Abstract
IsdB is a receptor on the surface of the bacterial pathogen Staphylococcus aureus that extracts heme from hemoglobin (Hb) to enable growth on Hb as a sole iron source. IsdB is critically important both for in vitro growth on Hb and in infection models and is also highly up-regulated in blood, serum, and tissue infection models, indicating a key role of this receptor in bacterial virulence. However, structural information for IsdB is limited. We present here a crystal structure of a complex between human Hb and IsdB. In this complex, the α subunits of Hb are refolded with the heme displaced to the interface with IsdB. We also observe that atypical residues of Hb, His58 and His89 of αHb, coordinate to the heme iron, which is poised for transfer into the heme-binding pocket of IsdB. Moreover, the porphyrin ring interacts with IsdB residues Tyr440 and Tyr444. Previously, Tyr440 was observed to coordinate heme iron in an IsdB·heme complex structure. A Y440F/Y444F IsdB variant we produced was defective in heme transfer yet formed a stable complex with Hb (Kd = 6 ± 2 μm) in solution with spectroscopic features of the bis-His species observed in the crystal structure. Haptoglobin binds to a distinct site on Hb to inhibit heme transfer to IsdB and growth of S. aureus, and a ternary complex of IsdB·Hb·Hp was observed. We propose a model for IsdB heme transfer from Hb that involves unfolding of Hb and heme iron ligand exchange.
Keywords: crystal structure, heme, hemoglobin, iron, Staphylococcus aureus (S. aureus)
Introduction
Staphylococcus aureus is most commonly found as a member of the normal human flora, colonizing primarily on the hands and in the nostrils (1). However, S. aureus is also one of the main agents of nosocomial infections (2) and can cause a range of diseases in humans, from mild skin infections such as boils and folliculitis, to severe, life-threatening bloodstream infections (3). Establishing infection requires effective iron scavenging systems because iron trafficking is tightly controlled within the human body, both to mitigate the toxicity of free iron and to limit microbial growth. The latter has been referred to as a form of innate immunity called “nutritional immunity” (4). S. aureus overcomes this nutritional immunity by employing two mechanisms to access iron sources: the secretion and uptake of iron-chelating siderophores to acquire iron from host proteins such transferrin and the expression of surface receptors for heme and hemoglobin. However, when presented with both heme and iron-bound transferrin as the only potential iron sources, S. aureus can acquire both forms of iron but preferentially utilizes heme (5).
S. aureus acquires heme using the Isd (iron-regulated surface determinant) system. This system consists of nine components: four surface proteins covalently anchored to the peptidoglycan that reversibly bind heme (IsdA, IsdB, IsdC, and IsdH); an ABC transporter (IsdF) with an associated lipoprotein (IsdE); and two intracellular heme-degrading enzymes (IsdG and IsdI) (6–8). The function of a predicted membrane protein, IsdD, remains unknown. Lastly, sortase B (SrtB) is encoded in a gene cluster with isdCDEFG and functions to anchor IsdC to the peptidoglycan, whereas the remaining Isd surface proteins (IsdABH) are anchored by sortase A, the housekeeping sortase of the cell (7). The expression of all Isd components appears to be classically regulated by the Fur regulator and is repressed in the presence of iron (8). IsdB and IsdH stand apart in the Isd system in that they are the only components capable of binding hemoproteins. IsdH can bind hemoglobin (Hb),2 haptoglobin (Hp), and the Hp·Hb complex. Hp is a high-abundance serum protein that binds free Hb with high affinity but does not, in itself, constitute an iron source. IsdB can bind Hb and the Hp·Hb complex but not Hp alone (9, 10). Thus, heme is stripped from Hb at the bacterial cell surface by IsdB or IsdH and is transferred in a unidirectional relay to IsdE via IsdA and IsdC (11–13). The heme is then imported into the cell by the permease IsdF for degradation by the homologous enzymes IsdG and IsdI to liberate iron for use by the cell (14, 15).
The four cell wall–anchored Isd proteins, IsdABCH, share three features: an N-terminal secretion signal, a C-terminal sortase signal for cell wall anchoring, and one to three copies of a NEAT (for near transporter) domain (16). IsdA and IsdC contain a single NEAT domain, which binds heme. IsdB and IsdH contain two and three NEAT domains, respectively, but only their C-terminal NEAT domains, IsdBN2 and IsdHN3, respectively, bind heme. The N-terminal and central NEAT domains of IsdH, recombinantly expressed individually as IsdHN1 and IsdHN2, are each able to bind Hb, Hp, and Hp·Hb (9, 17); however, the N-terminal IsdB domain alone, IsdBN1, does not bind Hb (18), despite >40% sequence identity between IsdBN1, IsdHN1, and IsdHN2 and the shared ability of the full-length proteins to bind Hb. Both IsdBN1 and IsdBN2 must be present and contiguous with the intervening “linker” region (represented as IsdBN1N2) for high affinity Hb binding. Moreover, IsdBN1N2 removes heme from oxidized Hb, known as methemoglobin (metHb), the form of Hb produced upon red blood cell lysis in the bloodstream. By contrast, IsdBN1N2 does not remove heme from oxyHb, the reduced, oxygen-bound form of Hb that is present in intact red blood cells, demonstrating specificity toward the probable biologically relevant form of Hb encountered by S. aureus during infection (18).
Structures of the Isd proteins have been elucidated by X-ray crystallography or NMR, revealing the mode of heme coordination within NEAT domains (17, 19–25). Recently, crystal structures of various portions of IsdH in complex with Hb have also been reported: IsdHN1·metHb (17), IsdHN2·metHb (19), and a heme transfer–deficient variant of IsdHN2N3 (Y642A) complexed with metHb (19, 26). These complex structures provide insight into the extensive interactions between IsdH and Hb. In these structures, the heme remains largely encapsulated by metHb with minor or no distortions of the αHb heme binding pocket. Therefore, these structures only provide insight into the initial step of the heme transfer process. These insights likely extend to IsdB·Hb, given the similarities between the two surface receptors. Nevertheless, IsdB is more important than IsdH in both in vitro growth on Hb and a mouse abscess model of infection (10, 27). IsdB is also a dominant antigen that is highly up-regulated in blood, serum, and a cage model of tissue infection (28–31), corroborating the key role of this receptor.
Herein, we report the crystal structure of an IsdBN1N2·Hb complex in which the heme is positioned between the two proteins, consistent with an intermediate state of heme transfer. Two point mutations were then introduced into IsdBN1N2 to trap a similar species in solution, which was characterized spectroscopically. In concordance, kinetic analyses of heme transfer revealed a multistep transfer process. Furthermore, we demonstrate that although IsdBN1N2 can bind to the Hp·metHb, as was previously demonstrated for IsdH, it is unable to remove heme from the complex. This finding correlates with our observation that S. aureus did not utilize Hp·Hb as a sole iron source.
Results
Crystal structure of an IsdBN1N2·Hb complex
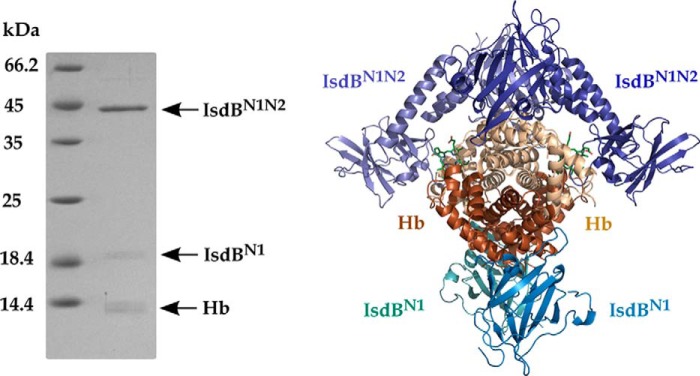
Because IsdBN1N2 does not extract heme from oxyHb (18), this form of Hb was used to obtain an IsdBN1N2·Hb complex for crystallization. Crystals of an IsdBN1N2·Hb complex were grown from solutions of citric acid or malonic acid, pH 5.0–5.5, and 2.1–2.4 m ammonium sulfate. Small crystals (∼0.05–0.1 μm) appeared after 2–4 weeks and contained both proteins (Fig. 1, left panel). A 3.6 Å resolution data set from a single crystal was collected, and the structure was solved by molecular replacement of the individual components. The structure revealed a central Hb a2b2 tetramer surrounded by four IsdB molecules in the asymmetric unit (Fig. 1, right panel). An intact copy of the recombinant IsdBN1N2 construct is bound to each αHb subunit; however, only the first NEAT domain of IsdB (IsdBN1) is observed bound to each βHb subunit. The 19-kDa band observed in the left panel of Fig. 1 is consistent with proteolysis of IsdBN1N2 during crystallization and is presumed to be IsdBN1. Inspection of crystal packing within the unit cell revealed that modeling the missing components of IsdB would result in a large scale steric clash with αHb-bound IsdBN1N2, suggesting that the partial proteolysis allowed for crystallization of the complex.
Figure 1.
The IsdBN1N2·Hb complex. Left panel, components of the IsdBN1N2·Hb crystal separated by SDS-PAGE. Bands of the expected molecular weight of Hb and IsdBN1N2 were detected along with a ∼19-kDa band presumed to be an IsdB fragment. Right panel, crystal structure of the overall IsdBN1N2·Hb complex. IsdB molecules are colored in shades of blue, αHb molecules are colored in beige, and βHb molecules are colored in dark orange. Heme moieties in the αHb chains are shown as green sticks.
