Abstract
Hepatitis C virus (HCV) infection is a major risk factor for the development of chronic liver disease. The disease typically progresses from chronic HCV to fibrosis, cirrhosis, hepatocellular carcinoma (HCC), and death. Chronic inflammation associated with HCV infection is implicated in cirrhosis and HCC, but the molecular players and signaling pathways contributing to these processes remain largely unknown. Interferon regulatory factor 5 (IRF5) is a molecule of interest in HCV-associated HCC because it has critical roles in virus-, Toll-like receptor (TLR)-, and IFN-induced signaling pathways. IRF5 is also a tumor suppressor, and its expression is dysregulated in several human cancers. Here, we present first evidence that IRF5 expression and signaling are modulated during HCV infection. Using HCV infection of human hepatocytes and cells with autonomously replicating HCV RNA, we found that levels of IRF5 mRNA and protein expression were down-regulated. Of note, reporter assays indicated that IRF5 re-expression inhibited HCV protein translation and RNA replication. Gene expression analysis revealed significant differences in the expression of cancer pathway mediators and autophagy proteins rather than in cytokines between IRF5- and empty vector–transfected HCV replicon cells. IRF5 re-expression induced apoptosis via loss in mitochondrial membrane potential, down-regulated autophagy, and inhibited hepatocyte cell migration/invasion. Analysis of clinical HCC specimens supports a pathologic role for IRF5 in HCV-induced HCC, as IRF5 expression was down-regulated in livers from HCV-positive versus HCV-negative HCC patients or healthy donor livers. These results identify IRF5 as an important suppressor of HCV replication and HCC pathogenesis.
Keywords: Hepatitis C virus (HCV), hepatocellular carcinoma, immunosuppression, interferon regulatory factor (IRF), tumor suppressor gene, viral immunology
Introduction
Hepatitis C virus (HCV)5 has emerged as a serious human pathogen of global public health relevance that replicates in hepatocytes, thus targeting the liver. HCV has a current health burden of ∼180 million chronically infected individuals worldwide and a reported 350,000 deaths annually (1, 2). Persistent infection can lead to chronic liver disease that appears as acute and chronic hepatitis, fibrosis, cirrhosis, and eventually hepatocellular carcinoma (HCC) over a lifetime infection. Approximately 40–60% of HCC cases are due to HCV infections (3). HCC is highly malignant, with current survival rates less than 1 year following diagnosis (3). The virus is a single-stranded positive RNA virus classified within the Flaviviridae family. The HCV genome is ∼9.7 kb in length and encodes a large polyprotein of about 3,000 amino acids from a single open reading frame consisting of HCV structural (core, E1, E2, and possibly p7) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (1, 3). HCV has an internal ribosome entry site (IRES) that initiates translation in the uncapped 5′-untranslated region (4). There are no prophylactic vaccines against HCV, and although the current standard of care, consisting of an all-oral, IFN-free, direct-acting antiviral treatment regimen targeting the HCV NS3, NS5A, and NS5B proteins, cures most HCV patients, there still exist limitations, including the evolution of drug-resistant HCV alleles, complications with co-morbidities, significant side effects, access to care, and cost of therapy (5). HCV infection activates the innate immune system, resulting in type I and III IFN expression (5, 6), and these IFNs play a central role in eliminating HCV by turning on the expression of numerous IFN-stimulated genes. Thus, HCV has evolved mechanisms to block innate antiviral immune response(s) to replicate and persist (5, 6).
Although the molecular mechanisms by which HCV inhibits type I and III IFN signaling are not extensively known, data in the past 10 years indicate that the family of interferon regulatory factors (IRFs) is a target of HCV proteins (7–12). IRFs are transcription factors that can be activated or induced by IFNs yet also regulate the expression of IFNs and IFN-stimulated genes (13, 14). The nonstructural HCV protein, NS5A, was found to influence HCV persistence by blocking IRF1 activation and disrupting a host antiviral pathway that suppresses virus replication (7). Subsequent studies showed that HCV infection or transfection of HCV core- or NS5A-expressing plasmid in hepatocytes resulted in a significant reduction of IRF1 mRNA and protein expression (8). HCV serine protease NS3/4A was shown to block the phosphorylation and effector function of IRF3 (9). Last, NS5A was shown to interact with IRF7, resulting in reduced IRF7 nuclear translocation and IFNA14 promoter regulation (10). Nandakumar et al. (11) showed that IRF5 may play a role in controlling HCV replication independent of type I IFNs because reconstitution of Irf5−/− mouse embryonic fibroblasts with constitutively active IRF5 protein inhibited HCV replication in the presence of type I IFN-depleting antibodies.
IRF5 is a key mediator of MyD88-dependent Toll-like receptor signaling (15, 16) and antiviral immunity through its ability to regulate numerous pro-inflammatory cytokines, including type I IFN (α and β), IL-6, IL-12, IL-1B, IL-23, and TNF-α (15–19). IRF5 also has important roles in cell proliferation, migration, apoptosis, and cell cycle, resulting in its identification as a human tumor suppressor gene (20–23). Similar findings were made in mice showing that Irf5−/− mouse embryonic fibroblasts transformed with c-Ha-Ras were resistant to undergoing DNA damage- and virus-induced apoptosis (17). In vitro colony formation and in vivo tumor cell growth were also found to be exacerbated in cells lacking Irf5 (17, 20, 21). Given these pleiotropic functions, it is not surprising that dysregulated IRF5 expression and function have been implicated in the pathogenic mechanisms of autoimmune diseases, such as systemic lupus erythematosus, and cancer. In this study, we investigated IRF5 expression and function in hepatocytes infected with HCV J6/JFH-1 chimeric virus, HCV replicon cells, and human primary tissue specimens from patients with HCV-positive and -negative HCC tumors. Our data identify IRF5 as a new negative effector of HCV replication and HCV-associated HCC pathogenesis.
Results
IRF5 expression is down-regulated in HCV replicon cells
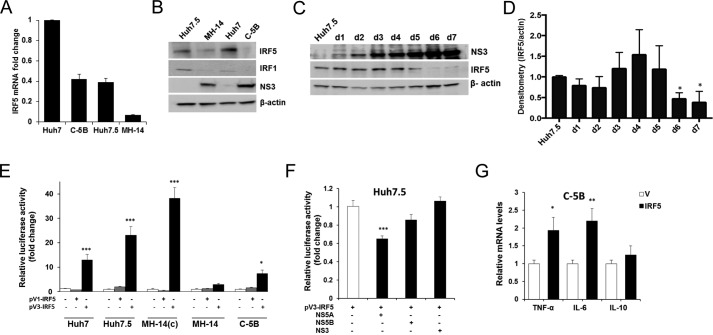
Although much is known of IRF5 expression and function in human lymphoid cells, little is known of its expression and function in normal hepatocytes or HCV-infected hepatocytes. IRF5 was shown to be required for Fas-induced apoptosis in murine hepatocytes (24–25), and basal IRF5 expression is detectable in healthy human liver (26). We examined endogenous IRF5 expression in cognate Huh cells (Huh7 and Huh7.5) and HCV replicon-bearing cells (MH-14 and C-5B). IRF5 expression was detected at both transcript and protein levels in Huh7 and Huh7.5 cell lines (Fig. 1, A and B). IRF5 transcript expression was highest in Huh7 and dramatically lower in the derivative, Huh7.5, which has a mutation in cytosolic retinoic acid–inducible gene I (RIG-I). Transcript levels similar to those detected in Huh7.5 were detected in C-5B, and levels were further decreased in MH-14. At the protein level, Huh7 and Huh7.5 showed similar IRF5 expression, whereas replicon-bearing MH-14 and C-5B had dramatically lower levels (Fig. 1B). Kim et al. (8) previously showed down-regulation of IRF1 expression in HCV-infected cells, and this was confirmed in our study. We were also unable to detect basal IRF7 expression in human hepatocytes, as reported by Ghosh et al. (27). We extended these findings to naive Huh7.5 cells infected with purified HCV J6/JFH-1 chimeric virus stock for a period of 7 days. Similar to HCV replicon-bearing cell lines, IRF5 expression was significantly reduced at the later stages of HCV chronic infection (days 2–7) (Fig. 1, C and D). HCV NS3 viral protein expression was confirmed to increase over the infection time (Fig. 1C).
Figure 1.
IRF5 expression and immunoregulation function are downregulated in HCV-infected hepatocytes and HCV replicon cell lines. A, endogenous IRF5 expression was examined in cognate control Huh7 and Huh7.5 cells, HCV replicon–bearing MH-14 and C-5B cells by real-time qPCR. -Fold change was determined after normalization to β-actin levels with relative comparison with Huh7 levels. B, representative data from Western blot analysis of endogenous IRF5, IRF1, and NS3 protein expression. C and D, HCV-infected cell culture supernatants were used to infect naive Huh7.5 cells for 7 days. Representative Western blot data (C) and densitometry analysis of IRF5 protein expression (D) is shown. Data are representative of four independent experiments. E, relative transactivation of IRF5 promoters, pV1-IRF5 and pV3-IRF5, in cognate and replicon-bearing cell lines. -Fold change is shown after normalization to Renilla. F, same as E except plasmids encoding HCV viral proteins were transfected with the pV3-IRF5 promoter. G, IRF5 induces transient proinflammatory cytokine expression in C-5B cells. Empty vector (V) or 80 ng of GFP-IRF5 was transiently transfected, and TNFA, IL6, and IL10 mRNA levels were measured by real-time qPCR at 36 h post-transfection. Data were normalized to β-actin and plotted relative to empty vector control. Data are representative of duplicate data points from three independent replicates; plotted values are means ± S.D. (error bars) (*, p < 0.05; **, p < 0.001; ***, p < 0.0001).
