Abstract
Background
Plants are frequently subjected to abiotic and biotic stresses, and WRKY proteins play a pivotal role in the response to such stress. OsWRKY11 is induced by pathogens, drought, and heat, suggesting a function in biotic and abiotic stress responses.
Results
This study identified OsWRKY11, a member of WRKY group IIc. It is a transcriptional activator that localized to the nucleus. Ectopic expression of OsWRKY11 resulted in enhanced resistance to a bacterial pathogen, Xanthomonas oryzae pv. oryzae; resistance was compromised in transgenic lines under-expressing OsWRKY11. Ectopic expression of OsWRKY11 resulted in constitutive expression of defense-associated genes, whereas knock-down (kd) of OsWRKY11 reduced expression of defense-associated genes during pathogen attack, suggesting that OsWRKY11 activates defense responses. OsWRKY11 bound directly to the promoter of CHITINASE 2, a gene associated with defense, and activated its transcription. In addition, ectopic expression of OsWRKY11 enhanced tolerance to drought stress and induced constitutive expression of drought-responsive genes. Induction of drought-responsive genes was compromised in OsWRKY11-kd plants. OsWRKY11 also bound directly to the promoter of a drought-responsive gene, RAB21, activating its transcription. In addition, OsWRKY11 protein levels were controlled by the ubiquitin-proteasome system.
Conclusion
OsWRKY11 integrates plant responses to pathogens and abiotic stresses by positively modulating the expression of biotic and abiotic stress-related genes.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0199-0) contains supplementary material, which is available to authorized users.
Keywords: OsWRKY11, Disease resistance, Drought tolerance, Cross-talk, Rice
Background
Plants in the field are often subjected to abiotic and biotic stresses simultaneously or successively and have therefore evolved elegant mechanisms to respond precisely to individual or combined stresses. Cross-talk between individual stress response signaling pathways is well documented (reviewed in Sharma et al. 2013; Kissoudis et al. 2014; Takatsuji 2017), and evidence has accumulated that the signaling pathways involved in biotic and abiotic stress responses interact either antagonistically or synergistically via the stress hormones salicylic acid, ethylene, jasmonic acid, and abscisic acid (Sharma et al. 2013; Kissoudis et al. 2014; Takatsuji 2017). In rice, many genes are involved at the points of intersection of abiotic and biotic stress signaling, including OsMPK5, OsMPK6, OsEIN2, and transcription factors (TF)s (Xiong and Yang, 2003; Sharma et al. 2013; Ueno et al., 2015).
Bioinformatics and functional analyses demonstrated that TFs, including NAC, MYB, AP2/ERF, and WRKY, are involved in biotic and abiotic stress responses (Atkinson and Urwin, 2012; Shaik and Ramakrishna, 2014). Of these, WRKY TFs are involved in various biological processes, including growth and development (Han et al. 2014; Dai et al. 2016), abiotic stress responses (Kim et al. 2016; Raineri et al. 2015; Yokotani et al. 2013), and biotic stress responses (Abbruscato et al. 2012; Choi et al. 2015; Han et al. 2013; Hwang et al. 2016; Lan et al. 2013; Shimono et al. 2007; Wang et al. 2015; Zhang et al. 2008). WRKY TFs are therefore likely candidates for proteins involved in the cross-talk between abiotic and biotic stresses in rice. Some of these proteins play opposing roles, either positive or negative, during biotic and abiotic stress responses. Involvement of OsWRKY13 gene in cross-talk between disease resistance and abiotic stress tolerance pathways has been intensively studied. OsWRKY13 enhances resistance to Xanthomonas oryzae pv oryzae (Xoo) and Magnaporthe oryzae and decreases tolerance to cold and salt stresses (Qiu et al. 2007, 2008). More recently, a molecular mechanism has been proposed by which the suppression of SNAC1 by OsWRKY13 enhances drought tolerance (Xiao et al. 2013). The OsWRKY45–2 TF confers broad-spectrum disease resistance in rice but reduces adaptation to salt, cold, and drought stresses (Tao et al. 2009, 2011). OsWRKY45–1 confers rice resistance to M. oryzae but reduces resistance to the bacterial pathogens Xoo and Xanthomonas oryzae pv oryzicola (Xoc) and to cold and drought stresses (Shimono et al. 2007; Tao et al. 2009, 2011; Goto et al., 2015). Interestingly OsWRKY62, one of the group IIa WRKY TFs, plays a positive role in pathogen defense together with OsWRKY45–1 but plays a negative role in pathogen defense under hypoxia stress by self dimerization (Fukushima et al., 2016). OsWRKY76 enhances susceptibility to M. oryzae and Xoo but increases cold tolerance (Seo et al. 2011; Yokotani et al. 2013).
Some OsWRKY TFs play positive roles in biotic and abiotic stresses. OsWRKY89 [renamed 104 by the Committee on Gene Symbolization, Nomenclature and Linkage (CGSNL)] enhances resistance to M. oryzae and UV-B irradiation (Rice WRKY working group, 2012; Wang et al. 2007). It is reported that over-expression of OsWRKY30 increases resistance to fungal pathogens and drought stress by two independent research groups (Peng et al. 2012; Shen et al. 2012). For OsWRKY71 two independent reports showed that OsWRKY71 enhances resistance to Xoo and tolerance to cold, respectively (Liu et al. 2007; Chujo et al., 2008; Kim et al. 2016).
Although evidence for cross-talk between biotic and abiotic stress responses is rapidly accumulating, the molecular mechanisms remain largely unknown, particularly with respect to positive cross-talk. We describe here the role played by OsWRKY11 in positive cross-talk between biotic and abiotic stress responses. OsWRKY11 is induced by heat, drought, combined heat/drought, and pathogen stresses (Shiroto et al. 2004; Ryu et al. 2006). Ectopic expression of OsWRKY11 under the control of the heat shock protein (HSP) 101 promoter enhances drought tolerance (Wu et al. 2009). These expression patterns suggest that OsWRKY11 is involved in biotic and abiotic stress responses. We analyzed the severity of disease in response to Xoo, drought tolerance, and expression of stress-related genes in OsWRKY11 over-expression (ox) and RNA interference (RNAi) lines, and demonstrated direct binding of OsWRKY11 to promoters of both biotic and abiotic stress-related genes.
