Abstract
Background
Ginsenoside F1 has been described to possess skin-whitening effects on humans. We aimed to synthesize a new ginsenoside derivative from F1 and investigate its cytotoxicity and melanogenesis inhibitory activity in B16BL6 cells using recombinant glycosyltransferase enzyme. Glycosylation has the advantage of synthesizing rare chemical compounds from common compounds with great ease.
Methods
UDP-glycosyltransferase (BSGT1) gene from Bacillus subtilis was selected for cloning. The recombinant glycosyltransferase enzyme was purified, characterized, and utilized to enzymatically transform F1 into its derivative. The new product was characterized by NMR techniques and evaluated by MTT, melanin count, and tyrosinase inhibition assay.
Results
The new derivative was identified as (20S)-3β,6α,12β,20-tetrahydroxydammar-24-ene-20-O-β-D-glucopyranosyl-3-O-β-D-glucopyranoside (ginsenoside Ia), which possesses an additional glucose linked into the C-3 position of substrate F1. Ia had been previously reported; however, no in vitro biological activity was further examined. This study focused on the mass production of arduous ginsenoside Ia from accessible F1 and its inhibitory effect of melanogenesis in B16BL6 cells. Ia showed greater inhibition of melanin and tyrosinase at 100 μmol/L than F1 and arbutin. These results suggested that Ia decreased cellular melanin synthesis in B16BL6 cells through downregulation of tyrosinase activity.
Conclusion
To our knowledge, this is the first study to report on the mass production of rare ginsenoside Ia from F1 using recombinant UDP-glycosyltransferase isolated from B. subtillis and its superior melanogenesis inhibitory activity in B16BL6 cells as compared to its precursor. In brief, ginsenoside Ia can be applied for further study in cosmetics.
Keywords: UDP-glycosyltransferase, ginsenoside F1, melanogenesis, ginsenoside Ia, B16BL6 cell line
1. Introduction
Glycosyltransferases (GTs; EC 2.4.x.y) consist of a ubiquitous family of enzymes that can catalyze glycosylation on a wide range of acceptor molecules [1], [2], which is paramount for the biosynthesis of oligosaccharides, polysaccharides, and glycoconjugates [3]. GTs are important in the development of complex natural glycosylated products. Based on their sequence, signature motif, stereochemistry of the glycoside linkage, and known target specificity, GTs are classified into 78 families [4] (http://afmb.cnrs-mrs.fr/CAZY/). In addition, GT Family 1 contains UDP-glucosyltransferases (UGTs), which have been found in bacteria, plants, and animals. UGTs can use UDP-activated sugar moieties such as UDP-glucose as a donor and some small molecules, such as flavonoids, antibiotics, and plant hormones, as sugar acceptors [5]. The Bacillus cereus genome was completely sequenced and found to contain four UDP-glycosyltransferases [6]. One of these, BcGT-1, was reported to carry out reactions on flavonoids [5]. However, there are few reports on the glycosylation of ginsenoside by UDP-glycosyltransferases [7]. Glycosylation plays a key biological role in many synthesis processes of natural saponins [5]. GTs are the most common enzymes used in glycosylation, and UDP-glucose is the necessary and specific sugar for the conversion of saponins.
Ginseng, the root of Panax ginseng Meyer, is a well-known traditional medicine widely used in China, Korea, Japan, and other Asian countries [8]. Among many other report pharmacological efficacies, ginseng has been used for more than 2,000 years to strengthen immunity [9], [10] and reduce fatigue. Ginsenosides, the principal components in ginseng, have been documented to possess various pharmacological activities, such as immune-modulatory [11], anti-inflammatory [12], antitumor [13], antidiabetic [14], and anti-aging effects [15]. More than 128 ginsenosides have been isolated from ginseng roots and are classified into three groups, namely protopanaxadiol (PPD), and protopanaxatriol, and oleanane [16]. Amidst the numerous ginsenosides, ginsenoside F1 is a minor ginsenoside with glucose in the 20-C position of protopanaxatriol aglycone. It is present naturally at low concentrations in Korean ginseng and can be produced by β-glucosidase-mediated transformation of ginsenosides Re and Rg1 in P. ginseng Meyer [17]. Ginsenoside F1 has been shown to flaunt anticancer [18], anti-aging, and antioxidant effects and has demonstrated competitive inhibition of CYP3A4 activity and weaker inhibition of CYP2D6 activity [19], [20]. Most interestingly, ginsenoside F1 has been shown to showcase an anti-skin cancer effect and the ability to block melanin production in B16BL6 cells [19]. However, its low water solubility hinders its cosmetic application in the human skin. Sugar conjugation of ginsenosides is considered essential for the coveted stability and water solubility [21]. Glycosylation enhances the pharmacological and bioavailability properties during drug development and in food additives [22], [23]. Compared with chemical synthesis, enzymatic biosynthesis is more selective and environmentally favorable. Recently, researchers have reported that recombinant enzymes have been used to transform ginsenosides at relatively high rates [24], [25].
Control of melanin biosynthesis is an important process in the treatment of abnormal skin pigmentation. Tyrosinase is a key enzyme in the production of melanin. Decrease in melanin synthesis and inhibition of tyrosinase activity are key factors for skin-lightening effects [26]. Earlier studies have claimed that several skin-whitening agents, such as hydroquinone, arbutin, azelaic acid, retinoic acid, and kojic acid, are effective in the treatment of hyperpigmentation disorders albeit with undesirable side effects, such as dermatitis, skin irritation, and melanocyte damage [27]. There is a need to identify new skin-lightening agents from natural sources with the aim of reducing or avoiding such side effects.
In this study, we report the cloning and expression of the GT gene, which encodes a UDP-glycosyltransferase derived from Bacillus subtilis KCTC1022. The new derivatives of ginsenoside were synthesized from ginsenoside F1 and UDP-glucose as the substrates using UDP-glycosyltransferase (BSGT1), which was already reported as ginsenoside Ia [28]. Finally, we aimed to study the effects of ginsenoside Ia on cell viability, melanin content, and tyrosinase inhibition in B16BL6 cells compared with those of F1 and arbutin.
