805–807. doi: 10.1067/S0022-3476(03)00495-5. [DOI] [PubMed] [Google Scholar]
- 26.Shaw GM, Rozen R, Finnell RH, Todoroff K, Lammer EJ. Infant C677T mutation in MTHFR, maternal periconceptional vitamin use, and cleft lip. Am J Med Genet. 1998;80:196–198. doi: 10.1002/(SICI)1096-8628(19981116)80:3<196::AID-AJMG2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 27.Gaspar DA, Pavanello RC, Zatz M, Passos-Bueno MR, Andre M, Steman S, et al. Role of the C677T polymorphism at the MTHFR gene on risk to nonsyndromic cleft lip with/without cleft palate: results from a case-control study in Brazil. Am J Med Genet. 1999;87:197–199. doi: 10.1002/(SICI)1096-8628(19991119)87:2<197::AID-AJMG15>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 28.Mills JL, Kirke PN, Molloy AM, Burke H, Conley MR, Lee YJ, et al. Methylenetetrahydrofolate reductase thermolabile variant and oral clefts. Am J Med Genet. 1999;86:71–74. doi: 10.1002/(SICI)1096-8628(19990903)86:1<71::AID-AJMG14>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Martinelli M, Scapoli L, Pezzetti F, Carinci F, Carinci P, Stabellini G, et al. C677T variant form at theMTHFR gene and CL/P: a risk factor for mothers? AmJ Med Genet. 2001;98:357–360. doi: 10.1002/1096-8628(20010201)98:4<357::AID-AJMG1108>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 30.Blanton SH, Patel S, Hecht JT, Mulliken JB. MTHFR is not a risk factor in the development of isolated nonsyndromic cleft lip and palate. Am J Med Genet. 2002;110(4):404–405. doi: 10.1002/ajmg.10496. [DOI] [PubMed] [Google Scholar]
- 31.Prescott NJ, Winter RM, Malcolm S. Maternal MTHFR genotype contributes to the risk of non-syndromic cleft lip and palate. J Med Genet. 2002;39:368–369. doi: 10.1136/jmg.39.5.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jugessur A, Wilcox AJ, Lie RT, Murray JC, Taylor JA, Ulvik A, et al. Exploring the effects of methylenetetrahydrofolate reductase gene variants C677T and A1298C on the risk of orofacial clefts in 261 Norwegian case-parent triads. Am J Epidemiol. 2003;157:1083. doi: 10.1093/aje/kwg097. [DOI] [PubMed] [Google Scholar]
- 33.Shotelersuk V, Ittiwut C, Siriwan P, Angspatt A. Maternal 677CT/1298AC genotype of the MTHFR gene as a risk factor for cleft lip. J Med Genet. 2003;40:e64. doi: 10.1136/jmg.40.5.e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Rooij LA, Swinkels DW, Blom HJ, Markus HM, Stegers-Theunissen RP. Vitamin and homocysteine status of mothers and infants and the risk of nonsyndromic orofacial clefts. Am J Obstet Gynecol. 2003;189:1155–1160. doi: 10.1067/S0002-9378(03)00592-1. [DOI] [PubMed] [Google Scholar]
- 35.Vieira AR, Murray JC, Trembath D, Orioli IM, Castilla EE, Cooper ME, et al. Studies of reduced folate carrier 1 (RFC1) A80G and 5,10-methylenetetrahydrofolate reductase (MTHFR) C677T polymorphisms with neural tube and orofacial cleft defects. Am J Med Genet. 2000;135A:220–223. doi: 10.1002/ajmg.a.30705. [DOI] [PubMed] [Google Scholar]
- 36.Frosst P, Bloom HJ, Milos R, Goyette P, Sheppard CA, Mattews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 37.van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hobbs CA, Sherman SL, Hopkins SE, Torfs CP, Hine RJ, Pogribna M, et al. Polymorphisms in genes involved in folate metabolism as maternal risk factors for down syndrome. Hum Genet. 2000;67:623–630. doi: 10.1086/303055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hobbs CA, Cleves MA, Karim MA, Zhao W, MacLeod SL. Maternal folate-related gene environment interactions and congenital heart defects. Obstet Gynecol. 2010;116:316–322. doi: 10.1097/AOG.0b013e3181e80979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hegele RA, Tully C, Young TK, Connelly PW. V677 mutation of methylenetetrahydrofolate reductases and cardiovascular disease in Canadian Inuit. Lancet. 1997;349:1221–1222. doi: 10.1016/S0140-6736(05)62414-2. [DOI] [PubMed] [Google Scholar]
