Abstract
Studies of virulence determinants in the bacterial phytopathogen Erwinia amylovora, the cause of devastating fire blight disease in apple and pear, have shown that HsvA, a putative amidinotransferase enzyme located in the Hrp pathogenicity island, is required for systemic infection in apple. However, the mechanism by which HsvA contributes to virulence is unclear. To investigate the role of HsvA in virulence, we carried out a series of biochemical and structural studies to characterize the amidinotransferase activity of HsvA. We found that HsvA displays a preference for linear aliphatic polyamines as the amidino acceptor substrate, especially for spermidine and putrescine (Km values of 33 μm and 3.9 mm, respectively). The three-dimensional structure, determined at 2.30 Å resolution using X-ray crystallography, revealed that the overall architecture of HsvA is similar to that of the human arginine–glycine amidinotransferase in the creatine biosynthesis pathway. The active site is located in the core of the protein at the base of a long, narrow substrate access channel. Specific amino acids near the entrance of the channel may serve as major determinants of the substrate specificity, including a glutamate residue at the rim of the channel entrance that appears to be positioned to interact with the distal primary amine in the putrescine substrate as well as the internal and distal amines in the spermidine substrate. These results suggest potential in vivo functions for HsvA as a virulence factor in fire blight and may also provide a basis for strategies to control fire blight by inhibiting HsvA activity.
Keywords: enzyme kinetics, polyamine, protein structure, virulence factor, X-ray crystallography, amidinotransferase, fire blight
Introduction
Fire blight is a devastating disease which affects a variety of plants in the Rosaceae family, with apple and pear being the hosts of greatest economic importance (1). Fire blight disease is caused by the Gram-negative bacterial pathogen Erwinia amylovora, a member of the Enterobacteriaceae. Although originally native to North America, the pathogen is now found in most regions of the world where apples and pears are grown commercially (2). This wide geographic spread, coupled with limited options for effective control, makes fire blight one of the most significant threats to commercial apple and pear production, an industry that was valued at nearly $4 billion in the United States in 2015 (3). Economic losses from fire blight in the United States are estimated to exceed $100 million annually (4).
Genome sequencing and genetic studies in E. amylovora have identified a number of genes required for pathogenicity, many of which are clustered within a ∼62-kb region of the genome that is designated as the Hrp pathogenicity island (5). Although most of the genes in the Hrp pathogenicity island encode a type III secretion system involved in the production and delivery of several essential virulence factors, one locus within the pathogenicity island contains five open reading frames, organized into two operons, which are predicted to encode enzymes. Expression of both of these operons is controlled by the alternative σ factor HrpL, which is specifically up-regulated during infection, suggesting a role for these two operons in virulence (5). Together, these two operons comprise a region designated as the Hrp-associated enzyme (HAE)2 locus. Genetic studies have shown that at least three of the genes in this locus, hsvA, hsvB, and hsvC, are required for full virulence in apple (5). Mutant strains of E. amylovora individually lacking either hsvA, hsvB, or hsvC exhibit significantly reduced virulence in apple shoots, suggesting that the encoded proteins may be part of a biochemical pathway operating during infection to synthesize a virulence factor that is required for systemic spread of the pathogen in apple.
HsvA is predicted to be an amidinotransferase enzyme on the basis of amino acid sequence similarity with other prokaryotic and eukaryotic amidinotransferases (5). Amidinotransferase enzymes catalyze the transfer of the amidino group from a donor molecule (usually arginine) to an acceptor molecule bearing a primary amine. Amidinotransferase enzymes are widespread in nature, occurring in essential metabolic pathways in eukaryotes as well as in biosynthetic pathways for antibiotics and virulence factors in prokaryotes.
In vertebrates, an arginine–glycine amidinotransferase (AGAT) catalyzes the first reaction in the creatine biosynthesis pathway, transferring the amidino group from arginine to glycine to form guanidinoacetate, an intermediate in the pathway, and ornithine, a by-product (6). In the three-dimensional structure of the human homolog (hAGAT), which displays 40% amino acid identity with HsvA, the active site lies in the interior of the protein at the base of a long, narrow channel and contains a catalytic triad composed of cysteine, histidine, and aspartic acid (7). Biochemical and kinetic studies clearly demonstrate a double-displacement (ping-pong) reaction mechanism involving temporary covalent modification of the active-site cysteine, which serves as a nucleophile to initiate the transamidination reaction upon binding of the amidino donor substrate (8, 9). The architecture of the active-site region also supports a double-displacement reaction mechanism, as the channel leading to the active site is too narrow to permit simultaneous binding of an amidino donor and acceptor substrate during catalysis (10).
In prokaryotes, amidinotransferase enzymes have been found in the biosynthetic pathways of two aminoglycoside antibiotics, including streptomycin (11) and bluensomycin (12), and the antimycobacterial peptide antibiotics pheganomycin and resorcinomycin (13). Amidinotransferase enzymes are also known to participate in the biosynthetic pathways of several bacterial toxins, including cylindrospermopsin (14, 15), saxitoxin (16), and phaseolotoxin (17, 18). HsvA displays 48% amino acid identity with AmtA, the amidinotransferase found in the biosynthetic pathway for phaseolotoxin, a derivatized tripeptide phytotoxin produced in Pseudomonas syringae pv. phaseolicola, the cause of bean halo blight. AmtA transfers an amidino group from arginine to lysine, forming homoarginine and ornithine, both of which are incorporated into the structure of the toxin (17).
Although genetic studies have shown that HsvA is necessary for full systemic virulence of E. amylovora in apple, the biochemical function of HsvA and the mechanism by which the protein contributes to virulence are unknown. We carried out a series of biochemical studies to characterize the predicted amidinotransferase activity of HsvA, and we also determined the atomic structure of HsvA at 2.30 Å resolution using X-ray crystallography. Our results show that HsvA is an amidinotransferase having novel acceptor substrate specificity, with a clear preference for linear polyamines, especially putrescine and spermidine, as the amidino acceptor substrate. The architecture of the active-site region in the three-dimensional structure provides insight into the structural basis for this substrate specificity. These results suggest some potential in vivo functions for HsvA as a virulence factor in fire blight and also provide a basis for the development of novel strategies to control fire blight through the inhibition of HsvA activity.
Results
Phylogenetic comparison of HsvA with other known or predicted amidinotransferase sequences
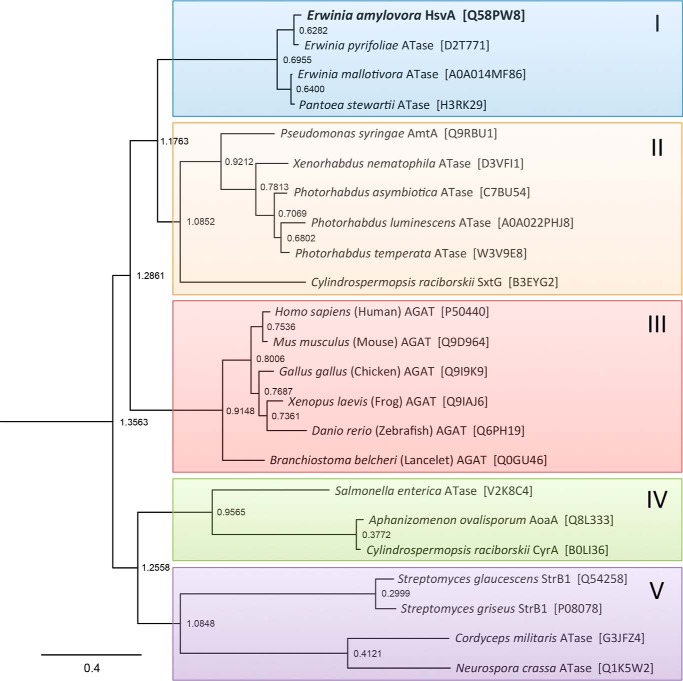
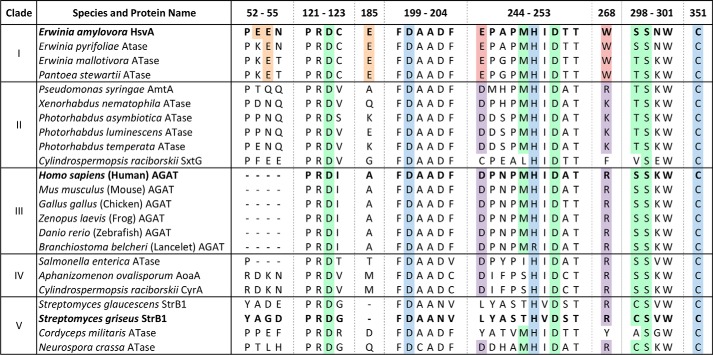
To gain insight into the relationship between HsvA and other amidinotransferases, the amino acid sequence of HsvA from E. amylovora was aligned with 22 other known or predicted amidinotransferase sequences from a variety of prokaryotes and eukaryotes (see Table S1 and Fig. S1), and a phylogeny tree was constructed (Fig. 1). The sequences used in phylogenetic analysis include several for which amidinotransferase activity has been confirmed experimentally (hAGAT and selected orthologs from other vertebrates, StrB1 from Streptomyces griseus, CyrA from Cylindrospermopsis raciborskii, AoaA from Aphanizomenon ovalisporum, and AmtA from P. syringaepv. phaseolicola). Phylogenetic analysis also included several predicted orthologs of HsvA from closely related pathogens, including Erwinia pyrifoliae, a cause of necrotic disease in Asian pear, Erwinia mallotivora, the cause of papaya blight, and Pantoea stewartii, the cause of corn leaf blight, a pathogen that was formerly classified within the Erwinia genus.
Figure 1.