2. Materials and methods
2.1. Materials
UDP-glucose was obtained from Sigma (St. Louis, MO). Ginsenoside F1 was obtained from the Ginseng Genetic Resource Bank (Suwon, Korea). The B16BL6 melanoma cell line was obtained from the Korean Cell Line Bank (Seoul, Korea). Arbutin was obtained from Abcam (Cambridge, UK), and α-melanocyte-stimulating hormone (MSH) was purchased from Sigma. Tyrosinase was purchased from Santa Cruz (Santa Cruz, CA).
2.2. Molecular cloning, expression, and purification of recombinant UDP-glycosyltransferase (BSGT1) gene from B. subtilis
Genomic DNA from B. subtilis was isolated using a genomic DNA extraction kit (GeneAll, Korea). The UDP-glycosyltransferase gene was amplified from genomic DNA using polymerase chain reaction (PCR) with Pfu DNA polymerase (GeneAll, Korea). The BSGT1 gene (GenBank accession number KU 500621) was amplified using the following primers (with NdeI and EcoRI restriction sites in boldface): BSGT1 F (5′-CGCATATGATGATTTACATGGTTGGTGAAA-3′) and BSGT1 R (5′-CGGAATTAAAAGAACGCTGATAATACTG-3′).
The amplified fragment was digested with NdeI and EcoRI and then inserted into a pMAL-c5X vector to generate a maltose-binding protein (MBP)-BSGT1 gene fusion using the EzFusion Cloning Kit (Enzynomics, Korea). The amplified gene was sequenced and confirmed at the Genotech facility (Daejeon, Korea). Escherichia coli BL21 (DE3) transformed with recombinant pMAL-BSGT1 were grown in Luria–Bertani medium with ampicillin at 37°C to an optical density of 0.4 at 600 nm, and protein expression was subsequently induced through the addition of 0.3mM isopropyl-β-D-thiogalactopyranoside (IPTG). The bacteria were incubated for additional 9 h at 20°C and harvested by centrifuging at 5,000g for 20 min at 4°C. The cells were washed twice with 20mM sodium phosphate buffer (pH 7.0, 1mM EDTA, and 1mM NaCl) and then resuspended in 20mM sodium phosphate buffer (pH 7.0). The cells were sonicated on ice, and debris was removed by centrifugation at 12,000g at 4°C for 20 min. The MBP-tagged fusion protein was purified in an amylose resin column. The supernatant was collected, and the homogeneity of the protein was assessed using 12% SDS-PAGE and Coomassie Blue staining. Initial biosynthesis experiments using ginsenoside F1 as the substrate revealed that the presence of MBP fused to BSGT1 did not affect the enzyme activity. Therefore, the MBP fusion protein was used to determine the specificity and selectivity of BSGT1 for the biosynthesis using UDPG as well as SDS-PAGE. The protein concentration was determined using bovine serum albumin as the standard according to the Bradford method.
2.3. Enzyme characterization
The Michaelis–Menten constant, Km (mol/L), and the kinetic parameters, Vmax (μmol/L*min), were measured by Michaelis–Menten plot using different substrate concentrations ranging from 0.5mM to 5mM UDP-glucose [29]. The values of Km and Vmax were calculated by the Lineweaver–Burk plot.
2.4. Biosynthesis of metabolites
Glycosylation ability was assayed with overexpressed BSGT1 enzyme and F1. The reaction mixtures contained 100 μL of 0.5mM F1 and 100 μL of 2.5mM UDP-glucose and 800 μL of purified enzyme (final concentration at 0.1 mg/mL) (pH 7.0). The mixtures were incubated at 30°C for 24 h. Moreover, three groups of controls were incubated under the same conditions: (1) control 1 (C1) consisted of ginsenoside F1 with BSGT1; (2) control 2 (C2) consisted of BSGT1 with UDP-glucose; and (3) control 3 (C3) consisted of ginsenoside F1 with UDP-glucose.
2.5. Analysis of optimal conditions for purified enzyme activity according to pH, temperature, and metal ion concentrations
Initial biotransformation experiments were determined in 1 mL of reaction mixture using 0.5mM ginsenoside F1 as a substrate. The activity of BSGT1 was assessed in the presence of 10mM of Co2+, Mg2+, Fe3+, Na+, Cu2+, NH4+, K+, Ca2+, and Zn2+ for 30 min at 30°C and subsequently compared with a control without metal ions. The effect of pH of enzyme activity was determined at 30°C using 0.5mM ginsenoside F1 as the substrate in 20mM glycine-HCl buffer (pH 3.0), citric acid-sodium citrate buffer (pH 4.0–5.0), sodium phosphate buffer (pH 6.0–7.0), Tris HCl buffer (pH 8.0–9.0), and glycine-NaOH buffer (pH 10.0) at optimum metal ion concentration. The effect of temperature on enzyme activity was determined at 20°C, 30°C, 37°C, 50°C, and 60°C at optimum pH and metal ion conditions for 24 h in the presence of 0.5mM ginsenoside F1. Samples were analyzed at regular intervals. An equal volume of water-saturated n-butanol was added to each sample to stop the reaction. The supernatant containing ginsenoside F1 and biotransformed products was analyzed using TLC, HPLC, and HR/MS.
2.6. TLC analysis of metabolites
The extract was collected and evaporated to dryness at room temperature. TLC analysis was performed using Silica gel 60 plates with the developing solvent CHCl3:CH3OH:H2O (65:35:10, v/v/v, lower phase). The spots on the TLC plates were detected by spraying 10% (v/v) H2SO4 followed by heating at 110°C for 10 min [30].