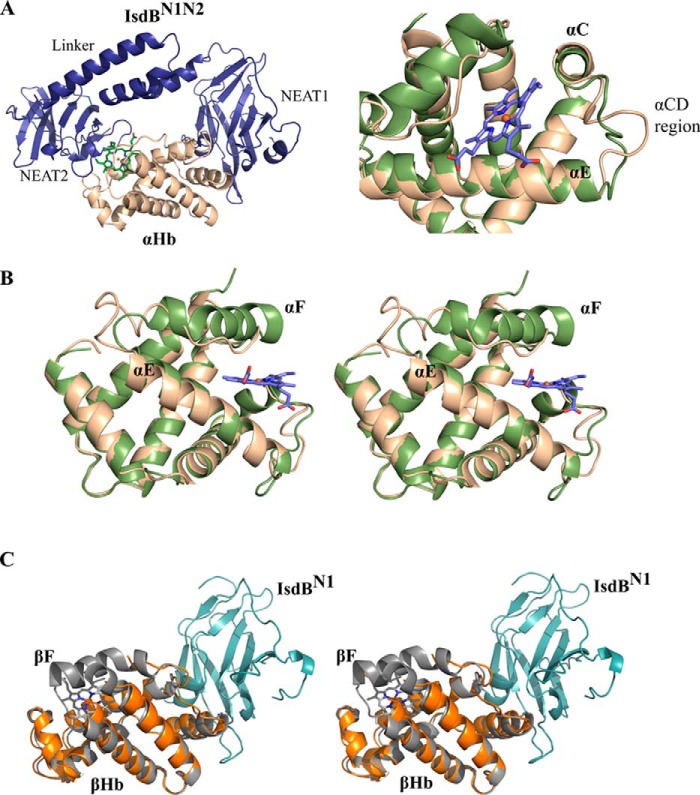
The structure of each IsdBN1N2 molecule resembles a dumbbell with the two NEAT domains joined by an α-helical linker (Fig. 2A, left panel), as was observed for IsdHN2N3 (19, 26). The loop between IsdBN1 and the linker region is flexible, as evidenced by poor electron density; conversely, well ordered density is observed between the linker and the second NEAT domain (IsdBN2), which includes a short 310 helix. The IsdBN1 and IsdBN2 domains of intact IsdBN1N2 each make numerous interactions with αHb (average interface area of 780 and 736 Å2, respectively), and similar interactions are observed for IsdBN1 bound to βHb (634 Å2). No interactions are observed between the linker region and Hb.
Figure 2.
Alterations in Hb polypeptide chain structure in the complex with IsdB. A, left panel, a single copy of αHb (beige) interacting with a single copy of IsdBN1N2 (dark blue). IsdBN2 directly interacts with the Hb heme and heme pocket. The linker is a three-helix bundle between the two NEAT domains. IsdBN1 binds onto the opposite face of αHb. Right panel, the conformation of the αHb C–D loop is significantly altered upon IsdB binding. B, stereo view of the interaction with IsdB, which results in the αHb F helix becoming highly unwound. The αHb chain of oxyHb (PDB code 2DN1; green) is overlaid to demonstrate the original state of the helix. The heme is illustrated in blue sticks with heme iron in red. C, stereo view of the interaction between βHb (orange) and IsdBN1 (cyan). The βHb chain of oxyHb with associated heme (PDB code 2DN1; gray) is overlaid. The orientation of the molecules is similar to that of A. Density for the βHb F helix and heme is absent, as are the IsdB linker and NEAT2 domains.
The Hb chains of the complex described here reveal major structural rearrangements as compared with the structure of isolated oxyHb (PDB code 2DN1). These changes are situated primarily in the two αHb chains of the IsdBN1N2·Hb structure, which superimpose poorly with the equivalent α chains in the free oxyHb structure (r.m.s.d. of 3.3 and 3.4 Å over all Cα) as compared with alignments between the βHb chains (r.m.s.d. of 1.4 and 1.3 Å over all Cα). In the α subunit of free human oxyHb, heme is bound in a pocket between the E and F helices, with His87 (F helix) coordinating directly to the heme iron and the distal His58 (E helix) forming a hydrogen bond to the heme-bound dioxygen molecule. To our knowledge, the heme iron axial residue is αHis87 in all previously reported αHb crystal structures regardless of the gaseous heme ligand. Upon binding to IsdBN1N2, a major deformation of the αHb heme pocket occurs, with the F helix and part of the E helix of each αHb chain (encompassing residues αAsp74 to αArg92) unwinding entirely (Fig. 2B). Clear electron density in this region indicated that heme (modeled at full occupancy) remains bound to αHb (Fig. 3A) but has been displaced 5 Å toward the heme-binding pocket of IsdBN2. This heme displacement was accompanied by the unexpected direct coordination of the heme iron by αHis58 and αHis89 (Fig. 3B). Through reorganization of the polypeptide chain, αHis87 has pivoted out of the heme pocket and points away from the heme iron (Nϵ2 translation of 13.4 Å). In its place, αHis89 (originally on the F helix) has moved from a solvent-exposed position to coordinate to the heme iron (13.2 Å translation in Nϵ2). Binding to IsdBN1N2 also resulted in the extension of the C-terminal H helix by another half turn, such that αTyr140 occupies the void left by the unwound F helix, with an accompanying Cα shift of 7.5 Å and the placement of the side chain phenol group within 3.2 Å from Nϵ2 of αHis87. Eleven residues between helices C and D (αTyr42 to αSer52) have also rearranged to accommodate the interaction with IsdBN1N2 (Fig. 2A, right panel). For the β chains of Hb, insufficient density was present to model most of the F helix (βLeu88 to βVal98 in chain B and βPhe85 to βVal98 in chain D), and no bound heme was observed (Fig. 2C). Otherwise, minimal differences were observed between the β chains of free and IsdB-complexed Hb.
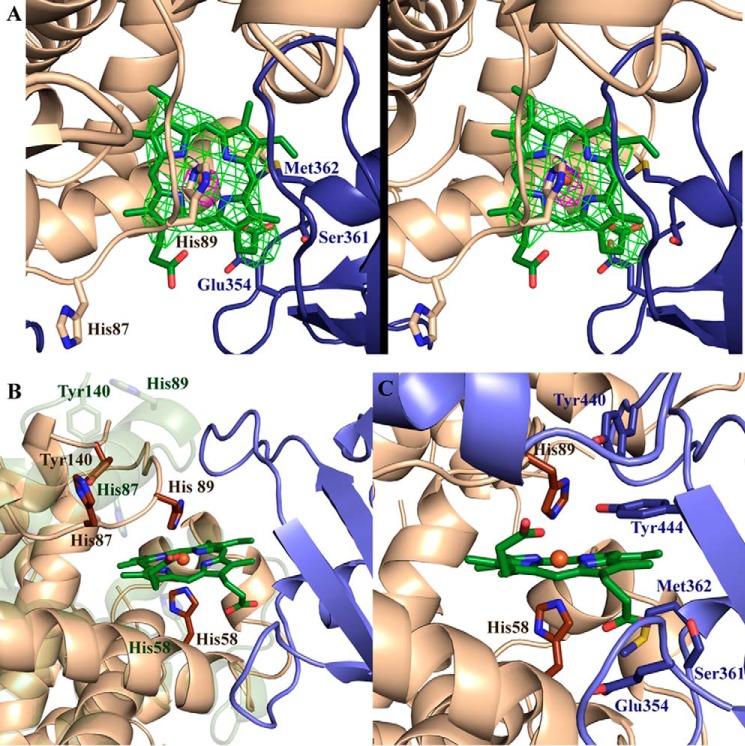
Figure 3.
Binding of IsdBN1N2 to Hb induces major changes in heme environment. A, stereo view of an Fo − Fc omit positive difference map (green) of the heme in the IsdBN1N2·αHb interface contoured at 3σ (refined with torsion-based simulated annealing). An anomalous density map (pink) contoured at 3σ is overlaid to unambiguously support correct placement of the heme. B, the complex structure is overlaid with the structure of the αHb chain from the oxyHb structure (PDB code 2DN1). The 2DN1 αHb structure is shown in green; αHb and IsdBN1N2 from the IsdBN1N2·Hb structure are shown in beige and blue, respectively. Relevant amino acid side chains are shown as sticks. Only the heme moiety from the structure presented here is shown (dark green). C, the IsdB heme pocket is positioned to accept αHb-heme. IsdB and αHb residues situated at the binding interface near the heme are shown as blue and beige sticks, respectively. The remainder of the structure is shown as cartoons. The αHb-heme is shown in green, with the heme iron (coordinated by αHis58 and αHis89) shown as an orange sphere.
The IsdBN2 domain interacts with the heme-binding site of αHb and superposed well with the structure of free IsdBN2 (20) (r.m.s.d. of 1.4 Å over 109 Cα). However, the β7-β8 loop (Val435 to Tyr440), which coordinates heme iron via Tyr440, is curved inward in the complex structure, making contacts with αHb, and is poised to receive the heme molecule (Fig. 3C). Tyr440 closely abuts αHis89 of Hb, whereas Tyr444 is positioned adjacent to the heme pyrrole ring, forming a π-stacking network of interactions. Interestingly, Tyr440 is positioned further back than Tyr444, suggesting that Tyr444 plays a role beyond simply stabilizing the position of Tyr440 in the heme pocket, as was previously proposed (20), and appears to be required for the heme transfer process. The second heme iron–coordinating residue of holo IsdBN2, Met362, has poor electron density in the complex structure and is likely conformationally flexible. Conversely, the IsdBN2 propionate-binding residue Ser361 remains engaged in hydrogen-bonding with the propionate, with Glu354 also participating in this interaction. The second heme propionate is not observed to form H-bonds to either Hb or IsdBN1N2.
Proteolyzed IsdBN1 (chains J and H) interact with βHb and are structurally similar to the IsdBN1 domains in intact IsdBN1N2 (chains E and F), which interact with αHb in the crystal structure (r.m.s.d. of 0.9–1.2 Å over 133 Cα). Minor differences are observed mainly in the β2-β3 and β7-β8 loops (numbered according to IsdAN1 (21)) and at the N and C termini. However, Hb-bound IsdBN1 differed substantially from the solution structure of IsdBN1 (25) (r.m.s.d. of ∼2.3 Å over all Cα of model 1). One significant structural difference was within the four-residue aromatic motif that is important for Hb-binding by IsdB and IsdH (27, 32). This motif, FYHY in IsdB (residues 164–167), was disordered in the solution structure but formed a short α-helix in the Hb-IsdBN1N2 structure. This α-helix forms a close contact with Hb, with Phe164 closely abutting αTrp14/βTrp15 in a T-shaped π-stacking interaction.
