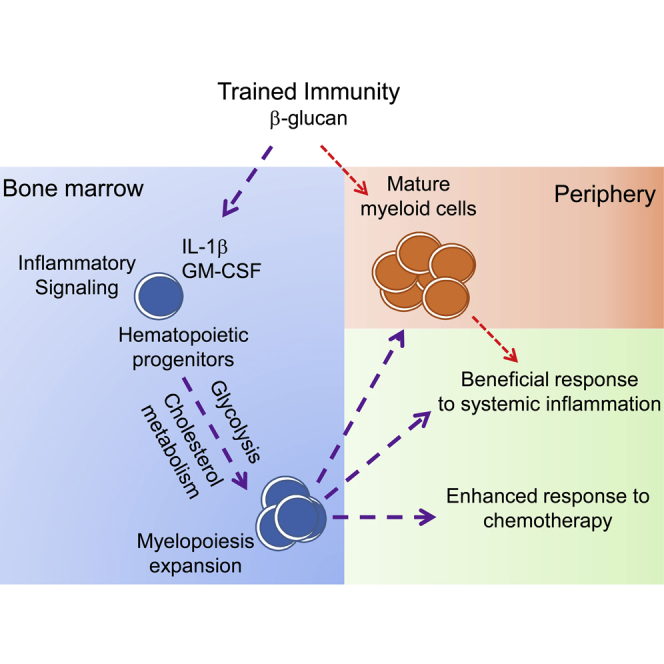
Summary
Trained innate immunity fosters a sustained favorable response of myeloid cells to a secondary challenge, despite their short lifespan in circulation. We thus hypothesized that trained immunity acts via modulation of hematopoietic stem and progenitor cells (HSPCs). Administration of β-glucan (prototypical trained-immunity-inducing agonist) to mice induced expansion of progenitors of the myeloid lineage, which was associated with elevated signaling by innate immune mediators, such as IL-1β and granulocyte-macrophage colony-stimulating factor (GM-CSF), and with adaptations in glucose metabolism and cholesterol biosynthesis. The trained-immunity-related increase in myelopoiesis resulted in a beneficial response to secondary LPS challenge and protection from chemotherapy-induced myelosuppression in mice. Therefore, modulation of myeloid progenitors in the bone marrow is an integral component of trained immunity, which to date, was considered to involve functional changes of mature myeloid cells in the periphery.
Keywords: trained innate immunity, β-glucan, innate immune memory, myelopoiesis, inflammation, interleukin-1β, myelosuppression, cholesterol biosynthesis, glycolysis, GM-CSF
Graphical Abstract
Highlights
-
•
Trained immunity (TI) modulates hematopoietic progenitors in bone marrow
-
•
TI is associated with adaptations in cell metabolism in progenitors
-
•
TI increases expansion of hematopoietic progenitors and myelopoiesis
-
•
TI promotes beneficial responses to systemic inflammation and chemotherapy
Modulation of hematopoietic stem and progenitor cells during trained immunity allows a sustained response of myeloid cells to a secondary challenge despite their short lifespan in circulation.
Introduction
Recent studies have shown that certain microbial challenges or vaccines promote a heightened response of myeloid cell populations to a subsequent infection with the same or even different pathogens. This process involves changes in cell transcription and is designated “trained immunity” (Goodridge et al., 2016, Jensen et al., 2015, Netea and van Crevel, 2014, Netea et al., 2016, Quintin et al., 2012). Pathogen-associated molecular patterns or cytokines induce a complex immunometabolic program in monocytes and macrophages, enabling a robust cellular response to re-stimulation, especially with regard to cytokine production (Arts et al., 2016a, Netea et al., 2016, Quintin et al., 2012). Transcriptomic and metabolomic analysis have implicated metabolic pathways, especially glucose metabolism, in the adaptation of monocytes and macrophages in the context of innate immune memory (Arts et al., 2016a, Arts et al., 2016b, Cheng et al., 2014, Netea et al., 2016). However, the long-term effects (up to months) of trained immunity on circulating monocytes are puzzling, as these cells have a relatively short lifespan in the circulation. We have, thus, hypothesized that the adaptive processes induced by trained immunity involve alterations to progenitors of the hematopoietic system, a concept that has not been addressed yet.
Hematopoiesis is a hierarchical system, in which hematopoietic stem and progenitor cells (HSPCs) are responsible for its maintenance via their differentiation into progressively committed progenitors and mature cells (Trumpp et al., 2010). HSPCs are able to respond to stress, such as severe infection, systemic inflammation, or iatrogenic myeloablation, by increasing their proliferation rate and hematopoiesis (Trumpp et al., 2010, Zhao and Baltimore, 2015). Myeloid lineage commitment and differentiation of hematopoietic and myeloid progenitors are orchestrated by myeloid-lineage-specific growth factors (Boettcher and Manz, 2017, Mossadegh-Keller et al., 2013, Sarrazin and Sieweke, 2011) and cytokines, such as interleukin (IL)-1β (Pietras et al., 2016). These mediators act on HSPCs and myeloid progenitors driving myelopoiesis via inducing transcriptional networks involving transcription factors (TFs) and expression of myeloid-lineage-specific genes (Boettcher and Manz, 2017, Rosenbauer and Tenen, 2007).
Cell metabolic pathways have been implicated in the regulation of HSPC function. Aerobic glycolysis promotes the quiescence—and, thus, maintenance—of long-term hematopoietic stem cells (LT-HSCs) (Simsek et al., 2010, Takubo et al., 2013, Wang et al., 2014), whereas oxidative phosphorylation (OXPHOS) is prominent in less primitive progenitors (Simsek et al., 2010). Accordingly, disruption of the glycolytic process impairs LT-HSC stemness, leading to loss of their self-renewal potential (Simsek et al., 2010, Takubo et al., 2013, Wang et al., 2014). In turn, fatty acid oxidation drives asymmetric division and differentiation (Ito et al., 2012), while cholesterol accumulation is associated with skewing of hematopoiesis toward myeloid lineage (Murphy et al., 2011, Yvan-Charvet et al., 2010).
Our present findings support the hypothesis stated earlier that trained immunity acts at the level of hematopoietic progenitors and specifically involves favorable adaptations in myelopoiesis. Indeed, we show that β-glucan administration induced the expansion of HSPCs, which was associated with IL-1β and GM-CSF (granulocyte-macrophage colony-stimulating factor) signaling as well as with changes in glucose and lipid metabolism. Importantly, these progenitor adaptations, as conferred by trained immunity, promoted a beneficial response to a secondary inflammatory challenge and gave protection from chemotherapy-induced myeloablation.
Results
β-Glucan-Induced Trained Immunity Acts in the Bone Marrow and Enhances Myelopoiesis
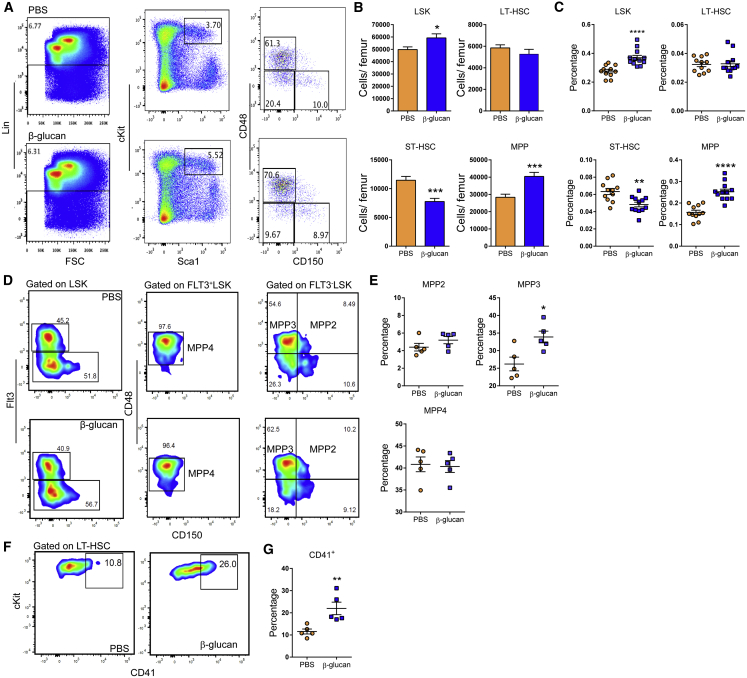
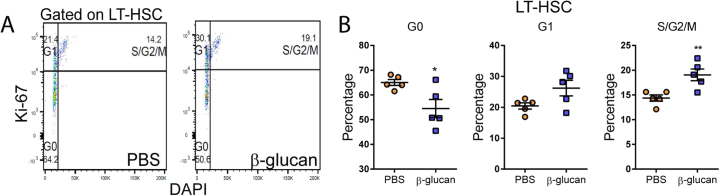
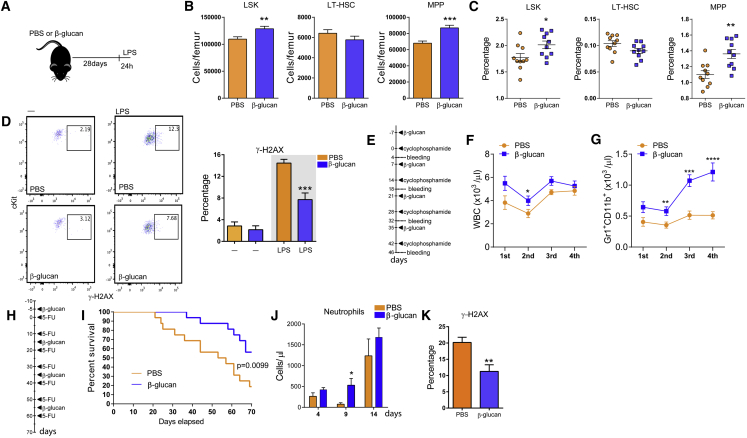
To study the response of the hematopoietic and myeloid progenitor pool to agonists implicated in innate immune training, mice received a single intraperitoneal injection of β-glucan. After 24 hr, β-glucan increased the numbers and frequency of hematopoietic progenitors (LSKs; Lin−cKit+Sca1+) and multipotent progenitors (MPPs; CD48+CD150−LSK) in the bone marrow (BM), as compared to injection of PBS (Figures 1A–1C). No difference was observed in LT-HSCs (CD48−CD150+LSK), while the numbers and frequency of short-term (ST)-HSCs (CD48−CD150−LSK) were decreased (Figures 1A–1C). Cell-cycle analysis in LT-HSCs revealed that β-glucan led to enhanced cell-cycle progression in HSPCs (Figure S1), thus suggesting enhanced differentiation-associated proliferation of LT-HSCs upon β-glucan administration. The β-glucan-induced increase in MPPs was linked to an increase in the frequency of the MPP3 subset (Flt3−CD48+CD150−LSK), which is biased toward the myeloid lineage (Pietras et al., 2015) (Figures 1D and 1E). In contrast, we found no significant difference in the lymphoid-biased MPP4 (Flt3+CD48+CD150−LSK) subset (Pietras et al., 2015) (Figures 1D and 1E). Moreover, β-glucan induced an increase in the frequency of CD41+ LT-HSCs (Figures 1F and 1G), which are considered the myeloid-biased LT-HSC subpopulation (Gekas and Graf, 2013), although the overall absolute numbers of LT-HSCs remained unaltered (Figure 1B).
Figure 1.
Administration of β-Glucan Drives Expansion of HSPC Subpopulations
WT mice were injected with β-glucan or PBS, and BM analysis was performed after 24 hr.
(A) Representative fluorescence-activated cell sorting (FACS) plots for the identification of hematopoietic progenitor cells. After gating for Lin− cells, LSK cells were characterized as cKit+Sca1+ cells. LSK cells subpopulations were further characterized as MPP (CD48+CD150−LSK), ST-HSC (CD48−CD150−LSK), and LT-HSC (CD48−CD150+LSK).
(B and C) Cell numbers of LSKs, MPPs, ST-HSCs, and LT-HSCs (B) and cell percentages of the same populations in total BM cells (C) of mice at 24 hr after the administration of PBS or β-glucan (ns = 11 and 12 mice).
(D) Representative FACS plots for the identification of MPP subpopulations. After gating for LSK cells, MPP4 cells are characterized as CD48+Flt3+CD150−LSK, MPP3 cells are characterized as CD48+Flt3−CD150−LSK, and MPP2 cells are characterized as CD48+Flt3−CD150+LSK.
(E) Frequency of MPP subpopulations in LSK cells in the BM of mice at 24 hr after the administration of PBS or β-glucan (n = 5 mice per group).
(F and G) Representative FACS plots for the identification of CD41+ LT-HSCs (F) and frequency of CD41+ LT-HSCs (in total LT-HSCs) (G) in the BM of mice at 24 hr after the administration of PBS or β-glucan (n = 5 mice per group).
Data are presented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
See also Figure S1.
Figure S1.