2.7. HPLC, NMR, and MS analysis of metabolite 1
The reaction mixture was extracted in n-butanol saturated with H2O and evaporated in a vacuum. HPLC was performed using an Agilent 1260 system (Agilent). HPLC-grade acetonitrile and water were purchased from SK Chemicals (Ulsan, Korea). Separation was performed on the C18 column (250 × 4.6 mm, ID 2.6 μm) using acetonitrile (solvent A) and distilled water (solvent B) mobile phases at 85% B for 5 min, 79% B for 20 min, 42% B for 55 min, 10% B for 12 min, and 85% B for 18 min at a flow rate of 1.6 mL/min. The sample was detected using UV (203 nm) absorbance [30]. The structure was identified using 1H-NMR and 13C-NMR. 1H-NMR and 13C-NMR spectra were obtained on a Bruker Av 600NMR spectrometer at 100 MHz with CD3OD as the solvent. MS analysis was performed on a Finnigan LCQ-Advantage mass spectrometer.
2.8. In vitro application in B16BL6 cell lines
2.8.1. Cell culture
B16BL6 cells were cultured in Dulbecco's modified Eagles medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in a humidified 95% air/5% CO2 atmosphere, as described previously [31].
2.8.2. Cell viability assay
Cell viability was determined for ginsenoside F1 and metabolite 1 using MTT (Life Technologies) conversion to formazan [32]. Cells were seeded at a density of 1 × 105 cells/well in a 96-well plate (Corning Costar), cultured for 24 h, and treated with various concentrations from 1μM to 200μM of F1 and metabolite 1 for 5 d [19]. Finally, 10 μL of MTT [5 mg/mL in phosphate-buffered saline (PBS)] was added to each well. Cells were incubated at 37°C for 3 h, and then dimethyl sulfoxide (DMSO, 100 μL) was added to dissolve the formazan crystals. The absorbance was measured at 570 nm with the reference wavelength of 630 nm using an ELISA reader (Bio-Tek Instruments, USA).
2.8.3. Melanin content measurement
The melanin content of the cultured B16BL6 cells was measured, as described previously with slight modifications [33], [34]. The cells were seeded at a density of 2 × 105 cells/well with α-MSH (1μM) in six-well plates. After 24 h, the cells were treated with 100μM F1, metabolite 1, or arbutin for 5 d. After the cells were washed with PBS, they were harvested by trypsinization. The cell pellet was solubilized in 500 μL of 1M NaOH containing 10% DMSO at 80°C for 1 h. The relative melanin content was determined by measuring the absorbance at 475 nm using an ELISA reader (Bio-Tek Instrument, USA). A standard curve for synthetic melanin (0–500 μg/mL) was prepared for each experiment. Melanin production was expressed as a percentage of untreated controls.
2.8.4. Tyrosinase activity assay
Tyrosinase activity was assayed indirectly via dopa oxidase activity [35], [36]. The cells were cultured in six-well plates at a density of 4 × 105 cells/well. After 24 h, the cells were treated with 100μM of F1, metabolite 1, or arbutin for 5 d. Next, the medium was removed, the cells were washed with ice-cold PBS, and subsequently lysed with phosphate buffer (pH 6.9) containing 1% Triton X-100. The mixture was freeze-thawed by incubating at −80°C for 15 min and then room temperature for 10 min. The samples were clarified by centrifugation at 12,000g for 15 min. After quantification of protein levels using the Bradford method and adjusting concentrations with lysis buffer, 10 μL of prewarmed freshly prepared substrate (15mM L-DOPA in 48mM sodium phosphate buffer pH 7.1) was added to 90 μL of supernatant and incubated at a 37°C for 1 h. The absorbance was then read at 475 nm using an ELISA reader (Bio-Tek Instrument, USA).
2.8.5. Statistical analysis
All data are presented as mean ± standard deviation (SD), and all experiments were independently performed in triplicate. The mean values of the treatment groups were compared with those of the untreated groups using Student t test. A p value of 0.05 or less was considered statistically significant.
3. Results and discussion
3.1. Heterologous expression of BSGT1 gene in E. coli BL21
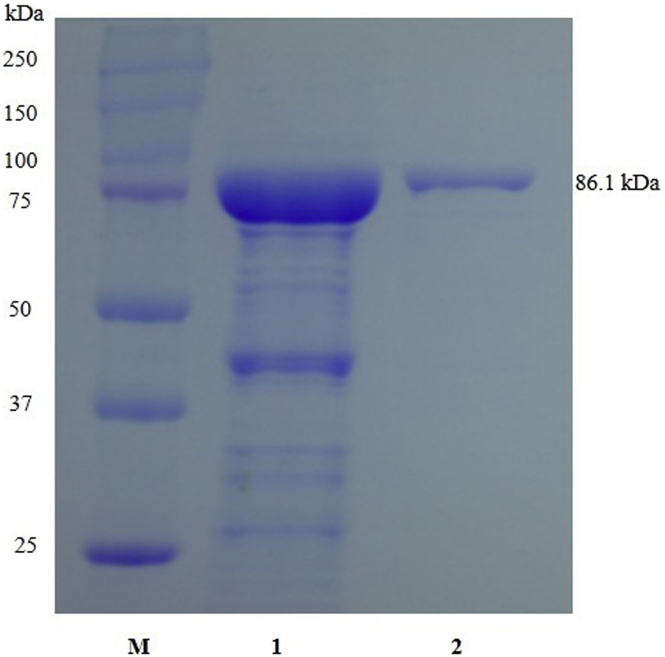
The BSGT1 gene was successfully cloned from the B. subtilis genome and consisted of 1,179 bp encoding 393 amino acid residues and was homologous to the protein domain of the GT family 1. The BSGT1 gene was inserted into a pMAL-C5X vector and then expressed in E. coli BL21 (DE3). Different conditions for protein induction were determined to maximize the yield of the fusion protein. Induction with 0.3mM IPTG at 20°C for 9 h produced the maximum level of soluble active fusion enzyme. The MBP-BSGT1 fusion protein was purified in MBP amylose resin, and then supernatant from cell lysates as well as purified protein was subjected to SDS-PAGE analysis [30]. The molecular weight of the recombinant MBP-BSGT1 calculated via an amino acid sequence was 86.12 kDa, which was similar to the mass of 86.1 kDa detected by SDS-PAGE (Fig. 1).
