Abstract
YidC/Oxa1/Alb3 are essential proteins that operate independently or cooperatively with the Sec machinery during membrane protein insertion in bacteria, archaea and eukaryotic organelles. Although the interaction between the bacterial SecYEG translocon and YidC has been observed in multiple studies, it is still unknown which domains of YidC are in contact with the SecYEG translocon. By in vivo and in vitro site-directed and para-formaldehyde cross-linking we identified the auxiliary transmembrane domain 1 of E. coli YidC as a major contact site for SecY and SecG. Additional SecY contacts were observed for the tightly packed globular domain and the C1 loop of YidC, which reveals that the hydrophilic cavity of YidC faces the lateral gate of SecY. Surprisingly, YidC-SecYEG contacts were only observed when YidC and SecYEG were present at about stoichiometric concentrations, suggesting that the YidC-SecYEG contact in vivo is either very transient or only observed for a very small SecYEG sub-population. This is different for the YidC-SRP and YidC-FtsY interaction, which involves the C1 loop of YidC and is efficiently observed even at sub-stoichiometric concentrations of SRP/FtsY. In summary, our data provide a first detailed view on how YidC interacts with the SecYEG translocon and the SRP-targeting machinery.
Introduction
The ability to transport proteins from their site of synthesis to their site of function is essential for prokaryotic and eukaryotic cells and involves universally conserved protein machineries that can transport a wide variety of potential substrates1–4. An example is the Sec translocon that is found as Sec 61αβγ complex in the endoplasmic reticulum membrane or as SecYEG complex in the bacterial cytoplasmic membrane5,6. SecY is composed of 10 transmembrane domains (TMs) and organized as two vestibules with a central constriction, called the pore ring, giving it an overall hour-glass like shape7. The constriction is further sealed on the periplasmic side by a small helix, called the plug, which likely helps to maintain the permeability barrier of the protein conducting channel during translocation8,9. During protein transport, SecY can open laterally towards the lipid phase by movement of TMs 2B, 3, 7 and 8, which constitute the so called lateral gate10–12. SecE in E. coli consists of three TMs, which surround the back of SecY and appears to stabilize SecY within the membrane13,14. The bacterial SecG contains two TMs and a connecting cytosolic loop and its function is primarily linked to the SecA-dependent translocation of secretory proteins6,15.
Although the trimeric SecYEG complex constitutes the minimal membrane embedded unit required for protein transport16,17, it associates with multiple partner proteins. This includes SecA, the membrane-associated receptor for secretory proteins18–22, and FtsY, the membrane-associated SRP receptor23–25. In addition to these membrane-associated partner proteins, SecYEG also interacts with membrane integral proteins, like YidC, the SecDFYajC complex, PpiD or YfgM15,26–28.
YidC contacts the four TMs of the lateral gate of SecY but also penetrates into the SecY channel interior where it interacts with pore ring residues29,30. The orientation of YidC at the lateral gate probably allows YidC to assist the release of substrates from the SecY channel into the lipid phase31,32. This involves conformational changes at the SecY-YidC interface, including the retraction of YidC from the SecY channel and a reorientation of YidC at the lateral gate29,30. YidC consists of a conserved five TM core, which forms a hydrophilic groove that is accessible from both the cytosol and the lipid phase4,33,34 (Fig. 1a). In E. coli, the five TM core is N-terminally preceded by an additional TM that is connected to TM2 via a large periplasmic domain (the P1 loop) of about 320 amino acids. Two short helices, the C1 loop, connect TM2 and TM3 and are required for the in vivo function of YidC35. The positively charged C2 loop, connecting TM4 and TM5 constitutes together with the C-terminus of YidC a composite ribosome binding site35. YidC is about five-times more abundant than SecYEG1 and functions both in complex with SecYEG27 but also as SecYEG-independent insertase for some membrane proteins36–38.
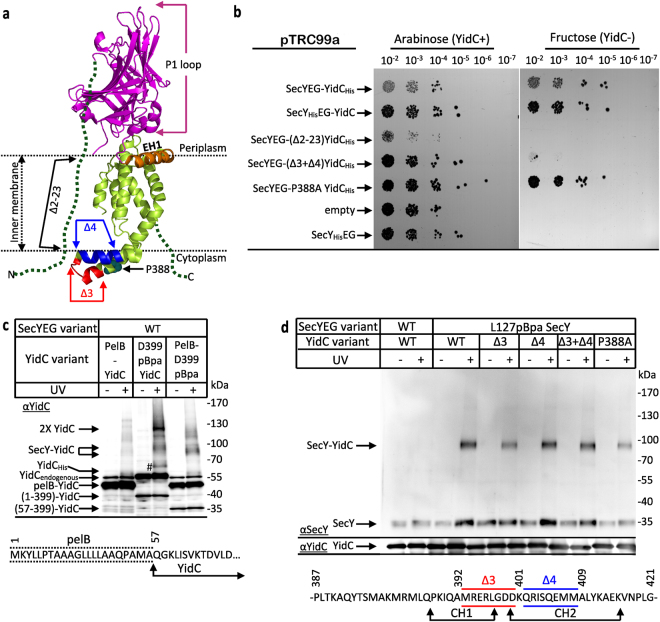
Figure 1.
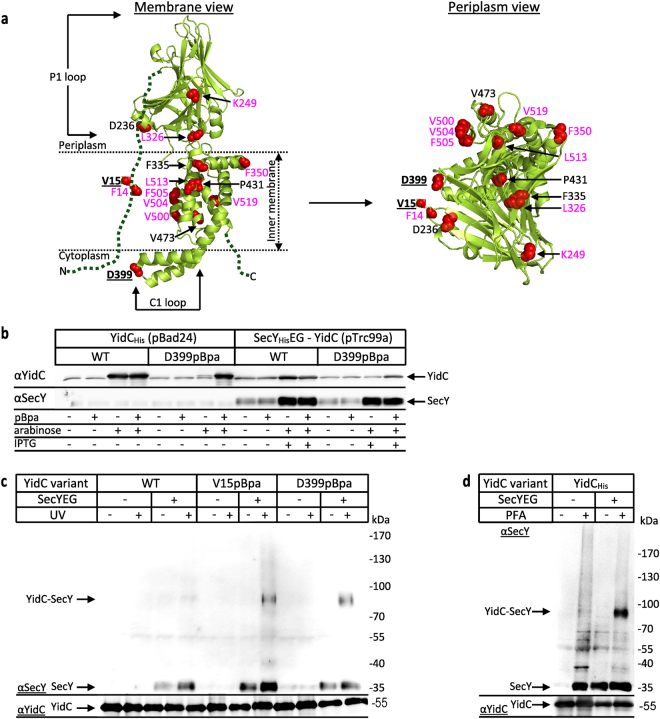
YidC contacts the SecYEG translocon via TM1 and the C1-loop in vivo. (a) The crystal structure of E. coli YidC (PDB accession no.: 3WFV) visualised from the membrane (left) or from the periplasmic side (right). The red spheres indicate the positions for pBpa insertion. Residues which show the strongest contacts to SecY are displayed in bold and underlined. Residues with weaker contacts are shown in black and residues that did not show significant cross-links are shown in magenta. The dashed green lines indicate TM1 and the C-terminus of YidC, which have not been crystallized so far. (b) The co-expression system shows balanced expression of YidC and SecYEG as revealed by western blotting. 1 × 108 E. coli BL21 cells expressing yidC under the arabinose promoter from plasmid pBad24 or co-expressing PlacsecYEG-yidC from plasmid pTrc99a were TCA precipitated, separated by SDS-PAGE and after western transfer decorated with the indicated antibodies. WT refers to wild type YidC and D399pBpa to a YidC variant with pBpa inserted at position 399, when pBpa was added to the growth medium. (c) In vivo photo-cross-linking performed with BL21 E coli cells expressing either yidC alone (-SecYEG) or co-expressing yidC and secYEG (+SecYEG). Either wild type YidC (WT) or YidC variants containing pBpa at position V15 or D399 were analysed. After UV-exposure, samples were purified via metal affinity chromatography using an N-terminal His-tag on YidC. A sample without UV-exposure served as a control. Samples were decorated with antibodies against SecY or YidC as indicated. The 95 kDa YidC-SecY cross-link is indicated. (d) In vivo para-formaldehyde (PFA) cross-linking with BL21 cells expressing either only YidC from plasmid pBad24 or YidC together with SecYEG from plasmid pTrc99a. All samples were treated as described in experimental procedures and YidC was further purified from the membrane fraction via its N-terminal His-tag and analyzed by western blotting using α-SecY antibodies for detecting SecY-YidC cross-links. α-YidC antibodies determined comparable amounts of YidC in all samples. Uncropped images are displayed in Supplementary Figure S4.
The contact between YidC and SecYEG is modulated by the SecDFYajC-complex39,40, although the SecY-YidC contact is maintained even in the absence of SecDF29,30. SecD and SecF are multi-spanning membrane proteins with large periplasmic loops41, while YajC is a small, single spanning membrane protein42. The SecDFYajC complex likely functions as proton-motive-force powered chaperone complex that aids ATP-independent steps during protein translocation41.
The available data indicate the presence of multiple SecYEG translocon assemblies within the membrane, which likely exist in a dynamic equilibrium4. The SecYEG-SecDFYajC-YidC complex is the most complex system and referred to as the holo-translocon (HTL)43,44. For some substrates it was shown that they are more efficiently transported by the HTL than by SecYEG alone44. However, considering the low abundance of SecDF in E. coli (approx. 50 copies versus approx. 500 SecYEG copies1), only a small number of HTLs can exist in E. coli, while there would be enough YidC (2500 copies) to attach to the majority of SecYEG complexes.
Our current understanding of how these accessory proteins connect to the SecYEG core is limited by the lack of high-resolution structures. A low resolution structure of the HTL points to the presence of a lipid-filled cavity at the SecYEG-YidC interface44, which could provide a shielded environment for membrane protein insertion and folding. However, it is still unknown which regions of YidC are in contact with SecYEG. Here, we mapped the YidC-SecYEG interface in E. coli and identified TM1 and the cytosolic C1 loop of YidC as major contact sites for the SecYEG translocon.
