Abstract
Highly concentrated radionuclide waste produced during the Cold War era is stored at US Department of Energy (DOE) production sites. This radioactive waste was often highly acidic and mixed with heavy metals, and has been leaking into the environment since the 1950s. Because of the danger and expense of cleanup of such radioactive sites by physicochemical processes, in situ bioremediation methods are being developed for cleanup of contaminated ground and groundwater. To date, the most developed microbial treatment proposed for high-level radioactive sites employs the radiation-resistant bacterium Deinococcus radiodurans. However, the use of Deinococcus spp. and other bacteria is limited by their sensitivity to low pH. We report the characterization of 27 diverse environmental yeasts for their resistance to ionizing radiation (chronic and acute), heavy metals, pH minima, temperature maxima and optima, and their ability to form biofilms. Remarkably, many yeasts are extremely resistant to ionizing radiation and heavy metals. They also excrete carboxylic acids and are exceptionally tolerant to low pH. A special focus is placed on Rhodotorula taiwanensis MD1149, which was the most resistant to acid and gamma radiation. MD1149 is capable of growing under 66 Gy/h at pH 2.3 and in the presence of high concentrations of mercury and chromium compounds, and forming biofilms under high-level chronic radiation and low pH. We present the whole genome sequence and annotation of R. taiwanensis strain MD1149, with a comparison to other Rhodotorula species. This survey elevates yeasts to the frontier of biology's most radiation-resistant representatives, presenting a strong rationale for a role of fungi in bioremediation of acidic radioactive waste sites.
Keywords: bioremediation, yeasts, radiation resistance, heavy metal resistance, pH minimum, temperature maximum, Rhodotorula taiwanensis, genome
Introduction
Between 1945 and 1986, immense volumes of radioactive waste were generated from the production of 46,000 nuclear weapons in the United States. This was a period of history when national security priorities often surmounted concerns over the environment. Many Cold War wastes contained mixtures of inorganic contaminants including radionuclides (e.g., U and Tc), heavy metals (e.g., Cr and Hg), and nitrate, which were disposed directly to the ground at 120 sites across the United States (Daly, 2000). As the processing of uranium ores involved dissolution and extraction with nitric acid, this led to large volumes of highly acidic radioactive waste, which were stored in subterranean holding tanks or ponds. Over the past six decades, low levels of widespread contamination originating from such waste sites have contaminated over 7.0 × 107 m3 of surface and subsurface soils, and over 3.0 × 1012 L of groundwater (McCullough et al., 1999; Daly, 2000). As a result of the chemical reprocessing of 1.1 × 108 kg of nuclear fuel at the Hanford Site (WA, USA) alone, 2.1 × 105 m3 of radioactive waste were produced at nine reactors and stored in 177 underground tanks. These storage tanks with a lifespan of 10–20 years have been used since 1943, and the first leaks were confirmed in 1959. The amount of waste leakage from the Hanford tanks continues to grow, with estimates in 2004 ranging from 2.3 to 3.7 × 106 L (Fredrickson et al., 2004). The scale of these waste environments leaves few options for cleanup other than bioremediation (Brim et al., 2000).
In 2000, more than 110 distinct aerobic heterotrophic bacteria were isolated from below Hanford tank SX-108, which has been leaking extremely radioactive waste since the 1960s (Fredrickson et al., 2004). Among the numerous bacteria identified, Arthrobacter spp. were the most prevalent and Deinococcus spp. the most radiation-resistant. Both bacterial genera are known for their ability to survive harsh environmental conditions and reduce a variety of metals, and for their dependence on Mn for growth and resistance (Daly et al., 2004; Fredrickson et al., 2004; Ehrlich and Newman, 2008). The isolation of Deinococcus radiodurans from sediments under tank SX-108 focused research on this extremophile: first, to engineer metal-reducing and organic toxin-degrading capabilities into this bacterium; and second, to test the ability of engineered D. radiodurans to reduce/immobilize different metals, and to couple those reactions to solvent degradation while growing under high-level chronic ionizing radiation (CIR). Metal reduction coupled to toluene degradation as a bioremediation strategy for radioactive sites was successfully demonstrated in D. radiodurans at near-neutral pH under CIR (60 Gy/h) (Brim et al., 2006). However, D. radiodurans strain R1 and its engineered counterparts cannot grow at pH values below 4.8 (unpublished results).
To determine whether or not radiation-resistant acidophilic microorganisms exist, we first screened approximately 60 different environmental samples (desert sands, acid mine drainages, soils) for microorganisms that are able to grow under 36 Gy/h at pH 2.3. This yielded the basidiomycetous yeast Rhodotorula taiwanensis MD1149, which can grow under 66 Gy/h at pH 2.3. Fungi play an important role in the biogeochemical cycling of manganese and other redox-active metals (Ehrlich and Newman, 2008; Culotta and Daly, 2013), which is related to their ability to survive radiation and other oxidative challenges (Gadd, 2007; Daly, 2009; Sharma et al., 2017). Nevertheless, any prospect of yeasts in bioremediation of radioactive waste sites has been neglected, mainly due to the lack of research in this nascent field of radiomycology; preliminary fungal isolates from beneath tank SX-108 were dismissed as contaminants (Fredrickson et al., 2004). We therefore screened 26 additional yeasts of the Microbial Culture Collection EX1. These EX yeast strains (EXF) and MD1149 were tested for their resistance to ionizing radiation (chronic and acute), heavy metal resistance, their pH minima and temperature maxima, and for their ability to form biofilms. From among the numerous CIR- and heavy metal-resistant yeasts identified, we judged MD1149 as the most suitable for bioremediation of acidic radioactive sites, therefore justifying its whole genome sequencing. We present a comparative analysis of MD1149 with three other Rhodotorula spp. Our analysis of the core metabolic and stress-resistance characteristics of MD1149, together with the identification of several yeasts capable of growth at low pH under high-level chronic γ-irradiation, strengthens the rationale for the important role of fungi in bioremediation of radioactive Cold War environmental waste sites.
Materials and methods
Radiological, chemical, and biological safety
All experimental work was performed under standard laboratory safety conditions, and all radiological, chemical, and biological safety precautions were observed following rules and regulations established for respective research institutions.
Strains, isolation of MD1149, and irradiations
The ascomycetous and basidiomycetous yeasts used in this study and their isolation sites are presented in Table 1.
Table 1.
Ranking of representative fungi by the survival index D10 together with other characteristics.
| Strain # | Name | Phylum | Isolated from | D10 [kGy] | Tmax [°C] | Topt [°C] | AM, pH 2.3, CIR | YPD, pH 7, CIR | AN, YPD | pHmin AM | pHmin YPD | AM, HgCl2 [μM] | AM, MER [μM] | AM, CrCl3 [μM] | AM, K2Cr2O7 [μM] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EXF-5294 | Saccharomyces cerevisiae | Ascomycota | Red wine, Slovenia | 3.2 | 40 | 30 | – | – | + | 3.0 | 2.5 | 25 | <500 | >500 | >10 |
| EXF-6408 | Metschnikowia fructicola | Ascomycota | Mofette, CO2 rich water, Slovenia | 3.0 | <40 | 23.5 | + | + | w | 2.5 | 2.5 | 10 | 1,000 | 500 | 100 |
| EXF-4909 | Saccharomyces bayanus x cerevisiae | Ascomycota | New wine, Slovenia | 3.0 | 40 | 26 | + | + | + | 2.5 | 2.5 | 25 | <1,000 | 500 | >500 |
| EXF-5281 | Saccharomyces cerevisiae | Ascomycota | Floor spilled with wine, Slovenia | 2.6 | 40 | 30 | + | + | – | 2.5 | 2.5 | 10 | >50 | >100 | >250 |
| MD1149 | Rhodotorula taiwanensis | Basidiomycota | Acid mine drainage, USA | 2.5 | 32 | 25 | + | + | – | 1.5 | 2 | 50 | >500 | 500 | 300 |
| EXF-6398 | Pichia kudriavzevii | Ascomycota | Mofette, Slovenia | 2.0 | 45 | 27 | + | + | + | 1.5 | 2.0 | 25 | 100 | 500 | 750 |
| EXF-308 | Rhodotorula rubra | Basidiomycota | Glacial ice in sea water, Svalbard | 2.0 | 37 | 30 | + | + | – | 2.0 | 2.0 | 25 | 1,000 | 750 | 100 |
| EXF-5293 | Saccharomyces bayanus | Ascomycota | Apple juice, Slovenia | 2.0 | 40 | 26 | + | + | + | 2.5 | 2.5 | 25 | 3,000 | >500 | >100 |
| EXF-6464 | Debaryomyces hansenii | Ascomycota | Water from slow moving creek showing CO2 release | 1.8 | <40 | 26 | + | + | – | 2.0 | 2.0 | 10 | 100 | 500 | 250 |
| EXF-7288 | Saccharomyces kudriavzevii | Ascomycota | Bark of Quercus ilex, Croatia | 1.5 | <40 | 24 | – | – | – | 2.5 | 2.5 | 10 | >50 | >100 | >250 |
| EXF-3501 | Rhodosporidium diobovatum | Basidiomycota | Ice, Svalbard | 1.4 | 37 | 24 | + | + | – | 2.0 | 2.0 | 25 | 1,000 | 750 | 100 |
| EXF-6402 | Kazachstania exigua | Ascomycota | Mofette, CO2 rich water, Slovenia | 1.2 | <40 | 27 | + | + | + | 2.0 | 2.0 | 10 | 100 | 750 | 50 |
| EXF-3697 | Rhodosporidium kratochvilovae | Basidiomycota | Ice, Svalbard | 1.2 | 37 | 30 | + | + | – | 2.5 | 1.5 | 50 | 1,000 | 500 | 250 |
| EXF-1534 | Rhodotorula lysinophila | Basidiomycota | Glacial ice in sea water, Svalbard | 1.1 | 37 | 25 | – | + | – | 3.0 | 3.0 | 25 | 1,000 | 500 | 250 |
| EXF-1529 | Rhodotorula minuta | Basidiomycota | Glacial ice in sea water, Svalbard | 1.1 | 37 | 25 | + | + | – | 2.5 | 2.0 | 25 | 1,000 | 500 | 250 |
| EXF-5557 | Rhodotorula slooffiae | Basidiomycota | Box of plasticizer in the washing machine, Slovenia | 1.1 | 37 | 30 | + | + | – | 2.0 | 2.0 | 25 | 100 | 500 | 250 |
| EXF-3409 | Cryptococcus liquefaciens | Basidiomycota | Glacier ice, Svalbard | 1.0 | <40 | 30 | + | + | – | 2.5 | >3 | 25 | 1,000 | 750 | 50 |
| EXF-6453 | Cyberlindnera saturnus | Ascomycota | Mofette, soil, Slovenia | 1.0 | <40 | 25 | + | + | + | 2.5 | 2.0 | 25 | 500 | 2,000 | >3,000 |
| EXF-1496 | Pichia guilliermondi | Ascomycota | Glacial ice, Svalbard | 1.0 | 43 | 25 | + | + | – | 2.0 | 2.0 | 25 | 250 | 750 | 1,500 |
| EXF-6094 | Rhodotorula calyptogenae | Basidiomycota | Dishwasher rubber, France | 1.0 | 40 | 26 | + | + | – | 2.5 | 7.0 | 25 | 1,000 | 500 | 100 |
| EXF-7210 | Saccharomyces kudriavzevii | Ascomycota | Bark of Quercus sp, Montenegro | 1.0 | <40 | 24 | – | – | + | 2.5 | 2.5 | 10 | >50 | >100 | >250 |
| EXF-7289 | Saccharomyces kudriavzevii | Ascomycota | Bark of Quercus ilex, Croatia | 1.0 | <40 | 24 | – | – | + | 2.5 | 2.5 | 10 | >50 | >100 | >250 |
| EXF-3800 | Rhodotorula benthica | Basidiomycota | Glacier ice with sediment, Svalbard | 0.9 | 25 | 25 | – | – | – | 2.5 | 2.0 | 10 | 1,000 | 500 | 75 |
| EXF-3909 | Rhodotorula laryngis | Basidiomycota | Sea water near the glacier, Svalbard | 0.9 | 25 | 20 | – | – | – | 2.5 | 2.5 | 25 | 1,000 | 750 | 25 |
| EXF-7964 | Wickerhamomyces anomalus | Ascomycota | Forest ditch water, Slovenia | 0.9 | 40 | 25 | + | + | + | 2.5 | 2.5 | 10 | 1,500 | >3,000 | 1,500 |
| EXF-6463 | Candida pseudolambica | Ascomycota | Mofette, CO2 rich water, Slovenia | 0.5 | <40 | 33 | + | + | – | 2.0 | 2.5 | 10 | 1,500 | >3,000 | 75 |
| EXF-589 | Debaryomyces hansenii | Ascomycota | By the Atlantic coast, Namibia | 0.3 | <40 | 23 | – | – | – | >3 | >3 | 10 | 100 | 250 | 100 |
Temperature maximum supporting growth (Tmax), ability to grow under CIR (36 Gy/h) on solid AM (pH 2.3) or YPD (pH 7). Growth under anaerobic conditions (AN), pH minimum (pHmin), and the highest heavy metal concentrations compatible with growth in medium supplemented with HgCl2 (Hg2+), merbromin (MER), CrCl3 (Cr3+), and K2Cr2O7 (Cr6+); growth (+), no growth (−), weak growth (w).