Fig. 1.
SDS-PAGE analysis of recombinant BSGT1. M, molecular mass marker; 1, crude extract of induced recombinant BL 21 (DE3) cells carrying pMAL-BSGT1; 2, pMAL-BSGT1 after purification by the maltose-binding protein-bound agarose resin.
3.2. Enzyme kinetic properties
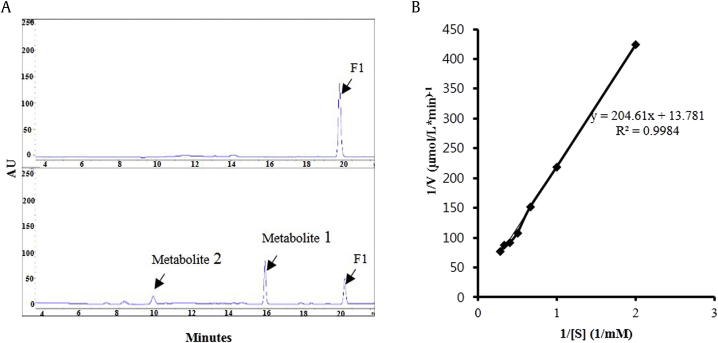
The kinetics of biosynthesis of metabolite 1 by BSGT1 were determined using the Michaelis–Menten model. The purified enzyme was incubated in the presence of a nonradiolabeled substrate UDP-glucose at concentrations varying from 0.5mM to 5mM. To measure the kinetic values, the product was detected by HPLC. The efficiency of BSGT1 is shown in Fig. 3A. Km and Vmax values were determined from the Lineweaver–Burk plot and were estimated to be 0.015 mol/L and 0.073 μmol/L*min, respectively.
Fig. 3.
(A) The Lineweaver–Burk plot of the recombinant BSGT1. (B) HPLC spectra of the transformation time course of ginsenoside F1 by recombinant BSGT1.
3.3. Biosynthesis of metabolites
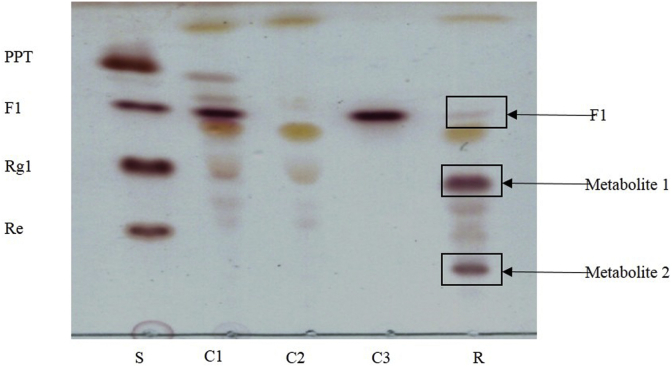
Transglycosylation is the formation of a glycosidic bond by transferring a glycosyl molecule from the donor to the acceptor. In this study, we aimed to synthesize a new transglycosylation metabolite from UDP-glucose as a donor and ginsenoside F1 as an acceptor using recombinant UDP-glycosyltransferase BSGT1. The mixtures were extracted by n-butanol, and a series of new spots were detected in TLC (Fig. 2). The new spots were not present in the control mixtures, which included ginsenoside F1 and enzyme without UDP-glucose, UDP-glucose and enzyme without ginsenoside F1, and UDP-glucose and ginsenoside F1 without enzyme. Two new spots appeared in TLC; metabolites 1 and 2 were detected by HPLC analysis (Fig. 3B). However, due to the low concentration of metabolite 2, we focused on the study of metabolite 1.
Fig. 2.
TLC analysis of transglycosylated products synthesized from ginsenoside F1 and UDP-glucose by BSGT1 enzyme. C1, enzyme and ginsenoside F1; C2, enzyme and UDP-glucose; C3, ginsenoside F1 and UDP-glucose; PPT, protopanaxatriol; S, standard; R, result of synthesis.
3.4. Optimal conditions of BSGT1 enzymes for metabolite production
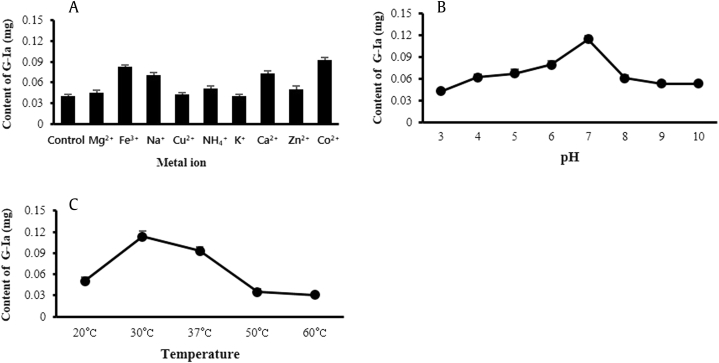
Based on the metal ion effect, the enzyme activity appeared to be significantly stimulated by Co2+, Mg2+, Fe3+, Na+, Cu2+, NH4+, K+, Ca2+, and Zn2+. Furthermore, the optimum metal ions with regard to enzyme activity were strongly affected by Co2+ (Fig. 4A). The enzyme showed optimal activity at pH 7 in 20mM sodium phosphate buffer and had more potent activity than those at other pH values (Fig. 4B). The optimum temperature range of BSGT1 was 20–37°C; however, the recombinant BSGT1 had the highest activity at 30°C (Fig. 4C).