Results
Generation of transgenic plants over-expressing and under-expressing OsWRKY11
We were most interested in investigating genes involved in the response to combined stresses such as drought and Xoo infection. The nine OsWRKY TFs (OsWRKY7, −10, −11, −30, −32, −67, −70, 83 (renamed 94 by CGSNL), −85 (renamed 96 by CGSNL) are previously reported on their induction upon the infection of an incompatible race of Xoo (Ryu et al. 2006). In the current study, we focused on OsWRKY11 (Os01g43650), whose expression enhances drought tolerance (Wu et al. 2009). More recently we reported that OsWRKY11 expression is increased in compatible and incompatible interaction but the level of its expression is more pronounced in the incompatible interaction than in the compatible interaction (Choi et al., 2017).
To elucidate the function of OsWRKY11 in defense signaling, we generated OsWRKY11-ox lines and OsWRKY11-knock-down (kd) lines by RNA interference approach in the rice cultivar, Nipponbare. We confirmed that OsWRKY11 was expressed at higher levels in transgenic lines #73 and #80 than in non-transgenic wild-type (WT) control plants (Fig. 1a; Additional file 1: Figure S2), and was compromised in OsWRKY11-kd lines #45 and #91 during pathogen attack (Fig. 1b; Additional file 1: Figure S2). Wu et al. (2009) reported that the heights of OsWRKY11-ox plants were similar to those of WT plants. Our results, however, were not consistent with this previous report. The heights of the OsWRKY11-ox plants, generated in this study and grown in a culture box (Fig. 1c) and a greenhouse (Fig. 1d), were about 60% those of the controls at the seedling stage, but had reached 80% of the controls’ height at the tillering stage (Fig. 1d). As plant height is an important agronomic trait, we examined heights of OsWRKY11-ox and OsWRKY11-kd plants grown in paddy fields. The heights of OsWRKY11-ox and OsWRKY11-kd plants were approximately 80% of the height of control plants under these conditions (Fig. 1e).
Fig. 1.
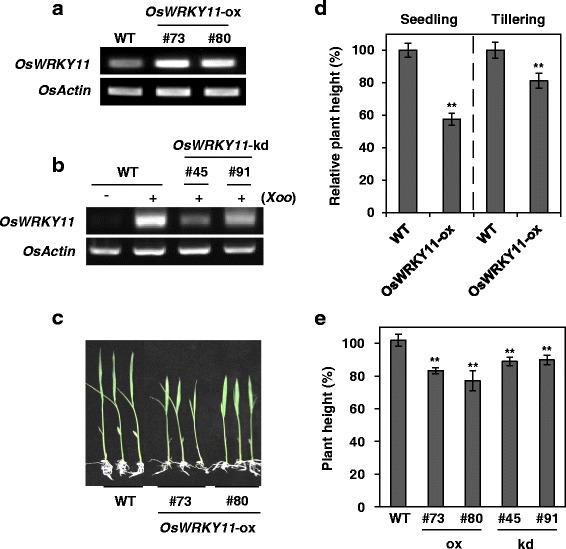
Analysis of OsWRKY11-ox and RNAi (kd) plants. OsWRKY11-ox lines and OsWRKY11-kd lines of Nipponbare cultivar were generated. Over-expression (a) and knock-down (b) of OsWRKY11 in these lines were confirmed by RT-PCR with gene-specific primers against OsWRKY11 (Table S1). Growth retardation of OsWKRY11-ox transgenic rice at the seedling and tillering stages. The photograph shows 10-day-old seedlings grown in MS medium(c). The relative heights of OsWKRY11-ox transgenic plants were obtained by comparing 10-day-old transgenic and WT seedlings grown in MS medium, and 10-week-old transgenic and WT plants at the tillering stage grown in a greenhouse (d). Growth retardation of OsWKRY11-ox and OsWRKY11-kd transgenic rice grown in rice paddy fields (e)
OsWRKY11 reduces the susceptibility to a bacterial pathogen, Xoo
To examine an effect of OsWRKY11 on bacterial blight disease caused by Xoo 10 plants from OsWRKY11-ox lines #73 and #80 and OsWRKY11-kd lines #45 and #91, respectively, were challenged with a compatible race of Xoo using the leaf-clip method. The areas of lesions on the OsWRKY11-ox lines #73 and #80 were smaller than those on the WT controls (Fig. 2a, b). By contrast, the areas of lesions on the OsWRKY11-kd lines were larger than those on the WT controls (Fig. 1e). This suggests that enhanced expression of OsWRKY11 results in a reduced susceptibility to the bacterial pathogen Xoo.
Fig. 2.
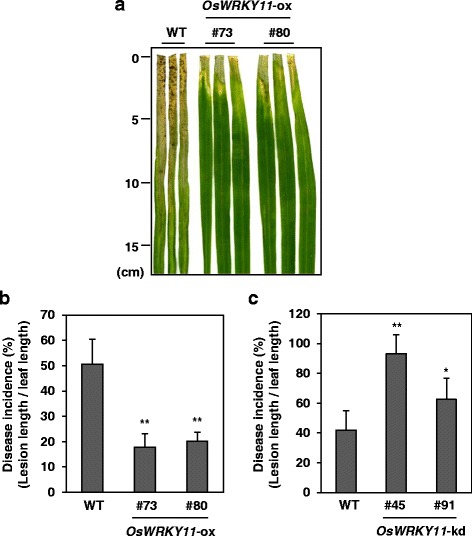
Analysis of OsWRKY11-ox and RNAi (kd) plants in response to Xoo infection. Wild-type (WT) and transgenic lines (T2) over-expressing or under-expressing OsWRKY11 were challenged with Xoo using the leaf-clip method and photographed at 14 dpi (a). Lesion lengths were measured at 14 dpi, and disease incidences for OsWRKY11-ox (b) and OsWRKY11-kd lines (c) were expressed as the percentage of lesion length/leaf length. Asterisks indicate significant differences from the WT (**: P < 0.01; *: P < 0.05)
OsWRKY11 increases the expression of defense-related genes
To elucidate the mechanism of OsWRKY11-mediated resistance to pathogens, we used RT-PCR to analyze the expression of defense-related genes. Levels of CHITINASE 2 (CHIT2), PATHOGENESIS-RELATED 10 (PR10), and Betv1 transcripts were higher in the OsWRKY11-ox plants than in WT plants before pathogen infection and increased transcript levels of these genes were maintained upon Xoo infection (Fig. 3a).
Fig. 3.