Administration of β-Glucan Promotes Cell Proliferation of LT-HSCs, Related to Figure 1
(A and B) Cell cycle analysis was performed in LT-HSC at 24h after the administration of PBS or β-glucan by staining for Ki67 and DAPI. (A) Representative flow cytometry plots and (B) frequency of LT-HSC at different phases of the cell cycle (n = 5 mice per group). Data presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
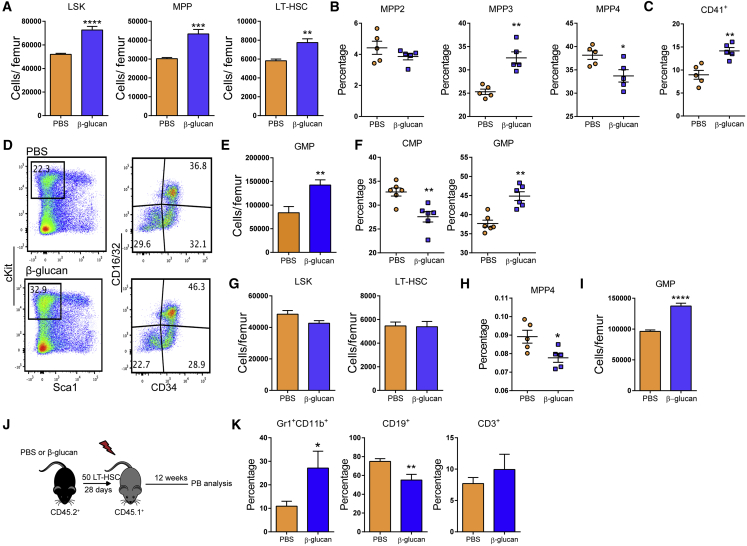
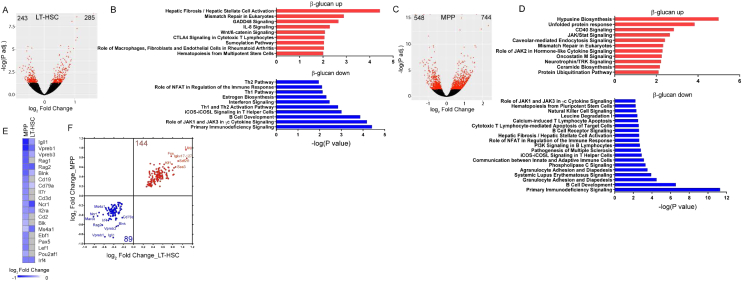
Seven days after β-glucan injection, we observed increased LSK, MPP, and LT-HSC cell numbers (Figure 2A), with an elevated frequency of myeloid-biased MPP3 cells, a corresponding decrease in the frequency of lymphoid-biased MPP4 cells (Figure 2B), and increased frequency of CD41+LT-HSCs (Figure 2C), as compared to findings with the injection of PBS. Analysis of myeloid progenitors (MyPs; Lin−cKit+Sca1−) at the 7-day time point revealed that β-glucan resulted in enhanced granulocyte macrophage progenitor (GMP; Lin−c-Kit+Sca1−CD16/32+CD34+) numbers and in an increased proportion of GMPs within the MyP population, accompanied by a corresponding reduction in the relative abundance of common myeloid progenitors (CMPs; Lin−c-Kit+Sca1−CD16/32−CD34+), as compared to control mice (Figures 2D–2F).
Figure 2.
Sustained Increase in Myelopoiesis upon β-Glucan Administration
(A–F) WT mice were injected with β-glucan or PBS, and BM analysis was performed after 7 days.
(A) LSK, MPP and LT-HSC cell numbers in the BM of mice on day 7 after administration of PBS or β-glucan (n = 6 mice per group).
(B) Frequency of MPP subpopulations in the LSK cells 7 days after β-glucan or PBS administration (n = 5 mice per group).
(C) Frequency of CD41+ LT-HSCs (in total LT-HSCs) on day 7 after the administration of PBS or β-glucan (n = 5 mice per group).
(D) Representative FACS plots for the identification of MyP subpopulations.
(E and F) GMP cell numbers (E) and frequency within the MyP pool of GMPs (Lin−c-Kit+Sca1−CD16/32+CD34+) and CMPs (Lin−c-Kit+Sca1−CD16/32−CD34+) (F) in the BM of mice on day 7 after the administration of PBS or β-glucan (n = 6 mice per group).
(G–I) WT mice were injected with β-glucan or PBS, and BM analysis was performed after 28 days.
(G) LSK and LT-HSC cell numbers (n = 5 mice per group).
(H and I) Frequency of MPP4 cells in total BM cells (H) and GMP cell numbers in the BM (I) of mice on day 28 after the administration of PBS or β-glucan (n = 5 mice per group).
(J and K) Transplantation.
(J) LT-HSCs (CD45.2+) were sorted 28 days after β-glucan or PBS administration and transplanted to lethally irradiated SJL/BL6 (CD45.1+) mice. CD45.1+ BM cells were co-transplanted in order to ensure the survival of recipients.
(K) Lineage output of donor LT-HSCs (CD45.2+) in peripheral blood of recipients at week 12 post-transplant (n = 10 recipient mice per group).
Data are presented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
See also Figure S2.
Innate immune training has a long-term adaptive effect on myeloid cells (Netea et al., 2016). We thus investigated whether the effects of β-glucan on hematopoiesis were sustained over a longer period. Although, 28 days after administration of β-glucan, the numbers of LSKs and LT-HSCs were not affected (Figure 2G), β-glucan induced a significant decrease in the frequency of lymphoid-biased MPP4 cells (Figure 2H) without altering the frequency of MPP3 cells (data not shown). In addition, a significant increase in the numbers of GMPs was observed (Figure 2I). To expand on these findings, we tested the β-glucan-induced effects on HSPCs through a transplantation experiment using the CD45.1/CD45.2 congenic system (Figure 2J). To this end, LT-HSCs were isolated from mice at 28 days after β-glucan or PBS administration and transferred to lethally irradiated recipient mice. Lineage output of donor-derived cells was studied 12 weeks post-transplantation. Peripheral blood analysis demonstrated that LT-HSCs from β-glucan-injected mice gave rise to an increased proportion of Gr1+CD11b+ myeloid cells with a corresponding reduction in the proportion of CD19+ B cells, as compared to LT-HSCs transplanted from PBS-treated mice (Figure 2K). No difference was observed in peripheral chimerism between LT-HSCs from PBS and β-glucan-injected mice, suggesting that the altered lineage output of the cells from the β-glucan group was not due to differences in reconstitution potential (Figure S2). Together, β-glucan induces a sustained increase in myelopoiesis.
Figure S2.
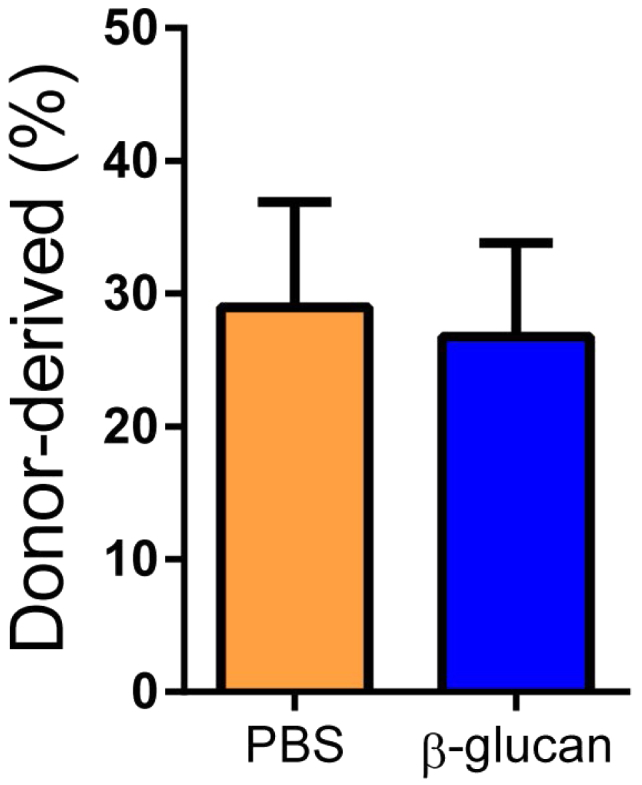
Peripheral Blood Chimerism in Recipients of LT-HSCs Isolated from β-Glucan-Administered or PBS-Injected Mice, Related to Figure 2
Briefly, LT-HSCs (CD45.2+) were sorted 28 days after β-glucan or PBS administration and transplanted to lethally irradiated SJL/BL6 (CD45.1+) mice. The percentage of donor-derived (CD45.2+) cells in peripheral blood of recipient mice at 12 weeks after transplantation is shown (n = 10 recipient mice per group). Data presented as mean ± SEM.
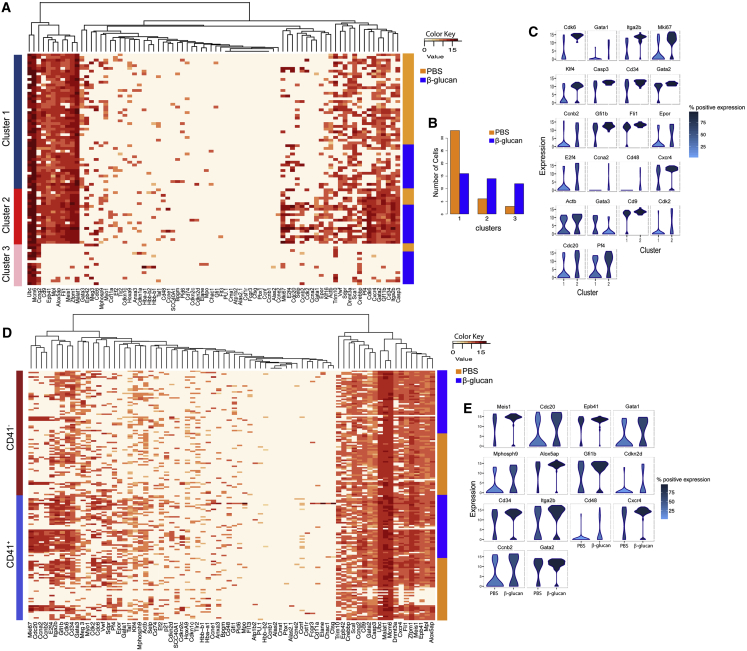
To dissect the effects of β-glucan, LT-HSCs isolated from mice 24 hr after β-glucan injection were analyzed by single-cell qPCR in terms of expression of genes linked to HSC maintenance, cell cycle, and differentiation. Hierarchical clustering analysis revealed that LT-HSCs clustered in three subpopulations, in accordance to previously published data (Yang et al., 2017). Relative to LT-HSCs from PBS-treated mice, LT-HSCs from β-glucan-treated mice showed higher association with a subpopulation (cluster #2; Figures 3A and 3B) that displayed an activated cell-cycle program, as indicated by enhanced expression of Mki67; the S/G2/M cyclins Ccna2, Ccnb2, and Cdc20; and the cyclin-dependent kinases Cdk2 and Cdk6 (Passegué et al., 2005, Yamada et al., 2013) (Figures 3A–3C). Moreover, cluster #2 showed increased expression of Itga2b (Cd41), which identifies myeloid-biased LT-HSCs (Gekas and Graf, 2013), and of the differentiation markers Cd34 and Cd48 (Wilson et al., 2008), while it showed decreased Gata3 expression, which regulates T cell-lineage development (Frelin et al., 2013, Hosoya et al., 2009) (Figures 3A–3C).
Figure 3.
Single-Cell Transcriptional Analysis in LT-HSCs upon β-Glucan Administration
(A–C) Single-cell qPCR in LT-HSCs isolated from mice at 24 hr after administration of PBS or β-glucan (n = 42 cells per condition).
(A and B) Hierarchical clustering analysis (A) and distribution of LT-HSCs in the three identified clusters (B) at 24 hr after the administration of PBS or β-glucan.
(C) Violin plots indicating genes with significantly altered expression between clusters 1 and 2. The y axis represents gene expression. The horizontal width of the plot shows the density of the data along the y axis. Color key represents the percentage of cells that express the specific gene.
(D and E) Single-cell qPCR was performed in CD41− and CD41+ LT-HSCs isolated from mice at 24 hr after the administration of PBS or β-glucan. Hierarchical clustering analysis (D) and violin plots indicating genes with significantly altered expression between CD41+ LT-HSCs from PBS and β-glucan-treated mice (E).
We next sorted CD41− and CD41+ LT-HSCs isolated from mice 24 hr after β-glucan or PBS injection and performed single-cell qPCR analysis. We found that the expression of the cell-cycle-associated genes Cdc20, Ccnb2, and Cdkn2d; of the TFs Meis1, Gata1, Gfi1b, and Gata2; and of the differentiation markers Itga2b, Cd34, and Cd48 was enhanced in CD41+ LT-HSCs (but not in CD41− LT-HSCs) from β-glucan-treated mice, as compared to CD41+ LT-HSCs from PBS control-treated mice (Figures 3D and 3E). These data suggest that β-glucan acts predominantly on myeloid-biased CD41+ LT-HSCs.