An IsdB heme pocket variant traps a bis-His metHb
Although the IsdBN1N2·Hb structure clearly showed the Hb-heme in a bis-His coordination state, a bis-His state involving the native proximal and distal heme ligands was previously observed in crystals of horse metHb grown at pH 5.4 as compared with pH 7.1 (33). Although the heme ligands differ, the low pH of our IsdBN1N2·Hb crystallization solution could have similarly induced bis-His coordination. However, incubation of metHb or oxyHb at pH 5.5 overnight, with or without IsdBN1N2, did not produce spectral changes associated with formation of a bis-His state (data not shown). Nonetheless, the observation of conformational changes in horse metHb to give a bis-His coordination state is a precedent for the conformational rearrangements observed in the IsdBN1N2·Hb structure.
A double mutant of IsdBN1N2 (IsdBYFYF) was created to prevent heme transfer by replacing Tyr440 (heme iron coordinating) and Tyr444 (H-bonds to Tyr440) with Phe residues in the heme pocket. In the complex structure, these two tyrosine residues are also juxtaposed to αHis89 of Hb, the heme-coordinating residue that is part of the unwound F helix. Upon addition of free heme to apo-IsdBYFYF, the spectra displayed a Soret peak at 403 nm, slightly blue-shifted relative to holo-IsdBN1N2, which peaks at 405 nm (Fig. 4A). The α/β region of the IsdBYFYF+heme spectrum was nearly featureless with a minor peak at ∼600 nm, similar to the spectrum of free heme in that region (Fig. 4B).
Figure 4.
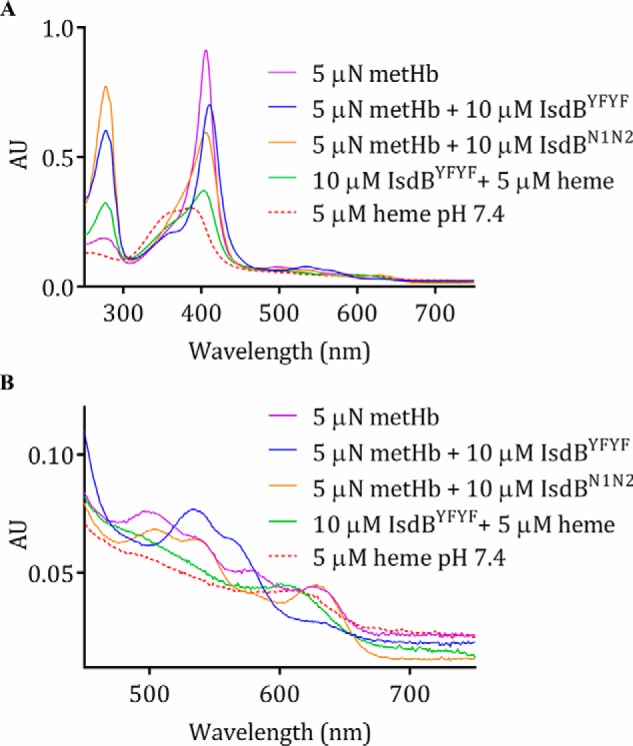
Electronic spectra of IsdBYFYF combined with heme and Hb. A, addition of excess IsdBYFYF to metHb results in a distinctly altered spectrum (blue); the heme spectrum alone (red), metHb spectrum alone (purple), the mixture of IsdBYFYF with heme (green), and reaction of wild-type IsdBN1N2 with metHb (orange) are shown for comparison. AU, absorbance units. B, a closer look at the visible region of the spectra presented in A.
As previously reported (18), the addition of IsdBN1N2 to metHb resulted in a large-scale shift in the intensity and shape of the Soret peak as heme was transferred, with minor changes in peak intensities in the visible region (Fig. 4A). Visible region peak wavelengths remained at ∼500 and 535 nm, with a charge-transfer band at 630 nm, indicating that the heme was a high-spin ferric species in both holo-IsdBN1N2 and metHb (34). Addition of IsdBYFYF to metHb also caused a large-scale shift in the Soret peak; however, the spectra was distinct from that formed upon heme addition to IsdBYFYF (Fig. 4A). The Soret peak became red-shifted, to 410 nm, and a distinct shoulder developed at ∼360 nm. Moreover, dramatic changes occurred in the α/β region (Fig. 4B); the charge-transfer band at 630 nm disappeared, and the highest absorption peak shifted to 533 nm with a shoulder peak at 564 nm. This spectrum is nearly identical to that of a ferric, low-spin, bis-His hemichrome form of human Hb observed when incubating purified α- or β-globin with heme (but not for heme addition to whole globin) (34). The change to a characteristic bis-His hemichrome spectrum upon addition of IsdBYFYF to 4 μn metHb was titratable and saturable (Fig. 5, A and B). A plot of the absorption change at a single wavelength (410 nm) as a function of added IsdBYFYF plateaued at ∼5 μm, indicating the stoichiometry of the reaction was ∼1:1 (Fig. 5C), as seen for interaction of IsdBN1N2 and Hb in the crystal structure.
Figure 5.
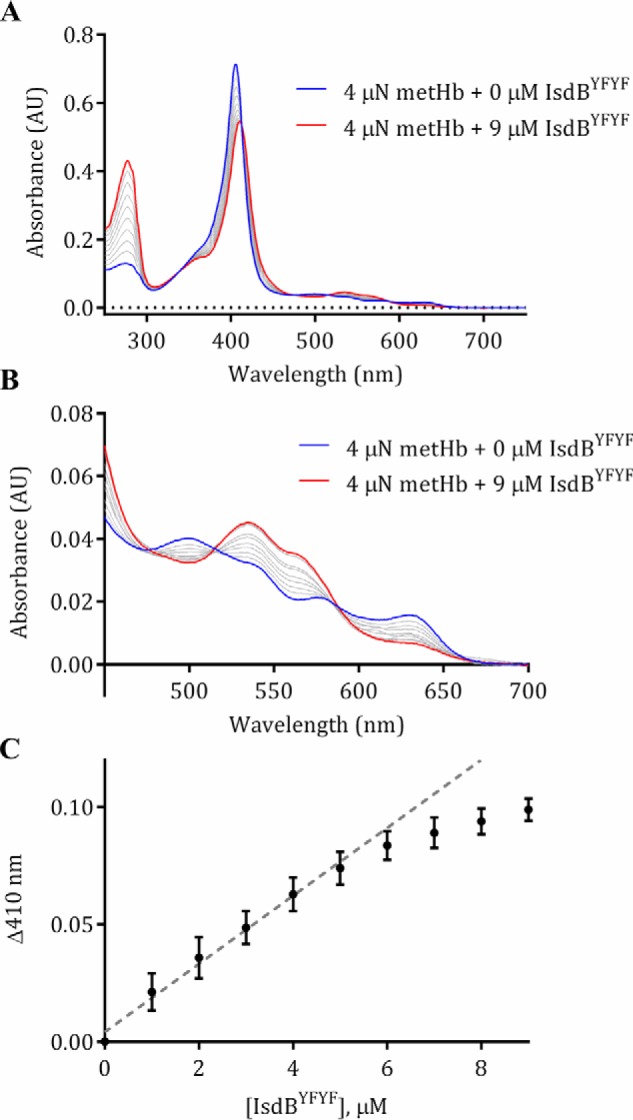
Titration of metHb with IsdBYFYF resulted in dose-dependent, saturable changes in electronic spectra. 4 μn metHb was titrated with increments of 1 μm of apo-IsdBYFYF from 1 to 9 μm at 22 °C; spectral changes were monitored in a conventional spectrophotometer. Spectra shown are the average of three independent replicates. A, overall spectral changes accompanying the titration of IsdBYFYF into metHb. The spectrum of metHb alone is shown in blue, the final titration spectrum is shown in red, and each gray line represents an intermediate titration spectrum in 1 μm increments. AU, absorbance units. B, expansion of spectra in the α/β region of the spectra shown in A. C, the change in absorption at 410 nm plotted against the concentration of IsdBYFYF for each titration point. Each point represents the mean and standard error of three replicates. The dotted line represents a linear fit to the first five titration points to indicate the concentration of IsdBYFYF where a plateau begins.
The binding of IsdBYFYF to metHb was confirmed by ITC. Titration of metHb into IsdBYFYF resulted in an exothermic reaction as observed by the negative change in enthalpy (Fig. S1). Analysis of the data with a one-site model gave a Kd of 6 ± 2 μm and a stoichiometry (N) of ∼0.5 (average of three runs), implying two IsdBYFYF molecules bound to one metHb monomer. Because Hb has two unequal subunits that may interact with IsdBN1N2 differently, the Kd and stoichiometry measurements are assumed to be the average of binding to αHb and βHb. A previous ITC study between IsdBN1N2 and carboxyhemoglobin reported a Kd of 0.42 ± 0.05 nm (18). The weaker interaction of IsdBYFYF for metHb may be due to a conformational change in the structure of metHb analogous to that observed in the IsdBN1N2·Hb crystal structure.