Phylogenetic comparison of HsvA with other amidinotransferase sequences. The amino acid sequence of HsvA from Erwinia amylovora was aligned with 22 predicted or confirmed amidinotransferase (ATase) sequences from a variety of prokaryotic and eukaryotic organisms, and a phylogram based upon the Jones–Taylor–Thornton probability model was constructed using PHYLIP. Colored shading was added to emphasize distinct clades. The short node-to-tip distances in the uppermost clade (I, highlighted in blue) show that HsvA and other predicted amidinotransferase sequences from the Erwinia genus (and one amidinotransferase sequence from the closely related genus Pantoea) cluster together in a unique clade, distinct from other bacterial amidinotransferase sequences. The UniProt accession number for each sequence is provided in brackets along with the name of the organism in which each sequence is found. The scale bar provides a measure of relative genetic distance along the branches. The node labels represent horizontal distance from the rightmost tip, a predicted amidinotransferase found in the model fungal organism N. crassa (Q1K5W2).
Using node-to-tip distances as an indication of sequence similarity, the amidinotransferase sequences in this phylogram appear to form five distinct clades. HsvA from E. amylovora aligns within the uppermost cluster of sequences, designated as group I, which also includes the predicted orthologs of HsvA from closely related pathogens in Erwinia and Pantoea. Group II is composed mostly of amidinotransferase sequences from various genera in the family Enterobacteriaceae, although one notable exception within this clade is SxtG from the cyanobacterium Cylindrospermopsis raciborskii, which functions as an amidinotransferase in the biosynthetic pathway for saxitoxin. Group III encompasses the well-characterized vertebrate AGAT sequences as well as an AGAT ortholog from lancelets, an invertebrate marine animal. Group IV includes two cyanobacterial sequences that are known to be arginine–glycine amidinotransferases (CyrA and AoaA), although they appear to have diverged fairly early in their evolutionary history from the lineage leading to the vertebrate AGAT sequences. The four amidinotransferase sequences included in group V form two clusters that are less closely related to one another, as judged from the longer node-to-tip distances observed in this portion of the phylogram; in addition to the well-studied StrB1 amidinotransferase from Streptomyces, this clade includes predicted amidinotransferases from two fungi, the model organism Neurospora crassa and the insect pathogen Cordyceps militaris.
Two observations emerge from this phylogenetic analysis. First, HsvA from E. amylovora and the predicted orthologs from E. pyrifoliae, E. mallotivora, and P. stewartii, form a unique clade that is distinct from amidinotransferase sequences found in a variety of other members of the family Enterobacteriaceae. Although Erwinia and Pantoea are also classified within the Enterobacteriaceae, the phylogram suggests that the amidinotransferase sequences found in these pathogens diverged relatively early from those found in other Enterobacteriaceae or that they are the result of a horizontal transfer event from a more distantly related organism. The possibility of horizontal transfer is supported by the observation that HsvA in E. amylovora is found on a ∼60-kb pathogenicity island that is bounded by sequences often associated with horizontal gene transfer in bacteria (5). Second, the four sequences in this clade appear to be quite closely related, as suggested by the comparatively short node-to-tip distances in the phylogram and the high degree of amino acid conservation that is seen among these four amidinotransferases in the sequence alignment shown in Fig. S1. The HsvA orthologs in these four plant pathogens are likely to have very similar functions.
Recombinant expression, purification, and biophysical characterization of HsvA
To investigate the enzymatic activity and determine the molecular structure of HsvA, the full-length hsvA gene from Erwinia amylovora strain Ea321 was cloned and expressed in Escherichia coli, and the protein was purified to homogeneity using affinity chromatography. Yields typically ranged between 25 and 50 mg/liter of bacterial culture.
Characterization of amidinotransferase activity
Amidino donor substrate specificity
Although amidinotransferases are generally able to use a variety of guanidine-containing molecules as amidino donor substrates, arginine is believed to be the primary amidino donor substrate in vivo, with reported Km values ranging from 0.74 mm in AoaA (15) to 2.0 mm in hAGAT (9), 3.5 mm in CyrA (14), and 7.7 mm in AmtA (19). To investigate the amidino donor substrate specificity profile of HsvA, we used a modified functional assay in which the amidino acceptor substrate is omitted, and thus only the first half of the double displacement reaction mechanism occurs as the amidino group is transferred from the donor substrate to the active-site cysteine. The observed product of this reaction is the portion of the amidino donor substrate that remains after the amidino group is removed. This low-efficiency arginase-like activity in the absence of an amidino acceptor substrate has been observed in other amidinotransferases (14, 20).
We applied this modified assay to screen for amidino transfer activity using arginine and several structural analogs, including homoarginine (an analog with one additional methylene in the side chain), agmatine (an analog without the carboxylate group), arcaine (an analog with an amidino group at both ends), and guanidinoacetate. As shown in Fig. S2, HsvA is able to convert arginine into ornithine, homoarginine into lysine, agmatine into putrescine, and arcaine into agmatine, indicating that these four guanidine-containing molecules can serve as donor substrates. HsvA did not appear to be able to convert guanidinoacetate into glycine, in contrast to hAGAT and CyrA, both arginine–glycine amidinotransferases that are able to use guanidinoacetate as an amidino donor (6, 21). Under conditions in which a suitable acceptor substrate was provided, we observed that HsvA is also able to use canavanine, a guanidinooxy analog of arginine, as an amidino donor substrate, as has been seen with hAGAT, StrB1, and AmtA (6, 11, 18).
Notably, agmatine and arcaine are positively charged at both ends of the molecule under physiological conditions, and, unlike arginine, homoarginine, or canavanine, neither has a negatively charged functional group. Thus, a common structural feature of all donor substrates observed to function in HsvA is the presence of a positive charge on the end of the molecule that is distal to the amidino group being transferred. This structure-activity relationship suggests that the active site may be designed to recognize donor substrates via interaction with the positively charged groups on both ends of the molecule. Taken together, these results indicate that the amidino donor substrate specificity profile for HsvA is distinct from other amidinotransferases for which comparable data is available.
Amidino acceptor substrate specificity
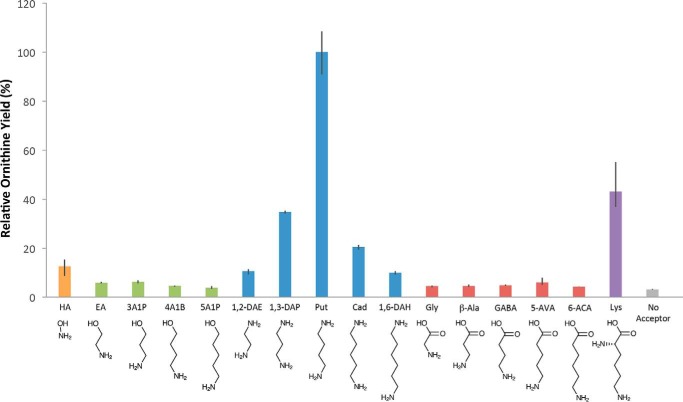
The functional role of an amidinotransferase is generally defined by the range of molecules that can be used as substrates for the second half of the amidinotransfer reaction, during which the amidino group is transferred from the nucleophilic cysteine in the active site to a primary amine on the acceptor substrate. To investigate the amidino acceptor substrate specificity for HsvA, we screened a panel of 16 candidate acceptor substrates in an assay that used arginine as a donor, detecting amidinotransferase activity by quantifying the amount of ornithine produced after 30 min.
The yield of ornithine observed in reactions containing each of the amidino group acceptor candidates is shown in Fig. 2. Putrescine was clearly the most efficient amidino acceptor among those evaluated in this panel. Other polyamines ranging in length from two carbons to six carbons showed varying levels of activity; the observed structure-activity relationship suggests that a chain length of four carbons provides the optimum separation between primary amines in the acceptor substrate. Lysine, which contains two primary amines separated by five carbons, also showed modest activity as an amidino acceptor substrate. As has been observed with other amidinotransferases, hydroxylamine also displayed limited activity as an acceptor substrate. In contrast, a series of amino alcohols and linear amino acids spanning a similar range of chain lengths displayed little to no activity. The yield of ornithine observed in connection with these candidate acceptor substrates was similar to the amount produced when no acceptor was provided, suggesting that the small quantity of ornithine accumulation in these reactions was due primarily to the basal-level arginase-like activity of the enzyme that is seen in absence of an acceptor (as indicated previously in Fig. S2).
Figure 2.
Relative efficiency of various short-chain candidate amidino acceptor substrates in HsvA. HsvA displays a clear preference for short polyamines as the amidino acceptor substrate, with four methylenes as the optimum separation between primary amine groups. Candidate acceptor substrates lacking a second primary amine are strongly disfavored. In these assays, HsvA was incubated with arginine and a panel of candidate amidino acceptor substrates at 30 °C for 30 min, and ornithine yield was quantified using HPLC. Relative ornithine yield is shown as a percentage of the ornithine accumulation observed when putrescine is provided as the amidino acceptor. Each bar represents the average of three trials, with error bars indicating the range of values observed. Negative control assays contained arginine but no acceptor substrate. HA, hydroxylamine; EA, ethanolamine; 3A1P, 3-amino-1-propanol; 4A1B, 4-amino-1-butanol; 5A1P, 5-amino-1-pentanol; 1,2-DAE, 1,2-diaminoethane; 1,3-DAP, 1,3-diaminopropane; Put, putrescine; Cad, cadaverine; 1,6-DAH, 1,6-diaminohexane; β-Ala, β-alanine; 5-AVA, 5-aminovaleric acid; 6-ACA, 6-aminocaproic acid.