- 41.Wilcken B, Bamforth F, Li Z, Zhu H, Ritvanen A, Renlund M, et al. Geographical and ethnic variation of the 677C > T allele of 5,10-methylenetetrahydrofolate reductase (MTHFR): findings from over 7000 newborns from 16 areas worldwide. J Med Genet. 2003;40:619–625. doi: 10.1136/jmg.40.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rai V, Yadav U, Kumar P, Yadav SK. Methyleletetrahydrofolate reductase polymorphism (C677T) in Muslim population of Eastern Uttar Pradesh, India. Ind J Med Sci. 2010;64(5):219–223. doi: 10.4103/0019-5359.98949. [DOI] [PubMed] [Google Scholar]
- 43.Rai V, Yadav U, Kumar P. Genotype prevalence and allele frequencies of 5,10-methylenetetrahydrofolate reductase (MTHFR) C677T mutation in two caste groups of India. Cell Mol Biol. 2012;58:OL1695–701. [PubMed] [Google Scholar]
- 44.Yadav U, Kumar P, Gupta S, Rai V. Distribution of MTHFR C677T gene polymorphism in healthy North Indian population and an updated meta-analysis. Ind J Clinical Biochem. 2016 doi: 10.1007/s12291-016-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tolarova MM, van Rooij IALM, Pastor M, van der Put NMJ, Goldberg AC, Hol F, et al. A common mutation in the MTHFR gene is a risk factor for non-syndromic cleft lip and palate anomalies. Am J Hum Genet. 1998;63:A27. [Google Scholar]
- 46.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 47.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 48.Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2004;24:1–15. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- 49.Zintzaras E, Hadjigeorgiou GM. The role of G196A polymorphism in the brain-derived neurotrophic factor gene in the cause of Parkinson’s disease: a meta-analysis. J Hum Genet. 2005;50:560–566. doi: 10.1007/s10038-005-0295-z. [DOI] [PubMed] [Google Scholar]
- 50.Higgins JP, Thompson SE. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 51.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zintzaras E, Koufakis T, Ziakas PD, Rodopoulou P, Giannouli S, Voulgarelis M. A meta-analysis of genotypes and haplotypes of methylenetetrahydrofolate reductase gene polymorphisms in acute lymphoblastic leukemia. Euro J Epidemiol. 2006;21:501–510. doi: 10.1007/s10654-006-9027-8. [DOI] [PubMed] [Google Scholar]
- 53.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 54.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bax L, Yu LM, Ikeda N, Tsuruta H, Moons KG. Development and validation of MIX: comprehensive free software for meta-analysis of causal research data. BMC Med Res Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaspar DA, Matioli SR, de Ca´ssia Pavanello R, Araujo BC, Alonso N, Wyszynski D, et al. Maternal MTHFR interacts with the offspring’s BCL3 genotypes, but not with TGFA, in increasing risk to nonsyndromic cleft lip with or without cleft palate. Eur J Hum Genet. 2004;12:521–526. doi: 10.1038/sj.ejhg.5201187. [DOI] [PubMed] [Google Scholar]
- 57.Wyszynski DF, Diehl SR. Infant C677T mutation in MTHFR, maternal periconceptional vitamin use, and risk of nonsyndromic cleft lip. Am J Med Genet. 2000;92:79–80. doi: 10.1002/(SICI)1096-8628(20000501)92:1<79::AID-AJMG14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 58.Grunert RR, Braune A, Schnackenberg E, Schloot W, Krause HR. Genetic differences in enzymes of folic acid metabolism in patients with lip-jaw-palate clefts and their relatives. Mund Kiefer Gesichtschir. 2002;6:131–133. doi: 10.1007/s10006-001-0361-4. [DOI] [PubMed] [Google Scholar]
- 59.Pezzetti F, Martinelli M, Scapoli L, Carinci F, Palmieri A, Marchesini J, et al. Maternal MTHFR variant forms increase the risk in offspring of isolated nonsyndromic cleft lip with or without cleft palate. Hum Mutat. 2004;24:104–105. doi: 10.1002/humu.9257. [DOI] [PubMed] [Google Scholar]
- 60.Brandalize AP, Bandinelli E, Borba JB, Felix TM, Roisenberg I, Schuler-Faccini L. Polymorphisms in genes MTHFR, MTR and MTRR are not risk factors for cleft lip/palate in South Brazil. Braz J Med Biol Res. 2007;40:787–791. doi: 10.1590/S0100-879X2006005000112. [DOI] [PubMed] [Google Scholar]
