Summary
Tissue regeneration requires dynamic cellular adaptation to the wound environment. It is currently unclear how this is orchestrated at the cellular level and how cell fate is affected by severe tissue damage. Here we dissect cell fate transitions during colonic regeneration in a mouse dextran sulfate sodium (DSS) colitis model, and we demonstrate that the epithelium is transiently reprogrammed into a primitive state. This is characterized by de novo expression of fetal markers as well as suppression of markers for adult stem and differentiated cells. The fate change is orchestrated by remodeling the extracellular matrix (ECM), increased FAK/Src signaling, and ultimately YAP/TAZ activation. In a defined cell culture system recapitulating the extracellular matrix remodeling observed in vivo, we show that a collagen 3D matrix supplemented with Wnt ligands is sufficient to sustain endogenous YAP/TAZ and induce conversion of cell fate. This provides a simple model for tissue regeneration, implicating cellular reprogramming as an essential element.
Keywords: intestinal stem cells, reprogramming, regeneration, YAP/TAZ, mechano-sensing
Graphical Abstract
Highlights
-
•
The repairing epithelium can be isolated based on Sca1 expression
-
•
Markers upregulated during tissue repair are expressed in the fetal intestine
-
•
Mechano-transduction via FAK, Src, and YAP/TAZ facilitate efficient tissue repair
-
•
YAP/TAZ activation is required and sufficient to induce cellular reprogramming
The mechanism that governs tissue regeneration following severe damage to the colonic epithelium remains poorly understood. Jensen and colleagues show that the colonic epithelium undergoes a profound reprogramming into a more primitive state with fetal-like properties. Moreover, they demonstrate that YAP and TAZ operate as essential mechano-sensors during tissue reprogramming.
Introduction
Intestinal epithelial stem cells ensure fast tissue replenishment. These adult stem cells reside at the bottom of crypts and express markers such as Lgr5, Olfm4, and Lrig1 (Barker et al., 2012). Upon tissue damage, cells that are distinct from adult intestinal stem cells contribute to wound repair, replenishment of lost stem cells, and restoration of tissue architecture (Blanpain and Fuchs, 2014). Yet, the cellular responses underlying this remarkable plasticity remain unclear.
Patients with inflammatory bowel disease, such as ulcerative colitis (UC) and Crohn’s disease, have impaired intestinal barrier function, and they experience recurrent severe inflammation. Current treatment strategies aim to reduce the intestinal inflammation burden in general, with only a limited focus on targeting the epithelium to promote tissue regeneration. Tissue regeneration is a complex process associated with pronounced changes to the environment at both the cellular and biophysical levels (Stappenbeck and Miyoshi, 2009). Sensing these environmental changes and spatial information is, however, essential during tissue regeneration. Two highly related transcriptional activators, YAP and TAZ, have recently emerged as primary sensors of the cellular microenvironment, integrating cell polarity and mechanical cues with growth factor signaling and inflammation (Piccolo et al., 2014). In vitro, activation of YAP/TAZ has recently been shown to dedifferentiate committed cells back to a progenitor and stem cell state (Panciera et al., 2016). YAP/TAZ-mediated signaling appears dispensable during steady-state homeostasis in the intestinal epithelium (Azzolin et al., 2014, Barry et al., 2013, Cai et al., 2010). Evidence does, however, suggest an important role of YAP during intestinal regeneration (Cai et al., 2010). Yet, it remains largely unknown how YAP/TAZ signaling is controlled during repair and which cellular processes are regulated by YAP/TAZ during tissue remodeling.
Here we examine wound repair in the colonic epithelium and identify markers for the repairing epithelium (RE). Characterization of the repairing epithelium demonstrates a strong and necessary response by the transcriptional regulators YAP/TAZ driven by environmental changes. We provide evidence that this can be recapitulated in vitro using defined cell culture conditions in a YAP/TAZ-dependent manner. Moreover, the changes associated with the repairing epithelium strongly suggest that the tissue undergoes injury-assisted transition into a primitive state with fetal-like properties. Importantly, the changes associated with the wound-induced reprogramming are reversible both in vivo and in vitro, allowing the tissue to regain its normal cellular architecture and adult-specific gene expression, when regeneration is complete. We believe that understanding the process of tissue repair and how this is orchestrated at the cellular and molecular levels will provide better solutions for enhancing regeneration.
Results
The Repairing Epithelium Represents a Distinct Transcriptional State
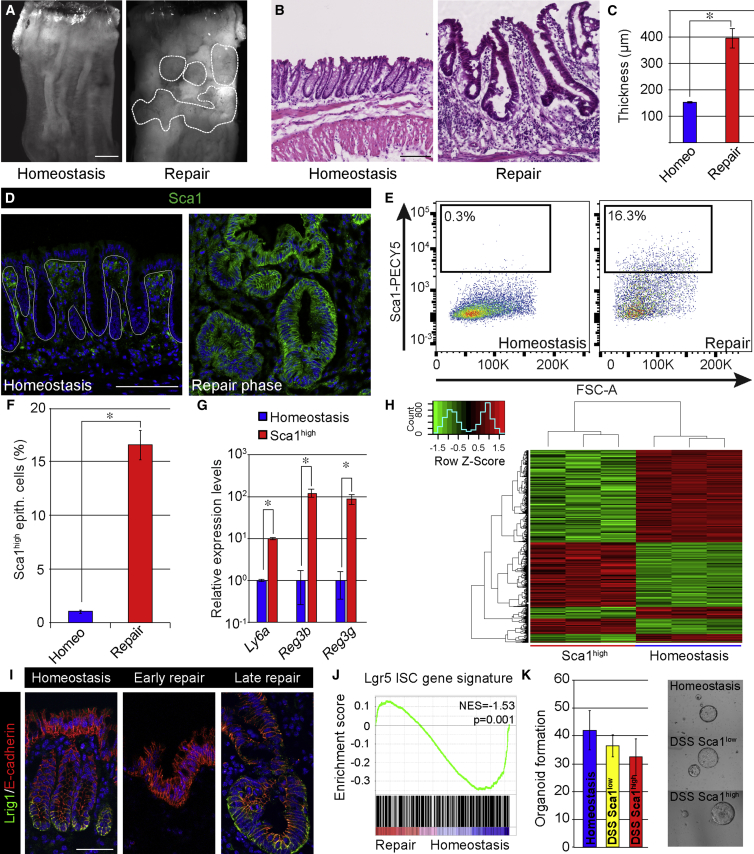
To investigate the mechanisms controlling tissue regeneration following severe injury, we took advantage of the dextran sulfate sodium (DSS) colitis model. Mice exposed to DSS for 5 days developed epithelial ulceration in the colon (Figures 1A, 1B, and S1A). Ulceration of the distal part of the colon was evident approximately 1 week after DSS administration (Figure S1A). This was followed by the formation of swelling islets (Figure 1A), which were covered by a thickened hyperplastic repairing epithelium (Figures 1B and 1C). At 1 month after the administration of DSS, tissue homeostasis was restored (Figure S1A).
Figure 1.
Repairing Intestinal Epithelium Has a Cellular and Molecular Profile Distinct from the Normal Epithelium
(A) Macroscopic images of the distal part of the colon in an untreated mouse (left) and a mouse 2 weeks post-administration of DSS (right). The demarcated area indicates regions undergoing active re-epithelialization. Scale bar, 2 mm.
(B) H&E staining of homeostatic tissue and tissue in the repair phase at 2 weeks following DSS administration. Scale bar, 100 μm.
(C) The mucosal/submucosal thickness at homeostasis and repair phase (2 weeks). Shown are mean distances ± SEM (n = 4 animals; p = 0.007 based on two-sided Student’s t test).
(D) Sca1 (green) expression in colonic epithelium in homeostatic and repair phase. Sections are counterstained with DAPI (blue). The demarcated line indicates the epithelial structure. Scale bar, 100 μm.
(E) Flow cytometric analysis of cells from the colonic epithelium during homeostasis and in repair phase (2 weeks post-administration of DSS). Diagrams show representative plots for Sca1-PECy5 in live CD45−veCD31−veEpcam+ve cell population.
(F) Quantification of the percentages of Sca1high cells in homeostasis and repair phase. Diagram shows average ± SEM (n = 3; p = 4 × 10−5 based on two-sided Student’s t test).
(G) qRT-PCR analyses in Epcam+Sca1high cells sorted via FACS from the repairing epithelium and in homeostatic epithelial cells (Epcam+). Bars represent average levels ± SEM (n = 3; Ly6a, p = 1.3 × 10−4; Reg3b, p = 9.7 × 10−3; Reg3g, p = 8.5 × 10−3 based on two-sided Student’s t test).
(H) Heatmap analysis of differentially expressed probe sets (fold change > 2.0; FDR < 0.05), comparing the expression profile of epithelial cells isolated from homeostatic tissue and Sca1high cells from the repairing epithelium.
(I) Detection of Lrig1 (green) in homeostasis, early and late repair phase counterstained for E-cadherin (red) with DAPI (blue). Scale bar, 50 μm.
(J) GSEA of repairing and homeostatic epithelium using Lgr5 intestinal stem cell gene signature.
(K) Organoid formation for Epcam+ cells isolated from normal homeostasis, as well as Epcam+ Sca1low and Sca1high cells during tissue repair showing representative images of formed organoids. Bars represent average number of organoids formed ± SEM (n = 3 animals).
Members of the LY6 superfamily of cell surface proteins are upregulated during colitis (Flanagan et al., 2008). Moreover, the LY6 member Stem Cell Antigen-1 (Sca1/Ly6a) has been used extensively in the hematopoietic field to enrich for murine hematopoietic stem cells (Morrison et al., 1995). We thus asked whether Sca1 could represent a marker for the repairing epithelium. Immunofluorescence time course analysis showed a dynamic regulation of Sca1, with initial elevated levels in the stroma and subsequently very high levels specifically associated with the swelling areas of the repairing epithelium (Figures 1D–1G and S1B). This pattern was mirrored by the secreted bactericidal C-type lectin Reg3b (Figure S1A). Further characterization of the repairing epithelium demonstrated an elevated number of proliferative cells (Ki67 positive) and fewer Muc2-expressing secretory cells than in homeostatic tissue (Figures S1C and S1D). Thus, the repairing epithelium expresses markers that distinguish it from the homeostatic epithelium, indicating that these cells have distinct properties.
To gain more insights into the molecular traits of the repairing epithelium, we performed expression profiling on epithelial cells purified from homeostatic tissue (EpCAMposSca1negCD45negCD31neg) and the repairing epithelium (EpCAMposSca1highCD45negCD31neg). As expected, a number of genes were differentially expressed between these purified epithelial cell populations (424 probe sets up and 585 probe sets down; fold change > 2; false discovery rate [FDR] < 0.05), reinforcing the notion that the repairing epithelium represents a cellular state profoundly different from the normal homeostatic epithelium (Figure 1H). Gene ontology (GO) analyses for molecular pathways illustrated that the Sca1high cells were specifically enriched in signaling cascades associated with chemokines and inflammation, whereas metabolic pathways were repressed (Figures S1E and S1F; Tables S1 and S2). Importantly, a number of markers specifically associated with the adult stem cell state were specifically repressed in the Sca1high state. This was evident at the protein level, where expression of the stem cell marker Lrig1 was reduced during the early phases of tissue repair (Figure 1I; Wong et al., 2012), as well as at the RNA level, where there was a general suppression of the gene signature associated with Lgr5-expressing intestinal stem cells (Figure 1J; Muñoz et al., 2012). Similarly, Sca1high cells were distinct from Sca1low cells following experimental colitis (Figure S1E). Despite the suppression of intestinal stem cell markers, Sca1high cells retained in vitro stem cell potential (Figure 1K). This demonstrates that the intestinal epithelium, as it progresses into the repair phase, undergoes extensive changes, including the suppression of normal markers of adult stem cell, without affecting stem cell potential.