Taken together, these results suggest that the active site is able to recognize amidino acceptor substrates having primary amines separated by 5–7 Å, a distance that is found in molecules having 3–5 methylene groups in the linear chain, with putrescine providing the ideal separation of ∼6 Å between primary amines. The absence of a second primary amine in a candidate substrate appears to prevent productive binding to the active site, as can be readily seen by comparing the activity of putrescine with that of the inactive analogs 4-amino-1-butanol and GABA, which otherwise have similar molecular shapes. The preference for amidino acceptor substrates having two primary amine groups, which would be positively charged under physiological conditions, is similar to the trend that was observed for amidino donor substrates, suggesting that the active site is designed to recognize both the donor and the acceptor substrates through similar enzyme–substrate interactions, probably involving the same amino acids in the active-site region.
Kinetic parameters
With arginine as an amidino donor and putrescine as an acceptor, HsvA obeyed Michaelis–Menten kinetics with a Km value of 0.47 mm for arginine and 3.9 mm for putrescine (see Table 1). The Km value for putrescine is similar to values reported for acceptor substrates used in other amidinotransferases, including glycine in hAGAT, CyrA, and AoaA (9, 14, 15) and lysine in AmtA (19), with reported Km values ranging between 2.0 and 6.9 mm. Although the structurally related molecules 1,3-diaminopropane, cadaverine, and lysine all displayed low to moderate amidino acceptor activity under the conditions used in the substrate specificity profile screen, we were not able to measure Km values for these acceptors, as we observed only very low levels of product formation within the time frame of our kinetic assays at acceptor substrate concentrations <30 mm (data not shown), suggesting that these molecules are relatively inefficient substrates.
Table 1.
Michaelis constants (Km) for arginine and various amidino acceptor substrates
The Km values for arginine and for various amidino acceptor substrates were measured in assays using arginine as the amidino donor in combination with various acceptors (ND, not determined). The Km for each substrate was determined with the other substrate present at saturating concentration.
| Acceptor substrate | Arginine Km: native HsvA | Acceptor substrate Km |
|
|---|---|---|---|
| Native HsvA | HsvA/E244Q | ||
| mm | mm | ||
| Putrescine | 0.47 | 3.9 | 7.9 |
| Spermidine | 1.5 | 0.033 | 3.8 |
| Lysine | ND | ND | 1.7 |
Putrescine, along with 1,3-diaminopropane and cadaverine, are common polyamines found in E. amylovora (22). Given that the E. amylovora genome encodes a homolog of spermidine synthase (SpeE), which catalyzes the formation of spermidine from putrescine and decarboxylated S-adenosylmethionine, we investigated whether HsvA could also use spermidine as an amidino acceptor substrate. We found that spermidine is a far more efficient acceptor than putrescine, with a Km value of 33 μm (see Table 1), significantly lower than any Km value previously reported for an amidinotransferase substrate. The products of the amidinotransfer reaction using spermidine as an acceptor would be either 2-[4-(3-aminopropylamino)butyl] guanidine (N1-aminopropylagmatine) or 2-[3-(4-aminobutylamino)propyl]guanidine (guanidinospermidine), depending upon whether the amidino group is attached to the terminal primary amine that is adjacent to four methylenes or three methylenes, respectively. Considering the greater efficiency of putrescine as an acceptor compared with 1,3-diaminopropane, the N1-aminopropylagmatine product is probably favored. It is also conceivable that diguanylspermidine (hirudonine) could be produced if both ends of spermidine undergo amidinylation in two successive rounds of catalysis. Interestingly, when spermidine is provided as the acceptor, rather than putrescine, the Km value for arginine increases from 0.47 to 1.5 mm. The slightly reduced affinity for arginine in the presence of a saturating concentration of spermidine suggests the likelihood of mild substrate inhibition by spermidine. Consistent with this possibility, we observed significant inhibition of HsvA activity at spermidine concentrations > 200 μm (data not shown).
Temperature and pH activity profile
To investigate the temperature and pH activity profile of HsvA, we used arginine and putrescine as substrates and quantified the production of ornithine under a range of temperature and pH conditions (Fig. S3). HsvA displayed maximum activity around a temperature of 35 °C and a pH of 8. The temperature and pH activity profiles are similar overall to those reported for other bacterial amidinotransferases (14, 15).
Three-dimensional structure of HsvA
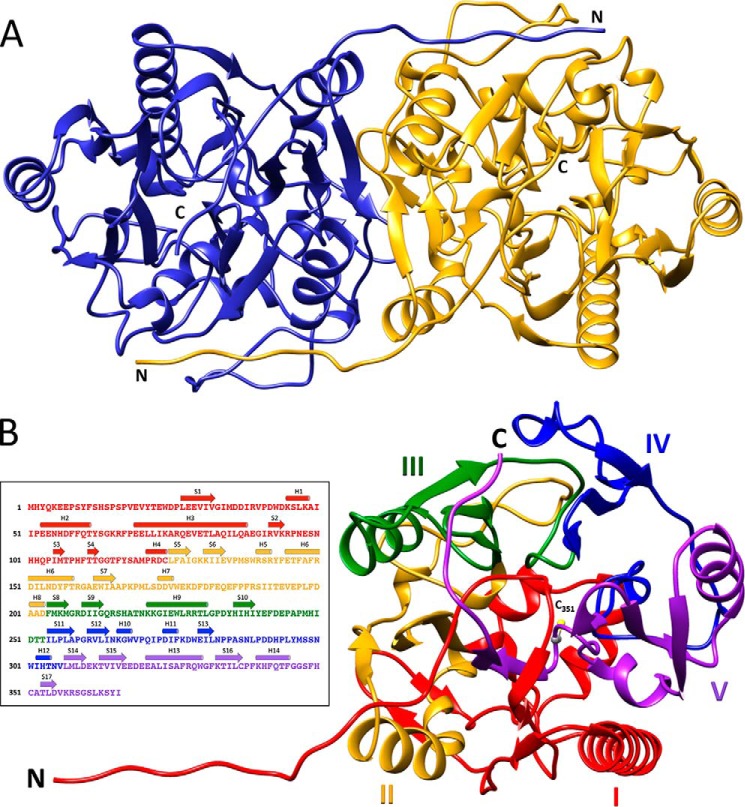
To investigate the structural basis for the novel substrate specificity of HsvA, we determined the three-dimensional structure using X-ray crystallography. HsvA crystallized in the trigonal space group P3221 and diffracted to 2.30 Å resolution. The asymmetric unit contains two molecules arranged as a dimer, with the two monomers related to each other through a noncrystallographic 2-fold symmetry axis along the dimer interface (Fig. 3A). The two HsvA monomers are identical and superimpose with a root mean square deviation of 0.4 Å. The model of each monomer begins with Tyr3 and extends continuously to the native carboxyl terminus at Ile366.
Figure 3.
Overall architecture of HsvA. A, ribbon diagram of the HsvA dimer. The monomers are related by a 2-fold symmetry axis at the dimer interface. The locations of the amino (N) and carboxyl (C) termini are indicated. B, domain organization of an HsvA monomer. The tertiary structure consists of five ββαβ modules arranged around a pseudo-5-fold axis, forming an α/β propeller. The modules are numbered I–V and are colored as follows: red, module I (residues 3–124); yellow, module II (residues 125–203); green, module III (residues 204–253); blue, module IV (residues 254–306); purple, module V (residues 307–366). The amino (N) and carboxyl (C) termini are identified, along with the position of the conserved nucleophilic cysteine in the active site (C351). The inset illustrates the position of secondary structural elements within the amino acid sequence, colored in the same manner as in the ribbon diagram. Helices (shown as cylinders) are numbered H1–H14, and β-strands (shown as arrows) are numbered S1–S17.
Overall architecture
The overall architecture of each HsvA monomer closely resembles the α/β propeller domain that is seen in hAGAT (PDB entry 1JDW) and StrB1 (PDB entry 1BWD) (7, 11), with root mean square deviations of 1.17 and 2.02 Å, respectively. Within each monomer, α-helices and β-strands are organized into five ββαβ modules arranged around a pseudo-5-fold symmetry axis (Fig. 3B), as is commonly seen in members of the guanidino group–modifying enzyme (GME) superfamily (23). The arrangement of ββαβ modules in this manner gives rise to a compact structure resembling a basket, as has been observed in hAGAT and StrB1. Four of the five modules contain an extended α-helix that is oriented roughly parallel to the pseudo-5-fold symmetry axis of the α/β propeller domain; these helices lie along the exterior of the basket and contribute significantly to the surface features of the monomer. Module I contributes several short helices and β-strands that define a lower surface of the basket, and several loops from modules I, IV, and V contribute an upper surface of the basket. Modules II and III contribute significantly to the dimerization interface, with additional intermolecular interactions coming from the first ∼12 amino acids in module I that are in an extended conformation, seen wrapping around the other monomer in the quaternary structure in Fig. 3A. The active site lies within the core of the monomer at the base of a long, narrow channel leading to the exterior of the protein. The features of this channel are defined by amino acids from each of the five ββαβ modules, with the majority coming from modules I, III, and IV. The position of the active-site cysteine (Cys351) within the context of the overall architecture of the monomer is shown in Fig. 3B.
Secondary structure conservation
All of the secondary structural elements defining the five ββαβ modules in hAGAT are highly conserved in HsvA, with two minor exceptions. One helix found in module II of hAGAT is not observed in HsvA; this helix consists primarily of a seven-residue insertion within a loop region that, in HsvA, is found on the exterior of the protein near the opening of the channel leading to the active site. Another minor difference in secondary structure arises in module IV, where HsvA contains a short helix (noted as H10 in Fig. 3B), also in the vicinity of the channel opening.