Fig. 4.
(A) Effect of metal ions on recombinant BSGT1 activity in the synthesis of ginsenoside Ia. (B) Effect of pH on recombinant BSGT1 activity in the synthesis of ginsenoside Ia. (C) Effect of temperature on the recombinant BSGT1 activity in the synthesis of ginsenoside Ia.
3.5. Biotransformation pathway of ginsenoside F1 by BSGT1
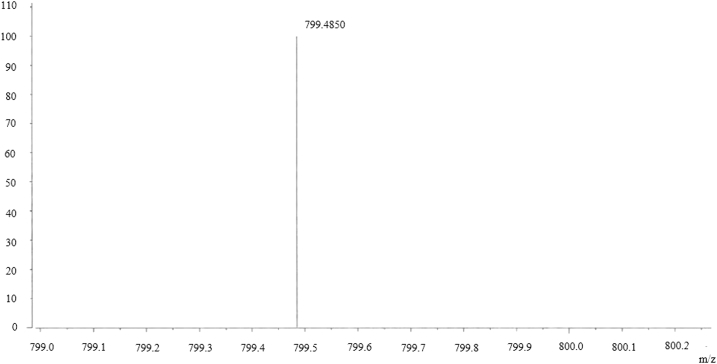
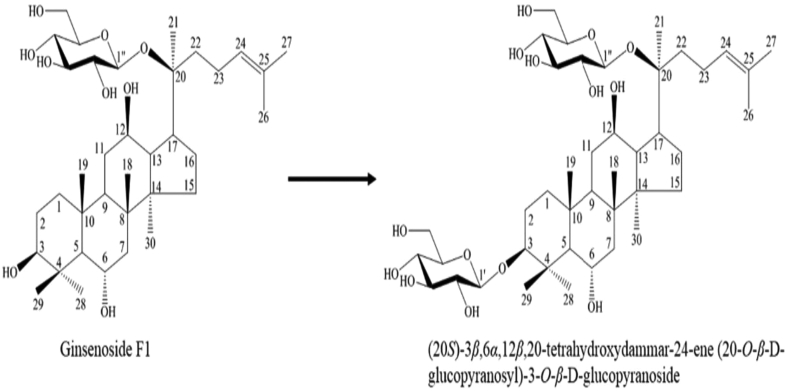
For large-scale production, final concentrations of 0.5mM ginsenoside F1, 2.5mM UDP-glucose, and 10mM Co2+ with BSGT1 enzyme (pH 7.0) were combined in 25 mL and reacted for 24 h at 30°C in a shaking incubator. The resulting metabolite 1 was purified by open silica column chromatography with a solvent system of CHCl3:CH3OH:H2O (65:35:10, v/v/v, lower phase); further separation and purification were achieved by repeated prep HPLC using a Nova-pak C18 column for further characterization [37]. The yield of metabolite 1 was 40.6%. Metabolite 1, a white amorphous powder, had the molecular formula C42H72O14 as deduced by a molecular ion peak at m/z 987.5532 ([M-H]–, calculated as C42H71O14 799.4844) (Fig. 5) in the negative HRFABMS and 13C-NMR (DEPT) spectra. The IR spectrum of metabolite 1 showed absorption bands at 3401 cm−1 and 1649 cm−1 due to an OH group and double bond, respectively. The 1H-NMR and 13C-NMR data were similar to those of ginsenoside F1 with the exception of an additional hexose sugar. The 13C-NMR signals of the sugar were observed as a hemiacetal (δC 107.1), four oxygenated methines (δC 78.2, 78.1, 76.4, 74.0), and an oxygenated methylene (δC 62.9), indicating the sugar to be β-glucopyranose. The β-configuration of the anomer hydroxyl group was confirmed from the coupling constant of the anomer proton signal (δH 4.31, d, J = 7.8 Hz). The sugar was linked to the C-3 hydroxyl group as observed from the chemical shift of C-3 (δC 90.8) downfield by 11.3 ppm compared to that of ginsenoside F1 owing to the glycosylation. Also, the positions of the two sugars were confirmed to be C-3 and C-20 from the HMBC spectrum. Two anomer proton signals H-1′and H-1″(δH 4.61, d, J = 7.8 Hz) showed cross peaks with the oxygenated methine carbon signal (δC 90.8, C-3) and the oxygenated quaternary carbon signal (δC 85.0, C-20), respectively. Taken together, these results indicate that metabolite 1 is (20S)-3β,6α,12β,20-tetrahydroxydammar-24-ene- 20-O-β-D-glucopyranosyl-3-O-β-D-glucopyranoside (Fig. 6), which is ginsenoside Ia (Supplementary Table 1) [28].
Fig. 5.
Mass spectrum of ginsenoside Ia after transformation by recombinant BSGT1.
Fig. 6.
Transformation reaction of ginsenoside F1 to (20S)-3β,6α,12β,20-tetrahydroxydammar-24-ene 20-O-β-D-glucopyranosyl-3-O-β-D-glucopyranoside by Bacillus subtilis.
Previously, ginsenoside Ia had been reported to be isolated from the leaves of P. ginseng Meyer [28]. In this work, we were successful to bioconvert ginsenoside F1 to rare ginsenoside Ia by means of recombinant UDP-glycosyltransferase isolated from B. subtillis. Gisenoside F1 has been reported to be able to modulate the clustering of B16 cells and the intracellular signaling cascade [19]. However, considering the poor solubility of ginsenoside and the absence of pharmacological activity of ginsenoside Ia in literatures, the glycosylation product Ia was expected to be applied as a novel skin cancer drug. Additionally, we aimed to conduct further in vitro study of ginsenoside Ia on B16BL6 skin cell line.