Results
The first transmembrane domain and the cytosolic loop of YidC constitute the primary contact site for SecY
By using site-directed cross-linking the SecY positions which are in contact with YidC have been mapped29,30. However, using the same approach and inserting the UV-sensitive amino acid derivative para-benzoyl-L-phenylalanine (pBpa)45 into different positions within YidC failed to identify by immune detection YidC residues that contact SecY. Only by combining cross-linking with mass spectrometry (MS), the C-terminus of YidC (position YidC540) was found to be in contact with SecY29. This was verified in the current study by using the amine-specific cross-linker Disuccinimidyl suberate (DSS) in combination with MS. The identification of the cross-linked peptides revealed that the C-terminus of YidC (residue K545) cross-links to the N-terminus of SecY (position A2) (Table 1). An additional cross-link to the N-terminus of SecY (position K3) was found from the cytosolic loop C1 of YidC (residue 413) (Table 1). These data demonstrated that the C-terminus of YidC and the cytosolic loop C1 (Fig. 1a) are in contact with the N-terminus of SecY. The data further showed that the N-terminal methionine residue of SecY is obviously cleaved, resulting in a free amino group of the succeeding alanine residue.
Table 1.
The N-terminus of SecY contacts the C-terminus and the cytosolic loop C1 of YidC.
| cross-link | cross-linked SecY peptide |
cross-linked YidC peptide |
calculated mass (Da) | mass deviation (ppm) | E-value |
|---|---|---|---|---|---|
| SecY(2)-YidC(545) | 2AKQPGLDFQSAK13 | 539GLHSREKK546 | 2380.2862 | 2.5 | 0.0012 |
| SecY(3)-YidC(413) | 2AKQPGLDFQSAK13 | 402QRISQEMMALYKAEK416 | 3251.6681 | 0.6 | 0.0247 |
E. coli BL21 cells carrying pTrc99a-SecYHisEG was used for SecYHisEG purification via metal affinity chromatography. The amine-specific cross-linker DSS was either added before SecYHisEG elution (on-column cross-linking) or after elution with elution buffer (solution cross-linking). Subsequently, samples were subjected to HPLC-MS. Cross-linked amino acids in the respective peptides are underlined and numbers in subscript indicate the position of the identified peptide within the protein. Identification of cross-linked peptides was performed essentially as described77 using the program pLink®79 for searching inter- or intra-molecular cross-links generated by amine-specific cross-linker DSS between peptides from SecY and YidC. The resulting set of peptide spectrum matches at a false discovery rate <0.05 was further filtered requiring a minimum E-value of 0.05 and identification of a pair of cross-linked residues in at least two independent experiments.
Attempts to verify these pBpa or DSS cross-links by immune detection were unsuccessful, suggesting that the experimental conditions allowed only for low-abundant YidC-SecYEG cross-links. This could reflect either a certain flexibility of YidC on the SecY surface, or alternatively, that the stoichiometry between both proteins prevented a more efficient cross-linking. In E. coli, YidC is about 5 times more abundant than the SecYEG complex1, but performing pBpa or DSS cross-linking with a plasmid-encoded SecYEG results in a surplus of SecYEG over YidC. On the contrary, using plasmid-encoded pBpa-containing YidC further increases the excess of YidC over SecYEG, resulting in only a small number of possible YidC-SecYEG complexes. To overcome this, a co-expression system was established that allowed simultaneous expression of YidC and SecYEG. This system was analyzed for both wild type YidC and for a YidC variant that contained pBpa at position 399 of the C1 loop. The amount of YidC in the co-expression system was lower than in the previously used single YidC expression system (pBad24-YidC), and required for YidC(D399pBpa) the addition of pBpa to the medium for efficient expression (Fig. 1b, compare lanes 7 and 8; and lanes 15 and 16). Without pBpa addition, translation stops at the amber stop codon and only a truncated YidC is synthesized. Importantly, the co-expression system allowed for a significantly increased and IPTG-dependent SecYEG production, compared to the wild type SecYEG levels present in the single expression system (Fig. 1b).
The consequences of simultaneous co-expression of YidC and SecYEG on the in vivo cross-linking were analyzed for two YidC variants, which contained pBpa either at position 399 or at position 15 within TM1 of YidC (Fig. 1a,c). TM1 was selected because it is unique to enterobacteria like E. coli and not part of the conserved YidC/Oxa core46. After UV-exposure of E. coli cells, the cross-linked material was purified via an N-terminal His-tag on YidC and probed with α-SecY antibodies. UV-dependent cross-linking products at approx. 95 kDa were detected for both YidC(V15pBpa) and YidC(D399pBpa), but only when SecYEG was co-expressed (Fig. 1c, upper panel). The observed 95 kDa band is in agreement with the previously observed SecY-YidC cross-linking product that was obtained when pBpa was inserted into SecY and which was verified by MS as SecY-YidC complex29. The same pBpa-containing YidC variants did not show detectable cross-linking products when expressed without SecYEG co-expression, i.e. at the endogenous wild type SecYEG levels (Fig. 1c). This was not related to significant differences in the amount of YidC (Fig. 1c, lower panel), but rather indicates that efficient SecYEG-YidC complex formation as visualized by cross-linking is only observed when SecYEG and YidC are present at about equal concentrations.
An intrinsic limitation of the position-dependent cross-linking is that even subtle changes in pBpa orientation can affect cross-linking. To overcome this limitation and to validate that YidC-SecY cross-links indeed require stoichiometric concentrations of both partner proteins, the amine-group specific, zero-length cross-linker para-formaldehyde (PFA) was used47. When cells expressing either YidC or SecYEG-YidC were treated with PFA in vivo and YidC was purified via its His-tag, the 95 kDa cross-link product was clearly detected by α-SecY antibodies in the co-expression system, but not when only YidC was expressed (Fig. 1d). This further verifies that about stoichiometric YidC/SecYEG amounts are required for efficient cross-linking. The 95 kDa band was not specifically recognized by the α-YidC antibodies (Supplementary Figure 4), because all YidC cross-linking products were purified via the His-tag purification, rendering a specific detection of a single cross-link species by the α-YidC antibodies unnecessary.
Incidentally, even though no SecY-YidC cross-link was observed without SecYEG co-expression, significant co-purification of native SecY with His-tagged YidC was noticed when PFA was added (Fig. 1d, compare SecY levels). This points to a PFA-stabilized SecYEG-YidC complex, in which SecY and YidC are not in direct contact but rather via accessory subunits of either SecYEG or YidC. In summary, the co-expression system revealed that YidC contacts SecY via TM1 and the C1 loop, which is also in agreement with the DSS cross-linking (Table 1).
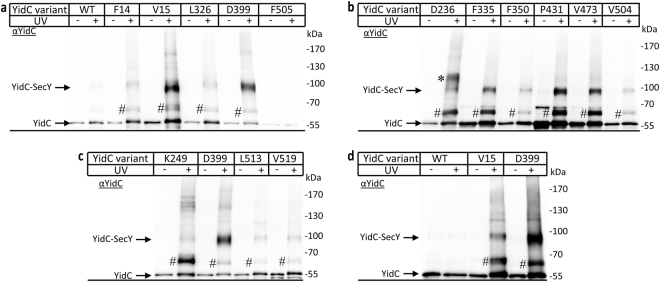
The co-expression system was then used for identifying additional YidC-SecY contacts. We selected surface exposed residues within the hydrophobic core and within the periplasmic domain of YidC (Fig. 1a). Compared to the strong cross-links at residues V15 and D399, only weak cross-links were observed for residues F14 within TM1, L326 within P1 and F505 in TM5 (Fig. 2a). However, in vivo UV exposure of cells expressing wild-type YidC (i.e. without pBpa insertion) also resulted in a very weak 95 kDa cross-linking product (Fig. 2a). This is probably the result of UV-dependent radical formation of aromatic amino acids that favors non-specific protein-protein and protein-nucleic acid cross-links48,49. Thus, the weak cross-linking products do not necessarily reflect site-specific contacts. Strong 95 kDa YidC-SecY cross-links were observed for residues F335, P431 and V473 and weaker cross-links for residues D236, F350 and V504 by α-YidC antibodies (Fig. 2b). Additional weak SecY contacts were observed for residue K249 in P1 and residues L513 and V519 within TM6 (Fig. 2c). The strong YidC-SecY cross-links were also detected by α-SecY antibodies (Supplementary Fig. S1), but the quality of western blots using α-SecY antibodies is generally lower, and requires extensive purification of the cross-linking products.
Figure 2.
YidC makes multiple contacts to SecY through its transmembrane and periplasmic domains. In vivo photo cross-linking was performed as described in Fig. 1 using the co-expression system with YidC variants containing pBpa at the indicated positions. After UV-exposure, samples were purified via the C-terminal His-tag on SecY and analysed on western blot with α-YidC antibodies (a–c). The 95 kDa YidC-SecY cross-links are indicated. For position D236, an additional strong band below the 130 kDa was recognized by α-YidC antibodies (*). In addition, α-YidC antibodies recognized a UV-dependent band at approx. 65 kDa (#). (d) In vitro photo cross-linking was performed using inner membrane vesicles carrying 10 μg of wild type YidC or YidC-pBpa variants co-expressed with SecYHisEG. Samples were purified by metal affinity chromatography using the His-tag on SecY and subsequently analysed on western blot with α-YidC antibodies. Uncropped images are displayed in Supplementary Figure S4.
Residue D236 within the P1 loop showed an additional cross-link product below the 130 kDa marker band (Fig. 2b,∗), which is likely identical to the about 130 kDa SecY-YidC cross-link product that was identified by MS when pBpa was incorporated at position 127 within TM3 of SecY29. It could correspond to a YidC-(SecY)2 complex, however, as the appearance of the 130 kDa cross-linking product appeared to be strain dependent, i.e. it is visible in strains C43 and BL21 but not in BL32529,30, this was not further analyzed. Finally, for all residues showing a clear YidC-SecY cross-linking product, an additional cross-link migrating below the 70 kDa marker band was recognized by α-YidC antibodies (Fig. 2#; see below for its characterization).