To begin to understand why HCV-infected Huh7.5 and HCV replicon cells express lower levels of IRF5 transcript and protein expression, IRF5 promoter reporter activity was examined in cognate and HCV replicon-bearing lines (Fig. 1E). Two IRF5 promoters known to have constitutive reporter activity, pV1-IRF5 and pV3-IRF5 (28), were utilized. Among other consensus sequences identified within the promoters that regulate IRF5 expression, pV1 was shown to have a functional IRF element, and pV3 was shown to have a functional interferon-stimulated response element (ISRE) (28). In all cell lines examined, pV3- and not pV1-IRF5 showed some level of constitutive reporter activity (Fig. 1E). Similar to protein levels detected in Fig. 1B, elevated reporter activity was found in cognate Huh7 and Huh7.5 cell lines and cured MH-14, with significantly decreased activity in replicon-bearing MH-14 and C-5B cells (Fig. 1E). To examine the effect of HCV proteins on pV3-IRF5 promoter reporter activity, plasmids encoding individual HCV proteins were transfected to Huh7.5 cells. It has previously been shown that NS3 and NS5A/B viral proteins inhibit the expression and/or activation of other IRF family members (7–10); thus, we examined their effect on IRF5 pV3 promoter activity. Interestingly, overexpression of NS5A and not NS5B or NS3 resulted in a significant reduction of pV3 transactivation (Fig. 1F). Similar to IRF1 and IRF7 (7–9), these data implicate NS5A in the mechanism(s) of IRF5 down-regulation in HCV-infected cells.
Ectopic IRF5 induces TNFA and IL-6 transcript expression in C-5B replicon cells
IRF5 has been previously shown to transactivate ISRE-containing promoters from a number of antiviral genes (15, 18, 19). To determine the effect of IRF5 on antiviral cytokine expression in HCV replicon cells, C-5B were transiently transfected with FLAG-tagged IRF5, and cytokine expression was measured by real-time quantitative PCR (qPCR). Overexpression of IRF5 resulted in significantly elevated TNFA and IL-6 transcript expression but not IL-10 (Fig. 1G). IRF5 is a direct regulator of IL-6 and TNFA expression (13, 15). Thus, re-expression of IRF5 in HCV-infected and/or HCV replicon-bearing cells triggers the activation of ISRE-containing promoters, such as those found in the antiviral genes TNFA and IL-6 (15). Enhanced expression of antiviral proteins, through IRF5 and other innate immune mediators, would lead to an antiviral response and negative regulation of HCV RNA replication. These data indicate that it is in the interest of HCV to suppress IRF5 expression.
Ectopic IRF5 negatively modulates HCV IRES-mediated protein translation and HCV RNA replication in HCV replicon cells
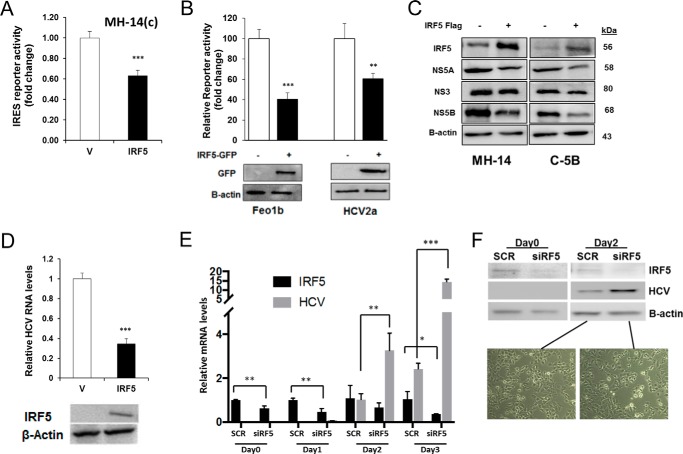
Because IRF5 is a key mediator of antiviral immunity through its regulation of proinflammatory cytokines (15, 18, 19), we examined the effect of IRF5 overexpression on HCV IRES-mediated protein translation and HCV RNA levels in HCV replicon cells. As shown in Fig. 2A, HCV IRES-mediated reporter activity was decreased ∼1.8-fold (40%) upon transient expression of IRF5 in MH-14(c) cells, indicating that IRF5 down-regulates HCV IRES-mediated protein translation. Similar levels of inhibition were found in Feo1b and HCV2a cells after IRF5 overexpression (Fig. 2B). Examination of endogenous HCV viral protein expression in replicon cells revealed that IRF5 overexpression decreased NS5A and NS5B levels (Fig. 2C). Analysis of HCV RNA in HCV replicon cells also showed that IRF5 overexpression decreased the levels of replicating HCV RNA by ∼2.7-fold (70%) (Fig. 2D). Conversely, knockdown of IRF5 by transient transfection of ON-TARGETplus IRF5 siRNA SMARTpools (Dharmacon) to Huh7 cells, followed by infection with purified virus stock for 3 days, revealed a significant reduction in IRF5 expression with a concomitant increase in HCV RNA levels (Fig. 2E). Knockdown efficiency and HCV viral protein levels were confirmed by Western blot analysis at days 0 and 2 postinfection (Fig. 2F). Representative Huh7 cell images after knockdown and viral infection are shown. Together, these data indicate that IRF5 regulates both HCV RNA replication and viral protein expression.
Figure 2.
IRF5 negatively modulates expression of HCV protein(s) and HCV RNA replication. A, MH-14(c) cells were co-transfected with 80 ng of GFP-IRF5 and the HCV IRES-Luc reporter. -Fold change in luciferase activity is shown after normalization to Renilla. B, same as in A except GFP-IRF5 and HCV IRES-Luc were transfected to Feo1b and HCV2a cells. A representative Western blot of GFP-IRF5 expression in transfected cells is shown. C, levels of HCV viral proteins in HCV replicon cells overexpressing IRF5 were determined by Western blot analysis. D, effect of ectopic IRF5 expression on replicating HCV RNA levels in C-5B cells. HCV RNA expression was analyzed by real-time qPCR 36 h after transfection of 0.1 μg of empty FLAG vector (V) or FLAG-IRF5 (IRF5). Data were normalized to β-actin levels and presented relative to empty vector control. E, Huh7 cells were transfected with scrambled (SCR) or IRF5 siRNAs (siRF5) for 48 h before infecting with purified virus stock for 3 days. IRF5 and HCV RNA levels were determined by real-time qPCR. F, representative Western blot from E showing IRF5 knockdown and HCV protein levels at days 0 and 2 of virus infection. Live cell images are from day 2 postinfection in Huh7 cells transfected with SCR or siIRF5. Images are representative of three independent transient transfections in each cell line. Images were taken at ×40 magnification. Data are representative of duplicate data points from three independent replicates; plotted values are means ± S.D. (error bars) (*, p < 0.05; **, p < 0.001; ***, p < 0.001).
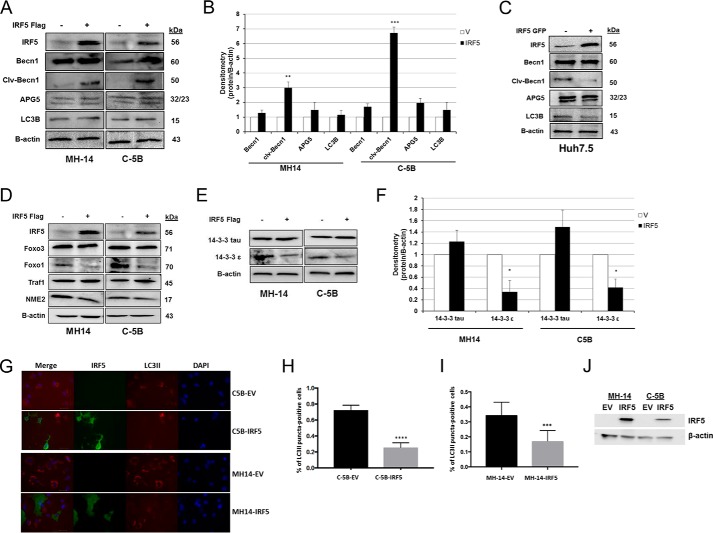
IRF5 impairs HCV-induced autophagy
Recent studies in the HCV field suggest that HCV exploits the autophagic process to accomplish translation, replication, assembly, and release of lipoviroparticles (29, 30). Multiple laboratories have shown that HCV-induced autophagy represses anti-HCV innate immune responses, whereas impairment leads to significant up-regulation of HCV-induced immune responses (29–32). Thus, further analysis of IRF5 function in HCV replicon cells led us to examine autophagy signaling. Results in Fig. 3 show specific alterations in key mediators of autophagy. In MH-14 and C-5B, ectopic IRF5 significantly increased the cleavage of Bcl-2–interacting protein-1 (Beclin-1) (Fig. 3, A and B), whereas no change was detected in IRF5-transfected Huh7.5 cells (Fig. 3C). No significant change was found in the expression of the long or short isoform of autophagy protein 5 (APG5/ATG5) or autophagy marker light chain 3 (LC3B) between IRF5- and empty vector–transfected cells. HCV can transcriptionally up-regulate Beclin-1 expression (30, 33), and cleavage is known to inactivate autophagy (34). Analysis of additional mediators of autophagy revealed that IRF5 overexpression specifically reduced FoxO1 and 14-3-3ϵ protein levels (Fig. 3, D–F). FoxO1, a forkhead O family protein, is a mediator of autophagy and is required for induction (35). 14-3-3ϵ forms a complex with phosphorylated Beclin-1 and dysregulates autophagy (36). To determine more clearly whether IRF5 alters HCV-induced autophagy, we measured LC3II-positive puncta in IRF5-expressing MH-14 and C-5 cells. Notably, we detected a significant reduction in the number of IRF5-expressing cells that contained LC3II-positive puncta (Fig. 3, G–J). Together, these data suggest that another mechanism by which IRF5 controls HCV infection is through inactivation of the autophagy pathway, leading to reduced HCV RNA replication and viral protein expression (Fig. 2).