Dimer interface
A total of 48 amino acids in each monomer form interactions with the other monomer at the dimer interface. Modules I–III contribute the majority of these interacting amino acids, although module IV contributes several as well. More than half of the interactions at the dimer interface appear to be hydrophobic in nature, with 34 different amino acids participating in the formation of nonpolar interactions with the other monomer. Further contributions to the dimer interface come from 46 intermolecular hydrogen bonds involving 28 different amino acids. Two electrostatic interactions are also observed, as Glu7 on one monomer interacts with Arg215 on the other monomer. The dimer interface involves 2,905 Å2/monomer of buried surface area, suggesting that the interaction between monomers is significantly stronger in HsvA than in hAGAT, where the dimer interface involves only 1,773 Å2/ monomer of buried surface area (7).
Active-site region
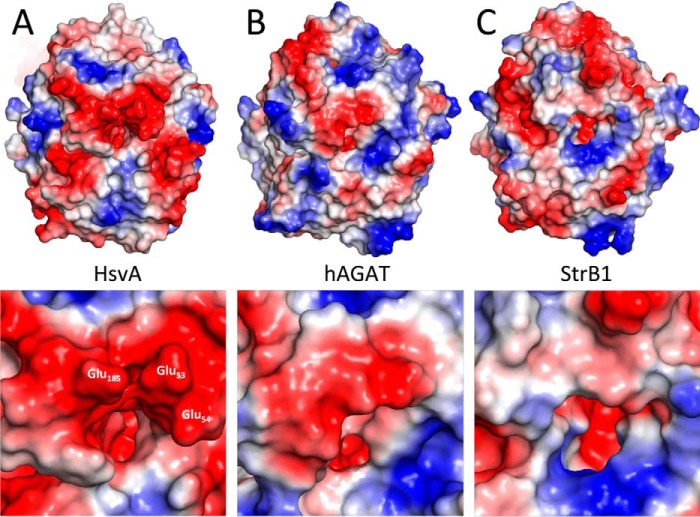
The active site of HsvA consists of the strictly conserved amino acids Cys351, His249, and Asp200 (Fig. 4). Together, these form a catalytic triad that is believed to function in a manner similar to the catalytic triad found in cysteine proteases (7). The side chains of Cys351 and His249 are oriented toward a small cavity that is found at the base of a long, narrow channel leading from the core of the protein to the exterior. A small region of weak electron density is seen within this long channel, but we were not able to model any ligand with confidence, suggesting that any bound ligands are probably present at low occupancy. Surrounding the opening of the channel on the exterior of the protein are three glutamate residues (Glu53, Glu54, and Glu185) that confer a significantly negative electrostatic potential to the surface immediately adjacent to the channel opening (Fig. 5A), a feature that could play a role in recruiting positively charged substrates into the channel. Altogether, ∼30 amino acids define the topology of this channel and the features of the protein surface adjacent to the channel opening. Around half of these amino acids are also found in hAGAT and slightly fewer in StrB1.
Figure 4.
Active-site region in HsvA. The side chains of selected amino acids in the active-site region are shown, with carbon shown in gray, oxygen in red, nitrogen in blue, and sulfur in yellow. Selected regions of secondary structure surrounding the active site and the substrate access channel are shown as well. In this cross-sectional view, the active-site region is oriented so that the channel is being viewed from the side, with the opening of the channel at the top of the diagram and the base of the channel near the bottom. The three amino acids forming the catalytic triad near the base of the channel (Cys351, His249, and Asp200) are labeled in boldface type. Glu244, which is believed to be a primary determinant of substrate specificity, is located just below the entrance of the channel, where the carboxylate is positioned to form an electrostatic interaction with the α-amino group in arginine or with a protonated amine in the polyamine acceptor substrate. Glu185 is located on the exterior of HsvA near the channel opening and, together with Glu53 and Glu54, confers a significantly negative electrostatic potential to the region surrounding the channel entrance.
Figure 5.
Electrostatic potential map of HsvA in comparison with hAGAT and StrB1. The molecular surface of HsvA (A), hAGAT (B), and StrB1 (C) is colored according to electrostatic potential, with red being negative, gray being neutral, and blue being positive. Each structure is oriented to show the region surrounding the channel leading to the active site. The inset provides an enlarged view of the region immediately adjacent to the channel entrance. In HsvA, three glutamate residues surrounding the channel entrance are labeled. The molecular surface of hAGAT was drawn from PDB entry 1JDW, and StrB1 was drawn from PDB entry 1BWD.
Substrate binding
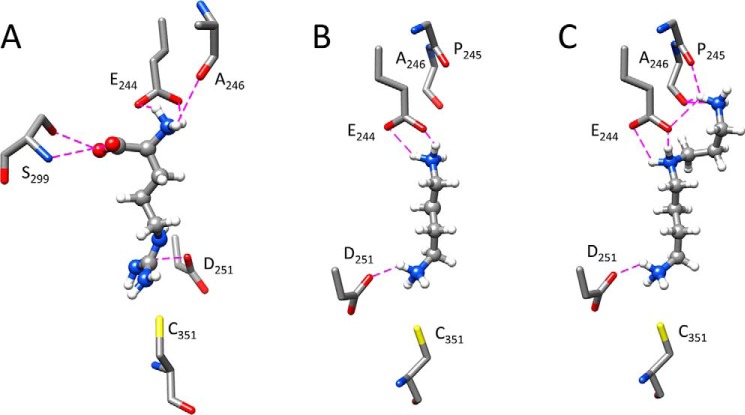
In hAGAT, seven amino acids in the active-site region play a direct role in binding the arginine substrate (7). Of these, six are found in nearly identical positions in the active-site region of HsvA, including Asp123, Met248, Asp251, Ser298, Ser299, and the active-site His249 (see Fig. 4). In hAGAT, an arginine residue located near the opening of the channel (designated as Arg322 in the hAGAT sequence) provides additional binding energy by forming an electrostatic interaction with the carboxylate group in the arginine substrate. This electrostatic interaction appears to be well-conserved among a broad range of amidinotransferases, which tend to have either an Arg or Lys at this position in the sequence, corresponding to position 268 in the HsvA sequence (see Fig. 7). In HsvA, the corresponding amino acid at this position near the opening of the channel is Trp268, which cannot form a comparable interaction with the arginine substrate. However, a negatively charged amino acid (Glu244) is located immediately adjacent to Trp268 at the channel opening. Computational modeling of arginine in the active-site channel suggests that the carboxylate in the Glu244 side chain is well-positioned to form an electrostatic interaction with the α-amino group in the arginine substrate; on the opposite wall of the channel, both the side-chain hydroxyl and main-chain amide of Ser299 are within hydrogen bonding distance of the carboxylate group in the arginine substrate (Fig. 6A). The side chain hydroxyl of Ser298 may also undergo repositioning to form a hydrogen bond with the carboxylate group in the arginine substrate upon binding.
Figure 7.
Amino acid sequence alignment of selected regions from known or predicted amidinotransferases. Selected regions from the 23 amino acid sequences used in the phylogram depicted in Fig. 1 were aligned using Clustal Omega and annotated manually (the full sequence alignment is given in Fig. S1). The clade designations from the phylogram in Fig. 1 are listed on the left. The UniProt accession numbers for the aligned sequences may be found in Table S1. The regions are numbered according to the amino acid positions in the HsvA sequence. The amidinotransferase proteins for which the molecular structure is known (HsvA, hAGAT, and StrB1) are highlighted in boldface type. Highlighted in blue are the strictly conserved amino acids forming the catalytic triad (Asp200, His249, and Cys351 in HsvA). Conserved amino acids outside of the catalytic triad that interact directly with the arginine substrate in hAGAT are highlighted in green (7). Highlighted in red are the Glu and Trp residues located at the opening of the substrate access channel in HsvA, along with conserved amino acids found in other homologs in clade I. The corresponding amino acids in the same positions in homologs from other clades are highlighted in purple. Glu residues on the surface of HsvA near the opening of the substrate access channel are highlighted in orange, along with conserved amino acids at corresponding positions in other homologs in clade I.
Figure 6.
Computational modeling of substrates in the HsvA active site. Selected amino acids predicted to play a role in binding arginine or various polyamine substrates in the active-site channel are shown, with carbon depicted in gray, oxygen in red, nitrogen in blue, sulfur in yellow, and hydrogen in white. Predicted hydrogen bonds are shown as dashed magenta lines. In these cross-sectional views, the catalytic Cys351 marks the location of the active site at the base of the channel, and Glu244, a major determinant of substrate specificity, marks the location of the channel entrance. Asp251 is positioned to form an electrostatic interaction with a positively charged functional group in each substrate close to the site of amidino transfer, and Glu244 is positioned to form an electrostatic interaction with a positively charged functional group at the opposite end of each substrate. The predicted mode of binding for arginine (A) suggests that the carboxylate in Glu244 and the main-chain carbonyl in Ala246 interact with the α-amino group in the substrate, whereas the side-chain hydroxyl and main-chain amide in Ser299 interact with the carboxylate in the substrate. Putrescine (B), with four methylenes separating two primary amines, has the optimum length to form electrostatic interactions with Glu244 and Asp251. With spermidine (C), the predicted binding model suggests that Glu244 forms electrostatic interactions with both the internal and distal amines, and the main-chain carbonyl groups in Pro245 and Ala246 both form hydrogen bonds with the distal primary amine. The orientation of the channel shown in A is the same as the orientation shown in Fig. 4, whereas the orientation shown in B and C is rotated ∼180°.
