Abstract
Copper-transporter ATP7B maintains copper homeostasis in the human cells and delivers copper to the biosynthetic pathways for incorporation into the newly synthesized copper-containing proteins. ATP7B is a target of several hundred mutations that lead to Wilson disease, a chronic copper toxicosis. ATP7B contains a chain of six cytosolic metal-binding domains (MBDs), the first four of which (MBD1-4) are believed to be regulatory, and the last two (MBD5-6) are required for enzyme activity. We report the NMR structure of MBD1, the last unsolved metal-binding domain of ATP7B. The structure reveals the disruptive mechanism of G85V mutation, one of the very few disease causing missense mutations in the MBD1-4 region of ATP7B.
Introduction
Membrane transporter ATP7B regulates the level and intracellular distribution of copper in human tissues1. At the basal copper levels, ATP7B is located in the trans-Golgi network, where it delivers copper to the newly synthesized copper-containing proteins, such as ceruloplasmin. At the elevated copper levels, ATP7B relocates to the cytosolic membrane vesicles and plasma membrane, where it exports copper from the cell. This function is particularly important in the liver, where copper excretion into the bile serves as a main route of excess copper disposal in the human body.
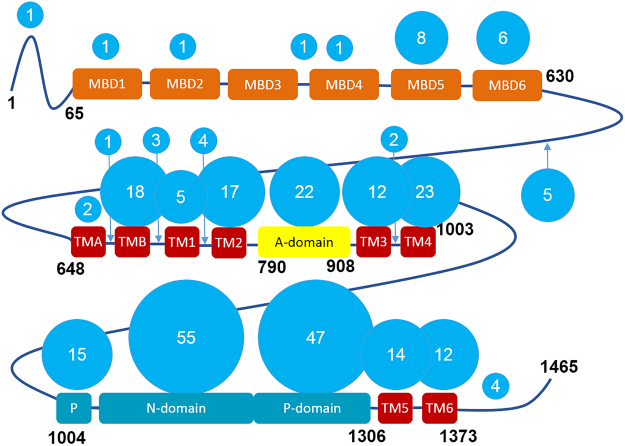
ATP7B uses the energy of ATP hydrolysis to translocate copper across the membrane. It belongs to the large family of P-type ATPases, and shares much of the domain composition and some fundamental features of the catalytic mechanism with the better studied members of the family, such as Ca-ATPase and Na,K-ATPase1–7. There is no high-resolution structure of ATP7B, but the structures of most cytosolic domains have been solved by NMR, and the overall structure of ATP7B has been modeled by homology5 using the X-ray structure of the bacterial copper ATPase CopA from Legionella pneumophila2 as a template. This model does not include the N-terminal chain of the six cytosolic metal-binding domains (MBDs) connected by flexible loops of various length, a unique structural feature of ATP7B and of the closely related copper transporter ATP7A (Fig. 1).
Figure 1.
Domain composition of ATP7B and the distribution of the Wilson disease mutations. ATP7B includes six cytosolic metal binding domains (MBD1-MBD6, orange), eight transmembrane helices (TMA-TM6, red), and the nucleotide-binding (N) and phosphorylation (P) domains (cyan), which together hydrolyze ATP, with the participation of the actuator (A) domain (yellow). The length of the interdomain linkers is not to scale. The number of known Wilson disease causing missense mutations in each domain and in the connecting loops, defined as distinct single amino acid substitutions, is shown in the blue circles. The list of mutations, as of 2010, was obtained from the Wilson disease mutation database (http://www.wilsondisease.med.ualberta.ca/database.asp)43. Except for the metal binding domains, a homology model of ATP7B5 based on the X-ray structure of the bacterial copper ATPase CopA2 was used to confirm domain assignment of the mutation sites.