3.6. In vitro cytotoxicity assay of ginsenoside Ia on B16BL6 skin cells
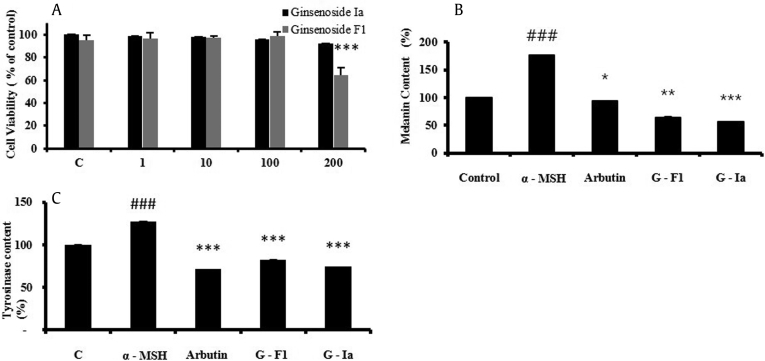
To evaluate the cytotoxic effects of ginsenoside Ia on melanoma cells, we performed the assay for over 5 d. The cell viability assay results are shown in Fig. 7A; the cell viabilities were greater than 90% even at the highest concentration of ginsenoside Ia (200μM). We compared these results to the MTT results for ginsenoside F1, which showed 68% cell viability at 200μM.
Fig. 7.
(A) Cell viability (%) of B16BL6 cells exposed to ginsenoside Ia and ginsenoside F1. Cell viability was determined by the MTT assay. Cells (1 × 105 cells/well) were incubated with various concentrations of ginsenoside Ia or F1 for 5 d. Data are expressed as a percentage of untreated control and reflect the mean ± standard deviation of two separate experiments. *** p < 0.0001 versus control. (B) Effect of ginsenoside Ia on melanin content. Cells (2 × 105 cells/well) were incubated with 100 μM of G-Ia, G-F1, or arbutin in the presence of 1μM of α-melanocyte-stimulating hormone (MSH) for 5 d. Melanin and protein content were determined as described in the Materials and methods section. Data are expressed as a percentage of α-MSH treated control and presented as mean ± SDM of two separate experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 versus α-MSH-treated control. (C) Effect of ginsenoside Ia on tyrosinase activity. Cells (4 × 105 cells/well) were incubated with 100μM of G-Ia, G-F1, or arbutin in the presence of 1μM α-MSH for 5 d. Tyrosinase activity in cellular lysates was determined as described in the Materials and methods section. Data are expressed as a percentage of α-MSH-treated control and presented as mean ± standard deviation of two separate experiments. * p < 0.05, ** p < 0.01, *** p < 0.001 versus α-MSH-treated control.
3.7. Inhibition effect of ginsenoside Ia on melanin and tyrosinase activity on B16BL6 skin cells
Melanin is chiefly responsible for the dark color of skin by UV irradiation [38]. Furthermore, the tyrosinase is the key enzyme for the formation of melanin. Thus, the inhibition of content of melanin and tyrosinase is critical for the reduction of skin darkness [39]. Previously Yoo et al [19] reported the whitening effect of ginsenoside F1 on B16 cells [19]. Based on the chemical similarity between ginsenoside F1 and ginsenoside Ia, we assay the effect of ginsenoside Ia on melanin synthesis and tyrosinase regulation in B16BL6 cells after stimulation with α-MSH. Fig. 7B shows that the treatment with α-MSH increases the melanin production in B16 cells, and this induction could be inhibited by ginsenoside Ia. Cellular melanin content was significantly decreased to 56 ± 0.02% at 100μM after treatment with ginsenoside Ia. However, cells treated with equal concentrations of arbutin and ginsenoside F1 showed that the melanin content decreased to 96 ± 0.02% and 65 ± 0.02%, respectively. Ginsenoside Ia decreased cellular tyrosinase activity in B16 cells (Fig. 7C); this result is in agreement with the decrease of melanin content. Tyrosinase enzyme is a key regulator in melanin production in cells. Ginsenoside Ia resulted in a greater decrease in the cellular tyrosinase activity in α-MSH-stimulated B16BL6 cells than F1, similar to that observed with an equal concentration of arbutin.
Overall, ginsenoside Ia showed better efficacy than F1 and arbutin in preventing pigmentation, and it can be applied as a newer and more potent skin-whitening agent in cosmetics.
4. Conclusion
In this study, the glycosyltransferase BSGT1 gene from B. subtilis was cloned and overexpressed in E. coli BL 21 (DE3). The recombinant enzyme was purified and characterized. The molecular weight of the purified MBP-BSGT1 was 86.1 kDa as determined by SDS-PAGE. Ginsenoside F1 was transformed into (20S)-3β,6α,12β,20-tetrahydroxydammar-24-ene-20-O-β-D-glucopyranosyl-3-O-β-D-glucopyranoside (ginsenoside Ia) by GT with high efficiency and specificity to the C-3 position of F1. Therefore, the enzyme is considered potentially useful for mass production of arduous ginsenoside Ia from F1.
Skin-whitening compounds suppress melanogenesis in B16BL6 melanoma cells through cytotoxic effects on melanocytes, direct tyrosinase inhibition, and melanin biosynthesis inhibition. In the present study, we investigated the inhibitory effect of enzymatically-synthesized ginsenoside Ia on melanogenesis in B16BL6 cells. Ginsenoside Ia had no significant cytotoxicity at 0–200μM. Furthermore, ginsenoside Ia significantly suppressed melanin synthesis and tyrosinase activity at 100μM, which demonstrated greater efficacy than ginsenoside F1 and arbutin at the same concentration. These results suggest that ginsenoside Ia decreases cellular melanin synthesis in B16BL6 cells through downregulation of tyrosinase activity.
In conclusion, we have identified a potential antimelanogenic ginsenoside Ia which decreases melanin production without reducing B16BL6 melanin cell viability. However, further study is needed to determine the molecular mechanism of melanin content reduction by ginsenoside Ia in B16BL6 cells.