Sixty environmental samples were collected between 2001 and 2015 as a part of a larger study. These samples represent desert sands (Arizona, Nevada, New Mexico); dried plant debris from deserts (Arizona, Nevada, New Mexico); water, sediments, and soil from abandoned mines and mine drainages (coal mine in Maryland, silver and gold mines in Colorado, mercury mine in Idrija, Slovenia); hot springs (Colorado; Radenci, Slovenia); water and sediments from acidic river (Rio Tinto, Spain); and radioactive waste storage tanks (Uniformed Services University of the Health Sciences, Maryland). One gram of each environmental sample was resuspended in 10 ml of MQ purified water and allowed to settle for 30 min. One milliliter of the supernatant was added to 10 ml of the oligotrophic medium AM (complex Acidiphilium Medium) (San Martin-Uriz et al., 2014) adjusted to pH 2.3 with HNO3, and incubated in a shaker incubator (200 rpm) at 25°C for 4 days. One hundred microliters were then spread on AM plates (pH 2.3) and incubated at 25°C under 36 Gy/h. After 3 days of continuous CIR, the plates were inspected for growth. Single colonies were re-inoculated on fresh AM solid medium.
Throughout this work, CIR exposures specified under 36 Gy/h (~22°C) were performed in a 137Cs irradiator (GammaCell 40, J. L. Shepard and Associates). For all other CIR exposures, we used a second adjustable dose rate 137Cs irradiator (Mark 1 Model 68 A, J. L. Shepard and Associates), also at ~22°C. Acute exposures were performed in a 60Co irradiator (10 kGy/h) (J. L. Shepard and Associates) at 0°C.
Phenotype characterization
The minimum pH and the highest Hg2+, merbromin, Cr6+, and Cr3+ concentrations supporting growth were determined in liquid AM and Yeast Extract-Peptone-Dextrose (YPD)2 medium. The overnight (O/N) cultures pre-grown at optimal temperatures were washed twice in sterile MQ and used to inoculate fresh liquid media adjusted for pH with HNO3, and/or supplemented with different concentrations of heavy metals to a final OD600 ~0.1. The strains were incubated in a shaker incubator, 200 rpm, at optimal temperatures. After inoculation, the OD600 was measured every 24 h for 1 week.
Maximum growth temperature and anaerobic growth were determined by observing colony formation on solid YPD medium incubated at various temperatures (25–65°C; temperature maxima); and for anaerobic growth, at a given strain's optimal temperature, in the presence or absence of atmospheric oxygen for 1 week.
Survival following acute forms of γ-radiation was determined on solid YPD medium by colony forming unit (CFU) assay as described previously (Daly et al., 2004). The ability of cells to grow under CIR on YPD pH 7.0 and AM pH 2.3 was monitored visually. The ability of a strain to form biofilms was tested in 96-well microtiter plates, as described by others (O'Toole, 2011), with eight replicate wells for every strain and each condition, and eight wells for blank controls. Pulsed-field gel electrophoresis (PFGE) with MD1149 genomic DNA was performed as described previously (Saracli et al., 2003).
Organic acid production by MD1149
The OD600 of an O/N culture of MD1149 in the Yeast Mold Broth (YM, Difco) was adjusted to 0.05 in modified Hommel's Minimal Salts (HMS; 0.3% (NH4)2SO4, 0.05% NaCl, 0.07% MgSO4, 0.04% Ca(NO3)2, 0.04% K2HPO4, 0.25% KH2PO4, 0.06% yeast extract, 5% glucose). At the indicated time (2, 4, 6, and 8 days), OD600 was assessed, and 10 ml of culture were centrifuged twice at 5,000 × g to obtain spent liquid medium (SLM).
Organic acids in SLM were identified and measured using a Waters Xevo G2-XS QTOF mass spectrometer (Waters Corporation, Milford, MA USA) coupled with a Waters Acquity H Class chromatography system. Organic acids were separated on a Waters Acquity UPLC HSS C18 1.8 μm 2.1 × 100 mm column using a modification of a previously published method (Fernández-Fernández et al., 2010). Mobile phases were methanol (solvent A) and water with 0.5% formic acid (solvent B). The separation method was as follows: initial, 90% B; 0.1 min, 90% B; 6 min, 70% B; 6.1 min, 90% B; 12 min, 90% B. The flow rate was 125 μl/min. The column compartment thermostat was set at 35°C, and the autosampler tray temperature was maintained at 4°C. Detection was accomplished by mass spectrometry with the electrospray ion source operating in negative ion, resolution mode. Data acquisition was performed using MassLynx Version 4.1 data acquisition software (Waters Corp.), with MSe data-independent centroid acquisition and leucine enkephalin lockmass correction.
Liquid chromatography mass spectrometry (LC-MS) and liquid chromatography tandem-mass spectrometry (LC-MS/MS) settings were as follows: Low Energy, 50–1000 Da, 6 V collision energy; High Energy, 50–1000 Da, collision energy ramp from 10–40 V; Scan time: 0.5 s; Source: 120°C; Desolvation, 450°C; Desolvation gas flow: 800 l/h; capillary voltage: 1.90 kV; Sampling cone voltage: 40 V. Experimental samples were compared with authentic standards (Sigma-Aldrich, St. Louis, MO) to identify organic acids present in cell culture media. Quantitation was performed with Waters TargetLynx software with quadratic calibration curve fitting.
MD1149 identification, DNA isolation, and genome analysis
MD1149 was first identified at the genus level based on micro- and macro morphology and assimilation test (YT MicroPlate™, BIOLOG Inc.), and then to the species level using genetic molecular identification (Mohamed et al., 2014).
Total DNA was isolated from MD1149 using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) and quantified by NanoDrop 2000 (Thermo Fisher Scientific).
The ITS1-5.8S rDNA-ITS2 and 18S rDNA sequences were matched to the GenBank non-redundant nucleotide database with the BLASTN algorithm (Altschul et al., 1990). MD1149 and related sequences were analyzed for similarity within the Geneious software package (Kearse et al., 2012) by using MUSCLE alignment (Edgar, 2004). The aligned sequences of representative strains were used to construct a phylogenetic tree with the PhyML 3.0 software (Guindon et al., 2010) with approximate likelihood-ratio test for branch supports, and with six substitution rate categories. The substitution model, alpha parameter of the gamma distribution and the proportion of invariable sites, was estimated by jModelTest 2.0 (Darriba et al., 2012).
The draft genome was generated using a combination of Illumina and 454 technologies. Two short-insert paired-end libraries, a fragment, 625-bp insert size (2 × 300 bp reads) and an overlapping fragment, 405-bp insert size (2 × 300 bp reads) were sequenced using version 3 chemistry on the MiSeq (Illumina, Inc., San Diego, CA, USA) (Bennett, 2004). Two large-insert paired-end libraries (8-kbp and 20-kbp insert size) were constructed and sequenced on the 454 GS FLX (Roche/454 Life Sciences, Branford, CT, USA) (Margulies et al., 2005). The draft data was assembled de novo with CLC Genomics Workbench v9.0 (QIAGEN Aarhus, Denmark). Repetitive sequences were identified using RepeatMasker (Smit et al., 2013–2015) and RepBase library (Jurka et al., 2005). The genome assembly completeness was evaluated with the Benchmarking Universal Single-Copy Orthologs (BUSCO 1.22) (Simão et al., 2015) software using the dataset for fungi.
For pairwise genome alignments, the following genomes were used: Rhodotorula sp. (Goordial et al., 2016), R. mucilaginosa (Deligios et al., 2015), R. glutinis (Paul et al., 2014), R. toruloides (Zhang et al., 2016), R. graminis (Firrincieli et al., 2015), Puccinia graminis (Duplessis et al., 2011). The genome alignments of contigs longer than 100 kbp were calculated with the PROmer algorithm, as implemented in MUMmer 3.23, and plotted with the MUMmerplot utility (Kurtz et al., 2004) as described by Hane et al. (2011).