In summary, these data revealed that YidC makes multiple contacts to SecY via TM1 (V15), the periplasmic domain (F335), the C1 loop (D399), TM3 (P431) and TM4 (V473) (Fig. 1a). Additional weak contacts were observed for F14 (TM1), D236/L326/F350 (P1), V504 (TM5) and L513/V519 (TM6). However the intensity of these weaker cross-links was identical or only slightly stronger than those observed for wild type YidC and thus do not necessarily reflect position-dependent cross-links. Making quantitative assessments based on non-equilibrium methods like cross-linking is difficult, but the intensity of the YidC-SecY cross-linking product from the different pBpa positions suggested that TM1 and the C1 loop are major contact sites for SecY. This was further verified by in vitro cross-linking using purified inner membrane vesicles (INV) of E. coli cells co-expressing SecYEG and YidC(pBpa), which also revealed strong contacts between SecY and YidC residues V15 and D399 (Fig. 2d).
Position dependent contacts of YidC with SecE and SecG
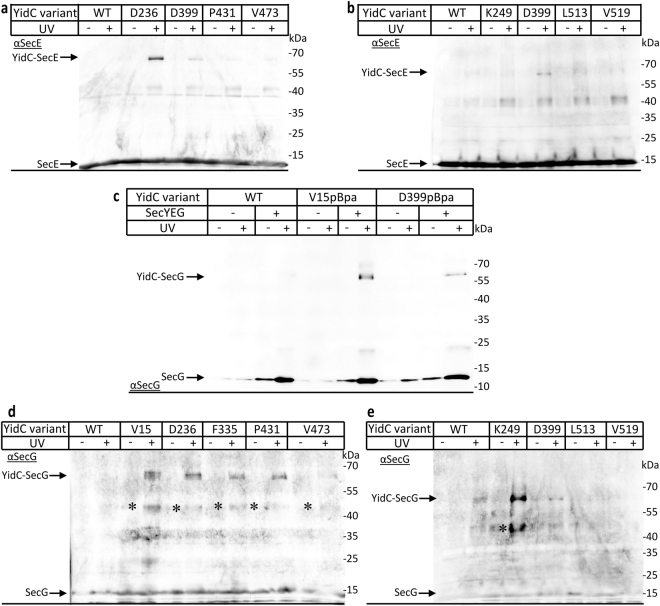
The α-YidC antibodies recognized not only the 95 kDa band but also a UV-dependent band at approx. 65 kDa (c.f. Fig. 2,#) and the intensity of this band largely coincided with the intensity of the 95 kDa SecY-YidC cross-link. We therefore reasoned that the 65 kDa band could reflect YidC cross-links to either SecE or SecG, which both have a MW of about 13 kDa13. α-SecE antibodies clearly recognized the 65 kDa band in the YidC(D236pBa) variant and weakly also for YidC(D399pBpa) (Fig. 3a). pBpa insertions at the other residues (F14, V15, L326, F335, F350, P431, V473,V504) did not show the 65 kDa band or only at the detection limit of the α-SecE antibodies (Fig. 3 and Supplementary Figure S2). These data demonstrate that the YidC-SecE contact primarily involves the periplasmic loop of YidC (D236) and weakly the cytoplasmic region C1, but no intra-membrane residues.
Figure 3.
YidC contacts the SecY associated proteins SecE and SecG. The cross-linked and purified material prepared for Figs 1c and 2a,c was decorated with antibodies against SecE (a,b) or SecG (c–e). (c) The samples were analysed with antibodies against SecG both at the endogenous SecYEG levels (-SecYEG) and in the co-expression system (+SecYEG). The cross-link products are indicated. The UV dependent 45 kDa bands (*) in Fig. 2d–e likely represent a proteolytic cleavage product of the YidC-SecG cross-links. Uncropped images are displayed in Supplementary Figure S4.
As the 65 kDa band was not recognized by α-SecE antibodies for all YidC(pBpa) variants, we assumed that for some YidC residues, it reflects a YidC-SecG cross-link. α-SecG antibodies recognized the 65 kDa band for V15 and also with lower intensity for D399 in the co-expression system, but not at the endogenous SecYEG levels (Fig. 3c). YidC-SecG cross-links were also observed for residues D236, K249, F335, P431 and (weakly) for V473 (Fig. 3d,e). Residues F350, V504, L513 and V519 gave no cross-linking product (Fig. 3e and Supplementary Figure S2). A UV-dependent band at 45 kDa was recognized by the α-SecG antibody in some samples (Fig. 3,*). This band could reflect a proteolytic fragment of the YidC-SecG cross-link product, but this was not further analyzed in the current study. Thus, YidC contacts SecG via TM1 (V15), the periplasmic loop (D236, F335, K249) and TM4 (P431), while no significant contacts were observed for pBpa insertions in TM5 and TM6.
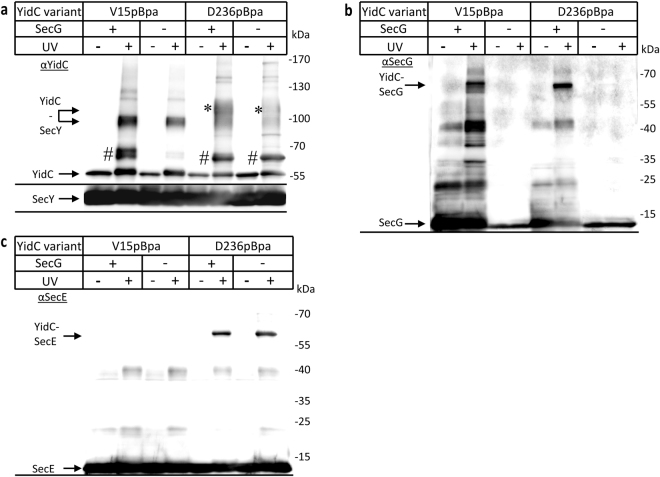
While SecE is essential for the stability of SecY14, SecG can be deleted50. This allowed us to further differentiate between YidC-SecE and YidC-SecG cross-links. YidC(pBpa) variants were either co-expressed with SecYEG or with just SecYE. In the absence of stoichiometric SecG amounts, the 95 kDa YidC-SecY cross-link was detected by α-YidC antibodies for YidC(V15pBpa), indicating that TM1 of YidC maintains contact to SecY even in the absence of SecG (Fig. 4a). This was different for YidC(D236pBpa), which showed a significant reduction of both the 95 kDa and the 130 kDa cross-linking products when SecG was not co-expressed (Fig. 4a). Thus, the presence of SecG appears to enhance the interaction of the P1 loop of YidC with SecY. The 65 kDa band that reflects either a YidC-SecE or a YidC-SecG cross-link was largely absent for YidC(V15pBpa), but unchanged for YidC(D236pBpa) when SecG was not co-expressed (Fig. 4a,#).
Figure 4.
The periplasmic loop of YidC contacts SecE and SecG, while TM1 is only in contact with SecG. In vivo photo-cross-linking was performed with a SecYHisEG-YidC or SecYHisE-YidC co-expression system with pBpa incorporated either in TM1 (position 15) or within the P1 loop (position 236) of YidC. After purification via the C-terminal His-tag on SecY, the samples were analysed using antibodies against YidC (a), SecG (b) or SecE (c). The 95 kDa YidC-SecY cross-link and the 65 kDa YidC-SecE/G cross-links are indicated (#). For pBpa-insertion at position 236 of YidC, in addition to the 95 kDa YidC-SecY cross-link, a second cross-link product below the 130 kDa marker was observed (*), as already shown in Fig. 2a. Uncropped images are displayed in Supplementary Figure S4.
α-SecG antibodies recognized the 65 kDa cross-link band only when SecG was present in stoichiometric amounts (Fig. 4b). In contrast, α-SecE antibodies recognized the 65 kDa band for position D236 both in the absence and presence of SecG (Fig. 4c), while they failed to detect a band at 65 kDa for position V15 in YidC. These results confirm that the 65 kDa cross-link band corresponds to both YidC-SecE and YidC-SecG complexes and reveal that TM1 of YidC makes exclusive contact to SecG, while the P1 loop is contact with both SecE and SecG.
The C1 loop is essential for YidC function but not essential for the interaction with SecYEG
The C1 loop of YidC is in contact with SecY and SecE and we therefore determined whether it is essential for the YidC-SecYEG interaction. The loop consists of two helices (CH1 and CH2), which are required for the in vivo function of YidC35,51. YidC loop mutants were generated in which residues 392–401 of CH1 (YidCΔ3) or residues 401–409 of CH2 (YidCΔ4) were deleted (Fig. 5a,d). In addition, a YidC mutant was generated which lacked residues 392–409 (YidCΔ3 + Δ4) and a mutant in which the conserved proline residue at position 388 was replaced by alanine. The latter is expected to change the orientation of the loop (Fig. 5a). The functionality of these YidC mutants was tested in the conditional YidC-depletion strain JS713136, which contains yidC under the control of the arabinose promotor and requires arabinose-supplementation for growth. When this strain was transformed with the empty vector (pTrc99a) or pTrc99a-SecYHisEG, it grew only on arabinose-supplemented plates (Fig. 5b, 6th and 7th row). In contrast, cells containing the co-expression plasmid pTrc99a-SecYEG-YidC grew both in the presence and absence of arabinose, validating that the co-expression system produced active YidC. Complementation was observed for both the His-tagged and non-His-tagged YidC (Fig. 5b, 1st and 2nd row), demonstrating that the N-terminal His-tag on YidC does not interfere with function. Arabinose-independent growth was also observed for the YidC(P388A) mutant (Fig. 5b, 5th row), indicating that this particular residue is not essential for YidC function. However, JS7131 cells expressing YidC(Δ3 + Δ4) were arabinose dependent, showing that the C1 loop is required for YidC function (Fig. 5b, 4th row), which is in agreement with previous studies51. We also constructed a YidC variant lacking TM1 (YidCΔ2-23) and JS7131 expressing this mutant was also strictly arabinose dependent (Fig. 5b, 3rd row), initially suggesting that TM1 is essential for YidC function. However, further proteolysis experiments revealed that YidCΔ2-23, although expressed (Supplementary Figure S3a), is not properly inserted into the E. coli membrane and therefore this mutant was not further characterized in the current study. Instead, we generated a YidC variant in which TM1 was replaced by the cleavable PelB signal sequence (Fig. 5c). When co-expressed with SecYEG, this construct complemented the JS7131 strain (Supplementary Figure S3b), confirming that TM1 is not essential under the conditions tested52. Immune-detection of these cells revealed expression of pelB-YidC and signal sequence cleavage, as indicated by the faster migration on SDS-PAGE (Fig. 5c). We then tested whether the YidC-SecY contact in the co-expression system was influenced by the absence of TM1. pBpa was inserted at position 399 of either YidC or pelB-YidC and cells were UV-exposed. Detection of the YidC-SecY cross-links was then performed without further purification, i.e. in crude cell extracts. The YidC-SecY cross-link was observed for pelB-YidC(D399pBpa), migrating slightly below the 95 kDa cross-link observed for YidC(D399 pBpa) (Fig. 5c). However, the 65 kDa SecE/SecG cross-link products (Fig. 5c,#) were not clearly detected with pelB-YidC. These data demonstrate that the YidC-SecY contact is maintained even when TM1 of YidC is missing, while contact to SecE and SecG is reduced.