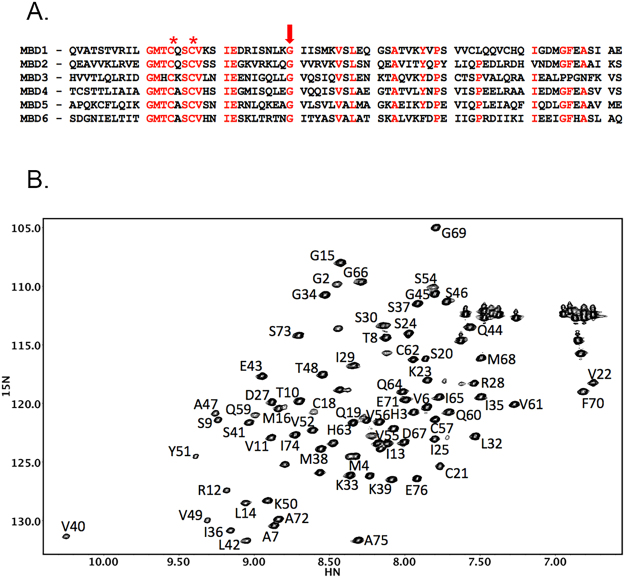
The structures of ATP7B metal-binding domains 2–6 have been solved previously by NMR8–10. Each of the six MBDs has a conserved ferredoxin-like fold and is approximately 70 amino acids long. The individual MBDs show significant sequence homology to each other, with the invariant GM(T/H)CxSCxxxIE motif responsible for binding copper(I) ions11,12 through the sulfur atoms of the two cysteine residues (Fig. 2A). Copper binding causes some changes in the local dynamics of the binding site, but does not alter overall conformation of the metal-binding domains13–16. Previous NMR studies show that MBD1-6 chain does not fold together into a compact structure, and the individual domains are highly mobile17–19.
Figure 2.
Amino acid sequence of the metal binding domains of ATP7B (A) and a fingerprint 1H,15N-HSQC spectrum of MBD1 (B). (A) Conserved residues are shown in red. Cysteine residues in the copper binding motif of the MBDs are marked by an asterisk. The invariant glycine, which is a target of the Wilson disease causing mutation in MBD1 (G85V), is shown by the arrow. (B) The sequential amino acid assignments in MBD1 are shown. In the protein construct used for structure determination, residues 1–4 are from the purification tag, and Q5 corresponds to Q56 in the full length ATP7B.
In the cell, ATP7B receives copper from a chaperone protein Atox1, which is structurally rather similar to the MBDs20,21. Presumably, copper is transferred from Atox1 to some or all of the MBDs, then to the copper-binding site in the transmembrane domain, and, finally, to an acceptor on the other side of the membrane. However, the exact path of copper transfer is unknown. MBDs 5–6, which are closest to the membrane, are required for activity, while MBD1-4 are believed to play a regulatory role22–24.
Mutations that impair transport activity or disrupt intracellular targeting of ATP7B cause Wilson disease, chronic copper toxicosis that primarily affects the liver and the brain. Wilson disease is an autosomal recessive disorder with highly variable symptoms, onset age and progression. This variability stems from the diverse effects of several hundred known disease mutations, further complicated by the fact that most Wilson disease patients are compound heterozygotes. While molecular basis of many Wilson disease mutations is known, disruptive effect of others is still unexplained, and, generally, there is no reliable correlation between the mutation type and disease symptoms and prognosis.
Analysis of the domain distribution of Wilson disease missense mutations reveals a striking difference between the first four metal binding domains, and the rest of the protein. Out of about 300 single amino acid substitutions known to be associated with Wilson disease, only five are located in the MBD1-4 region, and only one of those five, G85V, in MBD1 (Fig. 1). By comparison, MBD5 and MBD6, both very similar in size and structure to MBDs 1–4, are targets of eight and six disease causing missense mutations respectively, while the N-domain, which binds ATP, is a target of 55 such mutations. The paucity of missense disease mutations in MBD1-4 may reflect the fact that these domains play a regulatory role, and, unlike MBDs 5-6, are not strictly required for copper transport activity. The molecular basis of the few known mutations in MBD1-4 is therefore particularly interesting and may offer a clue to the individual functional roles of these domains in the native enzyme. To determine the disruptive mechanism of the G85V mutation, we have solved the structure of MBD1, the last unsolved metal-binding domain of ATP7B.
Results and Discussion
Structure of MBD1
The protein construct used for MBD1 structure determination produced high quality spectra. Using standard triple resonance experiments, we assigned backbone amides of all non-proline residues (Fig. 2B), with the exception of Q5, T17, and L58. These residues are located in flexible regions of the protein, and the backbone amide signals may not be observable due to the chemical exchange. The structure was calculated from 628 NOE restraints, 119 backbone dihedral angle constraints, with 40 hydrogen bond restraints added during structure refinement. The final structure ensemble had an RMSD of 0.59 Å for the backbone atoms, and 1.15 Å for all heavy atoms (Supplementary Table S1).