Significantly, the placement of a glutamate just below the opening of the channel imparts a negatively charged electrostatic potential to this region of the protein surface. Given the distance between this glutamate side chain and the active-site residues near the base of the channel (following amidinylation of the nucleophilic Cys), unbranched substrates having two positively charged functional groups separated by an optimal distance of four carbons, as is the case with putrescine, should be appropriately sized to form an electrostatic attraction with this negatively charged region near the channel opening (Fig. 6B). This structural feature provides an explanation for the substrate specificity profiles observed for both amidino donors and amidino acceptors, where the presence of two positively charged functional groups was a common feature seen in all molecules capable of serving as a donor or an acceptor substrate. These observations suggest that Glu244 serves as a primary determinant of substrate specificity and probably explain the preference of HsvA for polyamines as amidino acceptors, with putrescine serving as the most efficient substrate among the diamines surveyed. The channel appears to be too narrow to permit branched or cyclic polyamines to serve as substrates.
The significant difference in Km between the diamine putrescine (3.9 mm) and the triamine spermidine (33 μm) suggests that spermidine forms additional interactions with the enzyme upon binding. Computational modeling of spermidine binding in the active site (Fig. 6C) suggests that both the internal and distal amines in the triamine substrate are able to form electrostatic interactions with the carboxylate group in the side chain of Glu244. Furthermore, the main-chain carbonyls of Pro245 and Ala246 are also within hydrogen bonding distance of the distal primary amine in the spermidine substrate. The formation of additional electrostatic and hydrogen bonding interactions with spermidine during catalysis could explain the strong preference for spermidine over putrescine as an amidino acceptor substrate.
Altered substrate specificity in the HsvA/E244Q mutant
To investigate the role of Glu244 as a primary determinant of substrate specificity, we used site-directed mutagenesis to replace glutamate with glutamine at this position. The substitution of Gln for Glu replaces a charged carboxylate in the side chain with an uncharged carboxamide, eliminating the possibility of forming an electrostatic interaction with the substrate while preserving the potential for hydrogen bonding. This substitution was also chosen because it was expected to have little impact on the topology of the channel opening.
An examination of the amidino acceptor substrate specificity profile of HsvA/E244Q revealed that, whereas the mutant enzyme retained the ability to use spermidine, putrescine, and other linear diamines as substrates (data not shown), both putrescine and spermidine became less effective as amidino acceptors, as evidenced by increases in the Km values for these two substrates (Table 1). The reduced affinity of HsvA/E244Q for putrescine and spermidine is consistent with a decrease in binding energy that would result as an electrostatic interaction involved in substrate binding is replaced by a lower-energy hydrogen bond. These observed increases in Km support the proposed role of Glu244 as a determinant of substrate specificity.
Interestingly, whereas the elimination of a negative charge near the opening of the channel led to a reduction in the affinity for putrescine and spermidine, the replacement of this negative charge with an uncharged polar functional group led to a substantial increase in the affinity for lysine. In the native enzyme, the Km value for lysine was well above 30 mm (data not shown), whereas in HsvA/E244Q, the Km decreased to 1.7 mm (Table 1). The increase in affinity for lysine is probably the result of eliminating the electrostatic repulsion that would otherwise arise in native HsvA between Glu244 and the carboxylate in lysine during substrate recruitment.
Discussion
Genetic studies of virulence determinants in E. amylovora have identified HsvA, along with two other proteins encoded by the same operon in the HAE locus, HsvB and HsvC, as required for systemic infection in apple (5). To gain insight into the role of HsvA in the pathogenicity of E. amylovora, we carried out a series of biochemical and structural studies to characterize the predicted amidinotransferase activity of HsvA and to investigate the determinants of substrate specificity in the three-dimensional structure of the protein. Our results confirm that HsvA is an amidinotransferase, and we show that HsvA has novel substrate specificity, displaying a preference for linear aliphatic polyamines, especially putrescine and spermidine, as the amidino acceptor substrate.
An examination of the active-site region in the three-dimensional structure of HsvA reveals several features that explain the observed preference for putrescine and spermidine as amidino acceptor substrates. The active site lies at the base of a long, narrow channel that has dimensions well-suited to accommodate linear, but not branched or cyclic, substrates. A glutamate residue (Glu244) located just below the opening of the channel, with its side chain carboxylate ∼11 Å above the nucleophilic cysteine in the active site, is well-positioned to form an electrostatic interaction with the distal primary amine in putrescine, a four-carbon substrate, which, among the linear diamines, has the ideal degree of separation between primary amine groups (see Fig. 6B). When spermidine is bound, Glu244 is positioned to form electrostatic interactions with both the internal and the distal amines in this triamine substrate (see Fig. 6C). The potential for the formation of additional interactions with spermidine, in comparison with putrescine, could explain the lower Km that is observed for spermidine.
Given the preference of HsvA for substrates containing positively charged functional groups separated by 3–5 carbons, it is reasonable to suggest that agmatine, a linear polyamine containing a guanidine and a primary amine separated by four carbons, should be able to serve effectively as both an amidino donor and an acceptor substrate, producing putrescine or arcaine, respectively. Indeed, we observed a significant rate of putrescine formation when we provided HsvA with agmatine as a sole substrate, far greater than the rate of ornithine formation in the presence of arginine as a sole substrate (data not shown). The production of putrescine demonstrates the capability of agmatine to serve as an amidino donor, as shown in Fig. S2; likewise, the comparatively high rate of putrescine formation suggests that catalytic turnover is occurring relatively efficiently in the presence of agmatine as a sole substrate, which implies that agmatine is also able to serve as an amidino acceptor. Determination of Km values for agmatine as a donor and as an acceptor is difficult in light of the dual catalytic activities that occur simultaneously for this enzyme–substrate combination. Nevertheless, agmatine is a common polyamine in E. amylovora (22), and HsvA is probably able to use it as both an amidino donor and an acceptor in vivo with reasonable efficiency.
As seen in Fig. 7, the amino acids believed to play a direct role in substrate recognition in HsvA are highly conserved in the predicted orthologs of HsvA from E. pyrifoliae, E. mallotivora, and P. stewartii, all of which appear to form a unique clade in phylogenetic analysis (see Fig. 1). This sequence conservation includes Glu244, the amino acid believed to facilitate the preference for polyamine acceptor substrates. Additionally, two of three glutamate residues located on the surface of the protein near the entrance of the channel (corresponding to Glu54 and Glu185) are also conserved and could potentially be involved in the recruitment of polyamine substrates. These amidinotransferase orthologs probably have identical substrate specificity and perform a similar function in vivo during infection.
Comparison of substrate specificity with other amidinotransferases
The amidino acceptor substrate specificity of HsvA appears to be somewhat unique among the bacterial and vertebrate amidinotransferase homologs that have been characterized previously. In hAGAT and StrB1, two amidinotransferases for which the molecular structures are known, a conserved arginine (Arg322 in hAGAT and Arg246 in StrB1) is positioned near the entrance of the channel leading to the active site; this creates an area of positively charged electrostatic potential inside the channel near the rim, facilitating the formation of an electrostatic interaction with the carboxylate group in the arginine substrate (7, 11) during the first half of the amidinotransfer reaction. In StrB1, Arg246 is also predicted to form an electrostatic interaction with the phosphoryl group in the scyllo-inosamine 4-phosphate substrate during the second half of the amidinotransfer reaction. In HsvA, the corresponding amino acid at this position is Trp268, which is unable to form similar interactions with substrate. However, the adjacent Glu244 creates an area of negatively charged electrostatic potential near the opening of the channel, and the side chain of Glu244 is positioned to form an electrostatic interaction with the α-amino group in arginine during the first half of the amidinotransfer reaction and with an amine group in the polyamine substrate during the second half. Additionally, the molecular surface surrounding the entrance of the channel in HsvA has an electrostatic potential that is considerably more negative than the corresponding regions in hAGAT and StrB1 (Fig. 5); this probably also contributes to the novel acceptor substrate specificity seen in HsvA.
The AmtA amidinotransferase from P. syringae pv. phaseolicola, which is found in the biosynthetic pathway for phaseolotoxin and shares 48% amino acid identity with HsvA, uses arginine as a donor and lysine as an acceptor to produce ornithine and homoarginine, both of which are components of the toxin (17). Not surprisingly, the acceptor substrate specificity of AmtA bears some similarities with HsvA, with AmtA also having affinity for the lysine analog cadaverine and, to a lesser extent, putrescine (19). However, as seen in Fig. 7, the amino acids in AmtA that correspond to positions 244 and 268 in HsvA are Asp and Arg, as in hAGAT. The similarity with hAGAT at these positions suggests that the opening of the substrate access channel in AmtA is probably designed to interact with a negatively charged functional group in the substrate, as would be expected of an amidinotransferase designed primarily to bind amino acid substrates rather than polyamines. Likewise, none of the glutamate residues on the exterior surface of HsvA near the channel entrance that could potentially play a role in polyamine substrate recruitment are conserved in AmtA.
Amidinotransferase activity bearing some similarities to HsvA has been reported in two plant species, soybean and grass pea (Lathyrus sativus). Soybean amidinotransferase appears to have broad substrate specificity, preferring glycine as an amidino acceptor but also able to use agmatine, putrescine, and lysine to a limited extent (24). The amidinotransferase from L. sativus also appears to have broad substrate specificity, using lysine, ornithine, cadaverine, putrescine, and agmatine, but not glycine, as acceptor substrates, although the preferred acceptor is believed to be lysine (25). These enzymes are thought to play a role in modulating the levels of various guanidine-containing metabolites and structurally related polyamines during seedling development. Both enzymes were reported to be tetramers having molecular masses between 210 and 240 kDa. They are probably not homologs of the vertebrate or bacterial amidinotransferases, as we could not find any sequences matching hAGAT in a translated BLAST search of 108 sequenced plant genomes, including soybean. Given the absence of any detectable homology in plants, as well as the differences in oligomeric architecture when compared with the vertebrate and bacterial proteins, it is possible that these amidinotransferases in plants may be the result of convergent evolution. Interestingly, both plant enzymes are reportedly inactivated following exposure to thiol blockers, suggesting that the active site contains a reactive cysteine.