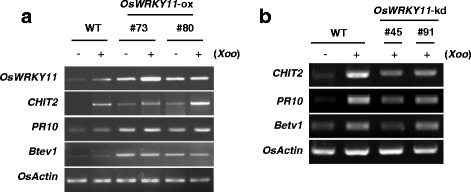
Expression analysis of defense-related genes in OsWRKY11-ox and OsWRKY11-kd lines. a Total RNA was isolated from WT plants and OsWRKY11-ox plants either infected with Xoo or non-infected. RT-PCR was performed using gene-specific primers for CHITINASE 2 (CHIT2), PR10, and Betv1. b OsWRKY11-kd plants were infected with Xoo using the toothpick inoculation method, and samples were collected at 24 hpi. Expression patterns of CHIT2, PR10, and Betv1 in OsWRKY11-kd lines were analyzed using RT-PCR with gene-specific primers. Transcript levels of OsACTIN were used as internal controls. These experiments were repeated more than twice
By contrast, induction of defense-associated genes during Xoo infection was more highly compromised in OsWRKY11-kd line #45 than in line #91. This indicated that OsWRKY11 regulated the induction of defense-associated genes such as CHIT2, PR10, and Betv1. Together with the data obtained from OsWRKY11-ox lines, these results show that OsWRKY11 regulates the expression of defense-related genes, resulting in reduction of disease susceptibility to Xoo.
OsWRKY11 trans-activates the CHIT 2 promoter by direct binding in vivo
Since subcellular localization of transcription factor is important to predict its function subcellular localization of OsWRKY11 was assessed. 35S::YFP-OsWRKY11 was generated and introduced into rice protoplasts along with 35S::NLS-RFP, a nuclear localization marker that contains a classical NLS (Choi et al. 2015). YFP::OsWRKY11 was localized exclusively in the nucleus, together with the nuclear marker NLS-RFP (Additional file 1: Figure S1a), suggesting that OsWRKY11 functions in nucleus.
Since defense-related genes were highly expressed in OsWRKY11-ox lines but their induction by pathogen infection was compromised in OsWRKY11-kd lines, we predicted that OsWRKY11 was a transcription activator. To test this hypothesis, a yeast vector (BD-OsWRKY11) was generated and transformed into yeast cells. Activity of a reporter gene was assessed (Additional file 1: Figure S1b). We found that OsWRKY11 was a transcription activator in yeast. To test this in plants, a promoter-reporter construct containing the CHIT 2 promoter upstream of a GFP-GUS fusion gene (pCHIT2::GFP-GUS) was generated. Agrobacterium-mediated transient assays were performed in N. benthamiana leaves (Fig. 4a; Additional file 1: Figure S2). Trans-activation activity of OsWRKY11 at the CHIT 2 promoter was assessed using GUS staining. GUS activity was stronger in leaves co-infiltrated with pCHIT2::GFP-GUS and 35S::OsWRKY11 than in leaves infiltrated with either pCHIT2::GFP-GUS or 35S::OsWRKY11 alone. We also performed a transient assay of promoter activity in rice protoplasts using PEG-mediated transformation (Additional file 1: Figure S3). Luciferase activity was about 2-fold higher in samples co-transformed with pCHIT2::LUC and 35S::OsWRKY11 than in those transformed with pCHIT2::LUC alone. These results suggest that OsWRKY11 trans-activates the CHIT 2 promoter in plants.
Fig. 4.
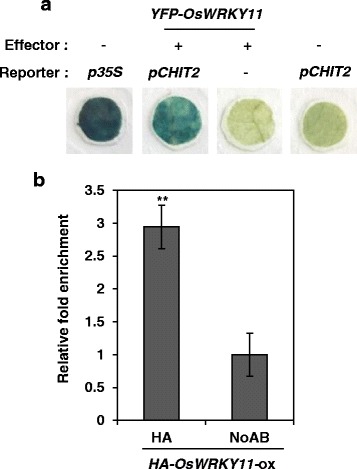
OsWRKY11 directly binds to and activates the CHITINASE 2 promoter. a Nicotiana benthamiana leaf discs were infiltrated with Agrobacterium carrying pCHIT2::GFP-GUS alone or with a mixture of Agrobacterium carrying 35S::OsWRKY11 and pCHIT2::GFP-GUS. Promoter activities in each sample were visualized 2 days post-infiltration using β-glucuronidase (GUS) activity staining. b Transgenic rice leaves expressing 35S::HA-OsWRKY11 were used for chromatin immunoprecipitation with anti-HA antibody. ChIP-PCRs were performed on genomic DNA fragments using promoter-specific primers against CHIT2 (Table S1). To normalize qPCR values, the value obtained from the sample with pre-immune serum (no antibody) was divided by the value obtained from the sample with 10% input of each sheared chromatin sample and arbitrarily set at 1. The value resulting from OsWRKY11 binding to the promoter was expressed as a relative ratio of the pre-immune serum sample. Asterisks indicate significant differences between treatments from the no-antibody control sample (**: P < 0.01; *: P < 0.05)
To examine direct binding of OsWRKY11 to the CHIT 2 promoter in vivo, transgenic lines over-expressing HA-OsWRKY11 were generated. In vivo binding of OsWRKY11 to the CHIT 2 promoter was assessed by chromatin immune-precipitation (ChIP) with anti-HA antibody followed by qPCR with primers binding near the W-box or WLE1 (Fig. 4b; Additional file 1: Figure S4). DNA binding was higher in IP samples treated with anti-HA antibody than in no-antibody samples, indicating that OsWRKY11 directly binds the CHIT 2 promoter in vivo.
OsWRKY11 positively regulates drought-responsive genes
OsWRKY11 was initially reported to be induced following infection with pathogens such as M. grisea and Xoo (Ryu et al. 2006). Later, Wu et al. (2009) reported that over-expression of OsWRKY11 under the control of a heat-inducible promoter such as HSP101 conferred enhanced tolerance of heat and drought stresses. Therefore, we performed a drought tolerance assay with the transgenic lines over-expressing OsWRKY11 under the control of the 35S constitutive promoter (Additional file 1: Figure S5a). Water was withheld from 6-week-old plants for 10 days to induce drought conditions. Transgenic lines #73 and #80, which over-expressed OsWRKY11, showed reduced leaf wilting after 8 days of drought treatment compared with WT plants (Additional file 1: Figure S5a). After 5 days of re-watering, OsWRKY11-ox plants showed a better recovery from wilting than WT plants. Our results are consistent with a previous report (Wu et al. 2009); however, those authors did not investigate the drought tolerance of OsWRKY11-kd lines. We performed a drought tolerance assay of OsWRKY11-kd lines by withholding water from 6-week-old plants for 7 days to induce drought conditions (Additional file 1: Figure S5b). After 5 days of re-watering, OsWRKY11-kd plants showed a lower recovery from wilting than WT plants. Tolerance to drought was compromised in OsWRKY11-kd lines (Additional file 1: Figure S5b). Our kd approach therefore further confirmed the effect of OsWRKY11 on drought tolerance.