Rapid heme transfer from metHb to IsdBN1N2
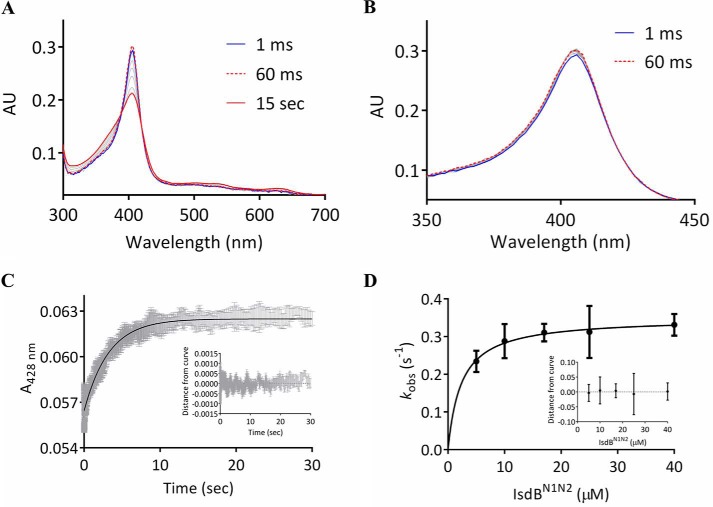
To identify potential intermediates in the heme transfer pathway, the kinetics of heme transfer from metHb to IsdBN1N2 was investigated by stopped-flow spectroscopy. Mixing of 2 μn metHb with 20 μm IsdBN1N2 resulted in spectral changes between 180 and 730 nm that were complete within 10 s, consistent with previous reports using a full-length IsdB construct (11). Coincident with heme transfer, the Soret peak underwent a large shift in intensity, particularly between ∼350 and 420 nm, along with spectral changes in the α/β region (Fig. 6A). Although the intensity of the Soret absorption peak increased during the first ∼60 ms (Fig. 6B), the overall effect was a reduction in intensity with broadening of the Soret band.
Figure 6.
Heme transfer kinetics from metHb to IsdBN1N2. A and B, electronic spectra collected with a stopped-flow spectrophotometer equipped with a photodiode array. A, spectra recorded over 15 s after mixing of 2 μn metHb with 20 μm IsdBN1N2. B, an increase in the Soret peak was observed over the first ∼60 ms, from ∼350–420 nm. AU, absorbance units. C, a representative single-wavelength (428 nm) stopped-flow spectroscopy experiment where 1 μn metHb was mixed with 17 μm IsdBN1N2. The gray bars represent the standard error of four replicates, and the black curve is a fit to a single exponential equation. The residuals for the experiment are in the inset. D, the observed transfer rate (kobs) from 1 μn metHb is plotted as a function of IsdBN1N2 concentration. The line is a hyperbolic fit assuming a two-step reaction model. Each point represents the mean, and the bars are the standard errors of four replicates. The residuals of the data to the model are in the inset.
Two wavelengths (406 and 428 nm) were chosen for single-wavelength stopped-flow spectroscopy under pseudo-first order conditions, with metHb held constant at 1 μn and IsdBN1N2 increasing from 5 to 40 μm. The kinetics at 428 nm were simpler (Fig. 6C), because this wavelength was outside the range that increased in the first 60 ms (∼350–420 nm). A single exponential fit yielded observed rate constants (kobs) that varied hyperbolically with IsdBN1N2 concentration (Fig. 6D), consistent with a two-step heme transfer mechanism (20) with a rate constant for heme transfer from metHb to IsdBN1N2 of 0.35 ± 0.02 s−1. At 406 nm, the kinetics were more complex. Curve fitting yielded four phases with differing amplitudes, rates, and concentration dependences (Fig. S2). In the first phase only, the rate was linearly dependent on IsdBN1N2 concentration (Table 1; see also Fig. S2C) and thus reflected concentration dependent collision events. Phases 2–4 did not display strong concentration dependence in their rates (Fig. S2, D–F), suggesting that they are associated with steps in the heme transfer process after IsdBN1N2·metHb complex formation.
Table 1.
Kinetics of heme transfer from metHb to IsdBN1N2 at 406 nm
The amplitude is given as a fractional quantity. All values represent the means and standard error of four replicates.
| IsdBN1N2 |
|||||
|---|---|---|---|---|---|
| 5 μm | 10 μm | 17 μm | 25 μm | 40 μm | |
| k1 | |||||
| kobs (s−1) | 11 ± 2 | 25 ± 2 | 32 ± 3 | 49 ± 7 | 108 ± 8 |
| Amplitude | 0.15 ± 0.02 | 0.13 ± 0.01 | 0.14 ± 0.01 | 0.12 ± 0.01 | 0.124 ± 0.003 |
| k2 | |||||
| kobs (s−1) | 1.9 ± 0.2 | 2.1 ± 0.2 | 5 ± 1 | 4 ± 1 | 4 ± 1 |
| Amplitude | 0.16 ± 0.02 | 0.19 ± 0.02 | 0.16 ± 0.02 | 0.17 ± 0.01 | 0.16 ± 0.02 |
| k3 | |||||
| kobs (s−1) | 0.41 ± 0.03 | 0.42 ± 0.05 | 0.5 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.1 |
| Amplitude | 0.48 ± 0.06 | 0.48 ± 0.06 | 0.47 ± 0.07 | 0.45 ± 0.07 | 0.50 ± 0.06 |
| k4 | |||||
| kobs (s−1) | 0.12 ± 0.02 | 0.13 ± 0.03 | 0.14 ± 0.04 | 0.15 ± 0.03 | 0.13 ± 0.04 |
| Amplitude | 0.20 ± 0.03 | 0.21 ± 0.06 | 0.22 ± 0.08 | 0.26 ± 0.08 | 0.22 ± 0.08 |
Hp prevents heme transfer from metHb to IsdBN1N2
Hp is a serum α2-sialoglycoprotein that binds metHb released from lysed erythrocytes to prevent oxidative damage (35, 36). The fundamental unit of human haptoglobin is a polymorphic αβ dimer composed of light chains (α) and heavy chains (β). The β chain is encoded by a single allele, whereas the α chains come in two forms: α1 and α2 (36). These alleles generate three possible phenotypes. Phenotype 1-1 (where both copies of the α gene are α1) is the simplest, with Hp forming a homodimer of two αβ heterodimers. Phenotypes 2-1 and 2-2 can form heterogeneous cyclical or linear multimers of increasing size (37). Within the human host, the normal plasma Hb concentration is less than 5 μn, whereas Hp is generally present in the range of 0.3–2 mg/ml (35) or roughly 6–50 μm depending on the phenotype.
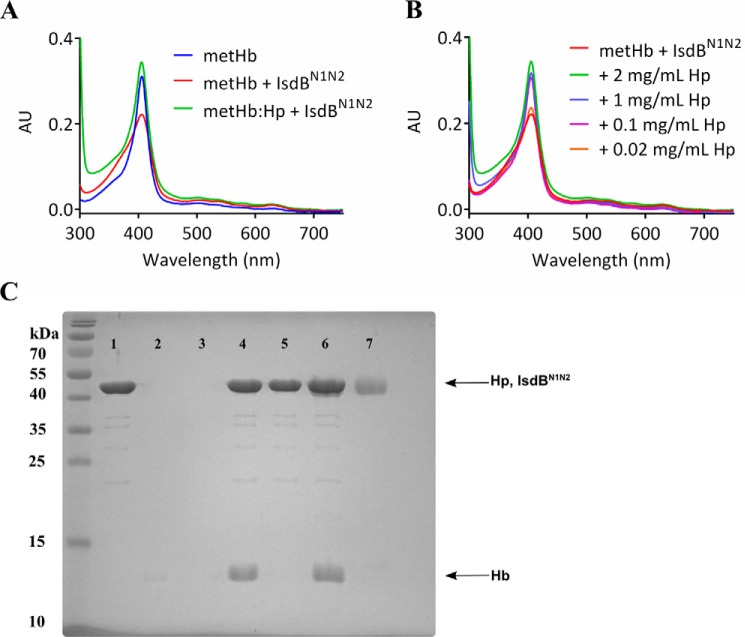
Binding of metHb by Hp has been proposed to make heme inaccessible to microbial pathogens (38). Therefore, to investigate the effect of Hp on heme uptake by IsdB from Hb, excess IsdBN1N2 was added to metHb preincubated with increasing concentrations of human Hp. A pooled mixed phenotype Hp was used to model normal human serum; however, the exact molar concentration was unknown. Therefore, Hp was used at the high end of the normal range for Hp serum concentration (2 mg/ml). Preincubation of 2 μn metHb with 2 mg/ml Hp resulted in electronic spectra that remained unchanged for 5 min after the addition of IsdBN1N2 (Fig. 7A). Only by decreasing the Hp concentration to 0.02 mg/ml or lower was a spectral change in the Soret region observed, implying heme transfer similar to that observed in the absence of Hp (Fig. 7B). Thus, Hp effectively blocked heme transfer from metHb to IsdBN1N2. Inhibition was not due to the inability of IsdBN1N2 to bind metHb when the latter is complexed to Hp because IsdBN1N2 was able to pull down metHb in both the presence and absence of Hp but did not interact with Hp alone (Fig. 7C). To elucidate the stoichiometry of the Hp inhibition, the heme transfer assay was repeated using Hp phenotype 1-1, which revealed a stoichiometric ratio of approximately one Hp 1-1 molecule (consisting of two αβ subunits) to one a2b2 metHb tetramer (Fig. S3).
Figure 7.
Inhibition of heme transfer from metHb by Hp. A, preincubating 2 μn metHb (blue) with 2 mg/ml mixed-serotype Hp resulted in metHb-like spectra (not shown). The addition of 10 μm IsdBN1N2 (green) resulted in a modest increase in the Soret peak. The reaction of 2 μn metHb with 10 μm IsdBN1N2 (in the absence of Hp) is shown for comparison (red). AU, absorbance units. B, 2 μn metHb was preincubated with decreasing amounts of Hp, as indicated, followed by the addition of 10 μm IsdBN1N2. Spectra for each reaction were recorded within 20 s of mixing and did not change within 5 min. C, SDS-PAGE separation of nickel-nitrilotriacetic acid bead pulldown of His6-IsdBN1N2, Hp, and metHb. 20 μm His6-IsdBN1N2 was used as bait to pull down 20 μn metHb and/or ∼20 μm Hp. His6-IsdBN1N2 could bind nickel beads alone (lane 1), whereas metHb and Hp could not (lanes 2 and 3, respectively). His6-IsdBN1N2 pulls down metHb (lane 4), but not Hp (lane 5). When metHb is added to nickel beads mixed with His6-IsdBN1N2 and Hp, all three species are pulled down (lane 6). 1 μg of Hp is shown in lane 7, for reference. Although Hp runs at nearly the same position on the gel as His6-IsdBN1N2, two separate bands in lane 6 are distinguished, largely because of their differential staining (Hp is glycosylated, affecting staining by Coomassie dye).