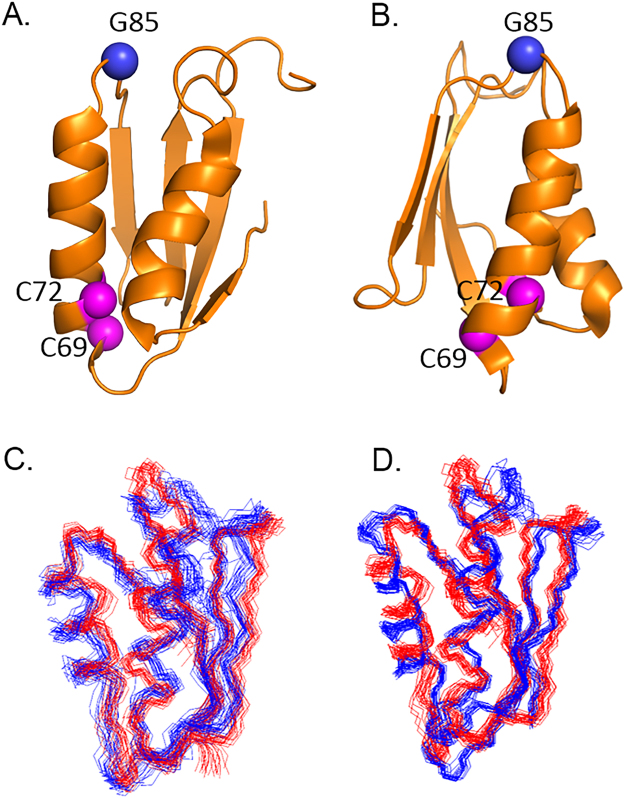
The MBD1 structure (Fig. 3A,B) shows the characteristic βαββαβ ferredoxin fold, similar to the other metal-binding domains of ATP7B, with the ensemble RMSD between the structures for the ordered core varying from 1.9 Å (MBD1 vs. MBD6, Fig. 3C) to 3.2 Å (MBD1 vs. MBD3, Fig. 3D). By comparison, the ensemble RMSD from MBD1 of ATP7A is 2.1 Å. The MBD1 rotational correlation time τc is 4.8 ns, as compared to 4.6 ns for MBD225, for example, and is consistent with the monomeric state of the protein. Similar to the other MBDs, the copper binding motif CxxC appears to experience complex dynamics (Fig. S1), while the rest of the folded core of the protein is well ordered, with some flexibility in some of the connecting loop regions.
Figure 3.
The structure of MBD1 of ATP7B. (A,B) The ribbon diagram of MBD1 structure with the invariant cysteines in the copper binding motif (C69 and C72 in the full-length protein, magenta), and G85 (blue) shown as spheres. Panels A and B are related by 90o rotation around the vertical axis. (C,D) MBD1 structure ensemble (PDB ID 2N7Y, red) aligned with the MBD6 (PDB ID 2EW9, C, blue) and MBD3 (PDB ID 2ROP, D, blue) ensembles by minimizing RMSD for the backbone atoms.
Structural effect of the G85V mutation
The structure of MBD1 explained the effect of G85V mutation, one of the very few known missense Wilson disease causing mutation in the MBD1-4 region of the protein. Although not among the most frequent, the G85V mutation has been found in various ethnic populations26–28. Biochemically, the G85V variant of ATP7B showed reduced interaction with the copper chaperone Atox129, but increased binding to COMMD1, a regulatory protein, involved in copper metabolism, among other cellular processes30. Previously, G85V substitution was also shown to cause the loss of ATP7B transport activity31, ATP7B retention in the endoplasmic reticulum, and rapid protein degradation30, indicative of protein misfolding. We were unable to purify G85V-MBD1 using the standard MBD purification procedure, presumably due to the degradation of the fusion protein in the bacterial cell. Taken together, these facts suggest that G85V mutation causes MBD1 misfolding.
The G85 residue is located in the loop connecting α1-helix and β2-strand. Interestingly, glycine in this position is conserved among all the MBDs of ATP7B (Fig. 2A), and, with one exception, all the MBDs of ATP7A. Yet, to date, only mutations of this glycine in MBD1 (G85V) and MBD6 (G591D) are known to cause Wilson disease. Analysis of the MBD local structure suggests an explanation of this distinct effect of the G85V mutation (Table 1).
Table 1.