Figure 3.
IRF5 impairs HCV-induced autophagy. Effect of IRF5 on autophagy regulation in HCV replicon cells. A and B, MH-14 and C-5B cells were transiently transfected with 0.1 μg of FLAG-IRF5 plasmid for 36 h. Expression of key proteins involved in autophagy was analyzed by Western blotting. B, results from densitometry analysis of mediators in A. C, same as A except expression was examined in Huh7.5 cells. D–F, similar to A except additional proteins involved in autophagy were examined and quantified by densitometry analysis. G, representative images of C5B-EV, C5B-IRF5, MH14-EV, and MH14-IRF5 cells stained for IRF5 and LC3II. Images were taken at ×50 magnification; scale bar, 20 μm. The percentage of cells positive for LC3 puncta was calculated from three independent experiments, and the results are shown in H for C-5B cells and I for MH-14 cells. J, representative Western blot of IRF5 expression in MH-14 and C-5B cells transfected with FLAG-IRF5 or empty vector (EV) plasmid. Data are representative of three independent replicates; plotted values are means ± S.D. (error bars) (*, p < 0.05; **, p < 0.001; ***, p < 0.0001).
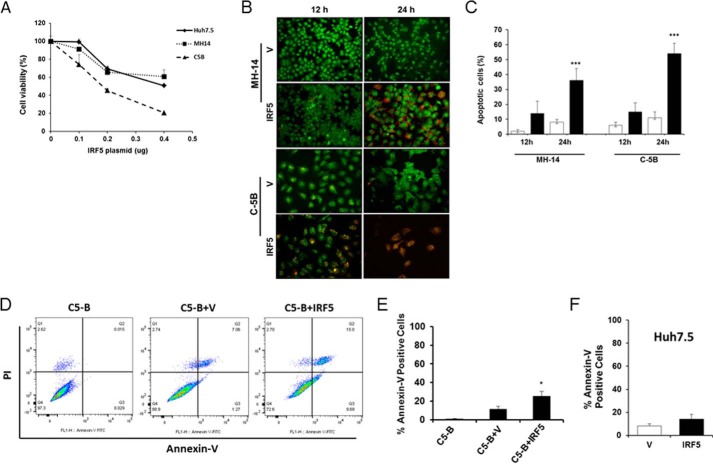
IRF5 enhances cell death in HCV replicon cells by triggering a loss in mitochondrial membrane potential
Inhibition of autophagy through the silencing of proteins involved in this pathway leads to apoptosis (30, 33). Because IRF5 is a known mediator of spontaneous and DNA damage–induced apoptosis (17, 20–22), and IRF5 expression enhanced Beclin-1 cleavage and significantly reduced LC3-GFP puncta (Fig. 3), we examined whether IRF5 alters cell viability and/or apoptosis. Viability was measured by an MTS assay in Huh7.5, MH-14, and C-5B cells transiently transfected with different amounts of FLAG-IRF5 plasmid. Results in Fig. 4A show a dose-dependent decrease in viability as IRF5 expression increased. At low concentrations of IRF5 (0.1 μg), C-5B was the most sensitive to IRF5-induced cell death and Huh7.5 the least sensitive; however, differences between cell lines were not statistically significant at the low concentration. Based on these data, all transfection experiments were performed with 0.08–0.1 μg of IRF5 plasmid to reduce the level of IRF5-induced cell death to <20% (Fig. 4A). Apoptosis was measured by two methods: acridine orange (AO)/EtBr double-staining and fluorescent microscopy and annexin V-FITC staining and flow cytometry. Representative images in Fig. 4B are from fluorescent microscopy analysis of AO/EtBr-stained cells with quantification of dual-stained cells shown in Fig. 4C. Dual-stained cells with condensed chromatin, as seen in IRF5-expressing C-5B cells, demarcate cells undergoing late apoptosis and not necrosis. Transient overexpression of IRF5 (black bars) in MH-14 and C-5B led to significant apoptosis at 24 h post-transfection as compared with empty vector (white bars) controls. These studies were complemented by flow cytometry to compare apoptosis in cognate Huh7.5 and C-5B cells. Percentage of annexin V-FITC single-stained cells was determined as a measure of early apoptosis (Fig. 4, D–F). At 12 h post-transfection, IRF5 overexpression induced significant yet low levels of spontaneous apoptosis in C5-B cells but not Huh7.5 (Fig. 4, D–F). Together, these data suggest that IRF5-mediated apoptosis in HCC cell lines is HCV-specific.
Figure 4.
IRF5 enhances cell death in HCV replicon cells. A, cell viability as determined by the MTS assay is shown for Huh7.5, MH-14, and C-5B cells. Cells were transiently transfected with the indicated concentrations of FLAG-IRF5 plasmid and harvested 24 h later. Data are means ± S.D. (error bars) of six independent replicates. B, representative fluorescent images of AO and EtBr co-staining in MH-14 and C-5B cells transfected with 0.1 μg of empty vector (V) or FLAG-IRF5 plasmid (IRF5). Images were taken at ×20 magnification. C, summary of data from B showing the percentage of EtBr-positive (apoptotic) cells 12 and 24 h post-transfection. White bars, cells transfected with empty vector; black bars, cells transfected with FLAG-IRF5 plasmid. D, representative dot plots from flow cytometry analysis of C-5B cells left untransfected or transiently transfected with 80 ng of empty vector (V) or FLAG-IRF5 (IRF5) plasmid. Apoptosis was determined from Annexin V-FITC single-stained cells 12 h later. E, summary of data from D showing mean percentage of single-stained cells ± S.D. Plotted data are duplicate data points from three independent replicates. F, same as E except data are from Huh7.5 cells. *, p < 0.05; ***, p < 0.0001.
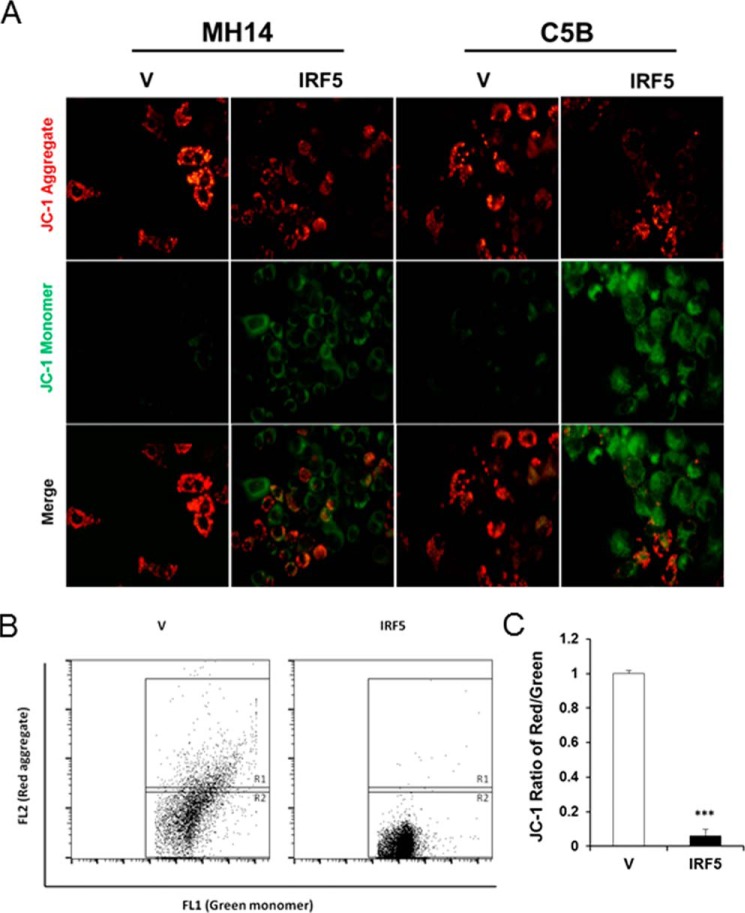
HCV has been shown to both associate with and alter mitochondrial function and signaling (37). This leads to important consequences for viral replication and the pathogenesis associated with chronic HCV, such as HCC. We examined loss of mitochondrial membrane potential (Δψm) via measurement of JC-1 aggregate/monomer ratio in IRF5- and empty vector–transfected HCV replicon cells. Mitochondrial depolarization is indicated by a decrease in the red (JC-1 aggregate)/green (JC-1 monomer) fluorescence intensity ratio. Representative images of JC-1 staining in MH-14 and C-5B cells either lacking (V) or expressing IRF5 revealed a clear increase in JC-1 monomers (green) with a concomitant decrease in JC-1 aggregates (red) in IRF5-expressing cells (Fig. 5A). Representative dot plots from flow cytometry analysis of JC-1 staining in C-5B cells are shown in Fig. 5B with quantitation in Fig. 5C. Data reveal a significant decrease in the ratio of red/green staining in IRF5-positive cells, indicating that IRF5 triggers a loss in Δψm resulting in enhanced apoptosis. These data support a key role for IRF5 in controlling HCV replicon cell viability whereby ectopic expression of IRF5 leads to apoptosis and cell death via loss in Δψm.