Training with β-Glucan Mediates a Favorable Response to Secondary Challenge and Protects from Chemotherapy-Induced Myelosuppression
We next continued to test whether training with β-glucan could improve the response of hematopoietic progenitors to a secondary stimulus that induces emergency myelopoiesis. LPS-mediated systemic inflammation induces hematopoietic progenitor expansion, which facilitates the restoration of BM cellularity and compensates for the increased need for mature myeloid cells (Mitroulis et al., 2017, Nagai et al., 2006, Takizawa et al., 2017). Therefore, mice were injected with a single dose of LPS 28 days after β-glucan or PBS administration, and BM analysis was performed after another 24 hr. Priming with β-glucan resulted in a more favorable response to the secondary LPS challenge 28 days later, as shown by more pronounced expansion of the LSK and MPP pools (Figures 4A–4C). Administration of LPS induces DNA damage in HSPCs due to replication stress, thereby leading to their functional decline (Takizawa et al., 2017). To address whether priming with β-glucan protects against replication stress induced by the secondary LPS challenge, we stained for phosphorylated H2AX (γ-H2AX), a marker of the DNA-damage response. Indeed, the frequency of γ-H2AX+ LT-HSCs at 24 hr after the secondary LPS challenge was significantly decreased in mice trained with β-glucan, as compared to mice that received PBS (Figure 4D). No difference in the frequency of γ-H2AX+ LT-HSCs was observed prior to the secondary LPS challenge in mice treated with β-glucan or PBS at 28 days after treatment (Figure 4D). Thus, β-glucan pre-treatment mediated a long-term protective response that mitigated DNA damage in LT-HSCs induced by the secondary acute LPS challenge 28 days after initial β-glucan administration.
Figure 4.
Training with β-Glucan Promotes a Beneficial Response to a Secondary Challenge
(A) WT mice were injected with β-glucan or PBS, and after 28 days, they received a secondary challenge with LPS.
(B and C) LSK, MPP, and LT-HSC cell numbers (B) and frequency of the same cells in total BM cells (C) at 24 hr after LPS injection (n = 10 mice per group).
(D) Representative FACS plots and frequency of γ-H2AX-positive LT-HSCs at 24 hr after LPS injection (n = 10 mice per group, right plots and gray background). The frequency of γ-H2AX-positive LT-HSCs at 28 days after β-glucan administration in mice not injected with LPS (—) is also shown; ns = 4 and 5, left plots and white background.
(E) Experimental protocol for the effect of β-glucan on the recovery of granulopoiesis after cyclophosphamide administration (4 rounds).
(F and G) Total white blood cell (WBC) (F) and granulocyte (Gr1+CD11b+) (G) counts in the peripheral blood (n = 10 mice per group).
(H) Experimental protocol for 5-FU administration.
(I) Survival curves of 5-FU-treated mice treated with β-glucan or PBS control (n = 16 mice per group). Comparison of survival curves was performed by log-rank (Mantel-Cox) test, and p value is shown.
(J and K) Mice were injected with β-glucan or PBS, and 7 days later, a single dose of 5-FU was administered.
(J) Neutrophil numbers in peripheral blood at different time points after the administration of 5-FU (n = 5 mice per group).
(K) Frequency of γ-H2AX-positive LT-HSCs 14 days after 5-FU administration (n = 10 mice per group).
Data are presented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
We subsequently studied the effects of β-glucan on the recovery of hematopoiesis in two models of chemotherapy-induced myeloablation (Trumpp et al., 2010, Wilson et al., 2008). Upon repeated rounds of cyclophosphamide administration, we observed that training with β-glucan resulted in increased numbers of white blood cells and, especially, Gr1+CD11b+ granulocytes in peripheral blood on day 4 after each cyclophosphamide round (Figures 4E–4G). Additionally, we used a 5-fluoracil (5-FU) suicide assay to study whether β-glucan can promote host survival upon cytotoxic stress (Figure 4H). Notably, β-glucan significantly enhanced mouse survival to repeated rounds of 5-FU administration (Figure 4I), suggesting that β-glucan improved chemoresistance of HSPCs. Moreover, training with β-glucan 7 days before administration of a single dose of 5-FU improved the recovery of neutrophil numbers in the circulation (Figure 4J). Furthermore, we found a decrease in the frequency of γ-H2AX+ LT-HSCs at day 14 after a single dose of 5-FU in mice that were pre-treated with β-glucan (Figure 4K). Together, β-glucan-induced trained immunity in mice mediates a protective hematopoiesis response to a secondary challenge represented by myeloablative chemotherapy.
β-Glucan-Induced Myelopoiesis Is Associated with Metabolic Changes in Progenitors
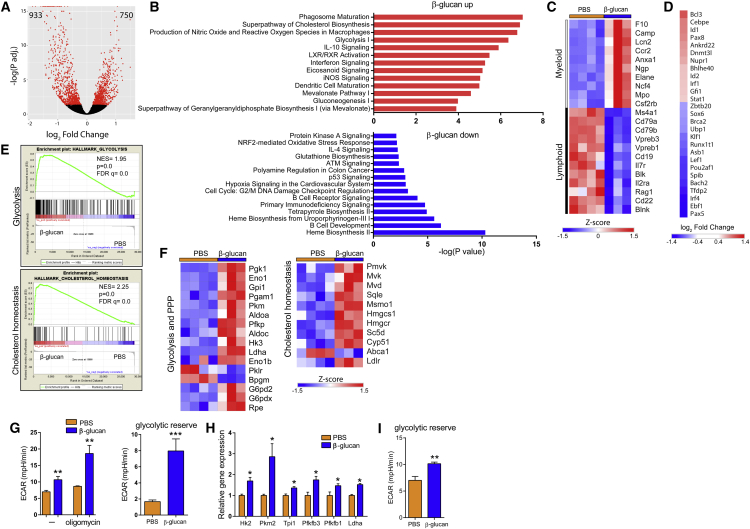
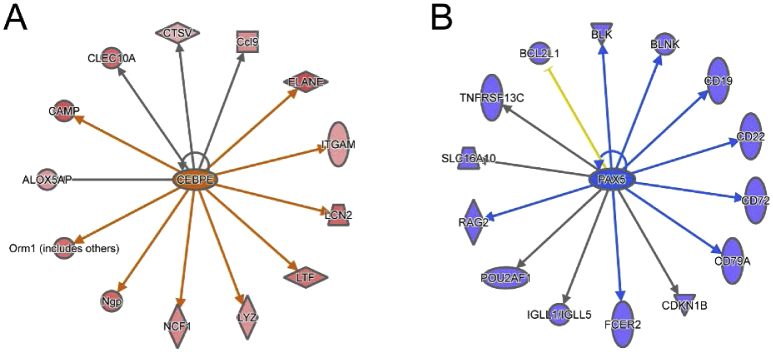
We next performed RNA sequencing (RNA-seq) of LT-HSCs from mice on day 7 after β-glucan or PBS administration. We identified 1,683 differentially expressed genes (false discovery rate [FDR = 0.05]) in cells from the β-glucan-injected group, compared to those from PBS-treated mice (Figure 5A). Ingenuity pathway analysis (IPA) demonstrated that pathways involved in innate immune function and pathways of cell metabolism, including glycolysis, cholesterol biosynthesis, and especially the mevalonate pathway, were overrepresented in the upregulated genes (Figure 5B). In contrast, pathways involved in lymphocyte development and function were overrepresented in the downregulated genes (Figure 5B). Upregulation of myeloid lineage markers, including Csf2rb, Elane, and Mpo, and downregulation of lymphoid lineage markers, such as Ms4a1, Il7r, Il2ra, Vpreb1, and Rag1, were also observed in LT-HSCs of β-glucan-treated mice (Figure 5C). Consistently, TFs promoting myelopoiesis (Cebpe, Id1, and Id2) were upregulated in LT-HSCs from β-glucan-treated mice, while lymphopoiesis-related TFs (Pax5, Ebf1, Irf4, Spib, and Lef1) were downregulated (Nimmo et al., 2015, Novershtern et al., 2011, Paul et al., 2015, Rosenbauer and Tenen, 2007) (Figure 5D). Additionally, genes regulated by the myeloid-lineage TF Cebpe and the lymphoid-lineage TF Pax5 were significantly up- and downregulated, respectively, in LT-HSCs from β-glucan-treated mice compared to LT-HSCs from PBS-treated mice (Figure S3). The sustained nature of the β-glucan-induced alterations in LT-HSCs and CD48+CD150− MPPs was further strengthened by transcriptomic analysis at 28 days post-β-glucan injection (Figure S4). IPA revealed a negative correlation between β-glucan-dependent transcriptomic alterations and lymphoid lineage gene signature in LT-HSCs and in MPPs (Figures S4A–S4D). Gene expression of the lymphoid cell markers Il2ra, Rag2, Vpreb1, Blnk, and Ms4a1 was downregulated in both cell populations in β-glucan-treated mice (Figures S4E and S4F).
Figure 5.
β-Glucan-Induced Alterations in LT-HSC Metabolic Pathways Revealed by Transcriptomic Analysis
(A–F) Transcriptome analysis in LT-HSCs sorted from mice on day 7 after β-glucan or PBS administration (n = 4 mice, PBS group; n = 3 mice, β-glucan group).
(A) Differential gene expression in LT-HSCs from β-glucan-treated mice as compared to PBS-treated mice. Volcano plot showing the distribution of the adjusted p values (−log(P-adj.)) and the fold changes (log2 fold change). Significant changes are indicated in red (FDR = 0.05).
(B) Top overrepresented canonical pathways showing upregulated (red) or downregulated (blue) genes in LT-HSCs from β-glucan-treated mice, as compared to PBS-treated mice.
(C) Heatmap of myeloid- and lymphoid-lineage-related genes.
(D) Heatmap depicting the differential gene expression of transcription regulators. Log2 fold change in cells derived from β-glucan-treated mice, as compared to PBS-treated mice, is indicated.
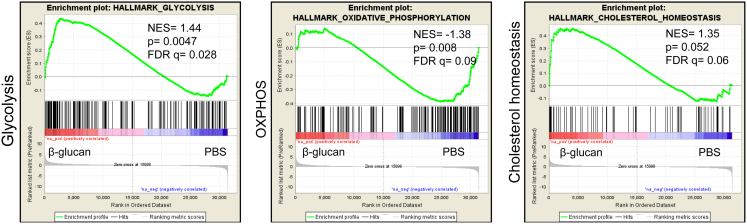
(E) GSEA for glycolytic genes and genes related to cholesterol homeostasis. NES, normalized enrichment score.
(F) Heatmap of genes involved in glycolysis and pentose phosphate pathway (PPP) and cholesterol homeostasis in LT-HSCs from β-glucan-treated mice compared to PBS-treated mice.
(G) Bioenergetic extracellular flux analysis (Seahorse) in Lin−cKit+ BM progenitors sorted from mice 24 hr after β-glucan or PBS administration (n = 5 mice per group). Basal and maximal ECARs (after oligomycin) (left) and glycolytic reserve (right), calculated as the difference between basal and maximal ECARs, are indicated.
(H) Glycolytic gene expression in LSKs from mice 24 hr after β-glucan or PBS administration using qPCR (ns = 4 and 5 mice).
(I) Bioenergetic extracellular flux analysis of Lin−cKit+ cells sorted from mice at 7 days after β-glucan administration; glycolytic reserve is shown (n = 5 mice per group).
Data are presented as mean ± SEM in (G)–(I). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
See also Figures S3 and S5.
Figure S3.
Transcriptomic Analysis of LT-HSCs on Day 7 after β-Glucan Administration, Related to Figure 5
(A and B) Upstream regulator analysis in transcriptomic data using IPA. Genes regulated by the TFs (A) Cebpe and (B) Pax5. Genes with increased expression in LT-HSC from mice injected with β-glucan (as compared to cells from PBS-treated mice) are depicted in red, whereas genes with decreased expression are shown in blue.
Figure S4.
Transcriptional Alterations in Hematopoietic Progenitors 28 Days after β-Glucan Administration, Related to Figure 5
LT-HSCs and MPPs were sorted from mice on day 28 after β-glucan or PBS administration and RNA sequencing was performed (n = 4 mice per group).
(A and C) Differential gene expression in (A) LT-HSCs and (C) MPPs; volcano plots depicting the distribution of the adjusted p values (-log(P-adj.)) and the fold changes (log2 Fold Change). Significant changes are colored red (FDR = 0.05).
(B and D) Overrepresented canonical pathways showing genes that were upregulated (red) or downregulated (blue) in (B) LT-HSCs and (D) MPPs from β-glucan–injected mice, as compared to PBS-treated mice.
(E) Heatmap depicting the expression of lymphoid lineage-related genes in LT-HSCs and MPPs from β-glucan injected mice, as compared to PBS treated mice. Transcripts that are not significantly altered are shown in gray.
(F) Differentially expressed genes in both LT-HSCs and MPPs. The number of upregulated (red) and downregulated (blue) genes is shown.