- 61.Mills JL, Molloy AM, Parle-McDermott A, Troendle JF, Brody LC, Conley MR, et al. Folate-related gene polymorphisms as risk factors for cleft lip and cleft palate. Birth Defects Res A. 2008;82:636–643. doi: 10.1002/bdra.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ali A, Singh SK, Raman R. MTHFR 677TT alone and IRF6 820GG together with MTHFR 677CT, but not MTHFR A1298C, are risks for non-syndromic cleft lip with or without cleft palate in an Indian population. Genet Test Mol Biomark. 2009;13:355–360. doi: 10.1089/gtmb.2008.0115. [DOI] [PubMed] [Google Scholar]
- 63.Sözen MA, Tolarova MM, Spritz RA. The common MTHFR C677T and A1298C variants are not associated with the risk of non-syndromic cleft lip/palate in northern Venezuela. J Genet Genomics. 2009;36:283–288. doi: 10.1016/S1673-8527(08)60116-2. [DOI] [PubMed] [Google Scholar]
- 64.Guo JZ, Song XM, Wang Y, Zhu WL, Li SQ, Li Y. Relationship between genetic polymorphisms of MTHFR C677T and nonsyndromic cleft lip with or without palate. Beijing Da Xue Xue Bao. 2009;41:432–436. [PubMed] [Google Scholar]
- 65.Chorna LB, Akopyan HR, Makukh HV, Fedoryk IM. Allelic polymorphisms in the MTHFR, MTR and MTRR genes in patients with cleft lip and/or palate and their mothers. Cytolo Genet. 2011;45:177–181. doi: 10.3103/S0095452711030029. [DOI] [PubMed] [Google Scholar]
- 66.Semiç-Jusufagiç A, Bircan R, Çelebiler O, Erdim M, Akarsu N. Elçioğlu1 NH. Association between C677T and A1298C MTHFR gene polymorphism and nonsyndromic orofacial clefts in the Turkish population: a case-parent study. Turkish J Pediatr. 2012;54:617–625. [PubMed] [Google Scholar]
- 67.Aslar D, Ozdiler E, Tastan H, Altug AB. Determination of Methylenetetrahydrofolate Reductase (MTHFR) gene polymorphism in Turkish patients with nonsyndromic cleft lip and palate. Int J Pediatr Otorhinolaryngol. 2013;77:1143–1146. doi: 10.1016/j.ijporl.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 68.Kumari P, Ali A, Shukla KK, Singh SK, Raman R. Lower incidence of nonsyndromic cleft lip with or without cleft palate in females: Is homocysteine a factor? J Biosci. 2013;38:21–26. doi: 10.1007/s12038-013-9298-7. [DOI] [PubMed] [Google Scholar]
- 69.Estandia-Ortega B, Velazquez-Aragon JA, Alcantara-Ortigoza MA, Reyna-Fabian ME, Villagomez-Martınez S, Gonzalez-del Angel A. 5,10-Methylenetetrahydrofolate reductase single nucleotide polymorphisms and gene–environment interaction analysis in analysis in non-syndromic cleft lip/palate. Eur J Oral Sci. 2014;122:109–113. doi: 10.1111/eos.12114. [DOI] [PubMed] [Google Scholar]
- 70.Rouget F, Monfort C, Bahuau M, Nelva A, Herman C, Francannet C, et al. Periconceptual folates and the prevention of orofacial clefts: role of dietary intakes in France. Rev Epidemiol Sante Publique. 2005;53:351–360. doi: 10.1016/S0398-7620(05)84617-6. [DOI] [PubMed] [Google Scholar]
- 71.Wilcox AJ, Lie RT, Solvoll K, Taylor J, Mcconnaughey DR, Abyholm F, et al. Folic acid supplements and risk of facial clefts: national population based case-control study. Br Med J. 2007;334:464. doi: 10.1136/bmj.39079.618287.0B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hernandez-Diaz S, Werler MM, Walker AM, Mitchell AA. Folic acid antagonists during pregnancy and the risk of birth defects. N Engl J Med. 2000;343:1608–1614. doi: 10.1056/NEJM200011303432204. [DOI] [PubMed] [Google Scholar]
- 73.Refsum H, Yajnik CS, Gadkari M, Schneede J, Vollset SE, Orning L, et al. Hyperhomocysteinemia and elevated methylmalonic acid indicate a high prevalence of cobalamin deficiency in Asian Indians. Am J Clin Nutr. 2001;74:233–241. doi: 10.1093/ajcn/74.2.233. [DOI] [PubMed] [Google Scholar]
- 74.Xiao WL, Wu M, Shi B. Folic acid rivals methylenetetrahydrofolate reductase (MTHFR) gene silencing effect on MEPM cell proliferation and apoptosis. Mol Cell Biochem. 2006;292:145–154. doi: 10.1007/s11010-006-9228-1. [DOI] [PubMed] [Google Scholar]