The expression of drought-responsive genes in transgenic plants over-expressing OsWRKY11 has not been previously investigated (Wu et al. 2009). We therefore analyzed expression of drought-responsive genes, including dehydrins such as DIP1, DHN1, and RAB21 (Fig. 5a). These three dehydrins were constitutively expressed in transgenic OsWRKY11-ox plants. By contrast, induction of dehydrins following drought stress was compromised in OsWRKY11-kd lines (Fig. 5b). All these results indicate that OsWRKY11 positively regulates the expression of drought-responsive genes.
Fig. 5.
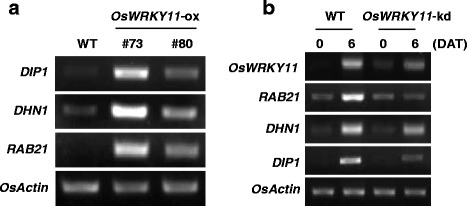
RT-PCR analysis of expression of drought-responsive genes in OsWRKY11-ox and OsWRKY11-kd plants. a Total RNA was isolated from leaves of OsWRKY11-ox plants from lines #73 and #80, and from WT controls. RT-PCR was performed using gene-specific primers against DIP1, DHN1, and RAB21. b OsWRKY11-kd and non-transgenic (WT) control plants were subjected to drought stress. Samples were collected 6 DAT. RT-PCR was performed using gene-specific primers against OsWRKY11, RAB21, DHN1, and DIP1. The transcript level of OsACTIN was used as an internal control. These experiments were repeated more than twice
OsWRKY11 trans-activates the RAB21 promoter by direct binding in vivo
We demonstrated that OsWRKY11 was a transcription activator and drought-responsive genes were up-regulated in OsWRKY11-ox lines but these effects were compromised in OsWRKY11-kd lines. To examine whether OsWRKY11 directly regulated drought-responsive genes as it did defense-related genes, we performed transient assays of promoter activity in N. benthamiana.
A promoter-reporter construct containing the RAB21 promoter upstream of a GFP-GUS fusion gene (pRAB21::GFP-GUS) was generated and transformed into Agrobacterium. Agrobacterium-mediated transient assays were performed in N. benthamiana leaves. Trans-activational activity of OsWRKY11 at the RAB21 promoter was assessed using GUS staining (Fig. 6a; Additional file 1: Figure S2). GUS activity was stronger in leaves co-infiltrated with pRAB21::GFP-GUS and 35S::OsWRKY11 than in leaves infiltrated with either pRAB21::GFP-GUS or 35S::OsWRKY11. This suggested that OsWRKY11 trans-activated the RAB21 promoter in planta.
Fig. 6.
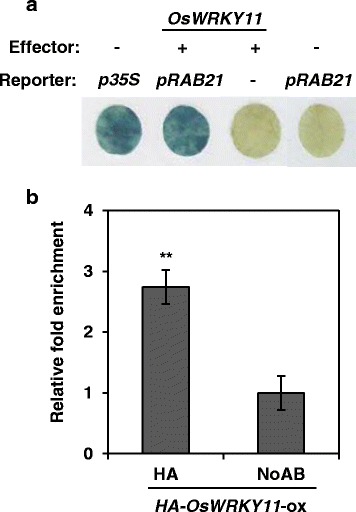
OsWRKY11 directly binds to and activates the RAB21 promoter. a Nicotiana benthamiana leaf discs were infiltrated with Agrobacterium carrying pRAB21::GFP-GUS alone or with a mixture of Agrobacterium carrying 35S::OsWRKY11 and pRAB21::GFP-GUS. Promoter activity in each sample was visualized 2 days post-infiltration using β-glucuronidase (GUS) activity staining. b Transgenic rice leaves expressing 35S::HA-OsWRKY11 were used for chromatin immunoprecipitation with anti-HA antibody. ChIP-PCRs were performed on genomic DNA fragments using promoter-specific primers against RAB21 (Table S1). To normalize qPCR values, the value obtained from the sample with pre-immune serum (no antibody) was divided by the value obtained from the sample with 10% input of each sheared chromatin sample and arbitrarily set at 1. The value obtained from OsWRKY11 binding to the promoter was expressed as a relative ratio of the pre-immune serum sample. Asterisks indicate significant differences between treatments from the no-antibody control sample (**: P < 0.01; *: P < 0.05)
In vivo binding of OsWRKY11 to the RAB21 promoter was also assessed in transgenic plants over-expressing HA-OsWRKY11 using ChIP with anti-HA antibody (Fig. 6b; Additional file 1: Figure S4). The signal obtained by PCR with primers binding sites near the W-box or WLE1 of the RAB21 promoter was higher in IP samples treated with anti-HA antibody than in no-antibody samples, suggesting that OsWRKY11 binds directly to the promoter .
OsWRKY11 protein is controlled by the Ubiquitin-Proteasome system
To prevent untimely activation of immune response transcription factors such as OsWRKY6 and OsWRKY45 are often controlled by the ubiquitin-proteasome system (UPS) (Choi et al., 2015; Matsushita et al., 2013). To examine whether OsWRKY11 was regulated by the UPS, we monitored the effect of the proteasome inhibitor MG132 on OsWRKY11 protein accumulation. GFP florescence was monitored in protoplasts transformed with 35S::YFP-OsWRKY11 (Fig. 7a). GFP florescence was stronger in protoplasts transfected with 35S::YFP-OsWRKY11 in the presence of MG132. This is suggesting that the level of OsWRKY11 may be controlled by the UPS. YFP-OsWRKY11 protein was detected by the immuno-blot analysis with a GFP antibody (Fig. 7b). OsWRKY11 protein levels were higher in the MG132-treated sample than in the mock-treated sample, while PAT protein levels were similar in both samples, again suggesting that the UPS controls levels of OsWRKY11 protein.
Fig. 7.
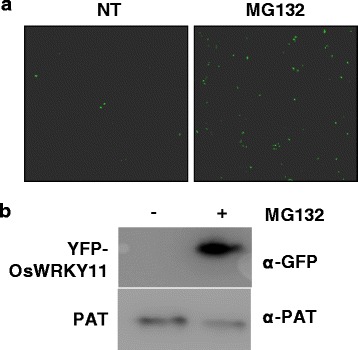
Degradation of OsWRKY11 protein by the UPS. a Rice (Oryza sativa) protoplasts were transformed with 35S::OsWRKY11-YFP. After 4 h, protoplasts were treated with 100 μM MG132 and buffer for 1 day. Fluorescence was observed using a confocal laser scanning microscope. Scale bars: 30 μm. NT: non-treated control. b Immunoblot analysis of total protoplast proteins in the absence (−) or presence (+) of 100 μM MG132. Blots were probed with green fluorescent protein (GFP) antibody. The PAT antibody was used as a control for transformation efficiency
Discussion
A number of studies show that WRKY TFs are involved in the integration of signaling pathways in biotic and abiotic stress responses negatively or positively (reviewed in Sharma et al. 2013; Kissoudis et al. 2014; Takatsuji 2017). WRKY TFs involved in positive cross-talk are thought to be good candidates for proteins conferring tolerance to abiotic and biotic stresses; however, there has been no consideration of the potential uses of these TFs in improving tolerance to abiotic and biotic stresses. This may be because the positive effects of those TFs were reported by independent research groups. Here, we report the involvement of OsWRKY11 in positive cross-talk between abiotic and biotic stress responses.