Besides typical pathways linked to innate immunity, cholesterol biosynthesis and glycolysis were among the highest enriched pathways at 7 days post-β-glucan injection (Figure 5B). Regarding metabolic pathways, gene set enrichment analysis (GSEA) using the Molecular Signatures Database (MSigDB) hallmark gene set collection (Liberzon et al., 2015) showed a significant positive correlation with glycolysis and cholesterol homeostasis gene sets in LT-HSCs from β-glucan-treated mice (Figure 5E; Table S1). Specifically, genes encoding key regulatory enzymes of the glycolytic pathway (Hk3, Pfkp, and Pkm) and the rate-limiting enzyme of the pentose phosphate pathways (G6pdx) were significantly upregulated (Figure 5F). Concerning cellular cholesterol homeostasis and the mevalonate pathway, there was a significant upregulation in the expression of several genes (Figure S5), including those encoding the rate-limiting enzyme of cholesterol biosynthesis HMG-CoA (coenzyme A) reductase (Hmgcr) and the low-density lipoprotein (LDL) receptor (Ldlr) that regulates LDL uptake, and a downregulation of the ATP-binding cassette transporter A1 (Abca1), which mediates cholesterol efflux (Figure 5F). These findings suggest an increased demand for cell cholesterol and enhanced cellular cholesterol biosynthesis and retention in LT-HSCs upon β-glucan treatment.
Figure S5.
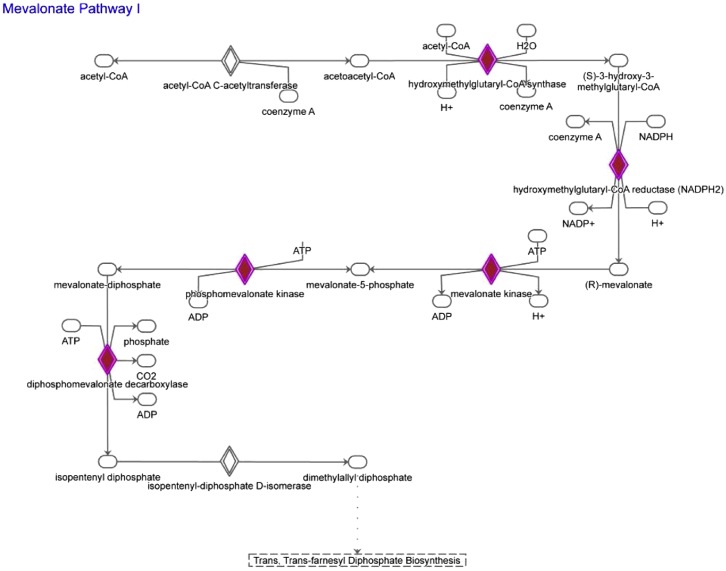
Mevalonate Pathway, Related to Figure 5
Schematic depiction of the mevalonate pathway with genes significantly upregulated in LT-HSC from β-glucan–injected mice (as compared to PBS-treated mice) shown in red.
We next assessed cellular bioenergetics in BM progenitors. We sorted cKit+ progenitors (Lin−cKit+), which include HSPCs and MyPs, from mice at 24 hr after β-glucan or PBS injection and performed bioenergetic flux analysis. Both under steady-state conditions and upon metabolic stress induced by the ATP synthase inhibitor oligomycin, we observed increased extracellular acidification rates (ECARs) in cells from β-glucan-treated mice (Figure 5G), thus suggesting the upregulation of glycolysis. These changes in energy metabolism were further associated with upregulated gene expression of glycolytic enzymes in LSK cells from β-glucan-treated mice (Figure 5H). The enhanced response of cKit+ progenitors to metabolic stress, as shown by the cellular response to oligomycin, was also found at 7 days after β-glucan administration (Figure 5I). These data suggest that the early proliferative phase after β-glucan administration involves a global increase in energy metabolism in BM progenitors.
Since cell metabolism in LT-HSCs is linked to LT-HSC maintenance (Ito et al., 2012, Murphy et al., 2011, Takubo et al., 2013, Wang et al., 2014, Yvan-Charvet et al., 2010), we investigated whether the β-glucan-induced metabolic changes in LT-HSCs were sustained also after 5-FU administration, thereby accompanying the beneficial response to chemotherapy-induced hematopoietic stress upon β-glucan injection (as shown in Figure 4). To this end, mice received a single injection of 5-FU on day 7 after β-glucan or PBS administration, and RNA-seq was performed in LT-HSCs isolated on day 14 after 5-FU injection. GSEA revealed a positive correlation between LT-HSCs from the β-glucan-trained group with the glycolysis and cholesterol homeostasis gene sets and a negative correlation with the OXPHOS gene set (Figure S6; Table S2). Thus, β-glucan-dependent enhanced glycolysis of LT-HSCs persists after severe hematopoietic stress.
Figure S6.
GSEA in LT-HSC after 5-FU Administration, Related to Figure 5
Mice were injected with β-glucan or PBS and 7 days later a single dose of 5-FU was administered. LT-HSCs were sorted from mice on day 14 after 5-FU administration and transcriptomic analysis was performed (n = 4 mice per group). GSEA for genes related to glycolysis, OXPHOS and cholesterol biosynthesis.
Trained Immunity Links Innate Immune Signaling to Metabolic Alterations in Progenitor Cells
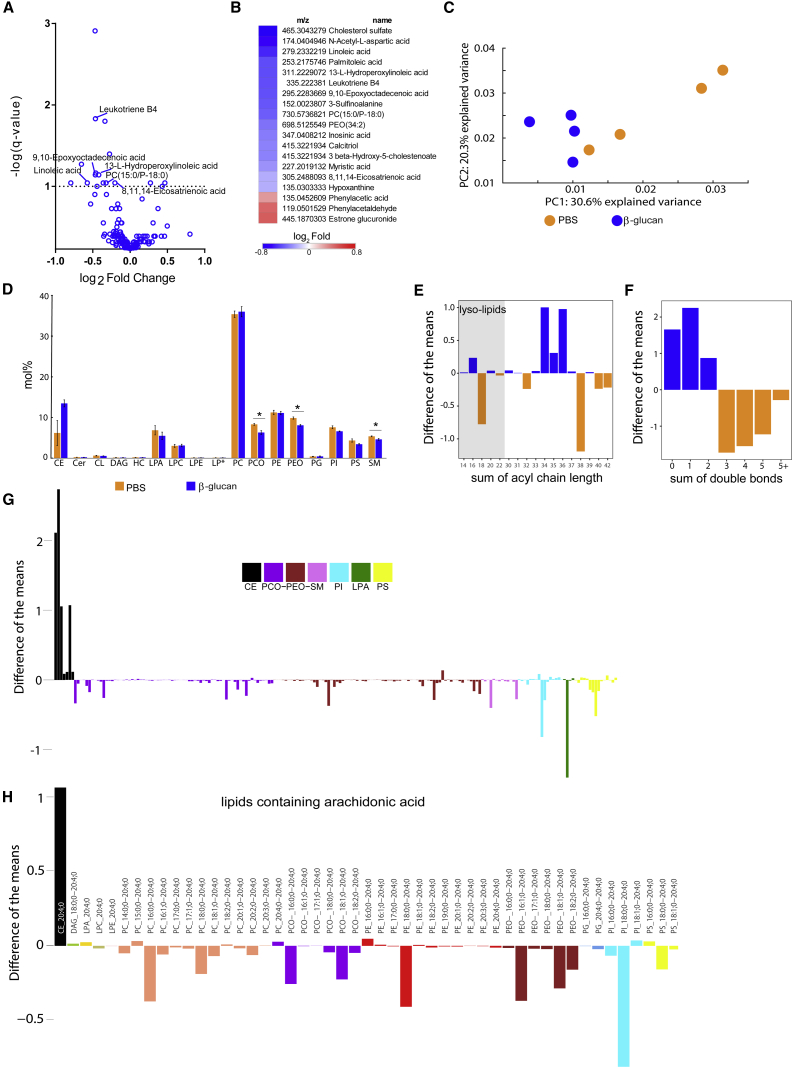
We next analyzed in more detail the β-glucan-induced metabolic changes by performing metabolomics and quantitative shotgun lipidomics of BM progenitor cells. Metabolomic analysis in cKit+ progenitors derived from mice at 24 hr after β-glucan injection revealed a reduction in metabolites involved in linoleic and arachidonic acid metabolism, compared to cells from the control group (Figures 6A and 6B). These findings, together with those of the transcriptomic analysis, which indicated alterations in cellular cholesterol biosynthesis pathways, prompted us to further assess lipid metabolism by lipidomic analysis in cKit+ progenitor cells from β-glucan- or PBS-treated mice at the 24-hr time point. The shotgun lipid mass spectrometry detects hundreds of lipid species present in the cells; however, in the present experimental approach, we did not perform an analysis of the free fatty acids, which had been covered by the metabolomic analysis. Principal-component analysis (PCA) of the lipidomic data suggested significant differences in the lipidomes of the BM progenitors from β-glucan-trained mice compared to the PBS group (Figure 6C). Significant differences were observed in certain lipid classes (Figure 6D). Whereas cells from β-glucan-trained mice contained substantially more lipids with shorter (34–36 carbon atoms) and more saturated acyl chains (having none, one, or two double bonds), cells from the PBS-treated control group contained lipids with longer acyl chains (38–42 carbon atoms) and increased levels of polyunsaturated fatty acids (≥3 double bonds) (Figures 6E and 6F). An accumulation of various species of cholesterol esters (CEs) and a decrease of a number of species of alkyl-ether-linked phosphatidylcholines (PCs) and phosphatidylethanolamines (PEs) (PC and PE plasmalogens; alkyl-ether-linked phosphatidylcholines [PCO]− and alkyl-ether-linked phosphatidylethanolamines [PEO]−, respectively), sphingomyelins, phosphatidylinositols, phosphatidylserines, and lysophospatidic acid was observed in cells from β-glucan-treated mice (Figure 6G). Consistent with the metabolomics data, we observed a decrease in the majority of lipids containing arachidonic acid (20:4) in cells from β-glucan-treated mice (Figure 6H). Therefore, lipid changes accompany trained-immunity-associated effects on hematopoietic progenitors.
Figure 6.
Alterations in Lipid Metabolism in BM Progenitor Cells upon β-Glucan Administration
Lin−cKit+ cells were sorted from mice 24 hr after β-glucan or PBS administration, and non-targeted metabolomic and lipidomic analyses were performed (n = 4 mice per group).
(A and B) Not-targeted metabolomics.
(A) Volcano plots depict the comparison of metabolite abundances between cells from β-glucan- and PBS-treated mice. Altered metabolites (q value < 0.1) involved in linoleate and arachidonic acid pathways (Kyoto Encyclopedia of Genes and Genomes [KEGG] pathways database) are indicated.
(B) Heatmap depicts the abundance of differentially regulated metabolites (q value < 0.1).
(C–H) Lipidomic analysis.
(C) Principal-components analysis shows that the two conditions segregate along the first dimension, PC1 (Mann-Whitney U test, p value = 0.028).
(D) Mol % abundance of lipid classes (CE, cholesterol esters; Cer, ceramides; CL, cardiolipin; DAG, diacylglycerols; HC, hexosyl ceramide; LPA, lysophosphatidic acid; LPC, lysophopshatidylcholines; LPE, lysophosphatidylethanolamine; LP∗, LPC and LPE plasmalogens; PC, phosphatidylcholines; PCO, PC plasmalogens; PE, phosphatidylethanolamines; PEO, PE plasmalogens; PG, phosphatidylglycerols; PI, phosphatidylinositols; PS, phosphatidylserines; SM, sphingomyelins). Data are presented as mean ± SEM. ∗p < 0.05.
(E) Difference between the mean abundance of lipid species (excluding CL and cholesterol esters) grouped by the number of carbon atoms: blue bars indicate lipids enriched in cells from β-glucan-treated mice, while orange bars indicate lipids enriched in cells from PBS-treated mice. Lysolipids containing a single fatty-acyl chain are highlighted by a gray background.
(F) Difference between the mean abundance of lipid species grouped by number of double bonds: blue bars indicate lipids enriched in cells from β-glucan-treated mice, while orange bars indicate lipids enriched in cells from PBS-treated mice.
(G) Relative abundance of lipid species belonging to PCO−, PEO−, SM, PI, LPA, PS, and cholesterol esters. Relative difference of lipid species is shown; negative bars represent species that are more abundant in the PBS group, while positive bars represent species that are more abundant in the β-glucan group.
(H) Lipid species containing arachidonic acid (fatty acid [FA] 20:4) are indicated; negative bars indicate species that are more abundant in the PBS group, while positive bars indicate species that are more abundant in the β-glucan group.