Novel mode of binding arginine substrate in HsvA
From the sequence alignment shown in Fig. 7, it appears evident that two distinct modes of interaction with the arginine substrate are represented among these diverse amidinotransferases. In most of the sequences clustered throughout clades II–V, a positively charged amino acid (Arg or Lys) is found at the position corresponding to residue 268 in HsvA, whereas an Asp is found at the position corresponding to residue 244 in HsvA, an arrangement that facilitates arginine binding via an electrostatic interaction with the carboxylate group in the substrate. This mode of arginine binding appears to be quite common among amidinotransferase sequences and indeed also among the wider GME superfamily, where an electrostatic interaction is often seen between an Arg or Lys in the enzyme and the carboxylate group of the arginine substrate; this trend is seen even in more distant amidinotransferase homologs, including dimethylarginine dimethylaminohydrolase, arginine deiminase, peptidyl-arginine deiminase, and succinylarginine dihydrolase enzymes (23).
In the amidinotransferase sequences clustered in clade I, Glu is found at position 244 and Trp at position 268, an arrangement that could facilitate arginine binding via an electrostatic interaction with the α-amino group in the substrate. This appears to represent a novel mode of interaction with arginine not previously seen among members of the GME superfamily. Although the origin of this alternate mode of arginine binding is unclear, it is possible that the Asp → Glu substitution at position 244, which moves the carboxylate group of the side chain closer to the location of substrate binding in the channel, arose to compensate for the interaction that is lost in the Arg → Trp substitution at position 268. Alternatively, it is also conceivable that the Asp → Glu substitution at 244 arose first, making possible the substrate specificity favoring polyamines, and then the Arg → Trp substitution at 268 was introduced to eliminate the possibility of a counterproductive ion-pair interaction between Glu and Arg at the opening of the channel, which would probably prevent substrate binding. Both amino acid substitutions may be achieved by single nucleotide substitutions in the corresponding codons.
Potential roles of HsvA as a virulence factor in E. amylovora
The demonstration of polyamine amidinotransferase activity in HsvA suggests some potential roles for this protein in the pathogenesis of E. amylovora. Because HsvA uses both arginine and polyamines as substrates, the protein has the capability to influence endogenous levels of these metabolites, potentially in ways that enhance the fitness of the pathogen during infection or support the production of other virulence factors through indirect means. It is also possible that the polyamine amidinotransferase activity of HsvA may play a direct role in the synthesis of an essential virulence factor that secures an advantage for the pathogen during infection in apple.
Polyamines play critical roles in a wide variety of processes in living systems through diverse mechanisms that continue to be elucidated (26). In bacteria, putrescine and spermidine are the most common polyamines, playing essential roles in cell growth, gene transcription and translation, detoxification of free radicals, and acid resistance, among other functions (27). In E. amylovora, cadaverine, putrescine, and other short polyamines serve as precursors for the synthesis of various desferrioxamine siderophores, which are essential virulence factors involved in iron scavenging (28). Studies from other pathogens in the Enterobacteriaceae have shown that putrescine and spermidine are specifically required for biofilm formation (29) and for the expression of proteins involved in type III secretion (30), although the extent to which polyamines are required for these processes in E. amylovora remains unknown.
Putrescine can be synthesized in a one-step process involving the action of ornithine decarboxylase (SpeC) or through a two-step process involving the conversion of arginine to agmatine via arginine decarboxylase (SpeA), followed by the removal of the amidino group via agmatinase (SpeB). Spermidine is synthesized from putrescine through the action of spermidine synthase (SpeE). The E. amylovora genome encodes homologs of all of these enzymes. Under conditions of excess agmatine and a shortage of putrescine, HsvA could contribute to the formation of putrescine by utilizing agmatine as an amidino donor substrate, mirroring the activity of SpeB. HsvA could also catalyze the direct formation of ornithine from arginine, mirroring the action of arginase, an enzyme activity that does not otherwise exist in E. amylovora. Ornithine could then be decarboxylated by SpeC to generate putrescine. In these ways, the amidinotransferase activity of HsvA may be able to contribute to virulence by helping to raise putrescine levels in support of endogenous metabolic and biosynthetic processes, such as those mentioned above, that are polyamine-dependent.
Although HsvA may support virulence indirectly by modulating endogenous levels of polyamines, a more direct role in pathogenicity is likely, given the significantly reduced virulence phenotype seen in mutant strains lacking hsvA or two of the other genes also predicted to encode enzymes in the HAE locus, hsvB and hsvC (5). All three of these genes have homologs in the phaseolotoxin biosynthesis pathway. Although no evidence of phaseolotoxin production has been found in E. amylovora (5), the overall similarities with components of the phaseolotoxin biosynthesis pathway suggest the possibility that a toxin or metabolite containing an amidinylated derivative of spermidine or putrescine (or another polyamine) as part of the structure could be synthesized during infection.
Such a polyamine-derived metabolite secreted from E. amylovora could potentially exert its effects by interacting with defense pathways in the plant that are polyamine-dependent. Polyamines are known to play a variety of essential roles in plant defenses, acting as signaling molecules in pathogen recognition and participating in a wide range of defense responses, including the production of H2O2 (required for the initiation of the hypersensitive response), reinforcement of the cell wall, and up-regulation of genes involved in systemic acquired resistance, to name a few (31). Some plant pathogens have been shown to manipulate polyamine metabolism in their hosts to weaken defense responses. A polyamine-derived metabolite from E. amylovora could conceivably act as an inhibitor of an enzyme in the plant that is required for polyamine metabolism or could interfere with polyamine-mediated signaling within the plant cell. Either of these scenarios could lead to impaired defense responses in the plant, providing an advantage for the pathogen.
Functional characterization of the other proteins in the HAE locus will lead to a better understanding of the mechanism by which this locus contributes to the pathogenicity of E. amylovora in apple. Investigation of changes in polyamine levels in susceptible and resistant cultivars of apple following exposure to E. amylovora may shed light on the possibility that E. amylovora could manipulate polyamine levels in the plant to its advantage during infection.
Experimental procedures
Phylogenetic analysis
Amino acid sequences for a variety of confirmed and predicted amidinotransferase enzymes were obtained from the UniProt database. Sequences were aligned with Clustal Omega (32). A phylogeny tree was constructed using PHYLIP Proml software, applying the Jones–Taylor–Thornton probability model (33), and manually annotated using FigTree (http://tree.bio.ed.ac.uk/software/figtree/)3 (34). The sequence alignment was annotated using Jalview (35).
Construction of pET28b-hsvA and pET28b-hsvA/E244Q expression plasmids
E. amylovora strain Ea321 (a generous gift from Dr. Steven Beer, Cornell University, Ithaca, NY) was grown in LB broth overnight at room temperature. Cells were recovered by centrifugation, resuspended in Tris-EDTA buffer, and lysed using a combination of SDS, proteinase K, and lysozyme. Following a brief incubation at 37 °C, protein was removed from the lysate by phenol-chloroform extraction. Nucleic acid was recovered by precipitation from the aqueous phase using 3 m potassium acetate (pH 5.5) and isopropyl alcohol, followed by several successive washes in ethanol. The purified genomic DNA was stored in Tris-EDTA buffer.
The native hsvA gene was amplified from genomic DNA using Phusion DNA polymerase (New England Biolabs) and primers containing the NdeI restriction site in the 5′-end (5′-GGGCATATGCATTATCAAAAAGAAGAGCCATC-3′) and XhoI in the 3′ end (5′-CCCCTCGAGTCAGATATAGCTTTTCAGGCTTCC-3′). The PCR product was digested with restriction enzymes and ligated into a modified pET28b vector, which encoded a tobacco etch virus protease cleavage site in place of the standard thrombin cleavage site. The nucleotide sequence of the cloned gene was verified by DNA sequence analysis.
A similar expression construct encoding HsvA with an E244Q mutation was assembled using a nested PCR strategy. A mutagenic primer pair encoding CAG in place of GAG at amino acid position 244 (5′-CATCCATATCTACGAGTTTGATCAGCCGGCTCCG-3′) was used in combination with terminal primers containing the NheI restriction site in the 5′-end and XhoI in the 3′-end.
Expression and purification of HsvA
The pET28b-hsvA plasmid was used to produce recombinant HsvA in BL21(DE3) E. coli cells. Expression was induced using 1 mm isopropyl β-d-1-thiogalactopyranoside, and the culture was incubated overnight at 25 °C. Cells were harvested by centrifugation at 10,000 × g, resuspended in an immobilized metal affinity chromatography binding buffer (20 mm sodium phosphate (pH 8), 500 mm NaCl, 20 mm imidazole, 1 mm DTT, and 10% (v/v) glycerol), and lysed using sonication. Clarified cell lysate was pumped over a 5-ml HisTrap immobilized metal affinity chromatography column (GE Healthcare) using an Äkta FPLC system (GE Healthcare). Protein was eluted using a buffer containing 20 mm sodium phosphate (pH 8), 200 mm imidazole, 1 mm DTT, and 5–10% glycerol. During elution, fractions were diluted into an equal volume of ultrapure water. Protein purity was confirmed using denaturing polyacrylamide gel electrophoresis, and the concentration was estimated by UV spectrophotometry, using a theoretical extinction coefficient of 72,880 m−1 cm−1 and a predicted molar mass of 45,065 Da.
For protein used in enzyme functional assays, the fractions containing pure HsvA following affinity chromatography were desalted into a buffer containing 20 mm HEPES (pH 7), 1 mm DTT, and 10% glycerol using a HiPrep 26/10 desalting column (GE Healthcare). The protein was flash-frozen in liquid nitrogen and stored at −80 °C. HsvA/E244Q protein was purified similarly.