OsWRKY11 is induced by heat and drought stresses, and over-expression of OsWRKY11 under the control of the HSP101 promoter enhances drought tolerance (Shiroto et al. 2004; Wu et al. 2009). In addition, OsWRKY11 expression is induced by pathogens such as M. grisea and Xoo (Ryu et al. 2006). There are, however, no further reports on the function of OsWRKY11 in pathogen-mediated defense signaling beyond these studies of OsWRKY11 expression. The expression pattern of OsWRKY11 prompted us to consider it a good candidate for playing a role in positive cross-talk between abiotic and biotic stresses.
OsWRKY11 is a member of group IIc (Xie et al. 2005), which contains 18 OsWRKY proteins (Berri et al. 2009). An Arabidopsis protein in this group, AtWRKY75, is involved in Pi stress as well as pathogen defense (Devaiah et al. 2007; Encinas-Villarejo et al. 2009; Choi et al. 2014). OsWRKY31 (OsWRKY23, according to CGSNL) confers enhanced resistance to Pseudomonas syringae when expressed heterologously in Arabidopsis (Jing et al. 2009; Berri et al. 2009). To assess the involvement of OsWRKY11 in pathogen-mediated defense signaling transgenic lines over- or under-expressing OsWRKY11 were therefore generated. OsWRKY11-ox lines displayed reduced susceptibility to bacterial leaf blight caused by a bacterial pathogen Xoo, while this reduced susceptibility was compromised in OsWRKY11-kd lines.
Defense-associated genes, such as PR10, Betv1, and CHIT2, were constitutively expressed in OsWRKY11-ox lines, while their expression was compromised in OsWRKY11-kd lines, indicating that OsWRKY11 was a positive regulator of expression of defense-associated genes (Peng et al. 2008). There are 125 WRKY TFs in rice (Rice WRKY Working Group, 2012), and, of these, OsWRKY03 (12 by CGSNL), 6, 13, 31 (23, by CGSNL), 45, 53, 62, 71, and 89 (104, by CGSNL) are known to be positive regulators of defense-associated genes (Liu et al. 2005; Li et al. 2006; Choi et al. 2015; Chujo et al. 2007; Liu et al. 2007; Shimono et al. 2007; Qiu et al. 2007; Zhang et al. 2008; Chujo et al. 2009; Hwang et al. 2011). Out of the OsWRKY proteins, only the functions of OsWRKY6, OsWRKY13, OsWRKY45, and OsWRKY62 (Fukushima et al., 2016) have been thoroughly investigated using kd or knock-out strategies. Our analysis of OsWRKY11-kd lines confirmed that OsWRKY11 functioned as a positive regulator in the expression of defense-associated genes, thereby conferring disease resistance, in a similar manner to the other WRKY factors described above.
A number of studies show that WRKY TFs act as transcriptional activators or repressors in pathogen-mediated defense signaling. Our trans-activation assay in yeast demonstrated that OsWRKY11 functioned as a transcriptional activator. The protein contains the consensus coactivator motif LXLL (L: leucine; X: any amino acid) (Xie et al. 2005). This motif may contribute to transcriptional activation of reporter genes. TFs mostly localize to the nucleus, either by themselves or with help from other protein(s). OsWRKY11 was localized in nuclei. The putative NLS may contribute to nuclear localization of OsWRKY11 (Xie et al. 2005). In addition, we used ChIP and transient assays of promoter activity to confirm that OsWRKY11 could bind directly to the CHIT2 promoter and induce transcriptional activation in vivo. We demonstrated that OsWRKY11 trans-activated and bound around the W box or WLE1 of the CHIT2 promoter in vivo. There was no report yet that OsWRKY TF directly regulates the expression of the chitinase in vivo in our knowledge. Previously we reported that OsWRKY6 directly regulates OsPR10a promoter (Choi et al. 2015).
It was previously reported that OsWRKY11 is induced by heat and enhances drought tolerance (Wu et al. 2009). However, this study did not investigate the downstream target genes of OsWRKY11 in terms of drought tolerance. We demonstrated that over-expressing OsWRKY11 under the control of the 35S promoter resulted in enhanced tolerance of drought, as previously reported (Wu et al. 2009). Moreover, we observed constitutive expression of drought-responsive genes in OsWRKY11-ox lines and that induction of drought-responsive genes, such as RAB21, DHN1, and DIP1, was compromised in OsWRKY11-kd lines during drought stress. Our ChIP and promoter transient assays showed that direct binding of OsWRKY11 to the RAB21 promoter led to transcriptional activation in vivo. Taken together, our data indicate that OsWRKY11 is a positive regulator of drought-responsive genes, and thereby enhances drought tolerance. Each TF positively or negatively regulates a subset of stress-related genes. In line with this concept, OsWRKY11 positively regulates drought-responsive genes, including several dehydrin genes.
Positive regulators of rice immunity, such as OsNPR1, OsWRKY6, and OsWRKY45, are often subject to control by the UPS to prevent untimely activation of the immune response (Choi et al. 2015; Liu et al. 2017; Matsushita et al. 2013). OsWRKY11 was also controlled by the UPS. Plant defense responses to abiotic and biotic stresses theoretically impart a cost of reduced growth and reproduction (recently reviewed in Karasov et al. 2017; Takatsuji 2017), and thus plants are likely to develop various means to minimize growth/defense trade-offs. Degradation of immune proteins by the UPS is considered one such strategy, and therefore degradation of OsWRKY11 by the UPS may underlie the beneficial phenotypes for aspects of plant performance observed in transgenic lines over-expressing OsWRKY11, such as plant height and yield.