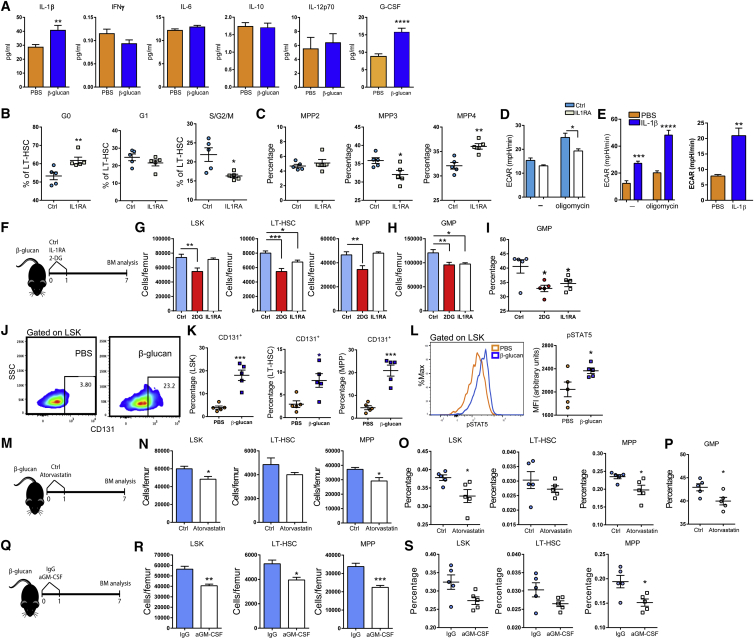
Since HSCs do not express the β-glucan receptor Dectin-1 (Yáñez et al., 2011), we sought to determine whether the effects of β-glucan on HSCs are mediated by cytokines induced in the BM in response to β-glucan administration. Cytokine analysis in the BM extracellular fluid at 24 hr following β-glucan administration revealed, in addition to an elevated concentration of G-CSF, enhanced levels of IL-1β, whereas the levels of other cytokines, such as interferon (IFN)γ, IL-6, IL-10, and IL-12p70, were not altered (Figure 7A). We thus next sought to identify the potential role of IL-1β-related BM inflammation in the β-glucan effects on BM progenitor cells.
Figure 7.
IL-1β, Glycolysis, Cholesterol Metabolism, and the GM-CSF/CD131 Axis Are Involved in β-Glucan-Dependent Training in the BM
(A) Cytokine and G-CSF concentrations in the BM extracellular fluid of mice at 24 hr after the administration of PBS or β-glucan (n = 10 mice per group).
(B and C) Mice were injected with β-glucan in the absence (vehicle control, Ctrl) or presence of IL1RA, and BM analysis was performed 24 hr later.
(B) Cell-cycle analysis in LT-HSCs.
(C) The frequency of MPP subpopulations in LSK cells in the BM is indicated (n = 5 mice per group).
(D) Bioenergetic extracellular flux analysis in Lin−cKit+ cells sorted from mice 24 hr after β-glucan administration in the absence (vehicle control, Ctrl) or presence of IL1RA (n = 5 mice per group). Basal and maximal ECAR are indicated.
(E) LSK cells were treated in vitro with IL-1β or PBS for 24 hr, and Seahorse analysis was performed (n = 5 cultures per group). Basal and maximal ECARs (left) and glycolytic reserve (right) are indicated.
(F–I) Mice were injected with β-glucan on day 0.
(F) Glycolysis and IL-1 were blocked by the administration of 2-DG and IL1RA, respectively, on days 0 and 1; PBS served as the vehicle control (Ctrl).
(G) LSK, MPP, and LT-HSC numbers in the BM at day 7 after β-glucan administration (n = 5 mice per group).
(H and I) GMP numbers in the BM (H) and frequency of GMPs within the MyP pool (I) at day 7 after β-glucan administration (n = 5 mice per group).
(J and K) Mice were injected with PBS or β-glucan, and BM analysis was performed 24 hr later; representative FACS plots (J) and frequency of CD131+ LSKs, CD131+ MPPs, and CD131+ LT-HSCs (K) (n = 5 mice per group) are indicated.
(L) Staining for pSTAT5 in LSK cells 24 hr after PBS or β-glucan administration. Representative FACS plots and median fluorescence intensity (MFI) are shown (n = 5 mice per group).
(M–P), As indicated in (M), mice were injected with β-glucan on day 0. Cholesterol metabolism was blocked by the administration of atorvastatin on days 0 and 1; Ctrl represents the vehicle control. (N) LSK, LT-HSC, and MPP cell numbers; (O) frequency of the same cells in total BM cells; and (P) frequency of GMPs within the MyP pool at day 7 after β-glucan administration (n = 5 mice per group).
(Q) Mice were injected with β-glucan on day 0. GM-CSF was blocked by specific anti-GM-CSF antibody on days 0 and 1. Immunoglobulin G (IgG) isotype served as the control.
(R and S) LSK, LT-HSC, and MPP cell numbers (R) and frequency of the same cells in total BM cells (S) at day 7 after β-glucan administration are indicated (n = 5 mice per group).
Data are presented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Pharmacologic inhibition of IL-1 by an IL-1 receptor antagonist (IL1RA; anakinra) prevented the increase in cell-cycle progression of LT-HSCs at 24 hr after the administration of β-glucan (Figure 7B). IL1RA also inhibited the β-glucan-induced increase in the frequency of MPP3 cells in the LSK population and resulted in a corresponding increase in the frequency of MPP4 cells (Figure 7C). Moreover, IL1RA prevented the β-glucan-dependent increase in glycolysis (ECAR) at 24 hr following β-glucan injection (Figure 7D). To investigate a possible direct link between IL-1β production and metabolic alterations in HSPCs, isolated LSK cells were treated ex vivo with IL-1β for 24 hr, and bioenergetic analysis was performed. In accordance with the in vivo findings, IL-1β induced an increase in ECAR and resulted in enhanced glycolytic reserve of LSK cells (Figure 7E). These data suggest that, upon β-glucan administration, IL-1 signaling promotes glycolysis and HPSC proliferation.
To more rigorously implicate IL-1β signaling and glycolysis in the β-glucan-induced innate immune-metabolic crosstalk within the BM, we studied the effects of IL1RA or of 2-DG, a pharmacologic inhibitor of the glycolytic enzyme hexokinase, on the β-glucan-dependent expansion of HSPCs observed on day 7 post-β-glucan injection. Inhibition of either IL-1 signaling or glycolysis reduced the numbers of HSPCs and GMPs in β-glucan-trained mice (Figures 7F–7H). Moreover, inhibition of IL1R or glycolysis in β-glucan-trained mice decreased the proportion of GMPs among MyPs, as compared to β-glucan administration alone (Figure 7I).
Previous studies have shown that disruption of cholesterol efflux drives myeloid cell expansion by increasing the surface expression of CD131, the common β subunit of the IL-3/GM-CSF receptor (IL-3Rβ) (Murphy et al., 2011, Yvan-Charvet et al., 2010). Since we observed enhanced cholesterol biosynthesis and reduction in gene expression of factors involved in cholesterol efflux in hematopoietic progenitors from β-glucan-trained mice, we next assessed the expression of CD131 in HSPCs. We detected a significant increase in CD131+ LSKs, CD131+ MPPs, and CD131+ LT-HSCs 24 hr after β-glucan injection (Figures 7J and 7K). We further assessed whether the increased expression of CD131 was associated with enhanced downstream signaling. Indeed, phosphorylation of STAT5 (pSTAT5) was increased in LSK cells from β-glucan-injected mice at 24 hr, as compared to control-treated mice (Figure 7L). Therefore, β-glucan-associated trained immunity causes changes in lipid metabolism associated with increased frequency of CD131-expressing HSPCs.
We also assessed the role of cholesterol biosynthesis in β-glucan-dependent effects on HSPCs. Inhibition of 3-hydroxy-3-methylglutaryl (HMG-CoA) reductase with atorvastatin in β-glucan-trained mice reduced the numbers of LSKs and MPPs, as compared to β-glucan administration alone (Figures 7M–7O), resulting also in decreased frequency of GMPs (Figure 7P). In line with a role of the GM-CSF/CD131 axis for the β-glucan-dependent effects on HSPCs, antibody-mediated blockade of GM-CSF in β-glucan-trained mice diminished the numbers of hematopoietic progenitors, as compared to β-glucan injection alone (Figures 7Q–7S). Together, the effects of β-glucan on HSPCs are associated with IL-1β signaling, key metabolic adaptations, and enhanced activation of the GM-CSF/CD131 axis.
Discussion
Preservation of immune homeostasis after infection is achieved not only through containment or eradication of the insulting pathogen but also through adaptations that limit or repair tissue damage or dysfunction and prepare the host against a secondary challenge. Although the latter property has been hitherto considered an exclusive hallmark of adaptive immunity, this dogma has been recently challenged. In this regard, innate immune training has been shown to cause long-term alterations in mature innate immune cells, enabling a robust innate host response to a secondary stimulus (Jensen et al., 2015, Netea and van Crevel, 2014, Netea et al., 2016). Because mature innate immune cells, such as monocytes, have a short lifespan in the circulation relative to the duration of trained immunity, the question arose as to whether trained immunity acts at the level of their cellular precursors. Herein, we show that β-glucan-induced trained immunity promotes HSPC expansion and enhances myelopoiesis, thereby conferring a protective response to a secondary challenge represented by systemic inflammation or chemotherapy-induced myeloablation. Thus, not only does trained immunity affect mature myeloid cells, but its effects may also be initiated by adaptations in the precursor cells of the innate immune system in the BM.
Administration of β-glucan increased the frequency of the myeloid-biased subsets of LT-HSCs and MPPs in the BM. This finding was in line with transcriptome analysis in LT-HSCs, which demonstrated enrichment of innate immune-related pathways at the expense of pathways related to lymphocyte development and function. The finding that β-glucan treatment induced increased transcription of cell-cycle-related genes in CD41+ LT-HSCs suggests the selective expansion of myeloid-biased cells as the likely target of trained immunity in the BM. Conceivably, trained immunity could also instruct primitive LT-HSCs toward myeloid differentiation, which is unclear at this point. Identifying the subsets of HSPCs that are responsive to signals related to innate immune training requires further investigation. Future studies, including additional transcriptional and epigenetic approaches at the single-cell level, may reveal whether the long-term impact of trained immunity on myeloid cells is initiated at the level of primitive HSCs or lineage-committed populations.
Additionally, β-glucan-induced trained immunity was associated with alterations in metabolic pathways in progenitor cells, such as glycolysis and cholesterol metabolism, which also accompany the induction of trained innate immunity in mature cells and are involved in innate immune cell adaptation to diverse stimuli (Arts et al., 2016b, Cheng et al., 2014, Mills and O’Neill, 2016, Norata et al., 2015). LT-HSCs depend on glycolysis for their maintenance (Simsek et al., 2010, Takubo et al., 2013, Wang et al., 2014), whereas the activation of OXPHOS is associated with their functional impairment, loss of self-renewal potential, and induction of DNA damage (Suda et al., 2011, Takizawa et al., 2017, Takubo et al., 2013, Walter et al., 2015). In β-glucan-treated mice, progenitor cells showed enhanced glycolysis. Importantly, this glycolytic signature was retained after chemotherapy and was associated with resistance to DNA damage upon hematopoietic stress, induced by systemic LPS challenge or chemotherapy. Hence, the metabolic alterations conferred by trained immunity may contribute to the protection from chemotherapy-induced myelosuppression.
Trained immunity mediated by β-glucan also induced changes in lipid metabolism in progenitors, such as increased gene expression of enzymes involved in cholesterol biosynthesis and decreased expression of Abca1, a transporter regulating cholesterol efflux (Yvan-Charvet et al., 2010). Importantly, atorvastatin-mediated blocking of cholesterol synthesis diminished the β-glucan-dependent expansion of HSPCs. Similar changes in lipid metabolism induced by blocking cholesterol efflux were previously shown to promote myeloid cell expansion in the BM via enhanced CD131 surface expression (Murphy et al., 2011, Yvan-Charvet et al., 2010). Consistently, β-glucan administration in mice not only upregulated CD131 expression in HSPCs but also activated downstream signaling, as revealed by STAT5 phosphorylation. The ability of β-glucan to increase the biosynthesis and retention of cholesteryl esters and substantially decrease glycerophospholipid-containing arachidonic fatty acid chains reflects the capacity of cells to actively remodel their lipidome and, thus, the physicochemical properties of their membranes. This adaptive response has direct implications in the lateral organization of cellular membranes (Sezgin et al., 2017) and in cell signaling by lipid-protein interactions (Coskun and Simons, 2011). In this regard, alterations in the cholesterol composition of the membrane can influence the localization and signaling capacity of CD131 (Kaul et al., 2016, Yvan-Charvet et al., 2010).
In emergency myelopoiesis, cell-fate decision already takes place at the level of non-committed progenitors (Boettcher and Manz, 2017). Myeloid-lineage priming is regulated by a complex interplay between myeloid-specific growth factors, cytokines, and TFs (Boettcher and Manz, 2017, Sarrazin and Sieweke, 2011). IL-1 promotes proliferation and myeloid differentiation in HSPCs by inducing a PU.1-based myeloid gene program (Pietras et al., 2016). Herein, we showed that β-glucan injection resulted in increased levels of IL-1β in the BM, presumably promoting a microenvironment tailored toward myelopoiesis. Our finding that β-glucan training promotes myelopoiesis recovery after chemotherapy is likely dependent on its ability to elevate IL-1β, which was previously associated with enhanced restoration of myelopoiesis after myeloablation (van der Meer et al., 1988, Nakai and Hirai, 1989).