RNA isolation and genome annotation
MD1149 total RNA was isolated, then pooled and sequenced from cells grown under the following conditions: YPD (25°C, O/N and 3 days), YNB + 2% glucose (25°C, O/N and 3 days), YNB + 2% glucose (6°C and 37°C, O/N), YNB + 2% glycerol (25°C, O/N), AM pH 2.3 (25°C, O/N, 0 and 36 Gy/h), and AM pH 7 (25°C, O/N, 0 and 36 Gy/h). The RNA was isolated from MD1149 using RiboPure RNA Purification Kit for Yeasts (Thermo Fisher Scientific). RNA integrity was assessed by Fragment Analyzer (Advanced Analytical Technologies Inc., Ankeny, IA, USA). Sequencing libraries were prepared from 500 ng of total RNA input using the TruSeq Stranded mRNA Library Preparation Kit (Illumina) with barcoded adapters. Sequencing libraries yield and concentration were determined using the Illumina/Universal Library Quantification Kit (KAPA Biosystems, Wilmington, MA, USA) on the Light Cycler 480 (Roche Diagnostics Co., Indianapolis, IN, USA). Library size distribution was determined using the Fragment Analyzer™ (Advanced Analytical Technologies Inc.). Clustering and sequencing was performed on the NextSeq 500 (Illumina) with paired-end reads of 75 bp length.
RNAseq reads were quality trimmed with Sickle (Joshi and Fass, 2011) and aligned to the assembled genome with TopHat 2.1.1 (Kim et al., 2013). The alignment was then used for the transcriptome assembly with Trinity 2.2.0 (Haas et al., 2013) in Genome Guided mode with jaccard clipping and a maximum intron length of 1,500 bp. Protein-coding and tRNA genes were annotated using MAKER 2.31.8 (Campbell et al., 2014). The complete Swissprot database was used as evidence, along with the database of BUSCO, a set of Basidiomycete fungal proteomes and the sequenced transcriptome of MD1149. Three gene predictors were used in the MAKER pipeline: SNAP (Korf, 2004; Campbell et al., 2014), GeneMark-ET (Lomsadze et al., 2014), and Augustus (Stanke and Waack, 2003). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PJQD00000000. The version described in this paper is version PJQD01000000.
Predicted protein sequences from the MD1149 genome were processed through automated functional annotation using the PSAT metaserver (Leung et al., 2016), which ran EFICAz 2.5 (Kumar and Skolnick, 2012), blastp against the KEGG, MetaCyc, BRENDA, and STRING databases (Caspi et al., 2014; Chang et al., 2015; Szklarczyk et al., 2015; Kanehisa et al., 2016), and Interproscan (Jones et al., 2014). Additionally, protein sequences were processed through online servers running SignalP 4.0 (Petersen et al., 2011) and TMHMM 2.0 (Krogh et al., 2001). The pepstats utility (EMBOSS suite) was used to calculate protein properties (Rice et al., 2000).
GO annotations were also quantified and then projected into the full GO hierarchy using slim-o-matic (Courtot et al., 2016) and reviewed using Protege (Munsen, 2015). High-level categories were selected, into which the MD1149 GO annotations were mapped (slimmed). Categories corresponding to GO molecular function and biological process were quantified and expressed as percent of annotations across the MD1149 genome.
Predicted proteins from MD1149, R. graminis, R. sp. JG-1b, and R. toruloides were compared by all-against-all blastp at identity cutoff 95% and query coverage ≥95% using CGP3, and post-processed to identify fasta sequences unique or in common among the species and putatively duplicated within each species. Sequence logos of conserved nucleotide positions in all introns of median length were drawn using WebLogo 3 (Crooks et al., 2004).
Results
Isolation of MD1149
In order to find a suitable candidate for bioremediation of acidic radioactive environmental waste sites, we first screened a variety of aquatic and terrestrial environments (desert sands, acid mine drainages, soils, and water samples) for strains that are both acid- and CIR-resistant. This strategy yielded only one strain, named MD1149, isolated from a sediment sample from an abandoned acid mine drainage facility in Maryland, USA (39°31′34.22″N, 79°1′12.16″W). MD1149 is a red-pigmented, unicellular, non-sporulating, ovoidal, obligately aerobic, budding yeast (Figure 1A, Table 1), which became pleomorphic under 36 Gy/h (Figure 1B). Phylogenetic analysis based on the internal transcribed spacer (ITS) and small subunit rRNA (SSU) sequences identified MD1149 as the basidiomycetous yeast R. taiwanensis [closely related to the type strain BCRC 23118(T) = CBS 11729(T)] (Figure 2), and confirmed by its micromorphological, macromorphological, and physiological characteristics (data not shown). Based on PFGE, the genome size of MD1149 was estimated to be >13 Mbp (Figure 1F). MD1149 was deposited with the Microbial Culture Collection EX as EXF-12971. Since other environmental samples screened did not yield additional acid- and CIR-resistant strains, we extended our study by including 26 distinct yeasts from the Microbial Culture Collection EX (Table 1).
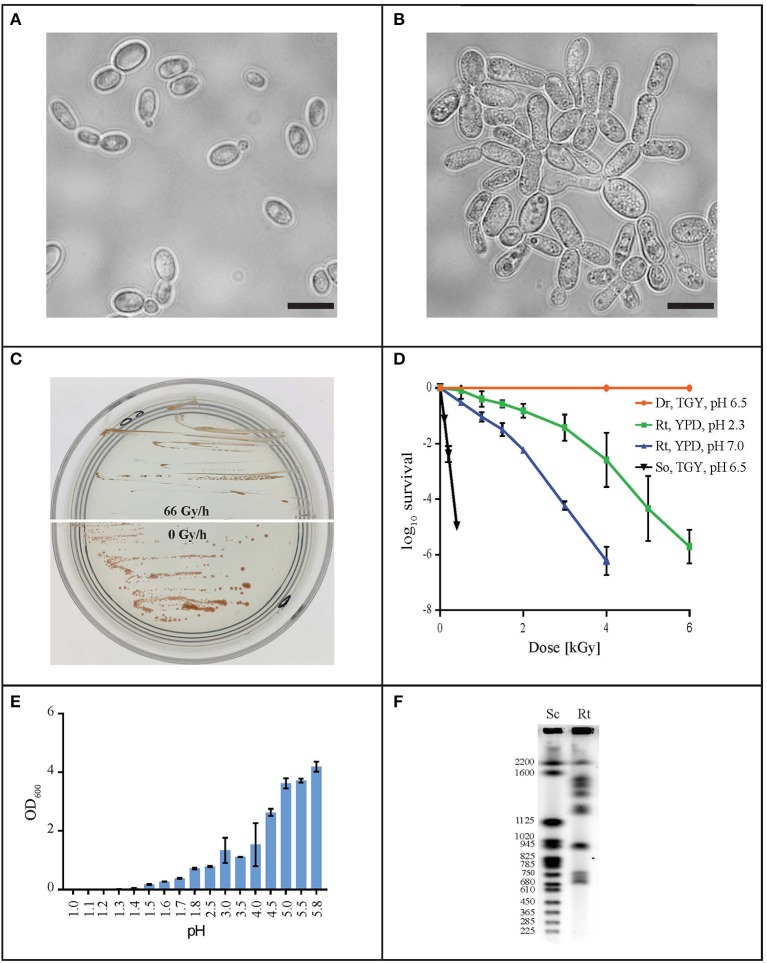
Figure 1.
Characterization of R. taiwanensis MD1149. Light microscopy of liquid O/N culture of MD1149 grown (A) without CIR and (B) under 36 Gy/h (137Cs). Scale bars: 5 μm. (C) Growth of MD1149 on AM plates at pH 2.3 under 66 Gy/h, and without CIR. (D) Survival after acute gamma irradiation (60Co) of MD1149 (Rt) pre-grown in and recovered on YPD at pH 2.3 and 7.0. Model bacteria D. radiodurans (Dr) and Shewanella oneidensis (So) were pre-grown in and recovered on TGY, pH 6.5. (E) pH-dependent growth of MD1149 in YPD (pH adjusted with HNO3) after 48 h. (F) Chromosomal partitioning in MD1149 by PFGE (Rt) and in S. cerevisiae size ladder (Sc).
Figure 2.
Phylogenetic analysis of R. taiwanensis MD1149 and related strains, and Sporobolomyces spp. as the root. The phylogenetic tree was constructed based on (A) ITS1-5.8S rDNA-ITS2 and (B) 18S rDNA sequences. GenBank accession number of each strain's sequence is in parentheses. TType strain.
Radiation resistance
Like most of the tested yeasts in this study, MD1149 was capable of growing luxuriantly at pH 2.3 and 7.0 under 36 Gy/h (Table 1). However, it was the only strain capable of growth under 66 Gy/h (Figure 1C).
Survival assays yield a radiation resistance metric named D10, which represents the acute radiation dose (Gy) giving 10% CFU survival (Daly et al., 2007, 2010; Sharma et al., 2017). Among tested strains, the most resistant was Saccharomyces cerevisiae EXF-5294 (D10, 3.2 kGy), and the most sensitive was Debaryomyces hansenii (D10, 0.3 kGy). The D10 of MD1149 is 2.5 kGy and ranks among the most radiation-resistant yeasts identified both for acute and chronic exposures (Table 1). Importantly, the radiation resistance of MD1149 increased with decreasing pH, from D10 0.8 kGy at pH 7.0 to D10 2.5 kGy at pH 2.3 (Figure 1D).
Temperature optima and maxima
Optimal and maximum growth temperatures for the strains are reported in Table 1. Pichia kudriavzevii was the most thermotolerant and could grow at 45°C. The temperature maximum of MD1149, which optimally grows at 20–25°C, was 32°C. The most temperature-sensitive strains were R. benthica and R. larynges, which could grow at 25°C or below.
pH minima
Over the course of 7 days, growth of strains in YPD and AM media adjusted to different pH values was montiored spectrophotometrically. Growth was considered as increasing when the OD600 rose above 0.1. The pH minima for growth of the yeasts are presented in Table 1. A full pH-dependent growth response curve for MD1149 is presented (Figure 1E). As shown in Table 1, the pH minima supporting growth of the yeast in rich (YPD) or oligotrophic (AM) media were very similar. Rhodotorula calyptogenae was the only strain that could not grow in YPD at low pH, whereas it grew well in AM at pH 2.5. Remarkably, growth responses of MD1149 and Pichia kudriavzevii in AM and Rhodosporidium kratochvilovae in YPD showed that their pH-minima approximate 1.5 (Figure 1E, Table 1).