Conclusions
Plants are normally exposed to abiotic and biotic stresses in their natural environment. Drought stress and diseases are major causes of reduced crop yields. To overcome both stresses efficiently, plants must establish elaborate cross-talk between signaling pathways at various levels. We found that OsWRKY11 was a positive regulator of plant defense responses to drought and pathogens, including the bacterium, Xoo. Many transgenic approaches have been performed to increase biotic and abiotic stress tolerance (Kissoudis et al. 2014). Transgenic rice cultivars with improved drought tolerance and resistance to two severe diseases can increase and stabilize crop yields in stressful environments. Our data indicated that OsWRKY11 is a good candidate for improving the yield of transgenic crops. Identification of upstream factor(s) regulating OsWRKY11 is required to elucidate OsWRKY11-mediated cross-talk between the abiotic and biotic stress response pathways.
Methods
Plant materials and treatments
Transgenic rice seedlings (Oryza sativa L. ssp. japonica cv. Nipponbare) were grown in a greenhouse for 3 weeks for RT-PCR analysis, and for 6 weeks for studies of disease severity. Xoo KACC10331, a compatible strain to the Nipponbare cultivar, were grown in peptone-sucrose-agar medium (10 g peptone, 10 g sucrose, 1 g sodium-glutamate, and 15 g agar per liter) at 28 °C for 2 days and resuspended in 10 mM MgCl2 to an OD600 of 0.5.
Transgenic rice seedlings (Nipponbare cultivar) were inoculated with Xoo using a toothpick at approximately 10 mm intervals. Samples were taken at the times indicated and stored at −80 °C prior to RNA isolation and RT-PCR analysis. For RT-PCR analysis of gene expression under drought conditions, 6-week-old seedlings were drought-treated, and samples were collected approximately 6 days after treatment (dat).
Vector construction and generation of transgenic rice plants
A full-length OsWRKY11 cDNA clone was obtained from cDNA reverse transcribed from total RNA extracted from rice leaves infected with Xoo by PCR (Additional file 1: Table S1). The PCR product was amplified using primers OsWRKY11-F and R, and cloned into pDONR221 using BP clonase (Invitrogen, Carlsbad, CA) to make an entry clone. This entry clone was confirmed by sequencing.
The 35S::OsWRKY11 and 35S::HA-OsWRKY11 constructs were produced using an LR reaction between the entry clone and the destination vectors pB2GW7 (Karimi et al. 2002) and pEarleygate201 (Earley et al. 2006), respectively. The 35S::OsWRKY11 RNAi construct was produced using an LR reaction between the entry clone pDONR201-OsWRKY11-RNAi and pB7GWIW2 (II) (Karimi et al. 2002).
The three constructs were introduced into rice using Agrobacterium-mediated transformation as previously described (Kim et al., 2009) with minor modifications. Agrobacterium LBA4404 carrying each construct was used to infect rice callus generated from Oryza sativa cv. Nipponbare. Transgenic lines were generated by transforming rice with the 35S::OsWRKY11-ox construct or the 35S::OsWRKY11 RNAi construct.
RT-PCR analysis
Leaf samples were ground to powder in liquid nitrogen, and total RNA was isolated for RT-PCR, as described (Hwang et al. 2008). RT-PCR analysis using M-MLV RTase (Promega, Madison, WI, USA) was performed using 1 μg total RNA according to the manufacturer’s instructions. Subsequent PCR was performed using 25–30 cycles and gene-specific primers (Additional file 1: Table S1). OsActin (XM469569) (Hwang et al. 2008) primers were used as a loading control. All gel images are generated by a program (Quantity one-4.5.2) in gel documentation system (Bio Rad Gel Doc XR system US170–8170) and trimmed around corresponding band for each gene in size.
Disease assays
For Xoo inoculation, the bacteria were grown on peptone-sucrose-agar plates at 28 °C for 2 days and resuspended in 10 mM MgCl2 to an OD600 of 0.5 (Hwang et al. 2011). OsWRKY11-ox lines (T1) and OsWRKY11 RNAi lines (T1) were challenged with a compatible strain of Xoo strain KXO85 (KACC10331) using the leaf-clip method (Kauffman et al. 1973). Lesion lengths in each plant were measured at 14 days post-inoculation (dpi). Disease severity was expressed as a percentage of lesion length (cm)/leaf length for individual plants (Tao et al. 2009).
Promoter transient expression assay
A 1.5 kb region of the RAB21 (Os11g26790) promoter were amplified using PCR with specific primers (Additional file 1: Table S1) and introduced into an entry vector, pENTR/d-TOPO (Invitrogen, Carlsbad, CA, USA). pRAB21::GFP-GUS constructs were made by an LR reaction between the entry clone containing the pRAB21 promoter and the destination vector pBGWFS7 (Karimi et al. 2002). A 2.0 kb region of the CHITINASE 2 (CHIT2; Os04g41620) promoter were amplified using PCR with specific primers in Additional file 1: Table S1 and used for entry vector. pCHIT2::GFP-GUS constructs were made by an LR reaction between the entry clone containing the pCHIT2 promoter and the destination vector pBGWFS7. Transient expression assays in Nicotiana benthamiana were performed using the protocol reported previously (Li, X. 2011). N. benthamiana was infiltrated with Agrobacterium carrying pCHIT2::GFP-GUS and pRAB21::GFP-GUS constructs alone, or with a mixture of Agrobacterium carrying 35S::OsWRKY11 and either pCHIT2::GFP-GUS or pRAB21::GFP-GUS. Infiltrated leaves were collected 2 days post-infiltration, and promoter activities in each sample were visualized using β-glucuronidase (GUS) activity staining. Leaf disks were immersed in a solution of 1 mM 5-bromo-4-chloro-3-indolyl-b-glucuronic acid in 100 mM sodium phosphate pH 7.0 plus 0.1% Triton X-100 and incubated at 37 °C before clearing in 70% ethanol.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were generally followed as previously described (Haring et al. 2007). Transgenic rice leaves over-expressing 35S::HA-OsWRKY11 were harvested and fixed with formaldehyde under a vacuum. The chromatin was sheared by sonication and mixed with anti-HA antibody (Abcam, Cambridge, UK). Protein A agarose (Thermo Scientific, Rockford, IL, USA) was added to precipitate the DNA fragments.
ChIP-PCRs were performed using promoter-specific primers against CHIT2 and RAB21 (Additional file 1: Table S1, see Supporting Information). qPCR values were normalized with the value obtained from the sample with pre-immune serum (no antibody) divided by the value obtained from the sample containing 10% input of each sheared chromatin was arbitrarily set at 1, and subsequent values resulting from OsWRKY11 binding to the promoter were expressed as relative ratios of the pre-immune serum sample. Chromatin samples precipitated with pre-immune serum were used as negative controls in all ChIP-qPCR reactions. Similar results were obtained in three independent experiments.