For native HsvA protein used in crystallization and structure determination, affinity-purified HsvA was desalted into a buffer containing 20 mm phosphate (pH 8) and 1 mm DTT using a HiPrep 26/10 desalting column. The protein was then injected onto a HiLoad 16/60 size exclusion column containing Superdex 200 medium (GE Healthcare) and eluted with a mobile phase buffer containing 50 mm sodium phosphate (pH 8), 50 mm NaCl, and 1 mm DTT. Fractions representing non-aggregated HsvA were pooled and desalted into a buffer containing 20 mm HEPES (pH 7) and 1 mm DTT. The protein was concentrated to ∼15 mg/ml using ultrafiltration, flash-frozen in liquid nitrogen, and stored at −80 °C.
Amidinotransferase activity assay
Standard assay conditions
An in vitro assay, coupled with HPLC, was used to investigate amidinotransferase activity. Unless otherwise noted, assays contained an amidino donor, an amidino acceptor, 100 mm HEPES (pH 8), and 1–3 μm HsvA. Either glutamate or tryptophan was included at 250 μm as an internal HPLC standard. Assays were incubated at 30 °C for a specific period of time and then stopped by the addition of HPLC-grade acetonitrile to precipitate the protein. Amidinotransferase assays were carried out in triplicate.
HPLC
The products of the amidinotransfer reaction were separated and quantified using an Agilent 1100 HPLC system as described previously (36), with several modifications. Mobile phase buffer A was composed of 10 mm sodium phosphate, 10 mm sodium borate, and 0.02% sodium azide (pH 8.0), and mobile phase buffer B was composed of methanol/acetonitrile/water in a 9:9:2 (v/v/v) ratio. Using an injector program with the autosampler, 1 μl of clarified supernatant from the amidinotransferase assay was combined with 2.5 μl of 0.4 n sodium borate (pH 10.2) and 2 μl of a freshly prepared derivatization reagent mixture containing 75 mm o-phthalaldehyde and 225 mm sodium 2-mercaptoethanesulfonate in 80% methanol. The use of sodium 2-mercaptoethanesulfonate as the thiol additive in the derivatization reaction facilitates efficient labeling of primary amines containing -CH2-CH2-NH2 (37). The reaction was mixed briefly, and then 32 μl of mobile phase buffer A was added as an injection diluent. The entire reaction mixture was injected onto a Zorbax Eclipse Plus C18 reverse phase column (4.6 × 150 mm, packed with 3.5-μm medium; Agilent) maintained at 40 °C. Derivatized amino acids were eluted using a gradient program and detected by UV spectrophotometry at 334 nm. The gradient program used a constant flow rate of 1.5 ml/min, with 2% B for the first 0.5 min, followed by a linear increase from 2% B at 0.5 min to 60% B at 16.0 min, and finally 100% B from 16.1 to 19.5 min. Between injections, the column was re-equilibrated into 2% B for ∼5 min. Amidinotransferase activity was quantified by estimating the yield of product (typically ornithine) from the peak area using a standard curve; the peak area of the internal standard was used to calculate the dilution factor for each reaction by comparison with the peak area of the internal standard obtained from a non-diluted protein-free control mixture.
Investigation of donor substrate specificity
To investigate the capability of arginine, homoarginine, agmatine, arcaine, and guanidinoacetate to serve as amidino donor substrates, a modified assay format was designed in which each candidate donor substrate was incubated with HsvA in the absence of an acceptor. Each reaction mixture contained 250 μm donor, 100 mm HEPES (pH 8), 250 μm glutamate, and 25 μm HsvA. Negative control reaction mixtures contained all components except for HsvA. Reactions were incubated at 30 °C for 120 min. HPLC was used to detect the expected product. Ornithine was measured in reactions containing arginine, lysine in reactions containing homoarginine, agmatine in reactions containing arcaine, putrescine in reactions containing agmatine, and glycine in reactions containing guanidinoacetate. To investigate the capability of canavanine to serve as an amidino donor, putrescine was provided as an acceptor, and both donor and acceptor substrates were included at 5 mm, with the reaction incubated at 30 °C for 10 min and agmatine measured as the product.
Investigation of acceptor substrate specificity
A variety of candidate amidino acceptor molecules, each containing at least one primary amine, were used in conjunction with arginine as the amidino donor in a series of amidinotransferase assays to investigate the structural requirements for the acceptor substrate in the amidinotransfer reaction. Reaction mixtures contained 10 mm arginine, 10 mm acceptor, 100 mm HEPES (pH 8), 250 μm glutamate, and 10 μm HsvA. Reactions were initiated with the addition of arginine and then incubated at 30 °C for 30 min. The yield of ornithine was measured using HPLC and compared with the yield of ornithine obtained using putrescine as the amidino acceptor under the same conditions.
Temperature and pH activity profiles
The temperature dependence of the amidinotransfer reaction was characterized using the standard assay with 10 mm arginine and 30 mm putrescine, incubating the reactions for 30 min at a variety of temperatures ranging between 15 and 50 °C and quantifying the yield of ornithine using HPLC. The effect of pH upon the amidinotransferase activity of HsvA was investigated by carrying out the standard assay with 10 mm arginine and 30 mm putrescine at pH values ranging between ∼5.5 and ∼9.5. The pH in each reaction was maintained with 100 mm buffer reagent; MES was used below pH 6.0, PIPES between 6.1 and 7.5, HEPES at pH 7.9, Bicine between pH 8.1 and 9.0, and sodium carbonate above pH 9.5. Reactions were incubated at 30 °C for 30 min, and the yield of ornithine was quantified using HPLC.
Measurement of Km for donor and acceptor substrates
Variations of the standard assay were used to measure the Km values for arginine and several amidino acceptor substrates. In each assay, one substrate (either arginine or an amidino acceptor) was maintained at a concentration at least 10-fold higher than its Km value, whereas the other substrate was varied between ∼0.2 × Km and 5 × Km. Each reaction also contained 100 mm HEPES (pH 8), 250 μm glutamate or tryptophan as an internal standard, and 1–3 μm HsvA. With all assays, reactions were initiated with the addition of arginine, and initial reaction velocities were estimated by quantifying the yield of ornithine after a 5-min incubation at 30 °C. The value of Km was estimated using nonlinear regression analysis to fit the kinetic data to a rectangular hyperbola (SigmaPlot version 12, Systat Software, Inc.).
Structure determination
Crystallization
HsvA crystallization trials were set up with commercially available crystal screens using the hanging-drop vapor diffusion method at 20 °C. HsvA protein (15 mg/ml) was mixed in a 1:1 ratio with the reservoir solution and set up by the nanoliter handling system Mosquito (TTP Labtech). Crystallization succeeded in a buffer containing 0.2 m MgCl2, 0.1 m HEPES, pH 7.5, and 30% (w/v) PEG 400. Crystals measuring 0.1–0.2 mm were obtained within a week. The crystals were cryoprotected in a reconstituted crystallization buffer containing 10% glycerol and flash-frozen in liquid nitrogen.
Diffraction, data collection, and structure determination
Diffraction data were collected using the home source RIGAKU FR-E+ (Rigaku) at Baylor College of Medicine (Houston, TX). Diffraction data were processed using d*TREK (Rigaku). The space group of the crystal was confirmed using Pointless (38) in the CCP4 suite (39). The initial electron density map was obtained by molecular replacement using Phaser (40), with the previously published structure of the human l-arginine–glycine amidinotransferase (PDB entry 1JDW) as the starting model. This was followed by ab initio automated model building and solvent addition using PHENIX AutoBuild (41) to reduce model bias. Further iterative cycles of refinement and model building were carried out based on the Fo − Fc difference maps. The programs phenix.refine (42) and Coot (43) were used throughout structure determination and refinement. Data collection and refinement statistics are provided in Table 2. The programs Chimera (44) and PyMOL (Schrödinger, LLC) were used to generate illustrations of the molecular structure.
Table 2.
Crystallographic data and refinement statistics
| Parameters | Valuesa |
|---|---|
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 127.54, 127.54, 92.23 |
| α, β, γ (degrees) | 90.00, 90.00, 120.00 |
| Resolution (Å) (range) | 38.03–2.30 (2.38–2.30) |
| No. of reflections | 230,012 |
| No. of unique reflections | 38,767 |
| Rmerge (%) | 11.7 (51.7) |
| I /σI | 7.9 (2.5) |
| Completeness (%) | 100 (100) |
| Redundancy | 5.93 (5.88) |
| Refinement | |
| Resolution (Å) (range) | 38.03–2.30 |
| Rwork/Rfree (%) | 17.01/22.59 |
| Ramachandran statistics (%) | |
| Favored | 95.30 |
| Outliers | 0 |
| No. of Atoms | |
| Protein | 5953 |
| Ligand/ion | 12 |
| Water | 282 |
| Average B-factor (Å2) | |
| Protein | 42.98 |
| Ligand/ion | 59.09 |
| Water | 42.57 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (degrees) | 0.879 |
a Numbers in parentheses correspond to values for the highest-resolution shell.
Substrate modeling in the active site
The amidino donor substrate arginine and the acceptor substrates putrescine and spermidine were computationally modeled into the active site using Chimera. Manual docking was carried out for the initial placement of each substrate within the active-site channel, and the position was optimized using the MMTK (Molecular Modeling Toolkit) energy minimization algorithms in Chimera (45), holding the HsvA atoms in place during refinement.