The Reprogrammed Repairing Epithelium Expresses Primitive Fetal-Associated Markers
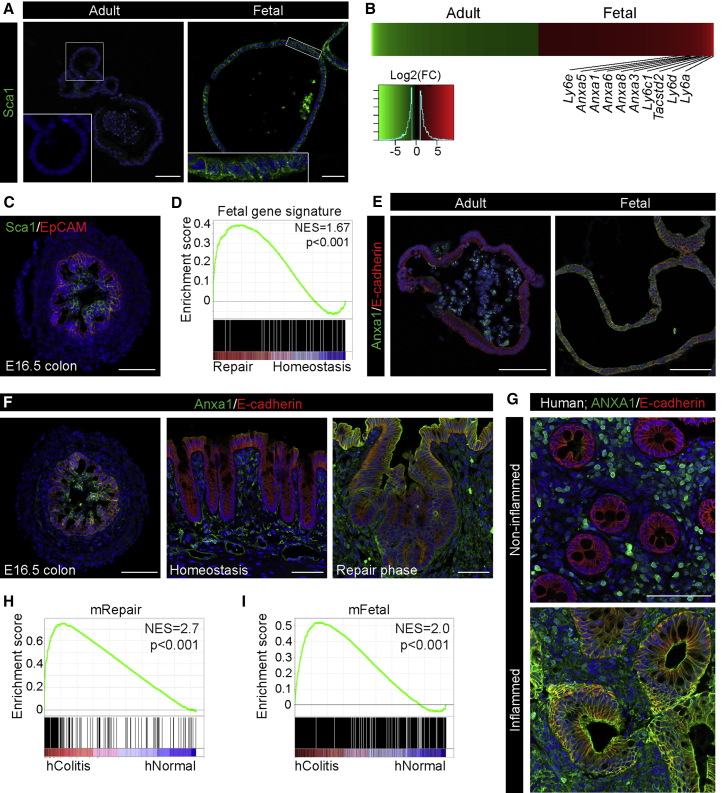
An assessment of the GO terms for biological processes associated with the repair phase revealed enrichment for transcriptional processes linked with morphogenesis and development (Figure S1F). We have previously demonstrated that the intestinal epithelium has distinct fetal and adult stem cells that can be propagated in vitro (Fordham et al., 2013). Expression analysis of cultures derived from the fetal and adult epithelium revealed that Sca1 was transcriptionally upregulated by fetal organoids along with a number of other Ly6 family members (Figures 2A and 2B). In vivo these observations were paralleled by the robust expression of Sca1 in the fetal colonic epithelium (Figure 2C), whereas it was absent in the adult homeostatic colonic epithelium (Figure 1D). All in all, this demonstrates that Sca1 is expressed by more primitive epithelia both in vitro and in vivo. Transcriptionally, gene set enrichment analysis (GSEA) of fetal signature genes (>2-fold; FDR < 0.05; fetal versus adult organoids) revealed a significant enrichment in the repairing epithelium when compared to the normal adult epithelium, strongly suggesting that the repairing epithelium represents an earlier developmental stage (Figure 2D). To further substantiate this observation, we assessed the expression of Annexin A1 (Anxa1), which was elevated in fetal organoids (Figures 2B and 2E), and Tacstd2/Trop2, which marks fetal progenitors (Mustata et al., 2013). Evidently, Anxa1 expression was upregulated both in the fetal and repairing epithelium (Figures 2F and S2A), and Tacstd2/Trop2 was also elevated during the repair phase and by fetal organoids (Figures S2A–S2C). These observations are consistent with the notion that, upon colonic injury, regeneration involves transcriptional reprogramming of epithelial cells to a more primitive state.
Figure 2.
The Repairing Epithelium Adopts a Fetal-like Expression Profile
(A) Sca1 (green) expression in organoids from adult animals (Adult) and in fetal organoids (Fetal) derived from the proximal small intestine. Insets show the enlarged view of the indicated region. Images are counterstained with DAPI (blue). Scale bar, 50 μm.
(B) Heatmap of differentially expressed probe sets showing genes upregulated more than 2-fold comparing adult organoids (green) and fetal organoids (red) (n = 3; fold change > 2; FDR < 0.05). Multiple members of Ly6 and Annexin gene families are upregulated in the fetal cells.
(C) Sca1 (green) expression in fetal colon at embryonic day (E)16.5 counterstained with EpCAM (red) and DAPI (blue). Scale bars, 50 μm.
(D) GSEA showing enrichment of the fetal intestinal gene signature in the repairing epithelium.
(E) Anxa1 (green) expression in adult organoids (Adult) and fetal organoids (Fetal) derived from the proximal small intestine. Images are counterstained with E-cadherin (red) and DAPI (blue). Scale bar, 100 μm.
(F) Anxa1 (green) expression in fetal colon at E16.5, colon in homeostasis and in the repair phase. Images are counterstained with E-cadherin (red) and DAPI (blue). Scale bars, 50 μm for fetal colon and 100 μm for adult colon.
(G) ANXA1 (green) in colonic biopsy from non-inflamed and inflamed regions obtained from a patient with ulcerative colitis. Images are counterstained with E-cadherin (red) and DAPI (blue). Scale bar, 100 μm.
(H and I) GSEAs showing enrichment of the transcriptional signatures for the mouse repairing epithelium (H, mRepair) and mouse fetal state (I, mFetal) in samples isolated from patients with active colitis (hColitis) compared to control individuals (hNormal).
We next evaluated human clinical material to address whether the observed reprogramming occurs following severe inflammation. As there is no human homolog of Sca1, we analyzed ANXA1 in intestinal specimens obtained by colonoscopy from non-inflamed areas and areas with active disease in the same patient with ulcerative colitis. Similar to our observations in mouse, ANXA1 was highly expressed in the areas of flaring disease, where the epithelium was in a state of regeneration as compared to the non-inflamed part (Figure 2G). Analyses of publicly available gene expression datasets from colonic tissue from patients with active ulcerative colitis and normal colon tissues (Van der Goten et al., 2014) further substantiated this finding, as genes upregulated by the repairing epithelium (>2-fold; FDR < 0.05; mRepair) and the fetal gene signatures (mFetal; Figures 2H and 2I; Table S3) were enriched in tissue from ulcerative colitis patients. We conclude that extensive transcriptional rewiring occurs upon regeneration in human ulcerative colitis and following DSS-induced colitis in mouse models.
The Repairing Epithelium Displays Elevated Integrin/FAK Signaling and YAP Activation
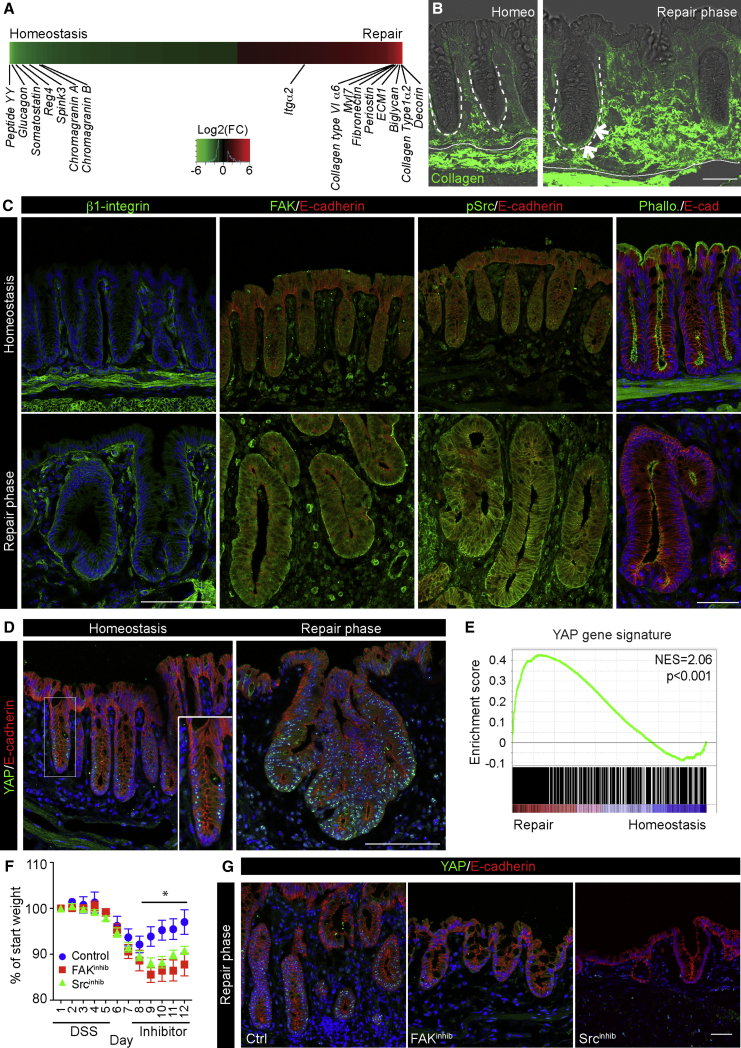
To investigate the molecular mechanism for the observed cellular reprogramming, we turned to the transcriptional profiles associated with the repairing epithelium, where we observed a profound repression of genes associated with secretory lineage. In contrast, several extracellular matrix components, including collagen type I, were highly elevated in the repairing epithelium (Figures 3A and S3A). This was also evident from GO analyses, where integrin signaling was among the most enriched pathways in the Sca1high population (Figure S1F). These gene responses correlated well with the pronounced changes in the wound bed, as detected by second harmonic generation microscopy, where collagen fibers accumulated around de novo-formed crypts (Figure 3B). Changes in the signaling between the extracellular matrix (ECM) and epithelial cells are sensed by transmembrane collagen receptors of the integrin family, activating focal adhesion kinase (FAK) and Src to control actin cytoskeleton rearrangement (Kim and Gumbiner, 2015). In line with the exposure to the wound-associated matrix, the repairing epithelium displayed elevated levels of the collagen receptor β1 integrin subunit and FAK at the basolateral cell surface, as well as enhanced levels of phosphorylated Src (pSrc) and loss of polarized F-actin filaments (Figures 3C and S3B). This reveals that the repairing epithelium is influenced by the altered environmental stimuli and mechanical properties of the dense collagen matrix present in the wound bed.
Figure 3.
The Repairing Epithelium Displays Features of Active Mechano-Transduction
(A) Differentially expressed probes ranked in heatmap comparing tissue homeostasis and repair phase (n = 3; 2-fold regulated; FDR < 0.05). Selected genes associated with lineage differentiation and extracellular matrix interactions are indicated.
(B) Second harmonic generation (Collagen; green) overlapped with bright-field microscopy in tissue sections from homeostasis and repair phase. Demarcated lines indicate crypts and muscularis mucosa. Arrows indicate collagen fibers encapsulating de novo-formed crypts. Scale bar, 50 μm.
(C) Detection of β1 integrin, FAK, pSrc, and Phalloidin (green) during the repair phase and homeostasis. Images are counterstained with E-cadherin (red) and DAPI (blue). Scale bars, 100 μm for β1 integrin, FAK, and pSrc and 50 μm for Phalloidin.
(D) Detection of YAP (green) during the repair phase and homeostasis. Inset shows enlarged view of a single colonic crypt during homeostasis. Sections were counterstained with E-cadherin (red) and DAPI (blue). Scale bars, 100 μm.
(E) GSEA showing enrichment of a YAP gene signature in repairing relative to normal colonic epithelium.
(F) Weight curves for animals treated with DSS and subsequently with either vehicle or FAK and Src inhibitors during tissue repair. Individual points represent the average weight relative to the starting point ± SEM (n = 5 for all groups). Significance was assessed using a two-way ANOVA with Bonferroni correction for multiple comparisons (Srcinhib versus Ctrl day 9, p = 0.02; day 10, p = 0.004; day 11, p = 0.03; and day 12, p = 0.01; FAKinhib versus Ctrl day 9, p = 4 × 10−4; day 10, p = 2 × 10−4; day 11, p = 10−4; and day 12, p < 10−4).
(G) Detection of YAP (green) during the repair phase (day 12) in vehicle and Srcinhibitor- and FAKinhibitor-treated animals. Sections were counterstained with E-cadherin (red) and DAPI (blue). Scale bars, 50 μm.
See also Figure S3.
YAP and TAZ are overarching mechano-transducers and key transcriptional regulators of cell fates (Dupont et al., 2011). In line with the altered accumulation and activation of components of the focal adhesion pathway, the number of cells with nuclear accumulation of YAP was highly elevated during the repair phase, when compared to the small subsets of positive cells in the normal epithelium (Figures 3D and S3C). Moreover, GSEA with a reported YAP/TAZ activity gene signature showed a highly significant enrichment in the repairing epithelium (Figure 3E; Gregorieff et al., 2015). The combination of YAP nuclear retention and YAP/TAZ transcriptional activation provides strong evidence that YAP/TAZ-mediated signaling is activated during tissue regeneration.