Figure 5.
IRF5 induces loss of Δψm in HCV replicon cells. A, representative images from JC-1 staining of C-5B and MH-14 cells overexpressing empty vector (V) or FLAG-IRF5 (IRF5) plasmid. Red and green fluorescence represent aggregated (non-apoptotic) and monomeric (apoptotic) JC-1 staining, respectively. Images were taken at ×40 magnification. B, similar to A except JC-1 staining was analyzed by flow cytometry. Representative dot plots from C-5B cells are shown illustrating gating of JC-1 red-positive (R1) and JC-1 green-positive (R2) populations; the ratio represents the JC1-positive population. C, graphical summary of data from B. Data are representative of duplicate data points from three independent replicates; plotted values are means ± S.D. (error bars) (***, p < 0.0001).
Ectopic IRF5 alters the expression of genes associated with mitochondrial apoptosis and HCC pathogenesis
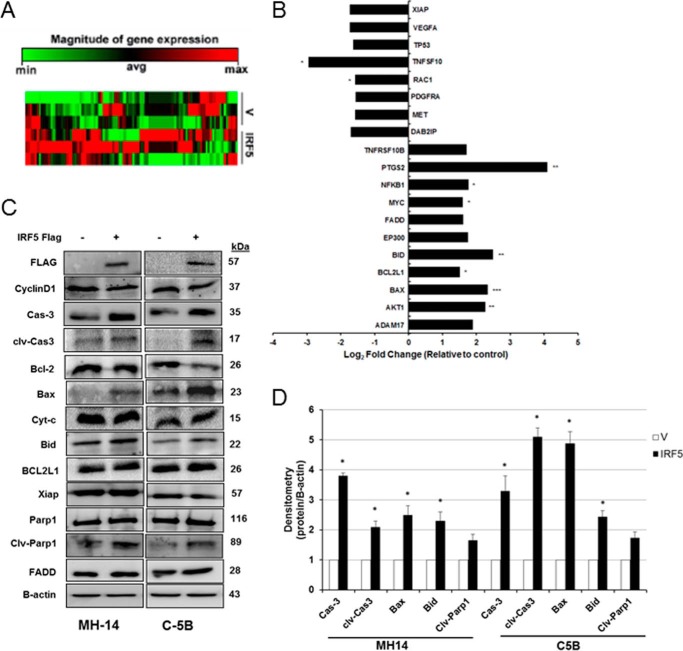
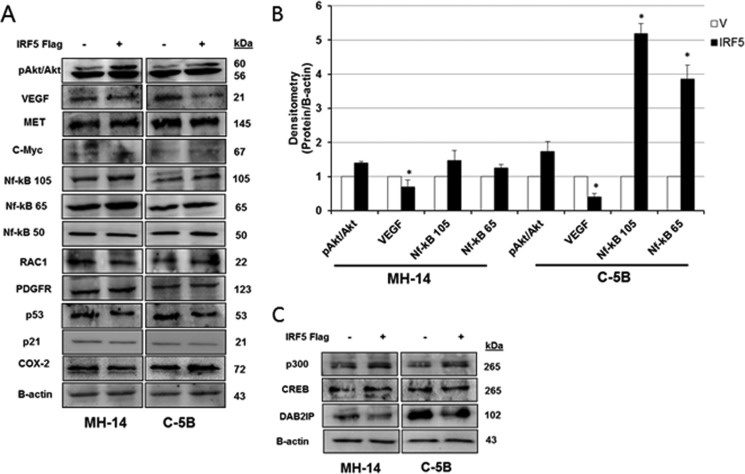
IRF5 tumor suppressor activity has been documented in many cancer cell types where its expression is down-regulated in cancer versus normal cells, and re-expression in cancer cells generally restores normal growth traits (20, 22, 23, 38). Because chronic HCV infection is a leading cause of HCC, and data herein indicate repression of IRF5 expression and function in HCV replicon cells, we examined IRF5 tumor suppressor function in HCV replicon cells. Using a liver cancer RT2 ProfilerTM PCR array, we assayed differential gene expression between IRF5- and empty vector–transfected C-5B cells. Distinct clusters of genes either up-regulated or down-regulated by IRF5 were identified (Fig. 6A and Table 1). Gene changes that were >1.5-fold over expression levels detected in vector control cells are shown in Fig. 6B. Expression was confirmed by Western blot analysis in C-5B and MH-14 cells (Figs. 6 and 7). Proteins important in the mitochondrial apoptotic pathway, such as Bax, Bid, and caspase-3, were found to be up-regulated in IRF5-positive cells, resulting in elevated cleaved caspase-3 and PARP (Fig. 6C). Quantification revealed significantly elevated caspase-3, cleaved caspase-3, Bax, and Bid in IRF5-positive MH-14 and C-5B cells (Fig. 6D). Elevated expression of NF-κB p105 and p65 was confirmed in IRF5-positive cells, along with E1A-binding protein p300 (Fig. 7). p300 functions as a histone acetyltransferase that regulates cell proliferation and differentiation. Conversely, vascular endothelial growth factor A (VEGFA) protein levels were significantly down-regulated in IRF5-positive cells, which correlates with enhanced apoptosis (Fig. 4). In addition to its pro-angiogenic function(s), VEGFA is anti-apoptotic and has been shown to foster tumor cell survival, proliferation, and vessel formation (39). Together, these data suggest pro-apoptotic and anti-tumorigenic functions for IRF5 in HCV replicon cells, suggesting that loss of IRF5 expression in HCV replicon cells may contribute to HCV-induced HCC pathogenesis.
Figure 6.
Ectopic IRF5 alters the expression of genes associated with mitochondrial apoptosis. A, C-5B replicon cells were transiently transfected with empty vector (V) control or GFP-IRF5 (IRF5) plasmid, and 36 h later, total RNA was isolated for analysis of gene expression using the liver cancer RT2 ProfilerTM PCR array. Data from three independent replicates is shown in a clustergram (heat map) representing relative expression levels for all samples and all genes included on the real-time qPCR array. Red, up-regulated expression; green, down-regulated expression. B, same as A except data are represented as -fold change compared with the control group after normalization to housekeeping genes. Genes with >1.5-fold change in expression between empty vector control and IRF5-overexpressing cells are shown. C, representative Western blot of genes in the liver cancer RT2 ProfilerTM PCR array. Data are representative of three independent replicates performed in each cell line. D, summarized results from densitometry analysis of C. Data are from three independent replicates; plotted values are means ± S.D. (error bars) (*, p < 0.05; **, p < 0.001; ***, p < 0.0001).
Table 1.
IRF5 alters the expression of genes known to play a role in HCC pathogenesis
Gene expression was analyzed in C-5B cells transiently transfected with empty vector or GFP-IRF5 using the liver cancer RT2 ProfilerTM PCR array from SABiosciences. Genes depicted are those that had >1.5-fold change between empty vector control cells and IRF5-overexpressing cells. Boldface values show statistically significant changes in gene expression between empty vector control cells and IRF5-overexpressing cells. NS, not significant.
| Gene symbol | Gene name | Regulation | p |
|---|---|---|---|
| -fold | |||
| ADAM17 | ADAM metallopeptidase domain 17 | 1.90 | NS |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 2.28 | p < 0.01 |
| BAX | BCL2-associated X protein | 2.35 | p < 0.001 |
| BCL2L1 | BCL2-like 1 | 1.51 | p < 0.05 |
| BID | BH3-interacting domain death agonist | 2.50 | p < 0.01 |
| DAB2IP | DAB2-interacting protein | −1.71 | NS |
| EP300 | E1A-binding protein p300 | 1.77 | NS |
| FADD | Fas (TNFRSF6)-associated via death domain | 1.62 | NS |
| MET | Met proto-oncogene (hepatocyte growth factor receptor) | −1.58 | NS |
| MYC | V-myc myelocytomatosis viral oncogene homolog (avian) | 1.60 | p < 0.05 |
| NFKB1 | Nuclear factor of κ light polypeptide gene enhancer in B-cells 1 | 1.78 | p < 0.05 |
| PDGFRA | Platelet-derived growth factor receptor, α polypeptide | −1.57 | NS |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 4.09 | p < 0.01 |
| RAC1 | Ras-related C3 botulinum toxin substrate1 (ρ family, small GTP binding protein Rac1) | −1.58 | p < 0.05 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | −2.95 | p < 0.05 |
| TP53 | Tumor protein p53 | −1.64 | NS |
| VEGFA | Vascular endothelial growth factor A | −1.74 | NS |
| XIAP | X-linked inhibitor of apoptosis | −1.73 | NS |
Figure 7.