- 75.Munger R. Maternal nutrition and oral clefts. In: Wyszynski D, editor. Cleft lip and palate: from origin to treatment. New York: Oxford University Press; 2002. pp. 170–192. [Google Scholar]
- 76.Rai V. Polymorphism in folate metabolic pathway gene as maternal risk factor for down syndrome. Int J Biol Med Res. 2011;2(4):1055–1060. [Google Scholar]
- 77.Rai V. Methylenetetrahydrofolate reductase C677T polymorphism and recurrent pregnancy loss risk in Asian population: a meta-analysis. Ind J Clin Biochem. 2016;32(4):402–413. doi: 10.1007/s12291-016-0554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yadav S, Hasan N, Marjot T, Khan MS, Prasad K, Bentley P, et al. Detailed analysis of gene polymorphisms associated with ischemic stroke in South Asians. PLoS One. 2013;8(3):e57305. doi: 10.1371/journal.pone.0057305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rai V. Association of methylenetetrahydrofolate reductase (MTHFR) gene C677T polymorphism with autism: evidence of genetic susceptibility. Metab Brain Dis. 2016;1:727–735. doi: 10.1007/s11011-016-9815-0. [DOI] [PubMed] [Google Scholar]
- 80.Yadav U, Kumar P, Gupta S, Rai V. Role of MTHFR C677T gene polymorphism in the susceptibility of schizophrenia: an updated meta-analysis. Asian J Psychiatr. 2016;20:41–51. doi: 10.1016/j.ajp.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Rai V. Genetic polymorphisms of methylenetetrahydrofolate reductase (MTHFR) gene and susceptibility to depression in Asian population: a systematic meta-analysis. Cell Mol Biol. 2014;60(3):29–36. [PubMed] [Google Scholar]
- 82.Rai V. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and alzheimer disease risk: a meta-analysis. Mol Neurobiol. 2016;54:1173–1186. doi: 10.1007/s12035-016-9722-8. [DOI] [PubMed] [Google Scholar]
- 83.Rai V. Methylenetetrahydrofolate reductase A1298C polymorphism and breast cancer risk: a meta-analysis of 33 studies. Ann Med Health Sci Res. 2014;4(6):841–851. doi: 10.4103/2141-9248.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar P, Yadav U, Rai V. Methylenetetrahydrofolate reductase gene C677T polymorphism and breast cancer risk: evidence for genetic susceptibility. Meta Gene. 2015;6:72–84. doi: 10.1016/j.mgene.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yadav U, Kumar P, Rai V. Role of MTHFR A1298C gene polymorphism in the etiology of prostate cancer: a systematic review and updated meta-analysis. Egypt J Med Hum Genet. 2016;17(2):141–148. doi: 10.1016/j.ejmhg.2015.06.005. [DOI] [Google Scholar]
- 86.Rai V. Evaluation of the MTHFR C677T polymorphism as a risk factor for colorectal cancer in Asian populations. Asian Pac J Cancer Prev. 2016;16(18):8093–8100. doi: 10.7314/APJCP.2015.16.18.8093. [DOI] [PubMed] [Google Scholar]
- 87.Verkleij-Hagoort A, Bliek J, Sayed-Tabatabaei F, Ursem N, Steegers E, Steegers-Theunissen R. Hyperhomocysteinemia and MTHFR polymorphisms in association with orofacial clefts and congenital heart defects: a meta-analysis. Am J Med Gene Part A. 2007;143A:952–960. doi: 10.1002/ajmg.a.31684. [DOI] [PubMed] [Google Scholar]
- 88.Rai V. Maternal methylenetetrahydrofolate reductase (MTHFR) gene A1298C polymorphism and risk of nonsyndromic Cleft lip and/or Palate (NSCL/P) in offspring: a meta-analysis. Asian J Med Sci. 2014;6(1):16–21. doi: 10.3126/ajms.v6i1.10281. [DOI] [Google Scholar]
- 89.Rai V. Meta-analysis of methylenetetrahydrofolate reductase (MTHFR) A1298C polymorphism and risk of orofacial cleft. J Med Genet Genomics. 2014;6:19–26. doi: 10.5897/JMGG2014.0075. [DOI] [Google Scholar]
- 90.Zhao M, Ren Y, Shen L, Zhang Y, Zhou B. Association between MTHFR C677T and A1298C polymorphisms and NSCL/P risk in Asians: a meta- analysis. PLoS One. 2014;9:e88242. doi: 10.1371/journal.pone.0088242. [DOI] [PMC free article] [PubMed] [Google Scholar]