Figure 5.
The C1-loop and TM1 form a composite binding site for SecY. (a) The positions of the partial YidC deletions used in (b) (Δ3 + Δ4 and Δ2-23) and (d) (Δ3, Δ4, Δ3 + Δ4) as well as the position of proline 388 which was mutated to alanine (P388A) (b,d) are indicated on the E. coli YidC crystal structure (PDB accession no. 3WFV). In addition, the amphipathic helix EH1 (orange) and the periplasmic loop 1 (magenta) are displayed. The dashed lines indicate TM1 and the C-terminus of YidC for which no structural information is available. (b) The conditional YidC depletion strain JS7131 was used to monitor the functionality of different pTrc99a-YidC-SecYEG co-expression systems. The presence of arabinose in the media results in expression of the endogenous YidC via the araBAD promoter while the absence of arabinose (replaced by fructose) induces YidC depletion. SecYHisEG-YidC corresponds to a co-expression system with a C-terminal His-tag on SecY, SecYEG-YidCHis to the co-expression system with a N-terminal His-tag on YidC; SecYEG-YidCHis-(Δ2-23)YidCHis to a co-expression system in which YidC lacked TM1 (residues 2–23), SecYEG-(Δ3 + Δ4)YidCHis to a co-expression system in which most of the C1 loop was deleted, SecYEG-(P388A)YidCHis to a co-expression system in which proline 388 of YidC was replaced by alanine. The empty plasmid (pTrc99a) or pTrc99a expressing just SecYHisEG served as control. Cells were sequentially diluted in LB medium and spotted onto LB plates with or without arabinose; cell growth was monitored after overnight incubation at 37 °C (upper panel). (c) In vivo photo-cross-linking was performed in the co-expression system with a YidC variant in which TM1 was replaced by the PelB signal sequence (PelB-YidC). When indicated pBpa was inserted at position 399 of either PelB-YidC (PelB-D399pBpa) or wild type YidC (D399pBpa-YidC). After UV-exposure of 1 × 108 cells, the material was not further purified but directly TCA precipitated and analysed by western blotting using α-YidC antibodies. Indicated are the cross-links (SecY-YidC), the endogenous YidC (YidCendogenous), PelB-YidC and the truncated YidC variants produced by incomplete suppression of the amber stop codon at position 399. In addition, a YidC dimer is indicated (2 × YidC). The N-terminal sequence of the PelB-YidC variant is displayed in the lower panel. The 65 kDa cross-link products between YidC and SecE/SecG are indicated (#) (d) In vivo photo-cross-linking was performed in the co-expression system with the indicated YidC variants. YidC was purified by metal affinity chromatography via an N-terminal His-tag on YidC and further analysed by western blotting with antibodies against SecY and YidC. The sequence information of the deletion mutants used in this experiment is displayed in the lower panel. CH1 and CH2 correspond to the two α-helices of the C1-loop. Uncropped images are displayed in Supplementary Figure S4.
The impact of the C1-mutants on the SecYEG-YidC interaction was also monitored in the co-expression system. Here, pBpa was inserted into position L127 of SecY, which is located in TM3 of SecY, one of the lateral gate helices that have been found to contact YidC29. For all C1 loop mutations, the SecY-YidC cross-linking band at 95 kDa was still observed (Fig. 5d), demonstrating that the YidC-SecYEG contact via the lateral gate of SecY is maintained even when the C1 loop of YidC is truncated. In summary, these data indicate that TM1 and the C1-loop constitute a composite binding site for SecY and that either one is sufficient to maintain the YidC-SecY interaction.
The cytosolic loop of YidC also binds SRP and the SRP receptor
The C1 loop seems to be dispensable for the insertion of some, but not all YidC substrates. As some substrates are targeted to YidC by the SRP system35,53, we analysed whether the C1 loop is involved in binding SRP or the SRP receptor FtsY. These cross-linking experiments were performed in the single expression system, i.e. without simultaneous SecYEG co-expression. The rationale here was to monitor only direct contacts to YidC and not those that were mediated by initial binding of SRP/FtsY to SecYEG in SecYEG-YidC complexes.
The cross-linked material from cells expressing YidC(D399pBpa) was first probed with antibodies against Ffh (48 kDa), the protein component of the bacterial SRP. A UV-dependent band was recognized by the α-Ffh antibody at approx. 110 kDa, in line with the predicted mass of a YidC-Ffh complex (Fig. 6a) and demonstrating that SRP interacts with the C1 loop of YidC. Whether the C1 loop was essential for the contact between YidC and SRP was analysed by using PFA cross-linking in vivo. After PFA cross-linking, YidC was purified and the material analysed by immune detection. α-Ffh antibodies recognized two PFA-dependent bands, the 110 kDa band that was also seen by pBpa-cross-linking and a second band at approx. 150 kDa (Fig. 6b). Different to pBpa, PFA can induce cross-links between multiple components and therefore the 150 kDa band likely reflects an Ffh-4.5 S RNA-YidC complex. Together with Ffh, the 35 kDa 4.5 S RNA forms the E. coli SRP54. The additional band would also be consistent with a SecY-YidC-Ffh cross-link. However, we consider this less likely, because the detection of a SecY-YidC complex by PFA cross-linking required the SecYEG-YidC co-expression and antibodies against SecY failed to detect a 150 kDa band (c.f. Fig. 1d), The PFA approach was then tested for YidC(Δ3 + Δ4), but we observed no significant reduction in the Ffh cross-linking efficiency compared to wild type YidC (Fig. 6c).
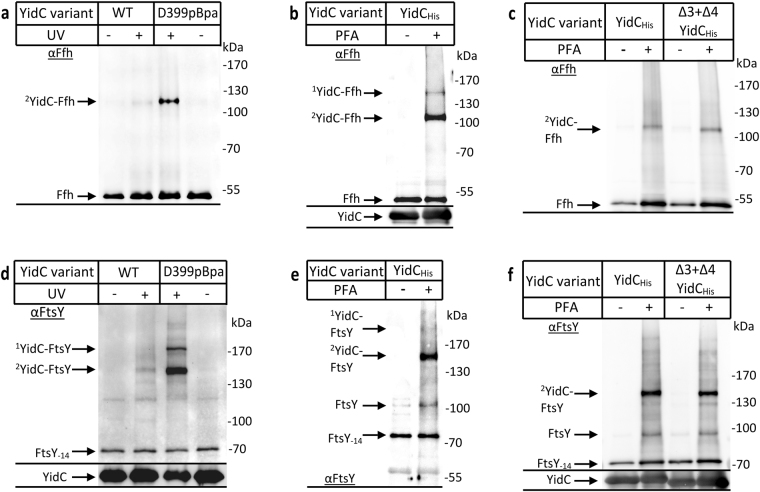
Figure 6.
The cytosolic loop 2 of YidC constitutes a binding site for SRP and its receptor FtsY. (a) In vivo photo-cross-linking was performed in the single-expression system using BL21 cells expressing yidC with pBpa at position 399. Subsequently YidC was purified via metal affinity chromatography and analysed on western blot with antibodies directed against Ffh, the protein subunit of the E. coli SRP. The 110 kDa YidC-Ffh cross-link is indicated. Note, a very weak, apparently also UV-dependent band was recognized by α-Ffh antibodies in the wild type. (b) BL21 cells expressing YidC were treated in vivo with p-formaldehyde (PFA) or buffer as a control. Subsequently, YidC was purified together with its cross-linking partners by metal affinity chromatography and analysed on western blot with α-Ffh antibodies. The 1YidC-Ffh species likely corresponds to a YidC-Ffh-4.5SRNA cross-link product and the 2YidC-Ffh species to the YidC-Ffh cross-link. (c) As in (b), but with cells expressing either wild type YidC or the YidC(Δ3 + Δ4) variant, which lacks most of the C1-loop. (d) The same material shown in (a) was decorated with α-FtsY antibodies, which revealed two cross-link products migrating at about 150 kDa and 180 kDa. FtsY-14 corresponds to the proteolytic cleavage product of FtsY, which is frequently observed. (e,f) The same material as in (b,c) was decorated with α-FtsY antibodies. Uncropped images are displayed in Supplementary Figure S4.