Ectopic IRF5 alters the expression of genes associated with HCC pathogenesis. A–C, same as in Fig. 6 except genes related to cell survival and growth were analyzed. B, summarized results from densitometry analysis of protein expression. Data are representative of three independent replicates from each cell line. Plotted values are means ± S.D. (error bars) (*, p < 0.05).
Re-expression of IRF5 in HCV replicon cells inhibits migration/invasion potential
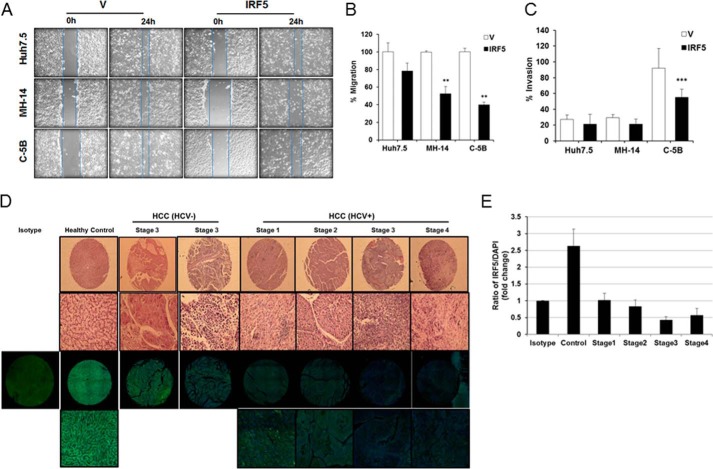
IRF5 was recently identified as a new regulator of epithelial cell migration (40). To further delineate a tumor suppressor role for IRF5 in HCV-associated HCC, IRF5-mediated cell migration and invasion was examined in cognate and HCV replicon–bearing cells (Fig. 8A). Summarized data in Fig. 8B show a significant reduction in wound-healing closure (i.e. migration) by IRF5-expressing HCV replicon cell lines only. Similar findings were made by a Matrigel invasion assay showing a significant reduction in IRF5-expressing C-5B cell invasion (Fig. 8C). These data support a role for IRF5 in HCV-infected hepatocyte migration and invasion, suggesting that loss of IRF5 expression in HCV-infected hepatocytes may contribute to a loss in hepatocyte growth control, HCC pathogenesis, and potentially metastasis to secondary organs.
Figure 8.
IRF5 inhibits the migration and invasion potential of HCV replicon cells. A, wound-healing assays were performed on Huh7.5, MH-14, and C-5B cells after transient transfection with empty vector (V) or GFP-IRF5 plasmid. Representative images are shown from 0 and 24 h after scratch. Lines demarcate visible wound borders. B, graphical representation of data from the 24-h time point is shown from four independent experiments performed in duplicate. Plotted values are means ± S.D. (error bars) (**, p < 0.001). C, similar to A except Matrigel invasion assays were performed on the indicated cell lines 16 h post-transfection. Graphical representation of data is shown from three independent experiments performed in duplicate. Plotted values are means ± S.D. (***, p < 0.0001). D, IRF5 expression is lost in the livers of HCV-positive HCC patients. Representative H&E and immunofluorescence staining of individual cores from a human liver cancer tissue array is shown. Representative images are from n = 20 healthy liver, n = 20 HCV- and HBV-negative HCC, and n = 18 HCV-positive HCC tissue cores. The top two panels show results from H&E staining; the top panel shows full core (×4 magnification), and the bottom panel shows a ×20 magnified image. The bottom two panels show immunofluorescence staining at the same magnifications; IRF5 is Alexa Fluor 488 (green), DAPI is blue. Left, single panel shows negative isotype control staining for IRF5 in healthy control core. E, summarized data of IRF5 expression is from n = 20 healthy liver and n = 38 HCC tissue cores stained in D. The ratio of IRF5 to DAPI staining is plotted for each disease stage.
Loss of IRF5 expression in human archived liver tissue specimens correlates with HCV-positive HCC
To further evaluate the contribution of IRF5 to HCV-associated HCC, we examined IRF5 expression in human clinical liver tissue specimens from patients with different stages of early, non-metastatic HCC that were either HCV-positive (n = 18) or HCV-negative (n = 20). Staging was based on the TNM (tumor, node, metastasis) system, and >95% of the samples analyzed were non-metastatic T2 or T3, N0, M0. Data in Fig. 8D reveal that endogenous IRF5 expression, while reduced in stage 3 HCV- and HBV-negative HCC as compared with healthy control liver, was further abrogated in livers of HCV-positive HCC patients. This loss in IRF5 protein expression occurred as early as stage 1 HCC, which is the earliest stage of HCC where the tumor has not spread to blood vessels, lymph nodes, or other parts of the body (T1, N0, M0), and was retained and further repressed as disease stage advanced (Fig. 8E). These data support a physiologic role for loss of IRF5 expression in the pathogenesis of HCV-associated HCC.
Discussion
HCC is the sixth most common cancer worldwide, and its incidence continues to increase because of the prevalence of chronic HCV infections (2, 41). Although HCV is now considered one of the major risk factors leading to HCC development, the precise mechanism(s) linking viral infection to HCC pathogenesis remains unclear (3). We present evidence implicating IRF5 in the mechanism(s) that links HCV infection with HCC pathogenesis. Data reveal that IRF5 expression is dramatically reduced in HCV-infected hepatocytes and HCV replicon–bearing cells as compared with uninfected, cognate cell lines. These findings are reminiscent of IRF1, whose expression was also found to be down-regulated in HCV-infected hepatocytes (8). However, this is not a shared feature for all IRF family members, as IRF3 expression was shown to be up-regulated by HCV infection, and IRF7 expression appeared unchanged by HCV infection, as expression was undetectable in human hepatocytes (27). Although the exact mechanism that HCV utilizes to attenuate IRF5 expression in replicon cells was not a focus of this first study, results from overexpression analysis of HCV proteins indicate that NS5A may be a candidate molecule for repressing IRF5 expression. Overexpression of NS5A but not NS5B or NS3 led to a significant reduction in IRF5 transcription from the pV3-IRF5 promoter (Fig. 1F). This finding is reminiscent of previous work showing modulation of IRF1 and IRF7 expression/function by NS5A for immune evasion (7, 10). Based on data presented herein and findings from Nandakumar (11), loss of IRF5 expression/function enables HCV replication, whereas retained expression restricts HCV RNA replication (Fig. 2). Among other functions identified for IRF5 in HCV replicon cells (Figs. 3–8), the virus has a clear and vested interest in shutting down IRF5 for the execution of a chronic infection state.
In addition to its well-established role(s) in lymphocytes to mediate innate and adaptive immune responses (13–19), results from this study provide the first insight into IRF5-mediated hepatocyte function. In the context of HCV, data indicate that IRF5 is necessary to limit hijacking of the autophagy pathway (Fig. 3). The effect of IRF5 on Beclin-1 cleavage is particularly interesting, as it occurs in a caspase-dependent manner and results in loss of function; it cannot induce autophagy or apoptosis (42). Simultaneously, it was found that a carboxyl-terminal cleaved fragment of Beclin-1 had pro-apoptotic function by inducing mitochondrion-mediated (intrinsic) apoptosis (34). Furthermore, up-regulated Bax induces the intrinsic apoptotic pathway and has been shown to cause caspase-mediated cleavage of Beclin-1 (43). Given that IRF5 re-expression in HCV replicon-bearing cells led to spontaneous apoptosis via the intrinsic apoptotic pathway (Figs. 4 and 5), these combined data suggest that IRF5 may mediate cross-talk between autophagy and apoptosis through its regulation of Bax. Bax functions in both pathways and was previously found to be regulated by IRF5, leading to enhanced caspase cleavage and apoptosis (17, 20, 22, 24). It remains to be determined whether the primary function of IRF5 in HCV replicon cells is to regulate gene transcription. This seems unlikely, however, because IRF5 cellular localization remained cytoplasmic (Fig. 3G). Nevertheless, in response to a productive in vivo HCV infection, we would expect IRF5 to become activated by post-translational modification(s), translocate to the nucleus, and induce a gene expression program that initiates functional host immunity to rid the body of infected hepatocytes.