To test whether signaling downstream of integrins is important for tissue repair and the localization of the mechano-sensor YAP, animals were treated with FAK and Src inhibitors during the transition from ulceration to the repair phase. We found that the activation of both pathway components was important for tissue repair, as mice treated with the inhibitors showed delayed repair following injury in line with previous reports (Cordero et al., 2014, Owen et al., 2011; Figure 3F). Moreover, large ulcerative regions remained in animals treated with both inhibitors (Figures S3D–S3G), and the remaining epithelium had very few cells with nuclear YAP and signs of active tissue remodeling (Figures 3G and S3H). This demonstrates that cell mechanics via the FAK pathway is important for colonic repair and the establishment of the highly proliferative repairing epithelium.
Rebuilding the Repairing Epithelium In Vitro Based on Defined Components
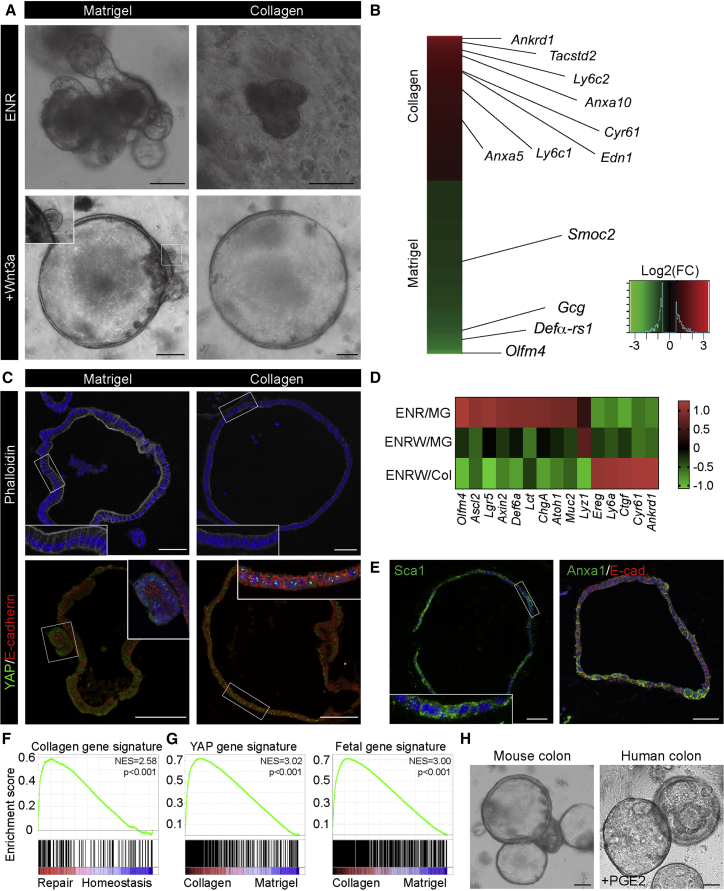
Based on the pronounced remodeling of the extracellular matrix during the repair phase, we hypothesized that these environmental changes have inductive properties. To test this, we took advantage of a 3D culture system for the adult small intestine (Sato et al., 2009). Unlike primary cells from the colon, small intestinal epithelial cells can be cultured as organoids under minimal conditions in the presence of epidermal growth factor (EGF), Noggin, and R-spondin1 (Sato et al., 2009, Sato et al., 2011). These organoids are typically cultured in Matrigel, consisting primarily of laminins and collagen IV.
Collagen type I was prominently upregulated during the repair phase (Figures 3B and S3A), but gels consisting of pure collagen type I did not support cultures of intestinal epithelial cells in medium supplemented with EGF, Noggin, and R-spondin1, whereas a combination of collagen type I and Matrigel did (Figures 4A and S4A). This suggests that collagen type I itself was not prohibiting expansion of small intestinal epithelial organoids and that additional factors should be incorporated to phenocopy the inductive microenvironment for the repairing epithelium. Recent evidence with artificially designed matrices indicates that conditions that sustain YAP activation can maintain primary epithelial cells in vitro (Gjorevski et al., 2016). Augmentation of canonical Wnt signaling has been connected to efficient tissue repair, and canonical Wnt factors cooperate with cell mechanics to sustain YAP/TAZ activation in normal and cancer cells (Azzolin et al., 2012, Panciera et al., 2016). This prompted us to investigate whether the combined stimulation of the canonical Wnt pathway together with collagen type I could install an repairing epithelium-like phenotype in vitro. Strikingly, we found that this combination supported the in vitro expansion of epithelial cells as spheroids for multiple passages in a transcriptionally distinct state (Figures 4A, 4B, and S4A). It was recently suggested that prostaglandin E2 (PGE2) acts via the activation of YAP in vivo (Kim et al., 2017); however, PGE2 cannot replace Wnt3a nor is the activity of the critical enzymes responsible for PGE2 production, Cox1 and Cox2, required for maintenance in collagen (Figure S4A). Importantly, the addition of Wnt3a to standard Matrigel cultures also promoted a spherical phenotype, but with occasional budding domains (Figures 4A and S4A).
Figure 4.
Rebuilding the Repairing Epithelium In Vitro Based on Defined Components
(A) Murine small intestinal epithelial cells cultured in the presence of EGF, Noggin, and R-spondin1 (ENR) or with the addition of Wnt3a (+Wnt3a) in either Matrigel or collagen type I. Scale bars, 100 μm.
(B) Heatmap of differentially expressed probe sets between culture conditions using MG/ENR+Wnt3a (ENRW, green) and COL/ENRW (red) (n = 6; fold change > 1.5; FDR < 0.1). Examples of differentially expressed genes are indicated.
(C) Detection of F-actin with Phalloidin (gray) counterstained with DAPI (blue) and YAP (green) counterstained with E-cadherin (red) and DAPI (blue) in Matrigel and collagen type I cultures from the small intestine. Insets show enlarged view of the indicated regions. Scale bars, 50 μm in the top panels and 100 μm in the bottom panels.
(D) Heatmap of Z score-transformed relative expression levels as determined by qPCR for selected YAP/TAZ target genes and markers of the fetal and adult states in cultures of small intestinal epithelial cells grown in either Matrigel (MG) or collagen type I (Col) in the presence of EGF/Noggin/Rspondin (ENR) with or without Wnt3a (W).
(E) Detection of Sca1 (green, left) and Anxa1 (green, right) in collagen type I cultures from the small intestine. Inset shows enlarged view of the region indicated. Images are counterstained with DAPI (blue) and image for Anxa1 is additionally counterstained with E-cadherin (red). Scale bars, 50 μm.
(F) GSEA showing enrichment of the collagen culture gene signature in the repairing epithelium.
(G) GSEA showing enrichment of YAP and fetal gene signatures in ENRW-Collagen relative to ENRW-Matrigel cultures.
(H) Mouse and human colonic organoids propagated in collagen type I. For human colonic crypts, Prostaglandin E2 (PGE2) is supplemented. Scale bar, 100 μm.
See also Figure S4.
Under collagen conditions, analogously to the in vivo colitis samples, the apical F-actin belt was lost and YAP was retained in the nuclei (Figures 4C and S4B). At the molecular level, stem cell (Lgr5, Olfm4, and Ascl2) and differentiation markers (Lyz1, Muc2, ChgA, Def6a, Lct, and Atoh1) were repressed, whereas the YAP/TAZ target genes (Ctgf, Ankrd1, Ereg, and Cyr61) as well as reprogramming marker Ly6a were induced (Figure 4D). Moreover, the epithelium started to express both Sca1 and Anxa1 (Figure 4E). Sca1 expression was retained after spheroid passaging, indicating that the Wnt/collagen condition installed a stable cellular state that recapitulated the cellular response during tissue repair. Transcriptional analysis confirmed that cells cultured under these conditions in vitro were remarkably similar to the gene profiles of the repairing epithelium in vivo (Figure 4F), including a prominent activation of YAP and fetal gene signatures (Figure 4G) and suppression of the Lgr5 stem cell signature (Figure S4C). Moreover, the conditions could be applied to both mouse and human colonic epithelial cells (Figure 4H), where overt transcriptional overlaps could be detected between adult colonic organoids cultured in collagen type I and fetal colonic organoids (Figure S4D). Importantly, inhibitors affecting signaling downstream of integrins via Rho kinase (C3 toxin; Mevastatin) and actin polymerization (Cytochalasin D) all significantly reduced growth in collagen type I (Figure S4E; Dupont et al., 2011, Sorrentino et al., 2014). Similarly, the FAK and Src inhibitors that affected tissue repair in vivo (Figure 3F) reduced growth in collagen type I (Figure S4E). This demonstrates the importance of matrix-mediated integrin signaling, and it suggests that mechano-transduction via YAP/TAZ is important for growth in collagen type I.
YAP and TAZ Promote Cellular Reprogramming
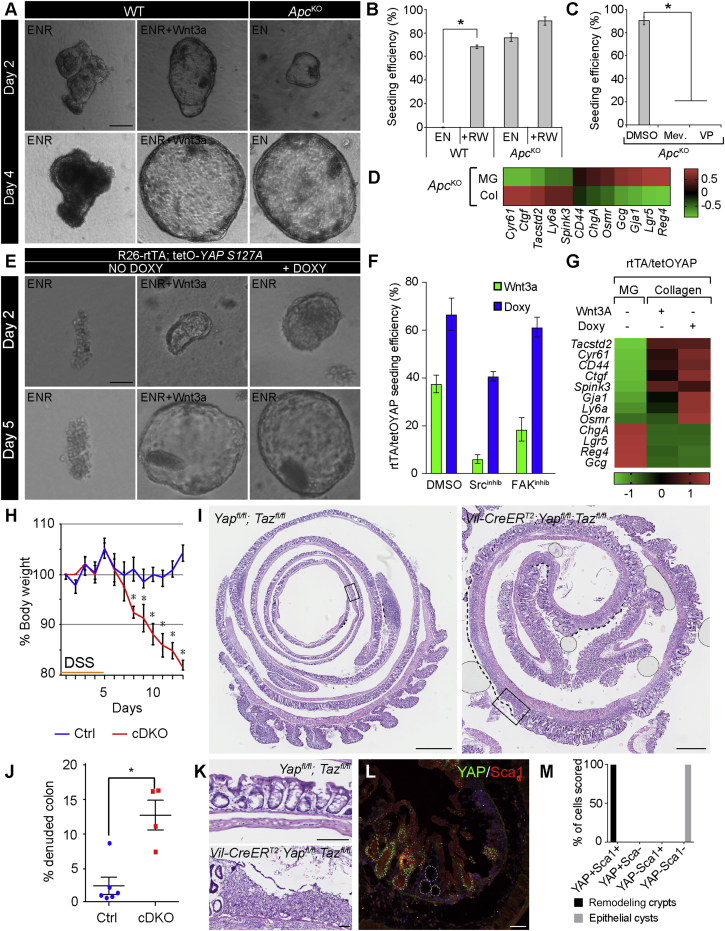
The above experiments suggest that YAP/TAZ act downstream of cell mechanics and Wnt signaling in cells of the repairing epithelium. Wnt signaling has previously been demonstrated to stimulate YAP/TAZ activation via the inhibition of Adenomatous Polyposis Coli (Apc)-mediated degradation (Azzolin et al., 2012, Azzolin et al., 2014). As expected, in collagen cultures Wnt3a could be substituted by the loss of Apc (Figures 5A and 5B). Moreover, ApcKO organoid growth could be blocked by treatment with the YAP/TAZ inhibitor verteporfin (Figure 5C). This demonstrates that Wnt via inhibition of the Apc complex supports growth in collagen type I via YAP/TAZ activation. This is similar to the reported requirement for YAP/TAZ during tumor formation upon the loss of Apc (Azzolin et al., 2014). Notably, blunting YAP/TAZ mechano-transduction by the inhibition of Rho signaling also opposed the growth of ApcKO organoids, a result consistent with the requirement of cell mechanics for YAP/TAZ activation in the repairing epithelium in vivo (Figure 5C). Importantly, even in the ApcKO model, cell mechanics were still causal for sustaining YAP/TAZ target genes (Cyr61 and Ctgf) and the associated appearance of markers associated with the fetal epithelium (Tacstd2 and Ly6a). This suggests that in our ex vivo-culturing conditions, activation of Wnt signaling and cell mechanics are additive signals for YAP/TAZ activation, but that cell mechanics remain essential for Wnt-induced YAP/TAZ activity.