The β-glucan-induced expansion of HSPCs resembles induction of an emergency myelopoiesis response (Boettcher and Manz, 2017). However, other types of emergency myelopoiesis, including LPS-induced inflammation, usually induce LT-HSC injury and exhaustion and, hence, myelosuppression (Chen et al., 2010, Takizawa et al., 2017, Zhang et al., 2016). Indeed, in the context of emergency myelopoiesis, LPS-induced cell stress inflicts DNA damage in LT-HSCs that is maintained for more than 14 days (Takizawa et al., 2017). Strikingly, this adverse effect is significantly reversed by priming with β-glucan as shown herein. Thus, β-glucan does not cause impairment of HSPC function and exhaustion but rather mediates a protective effect on hematopoiesis, which can thereby respond favorably to a secondary inflammatory challenge or chemotherapy. In conclusion, the actions of trained innate immunity on precursor cells of the innate immune system in the BM can be therapeutically exploited to counteract the adverse effects of chemotherapy-induced myelosuppression.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-mouse c-kit (CD117) | BioLegend | Cat# 105808; RRID: AB_313217 |
| Rat anti-mouse Sca-1 (Ly6-A/E) | BioLegend | Cat# 122514; RRID: AB_756199 |
| Mouse Lineage Antibody Cocktail | BD PharMingen | Cat# 558074/ 561301 |
| Biotin Mouse Lineage Panel | BD PharMingen | Cat# 559971 |
| Armenian Hamster anti-mouse CD48 | BioLegend | Cat# 103432; RRID: AB_2561463 |
| Rat anti-mouse CD150 | BioLegend | Cat# 115922; RRID: AB_2303663 |
| Rat anti-mouse CD34 | eBioscience | Cat# 11-0341; RRID: AB_465021 |
| Rat anti-mouse CD16/CD32 | eBioscience | Cat# 48-0161; RRID: AB_1272191 |
| Armenian Hamster anti-mouse CD3e | BioLegend | Cat# 100330; RRID: AB_1877170 |
| Rat anti-mouse CD19 | eBioscience | Cat# 11-0193; RRID: AB_657666 |
| Rat anti-mouse Gr1 (Ly-6G/C) | eBioscience | Cat# 48-5931; RRID:AB_1548788 |
| Rat anti-mouse CD11b | BD PharMingen | Cat# 552850; RRID: AB_394491 |
| Rat anti-mouse CD135 | BioLegend | Cat# 135310; RRID: AB_2107050 |
| Mouse anti-mouse CD45.1 | BD PharMingen | Cat# 553776; RRID: AB_395044 |
| Mouse anti-mouse CD45.2 | BD PharMingen | Cat# 558702 |
| Mouse anti-mouse/human H2A.X Phospho (Ser139) | BioLegend | Cat# 613403; RRID: AB_528918 |
| Rat anti-mouse CD41 | BioLegend | Cat# 133903; RRID: AB_1626237 |
| Mouse anti-mouse/human Phospho-STAT5 (Tyr694) | eBioscience | Cat# 17-9010-41; RRID: AB_2573272 |
| Rat anti-mouse Ki-67 | eBioscience | Cat# 11-5698-82; RRID: AB_11151330 |
| Rat anti-mouse CD131 | BD PharMingen | Cat# 740050 |
| LEAF Purified anti-mouse GM-CSF Antibody | BioLegend | Cat# 505408; RRID: AB_315384 |
| LEAF Purified Rat IgG2a, κ Isotype Ctrl Antibody | BioLegend | Cat# 400516 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Beta-glucan peptide (BGP) | Invivogen | Cat# tlrl-bgp |
| LPS-EB (LPS from E. coli O111:B4) | Invivogen | Cat# tlrl-eblps |
| Kineret (anakinra) | Anakinra, Amgen GmbH | NDC 66658-234-07 |
| 2-Deoxy-D-glucose | Sigma | Cat# D8375 |
| Atorvastatin | Calbiochem | Cat# 189291 |
| 5-Fluoruracil | Sigma | CAS# 51-21-8 |
| Cyclophosphamide | Sigma | CAS# 6055-19-2 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | ThermoFisher Scientific | Cat# D1306 |
| MyeloCult M5300 | StemCell Technologies | Cat# 05300 |
| Seahorse XF Base Medium | Agilent Technologies | Cat# 102353-100 |
| Recombinant Murine SCF | Peprotech | Cat# 250-03 |
| Recombinant Murine IL-1β | Peprotech | Cat# 211-11B |
| Critical Commercial Assays | ||
| Foxp3/TF Buffer Set | eBioscience | Cat# 00-5523-00 |
| G-CSF DuoSet ELISA | R&D systems | Cat# DY414 |
| V-PLEX Proinflammatory Panel 1 (mouse) kit | Meso Scale Discovery | Cat# K15048G |
| Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies | Cat# 103015-100 |
| RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 |
| iScript cDNA Synthesis Kit | BioRad | Cat# 1708891 |
| SsoFast EvaGreen Supermix | BioRad | Cat# 1725202 |
| qScript cDNA Supermix | Quantabio | Cat# 95048 |
| SMARTer Ultra Low Input RNA for Illumina Sequencing | Takara Bio | Cat# 634828 |
| Anti-Biotin MicroBeads | Milteniy Biotec | Cat# 130-090-485 |
| NEBNext Ultra DNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7370L |
| Multiplex PCR Kit | QIAGEN | Cat# 206143 |
| SsoFast EvaGreen Supermix with Low ROX | BioRad | Cat# 1725211 |
| Deposited Data | ||
| RNA sequencing data | This paper | GEO: GSE95617 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Janvier Labs | C57BL/6JRj |
| Mouse: C57BL/6 | The Jackson Laboratory | Stock# 000664 |
| Mouse: C57BL/6-CD45.1 B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | Stock# 002014 |
| Oligonucleotides | ||
| Single-cell qPCR primers | This paper | See Table S3 |
| qPCR primers | This paper | See Table S4 |
| Software and Algorithms | ||
| KNIME 2.11.2 | N/A | https://www.knime.com/ |
| GraphPad Prism 6 | Graphpad Software | N/A |
| R version 3.3.2 | R Development Core Team, 2016 | https://www.r-project.org/ |
| reshape2 package | Wickham, 2007 | https://cran.r-project.org/web/packages/reshape2/index.html |
| ggplot2 | Wickham, 2009 | http://ggplot2.org/ |
| LipidXplorer | Herzog et al., 2011, Herzog et al., 2012 | https://wiki.mpi-cbg.de/lipidx/Main_Page |
| MATLAB 9.0 | MathWorks | https://www.mathworks.com/products/compiler/matlab-runtime.html |
| Ingenuity Pathway Analysis | QIAGEN | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| GSEA software | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| Morpheus | Broad Institute | https://software.broadinstitute.org/morpheus/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/ release/bioc/html/DESeq2.html |
| GSNAP | Wu and Nacu, 2010 | http://research-pub.gene.com/gmap/ |
| Ensembl gene annotation version 81 | EMBL-EBI | https://www.ebi.ac.uk/ |
| featureCounts | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/ |
| RStudio Version 0.99.486 | N/A | https://www.rstudio.com/ |
| Fluidigm Real-Time PCR analysis software | Fluidigm | https://www.fluidigm.com/software |
| FlowJo version 10 | Tree Star | https://www.flowjo.com/solutions/flowjo |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, T. Chavakis (triantafyllos.chavakis@uniklinikum-dresden.de).
Experimental Model and Subject Details
Mice
C57BL/6-CD45.1 B6.SJL-Ptprca Pepcb/BoyJ (B6/SJL) mice were from Jackson Laboratories. C57BL/6 male mice (from Janvier Laboratories and Jackson Laboratories) were used at the age of 9-11 weeks. Mice were housed under specific pathogen-free conditions on a standard 12/12h light/dark cycle. Food and water was provided ad libitum. Animal experiments were approved by the Landesdirektion Sachsen, Germany and the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Method Details
Mouse experiments
To study the effect of β-glucan administration on hematopoietic progenitor cells, mice were injected intraperitoneally (i.p.) with 1mg of β-glucan peptide from Trametes versicolor (Invivogen) in 200 μL of PBS; as control, i.p. injections of PBS alone were performed; mice were euthanized 1, 7 or 28 days later. In other experiments, LPS was used as a secondary challenge. Specifically, LPS from E. coli O111:B4 (Invivogen; 35 μg per mouse) or PBS as control were injected i.p. into mice pretreated with β-glucan or PBS; mice were euthanized 24h after the LPS injection.
To assess lineage output of LT-HSC, the CD45.1/CD45.2 congenic system was used. Sorted LT-HSC (50 cells per recipient) isolated from CD45.2+ mice on day 28 post-administration of β-glucan or PBS were retro-orbitally transferred into lethally (9Gy) irradiated B6/SJL (CD45.1) recipients along with 2x105 CD45.1+ carrier cells (to ensure survival). The percentage of different CD45.2+ cell populations was assessed at 12 weeks post transplantation. Ιrradiated recipient mice were kept on antibiotic-containing water for 2 weeks after irradiation.
IL-1 inhibition was performed by i.p. administration of recombinant IL-1 receptor antagonist (IL1RA, Anakinra, Amgen GmbH, 100mg/kg body weight per dose) for one or two days (Cutando et al., 2013). Inhibition of glycolysis was performed by i.p. injection of 2-Deoxy-D-Glucose (2-DG) (Sigma; 500mg/kg body weight per dose) twice for two consecutive days. GM-CSF inhibition was performed by i.p. administration of LEAF purified anti-mouse GM-CSF antibody (Biolegend; clone MP1-22E9; 100 μg per dose) twice for two consecutive days. LEAF purified rat IgG2a, κ was used as isotype control (Biolegend; clone RTK2758; 100 μg per dose). Inhibition of cholesterol pathway was performed by administering atorvastatin (Calbiochem; 125 μg per dose) (Arts et al., 2016b) twice for two consecutive days.
Chemotherapy-induced myeloablation with 5-Fluoruracil (5-FU) (Sigma; 150 mg/kg body weight) was performed using two different protocols: (i) PBS or β-glucan were administered i.p. on day −7 and 5-FU was given once (day 0); blood cell population analysis was performed on days 4 and 9 post 5-FU injection and mice were euthanized on day 14 post 5-FU injection; peripheral blood analysis was performed using a Sysmex XT2000 blood analyzer (Sysmex Corporation). (ii) PBS or β-glucan peptide was injected i.p. once every 20 days starting on day −5. 5-FU was administered on day 0 and every 10 days thereafter for 7 rounds. Mice were observed every other day.
In the cyclophosphamide-induced myeloablation model, PBS or β-glucan were administered i.p. on day −7 and every 14 days thereafter and cyclophosphamide (Sigma) was injected at a dose of 200 mg/kg body weight i.p. on day 0 and every 14 days thereafter for a total of 4 rounds of injection (Winkler et al., 2012). Peripheral blood analysis was performed on day 4 after each round of cyclophosphamide administration, using a HemaVet automatic cell counter (Drew Scientific) for white blood cell measurement and flow cytometry for the quantification of Gr1+CD11b+ cells.
Flow cytometry and sorting
Cell analysis was performed by FACS Canto II or FACSCalibur flow cytometer (BD, Heidelberg, Germany). Cell sorting was performed using a FACS Aria II cell sorter (BD, Heidelberg, Germany). Enrichment of Lin— cells was performed prior to cell sorting by negative magnetic selection using MidiMACS Separator (Miltenyi Biotec). For this reason, cells were incubated with a biotin mouse lineage panel (Biotin-Conjugated Mouse Lineage Panel, BD PharMingen) and subsequently with anti-Biotin MicroBeads (Miltenyi Biotec). The analysis of BM cellularity was performed with MACSQuant (Miltenyi Biotec). For cell surface phenotype analysis, a lineage cocktail (Lin), including anti-CD3e (clone 145-2C11), anti-CD11b (clone M1/70), anti-Gr1 (clone RB6-8C5), anti-B220 (clone RA3-6B2), and anti-TER119 (clone TER-119), anti-Sca1 (clone E13-161.7), anti-cKit (clone 2B8), anti-CD135 (clone A2F10), anti-CD48 (clone HM48-1), anti-CD41 (clone MWReg30), anti-CD150 (clone TC15-12F12.2), anti-CD16/CD32 (clone 93), anti-CD34 (clone RAM34), anti-CD45.1 (clone A20), anti-CD45.2 (clone 104), anti-CD3e (clone 145-2C11), anti-CD19 (clone eBio1D3), anti-CD11b (clone M1/70), anti-Gr1 (clone RB6-8C5) and anti-CD131 (clone JORO50) were used. Data analysis was performed using FlowJo (Tree Star) software.
For cell cycle analysis, cells were stained for cell-surface markers as described in the previous paragraph, fixed and permeabilized using fixation / permeabilization buffer (Foxp3/TF Buffer Set; eBioscience) and stained with anti–Ki-67 (clone SolA15; eBioscience). After washing, cells were stained with DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) (ThermoFisher Scientific) and analyzed by flow cytometry. For analysis of phospholylated γ-H2AX and phospho-STAT5, cells were stained for cell surface markers, fixed and permeabilized using fixation / permeabilization buffer (Foxp3/TF Buffer Set; eBioscience) and then stained with the respective antibodies.