Heavy metal resistance
The most common metal contaminants at DOE sites are U, Sr, Cs, Tc, Cr, Pb, and Hg. Among these, U, Tc, Hg, and Cr are significantly less mobile when reduced, and are capable of being immobilized by microorganisms (Daly, 2000). We tested yeasts for their resistance to Hg and Cr: mercury in the form of HgCl2 (Hg2+) and merbromin (organo-Hg), and chromium in the form of CrCl3 (Cr3+) and K2Cr2O7 (Cr6+). Table 1 summarizes heavy metal tolerances for strains grown in oligotrophic medium (AM) instead of YPD; YPD contains phosphates and myriad small organic molecules (e.g., peptides) that can mask metal toxicity (e.g., Mergeay, 1995). As expected, Hg2+ in HgCl2 is considerably more toxic than Cr3+ and Cr6+. The two strains most resistant to Hg2+, Cr3+, and Cr6+, were MD1149 and R. kratochvilovae (Figure 3 and Table 1), which could grow in AM supplemented with 50 μM HgCl2, and at significantly higher concentrations of Cr3+ and Cr6+. In contrast, most strains were resistant to millimolar concentrations of Hg when added as merbromin. Wickerhamomyces anomalus and Candida pseudolambica could grow in liquid medium supplemented with 3 mM Cr3+, whereas MD 1149 was resistant only to 0.5 mM Cr3+ (Table 1). The growth responses of MD1149 in AM supplemented with increasing concentrations of HgCl2 or K2Cr2O7 were distinct. Whereas increasing the concentration of Hg2+ increased the length of the lag-phase before the onset of exponential growth, increasing the concentration of Cr6+ slowed the growth of MD1149 (Figures 3A,B). Furthermore, in contrast to Cr6+/Cr3+, we showed that Hg2+ had a significant detrimental effect on MD1149 growth under CIR or not (Figures 3C,D).
Figure 3.
Resistance of R. taiwanensis MD1149 to HgCl2 and K2Cr2O7. Growth in liquid AM supplemented with (A) HgCl2 and (B) K2Cr2O7. (C) Growth of diluted cell suspension (OD600 ~0.9) on solid AM with no metals added (control), with 100 μM K2Cr2O7 (Cr6+), and with 30 μM HgCl2 (Hg2+), no CIR. (D) As for Panel (C), under 36 Gy/h (+CIR). For corresponding CrCl3 (Cr3+) results, see Table 1.
Biofilm formation
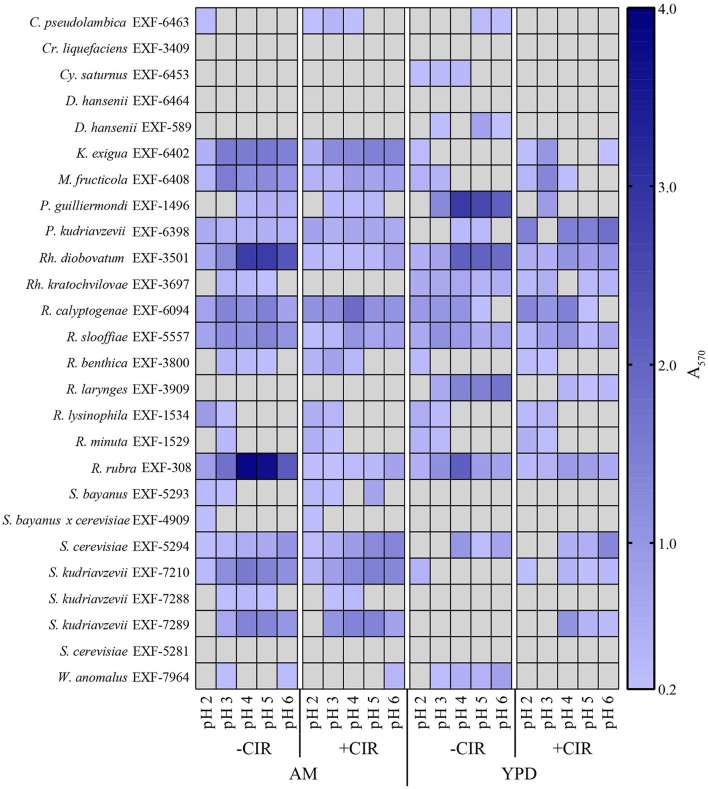
Biofilms are very important in bioremediation, since they offer sorption sites for many divalent cations that are toxic, and thus prevent their migration in the environment. The biofilm-forming capacity in yeasts was estimated with crystal violet assay (O'Toole, 2011) after 24 h incubation (in case of MD1149 it was additionally monitored over 5 days) at pH values 2–6, in the presence and absence of chronic gamma-radiation (36 Gy/h), and in oligotrophic (AM) and rich medium (YPD). This assay was performed on 8 parallels for each strain. The results are summarized in Figures 4, 5. The average absorbance of negative control (no inoculum) was subtracted from each measurement. Difference in A570 between the negative control and the sample above 0.2 was interpreted as an indication of biofilm formation.
Figure 4.
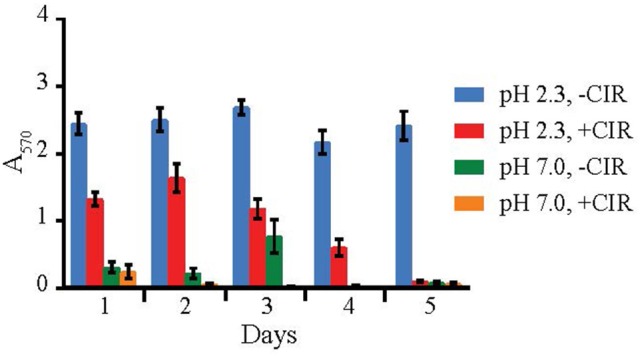
Biofilm formation by R. taiwanensis MD1149. Biofilm formation in YPD at pH 2.3 and 7.0 without CIR (-CIR) or under 36 Gy/h (+CIR) was quantified by crystal violet assay.
Figure 5.
Heat map showing biofilm formation in yeasts. Growth in liquid AM and YPD at pH 2.0, 3.0, 4.0, 5.0, and 6.0. Without CIR and under 36 Gy/h. Biofilms were stained with crystal violet and quantified. Gray area indicates the absence of detectable biofilm, determined by threshold-spectrophotometry at A570 < 0.2.
Out of 27 strains, 3 strains were unable to form biofilms: Cryptococcus liquefaciens, Debariomyces hansenii, and S. cerevisiae EXF-5281. For the remaining strains, biofilm forming capacity was strongly dependent on the species and physical parameters. For most strains, biofilm formation was inhibited by CIR. However, under specified conditions, biofilm formation in 7 species (C. pseudolambica, M. fruticola, P. kudriavzevii, R. benthica, S. bayanus, S. cerevisiae, and S. kudriavzevii) was moderately enhanced under CIR, based on A570 values (Figure 5). In the absence of CIR, pH values below 4 stimulated biofilm formation in 4 yeasts, but inhibited biofilms in the remainder. As pH values decreased to 2.3, in the presence or absence of CIR, MD1149 increasingly formed dense biofilms (Figure 4).
Organic acid production by MD1149
While monitoring the growth of MD1149 in YM medium (Figure 6A), we noted unusually high OD600 values in stationary phase cultures compared to growth in YPD or AM. This was not due to high cell concentrations in YM, but instead was caused by secretion of metabolites that absorb at 600 nm, which accompanied the drop in pH from 6 to 2.0–2.5 (Figure 6A). These metabolites are now being further investigated.
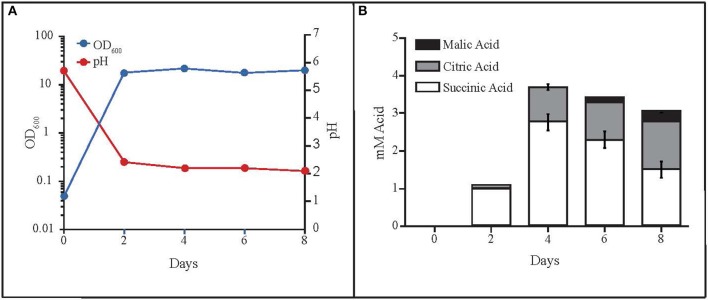
Figure 6.
Production of organic acids by R. taiwanensis MD1149. (A) Growth curve of MD1149 and the pH of the medium over an 8-day time period. (B) Quantitation of three organic acids in the SLM (spent liquid medium) for which authentic standards were available (citric, malic, and succinic acids).
The rapid pH drop suggested that MD1149 produced significant quantities of organic acids, theoretically in excess of 10 mM depending on pKa values of the acids present. Therefore, we analyzed the SLM by LC-MS to identify any excreted organic acids. We detected the presence of at least six organic acids by LC-MS and LC-MS/MS as well as elemental composition prediction using Waters MassLynx 4.1 software. These acids included citric, homoaconitic, homocitric/homoisocitric (constitutional isomers), malic, rhodotorulic, and succinic. It is noteworthy that organic acids with available reference spectra (citric, malic, and succinic) matched the precursor and product ions from LC-ESI-QTOF (liquid chromatography-electrospray ionization-quadrupole-time of flight-mass spectrometry) spectra published in MassBank (Horai et al., 2010). Furthermore, we procured standards for three of the six organic acids detected (citric, malic, and succinic) and quantitated their abundance (Figure 6B). The combined total concentration of these three acids was ~4 mM, consistent with the idea that these three acids contributed to the decrease in media pH, and that the other identified organic acids (which we were unable to characterize) contribute to the full pH change.
Sequencing, annotation, and analysis of the MD1149 genome
The estimated size of the genome assembly (without mitochondrial DNA) was 19.58 Mbp, and the final assembly of 181 scaffolds is based on 19.935 Gbp of draft sequence data, which provides 230× coverage of the genome. We identified a total of 26 scaffolds containing either 5′ or 3′ tandem DNA repeats with a sequence TTAGGG, which correspond to the most prevalent telomeric repeats (Teixeira and Gilson, 2005). Based on this result, we can conclude that the genome of MD1149 is organized in at least 13 chromosomes.
The size of the assembled mitochondrial genome was 38.20 kbp, slightly less than 40.39 kb reported by Zhao et al. (2013a). An alignment of both mitochondrial genomes showed that the sequences were largely syntenic (Figure 7D).