Author contributions
J. A. L. conceived and coordinated the study and wrote the paper. D. A. M. produced the expression construct for native HsvA, and J. H. produced the expression construct for the HsvA/E244Q mutant. S. S. crystallized the HsvA protein and determined the molecular structure. B. K. B. carried out the phylogenetic analysis. G. K. S., B. K. B., D. C., V. L. W.-K., and T. J. C. performed enzymatic assays. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Melvin Thomas for assistance with protein purification, and we thank Kristina Pansa, Edwin Ragwan, Ashley Beck, Chelsea Merkel, and Catherine Ruth for technical assistance with the development and optimization of the HsvA enzyme assay. We are grateful to Jeanne Bundens for helpful discussions about substrate accommodation within the structural model of the HsvA active site. We are also grateful to B. V. Venkataram Prasad for a critical reading of the manuscript.
This work was supported in part by Cottrell College Science Award 7714 from the Research Corporation for Science Advancement (to J. A. L.) and funding from the HBE Foundation, the VWR Charitable Foundation, the Eastern University Office of the Provost, and the Eastern University Department of Chemistry. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S3.
The atomic coordinates and structure factors (code 5WPI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- HAE
- Hrp-associated enzyme(s)
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- AGAT
- arginine-glycine amidinotransferase
- hAGAT
- human AGAT
- PDB
- Protein Data Bank.
References
- 1. Malnoy M., Martens S., Norelli J. L., Barny M. A., Sundin G. W., Smits T. H., and Duffy B. (2012) Fire blight: applied genomic insights of the pathogen and host. Annu. Rev. Phytopathol. 50, 475–494 [DOI] [PubMed] [Google Scholar]
- 2. Bonn W. G., and van der Zwet T. (2000) Distribution and economic importance of fire blight. In Fire Blight, the Disease and its Causitive Agent, Erwinia amylovora (Vanneste J. L., ed) pp. 37–54, CAB International, New York [Google Scholar]
- 3. National Agricultural Statistics Service (2016) Noncitrus Fruits and Nuts: 2015 Summary, United States Department of Agriculture, Washington, D. C. [Google Scholar]
- 4. Norelli J. L., Jones A. L., and Aldwinckle H. S. (2003) Fire blight management in the twenty-first century: using new technologies that enhance host resistance in apple. Plant Dis. 87, 756–765 [DOI] [PubMed] [Google Scholar]
- 5. Oh C.-S., Kim J. F., and Beer S. V. (2005) The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol. Plant Pathol. 6, 125–138 [DOI] [PubMed] [Google Scholar]
- 6. Walker J. B. (1979) Creatine: biosynthesis, regulation, and function. Adv. Enzymol. Relat. Areas Mol. Biol. 50, 177–242 [DOI] [PubMed] [Google Scholar]
- 7. Humm A., Fritsche E., Steinbacher S., and Huber R. (1997) Crystal structure and mechanism of human l-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis. EMBO J. 16, 3373–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grazi E., and Rossi N. (1968) Transamidinase of hog kidney. VII. Cysteine at the amidine-binding site. J. Biol. Chem. 243, 538–542 [PubMed] [Google Scholar]
- 9. Fritsche E., Humm A., and Huber R. (1997) Substrate binding and catalysis by l-arginine:glycine amidinotransferase: a mutagenesis and crystallographic study. Eur. J. Biochem. 247, 483–490 [DOI] [PubMed] [Google Scholar]
- 10. Humm A., Fritsche E., and Steinbacher S. (1997) Structure and reaction mechanism of l-arginine:glycine amidinotransferase. Biol. Chem. 378, 193–197 [PubMed] [Google Scholar]
- 11. Fritsche E., Bergner A., Humm A., Piepersberg W., and Huber R. (1998) Crystal structure of l-arginine:inosamine-phosphate amidinotransferase StrB1 from Streptomyces griseus: an enzyme involved in streptomycin biosynthesis. Biochemistry 37, 17664–17672 [DOI] [PubMed] [Google Scholar]
- 12. Jung Y.-G., Kang S.-H., Hyun C.-G., Yang Y.-Y., Kang C.-M., and Suh J.-W. (2003) Isolation and characterization of bluensomycin biosynthetic genes from Streptomyces bluensis. FEMS Microbiol. Lett. 219, 285–289 [DOI] [PubMed] [Google Scholar]
- 13. Ogasawara Y., Fujimori M., Kawata J., and Dairi T. (2016) Characterization of three amidinotransferases involved in the biosynthesis of ketomemicins. Bioorg. Med. Chem. Lett. 26, 3662–3664 [DOI] [PubMed] [Google Scholar]
- 14. Muenchhoff J., Siddiqui K. S., Poljak A., Raftery M. J., Barrow K. D., and Neilan B. A. (2010) A novel prokaryotic l-arginine:glycine amidinotransferase is involved in cylindrospermopsin biosynthesis. FEBS J. 277, 3844–3860 [DOI] [PubMed] [Google Scholar]
- 15. Barón-Sola A., Gutiérrez-Villanueva M. A., Del Campo F. F., and Sanz-Alférez S. (2013) Characterization of Aphanizomenon ovalisporum amidinotransferase involved in cylindrospermopsin synthesis. Microbiologyopen 2, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kellmann R., Mihali T. K., Jeon Y. J., Pickford R., Pomati F., and Neilan B. A. (2008) Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 74, 4044–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernández-Guzmán G., and Alvarez-Morales A. (2001) Isolation and characterization of the gene coding for the amidinotransferase involved in the biosynthesis of phaseolotoxin in Pseudomonas syringae pv. phaseolicola. Mol. Plant. Microbe Interact. 14, 545–554 [DOI] [PubMed] [Google Scholar]
- 18. Li M., Chen L., Deng Z., and Zhao C. (2016) Characterization of AmtA, an amidinotransferase involved in the biosynthesis of phaseolotoxins. FEBS Open Bio 6, 603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Markisch U., and Reuter G. (1990) Biosynthesis of homoarginine and ornithine as precursors of the phytoeffector phaseolotoxin by the amidinotransfer from arginine to lysine catalyzed by an amidinotransferase in Pseudomonas syringae pv. phaseolicola. J. Basic Microbiol. 30, 425–433 [Google Scholar]
- 20. Conconi F., and Grazi E. (1965) Transamidinase of hog kidney. I. Purification and properties. J. Biol. Chem. 240, 2461–2464 [PubMed] [Google Scholar]
- 21. Muenchhoff J., Siddiqui K. S., and Neilan B. A. (2012) Identification of two residues essential for the stringent substrate specificity and active site stability of the prokaryotic l-arginine:glycine amidinotransferase CyrA. FEBS J. 279, 805–815 [DOI] [PubMed] [Google Scholar]
- 22. Feistner G. J. (1994) Metabolism of polyamines and basic amino acids in Erwinia amylovora: application of liquid chromatography/electrospray mass spectrometry to proferrioxamine precursor feeding and inhibition studies. Biol. Mass Spectrom. 23, 793–803 [DOI] [PubMed] [Google Scholar]
- 23. Shirai H., Mokrab Y., and Mizuguchi K. (2006) The guanidino-group modifying enzymes: structural basis for their diversity and commonality. Proteins 64, 1010–1023 [DOI] [PubMed] [Google Scholar]
- 24. Lee G. T., Kim W. J., and Cho Y. D. (2002) Polyamine synthesis in plants: purification and properties of amidinotransferase from soybean (Glycine max) axes. Phytochemistry 61, 781–789 [DOI] [PubMed] [Google Scholar]
- 25. Srivenugopal K. S., and Adiga P. R. (1980) Partial purification and properties of a transamidinase from Lathyrus sativus seedlings: involvement in homoarginine metabolism and amine interconversions. Biochem. J. 189, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller-Fleming L., Olin-Sandoval V., Campbell K., and Ralser M. (2015) Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427, 3389–3406 [DOI] [PubMed] [Google Scholar]
- 27. Wortham B. W., Patel C. N., and Oliveira M. A. (2007) Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 603, 106–115 [DOI] [PubMed] [Google Scholar]
- 28. Feistner G. J., Stahl D. C., and Gabrik A. H. (1993) Proferrioxamine siderophores of Erwinia amylovora: a capillary liquid chromatographic/electrospray tandem mass spectrometric study. Organic Mass Spectrom. 28, 163–175 [Google Scholar]
- 29. Wortham B. W., Oliveira M. A., Fetherston J. D., and Perry R. D. (2010) Polyamines are required for the expression of key Hms proteins important for Yersinia pestis biofilm formation. Environ. Microbiol. 12, 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jelsbak L., Thomsen L. E., Wallrodt I., Jensen P. R., and Olsen J. E. (2012) Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS One 7, e36149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiménez-Bremont J. F., Marina M., Guerrero-González Mde L., Rossi F. R., Sánchez-Rangel D., Rodríguez-Kessler M., Ruiz O. A., and Gárriz A. (2014) Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 5, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sievers F., and Higgins D. G. (2014) Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 1079, 105–116 [DOI] [PubMed] [Google Scholar]
- 33. Jones D. T., Taylor W. R., and Thornton J. M. (1992) The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 [DOI] [PubMed] [Google Scholar]
- 34. Rambaut A. (2014) FigTree phylogenetic tree illustration tool, version 1.4.2 [Google Scholar]
- 35. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bartolomeo M. P., and Maisano F. (2006) Validation of a reversed-phase HPLC method for quantitative amino acid analysis. J. Biomol. Tech. 17, 131–137 [PMC free article] [PubMed] [Google Scholar]
- 37. Molnár-Perl I. (2011) Advancement in the derivatizations of the amino groups with the o-phthaldehyde-thiol and with the 9-fluorenylmethyloxycarbonyl chloride reagents. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 879, 1241–1269 [DOI] [PubMed] [Google Scholar]
- 38. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 39. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 45. Hinsen K. (2000) The molecular modeling toolkit: A new approach to molecular simulations. J. Comput. Chem. 21, 79–85 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.