S. aureus growth on Hb·Hp as sole iron source
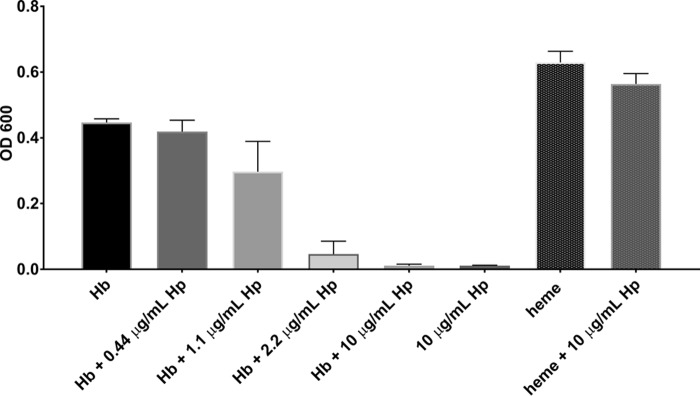
To examine the effects of Hp on Hb-heme utilization in vivo, the growth of S. aureus strain Newman was evaluated in iron-depleted RPMI media. As expected, S. aureus was able to grow in medium supplemented with 2 μn heme iron or 0.2 μn oxyHb as a sole iron source (Fig. 8; see also Fig. S4). A higher concentration of heme was required because 0.2 μn heme did not support growth of S. aureus (data not shown). OxyHb is expected to rapidly oxidize to metHb in culture. Assuming one Hp αβ dimer binds to one dimer of Hb, growth on Hb with the addition of 0.44–10 μg/ml Hp reduced the growth of S. aureus in a concentration-dependent manner. Conversely, addition of Hp to medium supplemented with heme did not reduce the growth of S. aureus, implying that Hp had a Hb-specific inhibitory effect.
Figure 8.
Growth of S. aureus for 16 h on iron-depleted RPMI medium supplemented with 200 nn Hb or 2 μn heme as the sole iron sources. Increasing concentrations of Hp inhibited growth of S. aureus on medium supplemented with Hb but not on heme. Each bar is the average of three independent growth experiments conducted on a Bioscreen C, each with three technical replicates.
Discussion
IsdB is the major Hb receptor functioning at the interface between the bacterial cell surface and the extracellular environment. The heme extraction function of IsdB is supported by the observed growth deficiency of S. aureus on nanomolar concentrations of Hb as a sole iron source upon deletion of IsdB (27) and more directly by heme transfer assays between metHb and IsdB (18). In contrast, IsdBN1N2 was unable to extract heme from oxyHb within hours of incubation, and this form of Hb was used in crystallization trials to obtain the structure of a complex between the two proteins.
Crystals of IsdBN1N2·Hb formed slowly over 2 weeks, sufficient time for oxyHb to be oxidized to metHb. Indeed, the heme groups from the βHb subunits are no longer observed in the complex structure and presumably were extracted by IsdBN1N2 that was subsequently degraded, leaving only the IsdB-N1 domain. The conformations of the αHb subunits in the complex with IsdBN1N2 are a large departure from all previously published structures of Hb. The large distortion of the polypeptide chain and the bis-His heme coordination state are accompanied by a shift of the heme from Hb to the interface with IsdB. The structure suggests that heme is removed from βHb before αHb, which may be a consequence of the kinetics or thermodynamics of crystal formation. The observed bis-His form of Hb observed in the crystal structure may be an artifact of crystallization and represent an off-path species. Attempts to identify this putative intermediate in solution by stopped-flow kinetics have not yet met with success, possibly because the putative intermediate is short-lived. However, spectroscopic analysis of the IsdBYFYF variant mixed with metHb supports the trapping of a bis-His heme complex in solution, thereby supporting the possible existence of a bis-His heme intermediate in the heme transfer pathway by the wild-type protein.
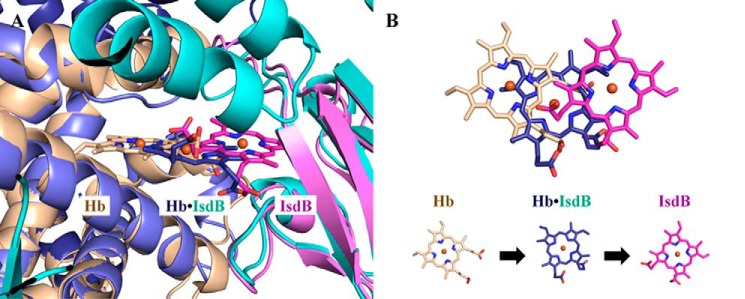
An attractive model of heme transfer from Hb to IsdB is suggested when the IsdBN1N2·Hb complex structure is overlaid with the structures of oxyHb (PDB code 2DN1; α chain) and holo-IsdBN2 (PDB code 3RTL) (Fig. 9A). In the IsdBN1N2·Hb structure presented here, the heme moiety is midway between the Hb and IsdB heme pockets (heme iron is ∼5 Å from either position). The motion of the heme is not solely translational, as one of the propionate groups in the complex is in nearly the same position as observed in the holo-IsdBN2 structure (Fig. 9). Instead, H-bond interactions between the propionate and both Ser361 and Glu354 appear to anchor the heme as it rotates ∼90° from the Hb heme pocket to the IsdB heme pocket, providing the most parsimonious route of heme transfer between the start and end states (as represented by the uncomplexed structures). Interestingly, the structures of oxyHb and our complex may also provide insight into the ambiguity observed in the electron density for the heme methyl/ethyl groups in the previously solved structure of holo-IsdBN2, which resulted in two possible heme conformations while maintaining the position of the propionate groups (20). Because it is unlikely that the heme face can flip over during transfer, one can argue that heme conformation A, as shown in Fig. 9B, is the biologically relevant heme binding mode when transferred from Hb.
Figure 9.
A model of the heme extraction pathway. A, heme positions observed in αHb, the IsdBN1N2·Hb complex, and isolated IsdB. The αHb chain of oxyHb (PDB code 2DN1; beige) overlaid on top of the complex αHb (dark blue) is used to represent the pretransfer heme position in uncomplexed, folded Hb. The heme-bound form of IsdBN2 (PDB code 3RTL heme conformation A; pink) overlaid on top of the complex IsdBN2 domain (cyan) represents the completed heme transfer reaction. B, top-down view of the positional changes of the heme molecule shown in A. The heme iron in the complex structure (dark blue) is ∼5 Å away from both the initial and final heme iron positions.
The interaction between IsdBN1 and βHb may represent an interaction after a successful heme transfer event, because the βHb heme and the IsdB NEAT1 and linker domains are not present. Additionally, helix F of βHb, which contains βHis92 (equivalent to axial heme-coordinating His87 of αHb) and βHis97, are disordered in the absence of bound heme. βHis97 is a single turn helix away from the equivalent position of αHis89 and may participate in the formation of a similar bis-His conformation in βHb. For the transfer to occur, βHb heme must have oxidized because IsdBN1N2 cannot remove ferrous heme from Hb (18). This slow oxidation is expected because the crystals of the complex took weeks to form. Subsequent to the oxidation, the heme is rapidly transferred to IsdBN2, and the flexible loop between IsdBN1 and the linker is cleaved to allow for growth of the crystal. Interestingly, under physiological conditions, αHb oxidizes seven to ten times more quickly than βHb, especially at acidic pH (our crystals were produced at pH 5.5) (39, 40). The observation of heme removal from βHb in the crystal structure may be explained by the differential positioning of αHis89 and βHis97 in αHb versus βHb, respectively, which may be a rate-determining factor in the heme uptake mechanism. The asynchronous heme extraction from αHb and βHb observed in the crystal structure is mirrored by the kinetic analysis of heme transfer monitored at 408 nm. The four distinct kinetic phases observed may correspond to steps in heme extraction from the each Hb domain; however, alternative models are possible because of the presence of different subunits and the complex allosteric nature of Hb. The complexity of the heme transfer kinetics is expected for a substrate (Hb) that is a tetramer with two non-equivalent sites where the rate of heme extraction and the spectral change would vary with the heme site and the number of heme molecules bound to the tetramer. This complexity may also be hindering our attempts to observe spectral features of the putative bis-His intermediate. Nonetheless, the rates observed are similar or faster than the overall rate of heme transfer observed under steady-state conditions with IsdA as the ultimate heme recipient (18).
Superposition of the αβ tetramer of Hb in complex with IsdBN1N2 upon the tetramers of oxyHb (PDB code 2DN1), deoxyHb (PDB code 2DN2), or metHb (PDB code 3P5Q) revealed an unusual quaternary structure. The IsdBN1N2-bound tetramer exhibited neither a T-like (deoxy) nor an R-like (oxy or met) state conformation. This quaternary structure was also observed in a crystal structure of a heme-transfer deficient mutant of IsdHN2N3 in complex with metHb (PDB code 4XS0) (26). However, the overall IsdHN2N3·metHb structure is more similar to that of oxyHb (αHb, r.m.s.d. of 1.2 Å; βHb, r.m.s.d. of 0.5 Å) than our IsdBN1N2·Hb structure and oxyHb (αHb, r.m.s.d. of 3.3–3.4 Å; βHb, r.m.s.d. of 1.3–1.4 Å). Dickson et al. (26) noted that the Hb dimer in IsdHN2N3·metHb structure was most similar to structures of unusual liganded hemoglobins in a T-like state, such as a human sickle-cell variant of embryonic Hb. Our structure of IsdBN1N2·Hb supports the hypothesis that distortion of the Hb quaternary structure into a liganded T-like state is a characteristic of the Hb-binding and heme-extraction process by Isd proteins.