Backbone conformation of the conserved glycine residue in the ATP7B metal binding domains (MBDs).
| MBD | PDB ID | Residue | φ | ψ | Wilson disease mutation |
|---|---|---|---|---|---|
| 1 | 2N7Y | G85 | 108 ± 10o | 3 ± 7o | G85V |
| 2 | 2LQB | G170 | −74 ± 16o | −57 ± 3o | — |
| 3 | 2ROP | G284 | −98 ± 19o | −16 ± 22o | — |
| 4 | 2ROP | G386 | 83 ± 11o | −15 ± 25o | —1 |
| 5 | 2EW9 | G515 | −72 ± 8o | −6 ± 19o | — |
| 6 | 2EW9 | G591 | −75 ± 4o | −34 ± 6o | G591D |
1Protein destabilization by G386V substitution observed in vitro.
In MBD1 and in MBD4, this residue adopts a backbone conformation that is only favorable for glycine, and is disallowed for valine: for G85 the backbone dihedral angle values are φ = 108 ± 10o and ψ = 3 ± 7o, and, for G386, φ = 83 ± 11o and ψ = −15 ± 25o. In the other MBDs, the corresponding glycine adopts backbone conformations falling into the universally allowed broad region of the Ramachandran plot surrounding the ideal α-helix values. Thus amino acid residues with bulky side chains will be disruptive at this position in MBD1 and MBD4, but not necessarily in the other MBDs. Analysis of the local dynamics is consistent with this interpretation. Residues 33–35 in MBD1, corresponding to K84-G85-I86 in the full-length ATP7B, all have high order parameter (S2) values over 0.98 indicating a rigid local structure, with very limited backbone flexibility (Fig. 1S,D).
Although, to the best of our knowledge, the G386V mutation in MBD4 has not been reported to cause Wilson disease, its effect has been tested in vitro. Consistent with the structural analysis, the G386V variant of MBD4 was shown to be unstable, with about 20oC lower midpoint temperature of thermal denaturation than the wild type protein32. The deleterious effect of G591D mutation in MBD6, where G591 has backbone dihedral angles of φ = −75 ± 4o and ψ = −34 ± 6o, cannot be explained by the constraints of the backbone conformation, and likely has other causes, such as disruption of domain-domain interactions, or local effects caused by the negative charge of the aspartate side chain. In fact, MBD6 is the closest to the membrane and likely forms a larger number of interdomain contacts than any other MBD.
Misfolding of G85V-MBD1 may have further structural consequences for ATP7B in the cell. Our recent work indicates that MBD1-3 interact with each other, forming a dynamically correlated domain group25. These interactions may be involved in the regulation of ATP7B activity and trafficking by copper24. MBD1 misfolding will disrupt MBD1-3 interactions, and interfere with proper ATP7B trafficking and activity regulation. This, in turn may be followed by protein degradation, as reported previously.
In summary, the structure of MBD1 taken together with the previously solved structures of the other MBDs explains the disruptive effect of a Wilson disease causing mutation located in the regulatory region of the protein, where very few disease causing mutations have been reported.
Methods
Protein expression and purification
MBD1 was expressed as fusion with the chitin-binding domain and intein using vector pTYB12 (New England BioLabs). The protein was isotopically labeled with 15N and 13C, and purified by chitin affinity chromatography combined with intein self-cleavage, essentially as described previously33. Prior to NMR experiments, MBD1 was dialyzed against 50 mM HEPES pH 7.4, 50 mM NaCl, 5 mM tris-(2-carboxyethyl)phosphine.
NMR experiments and structure determination
NMR experiments were performed on the 600 MHz and 750 MHz Bruker NMR spectrometers equipped with a CryoProbe. Data were processed using NMRPipe34 and analyzed using NMRView35. Backbone chemical shift assignments for MBD1 were made from HNCO, HNCA, HNCACB, HN(CA)CO, and HN(CO)CACB experiments36. Side-chain assignment experiments were done using H(CCO)NH, CC(O)NH, AND 1H, 15N-TOCSY. The backbone chemical shifts were assigned using CARA37. Hydrogen bond distance restraints were derived from H/D exchange measurements.
Peak tables and distance restraint generation from 3D 1H, 15N-NOESY and 3D 1H, 13C-NOESY experiments, as well structure calculation was done using PONDEROSA38. Peak tables and distance restraint generation from 3D 1H, 15N-NOESY and 3D 1H, 13C-NOESY experiments, as well structure calculation was done using PONDEROSA-C/S38 with XPLOR-NIH based calculation options39. NOE cross peaks were automatically assigned with AUDANA40 and manually verified by PONDEROSA Analyzer coupled with NMRFAM-SPARKY41 and PyMOL (Schroedinger LLC). Final structure calculation step was carried out with explicit water refinement option with validated restraints that selects best 20 from 100 calculated models with the lowest energy criteria. The structure has been deposited in RCSB as entry 2N7Y.