Figure 5.
YAP/TAZ Transcriptional Activation Is Required for Cellular Reprogramming
(A) Normal organoids (WT) and Apc-knockout (ApcKO) spheres derived from the small intestine cultured in collagen type I with the indicated cytokine cocktail. Time point of analysis is indicated in the left bar. Scale bar, 100 μm.
(B) Quantification of seeding efficiency for WT and ApcKO organoids in Noggin and EGF (EN) with or without R-spondin and Wnt3a (RW). Bars represent mean ± SEM (n = 3; EN versus +RW for WT, p = 4.8 × 10−6 based on two-sided Student’s t test).
(C) Quantification of seeding efficiency for ApcKO organoids in collagen with mevastatin (Mev) and verteporfin (VP). Bars represent mean ± SEM (n = 3; p = 1.8 × 10−5 based on two-sided Student’s t test).
(D) Heatmap of Z score-transformed relative expression levels as determined by qPCR for selected YAP/TAZ target genes and markers of the fetal and adult states in ApcKO organoid cultures in EGF/Noggin in either Matrigel (MG) or collagen type I (Col).
(E) Spheroids derived from the small intestine of Rosa26-rtTA;tetO-YAP-S127A (rtTA/tetOYAP), cultured in collagen type I under the indicated cytokine cocktail ± Doxycycline. Time point of analysis is indicated in the left bar. Scale bar, 100 μm.
(F) Quantification of seeding efficiency for organoids from rtTA/tetOYAP cultured with Wnt3a and doxycycline in collagen type I. Additionally, the efficiency is measured in the presence of either Src or FAK inhibitors. Bars represent mean ± SEM (n = 3).
(G) Heatmap of Z score-transformed relative expression levels as determined by qPCR for selected YAP/TAZ target genes and markers of the fetal and adult states in rtTA/tetOYAP organoids cultured in matrigel (MG) or collagen type I supplemented with either Wnt3a (no Dox) or Doxycycline (Dox) to induce YAP S127A expression.
(H) Weight curve for Yapfl/fl;Tazfl/fl (Ctrl) and Vil-CreERT2;Yapfl/fl;Tazfl/fl conditional double knockout (cDKO) animals following DSS administration. Mean ± SEM is presented (Ctrl n = 6; cDKO n = 4). The observed differences are significant from day 8 and onward based on two-sided ANOVA with Bonferroni correction for multiple comparisons (day 8, p = 6 × 10−4; day 9 onward, p < 10−4).
(I) Representative H&E staining of Swiss roll of DSS-treated colon from Ctrl and cDKO animals at 13 days following the administration of DSS. Scale bar, 1 mm.
(J) Quantification of the length of the denuded area in Ctrl and cDKO colons. Data are presented as mean ± SEM (Ctrl n = 6; cDKO n = 4; p = 0.019 with a Mann-Whitney exact two-sided test).
(K) Boxed areas from (I) shown in higher magnification. Scale bar, 100 μm.
(L) Detection of YAP (green) and Sca1 (red) during the repair phase in tissue from cDKO animals at day 13 following DSS administration. Demarcated lines indicate epithelial cysts without YAP and Sca1 expression. Scale bar, 100 μm.
(M) Quantification of YAP/Sca1 status in remodeling crypts and epithelial cysts observed in the cDKO tissue (n = 3 animals; 50–200 cells scored per condition; p < 10−4 based on two-way ANOVA).
See also Figure S5.
To directly assess the requirements for YAP/TAZ in the context of collagen-induced cellular reprogramming, we took advantage of both loss- and gain-of-function mutants. Using an inducible conditional double-knockout (cDKO) model for YAP/TAZ (Azzolin et al., 2014), it was evident that YAP/TAZ are required for growth in collagen type I matrices (Figure S5A). Intriguingly, overexpression of YAP in epithelial cells derived from the R26-rtTA;tetO-YAPS127A mouse model could compensate for the presence of Wnt in the culture medium (Figures 5E and 5F). Moreover, induction of exogenous YAPS127A expression in collagen rescued the detrimental effects of FAK and Src inhibition on growth (Figure 5F), demonstrating that YAP/TAZ activation lies downstream of the integrin-mediated mechano-transduction pathway. Importantly, the establishment of the repairing epithelium-like status in vitro via culturing in collagen type I, with either Wnt3a supplemented or by overexpression of YAP, led to a similar upregulation of markers of the fetal epithelium as well as a pronounced downregulation of markers associated with adult stem cells and differentiated lineages (Figure 5G). Collectively, gain- and loss-of-function lines of evidence concur at demonstrating that YAP/TAZ-mediated activation is required and sufficient for establishing the repairing epithelium-like state in vitro.
YAP and TAZ Are Required during Tissue Regeneration
To address the role of both YAP and TAZ in colonic tissue repair, we induced conditional loss of the two factors in the adult intestinal epithelium using the Villin-CreERT2 cDKO by administration of tamoxifen 2 weeks prior to the induction of colitis. As reported previously, loss of YAP/TAZ does not affect normal tissue homeostasis (Figures S5B and S5C; Azzolin et al., 2014). Upon the administration of DSS, at concentrations with minor effects on controls animals, cDKO animals rapidly lost weight, and ulcerated lesions spread throughout the colon similarly to the animals treated with either FAK or Src inhibitors (Figures 5H–5K). Importantly, the epithelium remaining in the YAP/TAZ cDKO could be divided into what appeared to be normal regenerating epithelium positive for YAP, demonstrating that these cells had escaped recombination, and thin-walled cysts found exclusively in the cDKO animals, where YAP and Sca1 were absent (Figures 5L, 5M, and S5E). This demonstrates that YAP and TAZ are required during the process of repair following DSS-mediated injury and tightly linked with establishing the Sca1-expressing repairing epithelium.
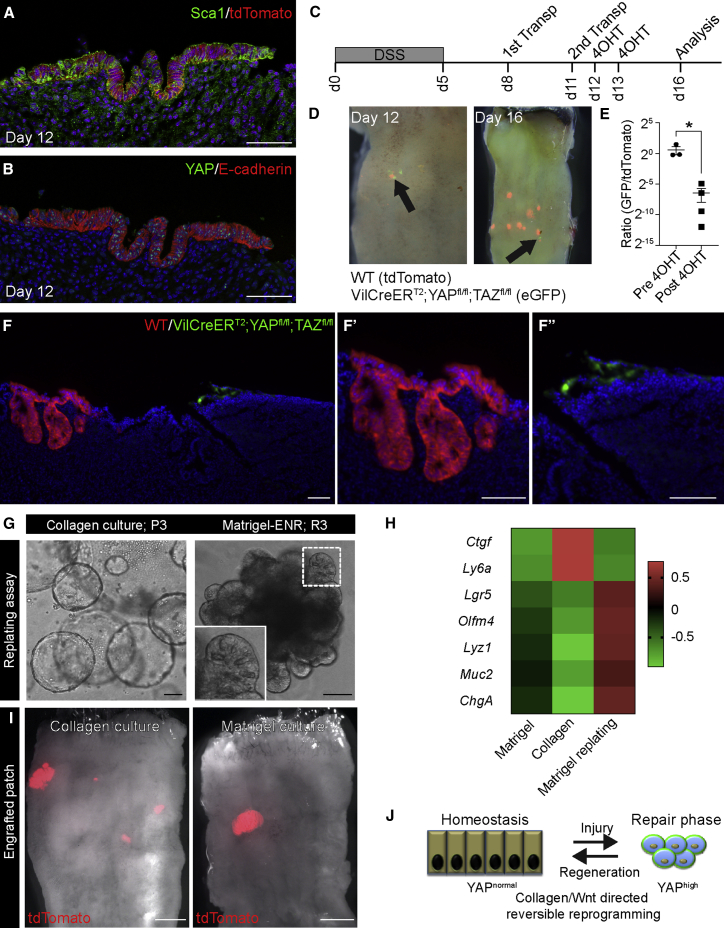
In experiments, where organoids grown in Matrigel are transplanted to the colon of animals by infusion (Yui et al., 2012), we also noticed that patches of engrafted cells displayed not only pronounced nuclear accumulation of YAP but also upregulation of Sca1 (Figures 6A and 6B). This illustrates that the cells transplanted into the epithelium, including those cultured in traditional Matrigel-based organoid medium, recapitulated the phenotypical changes associated with tissue regeneration. Next, we tested the requirement of YAP/TAZ in intestinal organoid engraftment into a damaged epithelium, a proxy of regenerative medicine application for ulcerative colitis. Crypts isolated from cDKO animals were labeled to express GFP, and control cells expressing tdTomato were expanded in vitro in Matrigel conditions. Organoid fragments from both control and cDKO organoids were infused into the colonic lumen following DSS-induced colitis, and YAP/TAZ deletion was induced by two applications of 4-hydroxy tamoxifen (4OHT; Figure 6C). Initially, the areas covered by GFP-expressing cDKO and tdTomato-expressing wild-type cells were equivalent (Figures 6D, 6E, and S6A). However, 3 days following the administration of 4OHT, it was evident that very few areas covered by cDKO cells remained, when compared to control cells (Figures 6D, 6E, and S6A). In sections, the control cells (red) formed large crypts, whereas the remaining cDKO (green) cells were found as small clusters of cells (Figure 6F, F’ and F”). This demonstrates that YAP and TAZ act as key effectors of the environmental signals that govern the maintenance of the repairing epithelium.
Figure 6.
Injury-Induced Cellular Reprogramming Is Reversible
(A and B) Serial sections of engrafted patches from Matrigel cultures analyzed for (A) tdTomato (red) and Sca1 (green) and (B) YAP (green) and E-cadherin (red) 1 day after transplantation (day 12). Scale bar, 100 μm.
(C) Diagram of the transplantation strategy using cells from the conditional YAP/TAZ cdKO cells and control animals. Indicated are the administration of DSS (day 0–5), time points for transplantation (days 8 and 11), administration of 4-hydroxy tamoxifen (days 12 and 13), as well as the final analysis (day 16).
(D) Whole-mount analysis of the colon for control (tdTomato+/red) and Villin CreER YAP/TAZ cDKO cells (eGFP+/green) before (day 12) and after (day 16) tamoxifen administration. Arrows illustrate areas of GFP+ cells.
(E) Quantification of the ratio of the area covered by GFP- versus RFP-expressing cells before and after tamoxifen treatment. Each dot represents independent animals, and data are presented as the mean ± SEM (p = 0.029 based on a Mann-Whitney exact one-sided test).
(F) Section of area with engrafted RFP- (red) and GFP- (green) expressing cells. The engraftments are shown at higher magnification in F’ and F”. Scale bar, 100 μm.
(G) Small intestinal epithelial cells cultured for 3 passages (P3) in collagen type I with ENRW grow as organoids, when replated in Matrigel (replating passage 3) in the presence of ENR. Inset shows secretory cells in an organoid. Scale bar, 100 μm.
(H) Heatmap of Z score-transformed relative expression levels as determined by qPCR for selected various markers expressed by fetal and adult epithelia grown in ENR in MG (Matrigel), then transferred into collagen and cultured in the presence of ENRW (Collagen), and lastly replated in Matrigel and cultured in ENR (Matrigel replating).
(I) Engrafted patches (red) from tdTomato-expressing cells cultured for 3 passages in either collagen type I or Matrigel. Scale bars, 2 mm.
(J) Model depicting how environmental changes control cellular identity during tissue repair via YAP/TAZ signaling.
See also Figure S6.
Injury-Induced Cellular Reprogramming Is Reversible
Regeneration is a reversible process orchestrated from residual cells leading to the formation of a de novo-formed tissue. It follows that the tissue architecture, which normally provides the framework and context for homeostatic cell behavior is temporarily lost. Thus, cell fate is dynamically regulated in vivo during tissue repair.