The same strategy was also used for monitoring possible interaction between the C1 loop and FtsY. α-FtsY antibodies recognized two UV dependent bands at approx. 150 kDa and 180 kDa for YidC(D399pBpa) (Fig. 6d). FtsY exists in two isoforms migrating at 100 kDa and 70 kDa, respectively55,56 and the two cross-link species probably reflect contacts of YidC with both isoforms, although only the proteolytically processed 70 kDa FtsY-14 isoform co-purified with YidC (Fig. 6d). The 180 kDa cross-link species would also fit in size with a SecY-YidC-FtsY complex, but SecY antibodies failed to recognize a 180 kDa complex and pBpa within YidC can only cross-link to one partner protein. PFA-cross-linking confirmed the presence of a strong 150 kDa and a weak 180 kDa YidC-FtsY complex (Fig. 6e), but as for Ffh this contact was maintained even in the YidC(Δ3 + Δ4) mutant (Fig. 6f). Thus, although the C1 loop is important for YidC interactions and essential for function, it is not essential for the interactions with SecYEG, FtsY, or SRP.
In contrast to the SecYEG-YidC interaction, which required their simultaneous co-expression to be detected, the interaction between Ffh/FtsY and YidC was detectable at the endogenous Ffh/FtsY levels. As Ffh is considered to exist in only about 200–300 copies per cell1, a significant portion of the cellular Ffh is obviously in direct contact with YidC. This interaction was still detectable when SecYEG was co-expressed, but became slightly weaker (Fig. 7a). Although direct contacts between SecYEG and Ffh have not been analysed so far, the likely explanation for the reduction is that a portion of Ffh is now recruited to SecYEG.
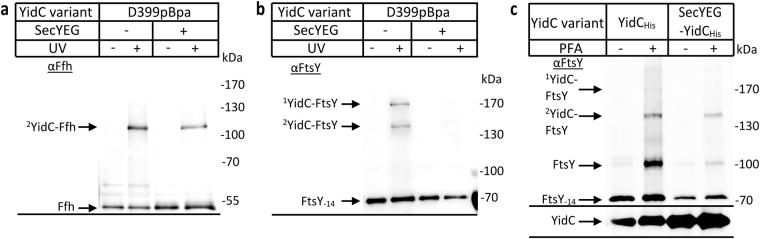
Figure 7.
The presence of SecYEG prevents binding of FtsY to the C1 loop of YidC. In vivo photo cross-linking was performed with BL21 cells expressing either just YidC or together with SecYEG. In both cases pBpa was present at position 399 of YidC. YidC was purified via its His-tag and analysed using antibodies against Ffh (a) or FtsY (b). Note that the YidC amounts which are present in (a) and (b) are displayed in Fig. 1c (α-YidC -bottom). (c) In vivo p-formaldehyde cross-linking was performed with BL21 cells expressing YidC only or together with SecYEG. The cross-linked material was analysed with α-FtsY (top) and α-YidC (bottom) antibodies. Uncropped images are displayed in Supplementary Figure S4.
Different to Ffh, FtsY is a well-studied partner protein of SecYEG23,25,57 and FtsY contact to position D399 of YidC was lost when SecYEG was co-expressed (Fig. 7b). Thus, in the presence of increased SecYEG concentrations, the majority of FtsY is obviously recruited to SecYEG. Alternatively, as the C1 loop also provides a binding site for SecYEG, the C1 loop could be re-oriented or shielded in the YidC-SecYEG complex, preventing cross-links to FtsY from position D399 of YidC. This was further analysed by PFA cross-linking, which revealed a significant reduction of the YidC-FtsY interaction when SecYEG was co-expressed (Fig. 7c). This position-independent cross-linking experiment supports the conclusion that the majority of FtsY is recruited to SecYEG in the co-expression system, suggesting that FtsY has a higher affinity for SecY than for YidC.
Discussion
Our study has revealed two unexpected aspects of the SecYEG-YidC assembly: (1) The non-conserved TM1 of E. coli YidC provides a major contact site for SecY and SecG and (2) efficient contacts between YidC and SecYEG as visualized by cross-linking are only observed when SecYEG is overexpressed. In addition, our data demonstrate that the C1 loop is an important hotspot for the interaction with SecYEG, SRP and FtsY.
YidC-like proteins facilitate protein transport in all three domains of life36,58–60 and they work autonomously for some substrates36, but also cooperate with the SecYEG translocon during membrane protein insertion27,43. In the resting SecYEG channel, YidC contacts the lateral gate and also penetrates deeply into the channel. Upon substrate insertion, YidC is expelled from the channel, but maintains contact to the lateral gate29,30, likely providing a sheltered substrate binding surface for TMs exiting SecY61,62. Our understanding on how YidC interacts with the SecYEG translocon is so far limited because the regions within YidC that contact the Sec translocon have not been identified, with the exception of a SecF binding site within the P1 loop40. Based on a genetic screen, residue G355 at the interface of P1 and TM2 and M471 within TM4 were predicted to contact SecY63. This is in line with the assumption that the conserved hydrophilic substrate binding groove of YidC probably faces the SecY lateral gate34. Our data now show that TM1 of YidC constitutes together with the C1 loop the major contact site for the SecYEG translocon. The exact localization of TM1 in E. coli YidC has not been resolved yet, but cysteine scanning mutagenesis has revealed that TM1 is involved in substrate binding, together with TMs 3–564,65. This indicates that TM1 is located close to the YidC core, although the connecting P1 loop likely provides some flexibility. TM1 is the most likely candidate to enter the interior of the SecY channel, because the other YidC TMs are tightly packed together at the periplasmic leaflet of the membrane62 and movements of these TMs would probably disrupt the hydrophilic substrate binding groove. The proposed flexibility of TM1 would also explain why cross-links to both SecY and SecG were observed. It is important to note that the in vivo cross-linking approach is not synchronized and thus monitors the YidC-SecY contact at different interaction states. This also explains why multiple surface exposed residues of YidC were found to contact SecY. Previous data had already demonstrated that YidC undergoes significant rearrangements at the SecY surface upon substrate insertion29,30 and this is likely represented by the number of cross-linking residues. As cross-linking reports on the proximity between two molecules, the weaker cross-link species could also result from a random collision between SecY and YidC. Still, the major contacts at TM1 and C1 were also confirmed by in vitro cross-linking using INVs, representing a resting state. TM1 and the P1 loop of E. coli YidC comprise 356 amino acids, yet they are largely dispensable for YidC function. Deleting TM1 and replacing it with a cleavable signal sequence or deleting most of the P1 loop, including the SecF binding site, does not inhibit YidC function40,66. The auxiliary TM1 and the periplasmic loop are also not present in YidC homologues from Gram-positive bacteria67, yet the B. subtilis YidC homologs can support SecY-dependent and –independent protein insertion in E. coli68. Chloroplast Alb3 also lacks TM1 but was still shown to interact with the chloroplast SecY59. Our data now show that YidC contains a conserved binding site for SecY within the C1 loop and a second, less-conserved in TM1. The latter appears to be a specific trait in Gram-negative bacteria. Detailed studies on the cooperation of SecYEG with YidC during membrane protein insertion have so far only been performed in E. coli and the identification of TM1 as SecY contact site could suggest that YidC-assisted protein insertion via SecYEG in E. coli is different from the analogous processes in Gram-positive bacteria or chloroplasts.
The C1 loop of YidC is essential for function, but its exact role is unknown. Deletion of the C1 loop interferes with the insertion of the Pf3 and M13 procoat proteins, but not with the insertion of Foc or CyoA35,69. We show here that the C1 loop is an important contact site for SecY, SRP and FtsY. We also demonstrate that the C-terminus of YidC, which is not resolved in the X-ray structure34, is in contact with SecY (Table 1). Together with previous data demonstrating that the C-terminus of YidC is also in contact with SRP, FtsY and ribosomes53 and that the C2 loop and the C-terminus form a composite ribosome binding site35, these data support the concept that the cytosolically exposed loops and the C-terminus of YidC constitute a composite docking site for multiple partner proteins. This is similar to the cytosolic loops of SecY, which are in contact with the ribosome, FtsY and SecA23,57.
Additional contacts to SecY were observed from YidC’s amphipathic helix EH1, which lies parallel to the plane of the membrane and which connects the P1 loop with TM234. EH1 and the P1 loop were also cross-linked to SecG, supporting the concept that the large periplasmic region of YidC serves as interface for the interaction with SecYEG. Finally, strong contacts to SecY were observed from TM3 and TM4, while cross-links from TM5 were weaker and no cross-links to TM6 were observed. TM3 and TM4 are part of the hydrophilic groove and their vicinity to SecY, which binds to YidC via its lateral gate29,30, provides evidence for the concept that substrate TMs are released from the SecY channel into the hydrophilic groove of YidC. TM5 is also part of the hydrophilic groove, but the weaker cross-links to SecY are probably explained by the fact that the selected residues 504 and 505 are located on the more tightly packed periplasmic side of the membrane, which makes these residues less accessible. These multiple contacts between YidC and SecY also explain why the truncation of the C1 loop or the replacement of the TM1 with an unrelated signal sequence does not preclude YidC-SecY contacts.
Although the results of our current and previous cross-linking experiments29,30 allow for a deeper insight into the YidC-SecYEG interaction, it is puzzling that SecYEG-YidC cross-links were only observed by immune-detection when SecYEG was overexpressed. This indicates that at the endogenous SecYEG concentrations, only a small number of SecYEG-YidC complexes exist that are not detected by cross-linking and immune-detection. On the other hand, we observed clear cross-links between YidC and the endogenous Ffh, although the cellular Ffh concentration is only half of the SecYEG concentration1. Thus, the YidC-SecYEG cross-linking efficiency is limited because most of the endogenous SecYEG complexes are either not accessible to YidC or because SecYEG only transiently associates with YidC during membrane protein insertion. Direct YidC-SecYEG interactions have so far primarily been observed when SecYEG was overproduced27,43,44; to our knowledge SecYEG-YidC complexes in native membranes were only detected by Blue-native studies26,70, and required the presence of a stalled membrane protein substrate70. This is in line with the sequential hand-over of a radioactively-labelled nascent membrane protein from SecY to YidC that was also observed with native membranes31,32,71. Thus, in native membranes YidC appears to interact with SecY only during membrane protein insertion. Whether SecYEG translocons not engaged in membrane protein insertion exist just as trimeric SecYEG core or in complex with other Sec translocon interacting proteins26,72 is currently unknown. The observation that YidC-SecYEG cross-links are only observed when SecYEG is overexpressed could indicate that under native conditions SecYEG is primarily in contact with partner proteins other than YidC. Only by overexpression sufficient SecYEG is available to form stable complexes with YidC, which otherwise would only transiently exist. The identification of the complete SecYEG interactome will probably reveal further partner proteins and extend the concept of a single holo-translocon43 to the presence of variable SecYEG assemblies, which transiently or stably recruit additional partner proteins for facilitating transport of the huge variety of exported proteins in bacteria.