Hp is present at 10 mg/ml in normal serum (37) and has a high affinity for dimeric Hb (Kd = ∼10−15 m (41)). Therefore, metHb present in serum is likely to exist as a complex with Hp. We have shown that heme transfer from metHb to IsdBN1N2 is inhibited by Hp, despite the ability of IsdB to bind Hp·Hb (9). In fact, under the conditions tested, heme transfer only occurred when the concentration of mixed-serotype Hp was decreased to 20 μg/ml, or ∼500-fold lower than the concentration of Hp in human serum. The data presented here are consistent with a recent study demonstrating Hp inhibition of heme transfer from metHb to a similar IsdBN1N2 construct but not to an analogous IsdH construct (42). Moreover, in contrast to IsdH, IsdB did not inhibit binding of Hb-Hp to CD163 (42), the macrophage receptor for recycling Hp·Hb complexes (43).
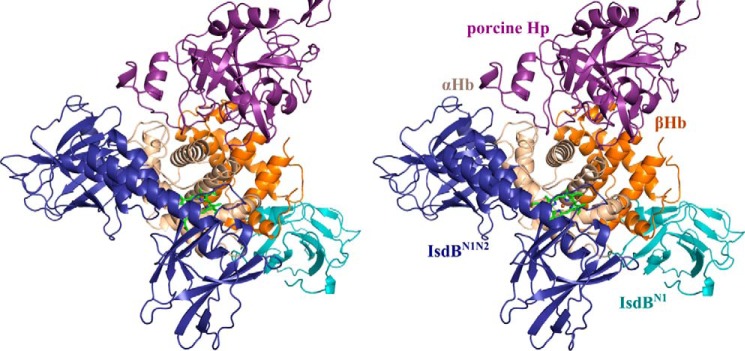
Binding of Hp to Hb has also been shown to stabilize the heme–Hb interaction (44). A structural alignment of our IsdB·Hb crystal structure with the porcine (PDB code 4F4O) and human (PDB code 4X0L) Hp·Hb structures revealed no steric clashes between IsdBN1N2 and Hp (Fig. 10). Thus, inhibition of heme uptake from the Hp·Hb complex may reflect modulated stability of heme in the Hb pocket rather than direct blocking of the IsdB binding site by Hp. More specifically, examination of the Hp·Hb structures reveals a Hp loop (Pro327–Gly329 in human Hp; Pro268–Gly270 in porcine Hp) that interacts with F helix at Pro78 of αHb. This interaction needs to be disrupted to unwind the F helix for subsequent heme transfer to IsdB.
Figure 10.
A stereo overlay of the Hp·Hb structure with the IsdBN1N2·Hb structure. Shown is superposition of porcine Hp·Hb (PDB code 4F4O) with one of the α/β Hb dimers of the IsdBN1N2·Hb structure. Hp binds at a distinct site from IsdB. Hp is shown in purple, and IsdBN1N2 and IsdBN1 are shown in dark blue and light blue, respectively. The αΗβ and βHb subunits are shown in beige and orange, respectively, with the αHb heme shown as bright green sticks.
The IsdBN1N2 construct could possibly lack regions required to remove heme from Hp·Hb complexes in vitro. However, S. aureus was not able to use Hb·Hp as a sole iron source, whereas nanomolar concentrations Hb alone supported S. aureus growth (Fig. 8). Previous studies have indicated that S. aureus can grow on Hb·Hp as sole iron source (45, 46); however, the quality of Hb was lower, and the concentration used in the growth medium was much higher than the more physiologically relevant concentration used here. In the work of Francis et al. (46), cells were grown in iron-rich tryptone and yeast extract, conditions under which the expression of the Isd system is repressed, and iron uptake was measured based on radioactivity of the cell pellet, a method that cannot distinguish between surface-bound and internalized iron. Recent studies have highlighted the importance of using human Hb from donors over commercial lyophilized Hb because of contamination with free heme (27, 47). During infection by S. aureus, hemolysin production and local hemolysis may be required to sufficiently increase the Hb concentration, allowing for excess Hb not bound to Hp.
Experimental procedures
Cloning
Plasmids were generated encoding residues 126–459 of IsdB with an N-terminal His6 tag and a thrombin cleavage site in pET28a(+)as previously described (18). Briefly, the IsdBN1N2 construct was subcloned from a GST-tagged construct cloned from S. aureus N315 chromosomal DNA (20) using a modified whole plasmid polymerase chain reaction method (48). A double variant of IsdBN1N2 was created by subcloning from the IsdBN1N2 clone using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). All clones were verified by sequencing (Agencourt, Beverly, MA).
Protein expression and purification
Recombinant protein was overexpressed in Escherichia coli BL21 (DE3) cells. A 2-liter bacterial culture was grown from 2 ml of overnight culture in LB broth supplemented with 25 μg/ml kanamycin at 30 °C to an A600 nm of 0.7–0.9 and then induced with 0.5 mm of isopropyl β-d-thiogalactopyranoside and grown for another 18 h at 25 °C. The cells were pelleted by centrifugation; resuspended in 20 ml of 50 mm Tris, pH 8.0, 100 mm NaCl; and then lysed at 4 °C using an EmulsiFlex-C5 homogenizer (Avestin, Ottawa, Canada). Insoluble material was removed by centrifugation; the soluble lysate contained a mixture of apo and holo His6 protein, and apo protein could be separated at 4 °C using a HisTrap nickel affinity column (GE Healthcare) by elution with an imidazole gradient. The apo protein was dialyzed against 50 mm Tris, pH 8.0, 100 mm NaCl and then cleaved with thrombin at a 1:500 ratio by weight of His6 protein to remove the His6 tag, leaving behind a two amino acid (Gly-Ser) N-terminal artifact. Recombinant protein was then dialyzed against 20 mm HEPES, pH 7.4, for cation exchange chromatography using a Source 15S column (GE Healthcare), and purified protein was obtained by elution with a NaCl gradient. The resulting pure (>95% by SDS-PAGE) apo protein was dialyzed against 20 mm HEPES, pH 7.4, 80 mm NaCl for all studies. Hemoglobin was prepared from fresh human blood as described previously.
Stopped-flow spectroscopic analysis of heme transfer from metHb to IsdB
An SX.18MV stopped-flow reaction analyzer (Applied Photophysics Ltd., Leatherhead, UK) was used to investigate the possibility of heme-coordination intermediates in the transfer process between metHb and IsdBN1N2. The temperature of the optical cell and drive syringe chamber was maintained at 25 °C using a circulating water bath. Multiple wavelength data from 180 to 730 nm were collected using the photodiode array detector of the system directly coupled to the xenon light source (the practical range was from 250 to 730 nm). 20 μm apo-IsdBN1N2 and 2 μn metHb samples were in a buffer composed of 20 mm NaH2PO4, pH 7.4, 50 mm NaCl. Each acquisition was 15 s long with spectra collected at logarithmic intervals; four acquisitions were averaged.
A monochromator was used to collect single-wavelength data at 406 and 428 nm for the reaction of metHb and IsdBN1N2 in 20 mm HEPES, pH 7.4, 80 mm NaCl. The reactions were carried out with 2 μn metHb in one syringe (final concentration, 1 μn) and concentrations of apo-IsdBN1N2 ranging from 10 to 80 μm (final concentrations, 5–40 μm) in the second syringe; a minimum 5-fold excess of the IsdB acceptor was used to attain pseudo-first order conditions. The temperature was again maintained at 25 °C, and five 30-s acquisitions were performed at each wavelength for each concentration pair. The first 3 ms were in the dead time of the instrument and thus were excluded from analysis. At least four acquisitions were averaged, and curve fitting was performed using the ProDataSX software.
Crystallization and data collection
Solutions of apo-IsdBN1N2 and oxyHb in an equimolar (by monomer) ratio were mixed together at 10 mg/ml and immediately used to set up crystal trays. The IsdBN1N2·Hb complex was crystallized in space group P21212 using reservoir solution containing 0.1 m malonic acid, pH 5.5, 2.2 m ammonium sulfate in 96-well sitting drop plates. Crystals appeared within a few days at room temperature but were allowed to grow to a sufficient size for ∼4 weeks. Crystals were cryoprotected with 30% sodium malonate, pH 7.0, and flash frozen in liquid nitrogen. A 3.60 Å resolution data set was collected at the Stanford Synchotron Radiation Lightsource on Beamline 7-1 (1.000 Å wavelength) at 100 K. The data were processed using HKL2000 (49) (Table 2). The data set was initially phased by molecular replacement using the oxyhemoglobin (PDB code 2DN1) and IsdBN2 (PDB code 3RTL) crystal structures and a SWISS-MODEL (50) generated IsdBN1 model (based on IsdHN1 PDB code 4IJ2) as search models with Phaser-MR (51) in the PHENIX suite of programs (52), which found the hemoglobin tetramer, three copies of IsdBN1 and three copies of IsdBN2. Manual inspection of the placement of IsdBN1 and IsdBN2 molecules, particularly at non-conserved bulky residues such as Tyr and Phe, revealed that electron density for one Phaser-placed copy of IsdBN1 was better fit by IsdBN2, and the model was thus corrected to include four copies of IsdBN2 and two copies of IsdBN1. SWISS-MODEL was also used to generate a model for the IsdB linker region based on the Hb-IsdH complex crystal structure (PDB code 4IJ2), which was added in a subsequent round of molecular replacement. Manual rebuilding with WinCoot (53) was used in all stages to complete the structure, and refinement was carried out using phenix.refine (54). Heme molecules were placed into omit difference maps, and the position of iron was confirmed by an anomalous difference map. The heme-iron ligand geometry was not restrained. The polypeptide chain has excellent stereochemistry, with 95.2% of residues in the favored region of the Ramachandran plots and 4.5% in allowed regions, and 0.3% were outliers.
Table 2.