To address whether cells can reversibly transit between a homeostatic and repair-like state, we took advantage of the collagen type I culture system. Intriguingly, epithelial cells that were propagated in collagen type I converted efficiently into adult-like organoids of substantially greater complexity, consisting of domains enriched in prospective stem and differentiated cells, when transferred to Matrigel (Figure 6G). Transcriptionally, the levels of the reprogramming marker Sca1, the YAP/TAZ target gene Ctgf, and stem cell (Olfm4) and lineage (Lyz1, Muc2, and ChgA) markers were reversibly regulated by the matrix (Figure 6H). This demonstrates the plasticity of cell fate and the fact that cells are indeed capable of reversible transitioning between different cellular states. Crucially, when transplanted into the colonic injury model, small intestine-derived cells cultured in collagen type I, similarly to cells cultured in Matrigel, retained the capacity to contribute to tissue regeneration (Figure 6I). The efficiency and sizes of engraftment were remarkably similar between Matrigel and collagen conditions (engraftment efficiencies: collagen 60% and Matrigel 50%). Analysis of the engrafted regions showed re-expression of appropriate secretory lineage markers and retention of markers associated with their original small intestinal identity (Figures S6B and S6C). Taken together, this not only demonstrates the reversible nature of cellular reprogramming of intestinal epithelial cells (Figure 6J) but also outlines essentially clinical compliant culture conditions relying on clinical grade collagen type I. This could complement Matrigel-based culturing systems, which when it comes to medical applications is undesirable, as the matrix is purified from animals and chemically undefined.
Discussion
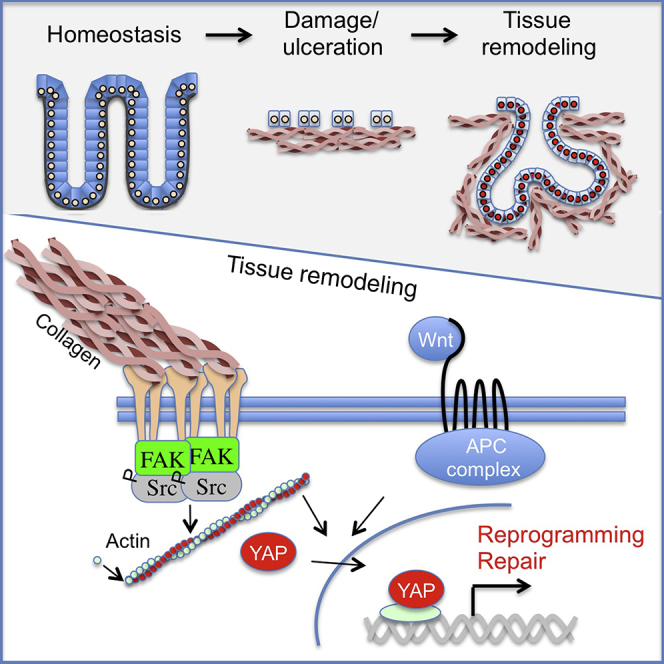
In summary, we reveal that the colonic epithelium during tissue repair is reprogrammed into a primitive state with fetal-like properties. This change of fate occurs downstream of signaling from the extracellular matrix during tissue repair. This mechano-sensory pathway entails activation of the focal adhesion pathway, cytoskeletal re-arrangement, and YAP/TAZ activation. We demonstrate that YAP and TAZ integrate mechanical, architectural, as well as soluble signals during intestinal epithelial repair to promote tissue regeneration. Reprograming into the highly proliferative repairing epithelium by YAP/TAZ is a reversible process that fuels regeneration. We propose that the ability to activate YAP/TAZ allows tissues to handle injuries by enabling cells to rapidly suppress differentiation of adult fates via transitioning into a more primitive fetal-like state (Figure 6G). This state is transient, persisting only until the return of the normal homeostatic microenvironment. Our findings point to a physiological role for YAP/TAZ in cellular reprogramming during tissue repair, and they extend previous observations using the forced expression of YAP (Panciera et al., 2016). This is potentially a broader phenomenon, since regeneration in other tissues also has been reported to involve some degree of reprogramming of resident cells (Fernandez Vallone et al., 2016, Yanger et al., 2013). Future studies will be needed to elucidate the role of YAP/TAZ in the transition into the primitive state observed during tissue repair, and YAP/TAZ’s role in the fetal epithelium.
Studies of colonic regeneration illustrate that the process is divided into discrete elements. First, a dormant wound-associated epithelium spreads over the wound in a Wnt5a-dependent manner (Miyoshi et al., 2017). Second, we now show that the epithelium enters a highly proliferative phase, where de novo crypt formation ensures that the tissue can regain prior functional features. In the absence of YAP/TAZ, a thin epithelium covers parts of the wounded regions, illustrating that defects lie in the establishment of the highly proliferative adaptive repairing epithelium. The phenotypic importance of integrin-mediated signaling has been implicated in the control of intestinal tissue repair following severe injury (Cordero et al., 2014, Owen et al., 2011, Taniguchi et al., 2015). Our data reveal that key signaling events downstream of integrins via FAK and Src control the mechano-sensors YAP/TAZ following injury. Similarly, previous studies have implicated YAP as an essential component in the response to colonic repair (Cai et al., 2010, Taniguchi et al., 2015). Our data support these observations, and they provide insight into how the pathway is activated via tissue remodeling and the consequences of YAP/TAZ activation. Moreover, we reveal that YAP and TAZ are essential during tissue repair, specifically in the establishment of the highly proliferative repairing epithelium, which governs regeneration.
One of the prominent components supporting cell behavior in vivo is the connective tissue, which provides a framework for appropriate organ functions. As with soluble growth factors, the scaffold consisting of various extracellular matrix components can be subjected to dynamic changes during tissue repair. Our results underscore the fact that the matrix is not only an inert scaffold but also an essential component that provides the context for specific signaling networks. In the case of the intestinal epithelium, changing the matrix into a collagen type I-enriched condition causes a completely reversible suppression of cellular identities. This is important to consider from a cancer perspective, where ulcerative colitis is a known risk factor for the development of colorectal cancer (Beaugerie and Itzkowitz, 2015). It is tempting to speculate that patients with ulcerative colitis, exhibiting uncontrolled inflammation and fibrosis, are at risk of developing cancer due to unchecked YAP/TAZ mechano-transduction. Interestingly, cohort studies revealed that the mutational spectrum in ulcerative colitis-associated colorectal cancer is distinct from that of sporadic cancer, including a lower prevalence of APC mutations (Robles et al., 2016, Yaeger et al., 2016). This implies that environmental stimulation of the YAP/TAZ pathway could bypass the requirements for loss of APC function.
Transcriptional networks have been the main focus as the framework that provides irreversible fate choices. The inductive potential of such networks are evident by their ability to reprogram otherwise terminally differentiated cells into either a more primitive state or a completely different cell lineage (Blanpain and Fuchs, 2014). Interestingly, recent work has illustrated that forced expression of the transcription factor YAP is capable of yet a different form of cellular reprogramming, that is, to convert differentiated cells from the mammary gland, pancreas, and neuronal lineage into the respective lineage-restricted, tissue-specific stem cell populations (Panciera et al., 2016). We speculate that the observed reprogramming into a fetal-like state upon tissue damage could represent a physiological counterpart to these in vitro observations, where tissue and germ layer identity is still maintained.
The in vitro model system for studying the intestinal epithelium using defined components in its repair phase or the transition between normal and repair is likely to have widespread impact on the field. First, it is based on off-the-shelf-components, which enable the in vitro expansion of clinical grade primary epithelial cells that would be suitable for clinical applications in patients with inflammatory bowel disease. Second, the fact that the culture conditions are fully defined enables systematic studies aimed at addressing how signaling networks operate in primary epithelial cells. Lastly, we highlight that the described in vitro model for tissue repair will improve our understanding of how tissue repair proceeds as well as the potential establishment of in vitro disease models (Schweiger and Jensen, 2016). Future studies should elucidate whether patients, who experience bowel inflammation/ulcerations, will benefit from therapies modulating either the YAP/TAZ levels directly or indirectly, including changing the biophysical properties of the wound bed.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti Sca1 (177228) | R&D systems | Cat #: BAM1226, RRID:AB_2070040 |
| Rabbit anti Anxa1 | Invitrogen | Cat #: PA5-27315, RRID:AB_2544791 |
| Mouse anti E-cadherin (36/E-cadherin) | BD Transduction Laboratory | Cat #: 610181, RRID:AB_397580 |
| Rat anti E-cadherin (ECCD-2) | Thermo Fisher Scientific | Cat #: 13-1900, RRID: AB_86571 |
| Rat anti b1 integrin (MB1.2) | Millipore | Cat #: MAB1997, RRID: AB_2128202 |
| Rabbit anti FAK | Millipore | Cat #: 06-543, RRID: AB_310162 |
| Rabbit anti pSrc (Tyr416) | Cell Signaling Technology | Cat #: 2101, RRID: AB_331697 |
| Rabbit anti YAP | Cell Signaling Technology | Cat #: 4912, RRID:AB_2218911 |
| Rabbit anti Lysozyme (EC.3.2.1.17) | Dako | Cat #: A0099, RRID:AB_2341230 |
| Rabbit anti Ki67 | Abcam | Cat #: ab15580, RRID: AB_443209 |
| Rabbit anti MUC2 (H-300) | Santa Cruz | Cat #: sc-15334, RRID: AB_2146667 |
| Rabbit anti Carbonic Anhydrase II (H-70) | Santa Cruz | Cat #: sc-25596, RRID: AB_2065996 |
| Goat anti Lrig1 | R&D systems | Cat #: AF3688, RRID: AB_2138836 |
| Rabbit anti EpCAM | B.N.G. Giepmans | Schnell et al., 2013 |
| Rat anti Reg3b antibody | R&D systems | Cat #: MAB5110, RRID: AB_2178585 |
| Goat and Trop2 antibody | R&D systems | Cat #: AF1122, RRID: AB_2205662 |
| Alexa Flour 488 Donkey anti Rat IgG (H+L) | Thermo Fisher scientific | Cat #: A-21208, RRID: AB_141709 |
| Alexa Flour 555 Donkey anti Rat IgG (H+L) | Abcam | Ab-150154 |
| Alexa Flour 488 Donkey anti Rabbit IgG (H+L) | Thermo Fisher scientific | Cat #: A-21206, RRID: AB_141708 |
| Alexa Flour 555 Goat anti Mouse IgG2a (H+L) | Thermo Fisher scientific | Cat #: A-21137, RRID: AB_2535776 |
| Alexa Flour 488 Donkey anti Goat IgG (H+L) | Thermo Fisher scientific | Cat #: A-11055, RRID: AB_142672 |
| Anti-Mouse EpCAM APC (G8.8) | eBioscience | Cat #: 17-5791-80, RRID: AB_1659714 |
| Anti-Mouse CD31 PE-Cyanine7 (390) | eBioscience | Cat #: 25-0311-81, RRID: AB_469615 |
| PECy7 Rat anti-Mouse CD45 (30-F11) | BD PharMingen | Cat #: 561868, RRID: AB_10893599 |
| PE/Cy5 anti-mouse Ly6A/E (Sca1) (D7) | Biolegend | Cat #: 108109, RRID: AB_313346 |
| Biological Samples | ||
| Biopsies from human colon | Herlev hospital | O.H. Nielsen |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dextran Sulfate Sodium (DSS) | MP Biomedicals | 0216011090 |
| Tamoxifen | Sigma | T5648 |
| Growth factor reduced Matrigel | Corning | 356231 |
| Cellmatrix Type I-A | Nitta Gelatin | 631-00651 |
| human EGF | Peprotech | AF-100-15 |
| murine Noggin | Peprotech | 250-38 |
| human Noggin | Peprotech | 120-10C |
| mouse R-spondin1 | R&D systems | 3474-RS |
| human R-spondin1 | Sino | 11083-H08H |
| murine Wnt3a | Cell guidance systems | GFM77 |
| Bovine Serum Albumin | Sigma | A9576 |
| Nicotinamide | Sigma | 72340 |
| Prostaglandin E2 | Sigma | P5640 |
| Y-27632 dihydrochloride | Sigma | Y0503 |
| Jagged-1 | Anaspec | AS-61298 |
| CHIR99021 | Stemgent | 04-0004 |
| Doxycycline | Sigma | D9891 |
| C3 toxin (Rho inhibitor I) | Cytoskeleton | CT04-A |
| Mevastatin | Sigma | M2537 |
| Cytochalasin D | Sigma | C8273 |