Methods
Strains, growth conditions, plasmids and plasmid construction
The following E. coli strains and plasmids were used: DH5α73, BL21(Merck, Darmstadt, Germany), JS713136, MC410074, pEVol75, pBad24-YidC; pBad24-YidCHis; pTrc99a-SecYHisEG57 and pTrc99a-SecYEGHis-YidC. Cells were grown in LB medium at either 30 °C or 37 °C; supplemented with 0.2% arabinose for JS7131. For generating a His-tag free SecYEG, the coding sequence for the C-Terminal His-tag in pTrc99a-SecYHisEG was removed using the Phusion PCR Kit (NE Biolabs, Frankfurt, Germany) with 5′ phosphorylated oligonucleotides SecY_HisR_for and SecY_HisR_rev (Supplementary Table S1), resulting in plasmid pTrc99a-SecYEG. TAG stop codons were incorporated at the indicated positions of pTRc99a-SecY(His)EG or pBad24-YidCHis using the Phusion PCR Kit or the Pfu Ultra II High Fidelity Polymerase Kit (Agilent, Stratagene, Germany) with 5′-phosphorylated oligonucleotides. pBad24-YidC was constructed by PCR amplification of yidC using genomic E. coli MC4100 DNA and corresponding oligonucleotide primer (Supplementary Table S1) that inserted NcoI and Xba1 restriction sites. The PCR product was then cloned via NcoI/Xba1 into pBad2476. Next, the coding sequence for ten consecutive histidine residues, followed by a gly-ser-gly linker were inserted immediately after the ATG start codon of yidC by reverse PCR. This resulted in plasmid pBad24-YidCHis. pTrc99a-SecYHisEG was used as a template for the generation of pTrc99a-SecYHisEG-YidC and pTrc99a-SecYEG-YidCHis co-expression systems. The His10-Gly-Ser-Gly-yidC coding sequence insert was amplified from plasmid pBad24-YidCHis using primers that contained SalI (5′) and XbaI (3′) restriction sites and the restricted PCR fragment was cloned into pTrc99a-SecYHisEG. In the resulting pTrc99a-SecYHisEG-YidCHis plasmid, yidC contains its own ribosome binding site (AGGAGG) and is located 44 base pairs downstream of the secG stop-codon. Subsequently, the His-tag from SecYHis was removed using inverse PCR and the primers SecY_HisR_for and SecY_HisR_rev primers which restored the wild type SecY sequence. Alternatively, the His-tag from YidCHis was removed using the YidC_HisR_for and YidC_HisR_rev primers.
pTrc99a-SecYHisEG-YidC was used as a template for the generation of all YidC-pBpa derivatives used in Figs 1b, 2, 3, 4, S1 and S2, as well as for the construction of the SecG deletion used in Fig. 4. pTrc99a-SecYEG-YidCHis was used as template for the production of YidC-amber stop codon variants which were used in Figs 1c, 5d and 7. Insertion of the TAG stop codon at position 127 of SecY was made by reverse PCR using the primer SecY_I91_for and SecY_I91_rev with pTrc99a-SecYEG-YidCHis as template. The generation of C2 loop deletions Δ3, Δ4, Δ3 + Δ4, the ΔTM1 strain and the point mutation P388A was constructed by reverse PCR using the pTrc99a-SecY(L127TAG)EG-YidCHis as template DNA. The pTrc99a-SecYEG-YidCHis construct was also used as template for generating the pelB-YidC variant. The PelB-YidC-for and PelB-YidC-rev primers (Supplementary Table S1) containing the pelB coding sequence were used for replacement of the N-terminal His10-Gly-Ser-Gly region and the ATG start codon of His10-Gly-Ser-Gly-yidC via reverse PCR. Subsequently the ATG start codon was inserted in front of the pelB sequence via reverse PCR using the Rest.pelB-for and Rest.pelB-rev primers (Supplementary Table S1).
Activity assays for YidC derivatives
The functionality of YidC variants was determined by transforming the indicated plasmids into the conditional YidC-depletion strain JS713136. An overnight LB culture supplemented with 50 μg/μl ampicillin, 50 µg/µl spectinomycin, and 0.2% L-arabinose (for YidC expression) was used to inoculate fresh LB cultures containing 50 μg/μl ampicillin, 50 µg/µl spectinomycin and 0.2% arabinose. These cultures were grown until they reached an OD600 = 0.5 and serially diluted with fresh LB medium in multi-well plates. Subsequently, 20 µl of these dilutions were spotted on LB agar plates containing either 0.2% L-arabinose (for YidC expression) or 0.2% fructose (for YidC depletion) and were grown overnight at 37 °C. The functionality of pelB-YidC variant was also assessed by transformation of the pTrc99a_SecYEG-(pelB-YidC) plasmid in the conditional YidC depletion strain JS7131 and overnight growth at 37 °C on LB agar plates containing 50 µg/µl spectinomycin and 0.2% L-arabinose (YidC+) or 0.2% Fructose (YidC−).
In vivo pBpa cross-linking
BL21 cells containing the pEVol plasmid together with YidC-pBpa variants in pBad24 or pTrc99a plasmids were cultured overnight in LB medium at 37 °C. 10 ml of the overnight culture was further used for inoculation of 1 l LB medium containing 0.5 mM pBpa (in NaOH), 50 μg/μl of ampicillin and 35 μg/μl of chloramphenicol. The cultures were further incubated at 37 °C until they reached the early exponential growth phase (OD600 = 0.5–0.8) and induced with 0.02% L-arabinose (for pEVol and pBad24 plasmids) and/or 0.5 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG, for pTrc99a plasmids). After induction, the cultures were grown for 2 hours at 37 °C, cooled down on ice for 10 minutes and harvested by centrifugation at 5000 rpm in a JLA 9.1000 rotor for 10 minutes. The cell pellets were resuspended in 10 ml of PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.76 mM KH2PO4) and divided in two multi-well plates. One plate was exposed to UV light on ice for 20 minutes (UV chamber: BLX-365, from Vilber Lourmat) while the other plate was kept in the dark. After UV irradiation, the cell suspension was transferred to 50 ml Falcon tubes and cells were collected by centrifugation at 5000 rpm for 10 minutes in a table-top centrifuge. Each cell pellet was resuspended in 10 ml of resuspension buffer (50 mM Tris/HCl pH 7.5, 300 mM NaCl, 10 mM Mg(Ac)2) for subsequent YidC purification or 20 mM Tris/HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2 for subsequent SecY purification). Next, the samples were lysed by French pressing and the cell debris was removed by centrifugation at 15800 rpm for 30 minutes in a SS34 rotor. The supernatant was further centrifuged at 45000 rpm for 1.5 hours in a TLA 50.2 rotor for membrane sedimentation. Next, the membrane pellets were solubilized in 1% (w/v) n-dodecyl β-D-maltoside (DDM) dissolved in resuspension buffer supplemented with 10% (w/v) glycerol for 1 hour at 4 °C. YidC or SecY were purified via metal affinity chromatography using TALON® Metal affinity resin (Clontech, USA). The samples were incubated for 1 hour at 4 °C with 1 ml/1 l LB culture of pre-equilibrated TALON® Metal affinity resin, washed five times with 10 ml/sample of washing buffer (40 mM Imidazol, 10% glycerol (w/v), 50 mM Tris/HCl, pH 7.5, 300 mM NaCl, 10 mM Mg(Ac)2 and 0.03% DDM for YidC purification or 5 mM Imidazol, 10% (w/v) glycerol, 20 mM Tris/HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2 and 0.03% DDM for SecY purification) and eluted four times in a total volume of 2 ml elution buffer (200 mM Imidazole, 10% glycerol (w/v), 50 mM Tris/HCl pH 7.5, 300 mM NaCl, 10 mM Mg(Ac)2 and 0.03% DDM for YidC purification or 200 mM Imidazole, 10% (w/v) glycerol, 10 ml of 20 mM Tris/HCl pH 7.5, 300 mM NaCl, 5 mM MgCl2 and 0.03% DDM for SecY purification). Next the samples were precipitated with 1 volume of 10% trichloracetic acid (TCA), denatured at 56 °C for 10 minutes in 35 μl of TCA loading dye (prepared by mixing one part of Solution III (1 M Dithiothreitol) with 4 parts of Solution II (6.33% SDS (w/v), 0.083 M Tris-Base, 30% glycerol and 0.053% Bromphenol blue) and 5 parts of Solution I (0.2 M Tris, 0.02 M EDTA pH 8) and analysed on SDS Page by western blotting or mass-spectrometry.
In vivo para-formaldehyde cross-linking
BL21 cells carrying the pBad24-YidCHis or the pTrc99a-SecYEG-YidCHis plasmids were used for inoculation of two flasks containing 1 l of LB medium with 50 µg/ml ampicillin. Both cultures were grown at 37 °C until they reached early exponential growth phase (OD600 = 0.5–0.8) and were then induced with 0.02% L-arabinose (pBad24-YidCHis) or 0.5 mM IPTG (pTrc99a-SecYEG-YidCHis). The cells were further grown at 37 °C for 2 hours to allow expression. Next, 50 ml of freshly dissolved para-formaldehyde (PFA) in para-formaldehyde buffer (PFA buffer) (2.7 mM KCl, 1.8 mM KH2PO4, 10 mM Na2HPO4, pH 6.5, final PFA concentration 1 g PFA/50 ml PFA buffer) was added to one flask, while only PFA buffer was added to the other flask. Both samples were further incubated at 37 °C for 30 minutes, cooled down on ice for 10 minutes and harvested by centrifugation in a JLA 9.1000 rotor at 5.000 rpm for 10 minutes. Subsequently the cell pellets were dissolved in 10 ml of resuspension buffer (50 mM Tris/HCl pH 7.5, 300 mM NaCl, 10 mM Mg(Ac)2) and lysed by French pressing followed by clearance of cell debris at 15.800 rpm for 30 min in a SS34 rotor. Next, the supernatant was used to sediment the membranes by centrifugation at 45.000 rpm for 1.5 hours in a TLA 50.2 rotor. The membranes were further solubilized in 1% DDM dissolved in resuspension buffer supplemented with 10% glycerol. Next, YidC was purified via metal affinity chromatography using TALON® Metal affinity resin as described above and analyzed on SDS Page by western blotting. In order to preserve the PFA cross-links, which are temperature sensitive, the samples were denatured in TCA loading dye at 37 °C for 18 minutes.