X-ray data collection and refinement statistics for IsdBN1N2·Hb structure
| Data collection | |
| Space group | P21212 |
| Unit cell dimensions (Å) | |
| a | 140.81 |
| b | 209.11 |
| c | 70.43 |
| Resolution range (Å) | 49.79–3.60 (3.66–3.60) |
| Rmerge | 0.12 (0.89) |
| I/σI | 16.9 (2.1) |
| Completeness (%) | 99.8 (100) |
| Redundancy | 6.0 (6.0) |
| Unique reflections | 24903 |
| Wilson B (Å2) | 116.5 |
| Refinement | |
| Rwork/Rfree | 0.252/0.302 |
| No. atoms | 11827 |
| Protein/heme | 11827 |
| Water | 0 |
| Average B-factors (Å2) | |
| Hemoglobin | 102.2 |
| IsdB | 116.3 |
| Heme | 112.4 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.80 |
| PDB entry code | 5VMM |
Hb+IsdBN1N2 crystal composition
Crystals from a well of oxyHb+IsdBN1N2 were looped and soaked in well solution to remove adventitiously bound protein and then dissolved in 5 μl of 20 mm HEPES, pH 7.4, 80 mm NaCl. Five small crystals in total were dissolved together and separated on a 15% SDS-PAGE gel to visualize the components of the crystal.
Haptoglobin inhibition assay
Human Hp of mixed serotype purified from plasma was purchased as a lyophilized solid (Athens Research and Technology, Athens, GA). 1 mg of Hp was dissolved in 100 μl of 20 mm HEPES, pH 7.4, 80 mm NaCl to give a concentration of 10 mg/ml. Spectra of 2 μn metHb alone or combined with 10 μm IsdBN1N2 and 2 mg/ml Hp (precombined with metHb) were taken from 250–750 nm in a conventional spectrophotometer (Cary50) with an optical path length of 1 cm in a quartz cell at room temperature, 22 °C. Subsequently, the minimum inhibitory concentration of Hp was determined by adding decreasing amounts of Hp under the same conditions (beginning with 2 mg/ml as above) until the heme transfer reaction from metHb to IsdBN1N2 was observed to proceed. Spectra (250–750 nm) were recorded immediately after mixing, as before. The reaction was monitored by recording additional spectra at 2 and 5 min after the initial spectrum.
IsdBN1N2·Hb·Hp pulldown assay
The ability of nickel bead-bound His6-IsdBN1N2 to pull down metHb·Hp complexes was tested. Six 25-μl aliquots of nickel bead slurry (Chelating Sepharose Fast Flow, GE Healthcare; stored in 20% ethanol) were washed twice with 1 ml of dH2O, followed by a 1-ml wash of binding buffer, consisting of 20 mm HEPES, pH 7.4, 80 mm NaCl, 75 mm imidazole. The moderate amount of imidazole was a concentration at which His6-IsdB could bind the nickel beads but Hb could not. To each aliquot of washed beads, 50 μl of 20 μm His6-IsdBN1N2, metHb, or Hp (alone for controls or together for pulldowns, as indicated) were added to the beads and kept at room temperature on the benchtop. Samples were incubated for 15 min and gently agitated occasionally. After incubation, the 50-μl supernatant was removed, and beads with bound protein were washed twice with 500 μl of binding buffer and then eluted with 1 m imidazole, pH 7.5. 4 μl of each eluate was run on a 15% SDS-PAGE gel at 200 V for 1 h and 5 min and stained using Coomassie Blue.
Isothermal titration calorimetry (ITC)
ITC was conducted using a MicroCal ITC-200 instrument at 25 °C. Syringe and cell samples were co-dialyzed overnight against 20 mm HEPES, pH 7.4, 80 mm NaCl. Dialysis buffer was collected and sterile-filtered for diluting samples and washing the ITC cell. ITC experiments were conducted with 800 μn (200 μm) of metHb as the titrant in the syringe and 80 μm of IsdBYFYF in the cell. Binding isotherms were analyzed with MicroCal Origin 7.0 software using a one-site model. Three ITC experiments were conducted to obtain an average dissociation constant (KD) and stoichiometry (N), and standard deviations were calculated and reported as error estimates.
IsdBYFYF titration of metHb
Spectra (250–750 nm) of metHb, IsdB constructs, or mixtures thereof were taken in a conventional Cary50 spectrophotometer with an optical path length of 1 cm in a quartz cell at room temperature (22 °C). 5 μn metHb was mixed with 10 μm of apo-IsdBN1N2 or apo-IsdBYFYF, and spectra were immediately recorded; spectra did not change within 5 min of first recording. Additionally, 4 μn metHb in a total volume of 1000 μl was titrated with aliquots (3.7 μl) of apo-IsdBYFYF in 1 μm increments from 1 to 9 μm. Spectra (250–750 nm) were taken in a Cary50 with an optical path length of 1 cm in a quartz cell at 22 °C, with three replicates carried out. The initial absorbance for each replicate at 410 nm (i.e. 0 μm IsdBYFYF) was subtracted from each subsequent titration absorbance value to yield the Δ410 nm, which was plotted against IsdBYFYF concentration; a straight line could be fit through the first five titration points to indicate stoichiometry.
S. aureus growth experiments
RPMI media 1640 with l-glutamine and Na2HCO3 (Sigma–Aldrich) was supplemented with 1% (w/v) casamino acids (BD, Sparks, MD). Metal-depleted RPMI (NRPMI) medium was prepared by first adding 7% (w/v) Chelex-100 (Sigma–Aldrich) to RPMI medium and stirring overnight. Chelex-100 was then removed from the medium, and 25 μm ZnCl2, 25 μm MnCl2, 100 mm CaCl2, and 1 mm MgCl2 were added. NRPMI medium was adjusted to pH 7.4, filter-sterilized, and stored at 4 °C.
Single colonies of S. aureus str. Newman on tryptic soy agar were inoculated into RPMI medium with 0.5 mm ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA, LGC Standards GmbH, Teddington, UK). The cultures were incubated at 37 °C for 16–20 h on a shaker set to 200 rpm. Overnight cultures were centrifuged for 2 min at 11,000 × g, and the pellet was washed three times in NRPMI with 0.5 mm EDDHA. The cells were then normalized to OD ∼3 and subcultured 1:100 into 200 μl of NRPMI with 0.5 mm EDDHA. In addition to a no-iron control, cultures were supplemented with 200 nn Hb, 200 nn human Hb, and 4–400 μg/ml Hp, mixed serotype (Athens Research and Technology), or 2 μm of heme with and without 400 μg/ml haptoglobin. The cultures were grown in triplicate with constant shaking at fast-speed and high-amplitude settings in a Bioscreen C instrument (Growth Curve USA, Piscataway, NJ) at 37 °C. Growth experiments were repeated three times, and the average growth with standard deviations are plotted using GraphPad Prism 6 (GraphPad Software, La Jolla, CA).
Author contributions
C. F. M. B. and M. E. P. M. conceptualized the study and designed the experiments; C. F. M. B., A. C. K. C., E. J. W. L., and A. L. A. conducted the experiments; C. F. M. B., A. C. K. C., and E. J. W. L. analyzed the data; C. F. M. B. drafted the article, with critical revisions by A. C. K. C., L. D. E., and M. E. P. M. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
Use of the Stanford Synchrotron Radiation Lightsource, Stanford Linear Accelerator Center National Accelerator Laboratory is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research, and by the NIGMS, National Institutes of Health Grant P41GM103393.
This work was supported by Canadian Institutes of Health Research Grant MOP-49597 (to M. E. P. M.) and a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship - Doctoral scholarship (to C. F. M. B.). Support for laboratory infrastructure was provided by the Canada Foundation for Innovation (to M. E. P. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
The atomic coordinates and structure factors (code 5VMM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Hb
- hemoglobin
- Hp
- haptoglobin
- metHb
- methemoglobin
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank
- ITC
- isothermal titration calorimetry
- NRPMI
- metal-depleted RPMI
- EDDHA
- ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid).