| FAK inhibitor (in vitro) | Tocris | PF573228 |
| Src inhibitor (in vitro) | Sigma | Dasatinib |
| FAK inhibitor (in vivo) | Selleckchem | PND-1186 |
| Src inhibitor (in vivo) | Selleckchem | Saracatinib |
| Verteporfin | Sigma | SML0534 |
| Jagged-1 | Anaspec | AS-61298 |
| Indomethacin | Sigma | I7378 |
| Puromycin | Sigma | P8833 |
| 4-hydroxytamoxifen | Sigma | H6278 |
| Critical Commercial Assays | ||
| Lipofectamin 2000 | Thermo Fisher Scientific | 11668027 |
| Hematoxylin QS | Vector | H-3404 |
| Eosin Y Solution Aqueous | Sigma | HT110232 |
| Vector Red Alkaline Phosphatase (Red AP) Substrate Kit | Vector | SK-5100 |
| Alexa Fluor 647 Phalloidin | Molecular Probes | A22287 |
| PureLink RNA Micro-scale kit | Invitrogen | 12183016 |
| Superscript III reverse transcriptase | Invitrogen | 18080093 |
| Deposited Data | ||
| Raw and analyzed data from homeostatic and repairing epithelium | This paper | E-MTAB-5249 |
| Raw and analyzed data from adult and fetal culture | This paper | E-MTAB-5246 |
| Raw and analyzed data from Collagen and Matrigel culture | This paper | E-MTAB-5247 |
| Experimental Models: Cell Lines | ||
| Wnt3a-expressing HEK293 cells | H. Clevers | n/a |
| Experimental Models: Organisms/Strains | ||
| Rosa26mT/mG | A.G. Botton | Muzumdar et al., 2007 |
| tetO-YAP S127A | F. Camargo | Camargo et al., 2007 |
| Yap1fl/fl | D.J. Pan | Zhang et al., 2010 |
| Villin-CreERT2 | S. Robine | el Marjou et al., 2004 |
| Tazfl/fl | S. Piccolo | Azzolin et al., 2014 |
| RAG2−/− | Taconic | C57BL/6NTac |
| Rosa26-rtTAM2 | Jackson Laboratory | Stock No: 006965 |
| Oligonucleotides | ||
| sgRNA targeting exon 15 of Apc (GCACTCAAAACGCTTTTGA) | B.K. Koo | Schwank et al., 2013 |
| Recombinant DNA | ||
| Cas9 | Schwank et al., 2013 | Addgene #41815 |
| Software and Algorithms | ||
| NDP.view2 | Hamamatsu | U12388-01 |
| ImageJ | NIH image | version 2.0.0-rc-41/1.50d |
| Adobe Photoshop CS6 | Adobe | version 13.0.6 x64 |
| Gene set enrichment analysis | http://software.broadinstitute.org/gsea/index.jsp | n/a |
| Other | ||
| Costar 48 Well Clear TC-Treated Multiple Well Plates | Corning | 3548 |
| Histoacryl Topical Skin Adhesive | B.Braun/TissueSeal | TS1050071FP |
| OCT compound | CellPath | KMA-0100-00A |
| Disposable microtome blades, S35 | Feather | 207500000 |
Contact for Reagents and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kim Jensen (kim.jensen@bric.ku.dk)
Experimental Model and Subject Details
Mice
Unless otherwise specified, C57BL/6J mice (purchased from Taconic, Denmark) were used for the experiments. Transgenic murine lines used in the experiments have been previously described: Rosa26mT/mG (Muzumdar et al., 2007), tetO-YAP S127A (kindly provided by F. Camargo (Camargo et al., 2007)), Yap1fl/fl (kindly provided by DJ Pan; (Zhang et al., 2010), Villin-CreERT2 (kindly provided by S. Robine; el Marjou et al., 2004), Tazfl/f (Azzolin et al., 2014), RAG2−/− (Taconic, US) and Rosa26-rtTAM2 (stock #006965, Jackson Laboratory).
Cohorts of C57BL/6J animals (3-6 months old) used for experimental colitis were female, immune compromised Rag2−/− animals (3-6 month old) used for transplantation experiments were a mix of males and females, and a mix of male and female Yapfl/fl;Tazfl/fl (Villin-CreERT2 negative; controls) and Villin-CreERT2;Yapfl/fl;Tazfl/fl siblings (2 months old) on a C57Bl6 background were used for assessing the role of YAP/TAZ in tissue repair. For in vitro cultures cells were obtained from animals ranging from embryonic day 16.5 to late adulthood (12 months).
None of the animals used in these studies had been subjected to prior procedures and were drug and test naive. All animals were housed in SPF (specific pathogen free) animal facilities, in either open or individually ventilated cages always with companion mice, and cages were placed under a 12hr light-dark cycle. Food and water were provided ad libitum. National animal ethics committees in Denmark and Italy (OPBA of Padua and Italian Ministry of Health) reviewed and approved all animal experiments.
Experimental Colitis
Colitis was induced in C57/BL6J animals (3-6 months old) by administration of 2.5% DSS (MP Biomedicals; MW; 36.000-50.000) in drinking water for 5 days followed by normal water. RAG2−/− animals (3-6 months old) were subjected to 3.3% DSS. In Yapfl/fl;Tazfl/fl (Villin-CreERT2 negative; controls) and Villin-CreERT2;Yapfl/fl;Tazfl/fl siblings (2 months old), deletion of YAP/TAZ was induced by intraperitoneal injections of Tamoxifen (3 mg/body; dissolved in corn oil; Sigma) 2 weeks before induction of experimental colitis. In this case DSS (2.5%) was administered for 5 days. FAK inhibitor (PND-1186, Selleckchem, 150mg/kg) and Src inhibitor (Saracatinib, AZD0530, Selleckchem, 50mg/kg) were administered via oral gavage from day 8 to day 12 after DSS initiation and analyzed at day 12. Control animals were administered with solvent (0.5% w/v HPMC / 0.1% w/v Polysorbate 80 in water).
Transplantation
RAG2−/− mice were infused twice with cultured intestinal epithelial cells on day 8 and day 11 after DSS initiation (Yui et al., 2012). Donor cells from Rosa26mT/mG mice were cultured as described below in either Matrigel (Corning) or collagen type I gel (Cellmatrix Type I-A; Nitta Gelatin). At passage 3, epithelial cells were released from matrix, and dissociated into sheets of epithelial cells. Cell fragments from approximately 600 organoids were resuspended in 300μL of 5% Matrigel in PBS. A flexible catheter was inserted into the colon of mice under general anesthesia (Isoflurane; Piramal Healthcare), and the cell suspension was subsequently infused into the colonic lumen. The anus was sealed with surgical histoacryl glue (B. Brawn), which was removed after 3 hours. After transplantation, animals were carefully monitored during recovery. From 1 day to 2 weeks after the second transplantation, recipient mice were analyzed (see below). In competitive assay based on the deletion of YAP/TAZ, GFP positive organoids derived from Villin-CreERT2;Yapfl/fl;Tazfl/fl and those from Rosa26mT/mG mice were transplanted (500 organoids of each) after expansion in Matrigel. After transplantation, mice received intraperitoneal injections of Tamoxifen (same as above) at day 12 and 13. Mice were analyzed either at day 12, or at day 16.
Flow cytometry and colony forming assay
Colonic tissue was isolated in the repair phase at 12-16 days after DSS initiation, and the injured regions were isolated using a stereomicroscope. Identical parts of colon from untreated animals were used as controls. Cells from the dissected tissue were isolated as reported (Yui et al., 2012). Briefly, each colonic fragment was finely minced using razorblade, and subjected to enzymatic digestion at 37°C for 20 min with Collagenase type IX (Sigma, 0.45mg/mL) and Dispase I (Roche; 0.2U/mL) in 12.5mL DMEM in 50mL tube. During incubation, the tube was shaken by hand every 2 min. After incubation, the solution was subjected to vigorous mechanical dissociation. The total volume was adjusted to 40 mL with DMEM supplemented with 10% adult bovine serum (ABS) and cells were pelleted by centrifugation at 500 g for 3 min. The resulting cell pellet was resuspended in 30% Percoll in Hank’s balanced salt solution, transferred in a 15 mL-tube and spun at 800 g for 3 min. The resulting cell pellet was resuspended in 10 mL DMEM and filtered through a 70μm-cell strainer (Falcon). To obtain a single cell suspension, cells were pelleted at 500 g for 3 min, resuspended in 5 mL TrypLE Express Enzyme (GIBCO) and incubated at 37°C for 10 min. Cells were then pelleted, resuspended in DMEM and stained for flow cytometry using EpCAM-APC, CD31-PECy7, CD45-PECy7, and Sca1-PECy5 for 30 min on ice. After washing, 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI; Sigma; 1μM) was added. Cells were subsequently sorted on a FACSAria I (BD Bioscience) cell sorter. In in vitro organoids assay, each fraction (104 cells) were harvested in 25 μL dome shaped Matrigel supplemented with Jagged-1 (Anaspec, 1 μM) and cultured in advanced DMEM/F12 (Thermo Fisher Scientific) supplemented with GlutaMAX (Thermo Fisher Scientific, 1%) and Penicillin/Streptomycin (Thermo Fisher Scientific, 0.5%) in the presence of human EGF (Peprotech; 50ng/mL), murine Noggin (Peprotech; 100ng/mL), mouse R-spondin1 (R&D; 500ng/mL), murine Wnt3a (Cell guidance systems; 100ng/ml), Chir99021 (Stemgent; 3 μM), Prostaglandin E2 (PGE2; Sigma; 2.5μM), Nicotinamide (Sigma; 10 mM), N-2 supplement (Thermo Fisher Scientific; 1%) and B-27 supplement (Thermo Fisher Scientific; 2%). Y-27632 dihydrochloride (Sigma; 10μM) was used for first 3 days of culture, and medium was changed every 3 days. Organoids number formed in each condition was manually identified at day 10.
Primary cultures of mouse small intestinal and colonic epithelial cells in Matrigel or collagen Type I
Small intestinal crypts were harvested from the proximal part of the small intestine using EDTA (2mM, 30 min) in PBS. Colonic crypts were harvested using 10 mM EDTA and incubation time extended to 60 minutes. Harvested crypts were embedded in Matrigel or collagen type I gel. Advanced DMEM/F12 with GlutaMAX and Penicillin/Streptomycin was used as basal medium and supplemented with human EGF, murine Noggin or human Noggin (Peprotech; 100ng/mL), mouse R-spondin1 or human R-spondin1 (Sino; 500ng/mL). When indicated, murine Wnt3a was supplemented to ENR medium in combination with 1% Bovine Serum Albumin (BSA; Sigma) (ENRW). For efficient expansion of colonic crypts, Nicotinamide (Sigma; 10 mM) was supplemented. When indicated, Prostaglandin E2 or Indomethacin (Sigma, 10 μM) were supplemented in ENR or ENRW medium, and collagen Type I gel was mixed with the same volume of Matrigel in ENR or ENRW medium. The growth under different conditions was quantified as seeding efficiency at day 6.
Apc KO organoids were generated using Cas9 (Addgene #41815) and sgRNA (GCACTCAAAACGCTTTTGA) targeting exon 15 of Apc (plasmid backbone: Addgene #41819; plasmids were a kind gift from Dr. Bon-Kyoung Koo University of Cambridge, UK). Briefly, WT organoids (derived in Matrigel) were dissociated into single cells using mechanical disruption and trypsin (Life Technologies) incubation for 10-15 min. Single cells were transfected with 0.7μg of Cas9-expressing plasmid and 0.7μg of the plasmid expressing the gRNA targeting Apc, using Lipofectamin®2000 (ThermoFisher). After addition of the transfection medium, the plate was centrifuged at 600 g at 32°C for 1 hour, followed by another 4-hour incubation at 37°C. Cells were then collected, centrifuged and re-suspended in Matrigel containing Jagged-1 and cultured in ENR medium supplemented with CHIR99021, Y-27632 and Nicotinamide. Two passages after transfection the medium was changed to EN (no R-spondin) for selection. Selected clones were verified by Sanger sequencing. Parental wild-type and Apc KO organoids were both replated in collagen type I in EN or ENRW and growth under each condition was quantified as seeding efficiency at day 5. Apc KO organoids were treated with Mevastatin (Sigma; 3 μM) and Verteporfin (Sigma; 3 μM) in collagen Type I in EN medium at day 2 after seeding, and the effects were quantified 3 days later. Control cells were treated with Dimethyl Sulfoxide (DMSO; Sigma).