Conflicts of interest
The authors have no conflicts of interest to report.
Acknowledgments
This research was supported by the Next-Generation BioGreen 21 Program (SSAC, grant#: PJ0120342016), Rural Development Administration, Korea.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2016.12.009.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Hu Y., Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 2.Luzhetskyy A., Bechthold A. Features and applications of bacterial glycosyltransferases: current state and prospects. Appl Microbiol Biotechnol. 2008;80:945–952. doi: 10.1007/s00253-008-1672-2. [DOI] [PubMed] [Google Scholar]
- 3.Taniguchi N., Honke K., Fukuda M. Springer Science & Business Media; Tokyo: 2002. Handbook of glycosyltransferases and related genes. [Google Scholar]
- 4.Campbell J.A., Davies G.J., Bulone V., Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326:929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyung Ko J., Gyu Kim B., Joong-Hoon A. Glycosylation of flavonoids with a glycosyltransferase from Bacillus cereus. FEMS Microbiol Lett. 2006;258:263–268. doi: 10.1111/j.1574-6968.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- 6.Ivanova N., Sorokin A., Anderson I., Galleron N., Candelon B., Kapatral V., Bhattacharyya A., Reznik G., Mikhailova N., Lapidus A. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 7.Luo S.L., Dang L.Z., Zhang K.Q., Liang L.M., Li G.H. Cloning and heterologous expression of UDP-glycosyltransferase genes from Bacillus subtilis and its application in the glycosylation of ginsenoside Rh1. Lett Appl Microbiol. 2015;60:72–78. doi: 10.1111/lam.12339. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y.J., Jeon J.N., Jang M.G., Oh J.Y., Kwon W.S., Jung S.K., Yang D.C. Ginsenoside profiles and related gene expression during foliation in Panax ginseng Meyer. J Ginseng Res. 2014;38:66–72. doi: 10.1016/j.jgr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng CA Meyer. Acta Pharmacol Sin. 2008;29:1109. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 10.Sathishkumar N., Sathiyamoorthy S., Ramya M., Yang D.U., Lee H.N., Yang D.C. Molecular docking studies of anti-apoptotic BCL-2, BCL-XL, and MCL-1 proteins with ginsenosides from Panax ginseng. J Enzyme Inhib Med Chem. 2012;27:685–692. doi: 10.3109/14756366.2011.608663. [DOI] [PubMed] [Google Scholar]
- 11.Lee K.Y., Lee Y., Kim S., Park J.H., Lee S. Ginsenoside Rg5 suppresses cyclin E-dependent protein kinase activity via up-regulating p21Cip/WAF1 and down-regulating cyclin E in SK-HEP-1 cells. Anticancer Res. 1996;17:1067–1072. [PubMed] [Google Scholar]
- 12.Ahn S., Siddiqi M.H., Noh H.Y., Kim Y.J., Kim Y.J., Jin C.G., Yang D.C. Anti-inflammatory activity of ginsenosides in LPS-stimulated RAW 264.7 cells. Sci Bull. 2015;60:773–784. [Google Scholar]
- 13.Mathiyalagan R., Subramaniyam S., Kim Y.J., Kim Y.C., Yang D.C. Ginsenoside compound K-bearing glycol chitosan conjugates: synthesis, physicochemical characterization, and in vitro biological studies. Carbohydr Polym. 2014;112:359–366. doi: 10.1016/j.carbpol.2014.05.098. [DOI] [PubMed] [Google Scholar]
- 14.Ni H.X., Yu N.J., Yang X.H. The study of ginsenoside on PPARγ expression of mononuclear macrophage in type 2 diabetes. Mol Biol Rep. 2010;37:2975–2979. doi: 10.1007/s11033-009-9864-0. [DOI] [PubMed] [Google Scholar]
- 15.Cho W.C., Chung W.S., Lee S.K., Leung A.W., Cheng C.H., Yue K.K. Ginsenoside Re of Panax ginseng possesses significant antioxidant and antihyperlipidemic efficacies in streptozotocin-induced diabetic rats. Eur J Pharmacol. 2006;550:173–179. doi: 10.1016/j.ejphar.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 16.Cheng L.Q., Na J.R., Kim M.K., Bang M.H., Yang D.C. Microbial conversion of ginsenoside Rb1 to minor ginsenoside F2 and gypenoside XVII by Intrasporangium sp. GS603 isolated from soil. J Microbiol Biotechnol. 2007;17:1937–1943. [PubMed] [Google Scholar]
- 17.Park C.S., Yoo M.H., Noh K.H., Oh D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl Microbiol Biotechnol. 2010;87:9–19. doi: 10.1007/s00253-010-2567-6. [DOI] [PubMed] [Google Scholar]
- 18.Lee E.H., Cho S.Y., Kim S.J., Shin E.S., Chang H.K., Kim D.H., Yeom M.H., Woe K.S., Lee J., Sim Y.C. Ginsenoside F1 protects human HaCaT keratinocytes from ultraviolet-B-induced apoptosis by maintaining constant levels of Bcl-2. J Invest Dermatol. 2003;121:607–613. doi: 10.1046/j.1523-1747.2003.12425.x. [DOI] [PubMed] [Google Scholar]
- 19.Yoo D.S., Rho H.S., Lee Y.G., Yeom M.H., Kim D.H., Lee S.J., Hong S., Lee J., Cho J.Y. Ginsenoside F1 modulates cellular responses of skin melanoma cells. J Ginseng Res. 2011;35:86–91. [Google Scholar]
- 20.Liu Y., Ma H., Zhang J.W., Deng M.C., Yang L. Influence of ginsenoside Rh1 and F1 on human cytochrome p450 enzymes. Planta Med. 2006;72:126. doi: 10.1055/s-2005-873197. [DOI] [PubMed] [Google Scholar]