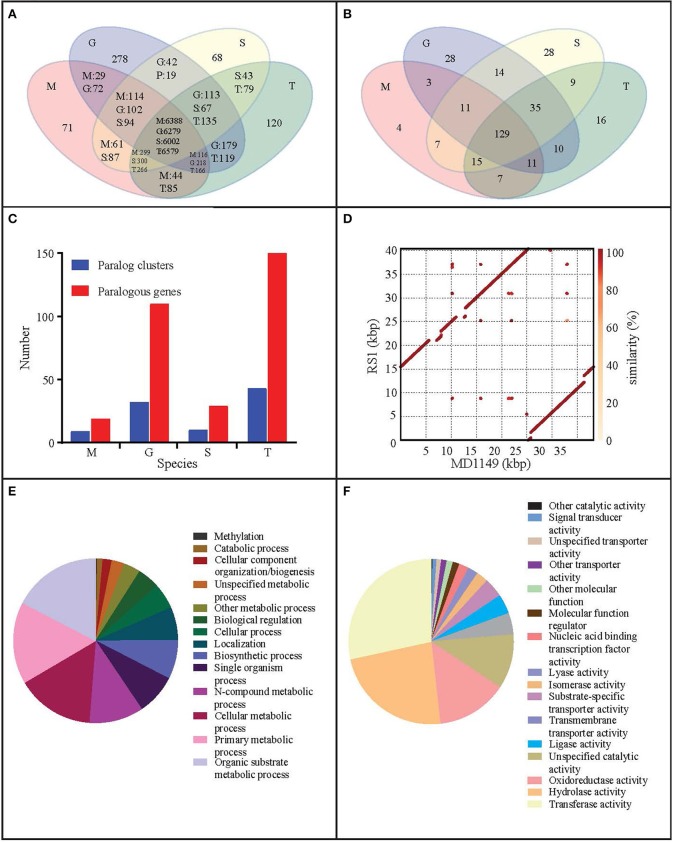
Figure 7.
Genome analysis of R. taiwanensis MD1149. Venn diagram representation of (A) shared/unique genes and (B) OrthoMLC groups in R. taiwanensis MD1149 (M), R. graminis (G), R. sp. JG-1b (S), and R. toruloides (T). (C) Numbers of genes/clusters determined to occur in at least two copies. (D) Alignment of mitochondrial DNA of MD1149 and R. taiwanensis RS1 (GeneBank: HF558455.1). Percentage of mapped GO annotation translated proteins of MD1149 belonging to two yeast GO-slim functional categories: (E) biological process and (F) molecular function.
The content of GC pairs was 40.85% in the mitochondrial genome and 61.69% in the nuclear DNA. This finding is comparable to R. glutinis (61.87%) and R. mucilaginosa (60.54%), but lower than in R. graminis (67.76%), which has one of the most GC-rich genomes among available fungal genomes. The number of repetitive sequences was relatively low at 1.49%.
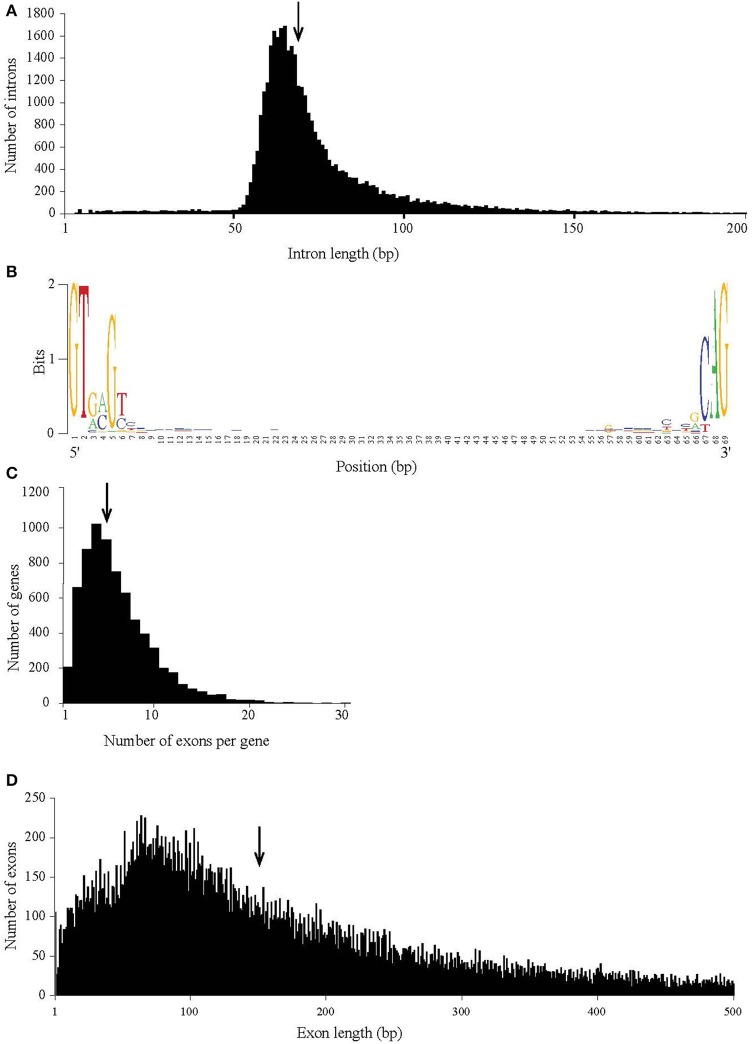
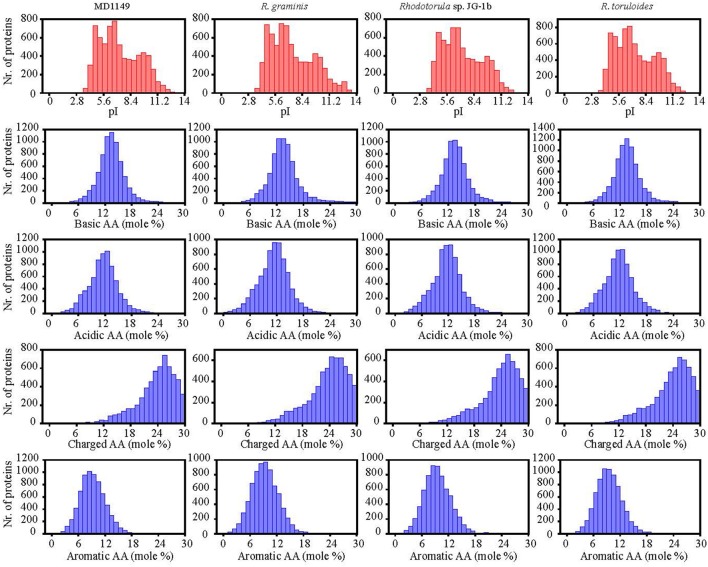
The number of genes annotated in the genome was 7,122. The genome completeness was estimated by searching the predicted proteome for 1,438 groups of BUSCO. We found 91% complete matches, 7% were fragmented and 2% were missing. More than 97% of MD1149 genes contained introns (Table 2), with an average of 6.2 exons and 5.2 introns per gene (Figure 8C, Table 2) (R. graminis: 6.2). The median length of introns was 69 bp (R. graminis: 101 bp), and they contained the typical 5′ and 3′ consensus sequences (Figures 8A,B). The median length of the exons was 151 bp (Figure 8D). The average length of the predicted proteins was 531, and their amino acid composition and isoelectric points were comparable to those of other Rhodotorula spp. (Figure 9).
Table 2.
Genome assembly and annotation statistics of R. taiwanensis MD1149.
| ASSEMBLY STATISTICS | |
| Assembly length (Mbp) | 19.58 |
| Mitochondrial genome size (kbp) | 38.2 |
| Number of contigs | 221 |
| Contig N50 | 18 |
| Contig L50 (kbp) | 345.82 |
| Number of scaffolds | 181 |
| Scaffold N50 | 17 |
| Scaffold L50 (kbp) | 388.69 |
| Percentage of scaffolds in gaps | 0.15% |
| Length of repeat-covered regions (bp) | 292515 |
| % of assembly covered by repeats | 1.49% |
| GC content | 61.69% |
| Mitochondrial GC content | 40.85% |
| GENE STATISTICS | |
| Number of genes | 7122 |
| Gene density (genes per kbp) | 0.36 |
| Protein length (amino acids, average) | 531 |
| Exon Length (bp, average) | 267 |
| Intron length (bp, average) | 80 |
| Intron length (bp, median) | 69 |
| Number of genes without introns | 208 |
| Percentage of genes without introns | 2.92% |
| Exons per gene (average) | 6.2 |
| Exons per gene (median) | 5 |
| Introns per gene (average) | 5.2 |
| GC content of CDS | 63.13% |
| GC content of introns | 59.81% |
| FUNCTIONAL ANNOTATIONS | |
| Genes with KEGG annotation | 2774 |
| Genes with Pfam domain | 2759 |
| Genes with Transmembrane domain | 1249 |
| Genes with SignalP peptide | 618 |
Figure 8.
Intron and exon statistics of R. taiwanensis MD1149. (A) The size distribution of introns. (B) The consensus sequence of all median-length introns. (C) The distribution of the number of exons per gene. (D) The size distribution of exons. The arrow indicates the median.
Figure 9.
Isoelectric points and amino acid composition of predicted proteins of R. taiwanensis MD1149 and related species with sequenced genome.
When compared to related species, the distributions of gene families were similar. Only 71 predicted MD1149 proteins and 4 OrthoMLC groups were unique (Figures 7A,B). Although the number of duplicated genes in MD1149 was similar to that in Rhodotorula sp. JG-1b, it was much lower than in R. graminis and R. toruloides (Figure 7C). GO-slim analysis revealed the expected distribution of functional categories (Figures 7E,F) for MD1149 genes. To better understand the remarkable radiation resistance of MD1149, we further analyzed the genome for the presence of genes involved in homologous DNA recombination, non-homologous end joining, oxidative stress response, Mn homeostasis, heavy metal resistance, and hydrolases; results are presented (Table 3). The set of genes and their copy number are comparable to other fungi.
Table 3.