Chronic HCV infection is a major contributor to HCC pathogenesis. Based on data presented herein, if the virus is successful in shutting down IRF5 expression/function in HCV-infected hepatocytes, these cells will gain additional survival and growth mechanisms to evade death and ultimately lead to HCC. Similar to other cancer cell types examined (20–23, 40), re-expression of IRF5 in HCV replicon cells led to spontaneous apoptosis and inhibition of hepatocyte cell migration and invasion (Figs. 4, 5, and 8). Although the mechanism(s) by which IRF5 regulates cell migration and invasion is not currently known, results from the current and previous studies suggest that VEGFA might be a valid target for regulation by IRF5 (40). VEGF is pivotal in the development of HCC by fostering tumor cell survival, proliferation, migration, and vessel formation (44). Serum VEGF concentrations are used as markers for HCC prognosis (45), and VEGF levels were found to be higher in HCV-infected HCC as compared with non-infected HCC patients (46). It is tempting to speculate that loss of IRF5 in HCV-infected hepatocytes leads to up-regulated VEGFA expression and HCC pathogenesis.
Last, our study provides the first clinical evidence that IRF5 expression is lost in HCC samples from HCV-infected individuals at early stages of disease development (Fig. 8). Whereas current data suggest that loss in IRF5 expression may be specific to HCV-associated HCC because expression was retained in stage 3 HCC (HCV- and HBV-negative), albeit to a lower extent than in healthy controls, additional samples will be necessary to confirm this finding. In conclusion, our study provides evidence that HCV-mediated loss of IRF5 expression in hepatocytes reduces their immune response(s) via reduced cytokine expression and enhances hepatocyte growth and survival. An understanding of the mechanisms by which HCV turns off critical immune mediators, such as IRF5, will have significant implications for therapeutic intervention of chronic infection and HCC development.
Materials and methods
Cell culture, treatments, and transfection
The human HCC cell line Huh7.5 was from the American Type Tissue Collection; Huh7 HCC cells were from Dr. C. Rice; and subgenomic HCV replicon-bearing MH-14 and cured MH-14 (MH-14(c)) were from Dr. Kunitada Shimotohno (47). Huh7/C-5B (from Dr. Stanley Lemon) and Huh7/Rep-Feo1b (from Dr. Naoya Sakamoto) cells contain the autonomously replicating HCV genotype 1b RNA; Huh7/Rep-Feo1b cells also contain firefly and Renilla luciferase reporter genes (48); Huh7.5-FGR-JC1-Rluc2A cells are similar to Feo1b cells except that they contain HCV genotype 2a RNA and were from Dr. Hangli Tang (49). All cell lines were grown similarly with 0.5 mg/ml G418, and experiments were performed at 70-80% confluence. All transfections were performed using LipoD293TM DNA in vitro transfection reagent (SignaGen) according to the manufacturer's instructions.
HCV infection
The pFL-J6/JFH-1 plasmid encoding the HCV J6/JFH-1 strain genome was transcribed and delivered into Huh7.5 cells by electroporation (50). The cell-free virus was propagated in Huh7.5 cell cultures and collected, as described (50). The expression of HCV NS3 protein in HCV-infected cells was confirmed by Western blotting. Purified viral supernatant was then used to infect naive Huh7.5 cells at a multiplicity of infection of 0.5 for 7 days at 37 °C and 5% CO2. Cells were harvested each day, and mRNA and protein expression was analyzed. In a complementary approach, IRF5 was knocked down with ON-TARGETplus IRF5 SMARTpool siRNAs (GE Dharmacon, catalog no. D-001810-10-05) before infection with purified virus stocks. Huh7 cells were transfected with either 80 nm ON-TARGETplus non-targeting control pool (GE Dharmacon, catalog no. D-001810-10-05), scrambled control, or IRF5 siRNAs in 6-well plates using LipoJet Transfection Kit II (SignaGen Laboratories) in accordance with the manufacturer's instructions. 48 h post-transfection, cells were infected with purified HCV as before. At the indicated time points, cells were harvested, and total RNA was isolated for real-time quantitative PCR analysis.
Western blotting
Cells were harvested in radioimmune precipitation assay lysis buffer (EMD Millipore) as described (28, 40). An equal amount of protein samples were resolved on 8–12% SDS-polyacrylamide gradient gels and transferred to 0.4-μm nitrocellulose membrane. After blocking, membranes were probed with primary antibodies overnight at 4 °C and HRP-conjugated secondary antibodies (Jackson Laboratory) at 1:5,000 dilution (40). Proteins were visualized using the enhanced ECL kit (Bio-Rad) and quantified on a FluorChem imaging system (ProteinSimple). β-Actin served as a loading control. Caspase-3, cleaved caspase-3, Bax, Bcl-2, Foxo3, Foxo1, p21, Bid, and Akt1 antibodies were from Cell Signaling Technology (Beverly, MA). FLAG antibodies were from Sigma-Aldrich; 14-3-3ϵ, β-actin, cyclin D1, cyto-c, PARP-1, c-Myc, NFκBp105, NFκBp50, p53, Beclin-1, APG5, MAP LC3β, and GFP antibodies were from Santa Cruz Biotechnology, Inc. (Dallas, TX). Antibodies to BCL2L1, XIAP, FADD, p300, CREB, DAB2IP, VEGF, Met, NFκBp65, Arc1, PDGFR, TRAF1, Nme2, and 14-3-3τ were from Fisher, and IRF5 and Cox2 antibodies were from Abcam (Cambridge, MA). Rabbit polyclonal antibody against NS5B was developed at Covance (Denver, PA) using recombinant NS5BCΔ21 as immunogen; anti-NS5A antibody was a kind gift from Dr. Craig Cameron; and monoclonal antibodies against NS3 were from Virogen (Watertown, MA). Protein band density was quantified using ImageJ software (National Institutes of Health).
Real-time qPCR
Total RNA was isolated and prepared as described (28, 40). Real-time qPCR was carried out using SYBR Green master mix (Applied Biosystems). All reactions were conducted in triplicate in a StepOneTM real-time PCR system (Applied Biosystems). The following primers were used for transcript detection: IRF5-F, 5′-GGGCTCATCCTCCAGCTAC-3′; IRF5-R, 5′-ACAAGGCCCGCTCCAGAA-3′; HCVRNA-F, 5′-CGGGAGAGCCATAGTGG-3′; HCVRNA-R, 5′-AGTACCACAAGGCCTTTCG-3′; IL6F, 5′-AGACAGCCACTCACCTCTTCAG-3′; IL6R, 5′-TTCTGCCAGTGCCTCTTTGCTG-3′; IL10F, 5′-TCTCCGAGATGCCTTCAGCAGA-3′; IL10R, 5′-TCAGACAAGGCTTGGCAACCCA-3′; TNFA-F, 5′-CTCTTCTGCCTGCTGCACTTTG-3′; TNFA-R, 5′-ATGGGCTACAGGCTTGTCACTC-3′. Gene expression was calculated relative to β-actin using the comparative 2−ΔΔCt method with StepOneTM software version 2.3.
Human liver cancer PCR array
Alterations in IRF5-mediated gene expression were examined in C-5B replicon cells transfected either with empty vector or IRF5 plasmid using the Liver Cancer RT2 ProfilerTM PCR array from SABiosciences (Frederick, MD) (catalog no. PHAS-133Z) (22, 51). This array profiles the expression of 84 genes involved in HCC progression. Total RNA was extracted and reverse-transcribed following the manufacturer's instructions. Four endogenous control genes (β-2-microglobulin (B2M), hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein large P0 (RPLP0), and β-actin (ACTB)) were used for data normalization with RT2 Profiler PCR array data analysis software version 3.5. Statistical significance was set at p < 0.05 and a mean difference ≥1.5-fold change in expression levels.
Dual-Luciferase assay
The IRF5 and ISRE promoter reporters have been described (18, 28). FLAG-tagged IRF5 contains a single FLAG tag at the amino terminus (18). Cells were seeded on 24- or 48-well plates and co-transfected with the indicated plasmids: IRF5 promoter V1 (pV1-IRF5-Luc), pV3-IRF5-Luc (150 ng/well), 350 ng/well viral proteins (NS5A, NS5B, and NS3 from HCV genotype 1b), or ISRE-Luc and FLAG-tagged IRF5 with Renilla luciferase (RL-SV40). HCV IRES-mediated translation was examined by co-transfection of FLAG-tagged IRF5 plasmid (80 ng/well) with Clneo-Rluc-IRES-Fluc (200 ng/well), in which Rluc is translated in a cap-dependent manner and Fluc is translated via HCV IRES-mediated initiation (52). Reporter activity was measured 36 h post-transfection with the Dual-Luciferase® reporter assay system (Promega, Madison, WI) (28). Levels of reporter firefly luciferase activity were normalized to Renilla luciferase activity, and -fold change was attained by comparing with empty vector control.
Acridine orange and ethidium bromide staining
Cells were seeded on cover slides at 70% confluence and transfected with empty vector or FLAG-tagged IRF5 (0.1 μg/well). At the indicated time points, cells were fixed with 2% paraformaldehyde in PBS for 15 min at 4 °C and incubated with 1 μg/ml AO and 1 μg/ml EtBr (Sigma-Aldrich) in PBS for 15 min at room temperature. After incubation, AO and EtBr were removed, and slides were washed three times with PBS. Slides were covered with anti-fade mounting medium and examined under a Nikon fluorescence microscope at ×20 magnification. Results are shown as the percentage of apoptotic cells by counting the number of dual AO/EtBr orange-stained cells within 500 cells. By this assay, dual-stained cells represent those undergoing late apoptosis and/or necrosis.