Protein accumulation assay for OsWRKY11 in rice protoplasts
Protoplasts were prepared from dark-grown rice seedlings for 10 days. PEG-mediated transformation was performed as previously described (Bart et al. 2006). The vector 35S::YFP-OsWRKY11 was introduced into rice protoplasts as described previously. After 4 h, 100 μM MG132, a proteasome inhibitor was added to the protoplasts and incubated for 1 day. Green fluorescent protein (GFP) fluorescence was examined under a confocal laser scanning microscope (Olympus FV300; Olympus, Germany).
Proteins extracts were prepared in ice-cold extraction buffer [150 mM NaCl, 50 mM Tris-Cl (pH 7.5), 1 mM EDTA, 0.5% NP-40, 1 mM PMSF, 50 μM MG132, and one protease inhibitor cocktail tablet per 10 ml; Roche, Mannheim, Germany]. Proteins were separated using 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Immunoblot analysis was performed with GFP antibody (Abcam; code: ab6556). Immunoblot analysis with phosphinothricin acetyltransferase (PAT) antibody (Abcam; code: ab1791) was used as a loading control.
Acknowledgments
This work was supported in part by two grants, #PJ01087001 and #PJ01246302 from the National Institute of Agricultural Science, the Rural Development Administration to Duk-Ju Hwang.
Availability of data and materials
All relevant data are provided as Tables within the paper and in the Supporting.
Information files.
Additional file
Primers used in this study. Figure S1. Subcellular localization and transcriptional activity of OsWRKY11. Figure S2. Schematic maps of constructs used for transient assays of promoter activity. Figure S3. Activation of the CHIT 2 promoter by OsWRKY11 in rice. Figure S4. W-box and W-box-like element 1 (WLE1) depicted in CHIT2 and RAB21 promoter. Figure S5. Drought response assays for plants over (ox)- or under-expressing (RNAi-knock-down; kd) OsWRKY11. Supplementary experimental procedures. (DOCX 130 kb)
Authors’ contributions
DJH developed ideas, cordinated the experiments, and wrote the manuscript. HL, JL, CC, and CC are carried out the experiments that analyzed transgenic plants. SRP and HJ carried out disease assays. SL produced transgenic rice plants. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Heyoung Lee and Jooyoung Cha contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0199-0) contains supplementary material, which is available to authorized users.
References
- Abbruscato P, Nepusz T, Mizzi L, Del Corvo M, Morandini P, Fumasoni I, Michel C, Paccanaro A, Guiderdoni E, Schaffrath U, Morel J, Piffannelli P, Faivre-Rampant O. OsWRKY22, a monocot WRKY22 gene, plays a role in the resistance response to blast. Mol Plant Pathol. 2012;13:828–841. doi: 10.1111/j.1364-3703.2012.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N, Urwin P. The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot. 2012;63:3523–3544. doi: 10.1093/jxb/ers100. [DOI] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods. 2006;2:13. doi: 10.1186/1746-4811-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro AC, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè ME, Piffanelli P. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Hwang SH, Fang IR, Kwon SI, Park SR, Ahn I, Kim JB, Hwang DJ. Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 2015;208:846–859. doi: 10.1111/nph.13516. [DOI] [PubMed] [Google Scholar]
- Choi C, Park Y, Kwon S, Yun C, Ahn I, Park S, Hwang D. Identification of AtWRKY75 as a transcriptional regulator in the defense response to Pcc through the screening of Arabidopsis activation-tagged lines. Plant Biotechnol Rep. 2014;8:183–192. doi: 10.1007/s11816-013-0308-x. [DOI] [Google Scholar]
- Choi NY, Lee E, Lee SG, Choi CH, Park SR, Ahn I, Bae SC, Hwang CH, Hwang DJ. Genome-wide expression profiling of OsWRKY superfamily genes during infection with Xanthomonas oryzae pv. oryzae using real-time PCR. Front in. Plant Sci. 2017;8:1628. doi: 10.3389/fpls.2017.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Kato T, Yamada K, Takai R, Akimoto-Tomiyama C, Minami E, Nagamura Y, Shibuya N, Yasuda M, Nakashita H, Umemura K, Okada A, Okada K, Nojiri H, Yamane H. Characterization of an elicitor-induced rice WRKY gene, OsWRKY71. Biosci Biotechnol Biochem. 2008;72:240–245. doi: 10.1271/bbb.70553. [DOI] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, Umemura K, Okada A, Okada K, Nojiri H, Yamane H. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta. 2007;1769:497–505. doi: 10.1016/j.bbaexp.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Dai X, Wang Y, Zhang W. OsWRKY74, a WRKY transcription factor, modulatestolerance to phosphate starvation in rice. J Exp Bot. 2016;67:947–960. doi: 10.1093/jxb/erv515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007;14:1789–1801. doi: 10.1104/pp.106.093971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Jeremy RH, Olga P, Kristen O, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Encinas-Villarejo S, Maldonado AM, Amil-Ruiz F, de los Santos B, Romero F, Pliego-Alfaro R, Mun˜oz-Blanco J, Caballero JL. Evidence for a positive regulatory role of strawberry (fragaria 9 ananassa) FaWRKY1 and Arabidopsis AtWRKY75 proteins in resistance. J Exp Bot. 2009;60:3043–3065. doi: 10.1093/jxb/erp152. [DOI] [PubMed] [Google Scholar]
- Fukushima S, Mori M, Sugano S, Takatsuji H. Transcription factor WRKY62 plays a role in pathogen defense and hypoxia-responsive gene expression in rice. Plant Cell Physiol. 2016;57:2541–2551. doi: 10.1093/pcp/pcw185. [DOI] [PubMed] [Google Scholar]
- Goto S, Sasakura-Shimoda F, Suetsugu M, Selvaraj MG, Hayashi N, Yamazaki M, Ishitani M, Shimono M, Sugano S, Matsushita A, Tanabata T, Takatsuji H. Development of disease-resistant rice by optimized expression of WRKY45. Plant Biotech J. 2015;13:753–765. doi: 10.1111/pbi.12303. [DOI] [PubMed] [Google Scholar]
- Han M, Kim CY, Lee J, Lee SK, Jeon JS. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in Rice. Mol Cells. 2014;37:532–539. doi: 10.14348/molcells.2014.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Ryu HS, Kim CY, Park DS, Ahn YK, Jeon JS. OsWRKY30 is a transcription activator that enhances rice resistance to the Xanthomonas oryzae pv. oryzae. J Plant Biol. 2013;56:258–265. doi: 10.1007/s12374-013-0160-0. [DOI] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:1–12. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Kwon SI, Jang JY, Fang IR, Lee H, Choi C, Park SR, Ahn I, Bae S, Hwang DJ. OsWRKY51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas oryzae pv. oryzae. Plat Cell Rep. 2016;35:1975–1985. doi: 10.1007/s00299-016-2012-0. [DOI] [PubMed] [Google Scholar]