NMR relaxation measurements, and data analysis using TENSOR 2.042 were performed essentially as described previously25, except that HSQC rather than TROSY versions of NMR experiments were used.
Electronic supplementary material
Acknowledgements
This research was supported by NSERC Discovery grant to O.Y.D. NMR data were collected at the National Magnetic Resonance Facility at Madison (NMRFAM), which is supported by NIH grant P41 GM103399 and by the University of Wisconsin-Madison.
Author Contributions
C.H.Y. cloned and purified the protein and, together with W.L., solved the structure. S.N. performed NMR experiments. O.Y.D. analyzed results and wrote the paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Corey H. Yu and Woonghee Lee contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-18951-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lutsenko S, Barnes NL, Bartee MY, Dmitriev OY. Function and regulation of human copper-transporting ATPases. Physiol Rev. 2007;87:1011–1046. doi: 10.1152/physrev.00004.2006. [DOI] [PubMed] [Google Scholar]
- 2.Gourdon P, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 3.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 4.Petrukhin K, et al. Characterization of the Wilson disease gene encoding a P-type copper transporting ATPase: genomic organization, alternative splicing, and structure/function predictions. Hum. Mol. Genet. 1994;3:1647–1656. doi: 10.1093/hmg/3.9.1647. [DOI] [PubMed] [Google Scholar]
- 5.Schushan M, Bhattacharjee A, Ben-Tal N, Lutsenko S. A structural model of the copper ATPase ATP7B to facilitate analysis of Wilson disease-causing mutations and studies of the transport mechanism. Metallomics. 2012;4:669–678. doi: 10.1039/c2mt20025b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 7.Toyoshima C, Inesi G. Structural basis of ion pumping by Ca2 + -ATPase of the sarcoplasmic reticulum. Annu. Rev. Biochem. 2004;73:269–292. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- 8.Achila D, et al. Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake. Proc. Natl. Acad. Sci. USA. 2006;103:5729–5734. doi: 10.1073/pnas.0504472103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banci L, Bertini I, Cantini F, Rosenzweig AC, Yatsunyk LA. Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry. 2008;47:7423–7429. doi: 10.1021/bi8004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolgova NV, Nokhrin S, Yu CH, George GN, Dmitriev OY. Copper chaperone Atox1 interacts with the metal-binding domain of Wilson's disease protein in cisplatin detoxification. Biochem. J. 2013;454:147–156. doi: 10.1042/BJ20121656. [DOI] [PubMed] [Google Scholar]
- 11.Arnesano F, et al. Metallochaperones and metal-transporting ATPases: a comparative analysis of sequences and structures. Genome Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- 12.Wernimont AK, Huffman DL, Lamb AL, O'Halloran TV, Rosenzweig AC. Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nat. Struct. Biol. 2000;7:766–771. doi: 10.1038/78999. [DOI] [PubMed] [Google Scholar]
- 13.Banci L, Bertini I, Del Conte R, D'Onofrio M, Rosato A. Solution structure and backbone dynamics of the Cu(I) and apo forms of the second metal-binding domain of the Menkes protein ATP7A. Biochemistry. 2004;43:3396–3403. doi: 10.1021/bi036042s. [DOI] [PubMed] [Google Scholar]
- 14.Banci L, et al. Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein. J. Biol. Chem. 2006;281:29141–29147. doi: 10.1074/jbc.M603176200. [DOI] [PubMed] [Google Scholar]
- 15.DeSilva TM, Veglia G, Opella SJ. Solution structures of the reduced and Cu(I) bound forms of the first metal binding sequence of ATP7A associated with Menkes disease. Proteins. 2005;61:1038–1049. doi: 10.1002/prot.20639. [DOI] [PubMed] [Google Scholar]
- 16.Gitschier J, Moffat B, Reilly D, Wood WI, Fairbrother WJ. Solution structure of the fourth metal-binding domain from the Menkes copper-transporting ATPase. Nat. Struct. Biol. 1998;5:47–54. doi: 10.1038/nsb0198-47. [DOI] [PubMed] [Google Scholar]
- 17.Banci L, et al. The different intermolecular interactions of the soluble copper-binding domains of the menkes protein, ATP7A. J. Biol. Chem. 2007;282:23140–23146. doi: 10.1074/jbc.M700695200. [DOI] [PubMed] [Google Scholar]
- 18.Banci L, et al. An NMR study of the interaction of the N-terminal cytoplasmic tail of the Wilson disease protein with copper(I)-HAH1. J. Biol. Chem. 2009;284:9354–9360. doi: 10.1074/jbc.M805981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatemi N, Korzhnev DM, Velyvis A, Sarkar B, Forman-Kay JD. NMR characterization of copper-binding domains 4-6 of ATP7B. Biochemistry. 2010;49:8468–8477. doi: 10.1021/bi1008535. [DOI] [PubMed] [Google Scholar]