Cell viability and apoptosis
Cell viability and proliferation were analyzed using the CellTiter96® AQueous One Solution cell proliferation assay (MTS) (Promega). Cells were seeded at a concentration of 2 × 104 cells/well in 96-well polystyrene tissue culture plates and transfected with different concentrations of IRF5 (0.1, 0.2, and 0.4 μg) or empty plasmid. 24 h post-transfection, 15 μl of MTS solution (1.9 mg/ml) was added, and absorbance was read at 490 nm. For apoptosis, cells were transiently transfected with 0.08 μg of IRF5 plasmid, and apoptosis was detected 12 h later with the BD PharmingenTM FITC annexin V apoptosis detection kit on a BD FACscan. Data were analyzed using CELLQuest software (BD Biosciences) (22, 40).
JC-1 assay
Mitochondrial membrane potential was measured using the JC-1 assay kit (BD Pharmingen). Cells were transiently transfected with 0.08 μg of FLAG-tagged IRF5 for 24 h, grown on glass coverslips, and examined by fluorescence microscopy. For flow cytometry analysis, cells were harvested according to the manufacturer's protocol and analyzed on a FACScan. JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide) is a lipophilic fluorochrome used to evaluate Δψ. At low membrane potentials, JC-1 exists as a monomer and produces a green fluorescence. At high membrane potentials or concentrations, JC-1 forms J aggregates and produces a red fluorescence. Results are shown as the ratio of red/green fluorescence.
Wound-healing and migration/invasion assays
Wound-healing assays were performed as described (40). Cells were transfected with empty vector or GFP-tagged IRF5 (0.1 μg/well), plated, and grown to confluence. 12 h post-transfection, a disruption in the monolayer was created with a sterile p-10 pipette tip (40). After a 24-h incubation, migrated cells were photographed under a phase-contrast inverted microscope, and wounds were measured using ImageJ software. Wounds were measured by width at three points and averaged. Average width was then used to calculate relative percentage of migration. Migration/invasion assays were performed using BD 24-well Boyden chambers (8-μm pore size) as described (40). Briefly, 1.5 × 105 cells were added to the top insert, and 750 μl of 5% FBS-containing medium was added to the bottom chamber. After a 16-h incubation, cells that migrated to the lower surface of the membrane were fixed, stained with a Diff-Quick stain kit (Dade Behring, Inc., Westwood, MA), and counted (40).
LC3II staining and microscopy
MH-14 and C-5B cells were transfected with 0.1 μg of FLAG-IRF5 or empty vector in 6-well plates using LipoJet Transfection Kit II. 48 h post-transfection, cells were fixed with 100% methanol at −20 °C for 15 min. Cells were then washed three times with 1× PBS and then blocked for 60 min at room temperature in blocking buffer (PBS, 5% goat serum, 0.3% Triton X-100). Cells were then incubated with mouse anti-IRF5 (Abcam) and rabbit anti-LC3II antibodies, followed by Alexa Fluor 488–conjugated goat anti-mouse (Invitrogen) and Cy3-conjugated donkey anti-rabbit antibodies (Jackson ImmunoResearch Laboratories). After washing in 1× PBS, cells were mounted with VECTASHIELD antifade mounting medium with DAPI (Vector Laboratories), which counterstained the nuclei. Images were captured on a Zeiss fluorescence apotome microscope. For tissue staining, purchased tissue arrays (T031a and LV8013, US Biomax, Rockville, MD) containing either 24 or 80 tissue cores were deparaffinized according to the manufacturer's instructions. Antigen retrieval was done in sodium citrate buffer (pH 6.0) as described (23, 51, 53). After blocking, slides were stained with mouse anti-human IRF5 (1:100; Novus) diluted in 4% BSA and secondary antibodies (1:1,000) applied at room temperature for 2 h. Isotype mouse and rabbit IgG controls and their corresponding secondary antibodies were used as negative controls. Slides were counterstained and mounted with DAPI aqueous mounting medium (Vectamount). Tissue arrays were scanned on a Nikon AIR confocal microscope (Nikon Instruments, Melville, NY). Data were analyzed using the ImageJ program (National Institutes of Health) by calculating the ratio of IRF5/DAPI staining.
Scoring
To determine IRF5-positive and -negative staining, images were compared with tissue cores stained only with IgG control or secondary antibody (23, 51). To be considered IRF5-positive, >50% of tumor cells had to show intensities above negative control samples (23, 51). Two independent reviewers unaware of the clinical data scored each tissue core as positive or negative for IRF5 (23). Tissue cores consisting of mostly ducts, adipose tissue, or blood vessel sections were not counted. Once the samples were scored, data were compared with HCV status and HCC stage.
Statistical analysis
Experimental data are presented as the mean ± S.D. from at least three independent experiments performed in triplicate. Differences between groups were analyzed by Student's t test. Statistical analysis was performed using GraphPad Prism version 5.0 software. Statistical significance is defined as follows: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.0001.
Author contributions
O. C., E. B., D. L., D. M., and E. M. P. carried out all experimental procedures, analyzed data, and generated figures. G. W. performed primary infections with HCV chimeric virus. O. C., D. L., G. W., B. J. B., and N. K.-B. prepared the manuscript; B. J. B. and N. K.-B. conceived and designed the study and drafted the manuscript.
Acknowledgments
We are grateful to Dr. Kunitada Shimotohno for providing the cured MH-14 and MH-14 cell lines and Dr. Stanley Lemon for the C-5B cells. Plasmids pCIneo-Rluc-IRES-Fluc and Huh7/Rep-Feo1b were generously provided by Dr. Naoya Sakamoto. We thank Dr. Hengli Tang for Huh7.5-FGR-JC1-Rluc2A replicon reporter cells.
This work was supported by National Institutes of Health Grants CA153147 (to N. K.-B.) and CA191903 (to B. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HCV
- hepatitis C virus
- HCC
- hepatocellular carcinoma
- IRES
- internal ribosome entry site
- IRF
- interferon regulatory factor
- Δψm
- mitochondrial membrane potential
- ISRE
- interferon-stimulated response element
- AO
- acridine orange
- VEGFA
- vascular endothelial growth factor A
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt.
References
- 1. Hajarizadeh B., Grebely J., Dore G. J. (2013) Epidemiology and natural history of HCV infection. Nat. Rev. Gastroenterol. Hepatol. 10, 553–562 [DOI] [PubMed] [Google Scholar]
- 2. Mohd Hanafiah K., Groeger J., Flaxman A. D., and Wiersma S. T. (2013) Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57, 1333–1342 [DOI] [PubMed] [Google Scholar]
- 3. Arzumanyan A., Reis H. M., and Feitelson M. A. (2013) Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 13, 123–135 [DOI] [PubMed] [Google Scholar]
- 4. Beales L. P., Holzenburg A., and Rowlands D. J. (2003) Viral internal ribosome entry site structures segregate into two distinct morphologies. J. Virol. 77, 6574–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barth H. (2015) Hepatitis C virus: is it time to say goodbye yet? Perspectives and challenges for the next decade. World J. Hepatol. 7, 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wack A., Terczyńska E., and Hartmann R. (2015) Guarding the frontiers: the biology of type III interferons. Nat. Immunol. 16, 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pflugheber J., Fredericksen B., Sumpter R. Jr., Wang C., Ware F., Sodora D. L., and Gale M. Jr. (2002) Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. U.S.A. 99, 4650–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim H., Mazumdar B., Bose S. K., Meyer K., Di Bisceglie A. M., Hoft D. F., and Ray R. (2012) Hepatitis C virus-mediated inhibition of cathepsin S increases invariant-chain expression on hepatocyte surface. J. Virol. 86, 9919–9928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Foy E., Li K., Wang C., Sumpter R. Jr., Ikeda M., Lemon S. M., and Gale M. Jr. (2003) Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300, 1145–1148 [DOI] [PubMed] [Google Scholar]
- 10. Raychoudhuri A., Shrivastava S., Steele R., Dash S., Kanda T., Ray R., and Ray R. B. (2010) Hepatitis C virus infection impairs IRF-7 translocation and alpha interferon synthesis in immortalized human hepatocytes. J. Virol. 84, 10991–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nandakumar R., Finsterbusch K., Lipps C., Neumann B., Grashoff M., Nair S., Hochnadel I., Lienenklaus S., Wappler I., Steinmann E., Hauser H., Pietschmann T., and Kröger A. (2013) Hepatitis C virus replication in mouse cells is restricted by IFN-dependent and -independent mechanisms. Gastroenterology 145, 1414–1423.e1 [DOI] [PubMed] [Google Scholar]
- 12. Binder M., Kochs G., Bartenschlager R., and Lohmann V. (2007) Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology 46, 1365–1374 [DOI] [PubMed] [Google Scholar]
- 13. Ikushima H., Negishi H., and Taniguchi T. (2013) The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb. Symp. Quant. Biol. 78, 105–116 [DOI] [PubMed] [Google Scholar]
- 14. Zhao G. N., Jiang D. S., and Li H. (2015) Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim. Biophys. Acta 1852, 365–378 [DOI] [PubMed] [Google Scholar]