Immunoassays
To collect BM extracellular fluid, a femoral bone was flushed with ice cold PBS (500 μl; Life Technologies) and the supernatant was harvested after pelleting cells by centrifugation at 500g for 5min at 4°C. G-CSF was measured using a mouse G-CSF DuoSet ELISA (R&D), according to the manufacturer’s instructions. V-PLEX Proinflammatory Panel 1 (mouse) kit was used for cytokine measurement (Meso Scale Discovery) using a Meso QuickPlex SQ 120 instrument (Meso Scale Discovery), according to the manufacturer’s instructions.
In vitro LSK incubation
Sorted LSK cells were cultured in suspension culture 96 well plates (Greiner Bio-One) in MyeloCult medium supplemented with stem cell factor (20ng/ml, PeproTech). To study the effect of IL-1β on cell metabolism, LSK cells from untreated mice were incubated with 20 ng/ml IL-1β (Peprotech) for 24h. At the end of cell cultures, Seahorse extracellular flux analysis in LSK cells was performed.
Extracellular flux analysis
Real-time extracellular acidification rate (ECAR) of sorted Lin—cKit+ progenitors from mice treated with β-glucan or PBS and of sorted LSK cells, incubated for 24h as described under “in vitro LSK incubation,” was assessed using a Seahorse XFe96 Analyzer (Agilent Technologies). After washing with unbuffered Seahorse XF Base Medium (Agilent Technologies), sorted Lin—cKit+ progenitors were seeded in duplicate in 96-well plates (105 cells per well) for Seahorse pretreated with poly-L-lysine (Sigma). To perform real-time ECAR analysis in LSK cells, cells were washed with unbuffered Seahorse XF Base Medium (Agilent Technologies), and seeded in duplicate in 96-well plates (5x104 cells per well) for Seahorse pretreated with poly-L-lysine (Sigma). Basal ECAR was measured during three consecutive measurements in unbuffered Seahorse XF Base Medium (Agilent Technologies), containing 5.5 mM glucose (Sigma), 2 mM L-glutamine (Life Technologies) and 1 mM pyruvate (Life Technologies). After three basal measurements, three consecutive measurements were taken following the addition of 2 μM oligomycin (Agilent Technologies), to measure maximal ECAR. Glycolytic reserve was defined as the difference between maximal and basal ECAR. Equal cell numbers in the wells were assessed by measuring total DNA content using DAPI.
Single-cell qPCR
Gene expression profiles of single cells (cells were pooled from either 3 PBS-treated mice or from 3 β-glucan treated mice) were obtained as previously described (Bonifacio et al., 2015) with some modifications. cDNA was synthesized with Quanta qScript TM cDNA Supermix directly on cells. Total cDNA was pre-amplified for 20 cycles (1x 95°C 5′, 95°C 45”, 60°C 1’, 72°C 1.5′) and 1x 68°C 10’ using the Multiplex PCR Kit (QIAGEN, Hilden, Germany) in a final volume of 35 μl in the presence of primer pairs for the genes (listed in Table S3) at 25nM final for each primer. Pre-amplified cDNA (10 μl) was then treated with 1.2 U Exonuclease I and expression quantified by real time PCR on the BioMark HD System (© Fluidigm Corporation, CA, USA) using the 96.96 Dynamic Array IFC and the GE 96x96 Fast PCR+ Melt protocol and SsoFast EvaGreen Supermix with Low ROX (Biorad, CA, USA) and 5 μM primers for each assay.
RNA isolation and real-time PCR
RNA isolation was performed using RNeasy Plus Micro Kit (QIAGEN), according to manufacturer’s instruction. RNA was reverse-transcribed with the iScript cDNA Synthesis Kit (Bio-Rad, Munich, Germany). qPCR was performed by using the SsoFast EvaGreen Supermix (BioRad, Munich, Germany) and gene-specific primers (Table S4) in a CFX384 Real time PCR detection system (BioRad, Munich, Germany). Relative mRNA expression levels were calculated according to the ΔΔCt method upon normalization to β2 m.
RNA sequencing
RNA isolation was performed using RNeasy Plus Micro Kit (QIAGEN) and RNA was subjected to the amplification workflow of the SMARTer Ultra HV v2 kit (Takara Bio). Amplified cDNA was successively converted into short read sequencing libraries using the NEBnext Ultra DNA library preparation chemistry (New England Biolabs). Libraries were equimolarly pooled and sequenced on an Illumina HiSeq 2500, resulting in ∼29–45 million single end reads per library.
Metabolomics
For metabolomics, 2x 105 Lin—cKit+ cells per sample were sorted by FACS in 75 mM ammonium carbonate adjusted to pH 7.4 with acetic acid and snap frozen in liquid nitrogen. Metabolite extraction from cells was performed twice using 400 μL extraction solution (acetonitrile:methanol:water in a 40:40:20 ratio) at −20°C. Untargeted analysis of metabolites was performed by flow injection-time-of-flight mass spectrometry on an Agilent 6550 QTOF Instrument operated in negative mode 4 GHz, high resolution in a mass to charge (m/z) range of 50-1000, as described previously (Fuhrer et al., 2011). Ions were annotated to the ionic derivatives of the metabolites listed in the KEGG database for Homo sapiens allowing 0.001 atomic mass units tolerance.
Lipidomics
For lipidomics, 2x 105 Lin—cKit+ cells per sample were sorted. Mass spectrometry-based lipid analysis was performed at Lipotype GmbH (Dresden, Germany) as previously described (Sampaio et al., 2011). Lipids were extracted using a two-step chloroform/methanol procedure (Ejsing et al., 2009). Samples were spiked with internal lipid standard mixture containing: cardiolipin 16:1/15:0/15:0/15:0 (CL), ceramide 18:1;2/17:0 (Cer), diacylglycerol 17:0/17:0 (DAG), hexosylceramide 18:1;2/12:0 (HexCer), lyso-phosphatidate 17:0 (LPA), lyso-phosphatidylcholine 12:0 (LPC), lysophosphatidylethanolamine 17:1 (LPE), lyso-phosphatidylglycerol 17:1 (LPG), lyso-phosphatidylinositol 17:1 (LPI), lyso-phosphatidylserine 17:1 (LPS), phosphatidate 17:0/17:0 (PA), phosphatidylcholine 17:0/17:0 (PC), phosphatidylethanolamine 17:0/17:0 (PE), phosphatidylglycerol 17:0/17:0 (PG), phosphatidylinositol 16:0/16:0 (PI), phosphatidylserine 17:0/17:0 (PS), cholesterol ester 20:0 (CE), sphingomyelin 18:1;2/12:0;0 (SM), triacylglycerol 17:0/17:0/17:0 (TAG) and cholesterol D6 (Chol). After extraction, the organic phase was transferred to an infusion plate and dried in a speed vacuum concentrator. First step dry extract was re-suspended in 7.5 mM ammonium acetate in chloroform/methanol/propanol (1:2:4, V:V:V) and second step dry extract in 33% ethanol solution of methylamine in chloroform/methanol (0.003:5:1; V:V:V). All liquid handling steps were performed using Hamilton Robotics STARlet robotic platform with the Anti Droplet Control feature for organic solvents pipetting.
Samples were analyzed by direct infusion on a QExactive mass spectrometer (Thermo Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences). Samples were analyzed in both positive and negative ion modes with a resolution of Rm/z = 200 = 280000 for MS and Rm/z = 200 = 17500 for MSMS experiments, in a single acquisition. MSMS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1 Da increments (Surma et al., 2015). Both MS and MSMS data were combined to monitor cholesterol esters, DAG and TAG ions as ammonium adducts; PC, PC O-, as acetate adducts; and CL, PA, PE, PE O-, PG, PI and PS as deprotonated anions. MS only was used to monitor LPA, LPE, LPE O-, LPI and LPS as deprotonated anions; Cer, HexCer, SM, LPC and LPC O- as acetate adduct and cholesterol as ammonium adduct of an acetylated derivative (Liebisch et al., 2006).
Quantification and Statistical Analysis
Single cell qPCR
Raw data were analyzed using the Fluidigm Real-Time PCR analysis software. Pre-processing and data analysis of single cell expression profiles was conducted using KNIME 2.11.2 and RStudio Version 0.99.486 (Boston, MA, USA). To model the bi-modal gene expression, the Hurdle model, a semi-continuous modeling framework, was applied to the pre-processed data (McDavid et al., 2014). This allowed us to assess the differential expression profiles with respect to the frequency of expression and the positive expression mean via a likelihood ratio test.
RNA Sequencing Analysis
FastQC (http://www.bioinformatics.babraham.ac.uk/) was used to perform a basic quality control on the resulting reads. As an additional control, library diversity was assessed by redundancy investigation in the reads. Alignment of the reads to the mouse reference (mm10) was done with GSNAP (Wu and Nacu, 2010) and Ensembl gene annotation version 81 was used to detect splice sites. The uniquely aligned reads were counted with featureCounts (Liao et al., 2014) and the same Ensembl annotation. Normalization of the raw read counts based on the library size and testing for differential expression between conditions was performed with the DESeq2 R package (Love et al., 2014). Genes, which have an adjusted p value (padj) < 0.05 and counts > 50 were considered as differentially expressed. Pathway analysis and upstream regulator analysis of gene lists containing significantly differentially expressed genes (padj < 0.05, log2FC < −0.3 and > 0.3) was done with Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity). Morpheus software (Broad Institute) was used to generate heatmaps. Top canonical pathways derived from IPA are shown. To perform gene set enrichment analysis (GSEA), gene sets were ranked by taking the -log10 transform of the p value and signed as positive or negative based on the direction of fold change. GSEA pre-ranked analysis (1000 permutations, minimum term size of 15, maximum term size of 500) was then performed using the GSEA software (Broad Institute) (Musso et al., 2015). Annotated gene sets from Molecular Signatures Database (MSigDB) were used as input. Lineage specific genes were selected based on IPA analysis (Molecular and Cellular Functions analysis). For generation of heatmaps of genes involved in glycolysis and pentose phosphate pathway we used the KEGG database.
Metabolomics
Data were processed and analyzed with MATLAB.
Lipidomics
Data were analyzed with a lipid identification software based on LipidXplorer (Herzog et al., 2011, Herzog et al., 2012). Data post-processing and normalization were performed using an in house developed data management system. Only lipid identifications with a signal-to-noise ratio > 5, and a signal intensity 5-fold higher than in corresponding blank samples were considered for further data analysis. Principal Component Analysis (PCA) was computed in MATLAB 9.0 (R2016a) using the Singular Value Decomposition function after transforming raw data to mol% (each quantity was divided by the sum of the lipids in the sample and multiplied by 100). Descriptive statistics and histograms were generated using base R version 3.3.2 (R Development Core Team, 2016) and packages reshape2 (Wickham, 2007) and ggplot2 (Wickham, 2009). The difference between the means was calculated species by species on the mol-percent transformed dataset. For each lipid feature we calculated the mean in the two groups (PBS and β-glucan), and subtracted the mean of the treated samples from the mean of the control samples in order to show whether and in which condition lipid species were more abundant.
Statistical analysis
All data are presented as mean ± SEM. A two-tailed unpaired Student’s t test for parametric variables and a Mann-Whitney U test for non-parametric variables were used for the comparison of two groups. For comparisons of multiple groups, One-way ANOVA followed by Holm-Sidak multiple comparison tests was used. In vitro comparison of matched groups was performed with a paired two-tailed Student’s t test. Comparison of survival curves was performed by Log-rank (Mantel-Cox) test. All statistical analysis was performed using GraphPad Prism (GraphPad Inc., La Jolla, CA). Significance was set at p < 0.05.
Data and Software Availability
Data are available upon request to the Lead Contact. Sequencing data are available at the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GEO: GSE95617.
Acknowledgments
This work was supported by grants from the ERC (Consolidator grant DEMETINL to T.C.), the Deutsche Forschungsgemeinschaft (CH279/5-1 and TR-SFB127 to T.C. and SFB 854/B26N to B.I.), and from the NIH (DE024716 to G.H. and DE026152 to G.H. and T.C.). A.S., M.B., and J.L.S. are members of the Excellence Cluster ImmunoSensation. Ü.C. was supported by the German Federal Ministry of Education and Research grant to the German Center for Diabetes Research (DZD e.V.). M.G.N. was supported by an ERC Consolidator Grant (#310372) and a Spinoza Grant of the Netherlands Association for Scientific Research. MGN and LABJ were partly supported by an IN-CONTROL CVON grant from the Netherlands Heart Foundation (CVON2012-03).
Author Contributions
I.M. designed the project, performed experiments, analyzed and interpreted data, supervised research, and wrote the manuscript; K.R., B.W., L.-S.C., M.T., M.G., I.K., and A.C. performed experiments and analyzed data; T.G., A.E., A. Palladini, M.L., A. Petzold, I.H., and A.D. analyzed and interpreted data; A.S., M.B., L.A.B.J., B.I., B.W., J.L.S., P.M., Ü.C., and K.S. interpreted data and contributed to experimental design; N.Z. performed metabolomics and analyzed data; G.H. participated in project design, supervised research, interpreted data, and edited the paper; M.G.N. participated in project design, interpreted data, and edited the paper; and T.C. designed the project, interpreted data, supervised research, and wrote the manuscript.
Declaration of Interests
K.S. is CEO of Lipotype, a start-up company that specializes in lipid analysis.