- Hwang SH, Lee IA, Yie SW, Hwang DJ. Identification of an OsPR10a promoter region responsive to salicylic acid. Planta. 2008;227:1141–1150. doi: 10.1007/s00425-007-0687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Yie SW, Hwang DJ. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 2011;181:316–323. doi: 10.1016/j.plantsci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Jing SJ, Zhou X, Song Y, Yu DQ. Heterologous expression of OsWRKY23 gene enhances pathogen defense and dark-induced leaf senescence in Arabidopsis. Plant Growth Regul. 2009;58:181–190. doi: 10.1007/s10725-009-9366-z. [DOI] [Google Scholar]
- Karasov TL, Chae E, Herman JJ, Bergelson J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell. 2017;29:666–680. doi: 10.1105/tpc.16.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kauffman HE, Reddy APK, Hsieh SPV, Merca SD. An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Kim C, Vo K, Nguyen C, Jeong D, Lee S, Kumar M, Kim S, Park S, Kim JK, Jeon JS. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol Rep. 2016;10:13–23. doi: 10.1007/s11816-015-0383-2. [DOI] [Google Scholar]
- Kim JA, Cho K, Singh R, Jung YH, Jeong SH, Kim SH, Lee JE, Cho YS, Agrawal GK, Rakwal R, Tamogami S, Kersten B, Jeon JS, An G, Jwa NS. Rice OsACDR1 (Oryza sativa accelerated cell death and resistance 1) is a potential positive regulator of fungal disease resistance. Mol Cells. 2009;28:431–439. doi: 10.1007/s10059-009-0161-5. [DOI] [PubMed] [Google Scholar]
- Kissoudis C, Wiel C, Visser R, Linden G. Enhancing crop resilience to combined abiotic and biotic stress through the dissection of physiological and molecular crosstalk. Front Plant Sci. 2014;5:1–20. doi: 10.3389/fpls.2014.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan A, Huang J, Zhao W, Peng Y, Chen Z, Kang D. A salicylicacid-induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana. Plant Biol. 2013;15:452–446. doi: 10.1111/j.1438-8677.2012.00664.x. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Tapio PE. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006;46:477–491. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- Li X (2011) Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-protocol Bio101 e95. 10.21769/BioProtoc.95
- Liu Q, Ning Y, Zhang Y, Yu N, Zhao C, Zhan X, Wu W, Chen D, Wei X, Wang G, Cheng S, Cao L. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell. 2017;29:345–359. doi: 10.1105/tpc.16.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Bai XQ, Qian Q, Wang XJ, Chen MS, Chu CC. OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 2005;15:593–603. doi: 10.1038/sj.cr.7290329. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Bai XQ, Wang XJ, Chu CC. OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol. 2007;164:969–979. doi: 10.1016/j.jplph.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H. Nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J. 2013;73:302–313. doi: 10.1111/tpj.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. Constitutive expression of rice WRKY30 gene increases the endog-enous jasmonic acid accumulation, PR gene expression and resis-tance to fungal pathogens in rice. Planta. 2012;236:1485–1498. doi: 10.1007/s00425-012-1698-7. [DOI] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant. 2008;1:446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice diseasistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. MPMI. 2007;5:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol Plant. 2008;1:538–551. doi: 10.1093/mp/ssn012. [DOI] [PubMed] [Google Scholar]
- Raineri J, Wang S, Peleg Z, Blumwald E, Chan RL. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol Biol. 2015;88:401–413. doi: 10.1007/s11103-015-0329-7. [DOI] [PubMed] [Google Scholar]
- Rice WRKY Working Group Nomenclature report on rice WRKY’s-conflict regarding gene names and its solution. Rice. 2012;5:1–3. doi: 10.1186/1939-8433-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu HS, Han M, Lee SK, Cho JI, Ryoo N, Heu S, Lee YH, Bhoo SH, Wang GL, Hahn TR, Jeon JS. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25:836–847. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- Seo YS, Chern M, Bartley LE, Han M, Jung KH, Le I, Walia H, Richter T, Xu X, Cao P, Bai W, Ramanan R, Amoupant F, Arul L, Canlas P, Ruan R, Park C, Chen X, Hwang S, Jeon JS, Ronald P. Towards establishment of a rice stresss response interactome. PLoS Genet. 2011;7(4):e1002020. doi: 10.1371/journal.pgen.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaik R, Ramakrishna W. Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candi- date genes for broad resistance in rice. Plant Physiol. 2014;164:481–495. doi: 10.1104/pp.113.225862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, DeVleesschauwer D, Sharma MK, Ronald PC. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6:250–260. doi: 10.1093/mp/sss147. [DOI] [PubMed] [Google Scholar]
- Shen H, Liu C, Zhang Y, Meng X, Zhou X, Chu C, Wang X. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol Biol. 2012;80:241–253. doi: 10.1007/s11103-012-9941-y. [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY 45 plays a crucial role in benzothiadiazole-inducilde blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroto Y, Toriyama K, Kishitani S. Gene expression analysis of transcription factors in rice plants subjected to drought, heat or combination of both stresses. Breed Res. 2004;6(Suppl 2):129. [Google Scholar]
- Takatsuji H. Regulating tradeoffs to improve rice production. Front Plant Sci. 2017;8:171. doi: 10.3389/fpls.2017.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot. 2011;62:4863–4874. doi: 10.1093/jxb/err144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009;151:936–948. doi: 10.1104/pp.109.145623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Yoshida R, Kishi-Kaboshi M, Matsushita A, Jiang CJ, Goto S, Takahashi A, Hirochika H, Takatsuji H. Abiotic stresses antagonize the rice defense pathway through the tyrosine-dephosphorylation of OsMPK6. PLoS Pathog. 2015;11:e1005231. doi: 10.1371/journal.ppat.1005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007;65:799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Meng J, Peng X, Tang X, Zhou P, Xiang J, Deng X. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol Biol. 2015;89:157–171. doi: 10.1007/s11103-015-0360-8. [DOI] [PubMed] [Google Scholar]
- Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- Xiao J, Cheng H, Li X, Xiao J, Xu C, Wang S. Rice WRKY13 regulates crosstalk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 2013;163:1868–1882. doi: 10.1104/pp.113.226019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005;137:176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003;15:745–759. doi: 10.1105/tpc.008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y. OsWRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot. 2013;64:5085–5097. doi: 10.1093/jxb/ert298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008;18:508–521. doi: 10.1038/cr.2007.104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are provided as Tables within the paper and in the Supporting.
Information files.