Small intestinal crypts were harvested from Rosa26rtTAM2/+; tetO-YAP S127A (rtTA/tetOYAP) mice (between 2 and 3 months of age) and were cultured in collagen type I using ENR or ENRW conditions. To induce expression of the transgenic allele, doxycycline (doxy; 2μg/ml, Sigma) was added to the culture medium (ENR) on the first day of culture. Additionally, Src inhibitor (Dasatinib; Sigma; 10 μM) and FAK inhibitor (PF573228; Tocris; 10 μM) were supplemented in the medium (ENRW or ENR plus doxy) at day 2. The seeding efficiency was measured after additional 3 days.
Analysis of chemical compounds affecting downstream components of integrin signaling was performed on crypts derived from small intestine seeded in collagen type I gel/ENRW conditions. At day 2, C3 toxin (Cytoskelton), Mevastatin (Sigma), Cytochalasin D (Sigma), FAK inhibitor and Src inhibitor were supplied to the culture medium at the concentrations indicated. Sphere numbers were counted in each condition after 3 days of incubation. Control cells were cultured in the presence of DMSO.
Small intestinal crypts derived from Villin-CreERT2;Yapfl/fl;Tazfl/flmice were cultured in Matrigel/ENR condition. Fragments of organoids were replated in collagen Type I under ENRW condition. At day 3, 4-hydroxytamoxifen (4OHT; Sigma; dissolved in ethanol; 1 μM) was supplemented to the culture medium and sphere numbers counted after 3 days to determine the effect of YAP/TAZ on organoid growth. Control organoids were derived from Rosa26mT/mG mice. Organoids cultured in Matrigel/ENR condition were labeled with GFP by lentiviral transduction using an empty pTRIPZ vector for transplantation assay. Organoids were dissociated into cell fragments using TryPLE (37°C, 5 minutes) and cell fragments were further dissociated into smaller fragments by pipetting. Cell fragments was subsequently incubated for 4 hours at 37°C with lentiviral particles in full medium containing EGF, Noggin, R-spondin1, Chir99021, Nicotinamide, N-2, B-27 and Y-27632. After 4 hours, fragments were harvested into Matrigel dome, and cultured in the same condition for 1 week. GFP positive clones were selected by Puromycin (Sigma; 2 μg/ml).
For replating assay, small intestinal epithelial cells under collagen/ENRW plus Nicotinamide condition were sequentially passaged into Matrigel. For passaging, spheres were released from collagen type I gel by collagenase solution (0.63mg/mL collagenase Type XI in PBS) and were mechanically dissociated into fragments by pipetting 10 times using P1000 pipettes. Y-27632 was supplemented for the first 2 days after passaging. At Passage 3, cells were replated in Matrigel and cultured in ENR medium. Control samples were cultured in Matrigel/ENR medium.
In all experiments using organoids, cells were harvested from WT C57/BL6 animals (3-6 months old) unless otherwise stated. In primary phase, approximately 300 crypts were seeded in a dome-shaped 25μl extracellular matrix droplet and 250μl culture medium was added to each well in a 48-well plate (flat bottom; Corning). Medium was subsequently changed every 2-3 days. The day of harvest is defined as day 0. For the quantification of growth of organoids, seeding efficiency was manually identified as a percentage of successful sphere formation in all seeded fragments.
Primary culture of human colonic epithelial cells in collagen Type I
Human colonic crypts were harvested from clinical biopsies as described for mouse colon and cultured with ENRW plus Nicotinamide/PGE2. The human cultures were established from intestinal pinch biopsies obtained from individuals scheduled for colonoscopy at the Department of Gastroenterology, Herlev Hospital, where all examinations subsequently fell out normal. The biopsies are obtained under strict anonymity. Accordingly, gender identity and age of the subjects cannot be recorded. The Scientific Ethics Committee of the Copenhagen Capital Region approved the use of this material for research purposes, and informed consent was obtained from all patients.
Primary culture of fetal intestinal and colonic epithelial cells in Matrigel
Cultures were established from the proximal part of small intestine from E16.5 mouse fetuses. The intestine was opened longitudinally and processed by mincing into small pieces. Epithelial compartment was dissociated by incubation in 2mM EDTA. Colonic tissue was digested in collagenase solution (125μg/mL in PBS) for 45 min at 37°C with vigorous pipetting by P1000 pipettes every 15 min. Isolated epithelial units were embedded in Matrigel and cultured in ENR. Wnt3a conditioned medium (50% in volume) was additionally supplemented for colon.
Methods Details
Imaging and histology
Tissues and cells were fixed with 4% Paraformaldehyde (PFA) from 3 hours to overnight at 4°C. Samples were embedded in OCT compound (CellPath) and frozen. Eight μm-sections were prepared by cryotome (Leica CM3050S). Hematoxylin and Eosin (H&E) staining (Hematoxylin QS; Vector, Eosin Y Solution Aqueous; Sigma) and Alkaline Phosphatase staining (Vector Red Alkaline Phosphatase Substrate kit; Vector) were performed following manufactures’ protocols. Images were acquired using NDP Zoomer Digital Pathology slide scanner (Hamamatsu). Phalloidin (Alexa Flour 647 Phalloidin; Molecular Probes) staining was performed according to manufacture’s protocol. Immunofluorescence analysis was done using antibodies listed in the Key Resources Table. DAPI was used to counterstain nuclei. Fluorescence images were acquired using laser scanning confocal microscopes (Leica TSC SP8). Some images of engrafted patch were acquired using a Zeiss Imager.M2 microscope equipped with ORCA-R2 Digital CCD camera C10600 (Hamamatsu). Phase contrast images of organoids were acquired using a Leica DM IL LED Fluo microscope equipped with Leica MC120 HD camera, or a Leica DM IRB inverted microscope equipped with a CCD camera (Leica DFC 450C). Macroscopic images of colon were acquired using a Leica M165FC microscope equipped with Leica DFC310 FX camera. All images were subsequently analyzed in NDP.view2 software, ImageJ, version 2.0.0-rc-41/1.50d and Adobe Photoshop CS6, version 13.0.6 x64.
Thickness of mucosa
The distance between crypt tip and muscularis mucosa was measured in H&E staining using NDP.view2 software. 4 animals each from control and colitis at 2 weeks after DSS administration were analyzed. In each animal, the distance was measured at 12 different points in the distal region of the colon.
Quantification of denuded regions in colon
Harvested colon from Yapfl/fl;Tazfl/fl and Villin-CreERT2;Yapfl/fl;Tazfl/fl animals, or animals treated with FAK and Src inhibitors was embedded as Swiss-rolls and processed to frozen sections. Denuded region was identified in H&E staining. The length of denuded regions and the length of entire colon were measured using NDP.view2 software. The quantification of denuded regions was used to reflect persisting tissue damage.
Quantification of immunofluorescence
All images within each series were acquired under the same setting of scanning confocal microscope, and were converted into gray scale adapting the same pre-set. Intensity of β1 integrin/FAK/pSrc was measured in ImageJ using line function to measure mean gray value at the basolateral side of epithelium to perform 50 to 100 independent measurements. Max intensity of Phalloidin staining at apical surface was measured using line function to determine max gray value at the apical side of the epithelium in 100 to 200 independent measurements. Samples from 3 animals in homeostatis and 3 in the repair phase were analyzed in all quantification. Max intensity of Phalloidin staining at apical surface of in vitro sample was measured using plot profile function in ImageJ by drawing a line penetrating the entire longitudinal aspect of cells. Samples from 6 Matrigel and 8 collagen specimens were analyzed. In each independent specimen 10 to 40 independent measurements were performed depending on the size of samples. In all quantification, mean background intensity was determined in each image, and this value was subtracted from the measured value, and this corrected value was adapted as output.
Sequential sectioning of a grafted epithelial patch
After dissection of the colon from mouse, it was washed with PBS, opened longitudinally and imaged by stereomicroscopy. Engrafted patches were dissected manually from the colon and fixed with 4% PFA for 3 hours. Tissue fragments were embedded in OCT. This process preserve endogenous expression of tdTomato/GFP in sequential frozen sections obtained every 8μm.
Fibrillar collagen analysis
Fifteen μm frozen sections were prepared from mock-treated and DSS-treated colons. After washing with PBS for 5 min, sections were placed in a drop of water in a glass-bottom Mattek dishes. Sections were imaged using a HCX PL APO lambda blue 20x, 0.70NA IMM UV objective fitted on an inverted Leica SP5-X MP multiphoton Leica microscope connected to a Ti-Sapphire laser. Fibrillar collagen was visualized by acquiring the second harmonic generation (SHG) signal between 410-450 nm after 2-photon excitation at 860 nm. The SHG signal was collected in the backward direction using a hybrid detector (HyD SP). A regular bright field image of the tissue was taken simultaneously with the SHG image by means of Supercontinuum White Light Laser excitation at 603 nm. All images were captured with a resolution of 2048 × 2048 pixels, at 100 Hz.
RNA extraction and qRT-PCR
Total RNA was isolated using PureLink RNA Micro-scale kit (Invitrogen). cDNA was synthetized using Superscript III reverse transcriptase (Invitrogen) and random primers. cDNA was synthetized from 300ng total RNA for all in vitro cultured cells or 2μl of total RNA from sorted materials. qPCR was performed with QuantStudio 6 Flex Real-Time PCR System (Life technologies) in SYBR Green analysis with optimized primer pairs. CT value was normalized to Tubb5, using the ΔCt method. Heatmaps from qPCR data were generated from z-score transformed relative expression data.
Microarray
Illumina beadarrays
Expression analysis using Illumina bead arrays was performed as described previously (Wong et al., 2012). Briefly, RNA was quality controlled for concentration, purity and integrity using spectroStar omega (BMG labtech) and Bioanalyzer (Agilent). The amplification was performed using the TotalPrep 96-RNA Amplification kit (Ambion). Total RNA (∼300ng) was reverse transcribed into cDNA and amplified by in vitro transcription to generate biotin-labeled cRNA. cRNA (1500ng) was hybridized to whole genome bead arrays (MouseWG-6 v2.0 Expression BeadChip) according to the direct hybridization assay from Illumina and scanned using an Illumina BeadArray scanner.
Affymetrix arrays
RNA quality was assessed using a Bioanalyzer for integrity and concentration. For cultured material RNA (200ng) were processed and labeled for array hybridization using the Ambion WT Expression kit (Life Technologies). For sorted material, biotinylated cDNA was synthesized from 1.8ng of total RNA using the Nugen Ovation Pico WTA System V2 kit. Five μg of ssDNA from sorted material was subsequently fragmented and biotinylated (NuGen Encore Biotin Module). Labeled, fragmented cDNA (Affymetrix GeneChip® WT Terminal Labeling and Controls Kit) was hybridized to Mouse Gene 2.0 arrays for 16 hours at 45°C (at 60 rpm) (Affymetrix GeneChip® Hybridization, Wash, and Stain Kit). Arrays were washed and stained using the Affymetrix Fluidics Station 450, and scanned using the Hewlett-Packard GeneArray Scanner 3000 7G.
Preprocessing of microarray data and calling of differentially expressed genes
Two different array platforms, Illumina Mouse WG6 v2.0 Expression BeadChip and Affymetrix Mouse Gene 2.0 ST Array platform, were used in this work and they were processed separately.
∗Illumina beadarrays
The Illumina BeadChip arrays were analyzed in R using the beadarray package (Dunning et al., 2007). Data were extracted directly from the tiff images using the readIllumina function with the option “useImages=TRUE.” Gene annotations were obtained from the illuminaMousev2.db package. Quality control was performed as suggested in the beadarray documentation, including the investigation of spatial effects. Data were normalized with the normalizeIllumina function using the neqc method, also from the beadarray package. Differential expression was called in R using the limma package and following the procedures suggested in the documentation (Ritchie et al., 2015).