Preparation of material for mass spectrometric analyses
E. coli BL21 cells carrying plasmid pEVol and plasmid pTrc99a-SecYHisEG were cultured overnight at 37 °C in LB medium containing 50 µg/ml ampicillin and 30 µg/ml chloramphenicol. 10 ml of the overnight culture was further used for inoculation in 1 l minimal medium (1% w/v glycerol, 47.7 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 18.7 mM NH4Cl, 0.153 mM CaCl2, 0.59 mM MgSO4, 2.07 µM MoNa2O4, 4.198 µM CoSO4, 37.49 µM MnSO4, 4.35 µM ZnSO4, 6.29 µM FeCl2, 16.17 µM H3BO3, 40 mg/l L-leucine, 1 mg/ml D-biotin, 1 mg/ml thiamine, 7 mg/l chloramphenicol, 10 mg/l ampicillin) until cells reached the early exponential growth phase (OD600 = 0.5–0.8) and protein expression was then induced with 0.5 mM IPTG. After induction, the cultures were grown for 4 hours at 30 °C, were cooled down on ice for 10 minutes and harvested by centrifugation at 5.000 rpm in a JLA 9.1000 rotor for 10 minutes. Each cell pellet was resuspended in 10 ml resuspension buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 5 mM MgCl2) to which 0.5 mM PMSF and 2 pills/l cOmplete™ protease inhibitor cocktail (Roche®; Mannheim Germany) were added. Next, the samples were lysed by French pressing and cell debris was removed by centrifugation at 15.800 rpm for 30 minutes in an SS34 rotor. The supernatant was further centrifuged at 45.000 rpm for 1.5 hours in a TLA 50.2 rotor for membrane sedimentation. Next, the membrane pellets were solubilized in solubilisation buffer (20 mM HEPES, pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 1% DDM) for 1 hour at 4 °C. Non-solubilized membranes were separated by centrifugation for 10 minutes at 14.000 rpm at 4 °C. The supernatant was transferred onto TALON® Metal affinity resin, which was then washed twice with washing buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.03% DDM, 5 mM imidazole) and subsequently washed four times with imidazole-free washing buffer. Further, the material was washed four times with imidazole-containing washing buffer and eluted in 2 ml elution buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.03% DDM, 200 mM imidazole).
The chemical cross-linker disuccinimidyl suberate (DSS, ThermoFisher Scientific, Langenselbold, Germany) was dissolved in DMSO and incubated with the samples for 30 minutes at 25 °C (final concentration DSS: 1 mM) after the second-imidazole-free washing step (on-column cross-linking) or after the elution step (solution cross-linking). After the DSS incubation step, the reaction was quenched by adding Tris/HCl, pH 7.5 to a final concentration of 20 mM.
Protein samples for mass spectrometry analyses were precipitated with acetone at −20 °C over-night and subsequently centrifuged for 20 minutes at 4 °C and 14.000 rpm. After the supernatant was removed, the protein samples were dried and denatured in 10 µl 8 M urea (in 50 mM NH4HCO3). Then, 1.1 µl 50 mM Tris(2-carboxyethyl)phosphine (in 50 mM NH4HCO3) was added and the sample was incubated for 20 minutes at 37 °C with continuous shaking at 600 rpm. Subsequently, 1.23 µl 500 mM iodoacetamide (in 50 mM NH4HCO3) was added and incubated for 20 minutes in the absence of light. Finally, 3 µl of 100 mM dithiothreitol (in 50 mM NH4HCO3) and subsequently 25 µl of 50 mM NH4HCO3 were added. The protein samples were digested overnight with 0.125 µg/µl trypsin at 37 °C, and then acidified with 0.1% trifluoroacetic acid and centrifuged for 6 minutes at 12.000 rpm.
Peptide mixtures were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an Ultimate 3000 RSLCnano coupled to a Q Exactive Plus (Thermo Fisher Scientific) mass spectrometer as described77. Proteins were identified using the MaxQuant software package (version 1.5.4.0,78) searching against the UniProt E. coli proteome set sequence database (version 2016_03). Identification of cross-linked peptides was performed essentially as described77 using the program pLink®79 for searching inter- or intra-molecular cross-links generated by amine-specific cross-linker DSS between peptides from SecY, SecE, SecG and YidC. The resulting set of peptide spectrum matches at a false discovery rate <0.05 was further filtered requiring a minimum E-value of 0.05 and identification of a pair of cross-linked residues in at least two independent experiments.
Reproducibility statement
All experiments shown in this study were performed at least twice as independent biological replicates. Each replicate consisted of at least two technical replicates. Only representative western blots and agar plates are displayed. Uncropped images of all western blots and LB plates are provided in the supplementary information files (Supplementary Figure S4).
Data availability statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We thank Bettina Knapp for technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft to HGK (DFG grant KO2184/8-1) and BW (DFG grant FOR 1905), the Else-Kröner Fresenius Stiftung/Motivate MD college of the University Freiburg Medical School to BJ, a DAAD PhD fellowship to NAP, a FF-Nord fellowship to LF and by the Excellence Initiative of the German Federal and State Governments (EXC294 BIOSS Centre for Biological Signaling Studies to BW).
Author Contributions
N.A.P. performed the pBpa and P.F.A. cross-linking experiments together with A.H., I.S. and L.F. A.H. analyzed the functionality of the YidC derivatives. N.A.P., B.J. and L.F. generated the plasmid-encoded SecYEG and YidC variants. B.J. performed the SecYEG purification for MS and the DSS cross-linking. F.D., B.J. and B.W. designed, performed and analyzed the MS experiments. N.A.P. and H.G.K. designed the cross-linking experiments and wrote the manuscript. All authors analyzed the data.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-19019-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kudva R, et al. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Research in microbiology. 2013;164:505–534. doi: 10.1016/j.resmic.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Park E, Rapoport TA. Mechanisms of Sec. 61/SecY-mediated protein translocation across membranes. Annual review of biophysics. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann R, Eyrisch S, Ahmad M, Helms V. Protein translocation across the ER membrane. Biochimica et biophysica acta. 2011;1808:912–924. doi: 10.1016/j.bbamem.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 4.Dalbey R, Koch HG, Kuhn A. Targeting and Insertion of MembraneProteins. EcoSalPlus. 2017;7:1–28. doi: 10.1128/ecosalplus.ESP-0012-2016. [DOI] [PubMed] [Google Scholar]
- 5.Denks K, et al. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Molecular membrane biology. 2014;31:58–84. doi: 10.3109/09687688.2014.907455. [DOI] [PubMed] [Google Scholar]
- 6.Koch HG, Muller M. Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. The Journal of cell biology. 2000;150:689–694. doi: 10.1083/jcb.150.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 8.Park E, Rapoport TA. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature. 2011;473:239–242. doi: 10.1038/nature10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saparov SM, et al. Determining the conductance of the SecY protein translocation channel for small molecules. Molecular cell. 2007;26:501–509. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 10.Hizlan D, et al. Structure of the SecY complex unlocked by a preprotein mimic. Cell reports. 2012;1:21–28. doi: 10.1016/j.celrep.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischoff L, Wickles S, Berninghaus O, van der Sluis EO, Beckmann R. Visualization of a polytopic membrane protein during SecY-mediated membrane insertion. Nature communications. 2014;5:4103. doi: 10.1038/ncomms5103. [DOI] [PubMed] [Google Scholar]
- 12.Gogala M, et al. Structures of the Sec. 61 complex engaged in nascent peptide translocation or membrane insertion. Nature. 2014;506:107–110. doi: 10.1038/nature12950. [DOI] [PubMed] [Google Scholar]
- 13.Schatz PJ, Bieker KL, Ottemann KM, Silhavy TJ, Beckwith J. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. The EMBO journal. 1991;10:1749–1757. doi: 10.1002/j.1460-2075.1991.tb07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lycklama a Nijeholt JA, de Keyzer J, Prabudiansyah I, Driessen AJ. Characterization of the supporting role of SecE in protein translocation. FEBS letters. 2013;587:3083–3088. doi: 10.1016/j.febslet.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 15.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. The EMBO journal. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braig D, et al. Signal sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle. Mol Biol Cell. 2011;22:2309–2323. doi: 10.1091/mbc.E11-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Laan M, Nouwen N, Driessen AJ. SecYEG proteoliposomes catalyze the Deltaphi-dependent membrane insertion of FtsQ. The Journal of biological chemistry. 2004;279:1659–1664. doi: 10.1074/jbc.M306527200. [DOI] [PubMed] [Google Scholar]
- 18.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomkiewicz D, Nouwen N, van Leeuwen R, Tans S, Driessen AJ. SecA supports a constant rate of preprotein translocation. The Journal of biological chemistry. 2006;281:15709–15713. doi: 10.1074/jbc.M600205200. [DOI] [PubMed] [Google Scholar]
- 20.Karamanou S, et al. Preprotein-controlled catalysis in the helicase motor of SecA. The EMBO journal. 2007;26:2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S, Oliver DB. Mapping of the SecA.SecY and SecA.SecG interfaces by site-directed in vivo photocross-linking. The Journal of biological chemistry. 2011;286:12371–12380. doi: 10.1074/jbc.M110.182931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto G, Yoshihisa T, Ito K. SecY and SecA interact to allow SecA insertion and protein translocation across the Escherichia coli plasma membrane. The EMBO journal. 1997;16:6384–6393. doi: 10.1093/emboj/16.21.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn P, et al. Ribosome binding induces repositioning of the signal recognition particle receptor on the translocon. The Journal of cell biology. 2015;211:91–104. doi: 10.1083/jcb.201502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu CJ, Schuenemann D, Hoffman NE. Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. The Journal of biological chemistry. 1999;274:27219–27224. doi: 10.1074/jbc.274.38.27219. [DOI] [PubMed] [Google Scholar]
- 25.Angelini S, Deitermann S, Koch HG. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO reports. 2005;6:476–481. doi: 10.1038/sj.embor.7400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Götzke, H. et al. YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. Journal of Biological Chemistry, in press (2014). [DOI] [PMC free article] [PubMed]