- 21.Lairson L., Henrissat B., Davies G., Withers S. Glycosyltransferases: structures, functions, and mechanisms. Biochem. 2008;77:521. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 22.Shimoda K., Otsuka T., Morimoto Y., Hamada H., Hamada H. Glycosylation and malonylation of quercetin, epicatechin, and catechin by cultured plant cells. Chem Lett. 2007;36:1292–1293. [Google Scholar]
- 23.Weymouth-Wilson A.C. The role of carbohydrates in biologically active natural products. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]
- 24.Hong H., Cui C.H., Kim J.K., Jin F.X., Kim S.C., Im W.T. Enzymatic biotransformation of ginsenoside Rb1 and gypenoside XVII into ginsenosides Rd and F2 by recombinant beta-glucosidase from Flavobacterium johnsoniae. J Ginseng Res. 2012;36:418–424. doi: 10.5142/jgr.2012.36.4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quan L.H., Min J.W., Jin Y., Wang C., Kim Y.J., Yang D.C. Enzymatic biotransformation of ginsenoside Rb1 to compound K by recombinant beta-glucosidase from Microbacterium esteraromaticum. J Agric Food Chem. 2012;60:3776–3781. doi: 10.1021/jf300186a. [DOI] [PubMed] [Google Scholar]
- 26.Solano F., Briganti S., Picardo M., Ghanem G. Hypopigmenting agents: an updated review on biological, chemical and clinical aspects. Pigment Cell Res. 2006;19:550–571. doi: 10.1111/j.1600-0749.2006.00334.x. [DOI] [PubMed] [Google Scholar]
- 27.Wen K.C., Chang C.S., Chien Y.C., Wang H.W., Wu W.C., Wu C.S., Chiang H.M. Tyrosol and its analogues inhibit alpha-melanocyte-stimulating hormone induced melanogenesis. Int J Mol Sci. 2013;14:23420–23440. doi: 10.3390/ijms141223420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dou D., Wen Y., Pei Y., Yao X., Chen Y., Kawai H., Fukushima H. Ginsenoside-Ia: a novel minor saponin from the leaves of Panax ginseng. Planta Med. 1996;62:179–181. doi: 10.1055/s-2006-957849. [DOI] [PubMed] [Google Scholar]
- 29.Thuan N.H., Yamaguchi T., Lee J.H., Sohng J.K. Characterization of sterol glucosyltransferase from Salinispora tropica CNB-440: potential enzyme for the biosynthesis of sitosteryl glucoside. Enzyme Microb Technol. 2013;52:234–240. doi: 10.1016/j.enzmictec.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Quan L.H., Min J.W., Yang D.U., Kim Y.J., Yang D.C. Enzymatic biotransformation of ginsenoside Rb1 to 20(S)-Rg3 by recombinant beta-glucosidase from Microbacterium esteraromaticum. Appl Microbiol Biotechnol. 2012;94:377–384. doi: 10.1007/s00253-011-3861-7. [DOI] [PubMed] [Google Scholar]
- 31.Jung G.D., Yang J.Y., Song E.S., Par J.W. Stimulation of melanogenesis by glycyrrhizin in B16 melanoma cells. Exp Mol Med. 2001;33:131–135. doi: 10.1038/emm.2001.23. [DOI] [PubMed] [Google Scholar]
- 32.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 33.Hosoi J., Abe E., Suda T., Kuroki T. Regulation of melanin synthesis of B16 mouse melanoma cells by 1α, 25-dihydroxyvitamin D3 and retinoic acid. Cancer Res. 1985;45:1474–1478. [PubMed] [Google Scholar]
- 34.Lim Y.J., Lee E.H., Kang T.H., Ha S.K., Oh M.S., Kim S.M., Yoon T.J., Kang C., Park J.H., Kim S.Y. Inhibitory effects of arbutin on melanin biosynthesis of α-melanocyte stimulating hormone-induced hyperpigmentation in cultured brownish Guinea pig skin tissues. Arch Pharma Res. 2009;32:367–373. doi: 10.1007/s12272-009-1309-8. [DOI] [PubMed] [Google Scholar]
- 35.Chiang H.M., Chien Y.C., Wu C.H., Kuo Y.H., Wu W.C., Pan Y.Y., Su Y.H., Wen K.C. Hydroalcoholic extract of Rhodiola rosea L. (Crassulaceae) and its hydrolysate inhibit melanogenesis in B16F0 cells by regulating the CREB/MITF/tyrosinase pathway. Food Chem Toxicol. 2014;65:129–139. doi: 10.1016/j.fct.2013.12.032. [DOI] [PubMed] [Google Scholar]
- 36.Jeong Y.M., Oh W.K., Tran T.L., Kim W.K., Sung S.H., Bae K., Lee S., Sung J.H. Aglycone of Rh4 inhibits melanin synthesis in B16 melanoma cells: possible involvement of the protein kinase A pathway. Biosci Biotechnol Biochem. 2013;77:119–125. doi: 10.1271/bbb.120602. [DOI] [PubMed] [Google Scholar]
- 37.Mathiyalagan R., Kim Y.H., Kim Y.J., Kim M.K., Kim M.J., Yang D.C. Enzymatic formation of novel ginsenoside Rg1-alpha-glucosides by rat intestinal homogenates. Appl Biochem and Biotechnol. 2015;177:1701–1715. doi: 10.1007/s12010-015-1847-0. [DOI] [PubMed] [Google Scholar]
- 38.Wood J.M., Gibbons N.C., Schallreuter K.U. Melanocortins in human melanocytes. Cell Mol Biol (Noisy-le-grand) 2006;52:75–78. [PubMed] [Google Scholar]
- 39.Kim S.J., Son K.H., Chang H.W., Kang S.S., Kim H.P. Tyrosinase inhibitory prenylated flavonoids from Sophora flavescens. Biol Pharm Bull. 2003;26:1348–1350. doi: 10.1248/bpb.26.1348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.