∗Affymetrix arrays
Affymetrix microarray data were analyzed in R using the oligo and affycoretools packages (Carvalho and Irizarry, 2010, MacDonald, 2008). Annotation was supplied by Affymetrix, which included both gene- and exon-level annotation. Only the gene-level information was used in this work. Normalization and probe summarization was performed using the rma function in the oligo package with default settings. Differential expression was called in R using the limma package (Ritchie et al., 2015).
Gene set enrichment analysis (GSEA) and GO-analysis
Gene set enrichment analysis was performed using the Broad institute GSEA tool software.broadinstitute.org/gsea/index.jsp) with standard settings. Based on samples numbers, gene_set permutation was used. Gene sets include a YAP signature gene set based on genes that are upregulated > 2 fold between wt and YAP overexpressing intestinal epithelial cells (Gregorieff et al., 2015), intestinal stem cell signature based on genes that are upregulated > 2 fold in Lgr5 expressing cells (Muñoz et al., 2012), a collagen type I signature gene set, which represent genes upregulated > 1.5 fold in collagen type when compared Matrigel cultures (n = 6, FDR < 0.1), a fetal signature gene set representing genes that are upregulated > 2 fold in cultures of fetal versus adult intestinal epithelial cells (n = 3, FDR < 0.05) and a repair signature representing genes that are upregulated > 2 fold in the repairing epithelium (EpCAMposSca1highCD45negCD31neg) relative to the normal homeostatic epithelium (EpCAMposSca1negCD45negCD31neg) (n = 3, FDR < 0.05). GO-term enrichment analysis for unbiased gene ontology analysis was performed using PANTHER Version 12.0 (http://pantherdb.org/).
Quantification and Statistical Analysis
The number of biological and technical replicates and the number of animals are indicated in figure legends and text. All tested animals were included. Sample size was not predetermined. Experiments were performed without methods of randomization or blinding. For all experiments with error bars, the standard error of the mean (SEM) was calculated to indicate the variation within each experiment.
Specifically for Figure 1D: 4 animals per condition; Figures 1F–1H, 1K, 3A, S1E, S3A, and S3B: 3 animals per conditions; Figures 2B, 4D, 5B–5D, 5F, 5G, 6H, S4A, S4E, and S5A: 3 cultures obtained from independent animals per condition; Figure 3F: 5 animals per condition; Figure 4B: 6 cultures obtained from independent animals per condition; Figures 5H and 5J: 6 control animals and 4 cDKO animals; Figure 5M: 3 animals analyzed for the two phenotypes; Figure 6E: 3 and 4 animals analyzed at Day 12 and 16, respectively; Figure S3G: 5, 5 and 3 for Control, FAK inhibitor and Src inhibitor experiments, respectively; Figure S4B: 6 and 8 individual structures analyzed from Matrigel and collagen cultures, respectively. Significance was assessed using the indicated tests calculated in Prism (GraphPad) or in R.
Data and Software Availability
Microarray data have been deposited at https://www.ebi.ac.uk/arrayexpress/ under the accession numbers as follows. ArrayExpress: E-MTAB5246, ArrayExpress: E-MTAB5247, and ArrayExpress: E-MTAB-5249.
Author Contributions
Conceptualization, S.Y., L.A., S.P., and K.B.J.; Methodology, S.Y., L.A., T.N., M.W., S.P., and K.B.J.; Investigation, S.Y., L.A., M.M., S.L.H., H.L.L., M.R.P.A., J.G., M.T.P., R.P.F., C.D.M., and P.J.S.; Formal Analysis, M.T.P., C.F.R., and J.V.J.; Resources, Y.L. and O.H.N.; Writing – Original Draft, S.Y., L.A., S.P., and K.B.J.; Writing – Review & Editing, all authors; Supervision, S.P. and K.B.J.; Funding Acquisition, S.Y., J.G., O.H.N., C.D.M., S.P., and K.B.J.
Acknowledgments
We thank members of the Jensen and Piccolo lab for comments and suggestions; A. Grapin-Botton (Rosa26mT/mG), D.J. Pan (Yapfl/fl), F. Camargo (tetO-YAP S127A), and S. Robine (Villin-CreERT2) for gifts of mice; B.-K. Koo for plasmids; H. Clevers for Wnt3a-expressing HEK293 cells; B. Giepmans for the EpCAM antibody; the EMBL genomics core facility; A. Fossum in the fluorescence-activated cell sorting (FACS) and Y. Antoku in imaging core facilities at BRIC; and the Center for Advanced Bioimaging at University of Copenhagen for experimental support. Rosa26-rtTAM2 mice were purchased from The Jackson Laboratory, where they were deposited by R. Jaenisch. This work was supported by Worldwide Cancer Research (13-1216 to K.B.J.), Lundbeck Foundation (R105-A9755 to K.B.J.), the Danish Cancer Society (R56-A2907 and R124-A7724 to K.B.J.), the Carlsberg Foundation (CF14-0122 to K.B.J.), EMBO Young Investigator programme (to K.B.J.), AIRC Special Program Molecular Clinical Oncology “5 per mille” (to S.P.), an AIRC PI-Grant (to S.P.), Epigenetics Flagship projects (CNR-Miur grants to S.P.), the DFF mobilex programme (1333-00130B to to S.Y.), Marie Curie fellowship programme (625238 to S.Y. and 656099 to J.G.), Foundation of Aase and Ejnar Danielsen (10-001992 to O.H.N.), Axel Muusfeldts Foundation (2017-678 to O.H.N.), and The Ragnar Söderberg Foundation (N91/15 to C.D.M.). This project has received funding from the European Union’s Horizon 2020 research and innovation programme (grant agreements STEMHEALTH ERCCoG682665 and INTENS 668294 to K.B.J. and DENOVOSTEM 670126 to S.P.).
Published: December 14, 2017
Footnotes
Supplemental Information includes six figures and three tables and can be found with this article online at https://doi.org/10.1016/j.stem.2017.11.001.
Contributor Information
Stefano Piccolo, Email: piccolo@biouni.pd.it.
Kim B. Jensen, Email: kim.jensen@bric.ku.dk.
Supplemental Information
References
- Azzolin L., Zanconato F., Bresolin S., Forcato M., Basso G., Bicciato S., Cordenonsi M., Piccolo S. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Barker N., van Oudenaarden A., Clevers H. Identifying the stem cell of the intestinal crypt: strategies and pitfalls. Cell Stem Cell. 2012;11:452–460. doi: 10.1016/j.stem.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Barry E.R., Morikawa T., Butler B.L., Shrestha K., de la Rosa R., Yan K.S., Fuchs C.S., Magness S.T., Smits R., Ogino S. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaugerie L., Itzkowitz S.H. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 2015;372:1441–1452. doi: 10.1056/NEJMra1403718. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014;344:1242281. doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Zhang N., Zheng Y., de Wilde R.F., Maitra A., Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., Brummelkamp T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero J.B., Ridgway R.A., Valeri N., Nixon C., Frame M.C., Muller W.J., Vidal M., Sansom O.J. c-Src drives intestinal regeneration and transformation. EMBO J. 2014;33:1474–1491. doi: 10.1002/embj.201387454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning M.J., Smith M.L., Ritchie M.E., Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- el Marjou F., Janssen K.P., Chang B.H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Fernandez Vallone V., Leprovots M., Strollo S., Vasile G., Lefort A., Libert F., Vassart G., Garcia M.I. Trop2 marks transient gastric fetal epithelium and adult regenerating cells after epithelial damage. Development. 2016;143:1452–1463. doi: 10.1242/dev.131490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan K., Modrusan Z., Cornelius J., Chavali A., Kasman I., Komuves L., Mo L., Diehl L. Intestinal epithelial cell up-regulation of LY6 molecules during colitis results in enhanced chemokine secretion. J. Immunol. 2008;180:3874–3881. doi: 10.4049/jimmunol.180.6.3874. [DOI] [PubMed] [Google Scholar]
- Fordham R.P., Yui S., Hannan N.R., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjorevski N., Sachs N., Manfrin A., Giger S., Bragina M.E., Ordóñez-Morán P., Clevers H., Lutolf M.P. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- Gregorieff A., Liu Y., Inanlou M.R., Khomchuk Y., Wrana J.L. Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature. 2015;526:715–718. doi: 10.1038/nature15382. [DOI] [PubMed] [Google Scholar]
- Kim N.G., Gumbiner B.M. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 2015;210:503–515. doi: 10.1083/jcb.201501025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.B., Kim M., Park Y.S., Park I., Kim T., Yang S.Y., Cho C.J., Hwang D., Jung J.H., Markowitz S.D. Prostaglandin E2 Activates YAP and a Positive-Signaling Loop to Promote Colon Regeneration After Colitis but Also Carcinogenesis in Mice. Gastroenterology. 2017;152:616–630. doi: 10.1053/j.gastro.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, J.W. (2008). affycoretools: functions useful for those doing repetitive analyses with Affymetrix GeneChips. R package version 1.46.1.
- Miyoshi H., VanDussen K.L., Malvin N.P., Ryu S.H., Wang Y., Sonnek N.M., Lai C.W., Stappenbeck T.S. Prostaglandin E2 promotes intestinal repair through an adaptive cellular response of the epithelium. EMBO J. 2017;36:5–24. doi: 10.15252/embj.201694660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Uchida N., Weissman I.L. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4′ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustata R.C., Vasile G., Fernandez-Vallone V., Strollo S., Lefort A., Libert F., Monteyne D., Pérez-Morga D., Vassart G., Garcia M.I. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 2013;5:421–432. doi: 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Owen K.A., Abshire M.Y., Tilghman R.W., Casanova J.E., Bouton A.H. FAK regulates intestinal epithelial cell survival and proliferation during mucosal wound healing. PLoS ONE. 2011;6:e23123. doi: 10.1371/journal.pone.0023123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera T., Azzolin L., Fujimura A., Di Biagio D., Frasson C., Bresolin S., Soligo S., Basso G., Bicciato S., Rosato A. Induction of Expandable Tissue-Specific Stem/Progenitor Cells through Transient Expression of YAP/TAZ. Cell Stem Cell. 2016;19:725–737. doi: 10.1016/j.stem.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S., Dupont S., Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles A.I., Traverso G., Zhang M., Roberts N.J., Khan M.A., Joseph C., Lauwers G.Y., Selaru F.M., Popoli M., Pittman M.E. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology. 2016;150:931–943. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Schnell U., Kuipers J., Giepmans B.N. EpCAM proteolysis: new fragments with distinct functions? Biosci. Rep. 2013;33:e00030. doi: 10.1042/BSR20120128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Schweiger P.J., Jensen K.B. Modeling human disease using organotypic cultures. Curr. Opin. Cell Biol. 2016;43:22–29. doi: 10.1016/j.ceb.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Sorrentino G., Ruggeri N., Specchia V., Cordenonsi M., Mano M., Dupont S., Manfrin A., Ingallina E., Sommaggio R., Piazza S. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat. Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science. 2009;324:1666–1669. doi: 10.1126/science.1172687. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Wu L.W., Grivennikov S.I., de Jong P.R., Lian I., Yu F.X., Wang K., Ho S.B., Boland B.S., Chang J.T. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Goten J., Vanhove W., Lemaire K., Van Lommel L., Machiels K., Wollants W.J., De Preter V., De Hertogh G., Ferrante M., Van Assche G. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS ONE. 2014;9:e116117. doi: 10.1371/journal.pone.0116117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V.W., Stange D.E., Page M.E., Buczacki S., Wabik A., Itami S., van de Wetering M., Poulsom R., Wright N.A., Trotter M.W. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger R., Shah M.A., Miller V.A., Kelsen J.R., Wang K., Heins Z.J., Ross J.S., He Y., Sanford E., Yantiss R.K. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology. 2016;151:278–287.e6. doi: 10.1053/j.gastro.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Zhang N., Bai H., David K.K., Dong J., Zheng Y., Cai J., Giovannini M., Liu P., Anders R.A., Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.