Published: January 11, 2018
Footnotes
Supplemental Information includes six figures and four tables and can be found with this article online at https://doi.org/10.1016/j.cell.2017.11.034.
Contributor Information
Ioannis Mitroulis, Email: ioannis.mitroulis@uniklinikum-dresden.de.
Triantafyllos Chavakis, Email: triantafyllos.chavakis@uniklinikum-dresden.de.
Supplemental Information
NAME is the gene set name; SIZE is the number of genes in the gene set after filtering out those genes not in the expression dataset; ES is the enrichment score for the gene set; NES is the normalized enrichment score that accounts for size differences in gene sets; NOM p-val is the nominal p-value of ES significance based on permutation test; FDR q-val is the False Discovery Rate; FWER p-val is the family-wise error rate; RANK AT MAX is the position in the ranked list at which the maximum running enrichment score occurred.
Results of GSEA on RNA seq data from LT-HSC isolated on day 14 after 5-FU administration using MSigDB hallmark gene set. (NAME is the gene set name; SIZE is the number of genes in the gene set after filtering out those genes not in the expression dataset; ES is the enrichment score for the gene set; NES is the normalized enrichment score that accounts for size differences in gene sets; NOM p-val is the nominal p-value of ES significance based on permutation test; FDR q-val is the False Discovery Rate; FWER p-val is the family-wise error rate; RANK AT MAX is the position in the ranked list at which the maximum running enrichment score occurred.).
References
- Arts R.J.W., Carvalho A., La Rocca C., Palma C., Rodrigues F., Silvestre R., Kleinnijenhuis J., Lachmandas E., Gonçalves L.G., Belinha A. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep. 2016;17:2562–2571. doi: 10.1016/j.celrep.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts R.J.W., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., Rodrigues F., Silvestre R., Cheng S.-C., Wang S.-Y. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metab. 2016;24:807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher S., Manz M.G. Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 2017;38:345–357. doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Bonifacio E., Ziegler A.-G., Klingensmith G., Schober E., Bingley P.J., Rottenkolber M., Theil A., Eugster A., Puff R., Peplow C., Pre-POINT Study Group Effects of high-dose oral insulin on immune responses in children at high risk for type 1 diabetes: the Pre-POINT randomized clinical trial. JAMA. 2015;313:1541–1549. doi: 10.1001/jama.2015.2928. [DOI] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu Y., Zheng P. Mammalian target of rapamycin activation underlies HSC defects in autoimmune disease and inflammation in mice. J. Clin. Invest. 2010;120:4091–4101. doi: 10.1172/JCI43873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.-C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun U., Simons K. Cell membranes: the lipid perspective. Structure. 2011;19:1543–1548. doi: 10.1016/j.str.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Cutando L., Busquets-Garcia A., Puighermanal E., Gomis-González M., Delgado-García J.M., Gruart A., Maldonado R., Ozaita A. Microglial activation underlies cerebellar deficits produced by repeated cannabis exposure. J. Clin. Invest. 2013;123:2816–2831. doi: 10.1172/JCI67569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing C.S., Sampaio J.L., Surendranath V., Duchoslav E., Ekroos K., Klemm R.W., Simons K., Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelin C., Herrington R., Janmohamed S., Barbara M., Tran G., Paige C.J., Benveniste P., Zuñiga-Pflücker J.-C., Souabni A., Busslinger M., Iscove N.N. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 2013;14:1037–1044. doi: 10.1038/ni.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T., Heer D., Begemann B., Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- Gekas C., Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood. 2013;121:4463–4472. doi: 10.1182/blood-2012-09-457929. [DOI] [PubMed] [Google Scholar]
- Goodridge H.S., Ahmed S.S., Curtis N., Kollmann T.R., Levy O., Netea M.G., Pollard A.J., van Crevel R., Wilson C.B. Harnessing the beneficial heterologous effects of vaccination. Nat. Rev. Immunol. 2016;16:392–400. doi: 10.1038/nri.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M., Shevchenko A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12:R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R., Schuhmann K., Schwudke D., Sampaio J.L., Bornstein S.R., Schroeder M., Shevchenko A. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS ONE. 2012;7:e29851. doi: 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T., Kuroha T., Moriguchi T., Cummings D., Maillard I., Lim K.-C., Engel J.D. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D.E., Schafer Z.T., Evans R.M., Suda T., Lee C.-H., Pandolfi P.P. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.J., Larsen N., Biering-Sørensen S., Andersen A., Eriksen H.B., Monteiro I., Hougaard D., Aaby P., Netea M.G., Flanagan K.L., Benn C.S. Heterologous immunological effects of early BCG vaccination in low-birth-weight infants in Guinea-Bissau: a randomized-controlled trial. J. Infect. Dis. 2015;211:956–967. doi: 10.1093/infdis/jiu508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S., Xu H., Zabalawi M., Maruko E., Fulp B.E., Bluemn T., Brzoza-Lewis K.L., Gerelus M., Weerasekera R., Kallinger R. Lipid-free apolipoprotein A-I reduces progression of atherosclerosis by mobilizing microdomain cholesterol and attenuating the number of CD131 expressing cells: monitoring cholesterol homeostasis using the cellular ester to total cholesterol ratio. J. Am. Heart Assoc. 2016;5:e004401. doi: 10.1161/JAHA.116.004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G., Binder M., Schifferer R., Langmann T., Schulz B., Schmitz G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS) Biochim. Biophys. Acta. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A., Dennis L., Danaher P., Finak G., Krouse M., Wang A., Webster P., Beechem J., Gottardo R. Modeling bi-modality improves characterization of cell cycle on gene expression in single cells. PLoS Comput. Biol. 2014;10:e1003696. doi: 10.1371/journal.pcbi.1003696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., O’Neill L.A. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur. J. Immunol. 2016;46:13–21. doi: 10.1002/eji.201445427. [DOI] [PubMed] [Google Scholar]
- Mitroulis I., Chen L.-S., Singh R.P., Kourtzelis I., Economopoulou M., Kajikawa T., Troullinaki M., Ziogas A., Ruppova K., Hosur K. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J. Clin. Invest. 2017;127:3624–3639. doi: 10.1172/JCI92571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossadegh-Keller N., Sarrazin S., Kandalla P.K., Espinosa L., Stanley E.R., Nutt S.L., Moore J., Sieweke M.H. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497:239–243. doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.J., Akhtari M., Tolani S., Pagler T., Bijl N., Kuo C.-L., Wang M., Sanson M., Abramowicz S., Welch C. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J. Clin. Invest. 2011;121:4138–4149. doi: 10.1172/JCI57559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G., Mosimann C., Panáková D., Burger A., Zhou Y., Zon L.I., MacRae C.A. Generating and evaluating a ranked candidate gene list for potential vertebrate heart field regulators. Genom. Data. 2015;6:199–201. doi: 10.1016/j.gdata.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., Kincade P.W. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S., Hirai Y. The therapeutic potential of interleukin-1 beta in the treatment of chemotherapy- or radiation-induced myelosuppression and in tumor therapy. Biotherapy. 1989;1:339–354. doi: 10.1007/BF02171010. [DOI] [PubMed] [Google Scholar]
- Netea M.G., van Crevel R. BCG-induced protection: effects on innate immune memory. Semin. Immunol. 2014;26:512–517. doi: 10.1016/j.smim.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Netea M.G., Joosten L.A.B., Latz E., Mills K.H.G., Natoli G., Stunnenberg H.G., O’Neill L.A.J., Xavier R.J. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo R.A., May G.E., Enver T. Primed and ready: understanding lineage commitment through single cell analysis. Trends Cell Biol. 2015;25:459–467. doi: 10.1016/j.tcb.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Norata G.D., Caligiuri G., Chavakis T., Matarese G., Netea M.G., Nicoletti A., O’Neill L.A.J., Marelli-Berg F.M. The cellular and molecular basis of translational immunometabolism. Immunity. 2015;43:421–434. doi: 10.1016/j.immuni.2015.08.023. [DOI] [PubMed] [Google Scholar]
- Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Wagers A.J., Giuriato S., Anderson W.C., Weissman I.L. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Pietras E.M., Reynaud D., Kang Y.-A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., Passegué E. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 2015;17:35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.-M., Will B. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 2016;18:607–618. doi: 10.1038/ncb3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J., Saeed S., Martens J.H.A., Giamarellos-Bourboulis E.J., Ifrim D.C., Logie C., Jacobs L., Jansen T., Kullberg B.-J., Wijmenga C. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2016. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rosenbauer F., Tenen D.G. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- Sampaio J.L., Gerl M.J., Klose C., Ejsing C.S., Beug H., Simons K., Shevchenko A. Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. USA. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S., Sieweke M. Integration of cytokine and transcription factor signals in hematopoietic stem cell commitment. Semin. Immunol. 2011;23:326–334. doi: 10.1016/j.smim.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Sezgin E., Levental I., Mayor S., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T., Kocabas F., Zheng J., Deberardinis R.J., Mahmoud A.I., Olson E.N., Schneider J.W., Zhang C.C., Sadek H.A. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T., Takubo K., Semenza G.L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Surma M.A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D., Simons K., Masoodi M., Sampaio J.L. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 2015;117:1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H., Fritsch K., Kovtonyuk L.V., Saito Y., Yakkala C., Jacobs K., Ahuja A.K., Lopes M., Hausmann A., Hardt W.-D. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell. 2017;21:225–240.e5. doi: 10.1016/j.stem.2017.06.013. [DOI] [PubMed] [Google Scholar]
- Takubo K., Nagamatsu G., Kobayashi C.I., Nakamura-Ishizu A., Kobayashi H., Ikeda E., Goda N., Rahimi Y., Johnson R.S., Soga T. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Essers M., Wilson A. Awakening dormant haematopoietic stem cells. Nat. Rev. Immunol. 2010;10:201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- van der Meer J.W., Barza M., Wolff S.M., Dinarello C.A. A low dose of recombinant interleukin 1 protects granulocytopenic mice from lethal gram-negative infection. Proc. Natl. Acad. Sci. USA. 1988;85:1620–1623. doi: 10.1073/pnas.85.5.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- Wang Y.-H., Israelsen W.J., Lee D., Yu V.W.C., Jeanson N.T., Clish C.B., Cantley L.C., Vander Heiden M.G., Scadden D.T. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158:1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Reshaping data with the reshape package. J. Stat. Softw. 2007;21:1–20. [Google Scholar]
- Wickham H. Springer; 2009. ggplot2: Elegant graphics for data analysis. [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Winkler I.G., Barbier V., Nowlan B., Jacobsen R.N., Forristal C.E., Patton J.T., Magnani J.L., Lévesque J.-P. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 2012;18:1651–1657. doi: 10.1038/nm.2969. [DOI] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Park C.S., Lacorazza H.D. Genetic control of quiescence in hematopoietic stem cells. Cell Cycle. 2013;12:2376–2383. doi: 10.4161/cc.25416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez A., Megías J., O’Connor J.-E., Gozalbo D., Gil M.L. Candida albicans induces selective development of macrophages and monocyte derived dendritic cells by a TLR2 dependent signalling. PLoS ONE. 2011;6:e24761. doi: 10.1371/journal.pone.0024761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tanaka Y., Seay M., Li Z., Jin J., Garmire L.X., Zhu X., Taylor A., Li W., Euskirchen G. Single cell transcriptomics reveals unanticipated features of early hematopoietic precursors. Nucleic Acids Res. 2017;45:1281–1296. doi: 10.1093/nar/gkw1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvan-Charvet L., Pagler T., Gautier E.L., Avagyan S., Siry R.L., Han S., Welch C.L., Wang N., Randolph G.J., Snoeck H.W., Tall A.R. ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Rodriguez S., Wang L., Wang S., Serezani H., Kapur R., Cardoso A.A., Carlesso N. Sepsis induces hematopoietic stem cell exhaustion and myelosuppression through distinct contributions of TRIF and MYD88. Stem Cell Reports. 2016;6:940–956. doi: 10.1016/j.stemcr.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.L., Baltimore D. Regulation of stress-induced hematopoiesis. Curr. Opin. Hematol. 2015;22:286–292. doi: 10.1097/MOH.0000000000000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NAME is the gene set name; SIZE is the number of genes in the gene set after filtering out those genes not in the expression dataset; ES is the enrichment score for the gene set; NES is the normalized enrichment score that accounts for size differences in gene sets; NOM p-val is the nominal p-value of ES significance based on permutation test; FDR q-val is the False Discovery Rate; FWER p-val is the family-wise error rate; RANK AT MAX is the position in the ranked list at which the maximum running enrichment score occurred.
Results of GSEA on RNA seq data from LT-HSC isolated on day 14 after 5-FU administration using MSigDB hallmark gene set. (NAME is the gene set name; SIZE is the number of genes in the gene set after filtering out those genes not in the expression dataset; ES is the enrichment score for the gene set; NES is the normalized enrichment score that accounts for size differences in gene sets; NOM p-val is the nominal p-value of ES significance based on permutation test; FDR q-val is the False Discovery Rate; FWER p-val is the family-wise error rate; RANK AT MAX is the position in the ranked list at which the maximum running enrichment score occurred.).