R. taiwanensis MD1149 homologs of genes that are known from other fungi to be involved in DNA repair, oxidative stress, Mn homeostasis, resistance to heavy metals, and selected hydrolase genes.
| Representative KEGG genes | Function | Nr. of homologs in MD1149 | Homologs numbers (BMF94_) | |
|---|---|---|---|---|
| HOMOLOGOUS DNA RECOMBINATION | ||||
| RAD50 | DNA repair protein RAD50 | 1 | 4958 | |
| MRE11 | Double-strand break repair protein MRE11 | 1 | 1623 | |
| RAD57 | DNA repair protein RAD57 | 1, low similarity | 1476 | |
| RFA1 | Replication factor A1 | 1 | 6843 | |
| RAD51 | DNA repair protein RAD51 | 1 | 4285 | |
| RAD52 | DNA repair and recombination protein RAD52 | 1 | 1177 | |
| BRCA2 | Breast cancer 2 susceptibility protein | 1 | 4266 | |
| RAD54 | DNA repair and recombination protein RAD54 | 2 | 1178 | 0432 |
| POLD1 | DNA polymerase delta subunit 1 | 1 | 4972 | |
| BLM | Bloom syndrome protein, ATP dependent DNA helicase | 4 | 4946 | 4947 |
| low similarity | 6361 | 3460 | ||
| TOP3 | DNA topoisomerase III | 1 | 1732 | |
| MUS81 | Crossover junction endonuclease MUS81 | 1 | 5222 | |
| EME1 | Crossover junction endonuclease EME1 | 0 | 4418 | |
| NON-HOMOLOGOUS END-JOINING | ||||
| KU70 | ATP-dependent DNA helicase 2 subunit 1 | 1 | 5927 | |
| KU80 | ATP-dependent DNA helicase 2 subunit 2 | 1 | 4455 | |
| RAD50 | DNA repair protein RAD50 | 1 | 4958 | |
| MRE11 | Double-strand break repair protein MRE11 | 1 | 1623 | |
| POLL | DNA polymerase lambda | 1 | 4374 | |
| RAD2 | Flap endonuclease-1 | 1 | 1460 | |
| DNL4 | DNA ligase 4 | 1 | 5514 | |
| OXIDATIVE STRESS | ||||
| SOD2 (S. cerevisiae) | Fe-Mn family superoxide dismutase | 1 | 4448 | |
| CTA1, CTT1 (S. cerevisiae) | Catalase | 2 | 3212 | 0981 |
| CTT1 (S. cerevisiae) | Cytosolic catalase T | 2 | 3212 | 0981 |
| TSA1, TSA2 (S. cerevisiae) | Peroxiredoxin, thioredoxin peroxidase | 1 | 1596 | |
| TSA2 (S. cerevisiae) | Stress inducible cytoplasmic thioredoxin peroxidase | 1 | 1596 | |
| PRX1 (S. cerevisiae) | Mitochondrial peroxiredoxin, thioredoxin peroxidase | 1 | 1823 | |
| DOT5 (S. cerevisiae) | Nuclear thiol peroxidase | 1, low similarity to peroxiredoxins | 1737 | |
| GPX1, GPX2, GPX3 (S. cerevisiae) | Glutathione peroxidase | 1 | 0010 | |
| HYR1 (S. cerevisiae) | GPX3 Thiol peroxidase | 1 | 0010 | |
| GTT1 (S. cerevisiae) | ER associated glutathione S-transferase | 1 | 4445 | |
| GTO1, ECM4, GTO3 (S. cerevisiae) | Omega-class glutathione S-transferase | 1 | 2093 | |
| GRX1, GRX2 (S. cerevisiae) | Dithiol glutaredoxin | 1 | 4981 | |
| GRX3, GRX4, GRX5 (S. cerevisiae) | Monothiol glutaredoxin | 2 | 1659 | 3737 |
| GLR1 (S. cerevisiae) | Cytosolic and mitochondrial glutathione oxidoreductase | 1 | 0249 | |
| TRX1, TRX2, TRX3 (S. cerevisiae) | Thioredoxin | 5 | 6822 | 6320 |
| 0478 | 4390 | |||
| 7059 | ||||
| TRR2 (S. cerevisiae) | Mitochondrial thioredoxin reductase | 5997 | ||
| NCU05770 (N. crassa) | Cytochrome c peroxidase | 2 | 5958 | 4954 |
| NCU07386 (N. crassa) | Fe-Mn family superoxide dismutase | 1 | 2434 | |
| NCU05780 (N. crassa) | Theta-class glutathione S-transferase | 2 | 3517 | 1660 |
| NCU01320 (N. crassa) | Microsomal glutathione S-transferase | 1 | 2590 | |
| NCU03339 (N. crassa) | Glutathione-disulfide reductase | 1 | 249 | |
| Mn HOMEOSTASIS | ||||
| SMF2 (S. cerevisiae) | Divalent metal ion transporter involved in manganese homeostasis has broad specificity for di-valent and tri-valent metals | 0853 | ||
| PHO84 (S. cerevisiae) | Inorganic phosphate (Pi) transporter, also low-affinity manganese transporter | 2 | 2399 | 0582 |
| PMR1 (S. cerevisiae) | CaMn P-type ATPase transporter | 2 | 5512 | 2539 |
| BSD2 (S. cerevisiae) | Heavy metal ion homeostasis protein | 1 | 5777 | |
| CCC1 (S. cerevisiae) | Similar to putative vacuolar FeMn transporter | 1 | 3438 | |
| RESISTANCE TO HEAVY METALS | ||||
| PCA1 | Copper or cadmium transporting P-type ATPase | 1 | 6825 | |
| YCF1 | Proteins with high similarity to the yeast vacuolar glutathione S-conjugate transporter with a known role in detoxifying Cd, Hg and As | at least 3 | 6201 | 3559 |
| 3360 | ||||
| COT1, ZRC1 | Transporter of heavy metals | 1 | 0535 | |
| HYDROLASE FAMILIES | ||||
| GNAT family acetyltransferases | 23 | 5507 | 6265 | |
| 5757 | 2541 | |||
| 5786 | 7019 | |||
| 4676 | 7020 | |||
| 1320 | 0152 | |||
| 1591 | 0305 | |||
| 4302 | 0442 | |||
| 1061 | 0435 | |||
| 1055 | 1739 | |||
| 1059 | 3511 | |||
| 1060 | 1915 | |||
| 6163 | ||||
| NUDIX hydrolases | 15 | 5675 | 1605 | |
| 5713 | 3374 | |||
| 5980 | 0416 | |||
| 5976 | 4813 | |||
| 1054 | 1893 | |||
| 0871 | 6385 | |||
| 6480 | 5468 | |||
| 4228 | ||||
| A/B superfamily hydrolases | 25 | 5552 | 3298 | |
| 1350 | 3063 | |||
| 5239 | 3669 | |||
| 1589 | 2387 | |||
| 6322 | 4897 | |||
| 5970 | 4890 | |||
| 0382 | 0636 | |||
| 0738 | 0676 | |||
| 7045 | 1717 | |||
| 6437 | 3427 | |||
| 6642 | 3439 | |||
| 0295 | 1961 | |||
| 3113 | ||||
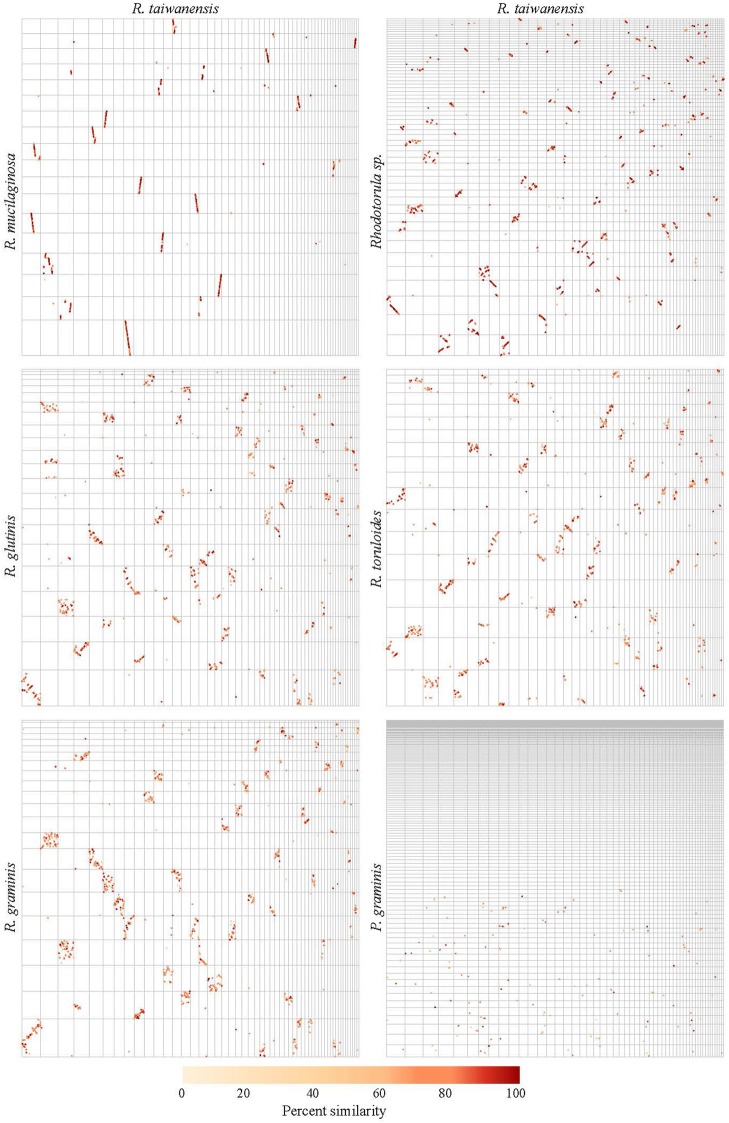
The pairwise genome alignments showed a high level of macrosynteny between MD1149, R. mucilaginosa and Rhodotorula sp. JG-1b (Figure 10). With R. toruloides, R. glutinis, and R. graminis the order of the alignable regions was mixed, but the genomic rearrangements appear to have occurred within the same DNA molecules and not between them (Figure 10). This form of evolution is known as mesosynteny, and it was previously thought to be restricted only to filamentous ascomycetes (Hane et al., 2011).
Figure 10.
Pairwise genome alignments of R. taiwanensis MD1149 and related species. Contigs longer than 100 kbp from the genomes of MD1149 (x-axes) and related species (y-axes) were ordered by length and aligned with Mummer software.
Discussion
The US Department of Energy (DOE) is the steward of the United States' nuclear waste legacy, comprised of immense volumes of long-lived radioactive environmental waste produced during the Cold War and stored at DOE sites. Over the last six decades, these radioactive wastes have been leaking into the environment, including mixtures of radionuclides, heavy metals and strong acids (e.g., HNO3) at levels (e.g., pH < 2.5) that exceed those tolerated by most microorganisms (Brim et al., 2000; Daly, 2000). Despite attempts to neutralize these acidic sites, low pH contamination zones persist, greatly diminishing the prospects for bioremediation at locations close to the originating leaks where the potential benefits are greatest, and where radiation levels are highest (Daly, 2000; Shelobolina et al., 2003).