- 20.Pufahl RA, et al. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 21.Rosenzweig AC, et al. Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution. Structure. 1999;7:605–617. doi: 10.1016/S0969-2126(99)80082-3. [DOI] [PubMed] [Google Scholar]
- 22.Cater MA, Forbes J, La Fontaine S, Cox D, Mercer JF. Intracellular trafficking of the human Wilson protein: the role of the six N-terminal metal-binding sites. Biochem. J. 2004;380:805–813. doi: 10.1042/bj20031804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huster D, Lutsenko S. The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson's disease protein. J. Biol. Chem. 2003;278:32212–32218. doi: 10.1074/jbc.M305408200. [DOI] [PubMed] [Google Scholar]
- 24.Yu CH, Dolgova NV, Dmitriev OY. Dynamics of the metal binding domains and regulation of the human copper transporters ATP7B and ATP7A. IUBMB. Life. 2017;69:226–235. doi: 10.1002/iub.1611. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, et al. Interactions between metal-binding domains modulate intracellular targeting of Cu(I)-ATPase ATP7B, as revealed by nanobody binding. J. Biol. Chem. 2014;289:32682–32693. doi: 10.1074/jbc.M114.580845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis D, et al. A study of Wilson disease mutations in Britain. Hum. Mutat. 1999;14:304–311. doi: 10.1002/(SICI)1098-1004(199910)14:4<304::AID-HUMU5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Gu YH, et al. Mutation spectrum and polymorphisms in ATP7B identified on direct sequencing of all exons in Chinese Han and Hui ethnic patients with Wilson's disease. Clin. Genet. 2003;64:479–484. doi: 10.1046/j.1399-0004.2003.00179.x. [DOI] [PubMed] [Google Scholar]
- 28.Loudianos G, et al. Further delineation of the molecular pathology of Wilson disease in the Mediterranean population. Hum. Mutat. 1998;12:89–94. doi: 10.1002/(SICI)1098-1004(1998)12:2<89::AID-HUMU3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 29.Hamza I, Schaefer M, Klomp LW, Gitlin JD. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc. Natl. Acad. Sci. USA. 1999;96:13363–13368. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Bie P, et al. Distinct Wilson's disease mutations in ATP7B are associated with enhanced binding to COMMD1 and reduced stability of ATP7B. Gastroenterology. 2007;133:1316–1326. doi: 10.1053/j.gastro.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huster D, et al. Diverse functional properties of Wilson disease ATP7B variants. Gastroenterology. 2012;142:947–956. doi: 10.1053/j.gastro.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar R, et al. Disease-causing point-mutations in metal-binding domains of Wilson disease protein decrease stability and increase structural dynamics. Biometals. 2017;30:27–35. doi: 10.1007/s10534-016-9976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dmitriev O, et al. Solution structure of the N-domain of Wilson disease protein: Distinct nucleotide-binding environment and effects of disease mutations. Proc. Natl. Acad. Sci. USA. 2006;103:5302–5307. doi: 10.1073/pnas.0507416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA, Blevins RA. Nmr View - A Computer-Program for the Visualization and Analysis of Nmr Data. Journal of Biomolecular Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 36.Gardner KH, Kay LE. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 37. Keller, R. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. Swiss Federal Institute of Technology, (2004).
- 38.Lee W, Kim JH, Westler WM, Markley JL. PONDEROSA, an automated 3D-NOESY peak picking program, enables automated protein structure determination. Bioinformatics. 2011;27:1727–1728. doi: 10.1093/bioinformatics/btr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn Reson. 2003;160:65–73. doi: 10.1016/S1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 40.Lee W, Petit CM, Cornilescu G, Stark JL, Markley JL. The AUDANA algorithm for automated protein 3D structure determination from NMR NOE data. J. Biomol. NMR. 2016;65:51–57. doi: 10.1007/s10858-016-0036-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/A:1008305808620. [DOI] [PubMed] [Google Scholar]
- 43.Kenney SM, Cox DW. Sequence variation database for the Wilson disease copper transporter, ATP7B. Hum. Mutat. 2007;28:1171–1177. doi: 10.1002/humu.20586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.