- 15. Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T. W., and Taniguchi T. (2005) Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434, 243–249 [DOI] [PubMed] [Google Scholar]
- 16. Schoenemeyer A., Barnes B. J., Mancl M. E., Latz E., Goutagny N., Pitha P. M., Fitzgerald K. A., and Golenbock D. T. (2005) The interferon regulatory 5, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 280, 17005–17012 [DOI] [PubMed] [Google Scholar]
- 17. Yanai H., Chen H. M., Inuzuka T., Kondo S., Mak T. W., Takaoka A., Honda K., and Taniguchi T. (2007) Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 104, 3402–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barnes B. J., Moore P. A., and Pitha P. M. (2001) Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon α genes. J. Biol. Chem. 276, 23382–23390 [DOI] [PubMed] [Google Scholar]
- 19. Barnes B. J., Kellum M. J., Field A. E., and Pitha P. M. (2002) Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol. Cell. Biol. 22, 5721–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barnes B. J., Kellum M. J., Pinder K. E., Frisancho J. A., and Pitha P. M. (2003) Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 63, 6424–6431 [PubMed] [Google Scholar]
- 21. Mori T., Anazawa Y., Iiizumi M., Fukuda S., Nakamura Y., and Arakawa H. (2002) Identification of the interferon regulatory factor 5 gene (IRF-5) as a direct target for p53. Oncogene 21, 2914–2918 [DOI] [PubMed] [Google Scholar]
- 22. Hu G., Mancl M. E., and Barnes B. J. (2005) Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res. 65, 7403–7412 [DOI] [PubMed] [Google Scholar]
- 23. Bi X., Hameed M., Mirani N., Pimenta E. M., Anari J., and Barnes B. J. (2011) Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis. Breast Cancer Res. 13, R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Couzinet A., Tamura K., Chen H. M., Nishimura K., Wang Z., Morishita Y., Takeda K., Yagita H., Taniguchi T., and Tamura T. (2008) A cell-type-specific requirement for IFN regulatory factor 5 (IRF5) in Fas-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alzaid F., Lagadec F., Albuquerque M., Ballaire R., Orliaguet L., Hainault I., Blugeon C., Lemoine S., Lehuen A., Saliba D. G., Udalova I. A., Paradis V., Foufelle F., and Venteclef N. (2016) IRF5 governs liver macrophage activation that promotes hepatic fibrosis in mice humans. JCI Insight 1, e88689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Courties G., Heidt T., Sebas M., Iwamoto Y., Jeon D., Truelove J., Tricot B., Wojtkiewicz G., Dutta P., Sager H. B., Borodovsky A., Novobrantseva T., Klebanov B., Fitzgerald K., Anderson D. G., et al. (2014) In vivo silencing of the transcription factor IRF5 reprograms the macrophage phenotype and improves infarct healing. J. Am. Coll. Cardiol. 63, 1556–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ghosh A. K., Majumder M., Steele R., Ray R., and Ray R. B. (2003) Modulation of interferon expression by hepatitis C virus NS5A protein and human homeodomain protein PTX1. Virology 306, 51–59 [DOI] [PubMed] [Google Scholar]
- 28. Mancl M. E., Hu G., Sangster-Guity N., Olshalsky S. L., Hoops K., Fitzgerald-Bocarsly P., Pitha P. M., Pinder K., and Barnes B. J. (2005) Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms: multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J. Biol. Chem. 280, 21078–21090 [DOI] [PubMed] [Google Scholar]
- 29. Vescozo T., Refolo G., Romagnoli A., Ciccosanti F., Corazzari M., Alonzi T., and Fimia G. M. (2014) Autophagy in HCV infection: keeping fat and inflamamtion at bay. Biomed. Res. Intern. 2014, 265353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dreux M., Gastaminza P., Wieland S. F., and Chisari F. V. (2009) The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 106, 14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rautou P. E., Cazals-Hatem D., Feldmann G., Mansouri A., Grodet A., Barge S., Martinot-Peignoux M., Duces A., Bièche I., Lebrec D., Bedossa P., Paradis V., Marcellin P., Valla D., Asselah T., and Moreau R. (2011) Changes in autophagic response in patients with chronic hepatitis C virus infection. Am. J. Pathol 178, 2708–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shrivastava S., Raychoudhuri A., Steele R., Ray R., and Ray R. B. (2011) Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 53, 406–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shrivastava S., Chowdhury J., Steele R., Ray R., and Ray R. B. (2012) Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J. Virol. 86, 8705–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wirawan E., Vande Walle L., Kersse K., Cornelis S., Claerhout S., Vanoverberghe I., Roelandt R., De Rycke R., Verspurten J., Declercq W., Agostinis P., Vanden Berghe T., Lippens S., and Vandenabeele P. (2010) Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 1, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao Y., Yang J., Liao W., Liu X., Zhang H., Wang S., Wang D., Feng J., Yu L., and Zhu W. G. (2010) Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 12, 665–675 [DOI] [PubMed] [Google Scholar]
- 36. Wang R. C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., and Levine B. (2012) Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brault C., Levy P. L., and Bartosch B. (2013) Hepatitis C virus-induced mitochondrial dysfunctions. Viruses 5, 954–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fresquet V., Robles E. F., Parker A., Martinez-Useros J., Mena M., Malumbres R., Agirre X., Catarino S., Arteta D., Osaba L., Mollejo M., Hernandez-Rivas J. M., Calasanz M. J., Daibata M., Dyer M. J., et al. (2012) High-throughput sequencing analysis of the chromosome 7q32 deletion reveals IRF5 as a potential tumour suppressor in splenic marginal-zone lymphoma. Br. J. Haematol. 158, 712–726 [DOI] [PubMed] [Google Scholar]
- 39. Ferrara N., Gerber H. P., and LeCouter J. (2003) The biology of VEGF and its receptors. Nat. Med. 9, 669–676 [DOI] [PubMed] [Google Scholar]
- 40. Pimenta E. M., and Barnes B. J. (2015) A conserved region within interferon regulatory factor 5 controls breast cancer cell migration through a cytoplasmic and transcription-independent mechanism. Mol. Cancer 14, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Attar B. M., and Van Thiel D. H. (2016) Hepatitis C virus, A time for decisions. Who should be treated and when? World J. Gastrointest. Pharmacol. Ther. 7, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H., Wang P., Yu J., and Zhang L. (2011) Cleaving Beclin 1 to suppress autophagy in chemotherapy-induced apoptosis. Autophagy 7, 1239–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luo S., Rubinsztein D. C. (2010) Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-XL. Cell Death Differ. 17, 268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bishayee A., and Darvesh A. S. (2012) Angiogenesis in hepatocellular carcinoma: a potential target for chemoprevention and therapy. Curr. Cancer Drug Targets 12, 1095–1118 [PubMed] [Google Scholar]
- 45. Kemik O., Sumer A., Kemik S. A., Purisa S., and Tuzun S. (2010) Circulating levels of VEGF family and their receptors in hepatocellular carcinoma. Bratisl. Lek. Listy 111, 485–488 [PubMed] [Google Scholar]
- 46. Chu J. S., Ge F. J., Zhang B., Wang Y., Silvestris N., Liu L. J., Zhao C. H., Lin L., Brunetti A. E., Fu Y. L., Wang J., Paradiso A., and Xu J. M. (2013) Expression and prognostic value of VEGFR-2, PDGFR-β, and c-Met in advanced hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 32, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Murata T., Ohshima T., Yamaji M., Hosaka M., Miyanari Y., Hijikata M., and Shimotohno K. (2005) Suppression of hepatitis C virus replicon by TGF-β. Virology 331, 407–417 [DOI] [PubMed] [Google Scholar]
- 48. Blight K. J., McKeating J. A., and Rice C. M. (2002) Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76, 13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Küçükgüzel I., Satilmış G., Gurukumar K. R., Basu A., Tatar E., Nichols D. B., Talele T. T., and Kaushik-Basu N. (2013) 2-Heteroarylimino-5-arylidene-4-thiazolidinones as a new class of non-nucleoside inhibitors of HCV NS5B polymerase. Eur. J. Med. Chem. 69, 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lindenbach B. D., Evans M. J., Syder A. J., Wölk B., Tellinghuisen T. L., Liu C. C., Maruyama T., Hynes R. O., Burton D. R., McKeating J. A., and Rice C. M. (2005) Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 [DOI] [PubMed] [Google Scholar]
- 51. Pimenta E. M., De S., Weiss R., Feng D., Hall K., Kilic S., Bhanot G., Ganesan S., Ran S., and Barnes B. J. (2015) IRF5 is a novel regulator of CXCL13 expression in breast cancer that regulates CXCR5(+) B- and T-cell trafficking to tumor-conditioned media. Immunol. Cell Biol. 93, 486–499 [DOI] [PubMed] [Google Scholar]
- 52. Kim K., Kim K. H., Kim H. Y., Cho H. K., Sakamoto N., and Cheong J. (2010) Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS Lett. 584, 707–712 [DOI] [PubMed] [Google Scholar]
- 53. Li D., De S., Li D., Song S., Matta B., and Barnes B. J. (2016) Specific detection of interferon regulatory factor 5 (IRF5): a case of antibody inequality. Sci. Rep. 6, 31002. [DOI] [PMC free article] [PubMed] [Google Scholar]