We studied 16 ascomycetous and 11 basidiomycetous yeasts isolated from diverse environments including arctic ice, acid mine drainage, red wine, and apple juice, as well as dry environments with elevated temperatures (Table 1). Whereas many yeasts and filamentous fungi are reported to be resistant to various extreme environments, there are no reports of yeasts being resistant to high-level CIR. All 27 yeasts were able to survive an acute exposure to gamma-rays over the range 0.3–3.2 kGy: 8 extremely resistant yeasts displayed D10 values between 2.0 and 3.2 kGy; 14 yeasts were moderately resistant with D10 values between 1–2 kGy; and 5 yeasts were relatively sensitive, with D10 values as low as 300 Gy, but still more resistant than many bacteria (Daly, 2012). For comparison, the D10 of the soil bacterium Shewanella oneidensis is 70 Gy (Daly, 2012). Thus, this survey elevates yeast to the frontier of biology's most radiation-resistant representatives (Daly, 2012). In the context of bioremediation of DOE sites, CIR resistance is most relevant: 18/27 strains were able to grow under 36 Gy/h at pH 2.3, comparable to dose rates and pH values reported for sediments beneath Hanford tank SX-108 (Fredrickson et al., 2004). Surprisingly, among the surveyed yeasts, we show that chronic and acute radiation responses are not always aligned: S. cerevisiae strain EXF-5294 (D10, 3.2 kGy) did not grow under 36 Gy/h, and similarly for S. kudriavzevii EXF-7288 (D10, 1.5 kGy). A special focus is placed on R. taiwanensis MD1149, isolated from an acid mine drainage facility. MD1149 is capable of growth under 66 Gy/h at pH 2.3 (Figure 1C).
The concentration of contaminant heavy metals at DOE sediments can reach 10–30 μM (Fredrickson et al., 2004). Many microorganisms are reported to resist the toxic effects of metals by immobilizing and/or transforming those metals to less toxic chemical states (Brim et al., 2000; Fredrickson et al., 2000). Far fewer microorganisms are known to be able to transform metals at low pH, and there have been no published reports on any organism capable of transforming metals at low pH under high-level CIR. We ranked the 27 yeasts for their resistance to two heavy metals that predominate at DOE waste sites: 1. ionic Hg2+ in the form of HgCl2, and Hg as an organo-Hg compound merbromin; and 2. chromium in the form of CrCl3 (Cr3+) and K2Cr2O7 (Cr6+), as presented in Table 1. Redox-active heavy metals propagate ROS in cells and typically are more toxic than their covalently-bound counterparts. Consistently, we show Hg2+ and Cr6+ were the most toxic, followed by Cr3+, then merbromin. The ability of many of the tested yeasts to grow in the presence of 50–100 μM concentrations of Hg or Cr thus elevates these radiation-resistant simple eukaryotes to the forefront of metal resistances encountered in the natural world: 14 of the strains were able to grow in the presence of 25 μM HgCl2; 2 strains, MD1149 and R. kratochvilovae, grew in 50 μM HgCl2; and 14 strains grew in 1 mM merbromin (Table 1). Unlike Hg0 and Hg2+, redox-active Cr can cycle between several oxidation states between +2 to +6, with the most stable forms in the environment being hexavalent Cr6+ and trivalent Cr3+. These oxidation states have different chemical properties. For example, Cr3+ is relatively insoluble in the environment and is far less toxic than Cr6+, which is highly soluble and generates ROS in cells (Viti et al., 2014). Indeed, most yeasts were able to grow at concentrations of 500 μM merbromin or CrCl3; and three species, W. anomalus, Cyberlindnera saturnus and C. pseudolambica grew at even higher concentrations of these heavy metals (Table 1).
In bioremediation, biofilm formation is a highly desirable characteristic because the polysaccharide/protein extracellular matrix can bind/adsorb cations and reduce their migration in the environment. In the past few decades, most research on biofilms has focused on medically important bacteria and a few yeasts (Niemira and Solomon, 2005). Importantly, we report that biofilm formation in some yeasts is facilitated by chronic gamma radiation (Figure 5). In particular, MD1149 is capable of forming biofilms and growing in the presence of heavy metals under 36 Gy/h (Figures 3, 4). We also show that MD1149 produces abundant carboxylic acids (e.g., succinic acid) (Figure 6), similarly to Rhodotorula glutinis (Glass and Bhattacharjee, 1971), which is expected to facilitate metal transformation and metal accumulation in biofilms formed at low pH under CIR, but more evidence is needed.
The yeast we judged most suitable for bioremediation of acidic radioactive DOE waste sites was MD1149. To further develop this basidiomycete as a bioremediating platform, we subjected MD1149 to whole genome sequencing, then compared the genome to three other Rhodotorula species (R. graminis, Rhodotorula sp. JG-1b, R. toruloides). The complete sequence of the MD1149 genome is organized into at least 13 chromosomes (Figure 1F). The sequence-based features are summarized (Table 2), and when compared to the other Rhodotorula spp., the genome is unremarkable with respect to its size and GC content. Moreover, compared to other basidiomycetes the genome and the predicted proteome are relatively small (Mohanta and Bae, 2015). Viewed from the perspective of radiation resistance, the MD1149 genome and the predicted proteome exemplify characteristics found in many other sequenced species across the tree of life (Paul et al., 2014; Deligios et al., 2015; Goordial et al., 2016; Zhang et al., 2016; Matrosova et al., 2017), viz. The predicted DSB homologous recombination and non-homologous end-joining repair functions of MD1149, as well as its enzymatic antioxidant enzymes, are unremarkable (Table 3). Further, MD1149 encodes numerous genes commonly implicated in generating low molecular weight (LMW) metabolites (e.g., orthophosphate). They include acetyltransferases of the GNAT family, Nudix hydrolases, a/b superfamily hydrolases and calcineurin family phosphoesterases, which are present in many fungi (Zhao et al., 2013b). For most of these predicted hydrolases, and phosphatases in particular, their substrate specificities are either unknown or the affinity of known substrates is extremely low. It is likely that these predicted MD1149 enzymes, similar to D. radiodurans, participate in the degradation of nucleic acids, proteins and lipids (Makarova et al., 2001). The prediction of so many hydrolase functions in MD1149, which also encodes systems for Mn accumulation (Table 3), is expected to give rise to high intracellular concentrations of low molecular weight Mn2+ antioxidants. The hydrolase genes may therefore play a role in MD1149's extreme radiation resistance, yielding high intracellular concentrations of the organic and inorganic ligand-precursors of Mn2+ antioxidants that maintain proteome functionality under oxidative stress (Daly et al., 2007, 2010; Sharma et al., 2017).
Physicochemical cleanup technologies that could be used to decontaminate the immense volumes of soils, sediments and groundwaters at DOE facilities are prohibitively expensive and dangerous. Thus, the use of microorganisms to stabilize and/or detoxify such waste environments may be a viable alternative (Prakash et al., 2013). A bioremediation strategy based on the basidiomycete MD1149 and other yeasts (this study; Chandran and Das, 2012) now offers a more promising path to stabilization of DOE sites than Deinococcus spp., which are intolerant of low pH and heavy metals. Remarkably, MD1149 is highly resistant to Hg, Cr and CIR, capable of forming biofilms under 36 Gy/h at pH 2.3, and surviving acute doses of 2.5 kGy at pH 2.3. Importantly, it is reported that Rhodotorula spp. are genetically tractable (Takahashi et al., 2014), and we anticipate that MD1149 could be a good candidate for fungal-based CRISPR/Cas9 technologies (DiCarlo et al., 2013). Thus, the proposed use of MD1149 and other fungi for treatment of environments where radiation, low pH, and heavy metals are the principle factors limiting microbial survival and function appears to be a realistic approach given these early data.
Author contributions
RT, MD, and LD: designed research; RT, VM, OG, RV, PK, EG, CZ, BS, ML, SM, BR, JS, CD, TH, KF, and NG-C: performed research; RT, CG, SM, MC, and CZ: analyzed data; and RT, LD, and MD: wrote the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the U.S. Department of Energy, Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 and at the Uniformed Services University of the Health Sciences under Contract DE-NA0002322/0006. The authors would also like to acknowledge Ms. Constance L. Loucks and Mr. Jaron Hawkins (Maryland Department of the Environment) for collecting samples, Mr. Michael E. Woolbert (USUHS) for technical support and maintenance of irradiation facilities, Dr. Tine Grebenc (Slovenian Forestry Institute, Slovenia) and Mr. John W. Hobson for proofreading, The American Genome Center (Bethesda, MD, USA) for services in library preparation and sequencing, and the Defense Threat Reduction Agency (HDTRA-18774-M), the Slovenian Research Agency (BI-US/12-13-003, BI-US/14-15-009, Infrastructural Centre Mycosmo, MRIC UL), the National Human Genome Research Institute (Gene Ontology Consortium P41, grant 5U41HG002273-14), and EMBL-EBI (core funds) for financial support.
The opinions expressed herein are those of the authors, and are not necessarily representative of those of the Uniformed Services University of the Health Sciences (USUHS), the Department of Defense (DOD); or, the United States Army, Navy, or Air Force. This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344.