- 27.Scotti PA, et al. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. The EMBO journal. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonoaea R, Fürst M, Nishiyama K-I, Müller M. The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry. 2008;47:5649–5656. doi: 10.1021/bi800233w. [DOI] [PubMed] [Google Scholar]
- 29.Sachelaru I, et al. YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. The Journal of biological chemistry. 2013;288:16295–16307. doi: 10.1074/jbc.M112.446583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachelaru I, et al. YidC and SecYEG form a heterotetrameric protein translocation channel. Scientific reports. 2017;7:101. doi: 10.1038/s41598-017-00109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck K, et al. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO reports. 2001;2:709–714. doi: 10.1093/embo-reports/kve154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houben EN, ten Hagen-Jongman CM, Brunner J, Oudega B, Luirink J. The two membrane segments of leader peptidase partition one by one into the lipid bilayer via a Sec/YidC interface. EMBO reports. 2004;5:970–975. doi: 10.1038/sj.embor.7400261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalbey RE, Kuhn A. How YidC inserts and folds proteins across a membrane. Nature structural & molecular biology. 2014;21:435–436. doi: 10.1038/nsmb.2823. [DOI] [PubMed] [Google Scholar]
- 34.Kumazaki K, et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 2014;4:7299. doi: 10.1038/srep07299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geng Y, et al. Role of the Cytosolic Loop C2 and the C Terminus of YidC in Ribosome Binding and Insertion Activity. The Journal of biological chemistry. 2015;290:17250–17261. doi: 10.1074/jbc.M115.650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samuelson JC, et al. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 37.Serek J, et al. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. The EMBO journal. 2004;23:294–301. doi: 10.1038/sj.emboj.7600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kol S, et al. YidC-mediated membrane insertion of assembly mutants of subunit c of the F1F0 ATPase. The Journal of biological chemistry. 2006;281:29762–29768. doi: 10.1074/jbc.M605317200. [DOI] [PubMed] [Google Scholar]
- 39.Nouwen N, Driessen AJ. SecDFyajC forms a heterotetrameric complex with YidC. Molecular microbiology. 2002;44:1397–1405. doi: 10.1046/j.1365-2958.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 40.Xie K, Kiefer D, Nagler G, Dalbey RE, Kuhn A. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry. 2006;45:13401–13408. doi: 10.1021/bi060826z. [DOI] [PubMed] [Google Scholar]
- 41.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. The EMBO journal. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulze RJ, et al. Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4844–4849. doi: 10.1073/pnas.1315901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Komar, J. et al. Membrane protein insertion and assembly by the bacterial holo-translocon SecYEG-SecDF-YajC-YidC. The Biochemical journal (2016). [DOI] [PMC free article] [PubMed]
- 45.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nature methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 46.Funes S, Kauff F, van der Sluis EO, Ott M, Herrmann JM. Evolution of YidC/Oxa1/Alb3 insertases: three independent gene duplications followed by functional specialization in bacteria, mitochondria and chloroplasts. Biological chemistry. 2011;392:13–19. doi: 10.1515/bc.2011.013. [DOI] [PubMed] [Google Scholar]
- 47.Lohmeyer E, et al. The ScoI homologue SenC is a copper binding protein that interacts directly with the cbb(3)-type cytochrome oxidase in Rhodobacter capsulatus. Biochimica et biophysica acta. 2012;1817:2005–2015. doi: 10.1016/j.bbabio.2012.06.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neves-Petersen MT, et al. Flash photolysis of cutinase: identification and decay kinetics of transient intermediates formed upon UV excitation of aromatic residues. Biophysical journal. 2009;97:211–226. doi: 10.1016/j.bpj.2009.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris ME, Christian EL. RNA crosslinking methods. Methods in enzymology. 2009;468:127–146. doi: 10.1016/S0076-6879(09)68007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugai R, Shimizu H, Nishiyama K, Tokuda H. Overexpression of gnsA, a multicopy suppressor of the secG null mutation, increases acidic phospholipid contents by inhibiting phosphatidylethanolamine synthesis at low temperatures. Journal of bacteriology. 2004;186:5968–5971. doi: 10.1128/JB.186.17.5968-5971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Soman R, Shanmugam S, Kuhn A, Dalbey R. The role of the strictly conserved positively charged residue differs among the Gram-positive, Gram-negative and chloroplast YidC homologs. J. Biol. Chem. 2015;289:35656–35667. doi: 10.1074/jbc.M114.595082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang F, et al. Defining the regions of Escherichia coli YidC that contribute to activity. The Journal of biological chemistry. 2003;278:48965–48972. doi: 10.1074/jbc.M307362200. [DOI] [PubMed] [Google Scholar]
- 53.Welte T, et al. Promiscuous targeting of polytopic membrane proteins to SecYEG or YidC by the Escherichia coli signal recognition particle. Mol Biol Cell. 2012;23:464–479. doi: 10.1091/mbc.E11-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koch HG, Moser M, Muller M. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Reviews of physiology, biochemistry and pharmacology. 2003;146:55–94. doi: 10.1007/s10254-002-0002-9. [DOI] [PubMed] [Google Scholar]
- 55.Luirink J, et al. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. The EMBO journal. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiche B, et al. A cleavable N-terminal membrane anchor is involved in membrane binding of the Escherichia coli SRP receptor. Journal of molecular biology. 2008;377:761–773. doi: 10.1016/j.jmb.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn P, et al. The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic (Copenhagen, Denmark) 2011;12:563–578. doi: 10.1111/j.1600-0854.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- 58.Borowska MT, Dominik PK, Anghel SA, Kossiakoff AA, Keenan RJ. Structure (London, England: 1993) 2015. A YidC-like Protein in the Archaeal Plasma Membrane; pp. 1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klostermann E, D G Helling I, Carde JP, Schunemann D. The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. The Biochemical journal. 2002;368:777–781. doi: 10.1042/bj20021291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. The EMBO journal. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimokawa-Chiba N, et al. Hydrophilic microenvironment required for the channel-independent insertase function of YidC protein. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:5063–5068. doi: 10.1073/pnas.1423817112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumazaki K, et al. Crystallization and preliminary X-ray diffraction analysis of YidC, a membrane-protein chaperone and insertase from Bacillus halodurans. Acta crystallographica. Section F, Structural biology communications. 2014;70:1056–1060. doi: 10.1107/S2053230X14012540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feltcher ME, Gibbons HS, Ligon LS, Braunstein M. Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain. Journal of bacteriology. 2013;195:672–681. doi: 10.1128/JB.02032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klenner C, Yuan J, Dalbey RE, Kuhn A. The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS letters. 2008;582:3967–3972. doi: 10.1016/j.febslet.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 65.Klenner C, Kuhn A. Dynamic disulfide scanning of the membrane-inserting Pf3 coat protein reveals multiple YidC substrate contacts. The Journal of biological chemistry. 2012;287:3769–3776. doi: 10.1074/jbc.M111.307223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang F, et al. Chloroplast YidC homolog Albino3 can functionally complement the bacterial YidC depletion strain and promote membrane insertion of both bacterial and chloroplast thylakoid proteins. The Journal of biological chemistry. 2002;277:19281–19288. doi: 10.1074/jbc.M110857200. [DOI] [PubMed] [Google Scholar]
- 67.Hennon SW, Soman R, Zhu L, Dalbey RE. YidC/Alb3/Oxa1 Family of Insertases. The Journal of biological chemistry. 2015;290:14866–14874. doi: 10.1074/jbc.R115.638171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saller MJ, Fusetti F, Driessen AJ. Bacillus subtilis SpoIIIJ and YqjG function in membrane protein biogenesis. J Bacteriol. 2009;191:6749–6757. doi: 10.1128/JB.00853-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y, Soman R, Shanmugam SK, Kuhn A, Dalbey RE. The role of the strictly conserved positively charged residue differs among the Gram-positive, Gram-negative, and chloroplast YidC homologs. The Journal of biological chemistry. 2014;289:35656–35667. doi: 10.1074/jbc.M114.595082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boy D, Koch HG. Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Molecular biology of the cell. 2009;20:1804–1815. doi: 10.1091/mbc.E08-08-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Houben EN, et al. YidC and SecY mediate membrane insertion of a Type I transmembrane domain. The Journal of biological chemistry. 2002;277:35880–35886. doi: 10.1074/jbc.M205556200. [DOI] [PubMed] [Google Scholar]
- 72.Sachelaru I, Petriman NA, Kudva R, Koch HG. Dynamic interaction of the Sec Translocon with the chaperone PpiD. J. Biol. Chem. 2014;289:21706–21715. doi: 10.1074/jbc.M114.577916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanahan. Studies on transformation of Escherichia coli with plasmids. Journal of molecular biology166 (1983). [DOI] [PubMed]
- 74.Alami M, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Molecular cell. 2003;12:937–946. doi: 10.1016/S1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 75.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. Journal of molecular biology. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 76.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of bacteriology. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan A, et al. Pex17p-dependent assembly of Pex14p/Dyn2p-subcomplexes of the peroxisomal protein import machinery. European journal of cell biology. 2016;95:585–597. doi: 10.1016/j.ejcb.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 78.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature protocols. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 79.Yang B, et al. Identification of cross-linked peptides from complex samples. Nature methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files) or are available from the corresponding author on reasonable request.