References
- 1. Wertheim H. F., Melles D. C., Vos M. C., van Leeuwen W., van Belkum A., Verbrugh H. A., and Nouwen J. L. (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762 10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 2. Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., and Edmond M. B. (2004) Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–317 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- 3. Waness A. (2010) Revisiting methicillin-resistant Staphylococcus aureus infections. J. Glob Infect. Dis. 2, 49–56 10.4103/0974-777X.59251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weinberg E. D. (1975) Nutritional immunity: host's attempt to withold iron from microbial invaders. JAMA 231, 39–41 10.1001/jama.1975.03240130021018,10.1001/jama.231.1.39 [DOI] [PubMed] [Google Scholar]
- 5. Skaar E. P., Humayun M., Bae T., DeBord K. L., and Schneewind O. (2004) Iron-source preference of Staphylococcus aureus infections. Science 305, 1626–1628 10.1126/science.1099930 [DOI] [PubMed] [Google Scholar]
- 6. Skaar E. P., and Schneewind O. (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6, 390–397 10.1016/j.micinf.2003.12.008 [DOI] [PubMed] [Google Scholar]
- 7. Mazmanian S. K., Ton-That H., Su K., and Schneewind O. (2002) An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298 10.1073/pnas.032523999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., Joachmiak A., Missiakas D. M., and Schneewind O. (2003) Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909 10.1126/science.1081147 [DOI] [PubMed] [Google Scholar]
- 9. Dryla A., Hoffmann B., Gelbmann D., Giefing C., Hanner M., Meinke A., Anderson A. S., Koppensteiner W., Konrat R., von Gabain A., and Nagy E. (2007) High-affinity binding of the staphylococcal HarA protein to haptoglobin and hemoglobin involves a domain with an antiparallel eight-stranded β-barrel fold. J. Bacteriol. 189, 254–264 10.1128/JB.01366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torres V. J., Pishchany G., Humayun M., Schneewind O., and Skaar E. P. (2006) Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188, 8421–8429 10.1128/JB.01335-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu H., Xie G., Liu M., Olson J. S., Fabian M., Dooley D. M., and Lei B. (2008) Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 283, 18450–18460 10.1074/jbc.M801466200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muryoi N., Tiedemann M. T., Pluym M., Cheung J., Heinrichs D. E., and Stillman M. J. (2008) Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 283, 28125–28136 10.1074/jbc.M802171200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grigg J. C., Vermeiren C. L., Heinrichs D. E., and Murphy M. E. (2007) Heme coordination by Staphylococcus aureus IsdE. J. Biol. Chem. 282, 28815–28822 10.1074/jbc.M704602200 [DOI] [PubMed] [Google Scholar]
- 14. Lee W. C., Reniere M. L., Skaar E. P., and Murphy M. E. (2008) Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283, 30957–30963 10.1074/jbc.M709486200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reniere M. L., Ukpabi G. N., Harry S. R., Stec D. F., Krull R., Wright D. W., Bachmann B. O., Murphy M. E., and Skaar E. P. (2010) The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol. Microbiol. 75, 1529–1538 10.1111/j.1365-2958.2010.07076.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Andrade M. A., Ciccarelli F. D., Perez-Iratxeta C., and Bork P. (2002) NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 3, RESEARCH0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krishna Kumar K., Jacques D. A., Pishchany G., Caradoc-Davies T., Spirig T., Malmirchegini G. R., Langley D. B., Dickson C. F., Mackay J. P., Clubb R. T., Skaar E. P., Guss J. M., and Gell D. A. (2011) Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH. J. Biol. Chem. 286, 38439–38447 10.1074/jbc.M111.287300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bowden C. F., Verstraete M. M., Eltis L. D., and Murphy M. E. (2014) Hemoglobin binding and catalytic heme extraction by IsdB near iron transporter domains. Biochemistry 53, 2286–2294 10.1021/bi500230f [DOI] [PubMed] [Google Scholar]
- 19. Dickson C. F., Kumar K. K., Jacques D. A., Malmirchegini G. R., Spirig T., Mackay J. P., Clubb R. T., Guss J. M., and Gell D. A. (2014) Structure of the hemoglobin-IsdH complex reveals the molecular basis of iron capture by Staphylococcus aureus. J. Biol. Chem. 289, 6728–6738 10.1074/jbc.M113.545566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaudin C. F., Grigg J. C., Arrieta A. L., and Murphy M. E. (2011) Unique heme-iron coordination by the hemoglobin receptor IsdB of Staphylococcus aureus. Biochemistry 50, 5443–5452 10.1021/bi200369p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grigg J. C., Vermeiren C. L., Heinrichs D. E., and Murphy M. E. (2007) Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 63, 139–149 10.1111/j.1365-2958.2006.05502.x [DOI] [PubMed] [Google Scholar]
- 22. Pilpa R. M., Fadeev E. A., Villareal V. A., Wong M. L., Phillips M., and Clubb R. T. (2006) Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus. J. Mol. Biol. 360, 435–447 10.1016/j.jmb.2006.05.019 [DOI] [PubMed] [Google Scholar]
- 23. Sharp K. H., Schneider S., Cockayne A., and Paoli M. (2007) Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J. Biol. Chem. 282, 10625–10631 10.1074/jbc.M700234200 [DOI] [PubMed] [Google Scholar]
- 24. Watanabe M., Tanaka Y., Suenaga A., Kuroda M., Yao M., Watanabe N., Arisaka F., Ohta T., Tanaka I., and Tsumoto K. (2008) Structural basis for multimeric heme complexation through a specific protein-heme interaction: the case of the third neat domain of IsdH from Staphylococcus aureus. J. Biol. Chem. 283, 28649–28659 10.1074/jbc.M803383200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fonner B. A., Tripet B. P., Eilers B. J., Stanisich J., Sullivan-Springhetti R. K., Moore R., Liu M., Lei B., and Copié V. (2014) Solution structure and molecular determinants of hemoglobin binding of the first NEAT domain of IsdB in Staphylococcus aureus. Biochemistry 53, 3922–3933 10.1021/bi5005188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dickson C. F., Jacques D. A., Clubb R. T., Guss J. M., and Gell D. A. (2015) The structure of haemoglobin bound to the haemoglobin receptor IsdH from Staphylococcus aureus shows disruption of the native α-globin haem pocket. Acta Crystallogr. D Biol. Crystallogr. 71, 1295–1306 10.1107/S1399004715005817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pishchany G., Sheldon J. R., Dickson C. F., Alam M. T., Read T. D., Gell D. A., Heinrichs D. E., and Skaar E. P. (2014) IsdB-dependent hemoglobin binding is required for acquisition of heme by Staphylococcus aureus. J. Infect. Dis. 209, 1764–1772 10.1093/infdis/jit817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allard M., Moisan H., Brouillette E., Gervais A. L., Jacques M., Lacasse P., Diarra M. S., and Malouin F. (2006) Transcriptional modulation of some Staphylococcus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 8, 1679–1690 10.1016/j.micinf.2006.01.022 [DOI] [PubMed] [Google Scholar]
- 29. Etz H., Minh D. B., Henics T., Dryla A., Winkler B., Triska C., Boyd A. P., Söllner J., Schmidt W., von Ahsen U., Buschle M., Gill S. R., Kolonay J., Khalak H., Fraser C. M., et al. (2002) Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 99, 6573–6578 10.1073/pnas.092569199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuklin N. A., Clark D. J., Secore S., Cook J., Cope L. D., McNeely T., Noble L., Brown M. J., Zorman J. K., Wang X. M., Pancari G., Fan H., Isett K., Burgess B., Bryan J., et al. (2006) A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect. Immun. 74, 2215–2223 10.1128/IAI.74.4.2215-2223.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Malachowa N., Whitney A. R., Kobayashi S. D., Sturdevant D. E., Kennedy A. D., Braughton K. R., Shabb D. W., Diep B. A., Chambers H. F., Otto M., and DeLeo F. R. (2011) Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6, e18617 10.1371/journal.pone.0018617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pilpa R. M., Robson S. A., Villareal V. A., Wong M. L., Phillips M., and Clubb R. T. (2009) Functionally distinct NEAT (NEAr Transporter) domains within the Staphylococcus aureus IsdH/HarA protein extract heme from methemoglobin. J. Biol. Chem. 284, 1166–1176 10.1074/jbc.M806007200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robinson V. L., Smith B. B., and Arnone A. (2003) A pH-dependent aquomet-to-hemichrome transition in crystalline horse methemoglobin. Biochemistry 42, 10113–10125 10.1021/bi030059t [DOI] [PubMed] [Google Scholar]
- 34. Rachmilewitz E. A., Peisach J., and Blumberg W. E. (1971) Studies on the stability of oxyhemoglobin A and its constituent chains and their derivatives. J. Biol. Chem. 246, 3356–3366 [PubMed] [Google Scholar]
- 35. Langlois M. R., and Delanghe J. R. (1996) Biological and clinical significance of haptoglobin polymorphism in humans. Clin. Chem. 42, 1589–1600 [PubMed] [Google Scholar]
- 36. Wobeto V. P. d. A., Zaccariotto T. R., and Sonati M. d. F. (2008) Polymorphism of human haptoglobin and its clinical importance. Genet. Mol. Biol. 31, 602–620 10.1590/S1415-47572008000400002 [DOI] [Google Scholar]
- 37. Cheng T. M., Pan J. P., Lai S. T., Kao L. P., Lin H. H., and Mao S. J. (2007) Immunochemical property of human haptoglobin phenotypes: determination of plasma haptoglobin using type-matched standards. Clin. Biochem. 40, 1045–1056 10.1016/j.clinbiochem.2007.04.018 [DOI] [PubMed] [Google Scholar]
- 38. Eaton J. W., Brandt P., Mahoney J. R., and Lee J. T. Jr. (1982) Haptoglobin: a natural bacteriostat. Science 215, 691–693 10.1126/science.7036344 [DOI] [PubMed] [Google Scholar]
- 39. Mansouri A., and Winterhalter K. H. (1973) Nonequivalence of chains in hemoglobin oxidation. Biochemistry 12, 4946–4949 10.1021/bi00748a020 [DOI] [PubMed] [Google Scholar]
- 40. Tsuruga M., Matsuoka A., Hachimori A., Sugawara Y., and Shikama K. (1998) The molecular mechanism of autoxidation for human oxyhemoglobin: tilting of the distal histidine causes nonequivalent oxidation in the β chain. J. Biol. Chem. 273, 8607–8615 10.1074/jbc.273.15.8607 [DOI] [PubMed] [Google Scholar]
- 41. Hwang P. K., and Greer J. (1980) Interaction between hemoglobin subunits in the hemoglobin·haptoglobin complex. J. Biol. Chem. 255, 3038–3041 [PubMed] [Google Scholar]
- 42. Sæderup K. L., Stødkilde K., Graversen J. H., Dickson C. F., Etzerodt A., Hansen S. W., Fago A., Gell D., Andersen C. B., and Moestrup S. K. (2016) The Staphylococcus aureus protein IsdH inhibits host hemoglobin scavenging to promote heme acquisition by the pathogen. J. Biol. Chem. 291, 23989–23998 10.1074/jbc.M116.755934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nielsen M. J., Andersen C. B., and Moestrup S. K. (2013) CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J. Biol. Chem. 288, 18834–18841 10.1074/jbc.M113.471060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bunn H. F., and Jandl J. H. (1968) Exchange of heme among hemoglobins and between hemoglobin and albumin. J. Biol. Chem. 243, 465–475 [PubMed] [Google Scholar]
- 45. Dryla A., Gelbmann D., von Gabain A., and Nagy E. (2003) Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol. Microbiol. 49, 37–53 10.1046/j.1365-2958.2003.03542.x [DOI] [PubMed] [Google Scholar]
- 46. Francis R. T. Jr., Booth J. W., and Becker R. R. (1985) Uptake of iron from hemoglobin and the haptoglobin-hemoglobin complex by hemolytic bacteria. Int. J. Biochem. 17, 767–773 10.1016/0020-711X(85)90262-9 [DOI] [PubMed] [Google Scholar]
- 47. Pishchany G., Haley K. P., and Skaar E. P. (2013) Staphylococcus aureus growth using human hemoglobin as an iron source. J. Vis. Exp. 72, e50072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. MacPherson I. S., Rosell F. I., Scofield M., Mauk A. G., and Murphy M. E. (2010) Directed evolution of copper nitrite reductase to a chromogenic reductant. Protein Eng., Des. Sel. 23, 137–145 10.1093/protein/gzp084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 50. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308,10.1107/S0108767312007234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.