Footnotes
References
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bennett S. (2004). Solexa Ltd. Pharmacogenomics 5, 433–438. 10.1517/14622416.5.4.433 [DOI] [PubMed] [Google Scholar]
- Brim H., McFarlan S. C., Fredrickson J. K., Minton K. W., Zhai M., Wackett L. P., et al. (2000). Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18, 85–90. 10.1038/71986 [DOI] [PubMed] [Google Scholar]
- Brim H., Osborne J. P., Kostandarithes H. M., Fredrickson J. K., Wackett L. P., Daly M. J. (2006). Deinococcus radiodurans engineered for complete toluene degradation facilitates Cr(VI) reduction. Microbiology 152, 2469–2477. 10.1099/mic.0.29009-0 [DOI] [PubMed] [Google Scholar]
- Campbell M. S., Holt C., Moore B., Yandell M. (2014). Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48, 4.11.1–39. 10.1002/0471250953.bi0411s48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R., Altman T., Billington R., Dreher K., Foerster H., Fulcher C. A., et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42, D459–D471. 10.1093/nar/gkt1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran P., Das N. (2012). Role of plasmid in diesel oil degradation by yeast species isolated from petroleum hydrocarbon-contaminated soil. Environ. Technol. 33, 645–652. 10.1080/09593330.2011.587024 [DOI] [PubMed] [Google Scholar]
- Chang A., Schomburg I., Placzek S., Jeske L., Ulbrich M., Xiao M., et al. (2015). BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 43, D439–D446. 10.1093/nar/gku1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtot M., Mitchell A., Scheremetjew M., Pi-ero J., Furlong L. I., Finn R., et al. (2016). Slim-o-matic: a semi-automated way to generate Gene Ontology slims, in Proceedings of SWAT4LS 2016 (Amsterdam: ). [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta V. C., Daly M. J. (2013). Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid. Redox Signal. 19, 933–944. 10.1089/ars.2012.5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. J. (2000). Engineering radiation-resistant bacteria for environmental biotechnology. Curr. Opin. Biotechnol. 11, 280–285. 10.1016/S0958-1669(00)00096-3 [DOI] [PubMed] [Google Scholar]
- Daly M. J. (2009). A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7, 237–245. 10.1038/nrmicro2073 [DOI] [PubMed] [Google Scholar]
- Daly M. J. (2012). Death by protein damage in irradiated cells. DNA Repair 11, 12–21. 10.1016/j.dnarep.2011.10.024 [DOI] [PubMed] [Google Scholar]
- Daly M. J., Gaidamakova E. K., Matrosova V. Y., Kiang J. G., Fukumoto R., Lee D. Y., et al. (2010). Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 5:e12570. 10.1371/journal.pone.0012570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. J., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Leapman R. D., et al. (2007). Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5:e92. 10.1371/journal.pbio.0050092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. J., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Venkateswaran A., et al. (2004). Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306, 1025–1028. 10.1126/science.1103185 [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L., Doallo R., Posada D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deligios M., Fraumene C., Abbondio M., Mannazzu I., Tanca A., Addis M. F., et al. (2015). Draft genome sequence of rhodotorula mucilaginosa, an emergent opportunistic pathogen. Genome Announc. 3:e00201-15. 10.1128/genomeA.00201-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., Church G. M. (2013). Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. 10.1093/nar/gkt135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplessis S., Cuomo C. A., Lin Y. C., Aerts A., Tisserant E., Veneault-Fourrey C., et al. (2011). Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. U.S.A. 108, 9166–9171. 10.1073/pnas.1019315108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich H. L., Newman D. K. (2008). Geomicrobiology of manganese, in Geomicrobiology, 5th Edn., eds Ehrlich H. L., Newman D. K. (Boca Raton, FL: CRC Press; ), 347–420. [Google Scholar]
- Fernández-Fernández R., López-Martínez J. C., Romero-González R., Martínez-Vidal J. L., Alarcón Flores M. I., Garrido Frenich A. (2010). Simple LC–MS Determination of Citric and Malic Acids in Fruits and Vegetables. Chromatographia. 72, 55–62. 10.1365/s10337-010-1611-0 [DOI] [Google Scholar]
- Firrincieli A., Otillar R., Salamov A., Schmutz J., Khan Z., Redman R. S., et al. (2015). Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Front. Microbiol. 6:978. 10.3389/fmicb.2015.00978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Kostandarithes H. M., Li S. W., Plymale A. E., Daly M. J. (2000). Reduction of Fe(III), Cr(VI), U(VI), and Tc(VII) by Deinococcus radiodurans R1. Appl. Environ. Microbiol. 66, 2006–2011. 10.1128/AEM.66.5.2006-2011.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Zachara J. M., Balkwill D. L., Kennedy D., Li S. M., Kostandarithes H. M., et al. (2004). Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the hanford site, Washington state. Appl. Environ. Microbiol. 70, 4230–4241. 10.1128/AEM.70.7.4230-4241.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd G. M. (2007). Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycol. Res. 111, 3–49. 10.1016/j.mycres.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Glass J., Bhattacharjee J. K. (1971). Biosynthesis of lysine in Rhodotorula: accumulation of homocitric, homoaconitic, and homoisocitric acids in a leaky mutant. Genetics 67, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goordial J., Raymond-Bouchard I., Riley R., Ronholm J., Shapiro N., Woyke T., et al. (2016). Improved high-quality draft genome sequence of the Eurypsychrophile Rhodotorula sp. JG1b, isolated from permafrost in the hyperarid upper-elevation McMurdo dry Valleys, Antarctica. Genome Announc. 4:e00069-16. 10.1128/genomeA.00069-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane J. K., Rouxel T., Howlett B. J., Kema G. H., Goodwin S. B., Oliver R. P. (2011). A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 12:R45. 10.1186/gb-2011-12-5-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., et al. (2010). MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714. 10.1002/jms.1777 [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H. Y., Fraser M., Li W., McAnulla C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N. A., Fass J. N. (2011). Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming tool for FastQ files. Available online at: https://github.com/najoshi/sickle
- Jurka J., Kapitonov V. V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. (2005). Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467. 10.1159/000084979 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. (2004). Gene finding in novel genomes. BMC Bioinformatics 5:59. 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kumar N., Skolnick J. (2012). EFICAz2.5: application of a high-precision enzyme function predictor to 396 proteomes. Bioinformatics 28, 2687–2688. 10.1093/bioinformatics/bts510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A. L., Smoot M., Shumway M., Antonescu C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung E., Huang A., Cadag E., Montana A., Soliman J. L., Zhou C. L. (2016). Protein Sequence Annotation Tool (PSAT): a centralized web-based meta-server for high-throughput sequence annotations. BMC Bioinformatics 17:43. 10.1186/s12859-016-0887-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A., Burns P. D., Borodovsky M. (2014). Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 42, e119. 10.1093/nar/gku557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Aravind L., Wolf Y. I., Tatusov R. L., Minton K. W., Koonin E. V., et al. (2001). Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79. 10.1128/MMBR.65.1.44-79.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Hazen T., Benson S. (1999). Bioremediation of Metals and Radionuclides: What It Is And How it Works. UC-Berkley, CA: Lawrence Berkeley National Laboratory. [Google Scholar]
- Matrosova V. Y., Gaidamakova E. K., Makarova K. S., Grichenko O., Klimenkova P., Volpe R. P., et al. (2017). High-quality genome sequence of the radioresistant bacterium Deinococcus ficus KS 0460. Stand Genomic. Sci. 12:46. 10.1186/s40793-017-0258-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M. (1995). Heavy metal resistances in microbial ecosystems, in Molecular Microbial Ecology Manual, eds Akkermans A. D. L., Van Elsas J. D., De Bruijn F. J. (Dordrecht: Springer; ), 439–455. [Google Scholar]
- Mohamed M. A., Abdelrazik A. B., Ibrahim S. A.-A. (2014). Identification of different species of Rhodotorula using Internal Transcribed Spacers. Open Sci. Reposit. Agric. e45011822 10.7392/openaccess.45011822 [DOI] [Google Scholar]
- Mohanta T. K., Bae H. (2015). The diversity of fungal genome. Biol. Proced. Online 17:8. 10.1186/s12575-015-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsen M. A. (2015). The Protégé project: A look back and a look forward. AI Matters 1, 4–12. 10.1145/2757001.2757003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemira B. A., Solomon E. B. (2005). Sensitivity of planktonic and biofilm-associated Salmonella spp. to ionizing radiation. Appl. Environ. Microbiol. 71, 2732–2736. 10.1128/AEM.71.5.2732-2736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. e2437. 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Magbanua Z., Arick M., II., French T., Bridges S. M., Burgess S. C., et al. (2014). Genome sequence of the oleaginous yeast Rhodotorula glutinis ATCC 204091. Genome Announc. 2:e00046-14. 10.1128/genomeA.00046-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8, 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Prakash D., Gabani P., Chandel A. K., Ronen Z., Singh O. V. (2013). Bioremediation: a genuine technology to remediate radionuclides from the environment. Microb. Biotechnol. 6, 349–360. 10.1111/1751-7915.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- San Martin-Uriz P., Mirete S., Alcolea P. J., Gomez M. J., Amils R., Gonzalez-Pastor J. E. (2014). Nickel-resistance determinants in Acidiphilium sp. PM identified by genome-wide functional screening. PLoS ONE 9:e95041. 10.1371/journal.pone.0095041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracli M. A., Sener K., Gonlum A., Yildiran S. T., Wickes B. L. (2003). Genotyping of clinical Rhodotorula mucilaginosa isolates by pulsed field gel electrophoresis. Mycoses 46, 487–491. 10.1046/j.0933-7407.2003.00925.x [DOI] [PubMed] [Google Scholar]
- Sharma A., Gaidamakova E. K., Grichenko O., Matrosova V. Y., Hoeke V., Klimenkova P., et al. (2017). Across the tree of life, radiation resistance is governed by antioxidant Mn2+, gauged by paramagnetic resonance. Proc. Natl. Acad. Sci. U.S.A. 114, E9253–E9260. 10.1073/pnas.1713608114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelobolina E. S., O'Neill K., Finneran K. T., Hayes L. A., Lovley D. R. (2003). Potential for in situ bioremediation of a Low-pH, High-Nitrate Uranium-contaminated groundwater. Soil Sediment Contaminat. Int. J. 12, 865–884. 10.1080/10588330390254928 [DOI] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R., Green P. (2013–2015). RepeatMasker Open-4.0. Available online at: http://www.repeatmasker.org/
- Stanke M., Waack S. (2003). Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl. 2), ii215–ii225. 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Okada H., Abe K., Kera Y. (2014). Genetic transformation of the yeast Rhodotorula gracilis ATCC 26217 by electroporation. Appl. Biochem. Microbiol. 50, 624–628. 10.1134/S0003683814110040 [DOI] [Google Scholar]
- Teixeira M. T., Gilson E. (2005). Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 13, 535–548. 10.1007/s10577-005-0999-0 [DOI] [PubMed] [Google Scholar]
- Viti C., Marchi E., Decorosi F., Giovannetti L. (2014). Molecular mechanisms of Cr(VI) resistance in bacteria and fungi. FEMS Microbiol. Rev. 38, 633–659. 10.1111/1574-6976.12051 [DOI] [PubMed] [Google Scholar]
- Zhang S., Skerker J. M., Rutter C. D., Maurer M. J., Arkin A. P., Rao C. V. (2016). Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol. Bioeng. 113, 1056–1066. 10.1002/bit.25864 [DOI] [PubMed] [Google Scholar]
- Zhao X. Q., Aizawa T., Schneider J., Wang C., Shen R. F., Sunairi M. (2013a). Complete mitochondrial genome of the aluminum-tolerant fungus Rhodotorula taiwanensis RS1 and comparative analysis of Basidiomycota mitochondrial genomes. Microbiologyopen 2, 308–317. 10.1002/mbo3.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Liu H., Wang C., Xu J. R. (2013b). Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14:274. 10.1186/1471-2164-14-274 [DOI] [PMC free article] [PubMed] [Google Scholar]